Background: Iron starvation triggers lipid droplet and triacylglycerol (TAG) accumulation in Chlamydomonas reinhardtii.
Results: The overall lipid profile shows a decrease in the absolute content of monogalactosyldiacylglycerol (MGDG) and an increase in saturated and monounsaturated fatty acids.
Conclusion: Iron starvation has an early and distinct effect on membrane lipids, before onset of chlorosis.
Significance: Iron deficiency affects distribution of lipid type as well as fatty acid profile.
Keywords: Algae, Iron, Lipid Metabolism, Plant Biochemistry, Triacylglycerol, Fatty Acid Desaturase
Abstract
Chlamydomonas reinhardtii cells exposed to abiotic stresses (e.g. nitrogen, zinc, or phosphorus deficiency) accumulate triacylglycerols (TAG), which are stored in lipid droplets. Here, we report that iron starvation leads to formation of lipid droplets and accumulation of TAGs. This occurs between 12 and 24 h after the switch to iron-starvation medium. C. reinhardtii cells deprived of iron have more saturated fatty acid (FA), possibly due to the loss of function of FA desaturases, which are iron-requiring enzymes with diiron centers. The abundance of a plastid acyl-ACP desaturase (FAB2) is decreased to the same degree as ferredoxin. Ferredoxin is a substrate of the desaturases and has been previously shown to be a major target of the iron deficiency response. The increase in saturated FA (C16:0 and C18:0) is concomitant with the decrease in unsaturated FA (C16:4, C18:3, or C18:4). This change was gradual for diacylglyceryl-N,N,N-trimethylhomoserine (DGTS) and digalactosyldiacylglycerol (DGDG), whereas the monogalactosyldiacylglycerol (MGDG) FA profile remained stable during the first 12 h, whereas MGDG levels were decreasing over the same period of time. These changes were detectable after only 2 h of iron starvation. On the other hand, DGTS and DGDG contents gradually decreased until a minimum was reached after 24–48 h. RNA-Seq analysis of iron-starved C. reinhardtii cells revealed notable changes in many transcripts coding for enzymes involved in FA metabolism. The mRNA abundances of genes coding for components involved in TAG accumulation (diacylglycerol acyltransferases or major lipid droplet protein) were increased. A more dramatic increase at the transcript level has been observed for many lipases, suggesting that major remodeling of lipid membranes occurs during iron starvation in C. reinhardtii.
Introduction
The fatty acid composition of the lipids in biological membranes is specific to the unique functions of individual membranes, and can adapt to changes in the environment. The most abundant constituents of chloroplast membranes are the non-phosphorus galactoglycerolipids, monogalactosyldiacylglycerol (MGDG)4 and digalactosyldiacylglycerol (DGDG). Another component of plastidic membranes is the sulfolipid sulfoquinovosyldiacylglycerol (SQDG). Genes encoding the enzymes involved in MGDG, DGDG, and SQDG have been found in Chlamydomonas reinhardtii based on orthology (MGD1, DGD1) or genetically confirmed (SQD1) (1, 2). A significant difference between plants and algae is that some algae, including C. reinhardtii contain betaine lipids like diacylglyceryl-N,N,N-trimethylhomoserine (DGTS), which is proposed to substitute for the phosphatidylcholine found in plants (3). C. reinhardtii membranes also contain phospholipids such as phosphatidylglycerol, phosphatidylinositol, and phosphatidylethanolamine (4–6).
Plants and algae also synthesize triacylglycerols (TAGs), non-membrane glycerolipids that can serve as a store of acyl moieties. The final step in TAG synthesis is catalyzed by diacylglycerol acyltransferases (DGATs). The C. reinhardtii genome codes for five type 2 DGATs (DGTT1–DGTT5) and one type 1 DGAT (DGAT1) (7). Recently, a cytosolic acyltransferase (DGAT3) has been shown to be involved in recycling of 18:2 and 18:3 FA into TAG in Arabidopsis thaliana (8). Another pathway to TAG formation is by an acyl-CoA-independent reaction, which is mediated by a phospholipid:DAG acyltransferase (PDAT) (7, 9). C. reinhardtii cells grown under conditions of macronutrient limitation (e.g. sulfur, phosphorus, and nitrogen deficiency) (7, 10, 11) or other growth-limiting conditions (high-salt) (12) accumulate TAGs in lipid droplets (7, 13, 14). Recently, it has been shown that limitation of micronutrients (e.g. zinc, iron) can result in formation of lipid droplets as well, as visualized by Nile Red staining (15–17).
C. reinhardtii cells, like other photosynthetic eukaryotes, become chlorotic (i.e. chlorophyll deficient) when grown under iron-deprived conditions. Their plastids are often smaller and less developed with the photosynthetic membranes strongly affected (18–20). Chlorosis results from decreased abundance of the photosynthetic machinery as a result of induced degradation, which is preceded by structural remodeling of photosystem I (20, 21). Although the transcriptome and proteome changes in iron-starved C. reinhardtii are well studied (22–24), the impact of poor iron nutrition on the lipid constituents of the membrane is under investigated. Studies with sugar beet have demonstrated that thylakoid membranes have a decreased ratio of MGDG/DGDG, but we do not know whether this is a general phenomenon or restricted to sugar beet and/or the land plant lineage (25).
In this study we investigated the impact of iron-starvation on membrane lipid composition and TAG accumulation in C. reinhardtii. To get insights into the changes in the transcript levels of genes coding proteins involved in FA metabolism, we surveyed the C. reinhardtii transcriptome from cells grown under iron-starved conditions. Our study indicates significant and rapid changes in glycerolipid composition, namely a decrease in MGDG content and an increase in TAG, along with changes in FA desaturation and FA composition of membrane lipids in C. reinhardtii exposed to iron deprivation.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
C. reinhardtii strain CC-4532 (2137 mt−) was grown in Tris acetate-phosphate (TAP) with Hutner trace elements (26, 27) at 24 °C and 50–100 μmol m−2 s−1 photon flux density. Iron nutritional stages were achieved by maintaining the strain in standard TAP medium (20 μm Fe-EDTA) followed by transfer to iron-free TAP supplemented with Fe-EDTA at the indicated concentrations as described in Refs. 20 and 22. Nitrogen-free medium was prepared by omitting the NH4Cl in TAP (7).
Iron Starvation Time Course
Two different iron-starvation experiments (0–48-h time course and 0–5-day time course) were performed to identify changes in lipid profiles in different stages of iron starvation. To induce iron starvation, cells were grown to mid-logarithmic phase (∼4 × 106 cells ml−1), collected by centrifugation (2,500 × g for 5 min at room temperature), washed twice in TAP medium lacking Fe-EDTA, and inoculated into TAP medium supplemented with 0 or 20 μm Fe-EDTA. For the 0–48-h time course samples were collected at 0, 0.5, 1, 2, 4, 8, 12, 24, and 48 h after transfer to TAP lacking iron and for the 0–5-day time course after 0, 1, 2, 3, 4, and 5 days in TAP medium without any Fe-EDTA for RNA and lipid analyses.
Nucleic Acid Analysis
Total RNA was extracted from exponentially grown C. reinhardtii cells as described previously (28). RNA quality was assessed on an Agilent 2100 Bioanalyzer and RNA hybridization as described in Ref. 29. The probe used for detection of CBLP was a 915-bp EcoRI fragment from the cDNA insert in plasmid pcf8-13 (30). For RNA-Seq, duplicate cDNA libraries were prepared from 4 μg of total RNA for each of the samples in the 0–48-h time course by means of the Illumina TruSeq RNA Sample Preparation kit version 1. Indexed libraries were pooled and sequenced on the Illumina HiSeq 2000 platform, at three libraries per lane, in 100-bp single end reads. Raw and processed sequence files are available at the NCBI Gene Expression Omnibus (accession number GSE44611).
Mapping the reads to the C. reinhardtii genome (version FM4 assembly, Augustus 10.2 annotation), calculation of transcript abundances in reads/kilobase of mappable transcript length/million mapped reads (RPKM), and fold-changes were performed as described previously (22). Differential expression analysis was performed using the DESeq package (31). p values obtained from DESeq were adjusted for multiple testing using Benjamini-Hochberg correction (32) to control false discovery rates.
Immunoblot Analysis
Proteins were separated by denaturing PAGE (7.5 to 15% acrylamide monomer), transferred to PVDF or nitrocellulose, and detected as described in Ref. 33. Primary antibody dilutions were: ferroxidase, 1:300; ferredoxin, 1:1,000; FAB2, 1 μg ml−1 of purified IgG; CTH1, 1:1,000; and CF1, 1:10,000. All antibodies were from Agrisera, except FAB2, which was obtained from John Shanklin.
Lipid and Fatty Acid Analysis
For quantitative total fatty acid analysis, lipids were extracted from lyophilized material and analyzed as previously described (7, 34, 35). The positional distribution of fatty acids was performed using Rhizopus arrhizus lipase (Sigma) as previously described in Ref. 36, with modifications according to Ref. 37.
Nile Red Staining and Microscopy
Cells were grown in triplicate to a density of 2 × 106 cells ml−1 in replete TAP medium containing a trace element mixture according to Hutner et al. (38). Each of the triplicates was split into two aliquots, which were washed twice in iron-deficient TAP medium before resuspension (1 × 106 cells ml−1) in iron-deficient or -replete medium, respectively. Samples were collected immediately after resuspension (= 0 h) and 4, 8, 12, 24, 48, 72, 96, and 120 h thereafter for staining with Nile Red (1 μg/ml final concentration, Sigma). Staining was performed for 15 min at room temperature under constant gentle agitation and cells were concentrated by centrifugation at 500 × g for 3 min. They were subsequently mixed with 2% low melting-agarose (Invitrogen) in phosphate buffer and observed by confocal microscopy on a Leica TCS SPE microscope with an ACS APO ×63 water objective lens (numerical aperture 1.15). The Nile Red signal was captured using a laser excitation line at 488 nm (intensity, 9%); and emission was collected between 554 and 599 nm (gain, 741; offset, 0). Chlorophyll autofluorescence was measured by excitation at 635 nm (intensity, 29%) and emission captured between 650 and 714 nm. Differential interference contrast images were acquired in the PM Trans channel (gain, 371; offset, 0). Images were colored using Leica confocal software.
RESULTS
Iron-limited C. reinhardtii Cells Accumulate TAG
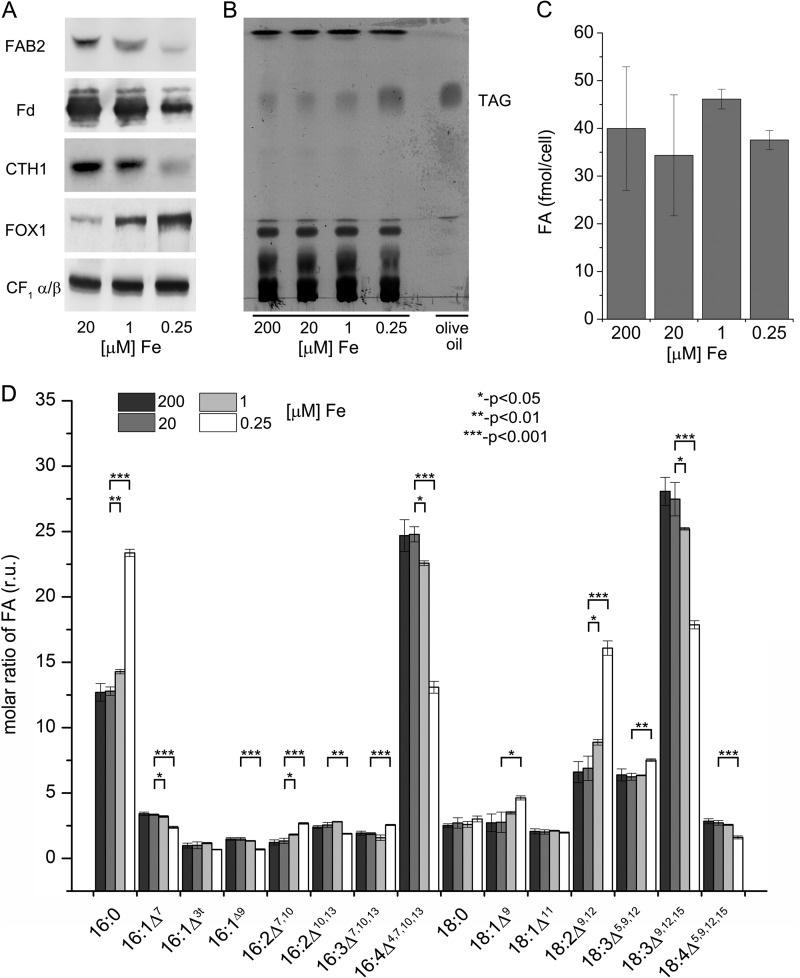
Previous studies performed in C. reinhardtii cells acclimated to iron limitation (22, 23) identified several differences in the abundance of transcript and proteins for enzymes involved in fatty acid biosynthesis and TAG accumulation. We took advantage of the availability of antibodies against one of the fatty acid desaturase enzymes (plastid acyl-ACP desaturase with a di-carboxylate diiron active site (FAB2)) and other iron-containing proteins (ferredoxin, ferredoxin with an Fe2S2 center) and copper target homolog 1, CTH1 (which is a diiron enzyme required for formation of the 5th ring in the Chl structure), also with a di-carboxylate diiron active site, to test their abundances as a function of iron nutrition. Immunoblot analysis confirmed and validated the previous proteomics results (23) (Table 1) indicating a lower amount in iron-limited conditions for each of the iron-containing proteins tested (Fig. 1A). The decrease in FAB2 abundance parallels the decrease in other iron-containing proteins. Because the iron cofactor is critical for activity of the protein, we wondered about the impact of iron deficiency on the profile of fatty acids in cellular lipids. In other words, is FAB2 rate-limiting for plastid fatty acid desaturation? A previous survey of nutritional stress in C. reinhardtii indicated accumulation of lipid droplets in iron-limited cells (16). A thin layer chromatogram (TLC) of lipids (Fig. 1B) indeed showed increased TAG content in iron-limited cells (0.25 μm Fe-EDTA) but no change in the total FA content (Fig. 1C), consistent with recycling of membrane fatty acids into storage neutral lipid. When we monitored the FA composition of the total lipid content, we observed a significant (p value <0.05, n = 3) increase in more saturated FAs such as 16:0 and 18:2 (number of carbons:number of double bonds) and a relative decrease in 16:4 and 18:3ω6 (Fig. 1D). Interestingly, the levels of 18:0 were not increased suggesting a distinct effect on specific FA desaturases. Together, these results indicate that iron limitation in C. reinhardtii leads to an altered FA profile, a relative increase in saturated FAs, and accumulation of TAG. This could be explained by the inactivation of fatty acid desaturases under these iron-limiting conditions, because they are diiron-containing enzymes (39).
TABLE 1.
Anti-correlated changes in protein and transcript abundances of plastid acyl-ACP desaturase (FAB2) in photoheterotrophic iron-limited cells
C. reinhardtii WT cells were grown in iron-replete, -deficient, or -limited photoheterotrophic conditions.
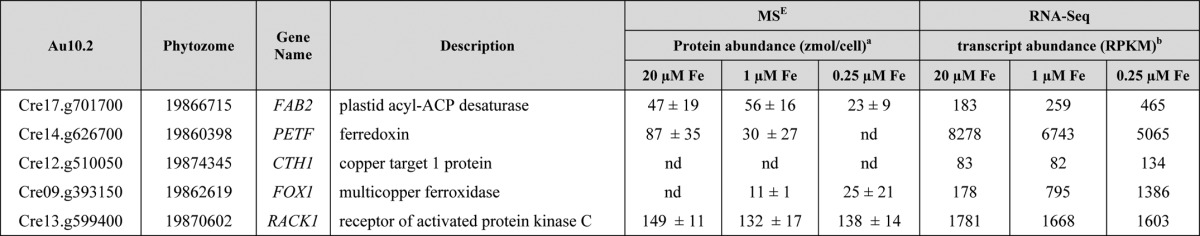
a Proteins were identified and quantified based on the signal intensity of the three most abundant peptides by MSE (35). Values represent the averages of three biological replicates; standard deviation is indicated.
b Changes in mRNA abundances as analyzed by RNA-Seq (22).
FIGURE 1.
Impact of iron limitation on TAG accumulation and FA desaturation in C. reinhardtii. A, protein abundances in response to iron nutrition. Twenty micrograms of total membrane protein were separated by denaturing PAGE and transferred to a PVDF or nitrocellulose membrane, which was probed with antibodies against several iron-containing proteins: plastid acyl-ACP desaturase (FAB2), ferredoxin (Fd), copper-target homolog 1 (CTH1), ferroxidase (FOX1), and CF1. B, thin layer chromatography for neutral lipids from iron excess (200 μm), iron (Fe)-replete (20 μm), iron-deficient (1 μm), and iron-limited (0.25 μm) cells stained with iodine vapor. 20 μg of total lipid were applied per lane. C, FA per cell in different iron nutritional conditions as determined by GC FID of FAMEs. FAMEs were prepared from lyophilized cells on glass fiber filters directly; cells were collected from iron-excess, -replete, -deficient, and -limited conditions. D, fatty acid composition in different iron nutritional states (expressed in relative units, r.u.), determined as FAMEs. Error bars represent the S.D. from three biological replicates. Fatty acids types are indicated as number of carbons: number and positions of double bonds (Δ). Asterisks indicate the values that are statistically different from the control (20 μm) (one asterisk, p < 0.05; two asterisks, p < 0.01, and three asterisks, p < 0.001; non-paired two sample Student's t test).
Iron Starvation Leads to TAG Accumulation
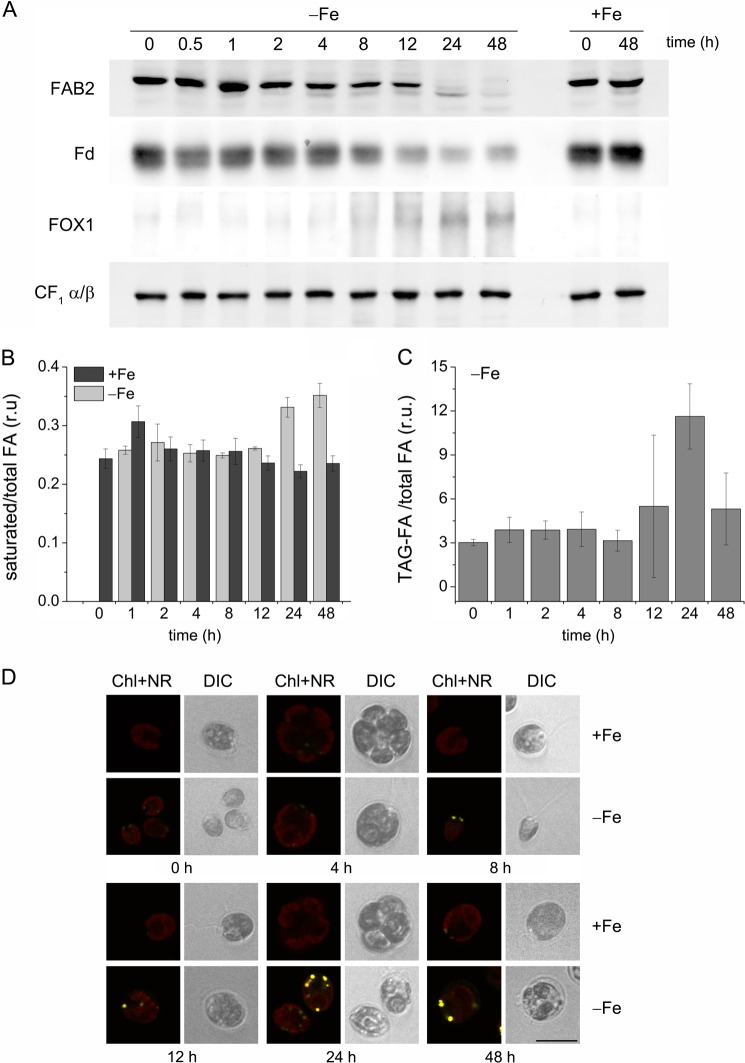
Naturally, iron starvation (0 μm Fe-EDTA) is a more damaging abiotic stress compared with iron limitation (0.25 μm Fe-EDTA), and indeed physiological parameters (growth rate) validate this point (not shown). To address the question of the timing of TAG accumulation during prolonged periods of iron deprivation, C. reinhardtii cells were grown in iron-replete conditions (20 μm Fe-EDTA) and then transferred to fresh iron-free medium (0 supplemental iron) or iron-replete medium (20 μm iron chelate, as a control) and samples were collected at time 0 h and every 24 h for 5 days. First, we monitored by immunoblotting the abundances of several iron-containing enzymes in C. reinhardtii cells deprived of iron for 5 days (Fig. 2). As observed previously (20, 40), iron starvation results in a rapid decrease in ferredoxin in the first 24 h, whereas CTH1, a diiron-containing enzyme was more slowly reduced. When the levels of FAB2 were monitored, we detected a similar profile as seen for ferredoxin with a dramatic decrease in the enzyme abundance in the first 24 h of iron starvation (Fig. 2A). The degree of impact of iron starvation is therefore not dependent on the type of iron cluster (diiron versus iron-sulfur). In this experiment, chlorophyll levels were changed significantly only after 48 h of iron starvation, chlorosis being visible only after this time. This is consistent with the slower impact on CTH1 (cyclase in Chl biosynthesis). Next, we determined the total FA and TAG levels during the time course of 5 days of iron starvation. Total FA levels did not change on a per cell basis in cells lacking iron compared with cells grown under iron-replete conditions (Fig. 2C). Nevertheless, the TAG content increased more than 5-fold after day 1 of iron starvation, but then slightly decreased over the next 4 days of the experiment (Fig. 2D). Next, we asked whether the increase in TAG after 24 h of iron starvation correlates with lipid droplet formation. C. reinhardtii cells grown under iron-deprived conditions were stained with Nile Red and lipid droplet formation was monitored by confocal microscopy (Fig. 2E). Indeed, we observed the accumulation of lipid droplets (yellow) after 24 h of iron starvation, which became relatively more abundant as the C. reinhardtii cells were grown further in medium lacking iron (Fig. 2E).
FIGURE 2.
Accumulation of TAG during an extended period of iron starvation. A, abundance of iron-containing proteins during 5 days of iron (Fe) starvation. Soluble or membrane fractions (20 μg) were separated by denaturing gel electrophoresis, transferred to PVDF or nitrocellulose membranes, and probed for the indicated proteins. B, chlorophyll content per cell in iron-starved C. reinhardtii cells. C, FA content on a per cell basis in iron-starved and control cells. D, TAG-FA content per cell during 5 days of iron starvation. E, formation of lipid droplets in C. reinhardtii exposed to 5 days of iron starvation. We used confocal microscopy with Nile Red® staining (yellow) to observe lipid accumulation and chlorophyll autofluorescence (red). Chl + NR, chlorophyll autofluorescence merged with Nile Red; DIC, differential interference contrast, scale bar, 10 μm. Error bars represent the S.D. from three biological replicates.
These results demonstrate that C. reinhardtii cells grown under iron-deprived conditions accumulate TAGs and show increased accumulation of lipid droplets after 24 h, whereas the total FA content remains relatively stable. This occurs before chlorosis is apparent. The slightly diminished TAG content after several days of iron starvation suggests the carbon stored in the form of TAGs can be used subsequently as an energy source to sustain growth during the following days of iron starvation.
Iron Starvation Results in More Saturated FAs
To more finely resolve the temporal changes in FA composition and TAG abundance in iron-starved cells, we sampled cells during a shorter time course (0, 1, 2, 4, 8, 12, 24, and 48 h) after transfer from iron-replete to fresh iron-free (0) or iron-replete (20 μm Fe-EDTA, control) medium. First, the abundance of FAB2, ferredoxin, and FOX1 were monitored as sentinels of iron status. FAB2 and ferredoxin decreased after 12–24 h of iron starvation, whereas the ferroxidase (involved in iron uptake) increased, indicative of selective changes in the proteome. Specifically, the increased abundance of FOX1 confirms that the cells were still biosynthetically active. The abundance of FAB2, ferroxidase, and FOX1 did not change under the control iron-replete conditions (Fig. 3A).
FIGURE 3.
Changes in FA saturation and TAG content occur during 12 and 24 h of iron (Fe) starvation. A, abundance of iron-containing proteins during 48 h of iron starvation. B, changes in the saturated versus unsaturated FAs distribution during the first 48 h of iron starvation. C, the fraction of total FAs in TAG during 48 h of iron starvation. D, formation of lipid droplets under iron starvation up to 48 h. See legend to Fig. 2. r.u., relative units.
Because the fatty acid desaturases are diiron enzymes, we expected that under conditions of complete iron deprivation, the levels of FA desaturation would be even lower than in the iron-limited cells. To answer that question, the FA composition was profiled over a 48-h period. As expected, we observed more saturated FAs (C16:0 and C18:0) in iron-deprived cells at 24 and 48 h (Fig. 3B). The increase in saturated FAs after 24–48 h of iron starvation correlated with the decrease in FA desaturase enzyme levels (e.g. FAB2). At the same time, we determined the content of TAG in C. reinhardtii cells during this 48-h iron starvation (Fig. 3C). The levels of TAGs increased 3–4-fold after 24 h of iron starvation compared with other time points. Interestingly, TAG levels dropped after 48 h by about 50%, but were still higher than at the 0-h time point. Next, we assayed lipid droplet formation by confocal microscopy using Nile Red staining to see if their numbers or size correlate with increased TAGs. The earliest detection of the accumulation of lipid droplets was noted after 8 h of iron starvation (Fig. 3D). The highest density of Nile Red-stained lipid droplets was observed when C. reinhardtii cells were iron starved for 24 h. This observation correlates with the amount of TAGs determined quantitatively at a similar time point (Fig. 3D), supporting the use of Nile Red staining as a proxy for measurement of TAG content in this work. No lipid droplets were detected under iron-replete conditions over the entire 48-h growth period.
Altogether, these results demonstrate that under iron starvation, C. reinhardtii cells accumulate more saturated FAs most likely due to defects in the enzymatic activities of FA desaturases. At the same time, the cells accumulate TAGs that are likely stored in the Nile Red-stained lipid droplets. All these effects appear after 24 h of iron starvation, prior to the onset of chlorosis and protein degradation.
Glycerolipid and FA Content Show Early Response to Iron Starvation
Besides TAGs, C. reinhardtii contains other glycerolipids (MGDG, DGTS, or DGDG) that are the major constituents of cellular membranes. We asked whether iron starvation would also influence the relative abundance of these glycerolipids and their FA profiles. We measured the FA composition of betaine-lipid (DGTS), a glycerolipid found mainly in extra plastidic membranes (Fig. 4A). The FA composition of DGTS changed progressively during the iron-starvation period, with an increase in saturated FA (C16:0). Significant changes (p < 0.05) appeared at later time points (12, 24, and 48 h) in C16:1Δ7, C18:1Δ11, C18:3Δ9,12,15, or C18:4Δ5,9,12,15. Overall, we observed an increase in more saturated FAs at the expense of unsaturated ones (C16:0 versus C16:1Δ7; C18:1Δ9 and C18:2Δ9, 12 versus C18:3Δ9,12,15 or C18:4Δ5,9,12,15) (Fig. 4A). This decrease in desaturated FA in DGTS composition might be the result of the loss of functional diiron FA desaturases during iron starvation.
FIGURE 4.

Iron starvation leads not only to changes in the fatty acids composition but also the types and amounts of membrane lipids. FA composition of DGTS (A) and MGDG (B). Lipids were isolated at different time points during the 48-h iron starvation, separated by thin layer chromatography, and quantified by GC as FAMEs. Molar ratios of FA composition are expressed in relative units (r.u.). C, FA content of individual lipids (TAG, MGDG, DGDG, and DGTS) per cell expressed in femtomole/cell at different time points during 48 h of iron starvation. Asterisks indicate the values that are statistically different from the control (0-h time point) (p < 0.05; non-paired two sample Student's t test).
In contrast to DGTS, the FA composition of MGDG, a plastidic-membrane glycerolipid remained relatively stable for the first 12 h of iron starvation (Fig. 4B). After 12 h of iron deprivation, we observed a decrease in desaturation of FAs in MGDG concomitant with the increase in saturated FAs (C16:2Δ7, 10 versus C16:4Δ4,7,10,13 and C18:2 versus C18:3Δ9,12,15). Interestingly, we detected rapid changes in the MGDG content, which starts to drop even after 2 h of iron starvation. The MGDG levels continue to decrease being about 3 times lower compared with 0 h (Fig. 4C). The absolute amounts of DGTS and DGDG decreased progressively during the 48 h of iron starvation, with levels around 2 times lower after 12 and 48 h. The amount of TAGs was considerably higher after 12 h (Fig. 4C).
The Diacylglycerol Moiety for TAG Biosynthesis Is of Chloroplast Origin
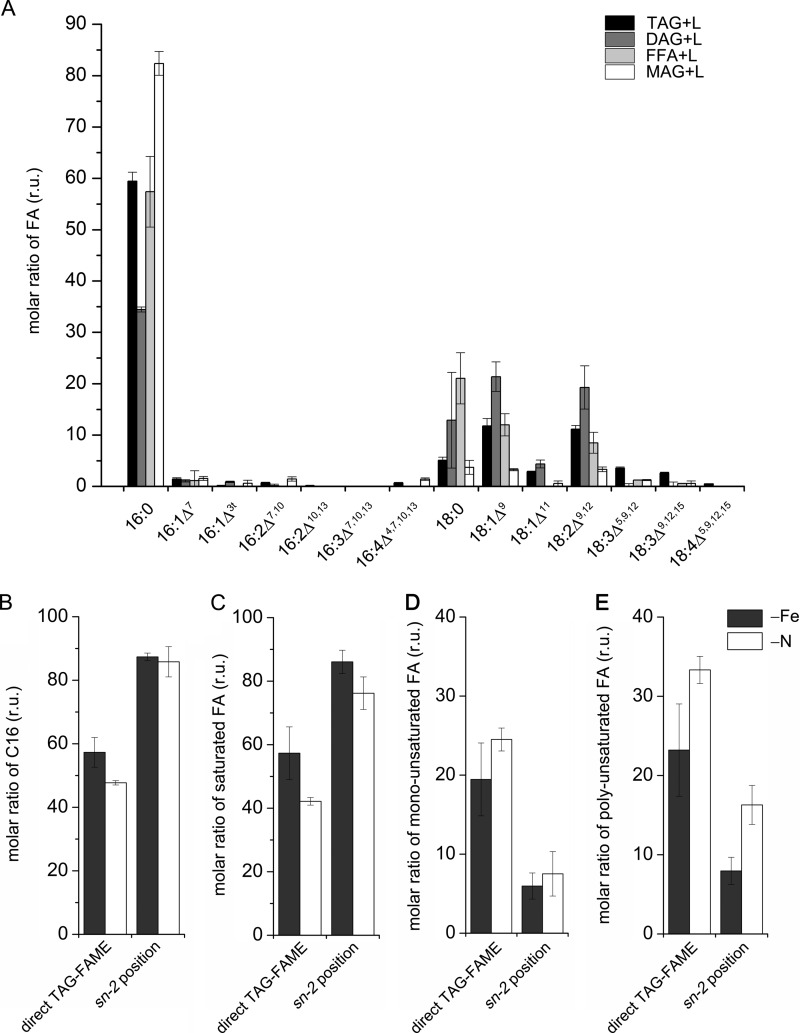
To investigate the origin of the DAG moiety in TAG, we performed positional analysis of acyl groups in TAGs isolated from 24-h iron-starved C. reinhardtii cells with R. arrhizus lipase. The lipase preferentially cleaves the acyl groups at sn-1/sn-3 positions of TAG (36). As previously shown (41), almost 90% of the FAs at the sn-2 position in TAG were C16 species (Fig. 5A). In contrast, all of the C18 acyl groups were predominately found at sn-1/sn-3 positions. This pattern is very similar to TAG from nitrogen-starved C. reinhardtii cells, where the amount of C16 FA at the sn-2 position is almost the same as in the iron-starved cells (Fig. 5B). Because in plants the enzyme responsible for attaching the acyl group at the sn-2 position has a preference for C16 FAs, this FA was most likely attached to the glycerol backbone inside the chloroplast, as shown for TAGs from nitrogen-deprived C. reinhardtii cells (41).
FIGURE 5.
Positional analysis of esterified fatty acids in TAG isolated from iron-starved C. reinhardtii cells compared with TAG from nitrogen-starved cells. A, partial digest on the TAG from C. reinhardtii cells after 24 h of iron (Fe) deprivation. The different products (tri-, di-, and monoacylglycerol (TAG, DAG, and MAG) and free fatty acids (FFA)) were separated by TLC and then analyzed as FAMEs by GC-FID. Direct TAG-FAME shows the results or TAG without lipase treatment as control. Molar ratio of C16 fatty acids (B), saturated fatty acids (C), monounsaturated fatty acids (D), and polyunsaturated fatty acids (E) at the sn-2 position separated from C. reinhardtii cells deprived for iron or nitrogen for 24 h. r.u., relative units.
We have shown that iron starvation results in an increase in saturated FAs for all glycerolipids investigated (TAG, DGTS, and MGDG). This could be a consequence of iron-containing desaturases being particularly sensitive to iron starvation and decreased intracellular iron or it may be a stress response. Therefore, we were interested to investigate whether other stresses (e.g. nitrogen starvation) will lead to increased levels of saturated FAs. TAGs isolated from iron- or nitrogen-starved C. reinhardtii cells for 24 h were analyzed by FAME or after R. arrhizus lipase treatment. The analysis showed that the impact on FA saturation is greater in iron-starved compared with nitrogen-starved cells. Specifically, we observed more saturated FAs and consequently less mono- and polyunsaturated FAs in TAG from iron-starved compared with TAG from nitrogen-deprived cells (Fig. 5, B–D).
Impact of Iron Starvation on FA Metabolism-related Genes
Iron starvation results in changes in FA desaturation levels, glycerolipid levels, and TAG accumulation. To investigate the changes in gene expression associated with these observations, we surveyed the C. reinhardtii transcriptome under iron-starved conditions. RNA was isolated from 0 to 48 h after transferring the cells from iron-replete (20 μm Fe-EDTA) to iron-deprived (0 μm Fe-EDTA) medium (supplemental Table S1). Several genes involved in de novo FA biosynthesis (BCX1, BCC1/2, BCR1, ACP1/2, MCT1, and PAP1/2) encoding β-carboxyltransferase, acetyl-CoA biotin carboxyl carrier 1/2, biotin carboxylase, acyl carrier protein 1, malonyl-CoA:ACP transacylase and phosphatidate phosphatase 1/2) were 2–3-fold down-regulated after 12 h of iron starvation. In contrast, a 3-ketoacyl-ACP-synthase (KAS2) and two putative β-ketoacyl synthases (KAS3 and Cre10.g438050) seemed to be more abundant after 0.5–4 h of iron starvation followed by a decrease to their initial levels (supplemental Table S1). Interestingly, PGPS3 encoding phosphatidylglycerolphosphate synthase was induced immediately after the cells were transferred to iron-deficient medium (0.5 h) and remained up-regulated for the entire 48-h time course. Our transcriptome analysis of iron-starved C. reinhardtii revealed changes in almost all FA desaturases (Table 2). The abundance of FAB2 transcripts was increased more than 2.5-fold after 8 h of iron deprivation and remained elevated for the entire 48-h time course. This is in contrast to decreased FAB2 polypeptide abundance observed after 24 h of iron starvation (Fig. 3A). Increased transcript levels were observed for other FA desaturases (e.g. FAD5b, FAD5c, FAD5d, DES6, and FAD2a), whereas FAD3, FAD7, FAD5a, Cre02.g103500 (coding for a putative sterol desaturase), and Cre16.g663950 (coding for a putative C-5 sterol desaturase) were mostly repressed after 48 h of iron deprivation (Table 2). In summary, we note that the transcript levels of all the FA desaturases, known to encode diiron enzymes, are changed. In contrast, fewer FA desaturases had changed transcript levels upon nitrogen starvation, and often they were changed in the opposite direction as observed upon iron starvation (see supplemental Table S1). Interestingly, two diacylglycerol kinases (KDG1 and KDG2) were induced under iron-deprived conditions (supplemental Table S1) indicating that under these conditions C. reinhardtii cells try to attenuate the DAG levels by converting it to phosphatidic acid.
TABLE 2.
Iron starvation has a major impact on fatty acid desaturases.
C. reinhardtii strain CC-4532 was grown photoheterotrophically under iron deprivation for 48 h. Samples were collected at various time points for RNA-Seq analysis. RPKM values represent the average from two biological replicates. Fold-changes were calculated relative to time point 0 h. Genes without names are indicates by Augustus 10.2 transcript IDs.
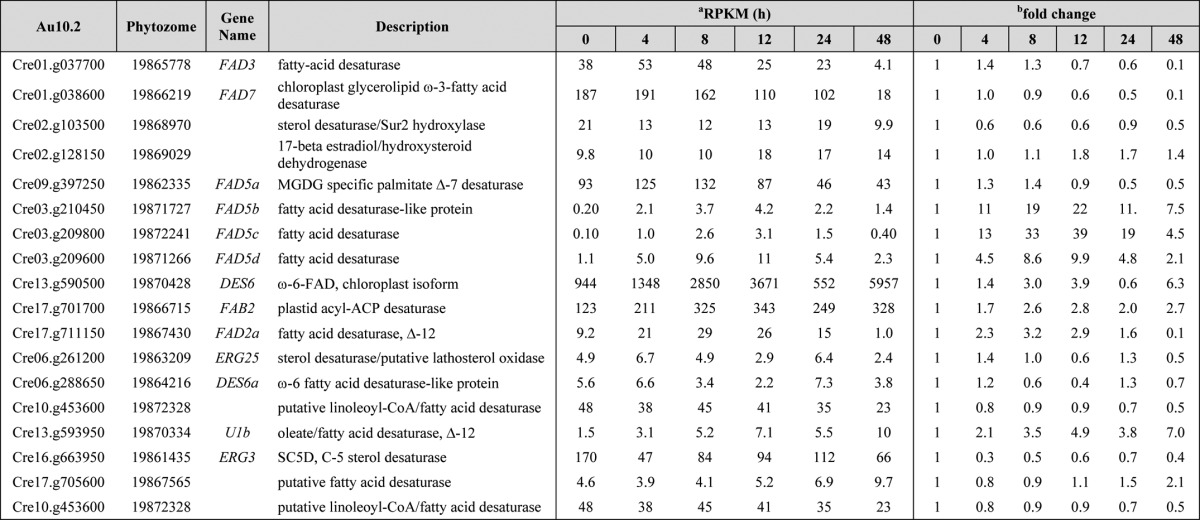
a RPKM, reads kilobase of mappable transcript length per million mapped reads.
b Fold-change relative to time 0 h and is derived from dividing the average of the two biological replicates for each time point.
We have demonstrated that the levels of glycerolipid MGDG were decreased as early as 2 h of iron starvation. The transcript levels of MGD1, coding for the enzyme responsible for the conversion of DAG to MGDG, were increased after 2 and 48 h of iron starvation, indicating a negative correlation between MGD1 mRNA levels and plastid MGDG content.
SAS1 (S-adenosylmethionine synthetase) and BTA1 (betaine lipid synthase) mRNAs coding for the two known enzymes involved in DGTS biosynthesis were strongly down-regulated (30- and 10-fold, respectively) (supplemental Table S1). These changes are consistent with the 2-fold decrease in DGTS levels observed after 48 h of iron deprivation.
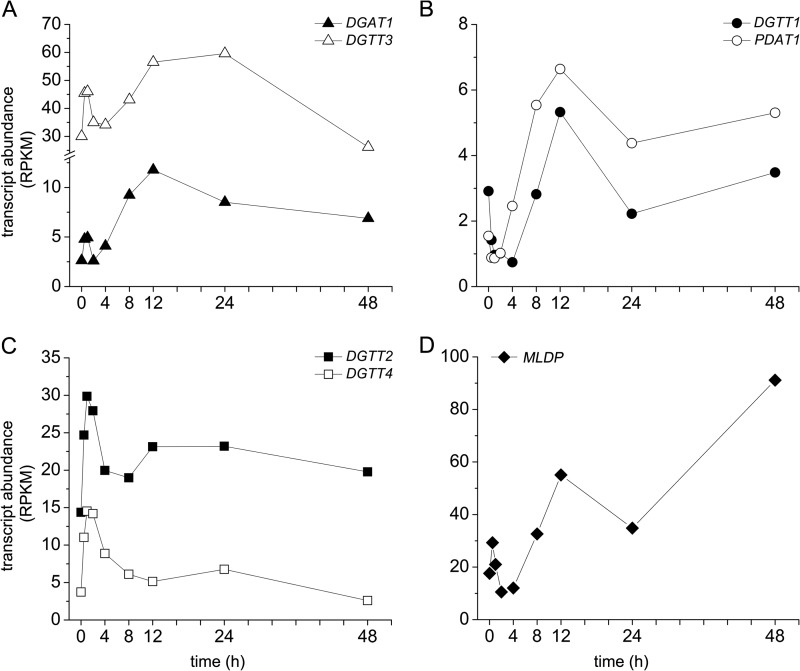
We have shown that C. reinhardtii cells accumulate TAGs after 24 h of iron deprivation, which are associated with the formation of lipid droplets. Therefore, we looked at transcript levels of DGATs in the 0–48-h iron-starvation experiment. The most abundant diacylglycerol acyltransferase is DGTT3, with transcript levels 2-fold higher after 24 h of iron deprivation compared with the 0-h time point (control). A similar trend of expression during the 0–48-h iron starvation was observed for DGAT1, but its overall abundance was lower (2.7 RPKM in 0 h control, which increased up to 11.5 RPKM at the 12 h time point) (Fig. 6A). DGTT1, a type 1 diacylglycerol acyltransferase and PDAT1 coding for a phospholipid diacylglycerol acyltransferase, showed similar patterns of expression. Both exhibited increasing transcript levels after 8 h and reached a maximum after 12 h of iron starvation (Fig. 6B). Noteworthy is that their overall transcript abundance was rather low and similar to DGAT1. Two other DGAT-encoding genes (DGTT2 and DGTT4) showed changes in transcript abundance only in the first 1–4 h of iron starvation (with a peak in expression at the 1-h time point). After 4 h of iron starvation, DGTT2 and DGTT4 mRNA levels decreased to almost the levels observed for the control (0 h) samples (Fig. 6C). DGTT5 was not expressed under iron starvation, similar to previous studies performed under nitrogen starvation (7). Moellering and Benning (42) showed that one major protein (MLDP) is associated with lipid droplet formation and MLDP mRNA repression affects lipid droplet size. Our RNA-Seq analysis of iron-deprived C. reinhardtii cells indicated an increase in MLDP transcript levels under iron deprivation (2.5-fold after 12 h and 6.5-fold after 48 h, respectively) (Fig. 6D and supplemental Table S1).
FIGURE 6.
Pattern of expression of diacylglycerol acyltransferases in response to iron starvation. mRNA transcript abundances are shown as quantified by RNA-Seq for DGAT1 and DGTT3 (A), DGTT1 and PDAT1 (B), and DGTT2 and DGTT4 (C). RPKM, reads/kilobase of mappable transcript length/million mapped reads.
Many putative lipase encoding genes showed an increase in their mRNA abundances, whereas for only 4–5 did we note decreased abundance (supplemental Table S1). Among the most highly induced of these are Cre05.g234800, Cre09.g407300, Cre12.g521650, Cre10.g422150, Cre06.g265850 (all coding for proteins of the esterase/lipase family), Cre08.g381250 (coding for a putative phospholipase A2), Cre12.g498750 and Cre07.g350000 (coding for putative triacylglycerol lipases). These putative lipase-encoding mRNAs were increased in abundance even after 0.5 h of iron starvation (supplemental Table S1). One of the genes most repressed in iron starvation is LIP1 (Cre02.g126050; 42-fold down-regulated after 48 h). LIP1 has been shown to be involved in TAG turnover in C. reinhardtii via degradation of DAG derived from TAG hydrolysis (43). The massive down-regulation of LIP1 correlates with the progressive increase in TAG accumulation and lipid droplet formation under iron deficiency. All these dramatic changes in the abundance of mRNAs coding for putative lipases suggest a remodeling of lipid membranes under conditions of iron deprivation.
DISCUSSION
Iron Starvation Has a Major Impact on Glycerolipid Content in C. reinhardtii
Previous studies indicated that C. reinhardtii accumulates TAGs when it is grown under conditions of macro (sulfur (11), phosphorus (10), and nitrogen (7, 13, 14)) and micro (zinc and iron (16, 17)) nutrient limitation or under conditions of abiotic stresses such as high salinity (12). Although the molecular details of the response to nitrogen starvation have received considerable attention (7, 14, 44) the biochemistry and molecular biology of other TAG-inducing conditions has not been explored.
Acyltransferases, type 1 DGAT1 or type 2 DGTTs, responsible for de novo TAG biosynthesis were identified previously in C. reinhardtii based on homology to A. thaliana (DGAT1 and DGAT2) and yeast (ARE1, ARE2, and DGA1, respectively) (45). In addition, plant and yeast phospholipid diacylglycerol transferases (PDAT1 and LRO1, respectively) have an ortholog in C. reinhardtii (PDAT1) (7, 45). In A. thaliana, PDAT1 and DGAT1 are both responsible for seed oil accumulation (46), and here we noted that C. reinhardtii PDAT1 and DGAT1 show a similar pattern of expression upon iron starvation as they do upon nitrogen starvation, validating the relevance of these enzymes for TAG accumulation.
The mRNAs encoding two type 2 diacylglycerol acyltransferases were also increased in abundance in iron-starved C. reinhardtii, DGTT1 and DGTT3. DGTT1 expression was increased also in nitrogen deprivation but no change in DGTT3 abundance was noted in those studies (7, 14). This is not a strain difference because the same strain was used here as compared with one of the previous studies. We conclude that there are clearly different signaling pathways controlling TAG biosynthesis and accumulation. The response to iron starvation is as fast as the one to nitrogen starvation, with mRNA abundances of DGAT1, DGTT1, DGTT3, and PDAT1 reaching the highest levels after 12–24 h of iron deprivation (Fig. 6). We conclude that the iron starvation-dependent TAG synthesis pathway relies also on both acyl-CoA-dependent and -independent acyltransferases. A recent study (9) has demonstrated that in addition to TAG synthesis, PDAT1 from C. reinhardtii mediates lipid turnover via its hydrolase activity using phospholipids, galactolipids, and TAG as substrates. Overexpression of plant DGAT1 and PDAT1 results in only a 2-fold increase in TAG content in A. thaliana (47), whereas overexpression of any one of three algal type 2 DGATs (DGTT1–DGTT3) in C. reinhardtii (48) did not increase TAG accumulation. However, in the C. reinhardtii work, it was not possible to ascertain whether the abundance of the proteins was increased in the overexpressing strains, because only mRNA levels were documented. Furthermore, the DGAT1 and PDAT1 genes (orthologs of the corresponding A. thaliana genes) were not tested. Nevertheless, it is likely that other factors, like metabolic regulation or substrate availability could limit TAG synthesis in C. reinhardtii.
Previous proteomic studies (42, 49) have identified a protein specific to algae to be associated with lipid droplets (MLDP). MLDP is induced under nitrogen deficiency in C. reinhardtii and RNAi-mediated depletion of MLDP increases lipid droplet size by about 40%, but does not increase the overall TAG content (42). Thus, MLDP has structural roles, but its specific molecular and biochemical functions remain to be shown. MLDP is highly induced in response to iron deprivation, and its transcript abundance correlates with the number and size of lipid droplets observed in response to iron starvation, supporting a role for this protein in lipid droplet formation (50).
One unforeseen finding was that iron starvation in C. reinhardtii leads to overall changes in relative glycerolipid abundance. The most dramatic impact was observed on MGDG levels that decreased very rapidly in response to iron starvation. On the other hand, the FA composition of MGDG does not change significantly, except for 16:2Δ7,10, 16:3Δ7,10,13, and 18:2Δ9,12, which were significantly elevated (p < 0.05). Because total FA levels remained stable over time upon iron starvation this suggests (a) comparable rates of synthesis and degradation and (b) that the more saturated MGDG species are metabolized very rapidly before we can detect them. Recently, it has been shown that PGD1, a MGDG-specific lipase can act predominantly on more saturated MGDG, and specifically on 16:0 and 18:1Δ9 from newly synthesized MGDG (51). PGD1 mRNA abundance in iron-starved cells is increased, suggesting that it might be involved in re-shuffling of saturated FA from MGDG to TAG.
The overall decrease in the MGDG levels correlates with a transient increase in MGD1 transcript levels after 2 h of iron starvation. It is likely that the increase in MGD1 mRNA indicates a compensatory change in response to decreased MGDG levels observed in iron starvation, suggesting a feedback regulation of MGD1 expression by MGDG levels. The more rapid response of MGDG levels versus chlorophyll-protein accumulation to iron starvation suggests that MGDG content may serve as a signal for changes in chlorophyll-protein content and assembly (20, 52).
The overall content of two other glycerolipids, DGDG and DGTS, decreased progressively over 48 h of iron starvation. Similar results were observed in iron-deficient pea (53) and peach (54) leaves, where the levels of the two major galactolipids, MGDG and DGDG, were decreased. Interestingly, this is in contrast to nitrogen starvation, where the total amount of membrane lipids was unchanged, except of DGDG, which increases (41, 51). One explanation for the decrease in the MGDG content upon iron starvation is that under these conditions photosynthetic membranes are remodeled and even degraded as previously shown to occur in iron-deficient C. reinhardtii (20, 21, 24). The acyl groups released from MGDG would then be recycled for TAG synthesis. However, TAGs do not contain considerable equal amounts of FAs typical for mature species of MGDG (i.e. 18:3 and 16:4) that would have to be recycled during degradation of the photosynthetic membrane. Alternatively, because cells are still dividing under these conditions and require the synthesis of membrane lipids, it seems possible that synthesis of MGDG and its modification by desaturases cannot keep up with demand explaining the lower levels. In fact, a recent study identified a FA desaturase in C. reinhardtii that is responsible for introducing the Δ4 double bond in 16:0 found almost exclusively at the sn-2 position in MGDG (55). Repression of the respective gene resulted not only in decreased cellular 16:4 content, but also in the overall reduction of MGDG, an essential lipid in the photosynthetic membrane. Although the transcript abundance of this MGDG-specific FA desaturase (FAD3) is marginally increased in the first 2 h of iron starvation, its mRNA abundance is 9-fold decreased after 48 h (Table 2 and supplemental Table S1). Thus, decreased desaturation of MGDG acyl groups due to iron deficiency could limit the assembly of the photosynthetic membrane.
In contrast to C. reinhardtii, the levels of TAG were dramatically decreased in response to iron deficiency in Saccharomyces cerevisiae (56). However, yeast cells grown under iron-deficient conditions show defects in ergosterol and sphingolipid biosynthesis, with impaired functions mostly in the steps catalyzed by iron-containing enzymes (Erg3, Erg25, Sur2, and Scs7) (56). Interestingly, Ole1, a Δ9-FA desaturase catalyzing the monodesaturation of C16:0 and C18:0 seems to be resistant to iron deficiency and correspondingly, no change in the levels of saturated and unsaturated FA has been observed (56).
C. reinhardtii cells have more saturated FA when completely deprived of iron. Because, TAG is naturally rich in saturated FA (16:0, 18:0), accumulation of TAG under iron starvation might account for the increase in saturated FA. Similarly, 20% more saturated FA were detected under nitrogen deficiency in C. reinhardtii. However, the total FA content in C. reinhardtii remained relatively stable under iron starvation conditions indicating that there is another mechanism that leads to elevated saturated FA content. The major catalytic step in FA desaturation is played by diiron enzymes, FA desaturases (57). Here we show that the amount of one of the most abundant FA desaturases in C. reinhardtii (FAB2) is reduced in response to iron starvation. Moreover, this decrease in FAB2 abundance correlates with the increase in TAG and lipid droplet accumulation, all occurring after 24 h of iron starvation. In contrast, mRNA abundance of FAB2 is increased in response to iron deprivation. This pattern of expression is often observed for iron-containing proteins, when the mRNA level is increased in an effort to make more protein, but the absence of iron prevents the formation of functional, mature protein (22, 23). However, the increase in saturated FA observed in iron-limited or -starved C. reinhardtii cells is more dramatic at the levels of C16:0, C16:2, and C18:2. Interestingly, the C18:0 content did not significantly increase upon iron limitation/starvation, suggesting that not all FA desaturases are equally affected under these conditions. Conversion of 18:2Δ9,12 to 18:3Δ9,12,15 is catalyzed by ω-3 desaturases, which are putatively encoded by FAD7 (plastid) and FAD3 (cytosol). Neither transcript abundance changed significantly under 24 h of iron starvation. Earlier desaturation steps are performed by DES6 (chloroplast) and FAD2 (cytosol) and these two mRNAs are increased upon iron starvation. Based on the RNA-Seq analysis, DES6 is the most abundant FA desaturase in C. reinhardtii and it could remain functional under iron starvation conditions by binding iron released, for example, by photosystem I degradation.
FA desaturation requires electrons from NADPH and reduced ferredoxin (57–59). Several studies, including the present one, demonstrated that under iron limitation and iron deprivation conditions, ferredoxin abundance decreases in C. reinhardtii (22, 23, 27, 40), suggesting that combined defects in all the iron-dependent enzymes required for FA desaturation have a negative impact on polyunsaturated FA levels. Noticeably, our study showed that ferredoxin and at least one FA desaturase (FAB2) have a similar pattern of protein expression and both have diminished abundance starting at 8–12 h of iron starvation. RNA-Seq analysis of iron-starved cells indicates that there are changes in the transcript abundance of several FA desaturases, reinforcing the idea that inactivation of FA desaturases in iron-starved C. reinhardtii cells is responsible for the decreased content of mono- and polyunsaturated FA. These observations indicate that FA desaturases are among the most sensitive targets of iron starvation and consequently all newly synthesized lipids will contain more saturated FAs.
Acknowledgment
We thank John Shanklin for kindly providing the FAB2 antibody (raised against purified protein from Ricinus communis).
This work was supported by the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy Grant DE-FD02-04ER15529 (to S. M.) and Air Force Office of Science Research Grants FA 9550-10-1-0095 (to M. P.) and FA9550-08-0165 (to C. B.).

This article contains supplemental Table S1.
- MGDG
- monogalactosyldiacylglycerol
- FA
- fatty acid
- TAG
- triacylglycerol
- DGDG
- digalactosyldiacylglycerol
- DGTS
- diacylglyceryl-N,N,N-trimethylhomoserine
- PDAT
- phospholipid:DAG acyltransferase
- CTH1
- copper target homolog 1
- DGAT
- diacylglycerol acyltransferase
- TAP
- Tris acetate-phosphate
- FAME
- fatty acid methyl ester
- DGTT
- DGAT type.
REFERENCES
- 1. Riekhof W. R., Ruckle M. E., Lydic T. A., Sears B. B., Benning C. (2003) The sulfolipids 2′-O-acyl-sulfoquinovosyldiacylglycerol and sulfoquinovosyldiacylglycerol are absent from a Chlamydomonas reinhardtii mutant deleted in SQD1. Plant Physiol. 133, 864–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Riekhof W. R., Sears B. B., Benning C. (2005) Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii. Discovery of the betaine lipid synthase BTA1Cr. Eukaryot. Cell 4, 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moore T. S., Du Z., Chen Z. (2001) Membrane lipid biosynthesis in Chlamydomonas reinhardtii. In vitro biosynthesis of diacylglyceryltrimethylhomoserine. Plant Physiol. 125, 423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang W., Mason C. B., Pollock S. V., Lavezzi T., Moroney J. V., Moore T. S. (2004) Membrane lipid biosynthesis in Chlamydomonas reinhardtii. Expression and characterization of CTP:phosphoethanolamine cytidylyltransferase. Biochem. J. 382, 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pineau B., Girard-Bascou J., Eberhard S., Choquet Y., Trémolières A., Gérard-Hirne C., Bennardo-Connan A., Decottignies P., Gillet S., Wollman F. A. (2004) A single mutation that causes phosphatidylglycerol deficiency impairs synthesis of photosystem II cores in Chlamydomonas reinhardtii. Eur. J. Biochem. 271, 329–338 [DOI] [PubMed] [Google Scholar]
- 6. Blouin A., Lavezzi T., Moore T. S. (2003) Membrane lipid biosynthesis in Chlamydomonas reinhardtii. Partial characterization of CDP-diacylglycerol:myo-inositol 3-phosphatidyltransferase. Plant Physiol. Biochem. 41, 11–16 [Google Scholar]
- 7. Boyle N. R., Page M. D., Liu B., Blaby I. K., Casero D., Kropat J., Cokus S. J., Hong-Hermesdorf A., Shaw J., Karpowicz S. J., Gallaher S. D., Johnson S., Benning C., Pellegrini M., Grossman A., Merchant S. S. (2012) Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287, 15811–15825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hernández M. L., Whitehead L., He Z., Gazda V., Gilday A., Kozhevnikova E., Vaistij F. E., Larson T. R., Graham I. A. (2012) A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued Arabidopsis seed oil catabolism mutants. Plant Physiol. 160, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoon K., Han D., Li Y., Sommerfeld M., Hu Q. (2012) Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 24, 3708–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weers P. M. M., Gulati R. D. (1997) Growth and reproduction of Daphnia galeata in response to changes in fatty acids, phosphorus, and nitrogen in Chlamydomonas reinhardtii. Limnol. Oceanogr. 42, 1584–1589 [Google Scholar]
- 11. Matthew T., Zhou W., Rupprecht J., Lim L., Thomas-Hall S. R., Doebbe A., Kruse O., Hankamer B., Marx U. C., Smith S. M., Schenk P. M. (2009) The metabolome of Chlamydomonas reinhardtii following induction of anaerobic H2 production by sulfur depletion. J. Biol. Chem. 284, 23415–23425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Siaut M., Cuiné S., Cagnon C., Fessler B., Nguyen M., Carrier P., Beyly A., Beisson F., Triantaphylidès C., Li-Beisson Y., Peltier G. (2011) Oil accumulation in the model green alga Chlamydomonas reinhardtii. Characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Z. T., Ullrich N., Joo S., Waffenschmidt S., Goodenough U. (2009) Algal lipid bodies. Stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell. 8, 1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miller R., Wu G., Deshpande R. R., Vieler A., Gärtner K., Li X., Moellering E. R., Zäuner S., Cornish A. J., Liu B., Bullard B., Sears B. B., Kuo M. H., Hegg E. L., Shachar-Hill Y., Shiu S. H., Benning C. (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154, 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen M., Tang H., Ma H., Holland T. C., Ng K. Y., Salley S. O. (2011) Effect of nutrients on growth and lipid accumulation in the green algae Dunaliella tertiolecta. Bioresour. Technol. 102, 1649–1655 [DOI] [PubMed] [Google Scholar]
- 16. Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S. S., Malasarn D. (2011) A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66, 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng X., Fei X., Li Y. (2011) The effects of nutritional restriction on neutral lipid accumulation in Chlamydomonas and Chlorella. Afr. J. Microbiol. Res. 5, 260–270 [Google Scholar]
- 18. Terry N. (1980) Limiting factors in photosynthesis. I. Use of iron stress to control photochemical capacity in vivo. Plant Physiol. 65, 114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stocking C. R. (1975) Iron deficiency and the structure and physiology of maize chloroplasts. Plant Physiol. 55, 626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moseley J. L., Allinger T., Herzog S., Hoerth P., Wehinger E., Merchant S., Hippler M. (2002) Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21, 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Busch A., Rimbauld B., Naumann B., Rensch S., Hippler M. (2008) Ferritin is required for rapid remodeling of the photosynthetic apparatus and minimizes photo-oxidative stress in response to iron availability in Chlamydomonas reinhardtii. Plant J. 55, 201–211 [DOI] [PubMed] [Google Scholar]
- 22. Urzica E. I., Casero D., Yamasaki H., Hsieh S. I., Adler L. N., Karpowicz S. J., Blaby-Haas C. E., Clarke S. G., Loo J. A., Pellegrini M., Merchant S. S. (2012) Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell 24, 3921–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsieh S. I., Castruita M., Malasarn D., Urzica E., Erde J., Page M. D., Yamasaki H., Casero D., Pellegrini M., Merchant S. S., Loo J. A. (2013) The proteome of copper, iron, zinc, and manganese micronutrient deficiency in Chlamydomonas reinhardtii. Mol. Cell. Proteomics 12, 65–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., Hippler M. (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7, 3964–3979 [DOI] [PubMed] [Google Scholar]
- 25. Nishio J. N., Taylor S. E., Terry N. (1985) Changes in thylakoid galactolipids and proteins during iron nutrition-mediated chloroplast development. Plant Physiol. 77, 705–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harris E. H. (2009) The Chlamydomonas Sourcebook, 2nd Ed., Academic Press, San Diego [Google Scholar]
- 27. Terauchi A. M., Peers G., Kobayashi M. C., Niyogi K. K., Merchant S. S. (2010) Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth. Res. 105, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quinn J. M., Merchant S. (1998) Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 297, 263–279 [DOI] [PubMed] [Google Scholar]
- 29. Allen M. D., del Campo J. A., Kropat J., Merchant S. S. (2007) FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii. Eukaryot. Cell 6, 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schloss J. A. (1990) A Chlamydomonas gene encodes a G protein β subunit-like polypeptide. Mol. Gen. Genet. 221, 443–452 [DOI] [PubMed] [Google Scholar]
- 31. Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate. A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 [Google Scholar]
- 33. Chen J. C., Hsieh S. I., Kropat J., Merchant S. S. (2008) A ferroxidase encoded by FOX1 contributes to iron assimilation under conditions of poor iron nutrition in Chlamydomonas. Eukaryot. Cell 7, 541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 35. Castruita M., Casero D., Karpowicz S. J., Kropat J., Vieler A., Hsieh S. I., Yan W., Cokus S., Loo J. A., Benning C., Pellegrini M., Merchant S. S. (2011) Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23, 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fischer W., Heinz E., Zeus M. (1973) The suitability of lipase from Rhizopus arrhizus delemar for analysis of fatty acid distribution in dihexosyl diglycerides, phospholipids and plant sulfolipids. Hoppe-Seylers Z. Physiol. Chem. 354, 1115–1123 [DOI] [PubMed] [Google Scholar]
- 37. Miquel M., Cassagne C., Browse J. (1998) A new class of Arabidopsis mutants with reduced hexadecatrienoic acid fatty acid levels. Plant Physiol. 117, 923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hutner S. H., Provasoli L., Schatz A., Haskins C. P. (1950) Some approaches to the study of the role of metals in the metabolism of microorganisms. Proc. Am. Philosophical. Soc. 94, 152–170 [Google Scholar]
- 39. Fox B. G., Shanklin J., Somerville C., Münck E. (1993) Stearoyl-acyl carrier protein δ9 desaturase from Ricinus communis is a diiron-oxo protein. Proc. Natl. Acad. Sci. U.S.A. 90, 2486–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Page M. D., Allen M. D., Kropat J., Urzica E. I., Karpowicz S. J., Hsieh S. I., Loo J. A., Merchant S. S. (2012) Fe sparing and Fe recycling contribute to increased superoxide dismutase capacity in iron-starved Chlamydomonas reinhardtii. Plant Cell 24, 2649–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fan J., Andre C., Xu C. (2011) A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 585, 1985–1991 [DOI] [PubMed] [Google Scholar]
- 42. Moellering E. R., Benning C. (2010) RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 9, 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li X., Benning C., Kuo M. H. (2012) Rapid triacylglycerol turnover in Chlamydomonas reinhardtii requires a lipase with broad substrate specificity. Eukaryot. Cell 11, 1451–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Msanne J., Xu D., Konda A. R., Casas-Mollano J. A., Awada T., Cahoon E. B., Cerutti H. (2012) Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169. Phytochemistry 75, 50–59 [DOI] [PubMed] [Google Scholar]
- 45. Merchant S. S., Kropat J., Liu B., Shaw J., Warakanont J. (2012) TAG, you're it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 23, 352–363 [DOI] [PubMed] [Google Scholar]
- 46. Zhang M., Fan J., Taylor D. C., Ohlrogge J. B. (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21, 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jako C., Kumar A., Wei Y., Zou J., Barton D. L., Giblin E. M., Covello P. S., Taylor D. C. (2001) Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 126, 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. La Russa M., Bogen C., Uhmeyer A., Doebbe A., Filippone E., Kruse O., Mussgnug J. H. (2012) Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J. Biotechnol. 162, 13–20 [DOI] [PubMed] [Google Scholar]
- 49. Nguyen H. M., Baudet M., Cuiné S., Adriano J. M., Barthe D., Billon E., Bruley C., Beisson F., Peltier G., Ferro M., Li-Beisson Y. (2011) Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii. With focus on proteins involved in lipid metabolism. Proteomics 11, 4266–4273 [DOI] [PubMed] [Google Scholar]
- 50. Liu B., Benning C. (2013) Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotechnol. 24, 300–309 [DOI] [PubMed] [Google Scholar]
- 51. Li X., Moellering E. R., Liu B., Johnny C., Fedewa M., Sears B. B., Kuo M. H., Benning C. (2012) A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24, 4670–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kobayashi K., Narise T., Sonoike K., Hashimoto H., Sato N., Kondo M., Nishimura M., Sato M., Toyooka K., Sugimoto K., Wada H., Masuda T., Ohta H. (2013) Role of galactolipid biosynthesis in coordinated development of photosynthetic complexes and thylakoid membranes during chloroplast biogenesis in Arabidopsis. Plant J. 73, 250–261 [DOI] [PubMed] [Google Scholar]
- 53. Abadia A., Ambard-Bretteville F., Remy R., Trémolièes A. (1988) Iron-deficiency in pea leaves. Effect on lipid composition and synthesis. Physiol. Plant. 72, 713–717 [Google Scholar]
- 54. Monge E., Pérez C., Pequerul A., Madero P., Val J. (1993) Effect of iron chlorosis on mineral nutrition and lipid composition of thylakoid biomembrane in Prunus persica (L.) Bastch. Plant Soil 154, 97–102 [Google Scholar]
- 55. Zäuner S., Jochum W., Bigorowski T., Benning C. (2012) A cytochrome b5-containing plastid-located fatty acid desaturase from Chlamydomonas reinhardtii. Eukaryot. Cell 11, 856–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shakoury-Elizeh M., Protchenko O., Berger A., Cox J., Gable K., Dunn T. M., Prinz W. A., Bard M., Philpott C. C. (2010) Metabolic response to iron deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 285, 14823–14833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shanklin J., Cahoon E. B. (1998) Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 611–641 [DOI] [PubMed] [Google Scholar]
- 58. Schmidt H., Heinz E. (1990) Desaturation of oleoyl groups in envelope membranes from spinach chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 87, 9477–9480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wada H., Schmidt H., Heinz E., Murata N. (1993) In vitro ferredoxin-dependent desaturation of fatty acids in cyanobacterial thylakoid membranes. J. Bacteriol. 175, 544–547 [DOI] [PMC free article] [PubMed] [Google Scholar]