Background: AtsR is a Burkholderia cenocepacia hybrid sensor kinase that negatively regulates quorum sensing.
Results: The AtsR phosphorelay mechanism includes the AtsT cognate response regulator.
Conclusion: AtsR regulation occurs through phosphorylation of AtsT, which is fine-tuned by the AtsR receiver domain.
Significance: The AtsR/AtsT phosphorelay pathway uncovers a major global regulator of B. cenocepacia pathogenicity.
Keywords: Bacterial Protein Kinases, Cystic Fibrosis, Histidine Kinases, Macrophages, Phosphorylation, Protease, Transcription Regulation, Virulence Factors
Abstract
AtsR is a membrane-bound hybrid sensor kinase of Burkholderia cenocepacia that negatively regulates quorum sensing and virulence factors such as biofilm production, type 6-secretion, and protease secretion. Here we elucidate the mechanism of AtsR phosphorelay by site-directed mutagenesis of predicted histidine and aspartic acid phosphoacceptor residues. We demonstrate by in vitro phosphorylation that histidine 245 and aspartic acid 536 are conserved sites of phosphorylation in AtsR, and we also identify the cytosolic response regulator AtsT (BCAM0381) as a key component of the AtsR phosphorelay pathway. Monitoring the function of AtsR and its derivatives in vivo by measuring extracellular protease activity and swarming motility confirmed the in vitro phosphorylation results. Together we find that the AtsR receiver domain plays a fine-tuning role in determining the levels of phosphotransfer from its sensor kinase domain to the AtsT response regulator.
Introduction
Phosphorylation cascades governed by two-component signal transduction systems provide key signaling mechanisms in bacteria, Archaea, simple eukaryotes, and higher plants, allowing them to translate signals into adaptive responses (1). These regulatory pathways consist of a transmembrane protein that responds to an environmental cue leading to autophosphorylation followed by the transfer of the phosphate to a cytoplasmic response regulator (RR).2 The sensor protein has a variable region dedicated to sensing a signal and a conserved histidine kinase (HK) domain. The RR typically consists of a conserved receiver domain (RD) and a variable effector domain that binds to DNA. Upon perceiving the signal, the HK becomes autophosphorylated on its conserved histidine (His) residue, and the phosphate is transferred to a conserved aspartic acid (Asp) residue on the RR. More complex phosphorelay systems require multiple phosphoryl transfer reactions involving a hybrid sensor kinase that contains an extra domain serving as a RD, a histidine-phosphotransfer protein, and response regulator proteins (1, 2). Because the mechanism of phosphotransfer is from His to Asp, histidine-phosphotransfer proteins shuttle phosphoryl groups between a hybrid sensor kinase and a RR, providing additional checkpoints of regulation in signaling pathways (3, 4).
Burkholderia cenocepacia is a Gram-negative opportunistic pathogen belonging to the Burkholderia cepacia complex that causes severe, chronic respiratory infections in patients with cystic fibrosis and other immunocompromised conditions (5, 6). Furthermore, B. cenocepacia can also be found in maize roots (7) and sewage (8) and cause banana fingertip rot (9) and onion rot (10). This bacterium is also virulent in zebrafish (11), alfalfa, nematodes, and insect infection models (12) and can survive within amoebae and macrophages (13, 14). We have previously identified AtsR (Adhesion and type six secretion system Regulator), which negatively regulates the expression of quorum sensing-regulated virulence factors in B. cenocepacia (15, 16). B. cenocepacia strains have two N-acylhomoserine lactone-dependent quorum-sensing systems, CepIR and CciIR, that coordinate the expression of ZmpA and ZmpB zinc metalloproteases and other virulence factors during infection (17–21). In the absence of atsR, expression of cepIR and cciIR is up-regulated and mediates early and increased N-acylhomoserine lactone production, suggesting that AtsR plays a role in controlling virulence gene expression by modulating the timing of quorum sensing signaling (16). AtsR also represses the expression of virulence genes by an N-acylhomoserine lactone-independent mechanism (16). Consequently, inactivation of atsR in B. cenocepacia also leads to increased biofilm formation, adherence to polystyrene and lung epithelial cells, extracellular protease secretion, and expression of a type 6 secretion system (T6SS). The latter is exemplified by actin cytoskeletal rearrangements with the formation of characteristic “pearls on a string-like structures around infected macrophages (15, 16, 22).
AtsR is a predicted membrane protein with two transmembrane domains and a sensor kinase region (containing the HK and ATPase domains) attached to an RD with conserved Asp residues. AtsR lacks a DNA binding motif found in canonical response regulator proteins, which suggests that AtsR does not bind directly to the promoter regions of target genes and is likely part of a multistep signal transduction pathway. Two genes near atsR encode two putative components of the AtsR phosphorelay pathway. One is BCAM0381 (herein designated atsT), a gene co-transcribed with atsR (15). AtsT is a putative cytoplasmic transcriptional regulator containing an N-terminal helix-turn-helix domain related to domains found in repressors (23) and a receiver domain with a conserved Asp at the C terminus. The other is BCAM0378, which locates immediately upstream of atsR and encodes a hypothetical protein containing His and Asp residues that form part of a conserved motif, UPF0047 (24). BCAM0378 might be functionally necessary as a histidine-phosphotransfer protein intermediate to transfer the phosphate from AtsR to the putative response regulator AtsT.
In this work we investigated the role of critical functional residues within the individual domains of AtsR using phosphorylation assays. We identified the conserved residues His-245 and Asp-536 as phosphoacceptor sites in AtsR. Furthermore, using in vivo and in vitro approaches, we demonstrate that AtsR is a hybrid sensor kinase that regulates downstream cellular activities through direct phosphorylation of AtsT.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria broth (8) (Difco) at 37 °C unless indicated otherwise. Escherichia coli cultures were supplemented as required with the following antibiotics (final concentrations): tetracycline 30 μg/ml, kanamycin 30 μg/ml, and trimethoprim 50 μg/ml. B. cenocepacia cultures were supplemented as required with trimethoprim 100 μg/ml, tetracycline 150 μg/ml, ampicillin 200 μg/ml, and polymyxin B 20 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain/plasmids | Relevant characteristicsa | Source/reference |
|---|---|---|
| Strains | ||
| B. cenocepacia | ||
| K56-2 | ET12 clone related to J2315, CF clinical isolate | BCRRCb (46) |
| K56-2 ΔatsR | Deletion of atsR | (34) |
| K56-2 ΔcepI | Deletion of cepI | (16) |
| K56-2 ΔatsRΔcepI | Deletion of cepI in K56-2 ΔatsR | (16) |
| K56-2 ΔatsR atsR+ | atsR integration in K56-2 ΔatsR | This study |
| K56-2 ΔatsRΔcepI atsR+ | atsR integration in K56-2 ΔatsRΔcepI | This study |
| K56-2 ΔatsRΔcepI atsRH245A+ | atsRH245A integration in K56-2 ΔatsRΔcepI | This study |
| K56-2 ΔatsRΔcepI atsRD536A+ | atsRD536A integration in K56-2ΔatsRΔcepI | This study |
| K56-2 ΔatsRΔcepI atsRΔRD+ | atsRΔRD integration in K56-2 ΔatsRΔcepI | This study |
| K56-2 ΔatsR atsRH245A+ | atsRH245A integration in K56-2 ΔatsR | This study |
| K56-2 ΔatsR atsRD536A+ | atsRD536A integration in K56-2ΔatsR | This study |
| K56-2 ΔatsR atsRΔRD+ | atsRΔRD integration in K56-2 ΔatsR | This study |
| K56-2 ΔatsRΔcepI +ΔatsT atsRD536A | Deletion of atsT in K56-2 ΔatsRΔcepI atsRD536A+ | This study |
| K56-2 ΔatsRΔcepI +ΔatsT atsRΔRD | Deletion of atsT in K56-2 ΔatsRΔcepI atsRΔRD+ | This study |
| K56-2 ΔatsR ΔatsT atsRD536A+ | Deletion of atsT in K56-2 ΔatsR atsRD536A+ | This study |
| K56-2 ΔatsR ΔatsT atsRΔRD+ | Deletion of atsT in K56-2 ΔatsR atsRΔRD+ | This study |
| K56-2 ΔcepI ΔatsT | Deletion of atsT in K56-2 ΔcepI | This study |
| K56-2 ΔcepI ΔatsT atsT + | atsT integration in K56-2 ΔcepI ΔatsT | This study |
| K56-2 ΔcepI ΔatsT atsTD208A+ | atsTD208A integration in K56-2 ΔcepI ΔatsT | This study |
| K56-2 ΔatsRΔhcp | Deletion of hcp in K56-2 ΔatsR | (34) |
| E. coli | ||
| DH5α | F− φ80lacZ M15 endA1 recA1 supE44 hsdR17 (rK− mK+) deoR thi-1 nupG supE44 gyrA96 relA1 Δ (lacZYA-argF)U169, λ− | Laboratory stock |
| GT115 | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80ΔlacZΔM15 ΔlacX74 recA1 rpsL (StrA) endA1Δdcm uidA(ΔMluI)::pir-116 ΔsbcC-sbcD | Invivogen |
| BL21 | F− dcm ompT lon hsdS (rB− mB−) gal λ (DE3) | Laboratory stock |
| Plasmids | ||
| pAtsRChr | atsR cloned in pMH447 for chromosomal complementation | This study |
| pAtsRH245AChr | atsRH245A cloned in pMH447 for chromosomal complementation | This study |
| pAtsRD536AChr | atsRD536A cloned in pMH447 for chromosomal complementation | This study |
| pAtsRΔRDChr | atsRΔRD cloned in pMH447 for chromosomal complementation | This study |
| pAtsTChr | atsT cloned in pMH447 for chromosomal complementation | This study |
| pAtsTD208AChr | atsTD208A cloned in pMH447 for chromosomal complementation | This study |
| pDAI-SceI-SacB | oripBBR1, TetR, Pdhfr, mob+, expressing I-SceI, SacB | (31) |
| pET28a(+) | Cloning vector, IPTG inducible for N-terminal His6 fusion, Kanr | Laboratory stock |
| pDelatsT | pGPI-SceI with fragments flanking atsT | This study |
| pDelM0378 | pGPI-SceI with fragments flanking bcam0378 | This study |
| pMK1 | pET28a (+) encoding B. cenocepacia bcam0378, His6: kanR | This study |
| pMK2 | pET28a (+) encoding B. cenocepacia atsT, His6: kanR | This study |
| pMK4 | pET28a (+) encoding B. cenocepacia atsR-RDD536A, His6: kanR | This study |
| pMK5 | pET28a (+) encoding B. cenocepacia atsRD536A, His6: kanR | This study |
| pMZ24 | pET28a (+) encoding B. cenocepacia atsR205–605, His6: kanR | M. Al-Zayer |
| pMZ25 | pET28a (+) encoding B. cenocepacia atsR, His6: kanR | M. Al-Zayer |
| pMZ33 | pET28a (+) encoding B. cenocepacia atsR-HK, His6: kanR | M. Al-Zayer |
| pMZ34 | pET28a (+) encoding B. cenocepacia atsR-RD, His6: kanR | M. Al-Zayer |
| pMZ36 | pET28a (+) encoding B. cenocepacia atsR atsRH245A, His6: kanR | M. Al-Zayer |
| pMH447 | pGPI-SceI derivative used for chromosomal complementation | (30) |
| pRK2013 | oricolE1, RK2 derivative, KanR, mob+, tra+ | (28) |
a kanR, kanamycin resistance; TetR, tetracycline resistance.
b B. cepacia Research and Referral Repository for Canadian CF Clinics.
General Molecular Techniques
DNA manipulations were performed as described previously (25). T4 DNA ligase (Roche Diagnostics) and Antarctic phosphatase (New England Biolabs, Pickering, Ontario, Canada) were used as recommended by the manufacturers. E. coli DH5α and E. coli GT115 were transformed by the calcium chloride method (26). Mobilization of complementing plasmids and mutagenesis plasmids into B. cenocepacia K56-2 was performed by triparental mating using E. coli DH5α carrying the helper plasmid pRK2013 (27, 28). DNA amplification by PCR was performed in a Bio-Rad C1000TM Thermal Cycler with Taq or HotStar HiFidelity DNA polymerases (Qiagen, Canada). The oligonucleotide primers are listed in Table 2. DNA sequencing was performed at the DNA sequencing Facility of York University, Toronto, Canada.
TABLE 2.
Oligonucleotide primers
NA indicates the absence of a restriction site.
| Primer no. | 5'-3' Primer sequencea | Restriction enzyme |
|---|---|---|
| 2836 | TTTGCTCGAGTTGTTGGCGGTGATGAGAT | XhoI |
| 2839 | TAGGAATTCAGGATCACGCCGTACTTGTC | EcoRI |
| 2840 | TTTTCTCGAGCACATCGTGTCGCACTACAA | XhoI |
| 2844 | TACGTCTAGAATCTTTAGGGCGATCGGGAAC | XbaI |
| 2869 | TTTCATATGTTTATCGTCGTCATCTTTGT | NdeI |
| 4009 | TTTTTCTAGACACCGAGCAACGCTACAC | XbaI |
| 4010 | TTTTCTCGAGCGTGATGGCCTGTTGCAT | XhoI |
| 4011 | TTTTCTCGAGGATATCGTGCTGCATCT | XhoI |
| 4012 | TTTTGAATTCACCTCGTCGTGCTCGATCT | EcoRI |
| 4632 | AAAACATATGACGCGGCGGCGATGGAAGAA | NdeI |
| 4633 | AAAAGCGGCCGCGGCGAGCAGTGTCTCGACGA | NotI |
| 4634 | AAAACATATGCGCACGCGCGACGACCT | NdeI |
| 4736 | AAAAGCGGCCGCTCAGGCGAGCAGTGTCTCGACGA | NotI |
| 4799 | AAAAGCGGCCGCTCATTCGACCGGCAGCGTCAC | NotI |
| 4800 | AAAACATATGGCGCTGGTGGTCGACGAC | NdeI |
| 4880 | GCGTTCCTCGGGATGGTCAGCGCCGAACTGCGCACGCCGCTG | NA |
| 4881 | CAGCGGCGTGCGCAGTTCGGCGCTGACCATCCCGAGGAACGC | NA |
| 5108 | AAAACATATGTCCACCACCGAGCAGGCCAA | NdeI |
| 5109 | AAAAGAATTCTCAGTTCGTCGCGGCCGCTG | EcoRI |
| 5798 | TTTTTTTCATATGCAACAGGCCATCACG | NdeI |
| 5799 | TTTTGCGGCCGCCTACTCGCCGAGCAGATG | NotI |
| 5866 | TTTTTCTAGAGCTTTGTTAGCAGCCGGATC | XbaI |
| 5885 | TTGATGGCGAGCGATTCTTC | NA |
| 5886 | CCAGTTCTTCAGCGTGACGA | NA |
| 5959 | CTCGTCGTGCTCGCGCTCGAACTGCCG | NA |
| 5960 | CGGCAGTTCGAGCGCGAGCACGACGA | NA |
| 6997 | GATGGCTACATCCTCGCCTGGATGCTCGGCG | NA |
| 6998 | GTCGCCGAGCATCCAGGCGAGGATGTAGCC | NA |
| 7020 | TTTTTTCATATGTGGAGACCCCATCGAGATG | NdeI |
| 7021 | TTTTTTCATATGGCATGCCTGCAGGTCGTCAC | NdeI |
a Restriction endonuclease sites incorporated in the oligonucleotide sequences are underlined.
Plasmid Construction and Chromosomal Complementation
Unmarked and nonpolar deletions were performed as described previously (16, 29). To delete atsT (BCAM0381) and BCAM0378, PCR amplifications of regions flanking these genes were performed individually using 2844–2836 and 2840–2839 primer pairs for atsT and 4009–41010 and 4011–4012 primer pairs for BCAM0378. The amplicons were digested with XbaI-XhoI and XhoI-EcoRI, respectively, and cloned into the mutagenic plasmid pGPI-SceI, also digested with XbaI and EcoRI, giving rise to pDelatsT and pDelM0378. To create His-tagged fusions of BCAM0378, AtsT, AtsR, and its truncated versions including AtsR-HK domain (residues 205–460), AtsR-RD receiver domain (residues 488–606), AtsRΔTM (residues 205–606), and AtsRΔRD, sequences were amplified from B. cenocepacia K56-2 genomic DNA using primers 5798-5799, 5108-5109, 4632-4633, 4799-4634, 4800-4736, 4634-4633, and 4632-4799. Amplicons were digested with the appropriate restriction enzymes and ligated into similarly digested pET28a cloning vector. pET28a (Novagen) was used to engineer C-terminal His-tagged proteins for expression in E. coli BL21. Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit from Stratagene (Santa Clara, CA) as recommended by the supplier. Primers were designed with 15–20 nucleotides flanking each side of the targeted mutation. Plasmid pMZ25 provided the template to create H245A and D536A using primers 4880–4881 and 5959–5960, respectively; pMK2 was the DNA template to create D208A using primers 6997–6998. The resulting PCR products were digested with DpnI and introduced into E. coli DH5α. All constructs and replacement mutants were confirmed by DNA sequencing.
Chromosomal complementations of ΔatsR, ΔatsRΔcepI, or ΔcepIΔatsT were performed using pMH447 (30, 31). Primers 5866 and 4632 were used to PCR amplify atsR and its mutated versions. The amplicon was digested with NdeI-XbaI and cloned into the similarly digested pMH447, giving rise to pAtsRChr, pAtsRH245AChr, pAtsRD536AChr, and pAtsRΔRDChr. Likewise, primers 7020–7021 were used to PCR-amplify atsT and its mutated version. The amplicons were digested with NdeI and cloned into the similarly digested pMH447, giving rise to pAtsTChr and pAtsTD536AChr.
Protein Expression and Purification
For overexpression and purification of recombinant proteins, a single colony was inoculated in LB broth supplemented with 30 μg/ml kanamycin and grown at 37 °C. Absorbance (A600) was monitored until it reached 0.6. The culture was then shifted to 30 °C, and 0.2 mm isopropyl-β-d-thiogalactopyranoside was added to induce the expression of proteins. Cultures were incubated for an additional 4 h. Samples were analyzed by SDS-PAGE and stained with Coomassie Blue to confirm protein expression. Cells were harvested by centrifugation at 8000 × g for 10 min. Cell pellets were resuspended in lysis buffer containing (50 mm sodium phosphate, pH 8, 0.3 m NaCl, 10 mm imidazole, pH 8.0, 10% glycerol, 0.25% Tween 20, and 1× of EDTA-free protease inhibitor mixture (Sigma)) and disrupted by One Shot cell disruptor (Thermo Scientific, Rockville, MD). After centrifugation at 27,000 × g for 30 min at 4 °C to remove the debris, the clarified cell lysate was loaded onto Ni2+-binding Sepharose beads and washed, and the His-tagged proteins were then eluted using increased gradient concentrations of imidazole (125–500 mm). Fractions were analyzed by SDS-PAGE and stained with Coomassie Blue to determine the integrity of the purified protein. Fractions were pooled and buffer-exchanged against dialysis buffer (100 mm Tris-HCl, pH 8, 50 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol (DTT), 10% glycerol) using Thermoscientific Slide-A-Lyzer mini dialysis devices. Proteins were concentrated in Amicon ultrafiltration devices (10-kDa molecular weight cutoff), and protein concentration was determined by standard Bradford assay (Bio-Rad). The purity of proteins was evaluated by SDS-PAGE followed by Coomassie Brilliant Blue staining (32).
In Vitro Phosphorylation Assay
For autophosphorylation and phosphotransfer reactions, 5 μmol of each protein was added to the phosphorylation buffer containing 50 mm Tris-HCl, pH 8.0, 50 mm KCl, 5 mm MgCl2, 1 mm DTT, and 5 μCi of [γ-33P]ATP (specific activity of 3000 Ci/mmol; 3.3 μm stock solution) (PerkinElmer Life Sciences) in a final volume of 10 μl. The reactions were carried out at room temperature for the desired time and were terminated by adding 3× sample buffer (32). Reaction products were separated by electrophoresis on 14% SDS-PAGE gels. Gels were fixed, exposed to a high resolution screen (Kodak), and analyzed using a PhosphorImager with ImageQuant software (Molecular Dynamics 5.0; Amersham Biosciences). For pulse-chase experiments, an excess of nonradioactive ATP (20 mm) was added to the reaction mixture after a 10-min preincubation with [γ-33P]ATP. Aliquots were taken before the addition of unlabeled ATP (time zero) and at various time points after the addition of cold ATP.
Chemical Stability of Phosphorylated His-245 and Asp-536
To probe the chemical stability of the phosphorylated residues, phosphorylated AtsR and AtsRD536A proteins were prepared as described above and treated with 1 m HCl, 1 m NaOH, or left untreated for 45 min at room temperature. The HCl reaction was neutralized with 0.25 volumes of 2 m Tris, pH 8, and analyzed by 16% SDS-PAGE followed by the PhosphorImager.
Protease, Swarming Motility, and Biofilm Formation
Protease assays were performed according to Aubert et al. (16). Briefly, 18-h cultures were normalized to an A600 of 1. The bacterial suspension (3 μl) was spotted onto dialyzed brain heart infusion (D-BHI) agar plates containing 1.5% carnation milk. The plates were incubated at 37 °C and examined for zones of clearing around the bacterial spots at 48 h. The protease activity was recorded by measuring the radius of the surrounding halo (from the outside of the spot to the edge of the halo). Swarming motility assays were performed as described previously (16). Three μl of overnight culture, adjusted to an A600 of 1, was spotted on a swarm plate (0.8% nutrient broth, 0.5% agar, 0.2% glucose). Plates were incubated at 37 °C for 20 h, and the diameters of swarming zones were measured. Biofilm formation assays were performed as described previously (15). Assays were done in triplicate and repeated independently three times.
Macrophage Infections and T6SS Activity
Infections were performed as previously described (15) using the C57BL/6 murine bone marrow-derived macrophage cell line ANA-1. Bacteria were added to ANA-1 cells grown on glass coverslips at a multiplicity of infection of 50. Coverslips were analyzed by phase contrast microscopy after 4 h of incubation at 37 °C. T6SS activity was recorded as the ability of the bacteria to induce the formation of characteristic ectopic actin nucleation around the macrophages (15, 22).
Western Blot Analysis
For E. coli, overnight bacterial cultures in 5 ml of LB were diluted to an initial A600 of 0.2 and incubated at 37 °C until reaching an A600 of 0.7. At this point, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.2 mm. Cells were incubated for 4 h at 30 °C and then harvested by centrifugation at 8000 × g for 10 min at 4 °C. The bacterial pellet was suspended in lysis buffer, and the suspension was lysed using a One-shot cell disrupter (Thermo Scientific). Cell debris were removed by centrifugation (15,000 × g for 15 min at 4 °C), and the clear supernatant was centrifuged at 40,000 × g for 30 min at 4 °C. The pellet, containing total membranes, was suspended in lysis buffer. The protein concentration was determined by the Bradford assay (Bio-Rad). Staining was performed with Coomassie Brilliant Blue. SDS-PAGE, protein transfers to nitrocellulose membranes, and immunoblots were performed as described (33). For detection of His6-tagged proteins, membranes were incubated with a 1:10,000 dilution of anti-His IgG2a monoclonal antibodies (Amersham Biosciences) and Alexa Fluor 680 anti-mouse IgG antibodies (Molecular Probes). For B. cenocepacia, overnight bacterial cultures in 5 ml of LB were diluted to an initial A600 of 0.2 and incubated at 37 °C for 8 h. His-tagged proteins were purified and detected by Western blot using an anti-His antibody as indicated above.
RESULTS
Deletion of atsT Causes the Same Phenotypes as ΔatsR Mutants
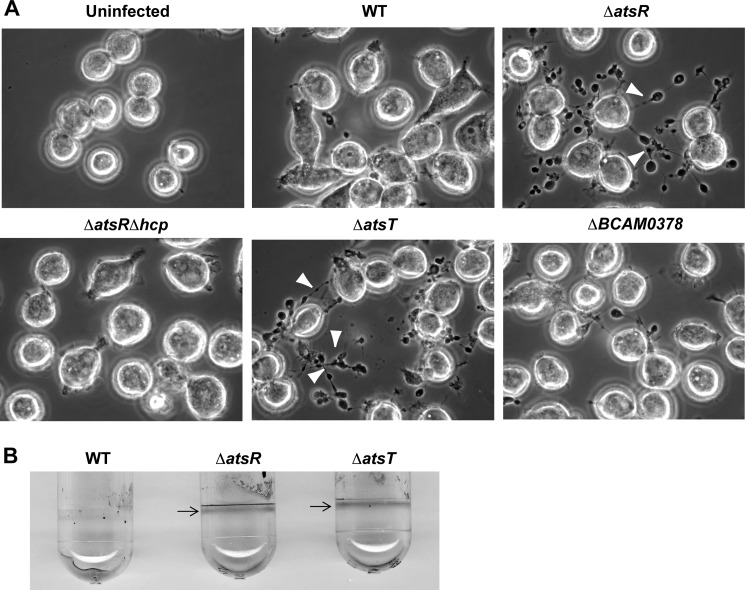
We reasoned that if AtsT (BCAM0381) was involved in the AtsR phosphorelay pathway, ΔatsT and ΔatsR mutants should have similar phenotypes. Therefore, ΔatsT was tested in the macrophage-infection model. Phase-contrast microscopy revealed that, in contrast to K56-2, ΔatsR and ΔatsT noticeably induce the formation of pearls on a string-like structures around infected macrophages (Fig. 1A, arrowheads). These structures depend on T6SS-mediated rearrangements of host actin and are characteristic of an up-regulated T6SS (15, 16, 22). B. cenocepacia ΔatsRΔhcp, a T6SS-defective mutant, was used as a negative control during the infections (34), and as expected, it did not mediate the pearls on a string phenotype in infected macrophages (Fig. 1A). Similarly to ΔatsR (15), ΔatsT showed increased biofilm formation as evidenced by a robust ring of biofilm at the air-liquid interface (Fig. 1B). Together, these results strongly suggest that AtsT and AtsR are in the same regulatory pathway, which is likely initiated by AtsR.
FIGURE 1.
T6SS activity and biofilm formation of B. cenocepacia K56-2 (WT) and its mutant derivatives. A, phase-contrast microscopy of infected ANA-1 murine macrophages to assess T6SS activity. Infections were performed at a multiplicity of infection of 50 for 4 h. White arrowheads indicate the presence of ectopic actin nucleation (pearls-on-a-string phenotype (15, 22)) extending from infected macrophages, which denotes T6SS activity. B. cenocepacia K56-2 ΔatsRΔhcp, a T6SS-defective mutant, was used as a negative control during the infections. Experiments consisted of three independent biological repeats where similar results were obtained. B, biofilm formation by parental strains, ΔatsR and ΔatsT mutants. B. cenocepacia K56-2 wild-type and derivative mutants were tested for biofilm formation by crystal violet staining. Arrows indicate the ring corresponding to the biofilm formation characteristic in ΔatsR (15) and ΔatsT mutants. The experiment was repeated three times in triplicate, and pictures were taken after 24 h of static incubation at 37 °C.
Identification of Phosphoacceptor Residues within AtsR
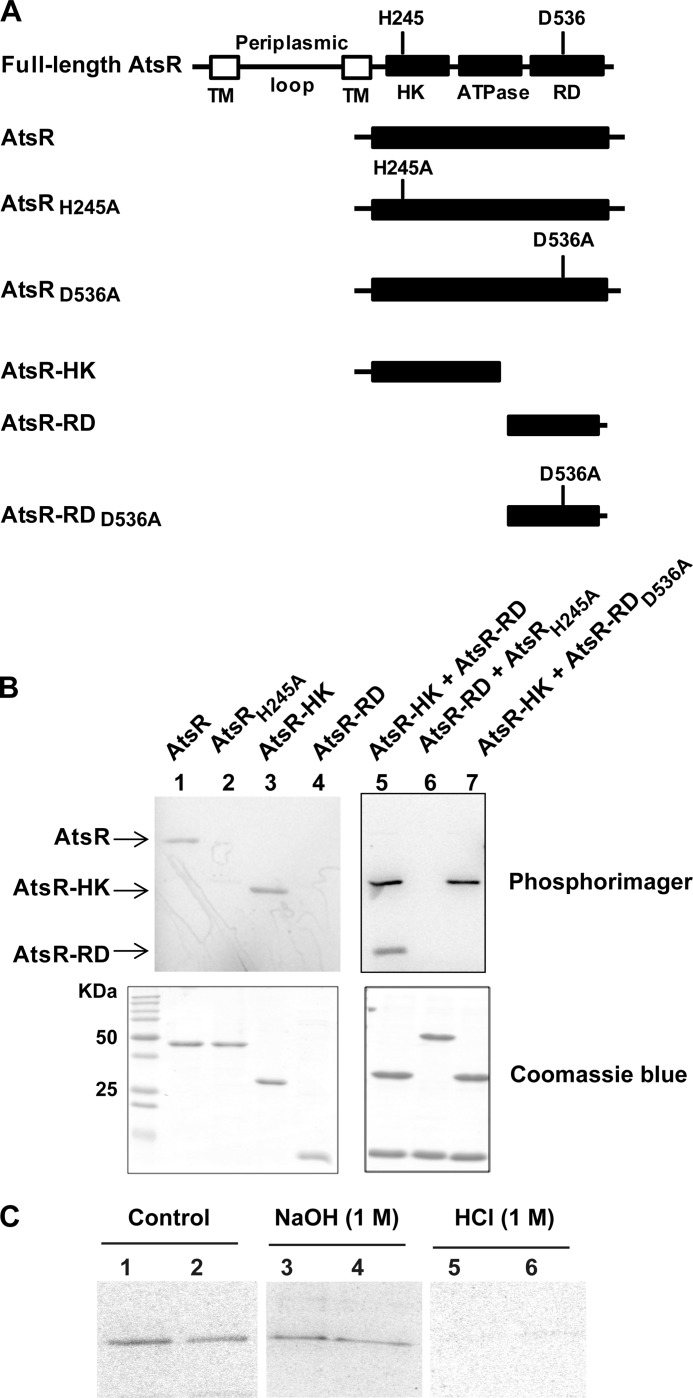
The region of AtsR corresponding to the HK domain (amino acids 233–457) was aligned with the HK domain of two well characterized hybrid sensor kinase proteins, ArcB from E. coli and RetS from Pseudomonas aeruginosa (3, 35–37). The amino acid sequence of AtsR (606 amino acids) is 34% identical and 50% similar to that of ArcB and 26% identical and 48% similar to that of RetS. The invariant His residue that is autophosphorylated in these proteins corresponds to His-245 in AtsR. Also, the AtsR-RD (amino acids 488–601) showed 52 and 50% similarity at the amino acid level to ArcB and RetS, respectively. This region has an invariant Asp at position 536, which corresponds to the site of phosphorylation in ArcB and RetS.
Truncated versions of AtsR spanning AtsR-HK and AtsR-RD were constructed and purified to analyze their biochemical properties (Fig. 2A). Residues 1–200, comprising the membrane-spanning domains, were removed to facilitate protein solubility and purification. Furthermore, to assess the contribution of conserved His and Asp residues to AtsR phosphorylation, His-245 and Asp-536 were individually replaced by alanine (Fig. 2A). These proteins were assessed for in vitro autophosphorylation using [γ-33P]ATP. Only polypeptides containing His-245 became autophosphorylated (Fig. 2B, lanes 1 and 3). Therefore, AtsR HK containing the native His-245 is necessary and sufficient for initiation of the phosphorelay pathway as the replacement of His with alanine at position 245 abolished autophosphorylation (Fig. 2B, lanes 2 and 6). Moreover, the AtsR-RD is unable to autophosphorylate in the absence of the HK domain (Fig. 2B, lane 4). The ability of AtsR-RD to accept the phosphoryl group from the AtsR-HK was tested in a phosphotransfer assay (Fig. 2B, lanes 5–7). Two bands were obtained after incubating AtsR-HK and AtsR-RD together in a reaction with [γ-33P]ATP (Fig. 2B, lane 5). These bands corresponded to the phosphorylated form of these proteins, indicating the transfer of phosphoryl group from the HK domain to the RD domain of AtsR. To test whether Asp-536 within AtsR-RD is the residue accepting the phosphate from His-245, AtsR-RDD536A was added to the reaction with AtsR-HK and [γ-33P]ATP. No band corresponding to AtsR-RDD536A was detected (Fig. 2B, lane 7), clearly indicating that Asp-536 is necessary for phosphorylation.
FIGURE 2.
Functional analysis of AtsR domains. A, schematic domain organization of AtsR and its derivatives (domains are not drawn to scale). The predicted sites of phosphorylation are His-245 (H245) and Asp-536 (D536). TM, transmembrane domain; HK, histidine kinase domain; ATPase, ATPase domain; RD, receiver domain; A, alanine. B, In vitro phosphorylation assays. Five μmol of purified AtsR, AtsRH245A, AtsR-HK, AtsR-RD, AtsR-HK and AtsR-RD, AtsRH245A and AtsR-RD, and AtsR-HK and AtsR-RDD536A were added in a standard phosphorylation mixture (100 mm Tris-HCl, pH 8, 50 mm KCl, 5 mm MgCl2, 1 mm DTT, 5 μCi [γ-33P]ATP) and incubated for 15 min at 25 °C. Reactions were terminated by adding 3× SDS-PAGE loading buffer and resolved by SDS-PAGE. The phosphorylated proteins were visualized using a PhosphorImager (top). Phosphorylated and non-phosphorylated proteins were detected by Coomassie Blue staining (bottom). The location of phosphorylated bands of the AtsR, AtsR-HK, and AtsR-RD proteins are denoted with arrows. C, chemical stability of phosphorylated proteins. Phosphorylated AtsR (lanes 1, 3, and 5) and AtsRD536A (lanes 2, 4, and 6) were treated with 1 m NaOH or 1 m HCl or were left untreated for 45 min at room temperature. The reactions were neutralized with 0.25 volumes of 2 m Tris, pH 8, and analyzed by PhosphorImager after SDS-PAGE.
Chemical Stability of Phosphorylated Proteins
Phosphoramidates, such as histidine or lysine phosphate, are stable in alkali conditions but extremely labile to acid. Acyl phosphates such as aspartate and glutamate, on the other hand, are labile to either acid or alkali treatment (38). To compare the chemical stability of the phosphorylation of AtsR and AtsRD536A proteins, phosphorylation reactions were subjected to either no treatment or treatment with acid or alkali. If AtsR is phosphorylated at both His-245 and Asp-536, whereas AtsRD536A is only phosphorylated at His-245, the two phosphorylated proteins should behave differently upon treatment with base. Indeed, AtsR and AtsRD536A were both labile to acid (Fig. 2C, lanes 5 and 6) and relatively stable to base (lanes 3 and 4), indicating that these two proteins contain amidylphosphates. The quantitative difference between phosphorylated AtsR before and after alkali treatment in Fig. 2C suggests that at least a portion of the wild-type protein may be phosphorylated at the Asp-536. In quantitative analysis by densitometry, the phosphorylated AtsR retained 82 ± 6% of the label after alkali treatment compared with the untreated control, whereas AtsRD536A retained 91 ± 5% of its label. These results suggest that the majority of the phosphate in both AtsR and AtsRD536A is in the amidylphosphate form.
AtsR and AtsT Form a Cognate HK-RR Pair
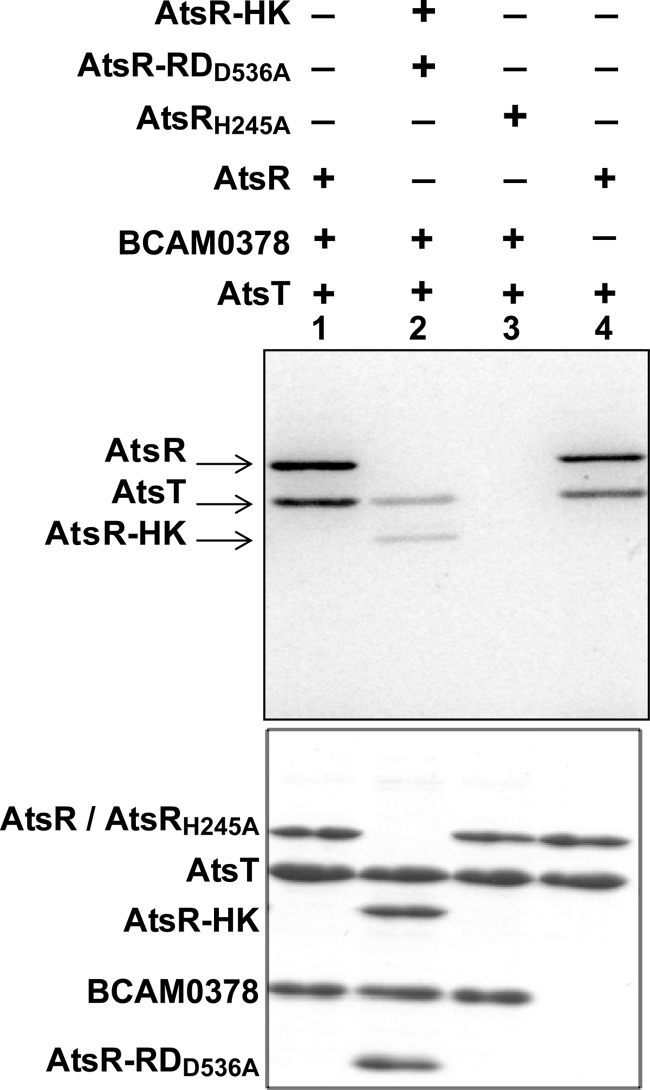
We initially hypothesized that BCAM0378 could be a histidine-phosphotransfer component of the AtsR phosphorelay based on the proximity of BCAM0378 to atsR (BCAM0379) and bioinformatics data indicating the presence of highly conserved His and Asp residues within this family of proteins. To investigate the relationship between AtsR, BCAM0378, and AtsT, we performed phosphotransfer assays employing combinations of these different proteins. The histidine-phosphotransfer candidate, BCAM0378, was not phosphorylated (Fig. 3, lanes 1–3). To rule out the possibility of a rapid phosphotransfer from AtsR to BCAM0378, we repeated the experiment at 1 and 5 min incubation times and increased the temperature to 30 °C. A phosphorylated form of BCAM0378 was never detected under any condition tested (data not shown). In contrast, AtsT was phosphorylated directly by AtsR and independently from BCAM0378 (Fig. 3, lane 4). To test whether AtsT autophosphorylates independently from AtsR, we incubated AtsT with AtsRH245A and BCAM0378 (Fig. 3, lane 3). No phosphorylated band was detected, indicating that AtsT cannot autophosphorylate in vitro without the native AtsR-phosphorylated HK domain. Together, these data suggest that AtsR acts as a conventional two-component signal transduction system that mediates signal transduction from a sensor kinase to a response regulator, and BCAM0378 is not involved in the transfer of the phosphate from AtsR to the AtsT under the conditions tested. This conclusion was supported by the lack of the pearls-on-a-string phenotype in macrophages infected with a ΔBCAM0378 mutant (Fig. 1A), strongly suggesting that BCAM0378 is not part of the AtsR regulatory pathway.
FIGURE 3.
Phosphotransfer from AtsR and its derivatives to the AtsT response regulator. Five μmol of AtsR, AtsRH245A, and AtsR-HK was preincubated in individual standard phosphorylation mixtures (100 mm Tris-HCl pH 8, 50 mm KCl, 5 mm MgCl2, 1 mm DTT, 5 μCi [γ-33P]ATP) for 15 min at 25 °C followed by the addition of 5 μmol of AtsT and/or BCAM0378. Reactions were terminated by adding 3× SDS-PAGE loading buffer after 15 min. Samples were resolved on 16% SDS-PAGE gel and stained with Coomassie Blue (bottom). Phosphorylated proteins were visualized by a PhosphorImager (Top). Phosphorylated bands corresponding to the expected masses of AtsR, AtsR-HK, and AtsT polypeptides are indicated by arrows.
Kinetics of AtsR Phosphorylation
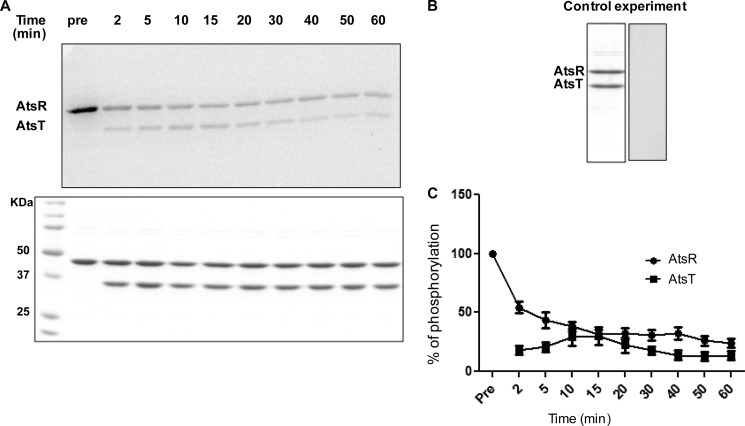
A pulse-chase experiment was performed to determine the stability of AtsR phosphorylation in the presence of AtsT and to determine whether phosphorylation of AtsT can take place independently from AtsR in excess of ATP. If AtsT is phosphorylated from the free nucleotide pool, then an excess of unlabeled ATP should compete with hot ATP for AtsT phosphorylation in a pulse-chase reaction. Conversely, if AtsR phosphorylates AtsT, then the label should be chased from AtsR to AtsT. AtsR was incubated with [γ-33P]ATP for 10 min, and an excess of unlabeled ATP (20 mm) and AtsT were simultaneously added to the reaction. Labeling of AtsT coincided with immediate loss of signal from phosphorylated AtsR (Fig. 4A). A decrease of phosphorylation of nearly 50% was observed between time 0 (preincubation) and 2 min (Fig. 4C). Phosphorylated AtsR was highly stable, and the signal remained relatively strong at 60 min. We repeated this experiment and chased phosphorylated proteins for up to 80 min, and phosphorylated bands were still visible (data not shown). To control the quality of unlabeled ATP, AtsR was incubated simultaneously with both labeled and unlabeled ATP for 10 min followed by the addition of AtsT (Fig. 4B). No labeling of AtsR or AtsT was detected in the control reaction, indicating that incorporation of γ-33P can be inhibited by an excess of unlabeled ATP, which competes with labeled ATP in the reaction. Thus, AtsR phosphorylation at His-245 is stable, and AtsT phosphorylation is resistant to competition from excess ATP, which confirms that AtsT acquires its phosphate directly from AtsR.
FIGURE 4.
Kinetics of phosphotransfer from AtsR to AtsT. A, 5 μmol of AtsR were preincubated with 5 μCi ([γ-33P]ATP) in a standard phosphorylation mixture (100 mm Tris-HCl, pH 8, 50 mm KCl, 5 mm MgCl2, 1 mm DTT) for 15 min, and then 5 μmol of AtsT and 20 mm ATP were simultaneously added to the reaction. The reaction was chased over time at 25 °C. Aliquots were removed before and after the chase at the times indicated. Reactions were terminated by adding SDS-PAGE loading buffer. Samples were run on 16% SDS-PAGE gel and stained with Coomassie Blue (bottom). Phosphorylated proteins were visualized by a PhosphorImager (top). The images shown here are the representatives of two independent repeats. B, 5 μmol of AtsR was incubated simultaneously with both labeled and unlabeled ATP for 10 min followed by the addition of 5 μmol of AtsT and incubated for 15 min. Samples were run on 16% SDS-PAGE gels and stained with Coomassie Blue (left) or visualized by a PhosphorImager (right). C, the y axis represents the percentage of normalized absorbance of densitometry readings from bands corresponding to phosphorylated proteins obtained from two independent experiments.
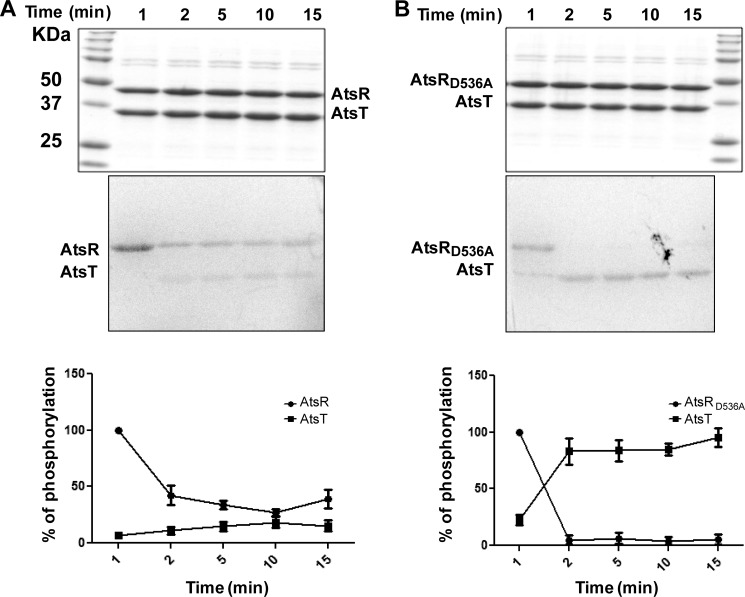
The Role of Asp-536 on AtsR-AtsT Phosphorylation Kinetics
To determine the effect of Asp-536 on phosphotransfer, AtsR and AtsRD536A were incubated with [γ-33P]ATP for 10 min in individual reactions to make a pool of phosphorylated proteins followed by the addition of AtsT to each reaction. The rate of phosphate incorporation to the RR was followed as a function of time. The D536A replacement did not abolish the ability of AtsR to autophosphorylate, but the phosphotransfer capabilities of the parental and AtsRD536A proteins were quite different. The transfer of phosphate from AtsR to AtsT took place not only slower but also at a lower rate than from AtsRD536A to AtsT. Indeed, >90% of the phosphate was transferred from AtsRD536A to AtsT after 90 s, and the amount of label gained by AtsT was almost equal to that lost by AtsRD536A (Fig. 5, A and B). The increase in AtsT phosphorylation correlates with the disappearance of the AtsR and AtsRD536A phosphorylation signals. In contrast to native AtsR, where the phosphorylation signal was maintained from 2 min until the end of the experiment, phosphorylation of AtsT by AtsRD536A was rapidly detected 1 min after the addition of RR to the reaction, and the signal of AtsRD536A had already disappeared after 2 min under the conditions tested (Fig. 5B). Together, these results indicate that replacing Asp-536 with alanine increases and accelerates the phosphotransfer reaction to the RR. It also might be due to the fact that AtsR has two sites of phosphorylation and only His-245 transfers phosphate to AtsT, and therefore, the persistence of the signal on wild-type protein is the result of Asp-536 phosphorylation.
FIGURE 5.
Kinetics of phosphotransfer from AtsR and AtsRD536A to AtsT. After 10 min of preincubation of 5 μmol AtsR (A) or AtsRD536A (B) in a standard phosphorylation mixture (100 mm Tris-HCl, pH 8, 50 mm KCl, 5 mm MgCl2, 1 mm DTT, 5 μCi [γ-33P]ATP), AtsT was added to the reaction, and aliquots were removed at the times indicated. The reaction was performed at 25 °C and terminated by adding SDS-PAGE loading buffer. Samples were run on 16% SDS-PAGE gel and stained with Coomassie Blue (top). Phosphorylated proteins were visualized with a PhosphorImager (middle). The images shown here are the representatives of two independent repeats. The y axis represents the percentage of normalized absorbance of densitometry readings from bands corresponding to phosphorylated proteins obtained from two independent experiments (bottom).
In Vivo Reconstitution of the AtsR Signaling Pathway
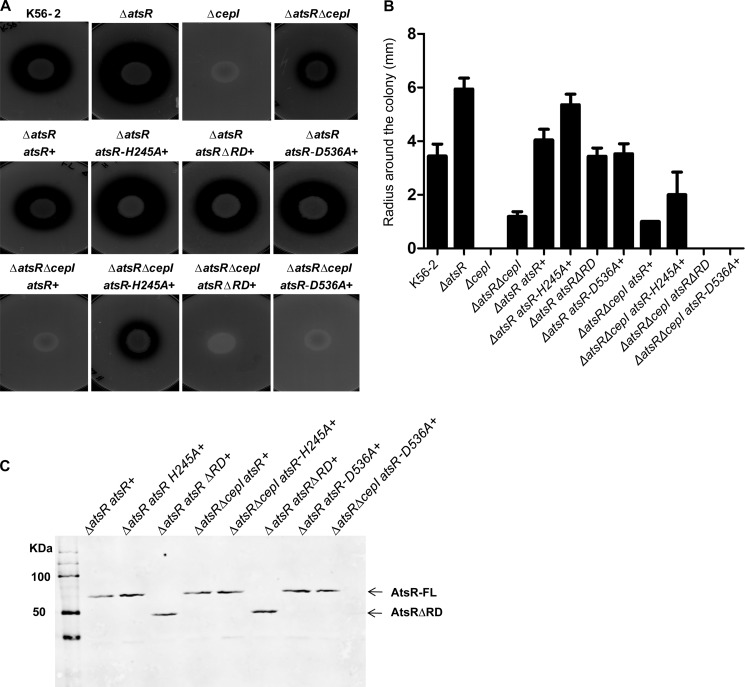
B. cenocepacia secretes two zinc metalloproteases, ZmpA and ZmpB (17–21). Deletion of atsR up-regulates the expression of several quorum-sensing regulated virulence factors, including swarming motility and the secretion of zinc metalloproteases, both of which are also positively regulated by the CepI/CepR quorum-sensing system (16). Moreover, whereas a ΔcepI mutant was protease-deficient at 48 h when spotted on D-BHI milk agar plates, further deletion of atsR in ΔcepI resulted in increased proteolytic activity, demonstrating that deletion of atsR up-regulates protease activity independently of quorum sensing (16). We took advantage of the protease and swarming motility phenotypes to investigate whether AtsR phosphorylation is required for its function in vivo. The mutant strains ΔatsR and ΔatsRΔcepI were complemented at the chromosomal level with full-length atsR or atsR variants encoding either AtsRH245A, AtsRD536A, or AtsR lacking the RD (AtsRΔRD), eliminating the putative effect of other conserved Asp residues adjacent to Asp-536. The ability of AtsRH245A, AtsRD536A, and AtsRΔRD to suppress the phenotypes of ΔatsR and ΔatsRΔcepI mutant backgrounds was first assessed using protease secretion assay (Fig. 6A). As expected, the radius of halo corresponding to casein degradation surrounding the ΔatsR spot was bigger compared with that of the wild type within 48 h of incubation, whereas no halo was present for ΔcepI (Fig. 6, A and B). Proteolytic activity was also detectable for ΔatsRΔcepI to a lesser extent compared with WT and ΔatsR (Fig. 6, A, top row, and B). Successful complementation was achieved when a chromosomal copy of the atsR gene was restored into ΔatsR and in ΔatsRΔcepI, leading to decreased protease activity to wild-type and ΔcepI levels, respectively (Fig. 6, A, middle and bottom row, and B).
FIGURE 6.
Proteolytic activity of B. cenocepacia K56-2 wild-type, ΔatsR, ΔatsRΔcepI, and ΔcepI mutants and complemented mutants at the chromosomal level in different genetic backgrounds. A, proteolysis was tested on D-BHI milk agar plates. The plates shown are representatives of three experiments performed in triplicate. Zones of clearing around the colonies were measured at 48 h of incubation at 37 °C. B, values are the average radius ± S.D. in millimeters of three experiments performed in triplicate. C, anti-His Western blot analysis of the His-tag-purified membrane pellet of AtsR, AtsRΔRD, AtsRD536A, and AtsRH245A in B. cenocepacia. Arrows indicate the positions of full-length AtsR (AtsR, AtsRD536A, and AtsRH245A) and AtsRΔRD. AtsR-FL, full-length AtsR.
Because Asp-536 significantly increases the phosphotransfer from His-245 to AtsT in vitro, one could have expected that complementation of ΔatsR with AtsRD536A would have a stronger inhibitory effect than atsR in vivo. However, similar results were obtained when either atsRΔRD or atsRD536A was introduced to ΔatsR or ΔatsRΔcepI, respectively. This suggests that Asp-D536 does not have a strong modulatory effect on the AtsR-AtsT phosphotransfer in vivo under the conditions tested or, alternatively, that additional components not identified in this study may be involved in modulating the AtsR-AtsT phosphorelay pathway.
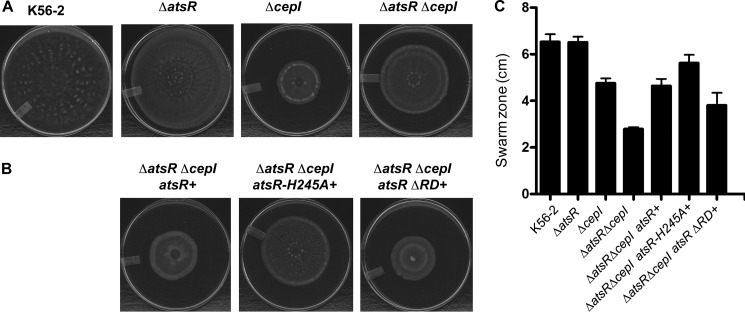
In contrast, the phosphorylation status of AtsR is critical for its role in expression and/or secretion of proteases, as complementation failed when atsRH245A was introduced. These strains were also tested in a swarming motility assay. In agreement with previous results, only complementation of ΔatsRΔcepI strains with atsR or atsRΔRD, but not atsRH245A, could restore swarming motility to ΔcepI levels (Fig. 7). Protein expression was confirmed by Western blot analysis of bacterial cell lysates prepared from K56-2 ΔatsR and ΔatsRΔcepI complemented with atsRH245A, atsRD536A, atsRΔRD, and atsR and demonstrated that the encoded AtsR variants were similarly expressed (Fig. 6C). Thus, a lack of complementation by atsRH245A was not due to a defect in protein expression. Together, these data confirm the in vitro results and suggest that His-245 is essential for the function of AtsR in vivo.
FIGURE 7.
Swarming motility. B. cenocepacia K56-2 wild type and derivative mutants (A) and ΔatsR ΔcepI mutant complemented by the integration of atsR, atsR-H245A, and atsRΔRD at the chromosomal level (B) were tested for swarming motility. The plates are representatives of at least three experiments performed in triplicate. The extent of the swarm zone was measured, and error bars represent the S.D. (C).
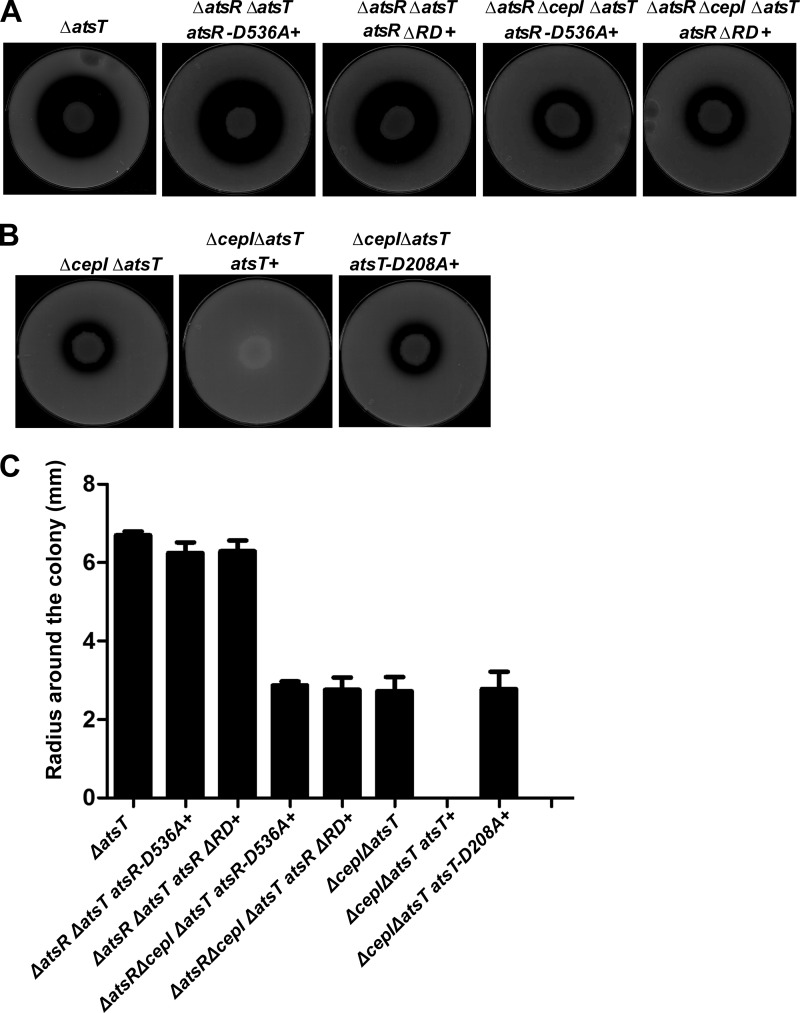
To confirm that AtsT is the cognate response regulator, which negatively controls the expression of protease activity when it is phosphorylated, atsT was deleted in ΔatsR, ΔatsRΔcepI, and ΔcepI strains, which were complemented with atsRD536A, atsRΔRD, atsT, or atsTD208A. The resulting strains were spotted on D-BHI-milk agar plates (Fig. 8, A and B), and the proteolysis was quantified by measuring the radius of clearing around the colonies (Fig. 8C). Although complementation of ΔatsR and ΔatsRΔcepI strains with atsRD536A or atsRΔRD reduced the proteolytic activity to WT and ΔcepI levels (Fig. 6), further deletion of atsT bypassed the atsRD536A or atsRΔRD complementation and resulted in an increase of proteolytic activity. Furthermore, ΔcepIΔatsT has the same phenotype as ΔatsRΔcepI, and as expected, by complementing ΔcepIΔatsT with atsT, proteolytic activity decreases to ΔcepI level, whereas atsTD208A was unable to complement. These results suggest that (i) AtsT indeed acts as a negative regulator and (ii) is a direct target of AtsR contributing to the regulatory role of this protein on proteolytic activity, and (iii) Asp-208 on AtsT is required for its function.
FIGURE 8.
Proteolytic activity of B. cenocepacia ΔatsT, ΔatsRΔatsT atsRΔRD+, ΔatsRΔcepIΔatsT atsRΔRD+, ΔatsRΔatsT atsRD536A+, and ΔatsRΔcepIΔatsT atsRD536A+. A, atsT was deleted from B. cenocepacia K56-2 ΔatsR atsRΔRD+, ΔatsRΔcepI atsRΔRD+, ΔatsR atsRD536A+, and ΔatsRΔcepI atsRD536A+ backgrounds. Mutants were tested on D-BHI milk agar plates. The plates shown are representatives of three experiments performed in triplicate. Zones of clearing around the colonies were measured at 48 h of incubation at 37 °C. B, atsT was deleted from B. cenocepacia K56-2 ΔcepI background, and the resulting strain was complemented with either atsT or atsTD208A at the chromosomal level. Mutants were tested on D-BHI milk agar plates. The plates shown are representatives of three experiments performed in triplicate. C, values are the average radius in millimeters of three experiments performed in triplicate.
DISCUSSION
The predicted structural features of AtsR suggested this protein could not directly control gene expression because it lacks an effector domain. We demonstrate in this study three key properties of AtsR: (i) upon autophosphorylation, AtsR transfers the phosphate to the response regulator AtsT without the participation of an intermediate histidine-phosphotransfer protein; (ii) AtsR function in vitro and in vivo depends on autophosphorylation of the His-245 residue, which is absolutely essential for initiation of signal transduction; (iii) the AtsR-RD and more specifically the Asp-536 to some extent plays a role in modulating the stability of phosphorylated AtsR.
Other studies have shown that the sequential phosphotransfer between residues within the same hybrid sensor kinase modulates the phosphotransfer to the cytosolic response regulator and the overall response by determining the specificity for the cognate RR or by regulating the autokinase activity (2, 3, 39–42). The stability of the phosphorylated AtsR and AtsT determined by the pulse-chase kinetic experiments is remarkable as these proteins can maintain the response memory for at least 60 min, which agrees with their physiological function as negative regulators of gene expression. Comparative studies have suggested that the in vitro assay of HK autophosphorylation reflects, to a certain extent, the in vivo situation (43). For example, ArcB, NarQ, and NtrB, with a high rate of phosphotransfer but a low level of autophosphorylation, are able to respond quickly to changes in the environment thereby returning to the steady-state levels after transient activation or repression by external stresses, whereas CheA and BaeS showed high levels of self-phosphorylation even though they had a slow signal transduction rate (43). In the case of AtsR, we find that the phosphate is transferred very quickly within 2 min from AtsR to AtsT, and both proteins are capable of prolonged phosphorylation. This may indicate that unlike some other systems (44, 45), AtsR might not have phosphatase activity, although further experiments are needed to validate this notion.
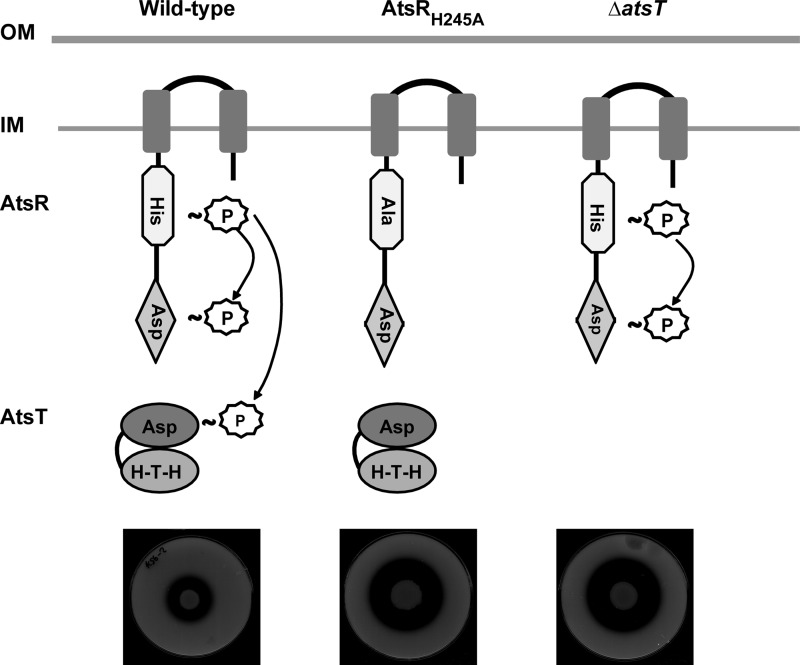
Based on the consistency of results obtained by in vitro and in vivo experimental approaches in this study, we propose a model (Fig. 9) in which AtsR autophosphorylates in vivo at His-245. The phosphate is then transferred to AtsT and partly to the AtsR receiver domain on Asp-536, as stable phosphorylation of the AtsR receiver domain was detected in vitro. Based on these findings, phosphorylation of AtsT in vitro correlates with repression of B. cenocepacia gene expression in vivo. Our data strongly support the notion that AtsR phosphorylation has a significant biological relevance as a global virulence regulator modulating the expression of proteases through AtsT. Experiments are under way to identify the genes that are specifically controlled by AtsT as well as the environmental signals that trigger activation or inactivation of the AtsR/AtsT phosphorelay.
FIGURE 9.
Diagram depicting a model for AtsR phosphorelay. OM, outer membrane; IM, inner membrane; HTH, helix-turn-helix; P, phosphate. Plates denote the proteolytic activity expected in each condition.
Acknowledgment
We gratefully thank M. Al-Zayer for the construction of some of the plasmids used in this study.
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada.
- RR
- response regulator protein
- D-BHI
- dialyzed brain heart infusion
- HK
- histidine kinase
- RD
- receiver domain
- T6SS
- type 6 secretion system.
REFERENCES
- 1. Wolanin P. M., Thomason P. A., Stock J. B. (2002) Histidine protein kinases. Key signal transducers outside the animal kingdom. Genome Biol. 3, REVIEWS3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krell T., Lacal J., Busch A., Silva-Jiménez H., Guazzaroni M. E., Ramos J. L. (2010) Bacterial sensor kinases. Diversity in the recognition of environmental signals. Annu. Rev. Microbiol. 64, 539–559 [DOI] [PubMed] [Google Scholar]
- 3. Georgellis D., Lynch A. S., Lin E. C. (1997) In vitro phosphorylation study of the arc two-component signal transduction system of Escherichia coli. J. Bacteriol. 179, 5429–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Uhl M. A., Miller J. F. (1994) Autophosphorylation and phosphotransfer in the Bordetella pertussis BvgAS signal transduction cascade. Proc. Natl. Acad. Sci. U.S.A. 91, 1163–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mahenthiralingam E., Baldwin A., Dowson C. G. (2008) Burkholderia cepacia complex bacteria. Opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104, 1539–1551 [DOI] [PubMed] [Google Scholar]
- 6. Vandamme P., Dawyndt P. (2011) Classification and identification of the Burkholderia cepacia complex. Past, present, and future. Syst. Appl. Microbiol. 34, 87–95 [DOI] [PubMed] [Google Scholar]
- 7. Bevivino A., Costa B., Cantale C., Cesarini S., Chiarini L., Tabacchioni S., Caballero-Mellado J., Dalmastri C. (2011) Genetic relationships among Italian and Mexican maize-rhizosphere Burkholderia cepacia complex (BCC) populations belonging to Burkholderia cenocepacia IIIB and BCC6 group. BMC Microbiol. 11, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McNeely D., Moore J. E., Elborn J. S., Millar B. C., Rendall J., Dooley J. S. (2009) Isolation of Burkholderia cenocepacia and Burkholderia vietnamiensis from human sewage. Int. J. Environ. Health Res. 19, 157–162 [DOI] [PubMed] [Google Scholar]
- 9. Lee Y. A., Chan C. W. (2007) Molecular typing and presence of genetic markers among strains of banana finger-tip rot pathogen, Burkholderia cenocepacia, in Taiwan. Phytopathology 97, 195–201 [DOI] [PubMed] [Google Scholar]
- 10. Burkholder W. H. (1950) Sour skin, a bacterial rot of onion bulbs. Phytopathol. 40, 115–117 [Google Scholar]
- 11. Vergunst A. C., Meijer A. H., Renshaw S. A., O'Callaghan D. (2010) Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infect. Immun. 78, 1495–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uehlinger S., Schwager S., Bernier S. P., Riedel K., Nguyen D. T., Sokol P. A., Eberl L. (2009) Identification of specific and universal virulence factors in Burkholderia cenocepacia strains by using multiple infection hosts. Infect. Immun. 77, 4102–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lamothe J., Huynh K. K., Grinstein S., Valvano M. A. (2007) Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacteria-containing vacuoles. Cell. Microbiol. 9, 40–53 [DOI] [PubMed] [Google Scholar]
- 14. Lamothe J., Thyssen S., Valvano M. A. (2004) Burkholderia cepacia complex isolates survive intracellularly without replication within acidic vacuoles of Acanthamoeba polyphaga. Cell. Microbiol. 6, 1127–1138 [DOI] [PubMed] [Google Scholar]
- 15. Aubert D. F., Flannagan R. S., Valvano M. A. (2008) A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect. Immun. 76, 1979–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aubert D. F., O'Grady E. P., Hamad M. A., Sokol P. A., Valvano M. A. (2013) The Burkholderia cenocepacia sensor kinase hybrid AtsR is a global regulator modulating quorum-sensing signalling. Environ. Microbiol. 15, 372–385 [DOI] [PubMed] [Google Scholar]
- 17. Baldwin A., Sokol P. A., Parkhill J., Mahenthiralingam E. (2004) The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72, 1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kooi C., Subsin B., Chen R., Pohorelic B., Sokol P. A. (2006) Burkholderia cenocepacia ZmpB is a broad-specificity zinc metalloprotease involved in virulence. Infect. Immun. 74, 4083–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Malott R. J., Baldwin A., Mahenthiralingam E., Sokol P. A. (2005) Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect. Immun. 73, 4982–4992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malott R. J., O'Grady E. P., Toller J., Inhülsen S., Eberl L., Sokol P. A. (2009) A Burkholderia cenocepacia orphan LuxR homolog is involved in quorum-sensing regulation. J. Bacteriol. 191, 2447–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sokol P. A., Sajjan U., Visser M. B., Gingues S., Forstner J., Kooi C. (2003) The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149, 3649–3658 [DOI] [PubMed] [Google Scholar]
- 22. Rosales-Reyes R., Skeldon A. M., Aubert D. F., Valvano M. A. (2012) The type VI secretion system of Burkholderia cenocepacia affects multiple Rho family GTPases disrupting the actin cytoskeleton and the assembly of NADPH oxidase complex in macrophages. Cell. Microbiol. 14, 255–273 [DOI] [PubMed] [Google Scholar]
- 23. Galperin M. Y. (2006) Structural classification of bacterial response regulators. Diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sambrook J., Russell D. W. (2006) The condensed protocols from molecular cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Cohen S. N., Chang A. C., Hsu L. (1972) Nonchromosomal antibiotic resistance in bacteria. Genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. U.S.A. 69, 2110–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Craig F. F., Coote J. G., Parton R., Freer J. H., Gilmour N. J. (1989) A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135, 2885–2890 [DOI] [PubMed] [Google Scholar]
- 28. Figurski D. H., Helinski D. R. (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Flannagan R. S., Linn T., Valvano M. A. (2008) A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol. 10, 1652–1660 [DOI] [PubMed] [Google Scholar]
- 30. Hamad M. A., Di Lorenzo F., Molinaro A., Valvano M. A. (2012) Aminoarabinose is essential for lipopolysaccharide export and intrinsic antimicrobial peptide resistance in Burkholderia cenocepacia. Mol. Microbiol. 85, 962–974 [DOI] [PubMed] [Google Scholar]
- 31. Hamad M. A., Skeldon A. M., Valvano M. A. (2010) Construction of aminoglycoside-sensitive Burkholderia cenocepacia strains for use in studies of intracellular bacteria with the gentamicin protection assay. Appl. Environ Microbiol. 76, 3170–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 33. Pérez J. M., McGarry M. A., Marolda C. L., Valvano M. A. (2008) Functional analysis of the large periplasmic loop of the Escherichia coli K-12 WaaL O-antigen ligase. Mol. Microbiol. 70, 1424–1440 [DOI] [PubMed] [Google Scholar]
- 34. Aubert D., MacDonald D. K., Valvano M. A. (2010) BcsK(C) is an essential protein for the type VI secretion system activity in Burkholderia cenocepacia that forms an outer membrane complex with BcsL(B). J. Biol. Chem. 285, 35988–35998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goodman A. L., Kulasekara B., Rietsch A., Boyd D., Smith R. S., Lory S. (2004) A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7, 745–754 [DOI] [PubMed] [Google Scholar]
- 36. Laskowski M. A., Osborn E., Kazmierczak B. I. (2004) A novel sensor kinase-response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa. Mol. Microbiol. 54, 1090–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ventre I., Goodman A. L., Vallet-Gely I., Vasseur P., Soscia C., Molin S., Bleves S., Lazdunski A., Lory S., Filloux A. (2006) Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U.S.A. 103, 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stock J. B., Ninfa A. J., Stock A. M. (1989) Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53, 450–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jourlin C., Ansaldi M., Méjean V. (1997) Transphosphorylation of the TorR response regulator requires the three phosphorylation sites of the TorS unorthodox sensor in Escherichia coli. J. Mol. Biol. 267, 770–777 [DOI] [PubMed] [Google Scholar]
- 40. Kwon O., Georgellis D., Lin E. C. (2000) Phosphorelay as the sole physiological route of signal transmission by the arc two-component system of Escherichia coli. J. Bacteriol. 182, 3858–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perraud A. L., Kimmel B., Weiss V., Gross R. (1998) Specificity of the BvgAS and EvgAS phosphorelay is mediated by the C-terminal HPt domains of the sensor proteins. Mol. Microbiol. 27, 875–887 [DOI] [PubMed] [Google Scholar]
- 42. Chang C. H., Zhu J., Winans S. C. (1996) Pleiotropic phenotypes caused by genetic ablation of the receiver module of the Agrobacterium tumefaciens VirA protein. J. Bacteriol. 178, 4710–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamamoto K., Hirao K., Oshima T., Aiba H., Utsumi R., Ishihama A. (2005) Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280, 1448–1456 [DOI] [PubMed] [Google Scholar]
- 44. Castelli M. E., García Véscovi E., Soncini F. C. (2000) The phosphatase activity is the target for Mg2+ regulation of the sensor protein PhoQ in Salmonella. J. Biol. Chem. 275, 22948–22954 [DOI] [PubMed] [Google Scholar]
- 45. Hsing W., Silhavy T. J. (1997) Function of conserved histidine 243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J. Bacteriol. 179, 3729–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mahenthiralingam E., Coenye T., Chung J. W., Speert D. P., Govan J. R., Taylor P., Vandamme P. (2000) Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38, 910–913 [DOI] [PMC free article] [PubMed] [Google Scholar]