Background: MyD88 is an adaptor protein that plays a crucial role in the immune response.
Results: We identified residues within the TIR domain of MyD88 required for protein self-association.
Conclusion: Interference with the surface of homodimerization identified by these residues inhibits MyD88 function.
Significance: The inhibition of MyD88 activity could be a good therapeutic strategy for inflammatory and autoimmune diseases.
Keywords: Cellular Immune Response, Mutagenesis, MyD88, Signal Transduction, Toll IL-1 Receptor (TIR) Domain, Toll-like Receptor (TLR)
Abstract
Myeloid differentiation factor 88 (MyD88) is an adaptor protein that transduces intracellular signaling pathways evoked by the Toll-like receptors (TLRs) and interleukin-1 receptors (IL-1Rs). MyD88 is composed of an N-terminal death domain (DD) and a C-terminal Toll/IL-1 receptor (TIR) domain, separated by a short region. Upon ligand binding, TLR/IL-1Rs hetero- or homodimerize and recruit MyD88 through their respective TIR domains. Then, MyD88 oligomerizes via its DD and TIR domain and interacts with the interleukin-1 receptor-associated kinases (IRAKs) to form the Myddosome complex. We performed site-directed mutagenesis of conserved residues that are located in exposed regions of the MyD88-TIR domain and analyzed the effect of the mutations on MyD88 signaling. Our studies revealed that mutation of Glu183, Ser244, and Arg288 impaired homodimerization of the MyD88-TIR domain, recruitment of IRAKs, and activation of NF-κB. Moreover, overexpression of two green fluorescent protein (GFP)-tagged MyD88 mini-proteins (GFP-MyD88151–189 and GFP-MyD88168–189), comprising the Glu183 residue, recapitulated these effects. Importantly, expression of these dominant negative MyD88 mini-proteins competed with the function of endogenous MyD88 and interfered with TLR2/4-mediated responses in a human monocytic cell line (THP-1) and in human primary monocyte-derived dendritic cells. Thus, our studies identify novel residues of the TIR domain that are crucially involved in MyD88 homodimerization and TLR signaling in immune cells.
Introduction
All living organisms counteract the invasion of pathogens by setting up an innate immune response mediated by members of the superfamily of the Toll-like and interleukin-1-like receptors (TLR/IL-1R)2 (1, 2) and by adaptor proteins that link them to downstream molecules involved in signaling pathways (3, 4). To date 10 human TLRs (TLR1–10) and 12 murine TLRs (TLR1–9, TLR11–13) have been identified (5). TLRs detect different pathogen-associated molecular patterns such as lipopeptides (TLR2), lipopolysaccharide (LPS) (TLR4), flagellin (TLR5), bacterial DNA (TLR9), and viral double- or single-stranded RNAs (TLR3 and TLR7/8, respectively) (6). TLRs are type I transmembrane proteins and comprise an extracellular domain, which mediates the recognition of pathogen-associated molecular patterns, a transmembrane region, and an intracellular Toll-IL-1 receptor (TIR) domain that activates downstream signaling pathways (5). Members of IL-1R family are characterized by the presence of extracellular immunoglobulin-like (Ig) domains and by an intracellular TIR domain (2, 3). The prototype of these receptors is IL-1RI, which heterodimerizes with the homologous IL-1R accessory protein to form a receptor complex for the IL-1α, IL-1β, and IL-1 receptor antagonist proteins (2, 3).
The adaptor protein family comprises five members, including the myeloid differentiation factor 88 (MyD88) (4). MyD88 was first shown to be essential for IL-1 and IL-18 signaling (7) and, subsequently, for signaling of most TLRs (4, 8, 9). MyD88 has a modular structure with a death domain (DD) at the N terminus, an intermediate linker domain (ID), and a TIR domain at the C terminus (10). The DD presents a fold that resembles a Greek key bundle of six antiparallel α-helices (11) and allows MyD88 oligomerization and its interaction with the respective DDs of the serine-threonine kinases IRAK1/2/4, thus resulting in a multimeric complex named Myddosome (12, 13). This complex propagates the signal and leads to activation of transcription factors, such as the nuclear factor κB (NF-κB), the activator protein 1, and the interferon-regulatory factors, and of mitogen-activated protein kinases (MAPKs), such as the stress kinase p38 and the extracellular signal-regulated kinases 1 and 2 (ERK1/2) (14). The ID is a short region involved with the DD in the interaction of MyD88 with IRAK4 (15). The TIR domain displays a globular form, consisting of five β-sheets (βA–βE) and four α-helices (αA–αC and αE), connected to the β-strands by surface-exposed loops (16). The structure of the TIR domain of MyD88 is similar to that of other TLR/IL-1R superfamily members, but it lacks an α helix in the region between the βD and βE strands (17–20).
TIR domains are crucial for TLR/IL-1R signal transduction as they mediate receptor-receptor, receptor-adaptor, and adaptor-adaptor interactions. For this reason, they have been the subject of numerous structural and functional studies aimed at identifying the residues participating to homo and hetero interactions (21–26). In the case of MyD88, it was demonstrated that the BB loop of the TIR domain plays a role in homodimerization (22, 27, 28). Nevertheless, current understanding of the nature of homotypic oligomerization of the MyD88-TIR domain is still limited. Here, we have performed an extensive structure-function analysis of the MyD88-TIR domain by mutagenesis. Our studies identify new residues (Glu183, Ser244, and Arg288) that are requested to ensure efficient self-association of MyD88. Modeling studies revealed that Glu183, Ser244, and a portion of the BB loop lie in an interface of dimerization between two contiguous TIR domains. Furthermore, we demonstrated that mutation of these residues also impairs the recruitment of IRAK1/4 and the activation of NF-κB. Importantly, forced expression of MyD88 mini-proteins encompassing the Glu183 residue competed with homodimerization of the endogenous MyD88 protein and attenuated TLR signaling in immune cells. Thus, our findings identified a novel region of the TIR domain of MyD88 that is crucial for TLR/IL-1R signal transduction.
EXPERIMENTAL PROCEDURES
Plasmids
Expression vectors for FLAG-tagged MyD88 or Myc-tagged MyD88 TIR, Myc-tagged MyD88, Myc-tagged IRAK1-kinase-dead (KD) (K239S), and Myc-tagged IRAK4-KD (KK213AA), were constructed as described previously (13, 27, 28). All constructs for the expression of mutated MyD88 TIR or MyD88 were generated by PCR using oligonucleotides containing the mutated residues. Constructs for GFP-MyD88 fusion proteins expression were obtained by subcloning cDNA encoding each protein into pCL-pCX vector. Lentiviral vectors for stable expression of GFP-MyD88 fusion proteins were constructed by subcloning cDNA encoding each protein into PCCLsin.PPT.hPGK vector. All constructs were confirmed by Cycle Sequencing (BMR Genomics, Padua, Italy). The vector for the expression of FLAG-tagged IL-1R was a kind gift from Dr. Alberto Mantovani (Humanitas Clinical and Research Center Rozzano, Italy). The NF-κB-luciferase reporter construct (pNiFty2-Luc) was purchased from InvivoGen, whereas Renilla luciferase construct (pRL-TK-Luc; Promega).
Cell Culture, Stimulation, and Transfections
Human embryonic kidney (HEK) 293T cells were cultured in Dulbecco's modified Eagle's medium, THP-1 cells were maintained in RPMI 1640 medium (Lonza), all supplemented with 10% fetal bovine serum (FBS; Lonza), penicillin and streptomycin (Invitrogen). Dendritic cells were generated from human monocytes of healthy donors. Peripheral blood mononuclear cells were isolated from buffy coats of healthy blood donor through density gradient centrifugation using Ficoll-Hypaque centrifugation (Amersham Biosciences). Monocytes were then positively separated using anti-CD14-labeled magnetic beads (Miltenyi) and resuspended in RPMI 1640 medium supplemented with 10% FBS, 2 mm l-glutamine, 10 mm Hepes, 10 mm sodium pyruvate, 10 mm nonessential amino acids (Lonza), penicillin and streptomycin (Invitrogen). Monocytes were cultured for 5 days in medium supplemented with 100 ng/ml GM-CSF (Miltenyi) and 40 ng/ml IL-4 (Miltenyi). This protocol leads to 98–99% of CD1a+/CD14− monocyte-derived dendritic cells (MDDCs). All cells were grown in a 37 °C humidified atmosphere of 5% CO2. For Western blot analysis, THP-1 cells were serum-starved overnight in medium containing 0.5% FBS and stimulated or not with Pam3CSK4 (1 μg/ml; InvivoGen) or TNF-α (10 ng/ml; Pepro Tech) for 30 min. At the end of incubation, cells were collected and lysed, and cell extracts were analyzed. For flow cytometry analysis the pro-monocytic THP-1 cells were differentiated into mature monocytic cells by treatment with phorbol 12-myristate 13-acetate (200 nm) for 24 h. Mature THP-1 cells and MDDCs were then stimulated with Pam3CSK4 (1 μg/ml; InvivoGen) or LPS (1 μg/ml; Sigma-Aldrich), respectively, for 6 h, in presence of brefeldin A (10 μg/ml) in the last 4 h to inhibit Golgi traffic. At the end of incubation, cells were collected, fixed, permeabilized, stained, and analyzed by flow cytometry. For co-immunoprecipitation assays, HEK293T cells were cultured in 6-cm-diameter dishes and transfected with the appropriate expression vectors by Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. MDDCs were transfected by electroporation with the Amaxa Nucleofector using the Human Dendritic Cell Nucleofector Kit (Lonza) according to the manufacturer's instructions.
Generation of Transduced THP-1
THP-1 stable cell lines were generated by transduction with lentiviral vectors encoding GFP or GFP-MyD88 fusion proteins. THP-1 cells (3 × 105) were infected with lentiviral particles at a multiplicity of infection of 40 in medium supplemented with Polybrene (6 μg/ml) for 24 h. At the end of incubation, cells were washed in PBS, resuspended in fresh medium, and the percentage of GFP-positive cells was assessed by FACS analysis.
Cell Extracts and Co-immunoprecipitation Assay
HEK293T cells were harvested 20 h after transfection, washed in ice-cold PBS, and lysed in buffer containing 50 mm Hepes, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 20 mm β-glycerophosphate, 2 mm DTT, 1 mm Na3VO4, protease inhibitor mixture (Sigma-Aldrich). After incubating for 10 min on ice, cell lysates were centrifuged at 10,000 × g for 10 min at 4 °C, and cytosolic fractions were collected for immunoprecipitation. Cell extracts were immunoprecipitated as described (13, 28) using 2 μg of mouse anti-FLAG M2 (Sigma-Aldrich) or rabbit anti-GFP antibodies (Molecular Probes). After three washes with lysis buffer, proteins were eluted in SDS sample buffer for Western blot analysis. THP-1 cells were collected 30 min after stimulation, washed in ice-cold PBS, and lysed in buffer containing 50 mm Hepes, pH 7.4, 15 mm MgCl2, 150 mm NaCl, 15 mm EGTA, 10% glycerol, 1% Triton X-100, 20 mm β-glycerophosphate, 1 mm DTT, 1 mm Na3VO4, protease inhibitor mixture (Sigma-Aldrich). Cells were pelleted by centrifugation at 10,000 × g for 10 min, and the resulting supernatants were diluted in SDS sample buffer for Western blot analysis.
Western Blot Analyses
Western blot analyses were performed as described (13, 28) using the following primary antibodies (overnight at 4 °C): mouse anti-Myc (1:1000; Santa Cruz Biotechnology), mouse anti-FLAG M2 (1:3000) and mouse anti-β-tubulin (1:1000; Sigma-Aldrich), rabbit anti-GFP (1:1000), rabbit anti-phospho-p38 and rabbit anti-phospho-p65 antibodies (1:1000; Cell Signaling Technology).
NF-κB Reporter Assay
HEK293T cells (5 × 105) were cultured in 24-well plates and transfected with 5 ng of NF-κB-dependent luciferase reporter gene and Renilla luciferase reporter gene (1 ng) as an internal control together with the constructs for the indicated MyD88 proteins by using the Lipofectamine 2000 reagent according to the manufacturer's instructions. THP-1 cells (1 × 106) were transfected by electroporation with the Amaxa Nucleofector using the Cell Line Nucleofector® Kit V (Lonza) according to the manufacturer's instructions. Twenty-four h after transfection, cells were stimulated or not with IL-1β (20 ng/ml), TNF-α (20 ng/ml), or Pam3CSK4 (1 μg/ml) for 6 h. At the end of incubation, cells were harvested and lysed, and luciferase activity was quantified with a biocounter luminometer using the dual-luciferase reporter assay system (Promega). Data were normalized for transfection efficiency by dividing firefly luciferase activity by that of the Renilla luciferase.
Flow Cytometry Analysis of Cytokine-producing Cells
THP-1cells and MDDCs were stained with live-dead fixable near-infrared (Invitrogen) to exclude dead cells from the subsequent analysis. After washing, cells were fixed with 4% paraformaldehyde for 5 min at 4 °C, permeabilized in phosphate-buffered saline (PBS) supplemented with 0.5% saponin, and stained for 15 min at 4 °C with PE-Cy7 conjugated anti-human TNF-α (clone MAb11; eBioscience) or Alexa Fluor 647 anti-human IL-1β (clone JK1B-1; Biolegend). The percentage of cytokine-producing cells was analyzed by flow cytometry (Cyan; Beckman Coulter).
Computational Analysis
NMR solutions of MyD88 TIR domain (PDB ID code 2Z5V) were extracted from the Protein Data Bank. Models of MyD88-TIR domain homodimer were generated using the algorithm PatchDock and sorted by shape complementarity criteria; the first 20 solutions were further refined by the FireDock algorithm using a restricted interface side chain rearrangement and a soft rigid-body optimization. Solutions were automatically ranked by a binding score that includes Atomic Contact Energy, softened van der Waals interactions, partial electrostatics, and additional estimations of the binding free energy.
Densitometry and Statistical Analysis
Densitometric analysis of the Western blots was performed using underexposed images from three to five experiments and analyzed by ImageQuant5.0 software. Statistical differences between mean values were determined using the two-tailed Student's t test.
RESULTS
Identification of New Residues within the MyD88-TIR Domain Required for Its Homodimerization
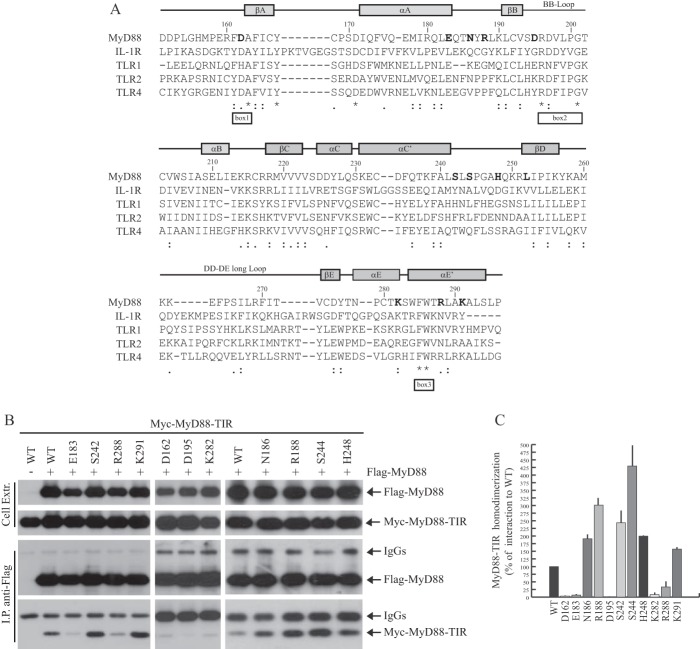
To identify the residues required for homotypic oligomerization of the TIR domain of MyD88, we performed an extensive mutational analysis. Relying on sequence comparison between different TIR domains (Fig. 1A), we selected several amino acids that might be sensitive to point mutations. We focused on residues present within the flanking regions of three highly conserved motifs denoted box 1, 2, and 3 (17) of the MyD88-TIR, with specific attention to charged/polar residues that are likely to be involved in protein-protein interactions (Fig. 1A). We also selected other residues within the AB and CD loops, as they are located in exposed regions that could play a role in the TIR-TIR interface of the MyD88 dimer. The amino acids shown in boldface type were substituted with alanine, with the exception of Lys282 (isoleucine), Arg288 (glycine), and Lys291 (methionine) that were substituted with the amino acid requiring the minimal change in codon usage. The effect of the introduced mutation on homodimerization of the TIR domain was then tested by co-immunoprecipitation assays in HEK293T cells co-transfected with FLAG-tagged wild-type MyD88 and wild-type or mutated Myc-tagged MyD88-TIR. Cell extracts were immunoprecipitated with anti-FLAG antibody, and homodimerization was tested by detecting Myc-MyD88-TIR in the immunoprecipitates. We found that several substitutions (D162A, E183A, D195A, K282I, and R288G) impaired MyD88-TIR homodimerization with respect to the wild-type TIR domain in this assay (Fig. 1, B and C). By contrast, other mutations (N186A, R188A, S242A, S244A, H248A, and K291M) were either ineffective or caused a reproducible increase of dimerization (Fig. 1, B and C).
FIGURE 1.
Mutation of select residues in MyD88 affect homodimerization of the TIR domain. A, sequence alignment of TIR domains of human MyD88, IL-1R, TLR1, TLR2, and TLR4 as the basis of homology modeling performed with ClustalW. Gray bars indicate secondary structure of the MyD88 TIR domain determined by solution NMR spectroscopy (16). White bars indicate box 1, 2, and 3, three highly conserved motifs within the TIR domains (17). The asterisks denote identical residues, whereas the double-dots and single-dots denote conservative or semi-conservative substitutions, respectively. In boldface type are shown the residues mutated for structure-function studies. B, Western blot analysis of the effect of site-specific mutations on homodimerization of MyD88-TIR domain. HEK293T cells were transfected with empty vector (first lane) or FLAG-MyD88 (other lanes) in combination with Myc-MyD88-TIR wild-type (WT) or mutated, as indicated. Cell extracts were immunoprecipitated (IP) with anti-FLAG antibody, and the immunoprecipitated proteins were then analyzed by with either anti-FLAG- or anti-Myc-specific antibodies to detect the interaction. C, densitometric analysis of the effect of MyD88-TIR mutants on homodimerization of TIR domain. Data are expressed as a percentage of the wild-type ± S.D. (error bars) from three separate experiments.
Glu183, Ser244, and Arg288 in the MyD88-TIR Domain Are Required for MyD88 Dimerization and IL-1 Signaling
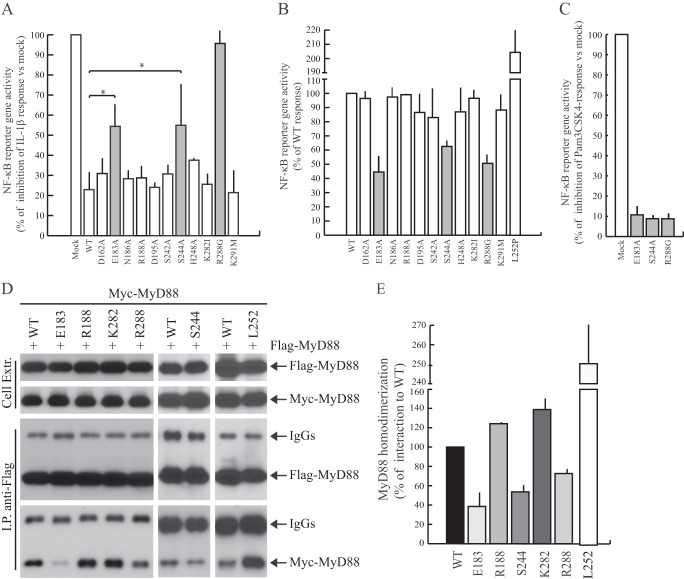
Homodimerization of the MyD88-TIR domain is required for propagation of TLR/IL-1R signaling (3, 4). Notably, overexpression of MyD88-TIR acts as a dominant negative inhibitor of signaling, presumably by competing for the interaction between endogenous MyD88 and TLR/IL-1Rs (16, 22, 24, 25). This feature allows the screening for loss-of-function mutations in the TIR domain by testing the recovery of IL-1-dependent activation of NF-κB in transfected cells (22). As expected, overexpression of the wild-type MyD88-TIR domain strongly suppressed IL-1β-dependent expression of an NF-κB reporter gene in transfected cells (Fig. 2A). Screening of the MyD88-TIR mutants by this assay revealed that MyD88-TIRE183A, MyD88-TIRS244A, and MyD88-TIRR288G were significantly defective (Fig. 2A), suggesting that Glu183, Ser244, and Arg288 are key residues in the MyD88-TIR domain.
FIGURE 2.
Functional analysis of MyD88 mutants. A, HEK293T cells were transfected with NF-κB reporter construct expressing firefly luciferase alone or in combination with wild-type or mutated Myc-MyD88-TIR. Twenty-four h after transfection, cells were stimulated or not with IL-1β (20 ng/ml) for 6 h and lysed for biocounter luminometer analysis. Data are normalized for transfection efficiency (see “Experimental Procedures”), and each column of the graph indicates the relative luciferase activity of the stimulated cells over the nonstimulated cells. Data are expressed as a percentage of inhibition of IL-1β response ± S.D. (error bars) from three separate experiments. Statistical significance was determined by Student's t test. *, p < 0.05. Gray bars indicate the mutants that exerted a significant increase of NF-κB activity compared with the wild-type protein. B, HEK293T cells were transiently transfected with wild-type (WT) or mutant MyD88 constructs together with NF-κB-luciferase reporter construct. Data were normalized for transfection efficiency as above and expressed as a percentage of wild type ± S.D. from three separate experiments. Gray bars indicate the defective MyD88 mutants. C, THP-1 cells were transfected with NF-κB reporter construct expressing firefly luciferase alone or in combination with wild-type or mutated Myc-MyD88. Twenty-four h after transfection, cells were stimulated or not with Pam3CSK4 (1 μg/ml) for 6 h and lysed for biocounter luminometer analysis. Data are normalized for transfection efficiency as described under “Experimental Procedures,” and each column of graph represents the relative luciferase activity of the stimulated cells over the nonstimulated cells. Data are expressed as a percentage of inhibition of TLR2 response ± S.D. from three separate experiments. D, representative Western blot analysis of the effect of selected mutations on MyD88 homodimerization is shown. HEK293T cells were transfected with FLAG-MyD88 in combination with wild-type (first and sixth lanes) or mutated (second through fifth and seventh lanes) Myc-MyD88. Cell extracts were immunoprecipitated (IP) with anti-FLAG antibody, and the immunoprecipitated proteins were then analyzed with either anti-FLAG or anti-Myc-specific antibodies to detect the interaction. E, densitometric analysis of the effect of MyD88 mutants used in C was performed on MyD88 homodimerization. Data are expressed as a percentage of the wild type ± S.D. from three separate experiments.
One surprising aspect of the above results was that although the D162A, E183A, D195A, K282I, and R288G mutations similarly reduced homotypic interaction of MyD88-TIR with full-length MyD88 (Fig. 1, B and C), only MyD88-TIRE183A and MyD88-TIRR288G displayed loss of a dominant negative effect on NF-κB activation (Fig. 2A). Moreover, MyD88-TIRS244A also lost a dominant negative effect in this assay, even though it strongly interacted with full-length MyD88 (Fig. 1, B and C). Thus, to further delineate the functional role of these residues in the TIR domain, we introduced the substitution in full-length MyD88. We also compared the effect of the novel mutations with that of the L252P substitution (based on the sequence of MyD88 variant 1 used in this study, equivalent to L265P in MyD88 variant 2), which was previously shown to confer oncogenic potential to MyD88 and to constitutively activate downstream signaling (29, 30). Overexpression of MyD88 per se induces forced protein oligomerization and consequent activation of NF-κB, even in the absence of exogenous stimuli (8). By using an NF-κB-dependent luciferase reporter assay in HEK293T cells, we observed that the MyD88E183A, MyD88S244A, and MyD88R288G mutants caused 40–60% lower activation of NF-κB than wild-type MyD88, whereas the other mutants activated NF-κB to a similar extent (Fig. 2B). Conversely, MyD88L252P potently activated NF-κB signaling (Fig. 2B). Importantly, expression of MyD88E183A, MyD88S244A, and MyD88R288G strongly interfered with activation of NF-κB by Pam3CSK4-induced TLR2 signaling pathway in monocytoid THP-1 cells (Fig. 2C). These results suggest that the MyD88E183A, MyD88S244A, and MyD88R288G mutants exerts a dominant negative effect on endogenous TLR pathways.
To assess whether the loss-of-function MyD88 mutants were impaired in their capacity to oligomerize correctly, we carried out co-immunoprecipitation assays. In line with the results of the NF-κB activation assays, we observed that the E183A, S244A, and R288G mutations impaired MyD88 homodimerization in HEK293T cells, whereas the other mutants were ineffective, and L252P enhanced it (Fig. 2, D and E, and Table 1).
TABLE 1.
Effect of mutations in MyD88 on protein homodimerization
The interaction of MyD88 mutants with MyD88 wild-type was analyzed by co-immunoprecipitation assays. The table reports a summary of the results obtained (yes: MyD88 mutants homodimerize with wild-type protein; no: MyD88 mutants are deficient in homodimerization).
| Mutation | Location | Homodimerization of MyD88 |
|---|---|---|
| D162A | βA | Yes |
| E183A | αA | No |
| N186A | AB loop | Yes |
| R188A | AB loop | Yes |
| D195A | BB loop | Yes |
| S242A | CD loop | No |
| H248A | CD loop | Yes |
| L252P | βD | Yes |
| K282I | αE | Yes |
| R288G | αE′ | No |
| K291M | αE′ | Yes |
Homodimerization-defective MyD88 Mutants Do Not Recruit IRAKs
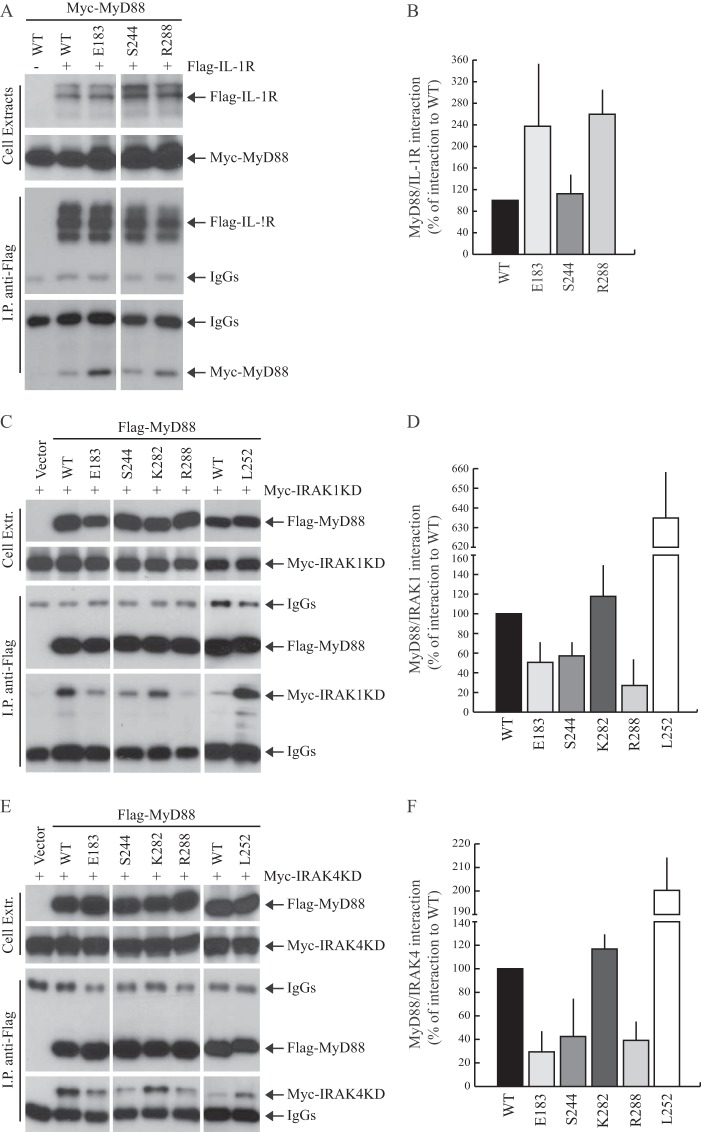
After the ligand-induced activation of IL-1R, MyD88 oligomerizes and interacts with the receptor through their respective TIR domains (31). Thus, we investigated whether the Glu183, Ser244, and Arg288 residues were also required for this heterotypic interaction. HEK293T cells were co-transfected with wild-type or mutated Myc-tagged MyD88 and FLAG-tagged IL-1R. Co-immunoprecipitation experiments indicated that the MyD88E183A, MyD88S244A, and MyD88R288G mutants are not compromised in their ability to associate with IL-1R (Fig. 3, A and B). On the contrary, the E183A and R288G mutations even increased interaction between the two proteins.
FIGURE 3.
MyD88 loss-of-function mutants interacts with IL-1R but fail to recruit IRAK1 and IRAK4. A, interaction of MyD88 mutants with IL-1R. HEK293T cells were transfected with empty vector (first lane) or FLAG-IL-1R (second through fifth lanes) in combination with Myc-MyD88 wild-type (WT) (first and second lanes) or mutated (third through fifth lanes). Twenty h after transfection, cells were collected, and interaction between MyD88 and IL-1R was evaluated by co-immunoprecipitation. Cell extracts were immunoprecipitated (IP) with anti-FLAG antibody, and the immunoprecipitated proteins were then analyzed by Western blotting with either anti-FLAG- or anti-Myc-specific antibodies to detect the interaction. B, densitometric analysis was done of the effect of MyD88 mutants used in A on its interaction with IL-1R. Data are expressed as a percentage of the wild type ± S.D. (error bars) from three separate experiments. C and E, Western blot analysis of the interaction of MyD88 mutants with IRAK1 (C) and IRAK4 (E). HEK293T cells were transfected with empty vector (first lane, C and E) or wild-type (second lane, C and E) or mutated (third through sixth lanes, C and E) FLAG-MyD88 in combination with Myc-IRAK1KD (C) or Myc-IRAK4KD (E). Cell extracts were immunoprecipitated with anti-FLAG antibody and the immunoprecipitated proteins analyzed with either anti-FLAG- or anti-Myc-specific antibodies to reveal the interaction. D–F, densitometric analysis was done of the interaction between MyD88 and IRAKs used in C and E. Data are expressed as a percentage of the wild type ± S.D. from three separate experiments.
Oligomerization of MyD88 is required for recruitment of IRAK1/2/4 (12). Thus, we set out to determine the ability of MyD88 loss-of-function mutants to recruit IRAKs. Because the interaction between MyD88 and these kinases is rapid and transient, it can be more reproducibly detected by expressing KD derivatives (IRAK1KD and IRAK4KD) (31, 32). Wild-type or mutated FLAG-MyD88 were co-expressed with either Myc-tagged IRAK1KD (Fig. 3C) or Myc-tagged-IRAK4KD (Fig. 3E), and cell extracts were immunoprecipitated with anti-FLAG antibodies. MyD88 efficiently associated with IRAK1KD and IRAK4KD, whereas MyD88E183A, MyD88S244A, and MyD88R288G were all impaired in the recruitment of IRAKs (Fig. 3, C–F). By contrast, MyD88K282I, which is not defective in its ability to oligomerize, interacted with IRAK1/4 like wild-type MyD88, and the constitutively active MyD88L252P was much more efficient than wild-type MyD88 in recruiting IRAKs (Fig. 3, C–F). These results suggest that the E183A, S244A, and R288G substitutions affect the recruitment of IRAKs by MyD88 as a consequence of their effect on MyD88-TIR dimerization.
Computational Modeling of MyD88-TIR Homodimer Suggests the Residues Involved in the Dimerization Surface
To evaluate the relevance of the identified residues in the dimerization interface, a computational approach was employed. The structure of the MyD88-TIR domain previously determined by solution NMR spectroscopy (16) was used as template for protein-protein interaction procedures (PDB code 2Z5V). We then employed the PatchDock algorithm (33, 34), which divides protein surfaces into patches according to the surface shape. Finally, the remaining dimers were ranked according to a geometric shape complementarity score. Twenty solutions from PatchDock were further refined using the FireDock algorithm (35, 36). Each dimer was refined by restricted interface side chain rearrangement and by soft rigid-body optimization. Following rearrangement of the side chains, the relative position of the docking partners was refined by Monte Carlo minimization of the binding score function. This score includes Atomic Contact Energy, softened van der Waals interactions, partial electrostatics, and additional estimations of the binding free energy. The refined homodimers were ranked by the binding score (supplemental Table S1). Analysis of the 20 top-scored solutions showed that only the first one contains Glu183 and Ser244 in the dimerization interface. Importantly, this solution scored as the most stable one, providing an independent confirmation of the relevance of Glu183 and Ser244 for MyD88-TIR homodimerization. These studies yielded a hypothetical model for MyD88-TIR homodimerization wherein Glu183 in the first monomer (blue) interacts with the αC' helix in the second monomer (yellow) (Fig. 4). Moreover, Ser244 in the second monomer interacts with the N-terminal fragment (amino acids 156–158) in the first monomer that is structured and exposed to the solvent.
FIGURE 4.
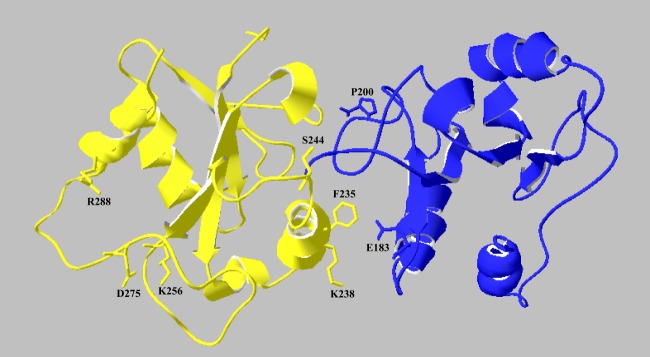
Model of MyD88-TIR domain homodimerization. Modeling of a MyD88-TIR dimer was performed by a computational approach using the available NMR structure as template (16) and the PatchDock and FireDock algorithms. Glu183 and Pro200 are shown on the first monomer (blue); Ser244 and Arg288 are shown on the second monomer (yellow).
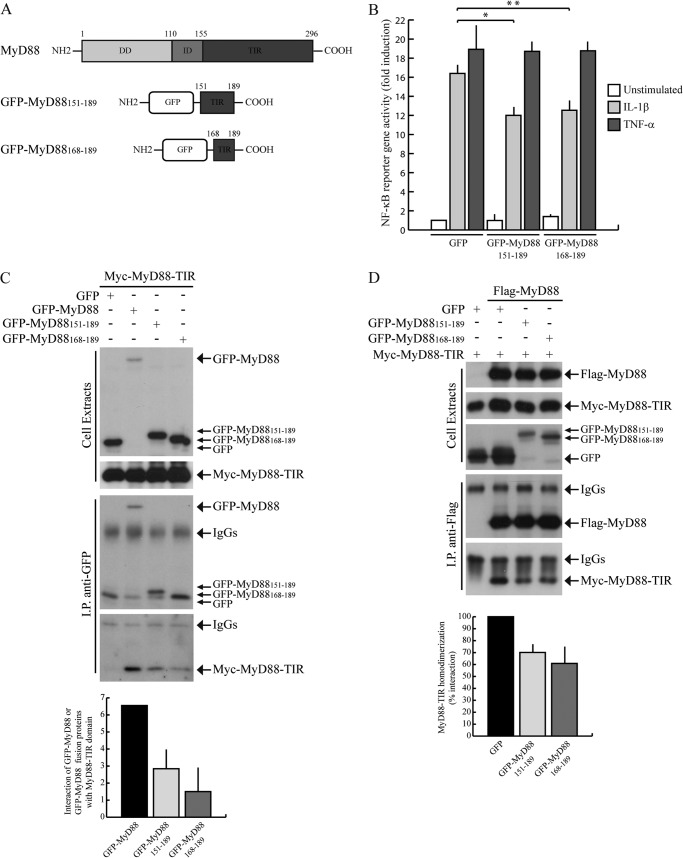
GFP-MyD88 Mini-proteins Encompassing the Glu183 Region Specifically Interfere with IL-1-dependent NF-κB Activation
The Glu183 residue is located at the junction between the αA helix and the loop that connects it to the BB loop of the TIR domain (Fig. 4), which was previously reported to influence MyD88 homodimerization and signaling (22, 27). Given the relevance of this region of the TIR domain for MyD88 function, we investigated it in further detail. First, to determine whether the expression of regions of MyD88-TIR encompassing the Glu183 residue can compete with the scaffold function of MyD88 and affect IL1-R signal transduction, we generated two green fluorescent protein (GFP)-tagged MyD88 fusion proteins containing different portions of this region: GFP-MyD88151–189 and GFP-MyD88168–189 (Fig. 5A). Using a reporter gene assay in HEK293T cells, we found that expression of the GFP-MyD88 MyD88151–189 and GFP-MyD88168–189 fusion proteins significantly reduced the IL-1R-dependent activation of NF-κB (by ∼30%, Fig. 5B). Importantly, the effect of these mini-proteins was specific, because they did not inhibit activation of NF-κB by the MyD88-independent TNF-α pathway (Fig. 5B).
FIGURE 5.
GFP-MyD88151–189 and GFP-MyD88168–189 interfere with IL-1β-induced NF-κB activation. A, schematic representation of GFP-MyD88 mini-proteins. B, NF-κB activity luciferase reporter assay. HEK293T cells were transfected with GFP, GFP-MyD88151–189, or GFP-MyD88168–189 together with NF-κB luciferase reporter construct and stimulated or not with either 20 ng/ml IL-1β (light gray bars) or 20 ng/ml TNF-α (dark gray bars) for 6 h. Data were normalized for transfection efficiency as described in Fig. 2 and expressed as mean -fold induction ± S.D. (error bars), compared with control expressing GFP from three experiments. Statistical significance was determined by Student's t test. *, p < 0.05; **, p < 0.01. C, GFP-MyD88151–189 and GFP-MyD88168–189 interaction with MyD88-TIR domain. HEK293T cells were transfected with Myc-MyD88-TIR in combination with GFP (first lane), GFP-MyD88 (second lane), or GFP-MyD88151–189 and GFP-MyD88168–189 (third and fourth lanes, respectively). Cell extracts were immunoprecipitated (IP) with anti-GFP antibody, and immunoprecipitated proteins were analyzed by Western blotting with either anti-GFP- or anti-Myc-specific antibodies. Densitometric analysis of the degree of interaction of Myc-MyD88-TIR with GFP-MyD88, GFP-MyD88151–189, or GFP-MyD88168–189 is shown in the bar graph. D, GFP-MyD88151–189 and GFP-MyD88168–189 interference with the homodimerization of MyD88-TIR domain. HEK293T cells were transfected with Myc-MyD88-TIR alone (first lane) or in combination with FLAG-MyD88 (second through fourth lanes) in the presence of GFP (first and second lanes), GFP-MyD88151–189 (third lane), or GFP-MyD88168–189 (fourth lane). Cell extracts were immunoprecipitated with anti-FLAG antibody, and immunoprecipitated proteins were analyzed by Western blotting with either anti-FLAG- or anti-Myc-specific antibodies. Densitometric analysis of MyD88-TIR homodimerization in the presence of GFP or GFP-MyD88 fusion proteins is shown and is expressed as percentage of interaction with respect to GFP.
To investigate whether the inhibitory effect of the GFP-MyD88 mini-proteins on IL-1β signaling was due to their ability to titrate out endogenous MyD88, first we tested whether they were able to interact with the MyD88-TIR domain by co-immunoprecipitation assays. GFP, GFP-MyD88, GFP-MyD88151–189, and GFP-MyD88168–189 were co-expressed in HEK293T cells with Myc-tagged MyD88-TIR. As expected, GFP-MyD88 interacted with MyD88-TIR (Fig. 5C). GFP-MyD88151–189 and GFP-MyD88168–189 also associated with the TIR domain, albeit with less efficiency (Fig. 5C). Despite the weaker interaction with MyD88-TIR, this experiment suggests that these GFP-MyD88 mini-proteins are folded correctly and maintain the original structure. Next, we investigated whether they were able to inhibit the activation of NF-κB by virtue of their interference with MyD88-TIR homodimerization. HEK293T cells were co-transfected with Myc-MyD88-TIR alone or in combination with FLAG-MyD88 in the presence of GFP, GFP-MyD88151–189, and GFP-MyD88168–189. Co-immunoprecipitation experiments showed that expression of the two MyD88 mini-proteins reduced MyD88-TIR homodimerization (Fig. 5D), whereas expression of GFP protein alone exerted no effect. Hence, these results indicate that the GFP-MyD88 mini-proteins impair IL-1R signaling by interfering with homodimerization of its TIR domain.
A MyD88 Mini-protein Encompassing the Glu183 Region Interferes with TLR Signaling in Monocytoid Cells
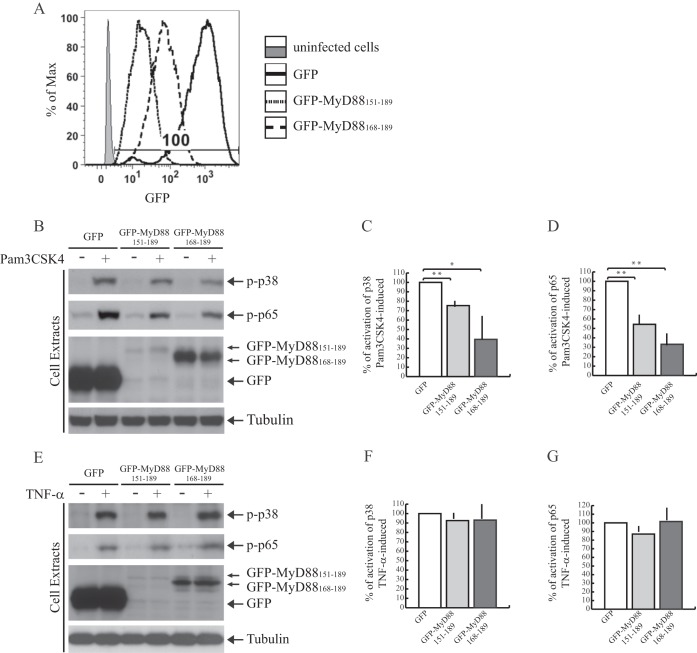
Hyperactivation of the TLR/IL-1R signal transduction pathway is involved in several autoinflammatory and autoimmune diseases (1, 2). Thus, agents that limit the activation of this pathway are of potential therapeutic interest. Given the dominant negative effect of the GFP-MyD88 mini-proteins on the IL-1β-signaling pathway, we asked whether they can also affect activation of TLR signaling in cells of relevance for immune system disorders. To test this possibility, we analyzed their effect on activation/phosphorylation of the endogenous p38 (14) and p65, a subunit of NF-κB (37), after stimulation of TLR2 in monocytoid cells (38). THP-1 cells were transduced by lentiviral particles to express GFP-MyD88151–189, or GFP-MyD88168–189, or GFP as control. Transduction efficiency (100%) was evaluated by flow cytometry analysis (Fig. 6A). Flow cytometry (Fig. 6A) and Western blot analysis (Fig. 6, B and E) of the transduced THP-1 cells showed that GFP was expressed at high levels, GFP-MyD88151–189 was expressed at low levels, whereas the expression of GFP-MyD88168–189 was detected at intermediate levels. Importantly, in THP-1 stimulated with the TLR2 ligand Pam3CSK4, expression of the two MyD88 mini-proteins caused a significant reduction of p38 (Fig. 6, B and C) and p65 phosphorylation (Fig. 6, B and D). GFP-MyD88168–189 was more efficient in inhibiting the TLR2 signaling than GFP-MyD88151–189, possibly due to its higher expression levels. Moreover, the inhibitory effect of two MyD88 mini-proteins on TLR2 signaling was specific because they did not interfere with phosphorylation of p38 and p65 induced by TNF-α (Fig. 6, E–G), which activates a MyD88-independent signaling pathway (37).
FIGURE 6.
GFP-MyD88151–189 and GFP-MyD88168–189 specifically interfere with TLR2 signaling in THP-1 cells. A, THP-1 cells transduced with lentiviral particles for stable expression of GFP or GFP-MyD88 mini-proteins (GFP-MyD88151–189 and GFP-MyD88168–189). The percentage of infected cells was determined by flow cytometry comparing the percentage of GFP fluorescent cells (solid, dotted, and dashed lines for GFP control, GFP-MyD88151–189, and GFP-MyD88168–189, respectively) to uninfected cells (gray solid line). B and E, Western blot analysis of phospho-p38 (p-p38) and phospo-p65 (p-p65) expression levels in pro-monocytic THP-1 cells. Cells were transduced with lentiviral particles for stable expression of GFP or GFP-MyD88 mini-proteins. After transduction, cells were serum-starved overnight in medium containing 0.5% FBS and stimulated or not with Pam3CSK4 (1 μg/ml) (B) or TNF-α (10 ng/ml) (E) for 30 min and analyzed with anti-phospho-p38 (top panel, B and E), anti-phospho-p65 (second panel, B and E), or anti-GFP-specific antibodies (third panel, B and E). C and D, densitometric analysis of TLR2-mediated p38 (C) and p65 (D) phosphorylation. F and G, densitometric analysis of TNF-α-mediated p38 (F) and p65 (G) phosphorylation. *, p < 0.05; **, p < 0.01. Tubulin staining was used as loading control.
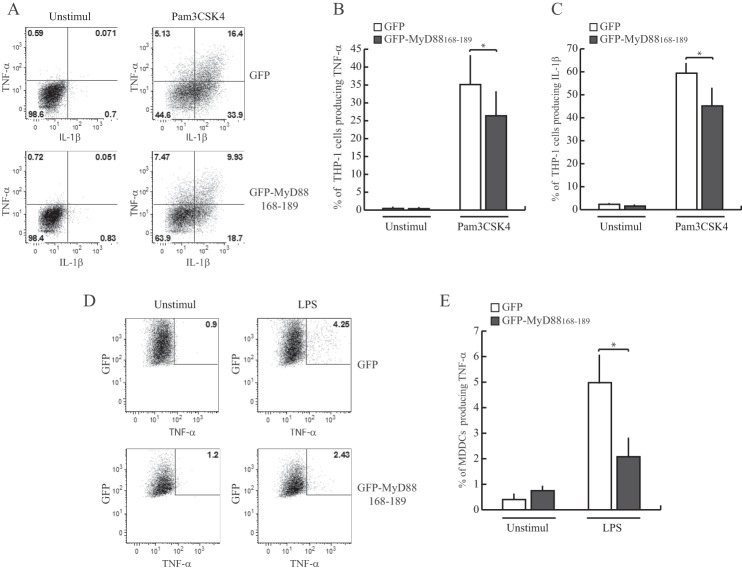
Activation of TLRs in monocytoid cells induces the secretion of cytokines such as TNF-α and IL-1β (1). Thus, we tested the ability of GFP-MyD88168–189 to interfere with production of TNF-α and IL-1β in THP1 stimulated with Pam3CSK4. We found that the percentage of cytokine-producing cells (within the alive GFP-positive cells) was significantly reduced in Pam3CSK4-stimulated THP-1 cells expressing GFP-MyD88168–189 (Fig. 7, A–C). Even more importantly, we observed that expression of GFP-MyD88168–189 reduced the frequency of TNF-α-producing cells in a primary human myeloid cell type, such as the MDDCs. In fact, the percentage of MDDCs that produce TNF-α after LPS stimulation was significantly lower in cells transfected with GFP-MyD88168–189 than in control GFP-expressing cells (Fig. 7, D and E). These results indicate that a MyD88 mini-protein encompassing the Glu183 residues interferes not only with IL-1R signaling but also with TLR pathways in live cells.
FIGURE 7.
GFP-MyD88168–189 interferes with the production of TNF-α and IL-1β induced by stimulation of TLR2 and TLR4 in human monocytoid cells. A, the pro-monocytic THP-1 cells stably expressing GFP or GFP-MyD88168–189 were differentiated into mature monocytic cells by phorbol 12-myristate 13-acetate treatment for 24 h and stimulated with Pam3CSK4 (1 μg/ml) for 6 h in presence of Golgi inhibitor brefeldin A (10 μg/ml). The percentage of TNF-α- or IL-1β-producing cells within GFP-positive cells was quantified by intracellular staining with the specific antibodies and flow cytometry analysis. B and C, bar graph represents quantitative data from three independent experiments (means ± S.E., error bars). *, p < 0.05 (paired t test). D, MDDCs transfected with constructs for transient expression of GFP or GFP-MyD88168–189 were stimulated with LPS (1 μg/ml) for 6 h in presence of Golgi inhibitor brefeldin A (10 μg/ml). At the end of incubation, the cells were harvested, and the percentage of TNF-α-producing cells within alive GFP-positive cells was quantified by flow cytometry. E, quantification of results as in D for three independent experiments was performed with cells from three individual donors. Data are represented as mean ± S.E. *, p < 0.05 (paired t test).
DISCUSSION
MyD88 is an adaptor protein that plays a pivotal role in immune responses mediated by TLR/IL-1Rs. Upon ligand binding, these receptors homo- or heterodimerize through their TIR domains and recruit MyD88, which in turn dimerizes via its DD or TIR domain to nucleate a macromolecular complex named Myddosome (12, 39). The TIR domain is a cytoplasmic domain typically composed of 135–160 residues, with sequence conservation between 20 and 30% in different proteins. The sequence and structure of different TIR domains determine the specificities of receptor and adaptor protein TIR-TIR interactions. A number of mutagenesis and docking studies have identified different TIR-TIR interfaces of interaction (16–27). The BB loop, which forms a protrusion on the surface of the TIR domain (17), has been particularly studied (16, 17, 22, 27, 28, 40–43). In the case of MyD88-TIR, the BB loop is involved in both adaptor-receptor and adaptor-adaptor interactions, as documented by its role in the association between MyD88 and TLR4 (16), IL-1R accessory protein (22), and TLR2 (24) as well as in MyD88 homodimerization (22, 27, 28). Nevertheless, given the plasticity and heterogeneity of complexes formed by this domain in MyD88, it is likely that additional regions of the TIR also play a key role in the interaction with TLR/IL1-R receptors and/or in the assembly of the Myddosome.
The identification of amino acidic stretches potentially involved in TIR-TIR interactions can be exploited to develop specific inhibitors of TLR/IL-1R signal transduction pathways. Such compounds would represent a valuable therapeutic tool to counteract the aberrant stimulation of these pathways that occur in a multitude of human diseases, such as cancer, autoimmune, chronic inflammatory or infectious diseases (44–48). We and others have shown previously that short amino acidic stretches can serve as templates to design decoy peptides and synthetic analogues (peptidomimetics) that interfere with TIR-TIR interactions and down-regulate TLR/IL-1R signaling (44–46). Regarding MyD88, a synthetic peptide encompassing the conserved residues of the BB loop was shown to compromise MyD88 self-association and to interfere with IL-1R-dependent signal transduction (27). Moreover, we developed a synthetic compound (ST2825) mimicking the BB loop peptide and demonstrated that it was efficient in inhibiting IL-1R and TLR9 signaling pathways (28, 49). Notably, because MyD88 plays a crucial role in the activation of the signaling pathways triggered by all TLR/IL-1Rs, with the sole exception of TLR3 (9), it represents a suitable target for diseases in which aberrant regulation of TLR/IL-1R signaling contributes to the pathology (44, 45). In this study, we have performed an extensive mutational analysis of the MyD88-TIR domain to identify new regions involved in MyD88 homodimerization. We identified several residues that affect the interaction of the isolated MyD88-TIR with full-length MyD88. Individual mutation of Asp162 (box 1), Glu183 (αA helix/AB loop), Asp195 (BB loop), Lys282 and Arg288 (flanking regions box 3) inhibited MyD88-TIR homodimerization in co-immunoprecipitation experiments. Notably, mutation of other residues, such as Asn186 (AB loop), Arg188 (AB loop), Ser242 (CD loop), Ser244 (CD loop), His248 (CD loop) and Lys291 (flanking region box 3), caused an increased association of the TIR domain with full-length MyD88 in the same assay. However, even if all of these residues affected in some way the homotypic interaction of the TIR domain, only the Glu183, Ser244, and Arg288 were required for IL-1R and TLR signal transduction in live cells. These results suggest that decreased or increased homodimerization in transfection experiments is not sufficient to explain the activity of MyD88. It is possible that the MyD88-TIR mutants that are still capable of supporting IL-1R/TLR signal transduction undergo conformational changes in the cell that make them behave like the wild-type MyD88-TIR, even though their homodimerization efficiency is altered. It is also worth noticing that these types of experiments are generally performed under overexpression conditions, which may represent a less sensitive system of detecting mild defects in protein functions. Nevertheless, these studies allowed us to identify Glu183, Ser244, and Arg288 as residues that are strictly required for the function of full-length MyD88. First, these amino acids were necessary for efficient homodimerization of the full-length protein, even though MyD88 homodimerizes mainly through the DD and ID (16). Moreover, mutation of these residues interfered with the ability of MyD88 to activate NF-κB in response to IL-1β and Pam3CSK4 stimulation, indicating that their effect on homodimerization was functionally relevant. Surprisingly, the S244A mutation induced an increase in homodimerization of the isolated MyD88-TIR, whereas it reduced self-association of the full-length protein, suggesting that in the context of full-length MyD88 the presence of the DD and ID might induce conformational rearrangements of the MyD88 dimer that affect the homotypic oligomerization of TIR domains and the specific contacts engaged by the exposed residues.
To fit our results in a three-dimensional model, we employed a computational approach based on the documented NMR structure of the TIR domain of MyD88. The resulting model provides some hints on how these three amino acids could stabilize TIR domain homodimerization. Indeed, in the lowest energy model Glu183 and Ser244 are directly involved in the dimerization interface. This model also predicts that residues Phe235 and Lys238, located in the αC′ helix, are involved in the interaction with αA helix containing Glu183. Moreover, amino acids at the N terminus of the monomer face Ser244. On the contrary, Arg288 is far away from this interface but lies within a very long and structured loop that could be stabilized by a salt bridge interaction between Arg288 and Asp275 or, alternatively, between Asp275 and Lys256, thus enhancing loop plasticity in respect to possible heterodimeric interactions. Noteworthy, our study identifies new functional surface sites in the MyD88-TIR that are important for homodimerization. The Glu183 and Ser244 residues are located at the end of the αA helix and in the CD loop, respectively. To our knowledge, this is the first evidence of an involvement of these portions in the homodimerization of MyD88-TIR. The Arg288 residue flanks the highly conserved box 3 and was reported to participate in TIR-TIR interactions between MyD88 and MAL (16), TLR2 (24) and TRAM (25). Here, we extend these findings by showing that the Arg288 residue is also involved in MyD88 homodimerization, suggesting that this extended loop plays a general role in TIR:TIR homo- and hetero-dimerization. Our model of the MyD88 TIR:TIR homodimer was generated using the experimental data reported in this work. Future studies by site-directed mutagenesis (for instance of Lys238 and Asp275) and the synthesis of small peptides derived by αΑ and αC′ helixes could be suitable approaches to identify the smallest active portion interfering with MyD88 signaling in live cells.
In our model, Glu183 is located in an accessible interface between the two MyD88-TIR monomers. To test this hypothesis, we engineered small MyD88 mini-proteins encompassing Glu183 (MyD88151–189 and MyD88168–189) and expressed them as fusions with GFP in eukaryotic cells. These mini-proteins were capable of interfering with MyD88 self-association, indicating that the N-terminal region of the MyD88-TIR domain, comprising the βA β-sheet, AA loop, αA helix, and AB loop (151–189 residues), is required for homotypic oligomerization. Moreover, the efficient competition exerted by these mini-proteins suggests that this interface region between two TIR domains is under a dynamic and exchangeable equilibrium, which is accessible to competing molecules. In support of this notion, we also found that these mini-proteins hampered IL-1β-induced activation of NF-κB in transfected cells, indicating that their inhibitory effect on MyD88 dimerization is functionally relevant. Furthermore, retroviral infection of these GFP-MyD88 mini-proteins in the human THP-1 monocytic cell line decreased the activation TLR2 signaling and down-regulated the production of TNF-α and IL-1β after stimulation of cells with Pam3CSK4. This observation is of considerable importance as TNF-α and IL-1β are pro-inflammatory cytokines involved in many autoinflammatory and autoimmune diseases, and modulation of their secretion from innate immune cells might represent a suitable target for therapeutic intervention (44–47). In this regard, we also tested the ability of the MyD88 mini-proteins to modulate TLR responses in primary immune cells of relevance for human diseases, such as monocyte-derived dendritic cells. Dendritic cells express a broad repertoire of TLRs through which they recognize different microbial compounds (50). After challenge with microbial stimuli, they undergo a complex process of maturation, including cytokine production, that results in the activation of an appropriate adaptive immune response (51). We found that the GFP-MyD88168–189 mini-protein is able to decrease the percentage of dendritic cells that produce TNF-α upon TLR2/TLR4 stimulation. Because dendritic cells play a key role in immune regulation, the observation that the GFP-MyD88168–189 mini-protein interferes with their TLR-induced response is potentially relevant for several diseases caused by excessive inflammatory responses, such as chronic diseases (i.e. atherosclerosis, Alzheimer disease) or autoimmune diseases (i.e. multiple sclerosis, Chron disease, psoriasis) (52). On the other hand, the identification of critical residues for the MyD88 function in dendritic cells might be of relevance for genetic diseases associated with microbial susceptibility. In this regard, our previous mutational study of the DD of MyD88 identified a crucial residue (Glu52) (13) that was found mutated in children affected by life-threatening recurrent pyogenic bacterial infections (53).
In conclusion, we provide evidence that Glu183, Ser244, and Arg288 are involved in the homotypic oligomerization of the TIR domain of MyD88. Our study also reveals that a short region in this domain encompassing Glu183 forms a critical surface in the interaction between two TIR domains. These results suggest that this amino acidic stretch could serve as template to design peptides and peptidomimetics that might behave as decoy molecules and compete with the endogenous MyD88, thus interfering with the activation of an aberrant TLR/IL-1R signal transduction in various pathological conditions (44–48).
Acknowledgments
We thank Dr. Alberto Mantovani and Dr. Gennaro Melino for the plasmids and reagents.
This work was supported by grants from Fondazione Italiana Sclerosi Multipla (FISM), Association for International Cancer Research (AICR) (to C. S.), and the Italian Ministry of Health (Progetto Giovani Ricercatori) (to E. V.).

This article contains supplemental Table S1.
- TLR
- Toll-like receptor
- DD
- death domain
- ID
- intermediate domain
- IL-1R
- inteleukin-1 receptor
- IRAK
- IL-1 receptor-associated kinase
- KD
- kinase-dead
- MDDC
- monocyte-derived dendritic cell
- MyD88
- myeloid differentiation factor 88
- TIR
- Toll/IL-1 receptor.
REFERENCES
- 1. Kawai T., Akira S. (2011) Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 [DOI] [PubMed] [Google Scholar]
- 2. Sims J. E., Smith D. E. (2010) The IL-1 family: regulators of immunity. Nat. Rev. Immunol. 10, 89–102 [DOI] [PubMed] [Google Scholar]
- 3. O'Neill L. A. (2008) The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol. Rev. 226, 10–18 [DOI] [PubMed] [Google Scholar]
- 4. O'Neill L. A., Bowie A. G. (2007) The family of five: TIR domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 5. Roach J. C., Glusman G., Rowen L., Kaur A., Purcell M. K., Smith K. D., Hood L. E., Aderem A. (2005) The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. U.S.A. 102, 9577–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sasai M., Yamamoto M. (2013) Pathogen recognition receptors: ligands and signaling pathways by Toll-like receptors. Int. Rev. Immunol. 32, 116–133 [DOI] [PubMed] [Google Scholar]
- 7. Muzio M., Ni J., Feng P., Dixit V. M. (1997) IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science 278, 1612–1615 [DOI] [PubMed] [Google Scholar]
- 8. Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., Janeway C. A. (1998) MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell 2, 253–258 [DOI] [PubMed] [Google Scholar]
- 9. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 10. Hardiman G., Rock F. L., Balasubramanian S., Kastelein R. A., Bazan J. F. (1996) Molecular characterization and modular analysis of human MyD88. Oncogene 13, 2467–2475 [PubMed] [Google Scholar]
- 11. Park H. H., Lo Y. C., Lin S. C., Wang L., Yang J. K., Wu H. (2007) The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu. Rev. Immunol. 25, 561–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin S. C., Lo Y. C., Wu H. (2010) Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signaling. Nature 465, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loiarro M., Gallo G., Fantò N., De Santis R., Carminati P., Ruggiero V., Sette C. (2009) Identification of critical residues of the MyD88 death domain involved in the recruitment of downstream kinases. J. Biol. Chem. 284, 28093–28103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brown J., Wang H., Hajishengallis G. N., Martin M. (2011) TLR-signaling networks: an integration of adaptor molecules, kinases, and cross-talk. J. Dent. Res. 90, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burns K., Janssens S., Brissoni B., Olivos N., Beyaert R., Tschopp J. (2003) Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J. Exp. Med. 197, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohnishi H., Tochio H., Kato Z., Orii K. E., Li A., Kimura T., Hiroaki H., Kondo N., Shirakawa M. (2009) Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 10260–10265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu Y., Tao X., Shen B., Horng T., Medzhitov R., Manley J. L., Tong L. (2000) Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 408, 111–115 [DOI] [PubMed] [Google Scholar]
- 18. Nyman T., Stenmark P., Flodin S., Johansson I., Hammarström M., Nordlund P. (2008) The crystal structure of the human Toll-like receptor 10 cytoplasmic domain reveals a putative signaling. J. Biol. Chem. 283, 11861–11865 [DOI] [PubMed] [Google Scholar]
- 19. Khan J. A., Brint E. K., O'Neill L. A., Tong L. (2004) Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. J. Biol. Chem. 279, 31664–31670 [DOI] [PubMed] [Google Scholar]
- 20. Valkov E., Stamp A., Dimaio F., Baker D., Verstak B., Roversi P., Kellie S., Sweet M. J., Mansell A., Gay N. J., Martin J. L., Kobe B. (2011) Crystal structure of Toll-like receptor adaptor MAL/TIRAP reveals the molecular basis for signal transduction and disease protection. Proc. Natl. Acad. Sci. U.S.A. 108, 14879–14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Radons J., Dove S., Neumann D., Altmann R., Botzki A., Martin M. U., Falk W. (2003) The interleukin 1 (IL-1) receptor accessory protein Toll/IL-1 receptor domain: analysis of putative interaction sites in vitro mutagenesis and molecular modeling. J. Biol. Chem. 278, 49145–49153 [DOI] [PubMed] [Google Scholar]
- 22. Li C., Zienkiewicz J., Hawiger J. (2005) Interactive sites in the MyD88 Toll/interleukin (IL) 1 receptor domain responsible for coupling to the IL1 signaling pathway. J. Biol. Chem. 280, 26152–26159 [DOI] [PubMed] [Google Scholar]
- 23. Bovijn C., Ulrichts P., De Smet A. S., Catteeuw D., Beyaert R., Tavernier J., Peelman F. (2012) Identification of interaction sites for dimerization and adaptor recruitment in Toll/interleukin-1 receptor (TIR) domain of Toll-like receptor 4. J. Biol. Chem. 287, 4088–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nada M., Ohnishi H., Tochio H., Kato Z., Kimura T., Kubota K., Yamamoto T., Kamatari Y. O., Tsutsumi N., Shirakawa M., Kondo N. (2012) Molecular analysis of the binding mode of Toll/interleukin-1 receptor (TIR) domain proteins during TLR2 signaling. Mol. Immunol. 52, 108–116 [DOI] [PubMed] [Google Scholar]
- 25. Ohnishi H., Tochio H., Kato Z., Kawamoto N., Kimura T., Kubota K., Yamamoto T., Funasaka T., Nakano H., Wong R. W., Shirakawa M., Kondo N. (2012) TRAM is involved in IL-18 signaling and functions as a sorting adaptor for MyD88. PLoS One 7, e38423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bovijn C., Desmet A. S., Uyttendaele I., Van Acker T., Tavernier J., Peelman F. (2013) Identification of binding sites for myeloid differentiation primary response gene 88 (MyD88) and Toll-like receptor 4 in MyD88 adaptor-like (Mal). J. Biol. Chem. 288, 12054–12066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loiarro M., Sette C., Gallo G., Ciacci A., Fantò N., Mastroianni D., Carminati P., Ruggiero V. (2005) Peptide-mediated interference of TIR domain dimerization in MyD88 inhibits interleukin-1-dependent activation of NF-κB. J. Biol. Chem. 280, 15809–15814 [DOI] [PubMed] [Google Scholar]
- 28. Loiarro M., Capolunghi F., Fantò N., Gallo G., Campo S., Arseni B., Carsetti R., Carminati P., De Santis R., Ruggiero V., Sette C. (2007) Pivotal advance: inhibition of MyD88 dimerization and recruitment of IRAK1 and IRAK4 by a novel peptidomimetic compound. J. Leukoc. Biol. 82, 801–810 [DOI] [PubMed] [Google Scholar]
- 29. Ngo V. N., Young R. M., Schmitz R., Jhavar S., Xiao W., Lim K. H., Kohlhammer H., Xu W., Yang Y., Zhao H., Shaffer A. L., Romesser P., Wright G., Powell J., Rosenwald A., Muller-Hermelink H. K., Ott G., Gascoyne R. D., Connors J. M., Rimsza L. M., Campo E., Jaffe E. S., Delabie J., Smeland E. B., Fisher R. I., Braziel R. M., Tubbs R. R., Cook J. R., Weisenburger D. D., Chan W. C., Staudt L. M. (2011) Oncogenically active MYD88 mutations in human lymphoma. Nature 470, 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Treon S. P., Xu L., Yang G., Zhou Y., Liu X., Cao Y., Sheehy P., Manning R. J., Patterson C. J., Tripsas C., Arcaini L., Pinkus G. S., Rodig S. J., Sohani A. R., Harris N. L., Laramie J. M., Skifter D. A., Lincoln S. E., Hunter Z. R. (2012) MyD88 L265P somatic mutation in Waldenstrom's macroglobulinemia. N. Engl. J. Med. 367, 826–833 [DOI] [PubMed] [Google Scholar]
- 31. Wesche H., Henzel W. J., Shillinglaw W., Li S., Cao Z. (1997) MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity 7, 837–847 [DOI] [PubMed] [Google Scholar]
- 32. Li S., Strelow A., Fontana E. J., Wesche H. (2002) IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc. Natl. Acad. Sci. U.S.A. 99, 5567–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duhovny D., Nussinov R., Wolfson H. J. (2002) in Proceedings of the 2nd Workshop on Algorithms in Bioinformatics (WABI) Rome, Italy, Lecture Notes in Computer Science 2452 (Guigò R., Gusveld D., eds) pp. 185–200, Springer Verlag [Google Scholar]
- 34. Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J. (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Andrusier N., Nussinov R., Wolfson H. J. (2007) FireDock: fast interaction refinement in molecular docking. Proteins 69, 139–159 [DOI] [PubMed] [Google Scholar]
- 36. Mashiach E., Schneidman-Duhovny D., Andrusier N., Nussinov R., Wolfson H. J. (2008) FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 36, W229–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Napetschnig J., Wu H. (2013) Molecular basis of NF-κB signaling. Annu. Rev. Biophys. 42, 443–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Remer K. A., Brcic M., Sauter K. S., Jungi T. W. (2006) Human monocytoid cells as a model to study Toll-like receptor-mediated activation. J. Immunol. Methods 313, 1–10 [DOI] [PubMed] [Google Scholar]
- 39. Burns K., Martinon F., Esslinger C., Pahl H., Schneider P., Bodmer J. L., Di Marco F., French L., Tschopp J. (1998) MyD88, an adapter protein involved in interleukin-1 signaling. J. Biol. Chem. 273, 12203–12209 [DOI] [PubMed] [Google Scholar]
- 40. Bartfai T., Behrens M. M., Gaidarova S., Pemberton J., Shivanyuk A., Rebek J., Jr. (2003) A low molecular weight mimic of the Toll/IL-1 receptor/resistance domain inhibits IL-1 receptor-mediated responses. Proc. Natl. Acad. Sci. U.S.A. 100, 7971–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 42. Horng T., Barton G. M., Medzhitov R. (2001) TIRAP: an adapter molecule in the Toll signaling pathway. Nat. Immunol. 2, 835–841 [DOI] [PubMed] [Google Scholar]
- 43. Fitzgerald K. A., Rowe D. C., Barnes B. J., Caffrey D. R., Visintin A., Latz E., Monks B., Pitha P. M., Golenbock D. T. (2003) LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the Toll adapters TRAM and TRIF. J. Exp. Med. 198, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Loiarro M., Ruggiero V., Sette C. (2010) Targeting TLR/IL-1R signalling in human diseases. Mediators Inflamm. 2010, 674363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Loiarro M., Ruggiero V., Sette C. (2013) Targeting the Toll-like receptor/interleukin 1 receptor pathway in human diseases: rational design of MyD88 inhibitors. Clin. Lymphoma Myeloma Leuk. 13, 222–226 [DOI] [PubMed] [Google Scholar]
- 46. Fekonja O., Avbelj M., Jerala R. (2012) Suppression of TLR signaling by targeting TIR domain-containing proteins. Curr. Protein Pept. Sci. 13, 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Verstak B., Hertzog P., Mansell A. (2007) Toll-like receptor signalling and the clinical benefits that lie within. Inflamm. Res. 56, 1–10 [DOI] [PubMed] [Google Scholar]
- 48. Basith S., Manavalan B., Yoo T. H., Kim S. G., Choi S. (2012) Roles of Toll-like receptors in cancer: a double-edged sword for defense and offense. Arch. Pharm. Res. 35, 1297–1316 [DOI] [PubMed] [Google Scholar]
- 49. Capolunghi F., Rosado M. M., Cascioli S., Girolami E., Bordasco S., Vivarelli M., Ruggiero B., Cortis E., Insalaco A., Fantò N., Gallo G., Nucera E., Loiarro M., Sette C., De Santis R., Carsetti R., Ruggiero V. (2010) Pharmacological inhibition of TLR9 activation blocks autoantibody production in human B cells from SLE patients. Rheumatology 49, 2281–2289 [DOI] [PubMed] [Google Scholar]
- 50. Iwasaki A., Medzhitov R. (2004) Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5, 987–995 [DOI] [PubMed] [Google Scholar]
- 51. Banchereau J., Steinman R. M. (1998) Dendritic cells and the control of immunity. Nature 392, 245–252 [DOI] [PubMed] [Google Scholar]
- 52. Tabas I., Glass C. K. (2013) Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science 339, 166–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. von Bernuth H., Picard C., Jin Z., Pankla R., Xiao H., Ku C. L., Chrabieh M., Mustapha I. B., Ghandil P., Camcioglu Y., Vasconcelos J., Sirvent N., Guedes M., Vitor A. B., Herrero-Mata M. J., Aróstegui J. I., Rodrigo C., Alsina L., Ruiz-Ortiz E., Juan M., Fortuny C., Yagüe J., Antón J., Pascal M., Chang H. H., Janniere L., Rose Y., Garty B. Z., Chapel H., Issekutz A., Maródi L., Rodriguez-Gallego C., Banchereau J., Abel L., Li X., Chaussabel D., Puel A., Casanova J. L. (2008) Pyogenic bacterial infections in humans with MyD88 deficiency. Science 321, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]