Background: Because CPZEN-45 is a promising antituberculous drug candidate, the identification of the target is required.
Results: CPZEN-45 inhibits the decaprenyl-phosphate–GlcNAc-1-phosphate transferase of Mycobacterium tuberculosis and the corresponding enzyme of Bacillus subtilis responsible for initiation of cell wall synthesis.
Conclusion: CPZEN-45 inhibits a novel target in cell wall assembly.
Significance: This study is critical for launching CPZEN-45 and for exploitation toward new antituberculous drugs.
Keywords: Antibiotics Action, Bacillus, Cell Wall, Teichoic Acid, Tuberculosis, CPZEN-45, TagO, WecA, Caprazamycin B, Mycolylarabinogalactan
Abstract
Because tuberculosis is one of the most prevalent and serious infections, countermeasures against it are urgently required. We isolated the antitubercular agents caprazamycins from the culture of an actinomycete strain and created CPZEN-45 as the most promising derivative of the caprazamycins. Herein, we describe the mode of action of CPZEN-45 first against Bacillus subtilis. Unlike the caprazamycins, CPZEN-45 strongly inhibited incorporation of radiolabeled glycerol into growing cultures and showed antibacterial activity against caprazamycin-resistant strains, including a strain overexpressing translocase-I (MraY, involved in the biosynthesis of peptidoglycan), the target of the caprazamycins. By contrast, CPZEN-45 was not effective against a strain overexpressing undecaprenyl-phosphate–GlcNAc-1-phosphate transferase (TagO, involved in the biosynthesis of teichoic acid), and a mutation was found in the tagO gene of the spontaneous CPZEN-45-resistant strain. This suggested that the primary target of CPZEN-45 in B. subtilis is TagO, which is a different target from that of the parent caprazamycins. This suggestion was confirmed by evaluation of the activities of these enzymes. Finally, we showed that CPZEN-45 was effective against WecA (Rv1302, also called Rfe) of Mycobacterium tuberculosis, the ortholog of TagO and involved in the biosynthesis of the mycolylarabinogalactan of the cell wall of M. tuberculosis. The outlook for WecA as a promising target for the development of antituberculous drugs as a countermeasure of drug resistant tuberculosis is discussed.
Introduction
Tuberculosis, caused by Mycobacterium tuberculosis, has long been among the most serious of infectious diseases. The World Health Organization estimated 8.7 million incident cases and 12 million prevalent cases worldwide in 2011 (1). The HIV epidemic exacerbates the problem; it is estimated that the number of deaths due to tuberculosis rose by 1.4 million due to HIV coinfection (1). In addition, the spreading of multidrug-resistant tuberculosis, which is resistant to at least, rifampicin and isoniazid, is now recognized as an additional complexity. Currently, there are an estimated 600,000 cases of multidrug-resistant tuberculosis worldwide, including extensively drug-resistant tuberculosis; extensively drug-resistant tuberculosis is additionally resistant to fluoroquinolones and one or more of the three injectable second-line anti-tuberculosis drugs (amikacin, kanamycin, and capreomycin). Moreover, cases of tuberculosis that are resistant to all currently used drugs, which are called totally drug-resistant tuberculosis, have been reported (2, 3). Clearly, new antituberculous drugs, especially those that have a novel mode of action, are urgently required.
As a response to these serious situations, we had explored antitubercular agents from the culture broth of actinomycetes and isolated promising candidate compounds named caprazamycins, which are a group of novel liponucleoside antibiotics containing many different alkyl side chains (Fig. 1A) (4, 5). Although caprazamycins themselves are effective against Gram-positive bacteria and mycobacteria, they were modified to optimize them for treatment of tuberculosis by acid hydrolysis to yield caprazene (Fig. 1B), and caprazene 4-butylanilide (CPZEN-45, Fig. 1C) (6) was finally chosen as the most promising derivative. CPZEN-45 showed excellent antitubercular activity against not only drug-sensitive strains but also some multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis strains (patented data (US2011237530 (A1)) as described previously (7). In addition, CPZEN-45 had a narrower spectrum, better water solubility, and higher resistance to biotransformation in mouse serum compared with caprazamycins. CPZEN-45 is in preclinical study now, and no problems such as cytotoxicity and mutagenicity have been observed up to this time.
FIGURE 1.
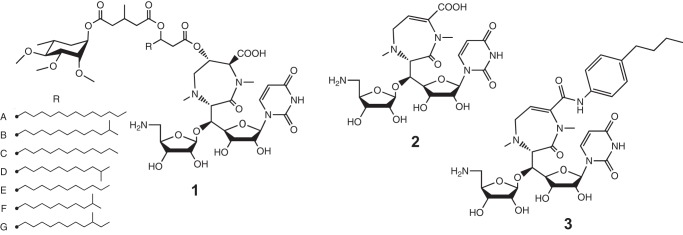
Structures of caprazamycin A-G (1); caprazene (2), which is a product of the acid hydrolysate of caprazamycins; and CPZEN-45 (3), which is 4-butylanilide of caprazene.
Caprazamycins are classified into the nucleoside antibiotics targeting bacterial translocase I (generally called MraY) involved in the biosynthesis of peptidoglycan; other such agents are liposidomycins, mureidomycins, muraymycins, and capuramycins (8–11). Because peptidoglycan is the main structural component of bacterial cell walls and is therefore essential for viability, MraY inhibitors, including caprazamycins, are effective against a broad spectrum of bacteria. In contrast, the spectrum of CPZEN-45 is narrower and is particularly effective against slowly growing mycobacteria, implying that CPZEN-45 has a different mode of action from caprazamycins.
In this report, the mode of action of both caprazamycin B, which is the most active analog among the caprazamycins, and CPZEN-45 were studied comparatively using Bacillus subtilis 168. With a combination of approaches such as evaluation of macromolecular biosynthesis, property of spontaneous drug-resistant strains or genetically modified strains, and enzymatic activity, the primary target of CPZEN-45 in B. subtilis was identified as TagO involved in the biosynthesis of teichoic acid, which is a major component of the cell wall of Gram-positive bacteria (12, 13). Furthermore, inhibition of WecA of M. tuberculosis, which is the ortholog of TagO of B. subtilis, by CPZEN-45 was also established.
EXPERIMENTAL PROCEDURES
Reagents
All analytical grade chemicals were purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan) or Sigma unless otherwise indicated.
Antibacterial Activity
The MICs2 of caprazamycin B and CPZEN-45 against bacteria shown in Fig. 2 were examined by a serial agar dilution method using Middlebrook 7H10 agar (BD Biosciences) with glycerol and OADC enrichment (BD Biosciences) for mycobacteria, or Mueller-Hinton agar (BD Biosciences) for the other microorganisms such as Staphylococcus aureus, Enterococcus faecalis, Streptococcus pneumoniae, B. subtilis, Micrococcus luteus, Escherichia coli, Shigella dysenteriae, Pseudomonas aeruginosa, and Klebsiella pneumoniae. MIC values were measured against mycobacteria strains after incubation for 2–21 days at 37 °C and against other bacteria after incubation for 18 h at 37 °C.
FIGURE 2.
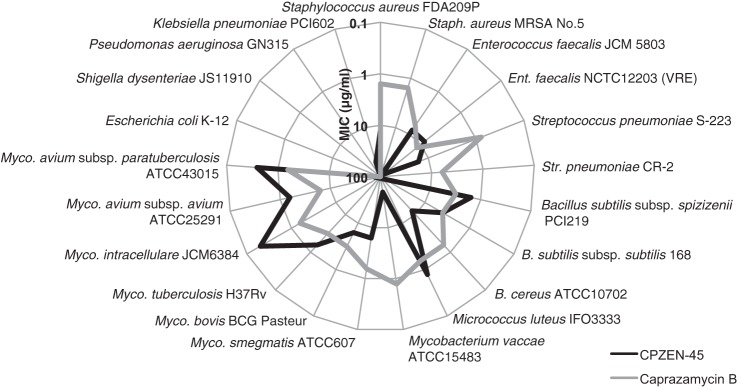
Radar chart of antibacterial activity of caprazamycin B (gray line) and CPZEN-45 (black line).
Inhibition of Macromolecular Synthesis
Inhibition of macromolecular synthesis was assayed by measurement of the incorporation of radiolabeled precursors into peptidoglycan, teichoic acid, fatty acid, DNA, RNA, and protein in the precipitate of a 10% trichloroacetic acid (TCA) extract of the cell as reported previously (14, 15). Log phase cell cultures (optical density at 600 nm (OD600) = 0.2) of B. subtilis 168 grown in nutrient broth were transferred to 96-well plates at 90 μl per well and then preincubated at 37 °C for 5 min with antibiotics. Nutrient broth consisted of 1% polypeptone (Nihon Pharmaceutical Co., Ltd., Tokyo, Japan), 1% nutrient (Kyokuto, Tokyo, Japan), and 0.2% NaCl in deionized water (pH was adjusted to 7.0 before sterilization). After preincubation, 10 μl of 10 nCi/μl N-acetyl-d-[1-14C]glucosamine, 1 nCi/μl [U-14C]glycerol, 100 nCi/μl [1-14C]acetate, 100 nCi/μl [methyl-3H]thymidine, 100 nCi/μl [5,6-3H]uridine, or 500 nCi/μl l-[4,5-3H]leucine were added to measure peptidoglycan, teichoic acid, fatty acid, DNA, RNA, and protein synthesis, respectively. All radiolabeled compounds were purchased from PerkinElmer Life Sciences. The mixtures were incubated for a further 10 min at 37 °C, and the reactions were stopped by adding an equal volume of 10% TCA. After 10 min of incubation at room temperature, the mixtures were transferred to 96-well filter plates (MultiScreenHTS, Millipore, Billerica, MA) and filter-washed three times with 5% TCA. After drying, the radioactivity remaining on the filter plate was counted by using a liquid scintillation counter (Tri-Carb 2800TR, PerkinElmer Life Sciences).
Construction of B. subtilis 168 Derivatives
B. subtilis strains and plasmids used are listed in Table 1. The antibiotic-resistant strains 168-BR and 168-45R1 were obtained by using continuous subculture. B. subtilis 168 cells were cultured in nutrient broth for 24 h at 37 °C with antibiotics at concentrations ranging from 0.125 to 8 times the MIC by 2-fold serial dilution; then the cells grown in the presence of one-quarter MIC of antibiotics were diluted 100-fold in nutrient broth and again cultured with various concentrations of antibiotics. This passage was repeated until the MIC had exceeded 128 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
The abbreviations used are as follows: ampR, ampicillin-resistant gene; tetR, tetracycline-resistant gene; cmlR, chloramphenicol-resistant gene; hygR, hygromycin-resistant gene; kanR, kanamycin-resistant gene; NIG, National Institute of Genetics, Japan.
| Strains or plasmids | Description | Origin |
|---|---|---|
| B. subtilis strains | ||
| 168 | trpC2 | ATCC |
| 168-BR | Caprazamycin B-resistant derivative obtained by continuous subculture with caprazamycin B | This work |
| 168-45R1 | CPZEZ-45-resistant derivative obtained by continuous subculture with CPZEN-45 | This work |
| 168-vec | 168 strain harboring control plasmid pHYcat | This work |
| 168-Yex | 168 strain harboring mraY-overexpression plasmid pHYcat-mraY | This work |
| 168-Oex | 168 strain harboring tagO-overexpression plasmid pHYcat-tagO | This work |
| M. smegmatis strains | ||
| mc2155 | ATCC 607 derivative, efficient plasmid transformation mutant | ATCC |
| 8a10 | mc2155 ΔwecA strain harboring p16Rkan-wecAMtb | This work |
| Plasmids | ||
| pHY300PLK | B. subtilis-E. coli shuttle vector, p15A and pAMα1 replicons, ampR, tetR | Takara Bio Inc. |
| pHT01 | B. subtilis expression vector, source of cmlR cassette | MoBiTec GmbH |
| pHYcat | p15A and pAMα1 replicons, cmlR | This work |
| pHYcat-mraY | pHYcat carrying an mraY fragment of B. subtilis 168 | This work |
| pHYcat-tagO | pHYcat carrying a tagO fragment of B. subtilis 168 | This work |
| p16R1 | Mycobacteria-E. coli shuttle vector, pAL5000 and pUC replicons, hygR | ATCC |
| pTH19kr | pSC101 replicon, source of kanR cassette | NIG |
| pUChphsacB-ΔwecAMsm | pUC replicon, hygR, sacB, wecA of M. smegmatis lacking the sequences between nucleotide positions 46 and 612 | This work |
| p16Rkan-wecAMtb | pAL5000 and pUC replicons, kanR, wecA of M. tuberculosis | This work |
B. subtilis 168 derivatives overexpressing mraY or tagO were constructed as follows. PCR fragments of mraY or tagO, including their own promoters, were inserted into the vector plasmid pHYcat, which was made from pHY300PLK (Takara Bio Inc., Shiga, Japan) and pHT01 (MoBiTec GmbH, Goettingen, Germany). The resulting plasmids pHYcat-mraY and pHYcat-tagO were introduced into B. subtilis 168, and the transformants obtained were designated 168-Yex and 168-Oex, respectively. The strain harboring pHYcat was also constructed as a vector control, named 168-vec. For amplification of mraY and tagO fragments, genomic DNA of B. subtilis 168 purified with the genomic DNA extraction kit (blood/bacteria/cultured cells) (RBC Bioscience Corp., New Taipei City, Taiwan) was used as the template, and the oligonucleotides listed in Table 2 were used as primers. PrimeSTAR GXL DNA polymerase, bacterial alkaline phosphatase, and T4 DNA ligase were purchased from Takara Bio Inc., and restriction enzymes were purchased from Takara Bio Inc. or New England Biolabs Inc. (Ipswich, MA). The transformation of B. subtilis was performed as described previously by Karamata and Gross (16). The MICs of the strains shown here were evaluated using the microbroth dilution method on LB medium (Wako Pure Chemical Industries, Ltd.). The strains were incubated for 18 h at 37 °C, and the MIC values were evaluated.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence | Description |
|---|---|---|
| 5′-mraY | AGGACATGAAACCTATCAGCAG | For amplification and sequencing of mraY gene of B. subtilis strain |
| 3′-mraY | CATAATGGTGATGAAGCGGAC | |
| 5′-tagO | CCGGACACAAGATTGGAATTGC | For amplification and sequencing of tagO gene of B. subtilis strain |
| 3′-tagO | AGCAGCACAAGCTCAAACAAC | |
| 5′-wecAMsm | CCGTCGCCATCGAACTGTTG | For amplification of wecA gene of M. smegmatis mc2155 |
| 3′-wecAMsm | TTTCGGGTTCTGCGAGCAC | |
| 5′-wecAMbo | GGCCGCTGCACCCGGTC | For amplification of wecA gene of M. bovis BCG |
| 3′-wecAMbo | GGCCGCACCGTACCACAAG | |
| wecAG1070T-F | GCATCGtTGCCTTCG | For mutation induction in wecA from M. bovis type to M. tuberculosis type (positions of mutation are indicated in lowercase letters) |
| wecAG1070T-R | GAAGGCAaCGATGCC |
Detection of the Mutation in B. subtilis 168 Derivatives
Genomic DNAs of B. subtilis strains 168, 168-BR, and 168-45R1 were isolated, and the DNA fragments containing mraY and tagO were amplified. The consequent fragments were sequenced using ABI3730XL (Invitrogen). Primers used for amplification and sequencing are listed in Table 2.
Evaluation of Enzyme Activity of MraY and TagO of B. subtilis
Enzyme activity of MraY and TagO was measured as described previously with some modifications (14, 17). The cell lysates of B. subtilis 168-Yex and 168-Oex were used for the enzyme sources of MraY and TagO, respectively. Log phase cell cultures (OD600 = 0.4) were collected by centrifugation for 10 min at 5000 rpm at 4 °C. Cells were washed twice with the same volume of ice-cold TMS buffer (50 mm Tris-HCl (pH 7.5), 625 mm sucrose, 10 mm MgCl2, 5 mm 3-mercapto-1,2-propanediol, 1 mm PMSF) and resuspended in 1/40 volume of TMS buffer. The cell suspensions were probe-sonicated 10 times for 30 s each with 30-s breaks on ice and centrifuged for 10 min at 3000 rpm to remove unbroken cells. These supernatants, cell lysates, were stored at −70 °C until use. For the evaluation of MraY activity, the reaction mixture, contained 20 ng of total protein/μl, 50 μm undecaprenyl phosphate, 0.02 μCi/μl [1-3H]undecaprenyl phosphate (American Radiolabeled Chemicals Inc., St. Louis, MO), 50 μm UDP-MurNAc-pentapeptide (UK Bacterial Cell Wall Biosynthesis Network, University of Warwick, Coventry, UK), 100 mm Tris-HCl, 20 mm MgCl2, 10 mm Triton X-100 in a total volume of 0.02 ml, followed by incubation for 20 min at 30 °C. The reaction was stopped by adding the same volume of 1-butanol, 6 m pyridine in acetic acid (2:1) and subsequent vigorous mixing, and the upper phase containing the radiolabeled substrate and the product was collected after centrifugation (15,000 rpm, 3 min). The resulting solution was chromatographed on a silica gel Kieselgel 60 F254 (Merck) by developing with CHCl3/MeOH/H2O, ∼14 m NH4OH (88:48:4:1) and subsequent autoradiography (Typhoon 9400, GE Healthcare). The intensity of the autoradiographic image was quantified with ImageJ software (National Institutes of Health).
For the evaluation of TagO activity, the reaction mixture used contained 250 ng of total protein/μl, 125 μm undecaprenyl phosphate, 20 μm UDP-GlcNAc, 0.05 μCi/μl UDP-[glucosamine-6-3H]GlcNAc (PerkinElmer Life Sciences), 100 mm Tris-HCl, 20 mm MgCl2, 1% CHAPS in a total volume of 0.02 ml, followed by incubation for 60 min at 30 °C. The reaction was stopped by adding 0.2 ml of CHCl3-MeOH (2:1), followed by subsequent vigorous mixing. The lower phase containing the radiolabeled product was collected by centrifugation (15,000 rpm, 3 min). The resulting solution was also chromatographed on silica gel plates by developing with CHCl3/MeOH/H2O, 14 m NH4OH (65:35:4:4) and subsequent autoradiography.
Construction of the M. smegmatis Strain That Lacks Its Own WecA and Is Expressing WecA of M. tuberculosis
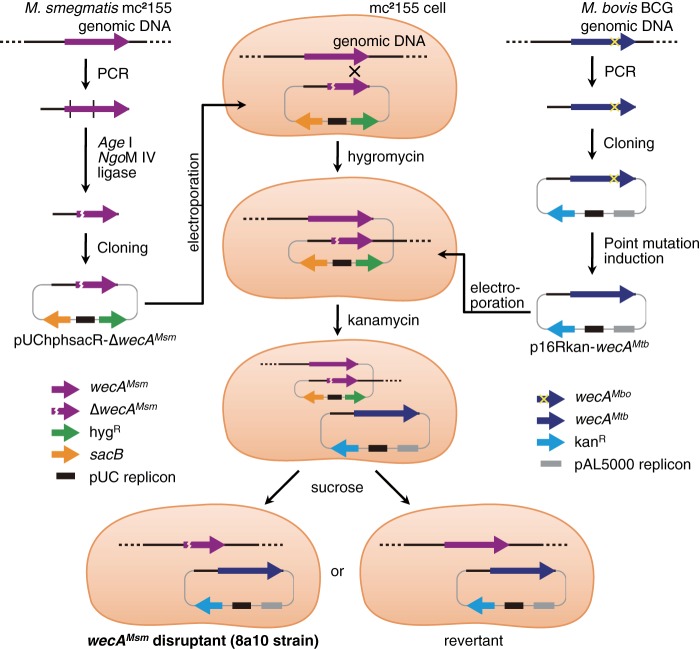
The M. smegmatis strain that lacks its original WecA and that expresses WecA of M. tuberculosis was constructed for the purpose of the detection of higher WecA activity and the avoidance of the biological hazard associated with M. tuberculosis. Mycobacterium smegmatis strains and the plasmids used are listed in Table 1; the oligonucleotides used for PCR amplification are shown in Table 2, and the strategy of the construction is illustrated in Fig. 3. First, plasmid containing the replication origin of pUC vectors, hygromycin-resistant gene, levansucrase gene sacB of B. subtilis, which confers bacterial cell sucrose sensitivity, and the partially deleted wecA gene of M. smegmatis was constructed. The partially deleted wecA gene was obtained with PCR amplification and subsequent restriction enzyme digestion with Age I and NgoM IV, and ligation. The consequent plasmid pUChphsacB-ΔwecAMsm was inserted into the genome of the mc2155 strain (18) using electroporation, as described previously with slight modifications (19). The resulting strain was transformed with p16Rkan-wecAMtb, which contains the sequences required for replication in E. coli and mycobacteria from p16R1 (20), the kanamycin-resistant gene from the plasmid pTH19kr (21), and the wecA gene of M. tuberculosis. The wecA fragment of M. tuberculosis was obtained through PCR amplification using the genomic DNA of Mycobacterium bovis BCG as the template and subsequent point mutation induction of the recombinant PCR product; there is only 1 base difference between the wecA of M. tuberculosis and M. bovis (nucleotide position 1070 is T in M. tuberculosis (encodes valine at position 357) but G in M. bovis (encodes glycine)). The resulting transformants were spread onto an agar plate containing 10% sucrose, and hygromycin-sensitive and sucrose-resistant strains were selected; those had the wild-type or disrupted wecA gene of M. smegmatis. The genotype of strains obtained was confirmed by genomic PCR, Southern blotting, transcription analysis by using RT-PCR, and subsequent sequencing. One of the resulting strains that had the disrupted wecA gene of M. smegmatis was designated M. smegmatis 8a10.
FIGURE 3.
Strategy for the construction of the M. smegmatis mc2155 derivative strain, containing the WecA of M. tuberculosis expressed from the multicopy plasmid instead of the original. The PCR fragment of wecA of M. smegmatis (wecAMsm) was amplified using the genomic DNA of the mc2155 strain as the template. The consequent DNA fragment was digested with restriction enzymes Age I and NgoM IV, and the 5′-end and 3′-end fragments were ligated to induce deletion from nucleotide positions 46–612 of wecAMsm. This fragment (ΔwecAMsm) was inserted into a plasmid containing the hygromycin-resistant gene (hygR), the levansucrase gene (sacB), and pUC replicon, and the resulting plasmid, pUChphsacB-ΔwecAMsm, was introduced into M. smegmatis mc2155 by electroporation. Because pUChphsacB-ΔwecAMsm does not have a replication origin for mycobacteria, only when the plasmid was integrated into the genome did the transformants show resistance to hygromycin. The resulting strain was transformed with another plasmid, p16Rkan-wecAMtb, by electroporation. This plasmid was constructed as follows: the PCR fragment of wecA of the BCG strain of M. bovis (wecAMbo) was amplified using the genomic DNA of BCG as the template. This fragment was cloned into a plasmid containing the kanamycin-resistant gene (kanR), the pAL5000 replicon for mycobacteria, and the pUC replicon. The resulting plasmid was used as template of another PCR to alter wecAMbo to wecA of M. tuberculosis (wecAMtb), which has 1 base difference from wecAMbo. Finally, the resulting strain, which is resistant to hygromycin and kanamycin, was challenged on agar plates containing 10% sucrose. Because the levansucrase encoded by the sacB gene leads to sucrose sensitivity, when integrated plasmid pUChphsacB-ΔwecAMsm was excluded from the genome, the transformant became sucrose-resistant again. The resulting sucrose-resistant and hygromycin-sensitive transformants had wild-type or disrupted wecA; thus, the genotype of each cell was analyzed by Southern blotting and transcription analysis using RT-PCR and subsequent sequencing.
Evaluation of Enzyme Activity of WecA of M. tuberculosis
Membrane fraction containing WecA of M. tuberculosis or M. smegmatis was prepared from M. smegmatis 8a10 as described by Mikusová et al. (22) with slight modifications. 10 g wet weight of M. smegmatis cells, which were grown to late log phase (2% inoculation, 24-h cultivation at 37 °C) in glycerol/alanine medium (23), were resuspended in 30 ml of buffer A (containing 50 mm MOPS (adjusted to pH 8.0 with KOH), 5 mm 3-mercapto-1,2-propanediol, and 10 mm MgCl2) and were disrupted in a French press (Thermo Spectronic French Pressure Cell Press; 16,000 p.s.i.) at 4 °C. The disrupted cells were centrifuged at 3000 × g for 10 min at 4 °C. The pellet was resuspended in buffer A to a final volume of 20 ml and again French pressed and centrifuged. The supernatants were combined and centrifuged at 27,000 × g for 12 min at 4 °C, and the membrane fraction was obtained by centrifugation of the supernatant at 100,000 × g for 60 min at 4 °C. The supernatant was carefully removed, and the precipitated membrane fraction was gently and superficially washed with buffer A and finally suspended in 0.4–0.5 ml of this buffer (protein concentration 15–20 mg/ml).
The reaction of WecA was carried out as follows: the reaction mixture, containing membrane fraction (40 μg of protein), 0.05 μCi/μl UDP-[glucosamine-6-3H]GlcNAc, 60 μm ATP in a total volume of 0.02 ml of buffer A, was incubated at 37 °C for 30 min. The reaction was stopped and analyzed by the same procedure as for the evaluation of TagO activity described above.
RESULTS
CPZEN-45 Showed an Antibacterial Spectrum Different from That of the Parent Compound Caprazamycin B
The radar chart of the antibacterial activity of caprazamycin B and CPZEN-45 is shown in Fig. 2. Caprazamycin B was uniformly effective against all tested Gram-positive bacteria, including mycobacteria with MICs ranging between 0.78 and 12.5 μg/ml. In contrast, CPZEN-45 was particularly effective against slowly growing bacteria such as M. tuberculosis H37Rv, Mycobacterium intracellulare JCM3684, Mycobacterium avium subsp. avium ATCC25291, and M. avium subsp. paratuberculosis ATCC43015 with MICs of 0.2–1.56 μg/ml, although it was not effective against pathogenic Gram-positive bacteria such as S. aureus strains FDA209P and MRSA No. 5, E. faecalis strains JCM 5803 and NCTC122103, and S. pneumoniae strains S-223 and CR-2 with MICs of 8–100 μg/ml. Neither caprazamycin B nor CPZEN-45 showed clear antibacterial activity against Gram-negative bacteria.
CPZEN-45 Inhibited Glycerol Incorporation Dominantly in B. subtilis
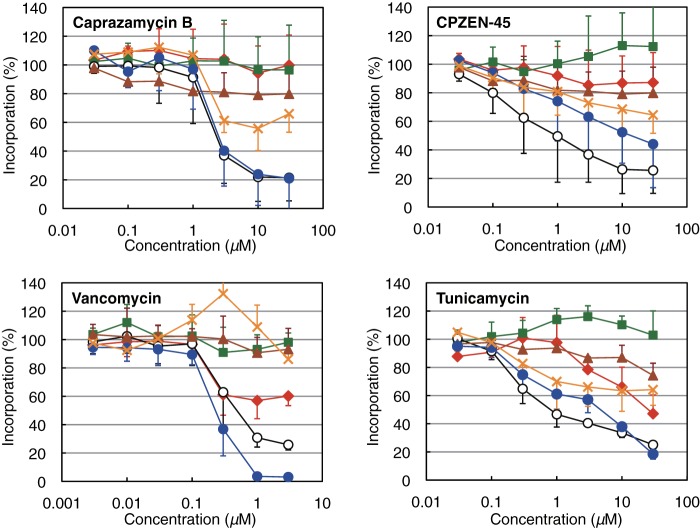
For the purpose of identification of the primary target of CPZEN-45, the effect of caprazamycin and CPZEN-45 on the incorporation of radiolabeled precursors into cellular macromolecules was evaluated in B. subtilis 168 (Fig. 4) as described (14, 15). Caprazamycin B inhibited the incorporation of both [1-14C]GlcNAc and [U-14C]glycerol with IC50 of less than 3 μm. In contrast, CPZEN-45 inhibited [U-14C]glycerol incorporation with IC50 of 1 μm, but the IC50 values for the inhibition of [1-14C]GlcNAc incorporation were more than 10 μm.
FIGURE 4.
Effects of caprazamycin B, CPZEN-45, vancomycin, and tunicamycin on the incorporation of radiolabeled precursors into the macromolecules of B. subtilis 168. The percentage of the incorporation of [methyl-3H]thymidine (for DNA, squares), [5,6-3H]uridine (for RNA, diamonds), l-[4,5-3H]leucine (for protein, triangles), [1-14C]acetate (for fatty acids, crosses), N-acetyl-d-[1-14C]glucosamine (for peptidoglycan, closed circles), and [U-14C]glycerol (for teichoic acid, open circles) are shown. Each plot and error bars represent the average and standard deviation of three independent trials.
The effect of vancomycin and tunicamycin was also observed. Vancomycin inhibits peptidoglycan biosynthesis by binding to lipid II, which contains a monomeric unit of peptidoglycan, and tunicamycin inhibits the biosynthesis of teichoic acid by inhibiting TagO (17, 24, 26). The pattern of inhibition curves of vancomycin was similar to caprazamycin B, whereas tunicamycin was similar to CPZEN-45.
Effects of Caprazamycin B and CPZEN-45 on the Viability of B. subtilis 168-derivatized Strains
Unlike caprazamycin B, CPZEN-45 inhibited the incorporation of glycerol only. In B. subtilis 168, glycerol was mainly incorporated into teichoic acid, suggesting that CPZEN-45 may inhibit the biosynthesis of teichoic acid. To evaluate this possibility, spontaneous mutants of B. subtilis 168 strain resistant to caprazamycin B and CPZEN-45, designated as 168-BR and 168-45R1, respectively, were obtained. The 168-BR strain was 32-fold more resistant to caprazamycin B than the parent strain but remained sensitive to CPZEN-45. In contrast, the 168-45R1 strain showed more than 32-fold higher resistance to CPZEN-45 but was still sensitive to caprazamycin B. The 168-BR and 168-45R1 strains showed 16- and 4-fold higher resistance to tunicamycin, respectively, but both strains showed similar vancomycin sensitivity to their parent 168 strain (Table 3).
TABLE 3.
Drug susceptibility and genotype of B. subtilis strains
| B. subtilis strains | MIC |
Genotype |
||||
|---|---|---|---|---|---|---|
| Caprazamycin B | CPZEN-45 | Tunicamycin | Vancomycin | mraY | tagO | |
| μg/ml | ||||||
| 168 | 4 | 4 | 0.25 | 0.25 | Wild type | Wild type |
| 168-BR | >128 | 4 | 4 | 0.25 | Wild type | Wild type |
| 168-45R1 | 4 | >128 | 1 | 0.5 | Wild type | T728G (I243S) |
| 168-vec | 4 | 8 | 1 | 0.5 | Wild type | Wild type |
| 168-Yex | >128 | 8 | 1 | 0.5 | Overexpression | Wild type |
| 168-Oex | 4 | 64 | 16 | 1 | Wild type | Overexpression |
In both resistant strains, the mraY and tagO genes, which encode translocase-I (generally called MraY) involved in biosynthesis of peptidoglycan, and undecaprenyl-phosphate GlcNAc-1-phosphate transferase (called TagO in B. subtilis) involved in teichoic acid biosynthesis, respectively, were sequenced and compared with their parent 168 strain. Although no mutation was found in the mraY gene from both strains, a nonsynonymous mutation of thymine-728 to guanine (T728G) in tagO, resulting in replacement of the isoleucine at position 243 by a serine (I243S), was detected only in the CPZEN-45-resistant 168-45R1 strain.
To confirm the involvement of tagO in the resistance to CPZEN-45, B. subtilis 168-derivatized strains overexpressing tagO and mraY, designated as 168-Oex and 168-Yex, respectively, were constructed using pHY300PLK-based plasmid pHYcat. The expression level of tagO mRNA in the 168-Oex strain and the level of mraY in the 168-Yex strain were 28- and 30-fold higher than in the vector control strain (designated as 168-vec), respectively. The 168-Oex strain was resistant to CPZEN-45 and tunicamycin but was still sensitive to caprazamycin B compared with the vector control strain 168-vec, and the 168-Yex strain was resistant to caprazamycin B but was sensitive to CPZEN-45 and tunicamycin (Table 3). These three strains displayed similar sensitivity to vancomycin. The results shown here implied that the target of caprazamycin B was MraY and that of CPZEN-45 was TagO.
Caprazamycin B Inhibited MraY and CPZEN-45 Inhibited TagO in B. subtilis
The effects of caprazamycin B and CPZEN-45 on the activity of MraY and TagO were evaluated using the sonicated cell lysate of 168-Yex and 168-Oex as the enzyme source and radiolabeled substrates, [1-3H]undecaprenyl phosphate for MraY and UDP-[glucosamine-6-3H]GlcNAc, for TagO enzyme activity estimations (Fig. 5). The IC50 values of caprazamycin B and CPZEN-45 against MraY enzyme activity were ∼50 and 400 ng/ml, respectively. In contrast, caprazamycin B and CPZEN-45 affected TagO with IC50 values of around 150 and 50 ng/ml, respectively. The effect of tunicamycin was similar to CPZEN-45, even though weaker. Vancomycin even at 4 mg/ml did not affect the activities of MraY and TagO (single trial; 100 and 108% residual activity of control, respectively).
FIGURE 5.
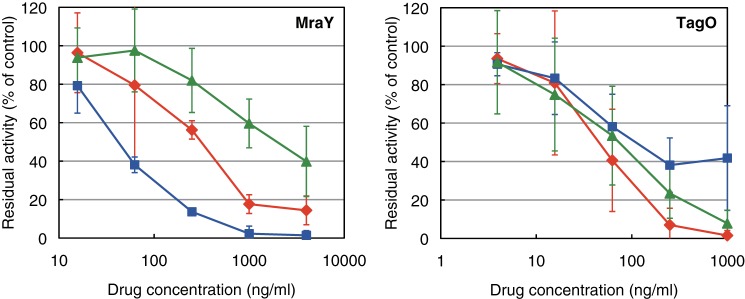
Effects of caprazamycin B, CPZEN-45, and tunicamycin on the activities of MraY and TagO in B. subtilis 168. The inhibitory curves of CPZEN-45 (diamonds), caprazamycin B (squares), and tunicamycin (triangles) on the activity of MraY and TagO are presented. Each plot and error bars represent the average and standard deviation of three independent trials.
CPZEN-45 Inhibited WecA of M. tuberculosis, Which Is the Ortholog of TagO in B. subtilis
We examined whether CPZEN-45 also inhibits WecA, which is the ortholog of TagO in B. subtilis. For this purpose, M. smegmatis cells lacking its original WecA but expressing WecA of M. tuberculosis was constructed as shown in Fig. 3. Subsequently, a membrane fraction of the strain was isolated, and the effect of CPZEN-45 on WecA activity was evaluated. WecA activity was strongly inhibited with IC50 values of ∼40 ng/ml (Fig. 6). Tunicamycin also inhibited WecA activity, but caprazamycin B was not effective compared with CPZEN-45 and tunicamycin. Vancomycin even at 4 mg/ml did not affect WecA activity (single trial; 90% residual activity of control). Additionally, CPZEN-45 also inhibited the activity of WecA of both M. smegmatis mc2155 (Fig. 7) and M. tuberculosis mc26230 (preliminary data) with an identical IC50 value of 4.4 ng/ml.
FIGURE 6.
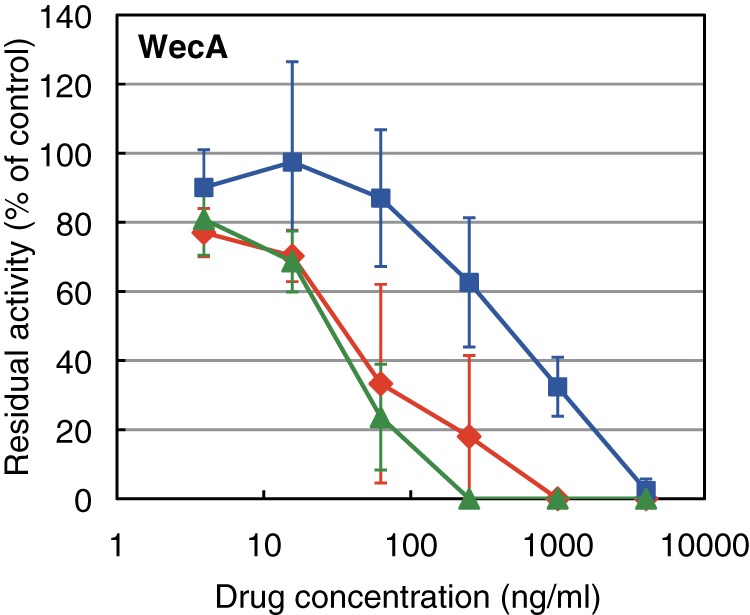
Effects of caprazamycin B, CPZEN-45, and tunicamycin on the activities of WecA of M. tuberculosis expressed in M. smegmatis 8a10. The inhibitory curves of CPZEN-45 (diamonds), caprazamycin B (squares), and tunicamycin (triangles) on the activity are presented. Each plot and error bars represent the average and standard deviation of three independent trials.
FIGURE 7.
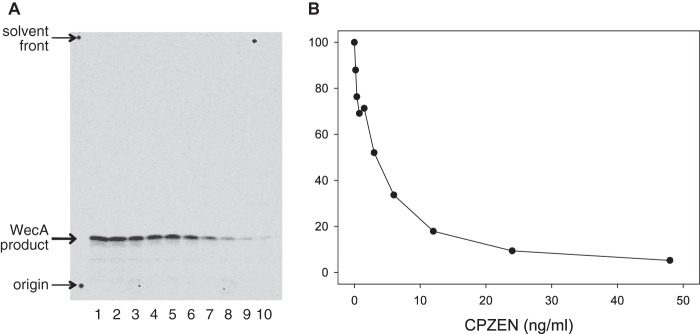
Effect of CPZEN-45 on the WecA activity of cytoplasmic membranes of M. smegmatis mc2155. A, TLC, developed as described above and subjected to radioautography, shows the effects of CPZEN-45 on the synthesis of the immediate product of WecA, the polyprenyl-P-P-GlcNAc. Preparation of membranes, cell-free assay conditions, including use of UDP-[U-14C]GlcNAc, were as described (22). B, CPZEN-45 was calculated to inhibit WecA of M. smegmatis strongly with an IC50 of 4.4 ng/ml. Data shown here are typical of three independent trials.
DISCUSSION
In this study, the mode of action of CPZEN-45, which is a promising antituberculous drug candidate, was studied by comparison with one of the parent compounds, caprazamycin B. In the first step to probe the comparative mechanisms of action of caprazamycin and CPZEN-45 on B. subtilis, the effects on macromolecular biosynthesis were evaluated (Fig. 4). Caprazamycin B inhibited incorporation of both radioactive GlcNAc and glycerol equivalently, whereas CPZEN-45 inhibited only glycerol incorporation. The inhibitory effect of caprazamycin B on the incorporation of GlcNAc and glycerol corresponded well to its MIC values against B. subtilis 168, demonstrating that the inhibition of the incorporation of GlcNAc and glycerol is directly linked to the antibacterial activities. In contrast, the MIC of CPZEN-45 was four times higher than the IC50 value of glycerol incorporation, implying that strong inhibition of glycerol incorporation is required for effective antibacterial activity of CPZEN-45.
Generally, in bacteria, GlcNAc is mainly incorporated into peptidoglycan of the bacterial cell wall, unlike glycerol that is mainly incorporated into the phospholipids of cellular membrane. However, in B. subtilis 168, glycerol is primarily incorporated into teichoic acid (24). Teichoic acids are polysaccharides or polymers of glycerol phosphate, ribitol phosphate, or sugar phosphates attached to peptidoglycan of the cell wall in many species of Gram-positive bacteria (27), and in the case of B. subtilis 168, the dominant teichoic acid is the polymer of glycerol phosphate (Fig. 8A). Because the biosynthesis of peptidoglycan and teichoic acid is thought to occur in concert (28), it is reasonable to think that the decrease of GlcNAc incorporation reflected in the inhibition of MraY, the resulting interruption of the biosynthesis of peptidoglycan, and the decrease in glycerol incorporation also occurred because de novo synthesis of peptidoglycan, to which teichoic acid is attached, was halted. The slope of the inhibitory curves in the case of vancomycin supported this hypothesis. In contrast, only inhibition of glycerol incorporation by CPZEN-45 and tunicamycin was observed because inhibition of the biosynthesis of teichoic acid occurred independently.
FIGURE 8.
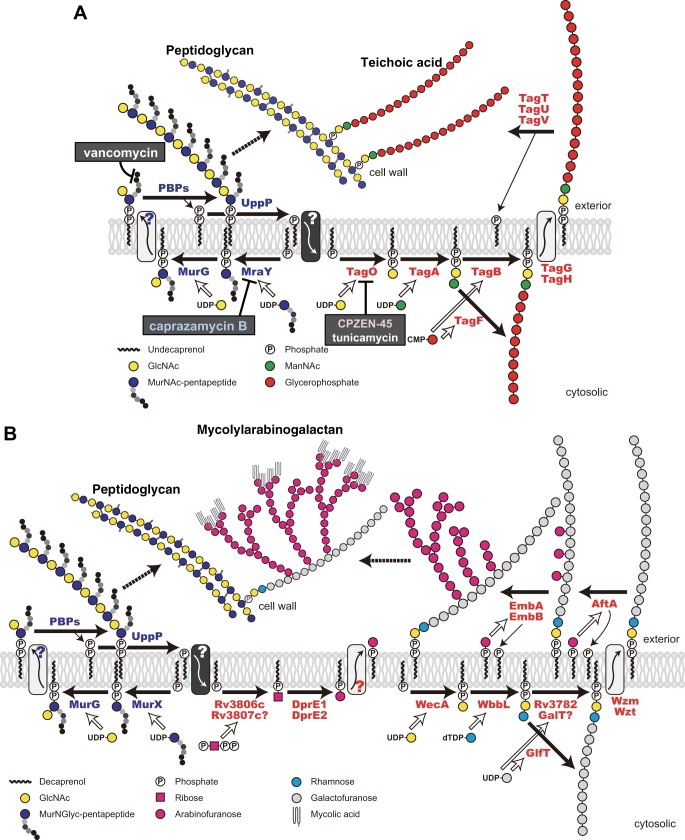
Overview of the biosynthetic pathway of peptidoglycan and teichoic acid in B. subtilis 168 (modified from Ref. 13) (A) and the biosynthetic pathway of peptidoglycan and mycolylarabinogalactan in M. tuberculosis (B). MurX and WecA are the orthologs of MraY and TagO in B. subtilis, respectively.
Considering that MraY is the target of caprazamycin B, TagO became the focal point as the target for the effects of CPZEN-45 on teichoic acid biosynthesis. TagO is a membrane protein, which mediates the first step of the biosynthesis of teichoic acid, that is the transfer of GlcNAc-1-phosphate from UDP-GlcNAc to undecaprenyl phosphate with the formation of undecaprenyl-P-P-GlcNAc. In contrast, MraY is the paralog of TagO and catalyzes the same reaction except its substrate is not UDP-GlcNAc but UDP-MurNAc pentapeptide (29). Thus, TagO, which is structurally and functionally similar to MraY, was worthy to be considered as the target of CPZEN-45, obviously a derivative of the MraY inhibitor caprazamycin B. This hypothesis was supported by the observation that tunicamycin, a known inhibitor of the TagO ortholog, showed a similar inhibitory curve as CPZEN-45 in macromolecular biosynthesis (Fig. 4).
To confirm this hypothesis, spontaneous mutants resistant to caprazamycin B and CPZEN-45 were obtained, and the genes encoding MraY and TagO were sequenced. Neither strain showed cross-resistance between caprazamycin B and CPZEN-45, implying different modes of action of these drugs (Table 3). Moreover, a nonsynonymous mutation of T728 to G in tagO was observed in the CPZEN-45-resistant strain, suggesting that the main target of CPZEN-45 is TagO. This mutation resulted in replacement of the isoleucine at position 243 by a serine, which is not located around the cytosolic active site but in the transmembrane region, implying that this mutation leads to structural alterations resulting in loss of the interaction between TagO and CPZEN-45 without any effect on the binding of the primary substrates. Additionally, the MraY- and TagO-overexpressing strains were resistant only against caprazamycin B and CPZEN-45, respectively (Table 3), further evidence in support of MraY and TagO as the targets of caprazamycin B and CPZEN-45, respectively.
Results of the effects of caprazamycin B and CPZEN-45 on the enzymatic activity of MraY and TagO supported these contentions. Predictably, MraY activity was inhibited by caprazamycin B rather than by CPZEN-45, whereas TagO was effectively inhibited by CPZEN-45 (Fig. 5). Additionally, caprazamycin B also inhibited TagO and CPZEN-45 inhibited MraY at these higher concentrations. Considering that CPZEN-45 is a derivative of caprazamycin B and that MraY and TagO are paralogs, this observation is expected. Some clinically successful antibiotics inhibit multiple targets (30). For instance, the β-lactams inhibit several penicillin-binding proteins; cycloserine inhibits alanine racemase and d-alanyl-d-alanine ligase; fluoroquinolones inhibit DNA topoisomerase IV and DNA gyrase; and the antituberculous drug ethambutol inhibits three of the arabinosyltransferases, EmbA, EmbB, and EmbC (31). Considering that CPZEN-45 partially inhibited the incorporation of radiolabeled GlcNAc (Fig. 4), it is likely that MraY inhibition is also partially involved in its antibacterial activity, likewise in the case of caprazamycin B.
Finally, the effect of CPZEN-45 on the enzymatic activity of WecA (decaprenyl-phosphate–GlcNAc-1-phosphate transferase, also called Rfe) of M. tuberculosis, which is the ortholog of the TagO of B. subtilis, was examined using the M. smegmatis strain, which had WecA of M. tuberculosis instead of its original one. As expected, WecA activity of the membrane fraction of the strain was inhibited by CPZEN-45 with a much lower concentration than the MIC against the M. tuberculosis H37Rv strain, indicating that the antibacterial activity of CPZEN-45 was due to the inhibition of WecA.
The fact that CPZEN-45 inhibited WecA of M. tuberculosis is the most important point in this report, suggesting that WecA is a novel target for antituberculous drugs. Mycobacteria do not contain teichoic acid but are characterized instead by mycolylarabinogalactan (Fig. 8B) (32). Although the structure of mycolylarabinogalactan is quite different from teichoic acid, the first step in its biosynthesis is identical; both WecA and TagO are polyprenyl-phosphate–GlcNAc-1-phosphate transferases. Because mycolylarabinogalactan is essential and some antituberculous drugs, such as ethambutol, and the new benzothiazinones, such as BTZ043 (33), target enzymes involved in mycolylarabinogalactan biosynthesis, WecA may be a promising target for a new generation of antituberculous drugs. In fact, WecA is apparently essential for the growth of M. smegmatis (34); however, WecA was initially reported to be nonessential in M. tuberculosis arising from a comprehensive study of a library of transposon insertion mutants (35, 36); more recently, however, WecA was reclassified as essential, through reevaluation of the transposon mutant library by deep sequencing and subsequent statistical analysis (37).
Some Gram-positive pathogenic bacteria were shown to be resistant to CPZEN-45 (Fig. 2). This apparent anomaly can be explained on the basis of the essentiality of the TagO/WecA ortholog. It is reported that the ortholog is nonessential in S. aureus (38) and E. faecalis (39) and that the ortholog is not required for biosynthesis of teichoic acid in S. pneumoniae (25), although it is essential in B. subtilis (12).
In conclusion, categorical statements can be made, namely that CPZEN-45, derived from caprazamycin B, an established antitubercular agent due to its inhibition of MraY, strongly inhibits TagO of B. subtilis and its WecA ortholog in M. tuberculosis. In addition, the TagO/WecA ortholog is nonessential for the major pathogenic bacteria with the exception of Myocbacterium spp., indicating that WecA provides a promising target in particular for exploitation toward new antituberculous drugs. Additional studies, such as genotyping of CPZEN-45-resistant strains of M. tuberculosis, kinetics of the interaction of WecA and CPZEN-45, and evaluation of its effect on MurX of M. tuberculosis (an ortholog of MraY of B. subtilis) will be required prior to full acceptability of the interaction of CPZEN-45 and WecA as the sole mode of action. Nevertheless, CPZEN-45 itself is a promising antituberculous drug candidate in light of its postulated site of action and lack of toxicity. Inhibitors of WecA, crucial in the initiation of the biogenesis of the cell wall core of M. tuberculosis, may become vital for the treatment of millions of patients with tuberculosis.
Acknowledgments
We thank the members of the Lilly TB Drug Discovery Initiative for helpful and fruitful discussions. We are grateful to Meiji Seika Pharma Co., Ltd., for facilitating the fermentation process toward the production of caprazamycins and to Dr. Seiichi Yasuda of National Institute of Genetics, Japan, for providing plasmid pTH19kr. We also thank Seiko Hattori and Dr. Hideki Hashizume of BIKAKEN for technical advice.
This work was supported, in whole or in part, by National Institutes of Health Grants AI049151 and AI018357 from NIAID.
- MIC
- minimum inhibitory concentration
- MurNAc
- N-acetyl-d-muramic acid.
REFERENCES
- 1. The World Health Organization (2012) Global Tuberculosis Report 2012, www.who.int/tb/publications/global_report/en/
- 2. Centers for Disease Control and Prevention (2006) Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs–worldwide, 2000–2004. MMWR Morb. Mortal Wkly. Rep. 55, 301–305 [PubMed] [Google Scholar]
- 3. Phillips L. (2013) Infectious disease: TB's revenge. Nature 493, 14–16 [DOI] [PubMed] [Google Scholar]
- 4. Igarashi M., Nakagawa N., Doi N., Hattori S., Naganawa H., Hamada M. (2003) Caprazamycin B, a novel anti-tuberculosis antibiotic, from Streptomyces sp. J. Antibiot. 56, 580–583 [DOI] [PubMed] [Google Scholar]
- 5. Igarashi M., Takahashi Y., Shitara T., Nakamura H., Naganawa H., Miyake T., Akamatsu Y. (2005) Caprazamycins, novel lipo-nucleoside antibiotics, from Streptomyces sp. II. Structure elucidation of caprazamycins. J. Antibiot. 58, 327–337 [DOI] [PubMed] [Google Scholar]
- 6. Takahashi Y., Igarashi M., Miyake T., Soutome H., Ishikawa K., Komatsuki Y., Koyama Y., Nakagawa N., Hattori S., Inoue K., Doi N., Akamatsu Y. (2013) Novel semisynthetic antibiotics from caprazamycins A–G: caprazene derivatives and their antibacterial activity. J. Antibiot. 66, 171–178 [DOI] [PubMed] [Google Scholar]
- 7. Engohang-Ndong J. (2012) Antimycobacterial drugs currently in phase II clinical trials and preclinical phase for tuberculosis treatment. Expert. Opin. Investig. Drugs 21, 1789–1800 [DOI] [PubMed] [Google Scholar]
- 8. Isono K., Uramoto M., Kusakabe H., Kimura K., Isaki K., Nelson C. C., McCloskey J.A. (1985) Liposidomycins: novel nucleoside antibiotics which inhibit bacterial peptidoglycan synthesis. J. Antibiot. 38, 1617–1621 [DOI] [PubMed] [Google Scholar]
- 9. Isono F., Inukai M. (1991) Mureidomycin A, a new inhibitor of bacterial peptidoglycan synthesis. Antimicrob. Agents Chemother. 35, 234–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McDonald L. A., Barbieri L. R., Carter G. T., Lenoy E., Lotvin J., Petersen P. J., Siegel M. M., Singh G., Williamson R. T. (2002) Structures of the muraymycins, novel peptidoglycan biosynthesis inhibitors. J. Am. Chem. Soc. 124, 10260–10261 [DOI] [PubMed] [Google Scholar]
- 11. Muramatsu Y., Ishii M. M., Inukai M. (2003) Studies on novel bacterial translocase I inhibitors, A-500359s. II. Biological activities of A-500359 A, C, D, and G. J. Antibiot. 56, 253–258 [DOI] [PubMed] [Google Scholar]
- 12. Soldo B., Lazarevic V., Karamata D. (2002) tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology 148, 2079–2087 [DOI] [PubMed] [Google Scholar]
- 13. Bhavsar A. P., Brown E. D. (2006) Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol. Microbiol. 60, 1077–1090 [DOI] [PubMed] [Google Scholar]
- 14. Hashizume H., Sawa R., Harada S., Igarashi M., Adachi H., Nishimura Y., Nomoto A. (2011) Tripropeptin C blocks the lipid cycle of cell wall biosynthesis by complex formation with undecaprenyl pyrophosphate. Antimicrob. Agents Chemother. 55, 3821–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sawa R., Takahashi Y., Hashizume H., Sasaki K., Ishizaki Y., Umekita M., Hatano M., Abe H., Watanabe T., Kinoshita N., Homma Y., Hayashi C., Inoue K., Ohba S., Masuda T., Arakawa M., Kobayashi Y., Hamada M., Igarashi M., Adachi H., Nishimura Y., Akamatsu Y. (2012) Amycolamicin: a novel broad-spectrum antibiotic inhibiting bacterial topoisomerase. Chem. Eur. J. 18, 15772–15781 [DOI] [PubMed] [Google Scholar]
- 16. Karamata D., Gross J. D. (1970) Isolation and genetic analysis of temperature-sensitive mutants of B. subtilis defective in DNA synthesis. Mol. Gen. Genet. 108, 277–287 [DOI] [PubMed] [Google Scholar]
- 17. Al-Dabbagh B., Mengin-Lecreulx D., Bouhss A. (2008) Purification and characterization of the bacterial UDP-GlcNAc:undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA. J. Bacteriol. 190, 7141–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Snapper S. B., Melton R. E., Mustafa S., Kieser T., Jacobs W. R., Jr. (1990) Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4, 1911–1919 [DOI] [PubMed] [Google Scholar]
- 19. Galamba A., Soetaert K., Wang X. M., De Bruyn J., Jacobs P., Content J. (2001) Disruption of adhC reveals a large duplication in the Mycobacterium smegmatis mc2155 genome. Microbiology 147, 3281–3294 [DOI] [PubMed] [Google Scholar]
- 20. Garbe T. R., Barathi J., Barnini S., Zhang Y., Abou-Zeid C., Tang D., Mukherjee R., Young D. B. (1994) Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology 140, 133–138 [DOI] [PubMed] [Google Scholar]
- 21. Hashimoto-Gotoh T., Yamaguchi M., Yasojima K., Tsujimura A., Wakabayashi Y., Watanabe Y. (2000) A set of temperature sensitive-replication/-segregation and temperature resistant plasmid vectors with different copy numbers and in an isogenic background (chloramphenicol, kanamycin, lacZ, repA, par, polA). Gene 241, 185–191 [DOI] [PubMed] [Google Scholar]
- 22. Mikusová K., Mikus M., Besra G. S., Hancock I., Brennan P. J. (1996) Biosynthesis of the linkage region of the mycobacterial cell wall. J. Biol. Chem. 271, 7820–7828 [DOI] [PubMed] [Google Scholar]
- 23. Takayama K., Schnoes H. K., Armstrong E. L., Boyle R. W. (1975) Site of inhibitory action of isoniazid in the synthesis of mycolic acids in Mycobacterium tuberculosis. J. Lipid Res. 16, 308–317 [PubMed] [Google Scholar]
- 24. Pooley H. M., Karamata D. (2000) Incorporation of [2-3H]glycerol into cell surface components of Bacillus subtilis 168 and thermosensitive mutants affected in wall teichoic acid synthesis: effect of tunicamycin. Microbiology 146, 797–805 [DOI] [PubMed] [Google Scholar]
- 25. Denapaite D., Brückner R., Hakenbeck R., Vollmer W. (2012) Biosynthesis of teichoic acids in Streptococcus pneumoniae and closely related species: lessons from genomes. Microb. Drug Resist. 18, 344–358 [DOI] [PubMed] [Google Scholar]
- 26. Campbell J., Singh A. K., Santa Maria J. P., Jr., Kim Y., Brown S., Swoboda J. G., Mylonakis E., Wilkinson B. J., Walker S. (2011) Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6, 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Swoboda J. G., Campbell J., Meredith T. C., Walker S. (2010) Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 11, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Atilano M. L., Pereira P. M., Yates J., Reed P., Veiga H., Pinho M. G., Filipe S. R. (2010) Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 107, 18991–18996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Price N. P., Momany F. A. (2005) Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology 15, 29R-42R [DOI] [PubMed] [Google Scholar]
- 30. Brötz-Oesterhelt H., Brunner N. A. (2008) How many modes of action should an antibiotic have? Curr. Opin. Pharmacol. 8, 564–573 [DOI] [PubMed] [Google Scholar]
- 31. Plinke C., Cox H. S., Zarkua N., Karimovich H. A., Braker K., Diel R., Rüsch-Gerdes S., Feuerriegel S., Niemann S. (2010) embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J. Antimicrob. Chemother. 65, 1359–1367 [DOI] [PubMed] [Google Scholar]
- 32. Berg S., Kaur D., Jackson M., Brennan P. J. (2007) The glycosyltransferases of Mycobacterium tuberculosis--roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology 17, 35R–56R [DOI] [PubMed] [Google Scholar]
- 33. Lechartier B., Hartkoorn R. C., Cole S. T. (2012) In vitro combination studies of benzothiazinone lead compound BTZ043 against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56, 5790–5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin Y., Xin Y., Zhang W., Ma Y. (2010) Mycobacterium tuberculosis Rv1302 and Mycobacterium smegmatis MSMEG_4947 have WecA function and MSMEG_4947 is required for the growth of M. smegmatis. FEMS Microbiol. Lett. 310, 54–61 [DOI] [PubMed] [Google Scholar]
- 35. Sassetti C. M., Boyd D. H., Rubin E. J. (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 36. Lamichhane G., Zignol M., Blades N. J., Geiman D. E., Dougherty A., Grosset J., Broman K. W., Bishai W. R. (2003) A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 100, 7213–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Griffin J. E., Gawronski J.D., Dejesus M. A., Ioerger T. R., Akerley B. J., Sassetti C. M. (2011) High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7, e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. D'Elia M. A., Pereira M. P., Chung Y. S., Zhao W., Chau A., Kenney T. J., Sulavik M. C., Black T. A., Brown E. D. (2006) Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 188, 4183–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Teng F., Singh K. V., Bourgogne A., Zeng J., Murray B. E. (2009) Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis. Infect. Immun. 77, 3759–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]