Background: OMA-1 is required for oocyte maturation and may function by regulating maternal mRNAs.
Results: OMA-1 binds with high affinity to UA(A/U) motifs and regulates glp-1 via its 3′-UTR in live worms.
Conclusion: OMA-1 is a sequence-specific RNA-binding protein that represses maternal mRNAs during oocyte maturation.
Significance: This work reveals the nature of OMA-1 RNA binding activity, which will help identify targets that contribute to the maturation defective phenotype.
Keywords: C. elegans, RNA, RNA-binding Protein, RNA-binding Proteins, RNA-Protein Interaction, Caenorhabditis elegans, OMA-1, RNA-binding Protein, glp-1, Post-transcriptional Regulation
Abstract
Maternally supplied mRNAs encode proteins that pattern early embryos in many species. In the nematode Caenorhabditis elegans, a suite of RNA-binding proteins regulates expression of maternal mRNAs during oogenesis, the oocyte to embryo transition, and early embryogenesis. To understand how these RNA-binding proteins contribute to development, it is necessary to determine how they select specific mRNA targets for regulation. OMA-1 and OMA-2 are redundant proteins required for oocyte maturation—an essential part of meiosis that prepares oocytes for fertilization. Both proteins have CCCH type tandem zinc finger RNA-binding domains. Here, we define the RNA binding specificity of OMA-1 and demonstrate that OMA-1/2 are required to repress the expression of a glp-1 3′-UTR reporter in developing oocytes. OMA-1 binds with high affinity to a conserved region of the glp-1 3′-UTR previously shown to interact with POS-1 and GLD-1, RNA-binding proteins required for glp-1 reporter repression in the posterior of fertilized embryos. Our results reveal that OMA-1 is a sequence-specific RNA-binding protein required to repress expression of maternal transcripts during oogenesis and suggest that interplay between OMA-1 and other factors for overlapping binding sites helps to coordinate the transition from oocyte to embryo.
Introduction
Post-transcriptional regulation of maternal mRNAs governs gene regulation during oogenesis and early embryogenesis in metazoans (1–3). Genetic studies have identified several RNA-binding proteins required for regulation of maternally supplied mRNAs during oogenesis, the oocyte to embryo transition, and early embryogenesis (4, 5). RNA-binding proteins are important during oocyte development because oocytes of metazoans are loaded with translationally repressed maternal RNAs (6–8). During the oocyte to embryo transition, RNA-binding proteins regulate their cognate RNA targets to coordinate events such as axis formation and cell fate specification (6).
Oocyte maturation is the complex process that prepares oocytes for fertilization (9–11). Metazoan sexual reproduction requires meiosis to produce fertile oocytes. Meiotic divisions in the oocytes must be completed before zygote formation. Therefore, precise regulation of meiosis during oocyte development is necessary to couple meiotic events to fertilization events. An evolutionarily conserved feature of oocyte development is meiotic arrest, which prepares the oocyte for fertilization (12). During oocyte maturation, meiotic arrest is released (9, 13), the nuclear envelope breaks down (9), and the cortical cytoskeleton rearranges morphologically (9). Caenorhabditis elegans provides a powerful system to study oocyte maturation because of its transparent body, established cellular lineage, and easy genetic manipulation (14). The oocyte proximal to the spermatheca receives a maturation signal from the sperm prior to ovulation and subsequent fertilization (15). This cycle is repeated approximately every 23 min (9, 16). Although the hallmark events of oocyte maturation are well understood morphologically, the molecular mechanisms governing these events are poorly understood.
During oogenesis, oocytes are loaded with translationally repressed RNAs. Expression of these RNAs must be coordinated in time and space to ensure correct patterning of the embryo. Genetic studies have identified several RNA-binding proteins required for regulation of maternally supplied mRNAs during oogenesis, the oocyte to embryo transition, and early embryogenesis (4, 5). To regulate expression of their cognate mRNA targets, these RNA-binding proteins must be capable of selecting their targets from a complex pool of mRNA sequences.
The putative RNA-binding proteins OMA-1 and OMA-2 are redundantly required for oocyte maturation (17, 18). They are expressed in maturing oocytes with the highest level present in the oocyte most proximal to the spermatheca. Their expression decreases rapidly following the first mitotic division of the one-cell embryo (17) (Fig. 1). Rapid turnover of OMA-1 and OMA-2 is required to prevent embryonic lethality (19, 20). Worms homozygous for oma-1 and oma-2 null alleles are sterile. They produce both sperm and oocytes but no embryos. The gonad arm fills with a higher number of oocytes as compared with wild-type worms. In addition, the oocytes of these worms are larger than wild-type oocytes (17).
FIGURE 1.
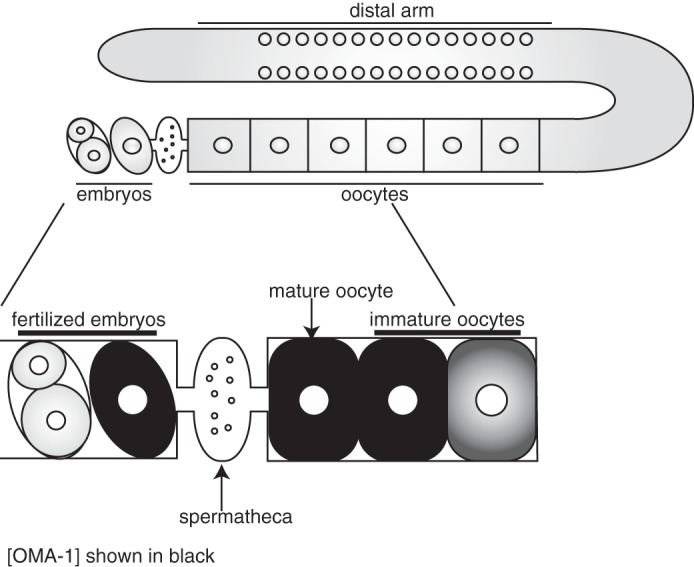
Schematic of the C. elegans germ line. Top, germ line of C. elegans. The syncytial region of nuclei in the distal arm of the gonad, the oocytes, and the embryos are shown. Bottom, oocyte maturation. The oocyte most proximal to the spermatheca matures first. Nuclear envelope breakdown is a hallmark event. The oocyte completes maturation, is ovulated, and is fertilized by sperm in the spermatheca. Embryos are then deposited in the uterus. Dark gray color in the oocytes denotes the abundance of OMA-1 and OMA-2.
OMA-1 and OMA-2 have two CCCH type tandem zinc finger domains typified by the mammalian homolog tristetraprolin (TTP).2 TTP has two CX8CX5CX3H motifs that bind to AU-rich elements (AREs) of the mRNA encoding the pro-inflammatory cytokine TNFα (21). Each finger binds one UAUU motif, and the binding event promotes the turnover of the mRNA and leads to regulation of the immune response (22). C. elegans expresses a number of tandem zinc finger proteins that regulate oogenesis (OMA-1, OMA-2, and MOE-3) (17, 18) or embryogenesis (MEX-5/6, POS-1, MEX-1, and PIE-1) (23–25). Of these, MEX-5 and POS-1 have been shown to bind to RNA with high affinity (26, 27). In contrast, MEX-1 and PIE-1 are proposed to function as transcription factors that bind to DNA (28–30).
OMA-1 and OMA-2 are proposed to function during oocyte maturation by regulating specific target maternal mRNAs at the oocyte to embryo transition. Consistent with this hypothesis, OMA-1 and OMA-2 are required to repress mei-1, zif-1, and nos-2 translation. The mei-1 gene encodes a katanin (a heterodimeric microtubule severing protein) subunit. Genetic studies showed that mei-1 is necessary for meiotic spindle formation; in the absence of mei-1 function, meiosis fails (31, 32). The zif-1 gene, on the other hand, encodes a subunit of the E3 ubiquitin ligase complex. ZIF-1 is required in embryos for proper asymmetric segregation of cell fate regulators through zif-1-dependent proteolysis (33, 34). nos-2 is Notch receptor homolog and is required for primordial germ cell development (35). OMA-2 was shown to repress a nos-2 3′-UTR reporter transgene in live worms (36). In addition, OMA-2 was shown in nonquantitative experiments to interact with the nos-2 3′-UTR via a UGCUAAUAAU sequence element. How OMA-1/2 represses mei-1, zif-1, and nos-2 mRNA translation, or whether OMA-1 regulates additional maternal transcripts, is not known. We set out to define the RNA recognition properties of OMA-1/2 in quantitative terms to gain insight as to how mRNA targets are selected for regulation.
EXPERIMENTAL PROCEDURES
OMA-1 Expression and Purification
The sequence encoding amino acids 1–182 of OMA-1 was cloned into pMal-ac (New England Biolabs). This construct was transformed into BL21(DE3) cells. The protein was then expressed after inducing the cells with 1 mm isopropyl 1-thio-β-d-galactopyranoside and 100 μm Zn(OAC)2 for 3 h, at 37 °C. The protein was expressed with an N-terminal maltose-binding protein (MBP) tag. The cells were then lysed in 200 mm NaCl, 50 mm Tris, pH 8.8, 2 mm DTT, 100 μm Zn(OAC)2, and EDTA-free protease inhibitor tablet. OMA-1 was then purified using an amylose (New England Biolabs) affinity column. Protein fractions were eluted in lysis buffer supplemented with 10 mm maltose. Fractions containing OMA-1 fusion were dialyzed into Q-column buffer (20 mm NaCl, 50 mm Tris, pH 8.8, 2 mm DTT, 100 μm Zn(OAC)2). After dialysis, purification was followed by HiTrap Q at 4 °C. Elution of the protein fractions was achieved by a salt gradient ranging from a low salt buffer (20 mm NaCl, 50 mm Tris, pH 8.8, 2 mm DTT, 100 μm Zn(OAC)2) to a high salt buffer (1 m NaCl, 50 mm Tris, pH 8.8, 2 mm DTT, 100 μm Zn(OAC)2). Final purification was done using a source 15Q (GE Healthcare) ion exchange column at 4 °C. Elution was achieved through the same salt gradient as in the HiTrap Q column purification. Pure fractions were determined by Coomassie-stained SDS-PAGE, and purified OMA-1 was dialyzed into storage buffer (25 mm Tris, pH 8.0, 25 mm NaCl, 2 mm DTT, 100 μm Zn(OAC)2) and stored at 4 °C.
In Vitro RNA Selection
RNA library design and in vitro selection protocols were adapted from a protocol described previously (37). The initial double-stranded DNA library was amplified from the template 5′-GGGAAGATCTCGACCAGAAG-(N30)-TATGTGCGTCTACATGGATCCTCA with a forward (5′-CGGAATTCTAATACGACTCACTATAGGGAAGATCTCGACCAGAAG-3′) and reverse (5′-TGAGGATCCATGTAGACGCACATA-3′) primer pair using three cycles of PCR. Binding reactions of the RNA pools to OMA-1 were performed in 200 μl of selection buffer (50 mm Tris, pH 8.0, 100 mm NaCl, 0.01 mg/ml tRNA, 0.01% Igepal CA-630, 2 mm DTT, 100 μm Zn(OAC)2). Between 5 and 800 nm of purified MBP-OMA-1(1–182) was equilibrated with the pool of RNA sequences in selection buffer for 1 h. Then OMA-1 was immobilized on amylose resin (New England Biolabs). At each round of selection, lowering the protein concentration from 800 nm to 200, 20, and 5 nm successively increased the stringency. OMA-1 bound to RNA was eluted from amylose resin with 10 mm maltose in selection buffer, at room temperature. Selected RNA was phenol/chloroform-extracted, ethanol-precipitated, and resuspended in 10 μl of TE buffer. RNA was then reverse transcribed and amplified with 15 rounds of PCR using the SuperScript III One-Step RT-PCR kit with Platinum Taq (Invitrogen). The new DNA pool was then in vitro transcribed to generate the next RNA pool that will enter the following round of selections. We performed four rounds of selection. The DNA was cloned using StrataClone PCR cloning kit (Stratagene).
Preparation of Fluorescently Labeled RNA
Synthesized oligonucleotides (Integrated DNA Technologies) were 3′-end labeled with fluorescein 5-thiosemicarbazide (Invitrogen) as previously described (38).
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift experiments and data analysis were carried out as previously described with a few modifications (26, 27, 38). Varying concentrations of purified OMA-1 were equilibrated with 3 nm of labeled RNA in equilibration buffer (0.01% Igepal, 0.01 mg/ml tRNA, 50 mm Tris, pH 8.0, 100 mm NaCl, 2 mm DTT, 100 μm Zn(OAc)2) for 3 h. Samples were loaded on a 5% native, slab polyacrylamide gel in 1× TB buffer (89 mm Tris and 89 mm boric acid, pH 8.3). The gels were run in 1× TB buffer for 120 min at 120 volts and at 4 °C. The gels were then scanned using a fluor imager (Fujifilm FLA-5000) with a blue laser at 473 nm.
oma-1;oma-2 RNAi Knockdown
We knocked down oma-1 and oma-2 using the RNAi feeding method (39). We cloned the oma-1 and oma-2 ORFs into the RNAi feeding vector construct L4440 by TA cloning, as previously described (39). These clones were then transformed into HT115(DE3) cells. Once these cells were grown to A600 = 0.4, the cells were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at a final concentration of 0.4 mm for 4 h. The cultures with the oma-1 RNAi feeding construct and oma-2 RNAi feeding construct were concentrated 10-fold and mixed at equal proportions. The mixed culture was the seeded onto nematode growth medium (1.7% (w/v) agar, 0.25% (w/v) peptone, 50 mm NaCl, 1 mm CaCl2, 5 μg/ml cholesterol, 25 mm KH2PO4, 1 mm MgSO4) plates that contain 1 mm isopropyl 1-thio-β-d-galactopyranoside and 100 μg/ml ampicillin. Worms were then bleached onto these plates and maintained at 25 °C for 2 days before imaging. As a control food, we used HT115 strain bacteria transformed with the empty RNAi feeding construct vector, L4440.
Imaging of Worm Strains
Worms at the appropriate age were picked on to 2% agarose pads in 0.4 mm levamisole. Differential interference contrast and GFP fluorescence images were taken using an oil immersion 40× objective on Zeiss Axioscope 2 plus microscope (Carl Zeiss, Jena, Germany). Confocal images were also taken using an oil immersion 40× objective on Leica DMIRE2 microscope (Leica, Wetzlar, Germany).
Quantifications of the GFP pixel intensities were performed as described previously (40, 41) with minor changes. We used a 20-pixel-wide segmented line that passes through the nuclei of the oocytes to determine the average pixel intensity across this line.
RESULTS
OMA-1 Binds with Limited Affinity to RNA Sequences Recognized by POS-1, MEX-5, and TTP
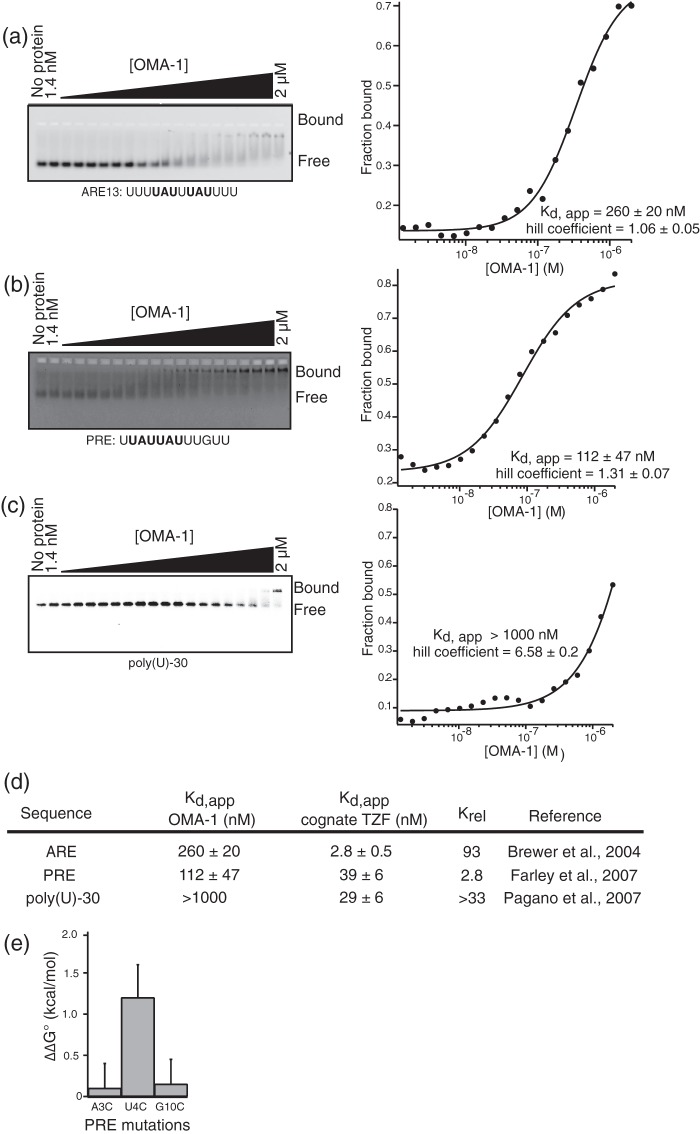
To assess OMA-1 RNA binding specificity, we first determined the ability of purified recombinant OMA-1 to bind sequences recognized by its mammalian homolog TTP and by two nematode family members MEX-5 and POS-1. We performed quantitative fluorescent electrophoretic mobility shift assays (F-EMSA) to measure the apparent binding affinity of purified recombinant OMA-1 to the fragment of an AU-rich element (ARE13) from the 3′-UTR of TNF-α mRNA (recognized by TTP) (22, 42), the POS-1 recognition element (PRE) from the 3′-UTR of mex-3 mRNA (recognized by POS-1) (27), and polyuridine-30 RNA (recognized by MEX-5) (26). OMA-1 binds with moderate affinity to the TNF-α ARE13 (Fig. 2a) and the PRE (Fig. 2b), and it binds weakly to polyuridine-30 RNA (Fig. 2c). However, the affinity of OMA-1 for all three sequences is weaker than the affinity of each sequence for its cognate RNA-binding protein (Fig. 2d). OMA-1 binds 90-fold more weakly to TNF-α ARE13 RNA compared with TTP, about 3-fold weaker to PRE compared with POS-1, and more than 30-fold weaker to polyuridine-30 compared with MEX-5 (Fig. 2d).
FIGURE 2.
OMA-1 is a sequence-specific RNA-binding protein. a, F-EMSA with the AU-rich element of TNF-a mRNA (ARE13) and OMA-1. The gel is shown with the bound and free RNA species labeled. Data are fit to the Hill equation. The values reported are the averages and standard deviation of three independent experiments. b, fluorescent electrophoretic mobility gel shift assay with the POS-1 binding sequence (PRE) and OMA-1. OMA-1 shows weak binding to this sequence. c, fluorescent electrophoretic mobility gel shift assay is done with the poly(U)-30, which binds MEX-5, and OMA-1, as described in a. d, table comparing the relative binding affinities of OMA-1 to the RNA sequences recognized by TTP, MEX-5, and POS-1 with respect to their cognate proteins. e, OMA-1 binds to variants of PRE differently than POS-1. Each bar shows the change in standard free energy change (ΔΔG°) caused by the mutation shown. The binding affinity of OMA-1 to these variants was measured by F-EMSA. This binding affinity is then compared with the binding affinity of OMA-1 to the PRE to calculate the ΔΔG°.
To determine whether OMA-1 binds RNA with identical specificity as POS-1, but with lower affinity, we measured OMA-1 binding to three PRE mutants (A3C, U4C, and G10C) that reduce POS-1 binding by >1 kcal/mol (27). OMA-1 binding is not affected by the A3C and G10C mutations. By contrast, the U4C mutant binds OMA-1 with reduced affinity (ΔΔG° = 1.2 kcal/mol) (Fig. 2e) We conclude that although OMA-1 is capable of binding to RNA sequences with variable affinity, its specificity is not the same as previously investigated members of the tandem zinc finger RNA-binding protein family.
OMA-1 SELEX
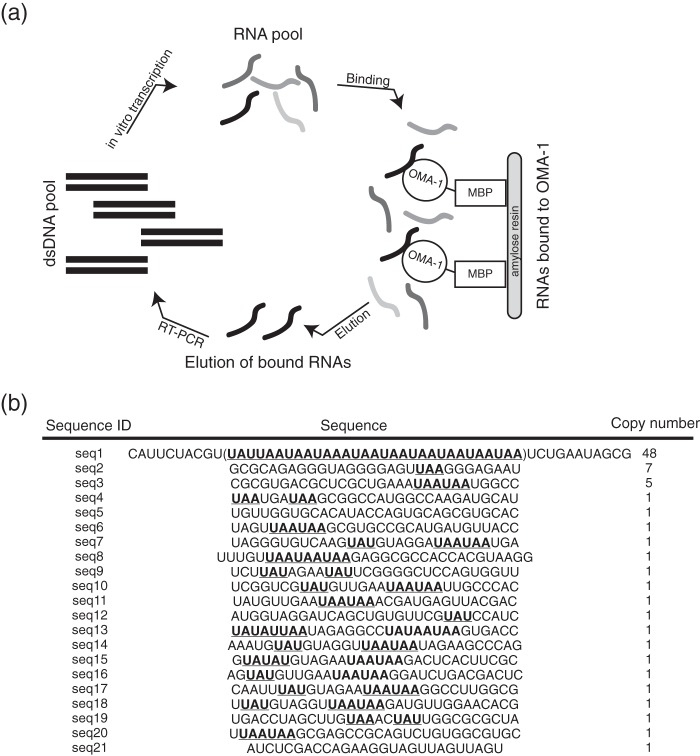
We hypothesized that OMA-1 binds to RNA with specificity that is different from MEX-5, POS-1, and TTP. To identify sequences that bind OMA-1 with high affinity, we performed an in vitro selection (systematic evolution of ligands by exponential enrichment (SELEX)) (43) using synthesized RNA sequences that contain 30 randomized bases, as described previously (37). In the first round of selection, we equilibrated the starting pool with a fragment of OMA-1 that includes the RNA-binding domain (amino acids 1–182) fused to an N-terminal MBP tag. This fusion protein was immobilized on an amylose resin, and unbound RNA sequences were washed away. The bound RNA sequences were eluted and amplified to generate a new library of RNA sequences for the next round of selection (Fig. 3a). F-EMSA was used to monitor the progress of selection. Our results reveal that RNA produced after the fourth round of selection is enriched for sequences that bind OMA-1 compared with the starting pool (data not shown). To identify the sequences within pool 4, we cloned cDNA generated from RNA sequences enriched in this pool and sequenced 69 clones. Of these, 48 contain extended repeats of motif UAA. These 48 sequences have variable lengths of UAA repeats. The sequence identified as seq1 in Fig. 4a is a representative of this class of recovered sequences. This sequence is longer than others, and this might be due to expansion of repeated sequences during the PCR amplification step of the SELEX procedure. Two additional sequences were recovered in multiple copies. Sequence 2 (seq2) was recovered in seven and sequence 3 (seq3) was recovered in five copies. These sequences also contain UAA elements. All but one of the remaining individual sequences also contained UAA elements. Eight of these sequences contain UAU motifs as well, which comprise a portion of the TTP and POS-1 recognition motifs (Fig. 3b) (27, 44).
FIGURE 3.
In vitro selection to identify high affinity binding sequences of OMA-1. a, schematic of SELEX. b, list of 69 DNA sequences that were recovered from the selection. The sequences and their respective copy number are also shown. These sequences are enriched in UAA and UAU repeats, which are shown in bold and underlined.
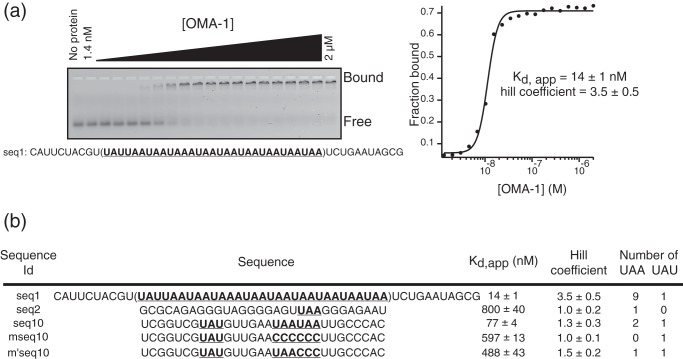
FIGURE 4.
UA(A/U) motifs are responsible for the binding of OMA-1 to the aptamer sequences. a, EMSA of OMA-1 with the highest ranking RNA sequence with respect to the copy number. This assay is analyzed as described in the legend to Fig. 1. The assay showed that seq1 binds with a high affinity to OMA-1, as shown by the plot on the right. b, a table of sequences that includes additional binding assay results with two more sequences recovered from the selection. seq2 is a sequence that has one UAA element. seq10 is a sequence that has three UA(A/U) elements. mseq10 is the sequence seq10 in which the UAA elements are replaced by CCC. m′seq10 binds with a similar affinity to mseq10.
To determine whether OMA-1 binds to the recovered aptamer sequences, we performed quantitative F-EMSA binding assays with RNA sequences that were recovered most frequently (Fig. 4a). Our results showed that OMA-1 binds with highest affinity to the aptamer sequence with the most UAA elements (Fig. 4b). Binding of OMA-1 to the RNA sequence with repeated motifs results in a Hill coefficient (nH) of 3.5 ± 0.5. This suggests that this interaction between OMA-1 and UAA-rich RNA sequences is cooperative. To test whether the UAA elements in these sequences are responsible for OMA-1 binding, we mutated the UAA motifs to CCC and tested the effect of this mutation on binding affinity. We chose seq10 as a representative sequence. Replacing the tandem UAA sequences to CCC in aptamer mseq10 led to a significant decrease in the binding affinity of OMA-1 (8-fold decrease). We also tested binding of OMA-1 to another variant of seq10 where a UAA element is retained in the center (Fig. 4b). This RNA binds with similar affinity to mseq10. Together, the data show that OMA-1 binds with high affinity to UAA-rich RNA.
OMA-1 Binds to Multiple Fragments of the glp-1 3′-UTR
GLP-1 is the C. elegans homolog of Notch. It is required for anterior cell fate specification in the early embryo and mitotic proliferation of progenitor cells in the germ line (45, 46). The mRNA that encodes glp-1 is found throughout the germ line, including oocytes, and in all cells of the early embryo (45). Several RNA-binding proteins have been shown to contribute to the asymmetric pattern of GLP-1 expression (40, 41, 47), but the identity of the factor that represses GLP-1 protein production in maturing oocytes is not known.
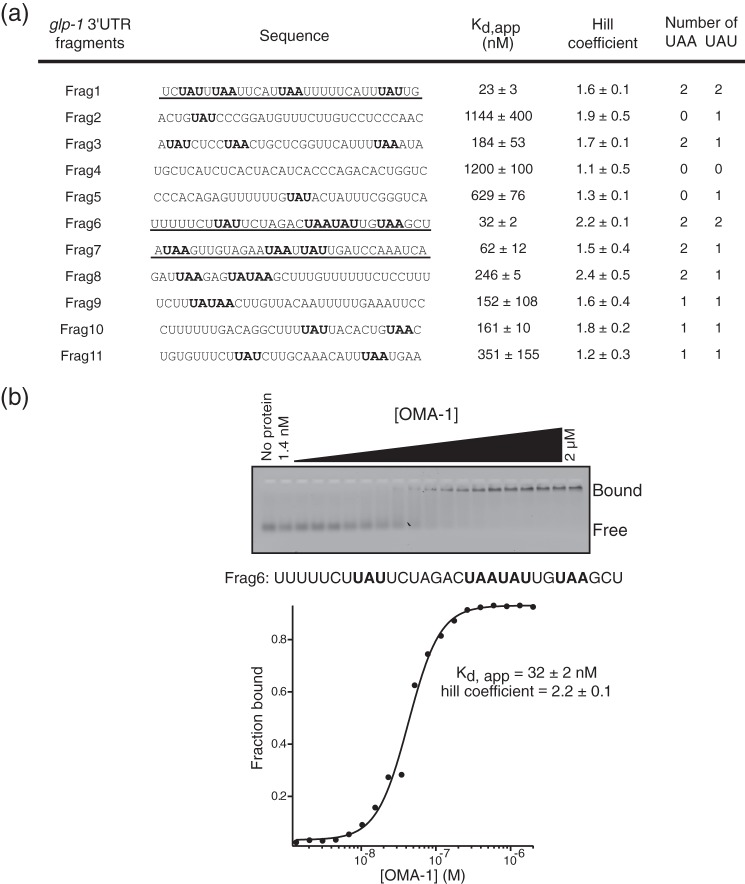
The glp-1 3′-UTR is densely packed with UA(A/U) motifs, suggesting that OMA-1 may bind to this transcript and repress its translation in oocytes. To determine whether OMA-1 binds to the glp-1 3′-UTR directly, we constructed nonoverlapping RNA fragments that span the 3′-UTR. Each RNA is ∼30 nucleotides in length. OMA-1 binds to multiple fragments of the glp-1 3′-UTR. OMA-1 binds to fragments 1, 6, and 7 with highest affinity, comparable to the affinity of OMA-1 for the selected aptamer sequences. Fragments 1 and 6 have four UA(A/U) motifs, whereas fragment 7 has three. OMA-1 binds with moderate affinity to fragments 3, 8, 9, 10, and 11, which contain two or three UA(A/U) motifs. Very weak binding is observed to fragments 2, 4, and 5, which have one or no motifs present. As such, the affinity of each fragment correlates with the number of UA(A/U) motifs present, with the highest affinity fragments containing four motifs (Fig. 5a). As with the selected aptamers, binding to the glp-1 3′-UTR fragments exhibits positive cooperativity when multiple UA(A/U) motifs are present (for example, fragment 6: nH = 2.2). The results are consistent with the SELEX results that suggest UA(A/U) motifs, which we now term OMA-1-binding motifs (OBMs), are required for high affinity OMA-1 binding.
FIGURE 5.
OMA-1 binds to the glp-1 3′-UTR. a, EMSA results for OMA-1 binding to fragments of RNA that span the glp-1 3′-UTR. Frag1, Frag6, and Frag7 (underlined) are the fragments of the glp-1 3′-UTR that bind OMA-1 with the highest affinities. b, gel image showing the binding of OMA-1 to Frag6, which is a conserved sequence of the glp-1 3′-UTR. Below the gel image is the plot of fraction bound of OMA-1 against the concentration of OMA-1. This plot is fit to the Hill equation.
Fragment 6 corresponds to a sequence that is evolutionarily conserved across nematode species and contains overlapping functional binding sites for POS-1 and GLD-1, RNA-binding proteins required for glp-1 silencing in embryos (40, 48, 49). As such, we decided to investigate the contribution of UA(A/U) motifs to OMA-1 binding to this fragment in more detail (Fig. 5b). We performed quantitative EMSA to determine the effect of mutating each UA(A/U) motif singly and in combinations to the OMA-1 binding affinity. Mutating each motif in isolation reduces the affinity by 3–5-fold (Table 1). Mutating three motifs causes a 15-fold decrease in the binding affinity. The two variants of the triple mutation (Triple1 and Triple2) show the same decrease in the binding affinity. Mutating all four of the motifs leads to a 25-fold decrease in OMA-1 binding affinity (Table 1). Our data show that the binding of OMA-1 to this sequence of RNA depends on the presence of UA(A/U) motifs. Plotting the apparent binding affinities against the number of UA(A/U) motifs show that the binding affinity improves with an increasing number of UA(A/U) motifs (Fig. 6), as expected.
TABLE 1.
Binding affinities of OMA-1 to variants of the glp-1 3′-UTR, where the OMA-1 binding motifs are mutated

FIGURE 6.
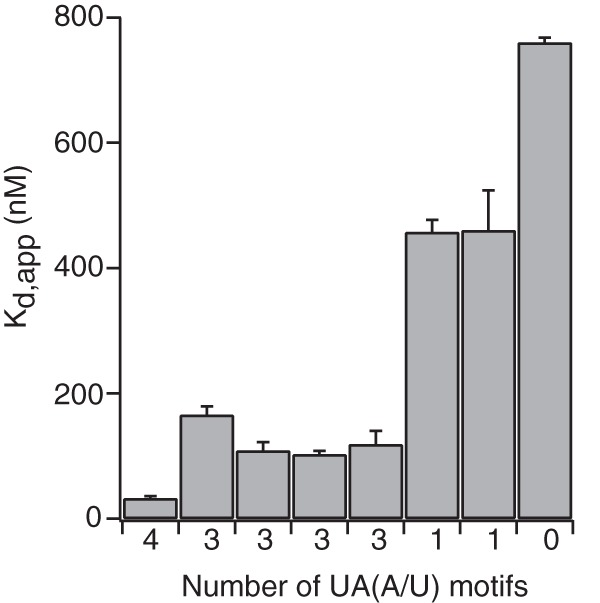
The binding affinity of OMA-1 to Frag6 sequence increases with the number of UA(A/U) motifs. The plot of Kd, app values against the corresponding number of UA(A/U) motifs shows that the more the number of these motifs in the RNA sequence, the better the binding affinity for OMA-1.
glp-1 Is a Regulatory Target of OMA-1
To test whether OMA-1 and OMA-2 contribute to the regulation of glp-1 mRNA in oocytes, we determined the effect of knocking down OMA-1/2 on the expression of a single-copy integrated green fluorescent protein reporter under the control of the glp-1 3′-UTR (40). When the reporter strain was treated with control food, GFP fluorescence was observed in the distal germ line and in embryos but was not observed in the proximal germ line or in oocytes, as has been previously reported (40). When the reporter strain was treated with oma-1, oma-2 RNAi food, we observed a strong increase in GFP fluorescence in the oocytes (Fig. 6; 88%, n = 27) (Fig. 7). We assessed knockdown effectiveness by verifying that embryos were not present, that a reduced number of eggs were laid, that oocytes were larger, and that there were greater numbers of oocytes stacked in the gonad arm, hallmarks of the oma-1, oma-2 phenotype. The data demonstrate that OMA-1 and OMA-2 are required to repress GLP-1 expression in the oocytes.
FIGURE 7.
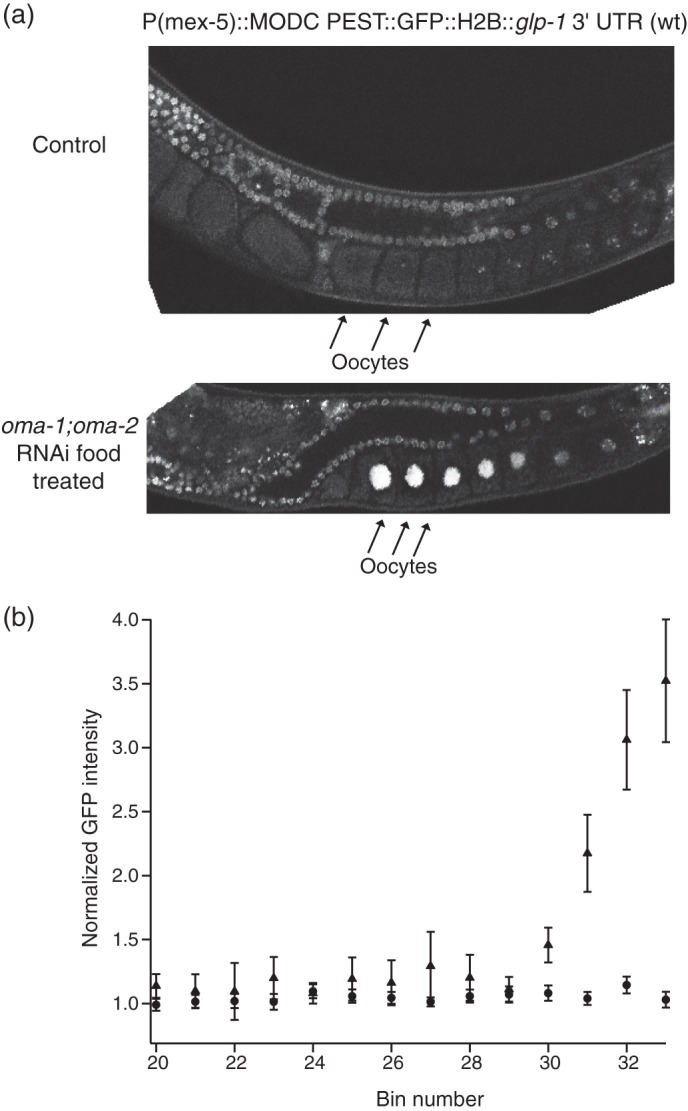
OMA-1 and OMA-2 contribute to repression of glp-1 in the oocytes. a, fluorescence images of single copy integrated strains that express GFP under the control of the wild-type glp-1 3′-UTR sequence. Top, worm grown on control RNAi food. Bottom, worm grown on oma-1;oma-2 RNAi food. Under oma-1;oma-2 knockdown conditions, glp-1 is derepressed in the oocytes. b, quantification of confocal images of the reporter strains under the same conditions described above. GFP intensity as normalized to the average intensity across the wild-type oocytes is plotted against the bin number. The intensities for oma-1;oma-2 RNAi food are denoted with triangles, and the intensities for the control food are denoted with circles.
DISCUSSION
In this study we demonstrated that the motif recognized by OMA-1 is different from those recognized by the related proteins TTP, POS-1, and MEX-5. From the in vitro selection, we showed that OMA-1 binds to UA(A/U) repeat sequences. This sequence is similar to the binding sequence of TTP, which is UAUUUAUU, yet the binding affinity of OMA-1 to this sequence is about 50-fold weaker. Similarly, OMA-1 binds weakly to POS-1 and MEX-5 motifs, revealing that its specificity is different from paralogs expressed in C. elegans.
It is likely that differences in primary sequence and structure account for the variance in RNA recognition properties. The NMR structure of the zinc finger domain of TIS11d, a mammalian tandem zinc finger protein, showed that each finger folds into a similar conformation that binds to UAUU. The RNA binding specificity was proposed to come from hydrogen bonding of the protein backbone to the Watson-Crick edges of the bases. In addition, side chains of conserved aromatic amino acids led to stacking interactions with the RNA bases, which are essential for RNA recognition (50).
It was reported that an amino acid in each finger, termed the “discriminator” residue, accounts for the difference specificity between TTP and MEX-5. In TTP, the discriminator residue is a glutamate in both fingers. In the NMR structure, the side chain carboxylate accepts a hydrogen bond from the N6-exocylic amine of an adenosine in the motif UAUU. In MEX-5, which binds to RNA with relaxed specificity, the corresponding amino acid is a lysine in finger 1 and an arginine in finger 2, predicted to form nonspecific backbone ionic interactions at the expense of the base specific hydrogen bonds found in Tis11D. Mutagenesis experiments confirm the importance of each amino acid to binding specificity (26, 51).
POS-1 has small hydrophobic residues at the corresponding positions and binds to RNA with different specificity compared with that of Tis11D and MEX-5 (PRE = UAU2–3RDN1–3G). It is not clear how the discriminator residues contribute to POS-1 RNA recognition. OMA-1 and OMA-2 have a basic residue in finger 1 and small hydrophobic residue in finger 2. Hence, a hybrid specificity between POS-1 and MEX-5 was expected (26). In line with this expectation, we showed that the RNA binding sequence specificity of OMA-1 is neither as relaxed as that of MEX-5 nor as specific as the POS-1 recognition element. The motif observed (UA(A/U)) bears some similarity to the 5′-portion of the PRE. It is not clear how a small hydrophobic residue would contribute to specificity. Perhaps van der Waals interactions help select for adenosine via interaction with the C2 carbon. More work, including structure determination of the OMA-1, POS-1, and MEX-5 RNA-bound complexes, is required to fully assess this hypothesis.
The relatively low information content of the OBM suggests that 1) many transcripts are regulated by OMA-1 or 2) additional factors may influence selection of its mRNA targets. In this study, we show that the apparent binding affinity of OMA-1 cooperatively increases as the number of OBMs increases, suggesting that multiple OBMs are required to achieve a high apparent binding affinity to mRNAs. It is possible that multiple OBMs are required to achieve regulation. Consistent with this hypothesis, mutation of sequences corresponding to OBM1, OBM3, and a double mutation of OBM1 and OBM3 in previous studies did not lead to activation of the glp-1 reporter in oocytes. There are 28 OBMs in the 3′-UTR of glp-1. Perhaps they function with some redundancy to ensure glp-1 repression.
Interestingly, analyzing the 3′-UTR of the putative mRNA targets nos-2, zif-1, and mei-1 revealed that there are 17 OBMs in the nos-2 3′-UTR, 27 OBMs in the zif-1 3′-UTR, and 9 OBMs in the mei-1 3′-UTR. These OBMs are densely clustered in the zif-1 3′-UTR but more scattered in the nos-2 and mei-1 3′-UTR. These OBMs could be the sites of regulation by OMA-1 in these mRNA targets.
glp-1 Regulation by OMA-1
Our data also show that OMA-1 regulates the translation of glp-1 in oocytes. Many RNA-binding proteins have been shown to regulate glp-1 mRNA post-transcriptionally. During oogenesis, PUF-5, PUF-6, and PUF-7 repress glp-1 in early stage oocytes (47). It was previously suggested that OMA-1 and OMA-2 might repress glp-1 in late stage oocytes because these proteins are abundant RNA-binding proteins in the maturing oocytes (47). Here we have shown that OMA-1 and OMA-2 do in fact repress glp-1 during oogenesis and oocyte to embryo transition. glp-1 gain of function mutation leads to a tumorous germ line because of excessive proliferation of mitotic germ cells (52). To prevent ectopic expression of GLP-1, the mRNA is tightly regulated by multiple RNA-binding proteins such as GLD-1, POS-1, PUF-5/6/7, and OMA-1/2. OMA-1 and OMA-2 repress glp-1 in late stage oocytes where the other RNA-binding proteins are not present. At the oocyte to embryo transition, OMA-1 is marked for degradation by phosphorylation. This leads to a rapid degradation of OMA-1 at one-cell stage embryo. Thus, as OMA-1 is degraded, it might hand off the regulation of glp-1 to embryonic RNA-binding factors. This is plausible because the POS-1 and GLD-1 binding sites that are overlapping with OBMs will be accessible upon OMA-1 and OMA-2 degradation.
The various phenotypes observed in oma-1;oma-2 mutants suggest both proteins regulate multiple targets. OMA-1 and OMA-2 likely prevent premature expression of mRNAs involved in embryonic cell fate pattering events prior to fertilization. The relatively relaxed RNA binding specificity of OMA-1 suggests that it binds to many mRNAs. As such, OMA-1 could be a general repressor of mRNA translation in oocytes. Alternatively, OMA-1 directed regulation could require additional factors that alter or enhance its RNA binding specificity. Future work will distinguish between these possibilities and define the mechanism of OMA-1-mediated repression.
Acknowledgments
We thank Dr. Ruth Zearfoss, Carina C. Clingman, and Neetha Makil for critical comments on the manuscript and advices on the project. We also thank Brian Farley for helpful discussions on the project. We thank Dr. Nicholas Rhind for sharing the fluorescence/differential interference contrast microscope. We also thank Lisa McCoig for providing the OMA-1 expression plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants GM081422 and GM098643 (to S. P. R.).
- TTP
- tristetraprolin
- OBM
- OMA-1 binding motif
- ARE
- AU-rich element
- MBP
- maltose-binding protein
- F-EMSA
- fluorescent EMSA
- PRE
- POS-1 recognition element
- SELEX
- systematic evolution of ligands by exponential enrichment.
REFERENCES
- 1. Moore M. J. (2005) From birth to death. The complex lives of eukaryotic mRNAs. Science 309, 1514–1518 [DOI] [PubMed] [Google Scholar]
- 2. Schier A. F. (2007) The maternal-zygotic transition. Death and birth of RNAs. Science 316, 406–407 [DOI] [PubMed] [Google Scholar]
- 3. Farley B. M., Ryder S. P. (2008) Regulation of maternal mRNAs in early development. Crit. Rev. Biochem. Mol. Biol. 43, 135–162 [DOI] [PubMed] [Google Scholar]
- 4. Colegrove-Otero L. J., Minshall N., Standart N. (2005) RNA-binding proteins in early development. Crit. Rev. Biochem. Mol. Biol. 40, 21–73 [DOI] [PubMed] [Google Scholar]
- 5. Lee M.-H., Schedl T. (2006) RNA-binding proteins. WormBook, 1–13, / 10.1895/wormbook.1.79.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Moor C. H., Meijer H., Lissenden S. (2005) Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin. Cell Dev. Biol. 16, 49–58 [DOI] [PubMed] [Google Scholar]
- 7. Spirin A. S. (1966) “Masked” forms of mRNA. Curr. Top. Dev. Biol. 1, 1–38 [PubMed] [Google Scholar]
- 8. Standart N. (1992) Masking and unmasking of maternal mRNA. Semin. Dev. Biol. 3, 367–379 [Google Scholar]
- 9. McCarter J., Bartlett B., Dang T., Schedl T. (1999) On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205, 111–128 [DOI] [PubMed] [Google Scholar]
- 10. Masui Y. (2001) From oocyte maturation to the in vitro cell cycle. The history of discoveries of maturation-promoting factor (MPF) and cytostatic factor (CSF). Differentiation 69, 1–17 [DOI] [PubMed] [Google Scholar]
- 11. Masui Y., Clarke H. J. (1979) Oocyte maturation. Int. Rev. Cytol. 57, 185–282 [DOI] [PubMed] [Google Scholar]
- 12. Greenstein D. (2005) Control of oocyte meiotic maturation and fertilization. WormBook, 1–12, / 10.1895/wormbook.1.53.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamamoto I., Kosinski M. E., Greenstein D. (2006) Start me up. Cell signaling and the journey from oocyte to embryo in C. elegans. Dev Dyn 235, 571–585 [DOI] [PubMed] [Google Scholar]
- 14. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller M. A., Nguyen V. Q., Lee M. H., Kosinski M., Schedl T., Caprioli R. M., Greenstein D. (2001) A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291, 2144–2147 [DOI] [PubMed] [Google Scholar]
- 16. Kimble J., Crittenden S. L. (2007) Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23, 405–433 [DOI] [PubMed] [Google Scholar]
- 17. Detwiler M. R., Reuben M., Li X., Rogers E., Lin R. (2001) Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev Cell 1, 187–199 [DOI] [PubMed] [Google Scholar]
- 18. Shimada M., Kawahara H., Doi H. (2002) Novel family of CCCH-type zinc-finger proteins, MOE-1, -2 and -3, participates in C. elegans oocyte maturation. Genes Cells 7, 933–947 [DOI] [PubMed] [Google Scholar]
- 19. Lin R. (2003) A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev. Biol. 258, 226–239 [DOI] [PubMed] [Google Scholar]
- 20. Nishi Y., Lin R. (2005) DYRK2 and GSK-3 phosphorylate and promote the timely degradation of OMA-1, a key regulator of the oocyte-to-embryo transition in C. elegans. Dev. Biol. 288, 139–149 [DOI] [PubMed] [Google Scholar]
- 21. Blackshear P. J. (2002) Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc Trans 30, 945–952 [DOI] [PubMed] [Google Scholar]
- 22. Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19, 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schubert C. M., Lin R., de Vries C. J., Plasterk R. H., Priess J. R. (2000) MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell 5, 671–682 [DOI] [PubMed] [Google Scholar]
- 24. Tabara H., Hill R. J., Mello C. C., Priess J. R., Kohara Y. (1999) pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126, 1–11 [DOI] [PubMed] [Google Scholar]
- 25. Mello C. C., Draper B. W., Krause M., Weintraub H., Priess J. R. (1992) The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell 70, 163–176 [DOI] [PubMed] [Google Scholar]
- 26. Pagano J. M., Farley B. M., McCoig L. M., Ryder S. P. (2007) Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J. Biol. Chem. 282, 8883–8894 [DOI] [PubMed] [Google Scholar]
- 27. Farley B. M., Pagano J. M., Ryder S. P. (2008) RNA target specificity of the embryonic cell fate determinant POS-1. RNA 14, 2685–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tenenhaus C., Subramaniam K., Dunn M. A., Seydoux G. (2001) PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev. 15, 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seydoux G., Mello C. C., Pettitt J., Wood W. B., Priess J. R., Fire A. (1996) Repression of gene expression in the embryonic germ lineage of C. elegans. Nature 382, 713–716 [DOI] [PubMed] [Google Scholar]
- 30. Guedes S., Priess J. R. (1997) The C. elegans MEX-1 protein is present in germline blastomeres and is a P granule component. Development 124, 731–739 [DOI] [PubMed] [Google Scholar]
- 31. Clark-Maguire S., Mains P. E. (1994) Localization of the mei-1 gene product of Caenorhaditis elegans, a meiotic-specific spindle component. J. Cell Biol. 126, 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clark-Maguire S., Mains P. E. (1994) mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics 136, 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guven-Ozkan T., Robertson S. M., Nishi Y., Lin R. (2010) zif-1 translational repression defines a second, mutually exclusive OMA function in germline transcriptional repression. Development 137, 3373–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeRenzo C., Reese K. J., Seydoux G. (2003) Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 424, 685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Subramaniam K., Seydoux G. (1999) nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126, 4861–4871 [DOI] [PubMed] [Google Scholar]
- 36. Jadhav S., Rana M., Subramaniam K. (2008) Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development 135, 1803–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pagano J. M., Farley B. M., Essien K. I., Ryder S. P. (2009) RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc. Natl. Acad. Sci. U.S.A. 106, 20252–20257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pagano J. M., Clingman C. C., Ryder S. P. (2011) Quantitative approaches to monitor protein-nucleic acid interactions using fluorescent probes. RNA 17, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Welchman D. P., Zipperlen P., Ahringer J. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 [DOI] [PubMed] [Google Scholar]
- 40. Farley B. M., Ryder S. P. (2012) POS-1 and GLD-1 repress glp-1 translation through a conserved binding-site cluster. Mol. Biol. Cell 23, 4473–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wright J. E., Gaidatzis D., Senften M., Farley B. M., Westhof E., Ryder S. P., Ciosk R. (2010) A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J. 30, 533–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Blackshear P. J., Phillips R. S., Lai W. S. (2005) Tandem CCCH zinc finger proteins in mRNA binding. Zinc Finger Proteins: From Atomic Contact to Cellular Function (Iuchi S., Kuldell N., eds) pp. 80–90, Landes Bioscience, Austin, TX [Google Scholar]
- 43. Tuerk C., Gold L. (1990) Systematic evolution of ligands by exponential enrichment. RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 [DOI] [PubMed] [Google Scholar]
- 44. Brewer B. Y., Malicka J., Blackshear P. J., Wilson G. M. (2004) RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin. Conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J. Biol. Chem. 279, 27870–27877 [DOI] [PubMed] [Google Scholar]
- 45. Evans T. C., Crittenden S. L., Kodoyianni V., Kimble J. (1994) Translational control of maternal glp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell 77, 183–194 [DOI] [PubMed] [Google Scholar]
- 46. Crittenden S. L., Troemel E. R., Evans T. C., Kimble J. (1994) GLP-1 is localized to the mitotic region of the C. elegans germ line. Development 120, 2901–2911 [DOI] [PubMed] [Google Scholar]
- 47. Lublin A. L., Evans T. C. (2007) The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Dev. Biol. 303, 635–649 [DOI] [PubMed] [Google Scholar]
- 48. Ryder S. P., Frater L. A., Abramovitz D. L., Goodwin E. B., Williamson J. R. (2004) RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat. Struct. Mol. Biol. 11, 20–28 [DOI] [PubMed] [Google Scholar]
- 49. Marin V. A., Evans T. C. (2003) Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 130, 2623–2632 [DOI] [PubMed] [Google Scholar]
- 50. Hudson B. P., Martinez-Yamout M. A., Dyson H. J., Wright P. E. (2004) Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 11, 257–264 [DOI] [PubMed] [Google Scholar]
- 51. Kaymak E., Wee L. M., Ryder S. P. (2010) Structure and function of nematode RNA-binding proteins. Curr. Opin. Struct. Biol. 20, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Berry L. W., Westlund B., Schedl T. (1997) Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124, 925–936 [DOI] [PubMed] [Google Scholar]