Background: Little is known about the very first steps of infection of bacterial viruses.
Results: We specifically access the structure of phage T5 receptor-binding protein bound to its membrane receptor using specific techniques in small angle neutron scattering.
Conclusion: There are no large conformational changes.
Significance: Signal transduction within T5 appears different from that of phages binding cell wall saccharides.
Keywords: Bacteriophage, Biophysics, Electron Microscopy (EM), Membrane Proteins, Viral Protein, Fluorinated Surfactant, Small Angle Neutron Scattering
Abstract
Within tailed bacteriophages, interaction of the receptor-binding protein (RBP) with the target cell triggers viral DNA ejection into the host cytoplasm. In the case of phage T5, the RBP pb5 and the receptor FhuA, an outer membrane protein of Escherichia coli, have been identified. Here, we use small angle neutron scattering and electron microscopy to investigate the FhuA-pb5 complex. Specific deuteration of one of the partners allows the complete masking in small angle neutron scattering of the surfactant and unlabeled proteins when the complex is solubilized in the fluorinated surfactant F6-DigluM. Thus, individual structures within a membrane protein complex can be described. The solution structure of FhuA agrees with its crystal structure; that of pb5 shows an elongated shape. Neither displays significant conformational changes upon interaction. The mechanism of signal transduction within phage T5 thus appears different from that of phages binding cell wall saccharides, for which structural information is available.
Introduction
Bacteriophages are the most abundant microorganisms in the biosphere. At the origin of major discoveries in modern genetics, their study is gaining increasing popularity in fields as diverse as ecology, genetics, phylogeny, and nanophysics (1, 2). Phagotherapy is also blooming with the increased multiresistance of human, animal, and plant pathogens to antibiotics (3, 4). Furthermore, deciphering the sophisticated but minimalist phage molecular machineries elaborated for host cell recognition and infection is fascinating. Structural data have revealed horizontal gene transfer, masked at the sequence level by rapid evolution, from phages to bacteria, of machineries developed for bacterial pathogenicity (e.g. Type VI secretion systems, reviewed in Ref. 5).
The vast majority of bacteriophages, Caudovirales, are tailed phages that have an icosahedral protein capsid containing a densely packed double-stranded DNA and a tail with an adsorption device at its end that serves to recognize the cell surface and deliver the phage genome into the bacterium cytoplasm. In the case of lytic phages, the biosynthetic host machinery is then hijacked to transcribe the viral genome and produce new virions. The interaction between the phage receptor-binding protein (RBP)4 and its bacterial receptor is the key signal that leads to capsid opening and cell wall perforation, resulting in the ejection of the DNA from the capsid into the cytoplasm, across the complex host envelope. Our aim is to describe the conformational rearrangements that initially take place between the RBP and its receptor. The coliphage T5 is an excellent model to study phage-host interactions for the following reasons: (i) both its RBP, pb5 (6), and its receptor, the E. coli outer membrane iron-ferrichrome transporter FhuA, have been identified and purified; (ii) the mere interaction between T5 and FhuA, either detergent-solubilized or reconstituted in liposomes, is enough to induce DNA ejection (7); and (iii) the interaction between purified pb5 and FhuA yields a highly stable, stoichiometric complex (8). Furthermore, this complex is currently the only complex biochemically available between an outer membrane receptor and a phage RBP. FhuA is composed of a 22-β-strand barrel, which is obstructed by a globular domain, or the “plug.” Large extracellular loops and loops of the plug form the ligand binding sites (9). pb5 appears to fold as a unique domain, which undergoes secondary structural changes and most probably a rigidification of its structure upon binding to FhuA (10). In parallel to crystallization attempts of the FhuA-pb5 complex, small angle neutron scattering (SANS) and electron microscopy (EM) have been chosen to investigate the conformational changes that occur in pb5 following FhuA binding.
SANS combined with contrast variation and specific deuterium labeling of proteins is a particularly powerful technique to monitor conformational changes undergone by each protein within a complex. The neutron scattering length of the hydrogen nucleus is negative, whereas that of deuterium (and of most of the atoms) is positive. The difference between the scattering of the macromolecule and that of the solvent is proportional to a contrast term that can be modulated by changing the hydrogen content of the macromolecule or of the solvent by deuterium labeling. Very importantly, the contrast term of proteins vanishes in a buffer containing an appropriate H2O/D2O mixture at the contrast match point (CMP), expressed in D2O %. Thus, the signal of a partner within a protein complex can be selectively highlighted or suppressed (11–13).
Purified membrane protein samples comprise at least one additional component, detergent, present as bound detergent and as free micelles. Thus, detergents need to be either matched or modeled, in order to obtain structural information from the non-matched protein partner. Fluorinated surfactants share with detergents their amphipathic structure (Scheme 1). They have been investigated as potential mild surfactants (14–18). F6-Diglu (19, 20) and the chemically related and easier to synthesize F6-DigluM (21) (Scheme 1A) are stabilizing toward membrane proteins and form homogeneous and globular micelles and well defined membrane protein complexes. The CMPs of F6-Diglu (20) and of F6-DigluM (11) are close to that of most proteins (20), making these fluorinated surfactants ideal candidates for SANS study of solubilized membrane protein complexes; both hydrogenated protein partner(s) and surfactant will be matched simultaneously, allowing us to focus on deuterated protein partner(s) only.
SCHEME 1.
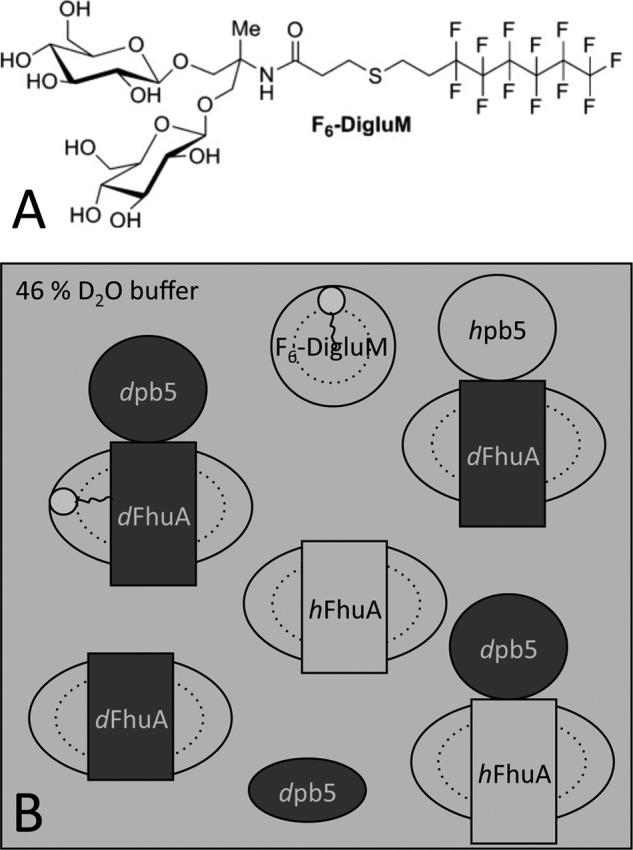
SANS contrast strategy. A, F6-DigluM chemical structure; B, SANS contrast strategy to highlight specific parts of the protein-detergent complexes. Partially deuterated proteins are represented in dark gray. Hydrogenated proteins and the F6-DigluM micelles are represented in light gray as is the 46% D2O buffer in which they are contrast-matched.
Here, we have analyzed the FhuA-pb5 complex solubilized in F6-DigluM by SANS. A contrast matching strategy shown in Scheme 1B was defined to separately resolve each partner alone and within the complex. Hydrogenated proteins and the F6-DigluM micelles were expected to both be contrast-matched at the same D2O %, the observed signal being due to the deuterated partner protein. FhuA low resolution structure in solution, determined by SANS, is in very good agreement with its crystal structure. It remains unchanged upon formation of the complex. pb5 is shown to be an elongated protein, whose structure, at the resolution of the technique, does not show significant conformational changes upon binding to FhuA. The SANS envelopes of the complex and the individual proteins are in excellent agreement with negative stain single particle EM. We propose that the mechanism of signal transduction from the RBP to the rest of the phage within phage T5, and thus other siphophages with straight fibers binding protein receptors, is different from that of siphophages binding cell wall saccharides, for which structural information is available.
EXPERIMENTAL PROCEDURES
Chemicals
F6-DigluM was synthesized by our general procedure consisting of one-pot reduction/alkylation of a thiol-based fluorinated chain onto an acrylamido-type polar head (19) with slight modifications. The 1H,1H,2H,2H-perfluorooctanethiol was directly used instead of its corresponding thioacetate (21), allowing the preparation of F6-DigluM in only three steps. F6DigluM was purified by flash chromatography on silica gel (AcOEt/MeOH/H2O, 7:2:1, v/v/v), followed by size exclusion chromatography on Sephadex LH 20 (MeOH). The solvent was removed under vacuum; the resulting powder was solubilized in MilliQ water and freeze-dried to give F6DigluM as a white foam. 300 mg of F6-DigluM was used in this study.
Adaptation, Overexpression, and Purification of dFhuA and hFhuA and of hpb5 and dpb5
FhuA was purified from the E. coli strain AW740 transformed with a plasmid encoding the fhuA gene in which a His tag has been inserted in the extracellular loop L5 (22). pb5 was purified from the E. coli strain BL21(DE3) carrying the oad gene encoding pb5 fused to an N terminus His tag in a pET-28 vector (8). Cells were grown in LB medium at 37 °C in the presence of 100 μm of the iron-chelating agent dipyridine for AW740 cells, and at 20 °C without induction for BL21(DE3) cells. To produce deuterated proteins, the two strains were first adapted to hydrogenated Enfors minimal medium for 7 days at 37 °C. Strains were then adapted to Enfors minimal medium in 80% D2O for 5 days. AW740 cells were then grown for 48 h at 37 °C in the presence of 100 μm dipyridine. BL21(DE3) cells were grown at 20 °C, and overexpression of dpb5 was induced at A600 nm = 0.9 with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 24 h. hFhuA, dpb5, and hpb5 purifications were carried out as described (10). For hFhuA, a molecular sieve step was added (SD200 10/300 GL; GE Healthcare; buffer: 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.05% N,N-dimethyldodecylamine N-oxide (LDAO)). dFhuA purification was performed as for hFhuA except that the anion exchange chromatography was omitted, because the deuterated protein remained adsorbed onto the matrix in the presence of 1 m NaCl. dpb5 and hpb5 were eluted from the cation exchange column (HTSP, 1 ml; GE Healthcare) in H2O or 46% D2O buffer and were used for data collection or complexation with FhuA.
Mass Spectrometry
LC/electrospray ionization-TOF-MS analysis was carried out on FhuA. The mass of hFhuA agrees with the sequence (79,965 versus 79,960 Da). The mass for dFhuA (82,930 Da) combined with the calculated mass (with deuteration of nonexchangeable H only, 84,061 Da) gives an estimate of 72% deuteration.
Surfactant Exchange, Formation of the FhuA-pb5 Complexes, and Quality Control of the Samples
First Series
SD200 fractions containing hFhuA or dFhuA solubilized in 0.05% LDAO were pooled and concentrated, desalted onto a PG10 column (3 ml; Bio-Rad), and loaded onto a 2-ml nickel-nitrilotriacetic acid home packed column (Qiagen resin) equilibrated in 20 mm Tris-HCl, pH 8.0, 0.05% LDAO. The flow-through, containing some unbound FhuA, was reloaded onto the column (6 cycles). The column was then washed with 10 ml of the equilibration buffer, and surfactant exchange was performed by washing the column with 20 ml of 20 mm Tris-HCl, pH 8.0, 2 mm F6-DigluM. The protein was eluted with 20 mm Tris-HCl, pH 8.0, 4 mm F6-DigluM, and 200 mm imidazole. To avoid excess of one of the partners, different FhuA-pb5 ratios were screened on small volumes and analyzed on SDS gels before formation of the complex on a preparative scale. Except for pb5, all samples were concentrated by ultrafiltration (Centricon 50) and finally dialyzed overnight against 2× 100 ml of 150 mm NaCl, 20 mm Tris-HCl, pH 8.0, 0.7 mm F6-DigluM, 46% D2O. Final protein concentrations were as follows: hFhuA, 4.5 mg/ml; dFhuA, 4.6 mg/ml; dFhuA-hpb5, 4.9 mg/ml; hFhuA-dpb5, 2.1 mg/ml; dFhuA-dpb5, 3.2 mg/ml. dpb5 was studied in H2O buffer at 0.5 mg/ml.
Second Series
dFhuA or hFhuA monomer fractions of gel filtration were concentrated by ultrafiltration (30,000 molecular weight cut-off; Millipore) to ∼8 mg/ml. Samples were supplemented with 10 mm + 2 g/g F6-DigluM/protein (total concentration, ∼32 mm) and incubated for 30 min. BioBeads (Bio-Rad) were then added (100 mg/ml), and the samples were incubated on a rotating wheel at room temperature for 1 h. BioBeads were removed, and the FhuA/LDAO/F6-DigluM mixture was submitted to size exclusion chromatography (SEC) (SD200 10/300 GL; buffer: 150 mm NaCl, 20 mm Tris-HCl, pH 8.0, 0.7 mm F6-DigluM, 46% D2O). Fractions of the second half of the monomer peak (to avoid any dimer contamination) were pooled and supplemented with 2 mm F6-DigluM and, for half of the samples, with stoichiometric amounts of dpb5 to form the complexes. dFhuA, hFhuA, hFhuA-dpb5, and dFhuA-dpb5 samples were concentrated by ultrafiltration (molecular weight cut-off 30,000 and 100,000 (Millipore) for FhuA and the complexes, respectively) before another SEC equilibrated in the same buffer. 320-μl fractions were collected, and the most concentrated fractions of the second half of the peak were supplemented with 2 mm F6-DigluM and directly used for SANS measurements. Final protein concentrations were as follows: hFhuA, 0.9 mg/ml; dFhuA, 0.8 mg/ml; hFhuA-dpb5, 1.0 mg/ml; dFhuA-dpb5, 1.2 mg/ml. dpb5 was also studied in 46% D2O at 0.4 mg/ml.
In the two series, surfactant exchange was controlled by thin layer chromatography (solvent chloroform/methanol/water, 65:35:5) stained by sulfuric acid and heating (300 °C). No residual LDAO was detected (detection limit <0.005%). To check for the homogeneity of the samples, AUC was performed on the SANS samples, sometimes after a 2-fold dilution in 150 mm NaCl, 20 mm Tris-HCl, pH 8.0, 46% D2O, using a Beckman-XLI ultracentrifuge at 42,000 rpm and 20 °C, in 3-mm optical path two-channel centerpieces, and analyzed as described (e.g. see Refs. 23 and 24). SEC was also performed for the second series. 10-μl aliquots were loaded onto an analytical SD200 5/150 column (GE Healthcare) in 150 mm NaCl, 20 mm Tris-HCl, pH 8.0, 0.7 mm F6-DigluM, 46% D2O.
Small Angle X-ray Scattering Experiments
Small angle x-ray scattering experiments were conducted on the ID14-3 beamline at the European Synchrotron Radiation Facility (Grenoble, France). The data were recorded using a two-dimensional Pilatus detector, with a monochromatic x-ray beam (λ = 0.931 Å) and a sample to detector distance of 2430 mm. The concentration of pb5 was 0.48 mg/ml in 25 mm MES buffer, pH 6.0, and 330 mm NaCl. 96 acquisitions (1 s/acquisition) were recorded during 300 s using 100 μl of sample in a continuous flow for both the protein and the buffer. The spectra were corrected individually for the response of the detector and scaled with respect to the incident beam intensity and the absorption of the samples. The averaged spectrum of the buffer was subtracted from the corrected averaged spectrum of pb5. A solution of bovine serum albumin at 4.94 mg/ml was used as a reference for the calibration of the spectra.
SANS Experiments
All samples were measured in Hellma quartz cells 100QS with 1-mm optical path length at the SANS instrument D22 at the Institute Laue-Langevin (Grenoble, France). The sample temperature was kept at 20 °C (first series) and 5 °C (second series) during the exposure times. Scattering data from all samples were recorded at two instrumental detector/collimator configurations, 2m/2m and 8m/8m, using a neutron wavelength of λ = 6 Å.
SANS Data Analysis
At each instrumental configuration, water, the H2O/D2O buffers, the empty beam, an empty quartz cell, and a boron sample (electronic background) were measured. Exposure times varied between 20 min (empty cell, boron) and 150 min (dpb5 in 46% D2O) according to the sample and instrument setup. Transmissions were measured during 2–3 min for each sample. The raw data were reduced (detector efficiency, electronic background, angular averaging) using a standard Institute Laue-Langevin software package (25) and normalized by the scattering of water. When applicable, the corrected scattered intensities from the two different Q ranges were merged (Q = (4π/λ)sinθ is the modulus of the scattering vector, with 2θ being the scattering angle), and in all cases, the respective buffer signals were subtracted. The radii of gyration (Rg) and the intensities scattered in the forward direction (I(0)) of all samples were extracted by the Guinier approximation, with RgQ ≤ 1.3, using the program Primus (26). The analysis of I(0) in terms of molar mass is done according to Ref. 27, I(0)/c = (k/NA)ΣM(∂ρN/∂c)2, where c represents the macromolecule weight concentration (g/liter), k is a geometrical factor, NA is Avogadro's number, Σ is for the different types of macromolecule in solution, and M and ∂ρN/∂c are their molecular mass and neutron scattering length density increment (cm/g). For a homogeneous macromolecule, ∂ρN/∂c = Σb/M − ρN°v̄, where Σb is the sum of the scattering lengths of the constituting atoms, v̄ is the partial specific volume (cm3/g) of the macromolecule, and ρN° is the solvent neutron scattering length density (cm−2). For a multicomponent complex comprising deuterated and hydrogenated macromolecules, we use the derived equation, I(0)/cd = (k/NA)Md (∂ρN/∂cd)2, where cd is the concentration of the deuterated partner; ∂ρN/∂cd = (∂ρN/∂c)d + Mh/Md (∂ρN/∂c)h, the second contribution being null at 46% D2O. Numerical values are in Ref. 11.
Low Resolution Models Determined by ab initio Analysis
We used the program DAMMIF (28) to generate low resolution envelopes of FhuA, of pb5 alone or within the complex, and of the complex from the SANS data, and of pb5 from the small angle x-ray scattering data. We used several variations of the input parameters in the default mode to model the data. Data up to Qmax = 0.15–0.2 Å−1 were included. The DAMMIF input files were generated using the program GNOM (29), imposing the restraints p(r = 0) = 0 and p(r = Dmax) = 0 for the pair distance distribution function p(r). For each sample, the values of Dmax were varied to obtain the best fit of the data and a smooth p(r) curve. As a quality control, the radii of gyration determined from the pair distribution analyses were compared with the ones determined by the Guinier analysis. 10–30 DAMMIF models were averaged using DAMAVER (30). The experimental curve for the dFhuA data was compared with the Protein Data Bank structure 2FCP of FhuA using CRYSON (31), applying a 72% deuteration degree of the protein, as determined by mass spectrometry.
Transmission Electron Microscopy and Image Processing
Samples were diluted at 0.01 mg/ml in 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.05% LDAO. Aliquots of 5 μl were adsorbed onto a glow-discharged carbon film-coated copper grid, washed with three droplets of pure water, and subsequently stained with 2% uranyl-acetate. Images were recorded using a Philips CM10 microscope operating at 80 kV on a Veleta 2000 × 2000 CCD camera (Olympus).
Reference-free alignment was performed on manually selected particles from digitized electron micrographs using the EMAN image processing package (32). After using a reference-free alignment procedure, particle projections were classified by multivariant statistical analysis. The class averages with the best signal/noise ratio were selected and gathered in a gallery.
RESULTS
F6-DigluM in SANS Can Be Matched Homogeneously in the Whole Q Range
F6-DigluM scattering was measured at 10.0 mg/ml in H2O and at 3.2, 6.4, and 9.7 mg/ml in 100% D2O. It displays a very neat, linear Guinier plot (Fig. 1A), indicative of well defined globular particles. The derived radii of gyration (Rg) are 1.87 ± 0.02 nm (H2O) and 2.05 ± 0.01 nm (D2O). An estimate of the critical micellar concentration (0.7 mm) in D2O can be obtained from the change of the forward scattering intensity I(0) with the total concentration, c, of surfactant. This value is close to the value determined from tensiometry and analytical ultracentrifugation (AUC) measurements (0.38 and 0.48 mm, respectively) (21). From an absolute calibration of I(0), a molecular mass of 64 ± 2 kDa can be calculated in D2O for F6-DigluM micelles, close to the value of 47 kDa from AUC, considering a globular shape, the difference being most probably related to the inaccuracy of the partial specific volume. A match point of 46% D2O is derived from the √((I(0)/(c − cmc)T) plot, with cmc the critical micellar concentration and T the sample transmission, close to the calculated one of 49% (11). The Rg, critical micellar concentration, molar mass, and match point are close to those measured by SANS for the very similar F6-Diglu (20). Neutron scattering was measured for F6-DigluM at 17.6 mg/ml (20 mm) in 150 mm NaCl, 20 mm Tris-HCl, pH 8.0, 46% D2O. Fig. 1B shows that the scattering of F6-DigluM is indistinguishable from that of the solvent. Thus, F6-DigluM not only is globally matched at the forward direction (Q = 0) but also displays no structural features in the investigated Q range. This is not common for surfactants, for which hydrophobic tail and hydrophilic head do not have the same chemical composition (11). The match point of F6-DigluM, as for F6-Diglu, corresponds roughly to that of hydrogenated proteins, which is highly interesting for SANS studies of membrane protein complexes in combination with specific deuteration.
FIGURE 1.
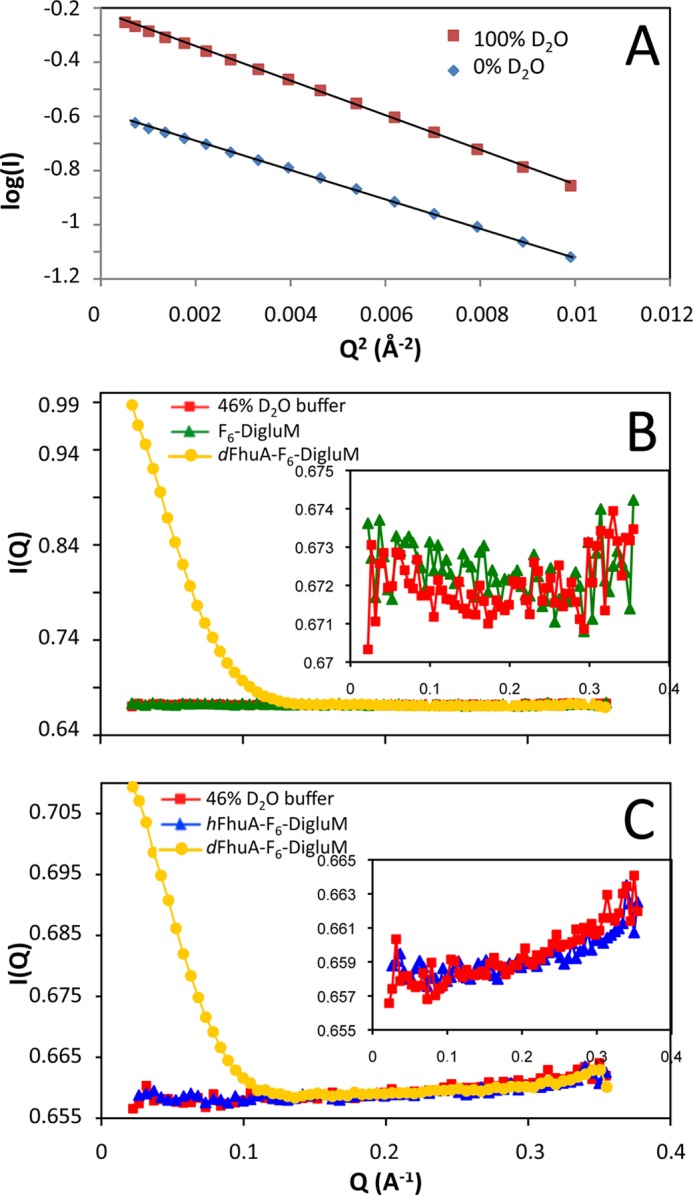
SANS of F6-DigluM and validation of SANS strategy. A, Guinier plot of F6-DigluM (10 mg/ml) in 0% (triangles) or 100% D2O (squares). B and C, scattering curves, before buffer subtraction, of F6-DigluM (17.6 mg/ml; B, triangles) and hFhuA (4.5 mg/ml; C, triangles) in F6-DigluM (8 mg/ml free micelles from AUC), measured in 46% D2O buffer, compared with the scattering curve of the 46% D2O buffer (squares in B and C) and dFhuA in F6-DigluM (circles in B and C). dFhuA was at 4.6 (B) and 0.8 (C) mg/ml, and AUC estimated 9 (B) and 2.3 (C) mg/ml free micelles. Insets, enlargements of the buffer curves.
Production and Quality Control of the SANS Samples
In view of our strategy (Scheme 1B), pb5 and FhuA were individually overproduced and purified, according to published protocols for the hydrogenated protein. Deuterated proteins were purified from E. coli stains adapted to 80% D2O in minimal medium, with slight modifications in the purification (see “Experimental Procedures”), leading to the purification of 1.5 mg of dpb5 and 2 mg of dFhuA per liter of culture. The deuteration rate, estimated for dFhuA using mass spectrometry, was ∼72%.
Two series of SANS experiments were performed with surfactant and solvent exchange following two different protocols: (i) affinity chromatography, mixture of the partners, dialysis, and concentration, and (ii) incubation with BioBeads, SEC, concentration, mixture of the partners, and SEC. These protocols led to sample concentrations of 2–4 and ∼1 mg/ml, respectively. In the first protocol, some of the samples contained aggregates, detected by SANS and AUC, and satisfactory samples were found only for hFhuA, dFhuA, and dFhuA-hpb5. The second protocol, avoiding final concentration steps, was thus designed to fulfill the requirements of full surfactant exchange, taking into account the limited availability of F6-DigluM, control of the D2O content, and sample homogeneity, with protein concentrations compatible with SANS analysis. Sample homogeneity was checked after SANS experiments by AUC and analytical SEC. Fig. 2 shows sedimentation velocity AUC analysis for dFhuA. F6-DigluM micelles are detected mainly using the interference optics at 5.3 ± 0.2 S, at a concentration, from fringe shifts, of ∼3 mm (for the second protocol) in the SANS samples. The main species is detected at 10.7 S for dFhuA. Similar results were obtained for hFhuA, hFhuA-dpb5, dFhuA-hpb5, and dFhuA-dpb5, with main peaks obtained at 10.4, 11.5, 11.8, and 11.9 ± 0.2 S, the variation in the s values being logically related to the deuteration level, deuterated molecules being more dense than hydrogenated ones. The combined analysis of the absorbance and interference signals gave an identical amount, in mol/mol, of bound F6-DigluM of 130 ± 15 and 130 ± 45 for FhuA and FhuA-pb5, respectively. The derived frictional ratio of 1.28 for dFhuA and 1.35 for the complexes considered as monomer are close to that of 1.25 expected for a globular compact shape. Larger species were also detected in limited amounts (∼5%) in both protocols, the second one, however, leading to more homogeneous samples. In the case of the complexes, aggregates are detected in AUC in larger amounts, a feature probably related to pb5 sensitivity to pressure and propensity to aggregate at a concentration above 0.5 mg/ml, because they were undetected in SEC (Fig. 2D) and in the SANS analysis.
FIGURE 2.
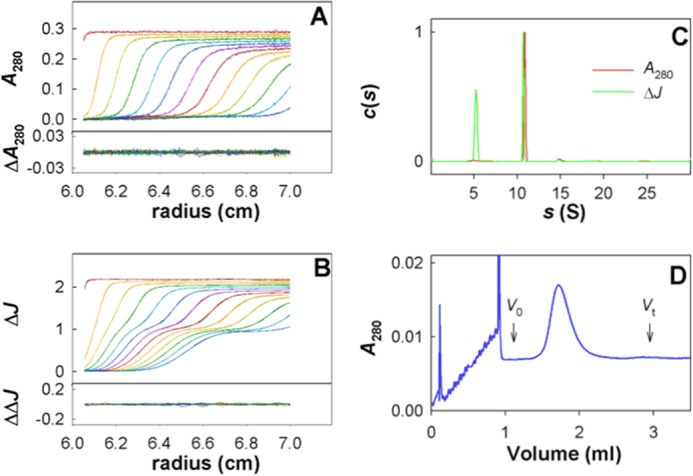
AUC and SEC quality control for dFhuA and dFhuA-dpb5 sample following SANS. A and B, superposition of experimental and fitted sedimentation velocity profiles obtained in 3-mm optical path length cells during 2 h at 42,000 rpm at 20 °C (top subpanels) and their differences (bottom subpanels), at 284 nm (A) and using interference optics (B), for dFhuA SANS sample in 46% D2O (second protocol) after 4-day storage at 4 °C. C, Sedimentation coefficient distributions c(s). The distributions are normalized to the main protein peak value. D, SEC of the dFhuA-dpb5 SANS sample (second protocol), loaded after the SANS measurements onto an SD200 5/150 column in 20 mm Tris-HCl, pH 8, 150 mm NaCl, 0.7 mm F6-DigluM, 46% D2O, run at 0.45 ml/min. Void volume (V0) = 1.12 ml; total volume (Vt) = 2.83 ml.
Hydrogenated Proteins Are Masked in SANS at the CMP of F6-DigluM
Because the CMP of F6-DigluM is close to that of most proteins (∼44% D2O), it is expected that both surfactant and hydrogenated proteins can be matched in the same buffer condition. Fig. 1C shows the scattering curve of hFhuA solubilized in F6-DigluM at 46% D2O. It clearly shows that both hFhuA and F6-DigluM are indeed indistinguishable from the buffer, showing no structural feature in the investigated Q range.
The Low Resolution SANS Structures of FhuA Alone and in Complex with pb5 Are Identical to the Crystal Structure
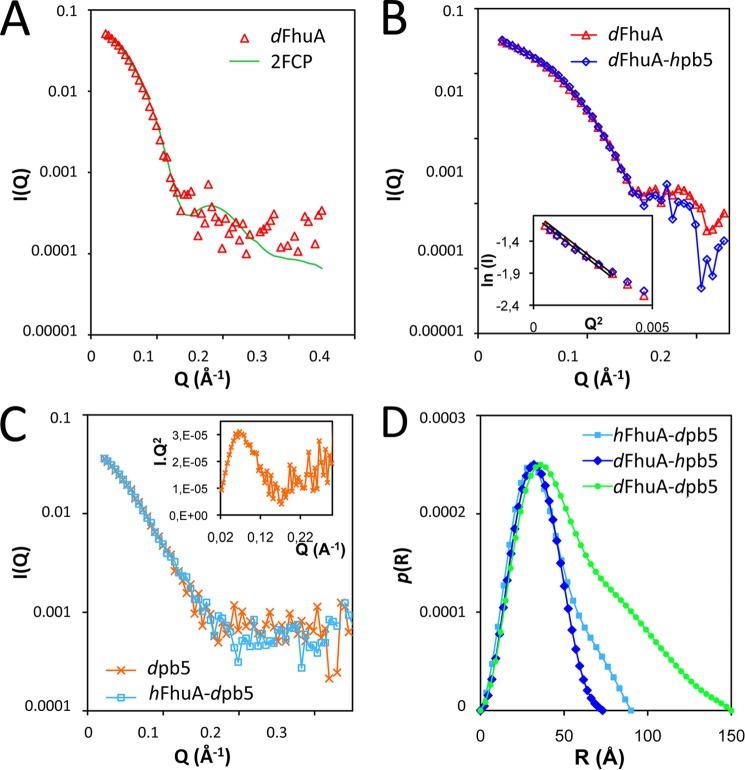
Fig. 3A shows the scattering curve of isolated dFhuA, solubilized in F6-DigluM, at 46% D2O. The measured data fit perfectly well with the calculated scattering curve of FhuA (Protein Data Bank entry 2FCP).
FIGURE 3.
SANS analysis of FhuA, pb5, and complex. A, calculated and measured scattering curve of dFhuA solubilized in F6-DigluM (triangles) at 46% D2O. The 2FCP curve was calculated with the program Cryson, considering a 46% D2O buffer and a 72% deuterated protein (solid line). χ2 = 1.173 on the whole Q range. B, experimental scattering curves of isolated dFhuA solubilized in F6-DigluM (triangles) or in complex with hpb5 (diamonds). Inset, Guinier analysis of the two data sets. C, scattering curves of dpb5 alone (crosses) or in complex with hFhuA solubilized in F6-DigluM (squares) measured in 46% D2O. Inset, Kratky representation of the dpb5 sample. D, pair distance distribution function, p(r), calculated using Gnom, of hFhuA-dpb5 (squares), dFhuA-hpb5 (diamonds), and dFhuA-dpb5 (circles) samples measured in a 46% D2O buffer.
In order to detect conformational changes induced upon formation of a complex with pb5, scattering data of dFhuA solubilized in F6-DigluM in complex with hpb5 were recorded. As shown in Fig. 3B, the data are superimposable, suggesting that the FhuA structure remains unchanged upon binding of pb5. The Guinier analyses of the two data sets (Fig. 3B, inset) allow the extrapolation of the forward intensities, which, normalized by FhuA concentration, are insignificantly different for dFhuA and for dFhuA-hpb5 samples, and correspond to FhuA as a monomer with 85% deuteration. The derived Rg are 28.1 ± 0.3 and 27.8 ± 0.2 Å for dFhuA and for dFhuA-hpb5, respectively. Both results corroborate the complete masking of hpb5 and the absence of conformational changes in the structure of FhuA upon formation of the complex. As expected from the nice superpositions of the curves, ab initio modeling of the dFhuA and dFhuA-hpb5 at 46% D2O shows very similar cylindrical volumes, ∼75 Å long and ∼50 Å wide, in which the Protein Data Bank structure of FhuA fits very well (Fig. 4A).
FIGURE 4.
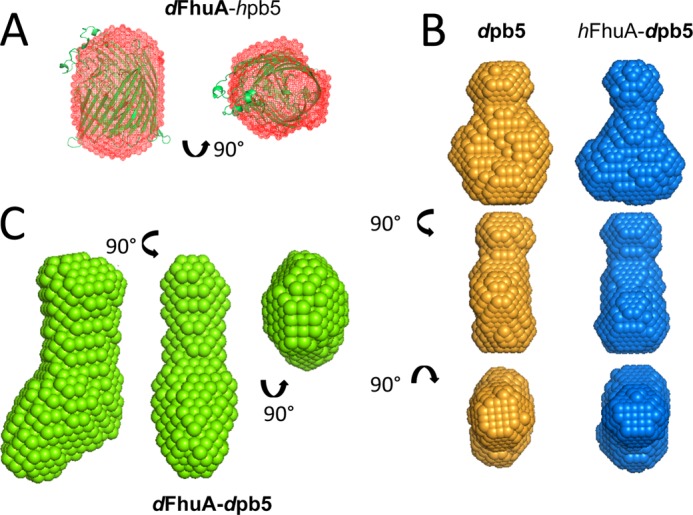
Ab initio envelopes from SANS. Ab initio envelopes (at 46% D2O) of dFhuA-hpb5 (red mesh), superimposed with the crystal structure of FhuA (2FCP, green) (A); dpb5 (orange) and hFhuA-dpb5 (blue) (B); and dFhuA-dpb5 (C). 10 (dFhuA-hpb5) and 20 (dpb5, hFhuA-dpb5, and dFhuA-dpb5) independent, very similar structures, calculated using Dammif, were averaged using DAMAVER. Side and top views are shown.
SANS Structure of pb5 Alone and in Complex with FhuA
pb5 has a strong tendency to aggregate whenever concentrated or dialyzed or at concentrations above 0.5 mg/ml. Thus, sample preparation had to be optimized without protein concentration, and the most concentrated fraction of the cation exchange chromatography was recovered and immediately diluted to 0.5 mg/ml if exceeding this concentration. However, even at these relatively low concentrations, interpretable SANS data of the dpb5 samples could be recorded (Fig. 3C), in particular for dpb5 in H2O buffer, where contrast is maximal. A Kratky representation of the data displays a bell shape, suggesting that pb5 is a compact protein, showing no sign of unfolding or structural disorder (Fig. 3C, inset) (33). Neutron diffraction curves of dpb5 measured either in 46% D2O or in H2O or x-ray diffusion curves of pb5 were superimposable, modulo the hydration shell, and so were their derived pair distance distribution functions (not shown). Ab initio modeling reveals an elongated, scamorza-shaped molecule (Fig. 4B), with a maximum distance of 90 Å and a width of ∼20–60 Å. Fig. 3C shows the scattering curve of dpb5, alone or in complex with hFhuA, both in a 46% D2O buffer, where hFhuA and F6-DigluM are masked. The two curves, like their derived pair distribution functions (not shown), are very similar, indicating that pb5 does not undergo important conformational changes upon binding of FhuA. Radii of gyration of 28.0 ± 1.2 and 29.6 ± 0.9 Å were extracted for dpb5 and hFhuA-dpb5, respectively, suggesting a slight elongation of pb5 within the complex. However, it cannot be excluded that this small difference is due to residual contributions from the hFhuA-F6DigluM moiety of the complex.
SANS Structure of the Complex
The overall structure of the complex can be obtained by measuring SANS data on a dpb5-dFhuA complex solubilized in F6-DigluM in a 46% D2O buffer. A molar mass of 196 ± 41 Da (i.e. compatible with a 1:1 complex of 151 kDa) and an Rg of 49.2 ± 0.9 Å are extracted from the Guinier plot, and GNOM allows determination of Dmax = 150 Å (Fig. 3D). Ab initio modeling reveals an elongated particle, with a socklike shape (Fig. 4C). The shape and dimensions of the dFhuA-dpb5 complex are compatible with a longitudinal organization of the individual FhuA and pb5 proteins. Without further constraints in the SANS data, the resolution (∼30 Å) is, however, too low to allow fitting of the individual proteins within the envelope.
EM Negative Stain Data Structure of the FhuA-pb5 Complex
FhuA, pb5, and the complex were analyzed by EM and single particle analysis of negatively stained sample, the particles being too small to be visualized by cryo-EM. Fig. 5 shows the overall projection structure of FhuA, pb5, and the complex. The dimensions of FhuA are in agreement with the crystal structure (height of 70 Å). pb5 protein has an elongated shape, 80 Å long and 50 Å wide, and presents a slightly sharp extremity. There is a good agreement between pb5 structures determined by SANS and by EM (Fig. 6). The dimensions and overall shape of the complex are also similar between SANS and EM data, with an elongated shape of 150 Å long, even if the reconstituted volumes do not perfectly coincide. The slight differences may be attributed to the differences in the techniques (see “Discussion”). The localization of each protein within the EM complex projection is facilitated by the substructures within each protein (Fig. 5B). The peaked side of pb5 is recognizable at the extremity of the complex. Even if the different partners within the complex are not distinguished individually, the EM data strongly suggest that, at this resolution and in agreement with SANS data, there are no noticeable conformational changes of pb5 upon complex formation.
FIGURE 5.
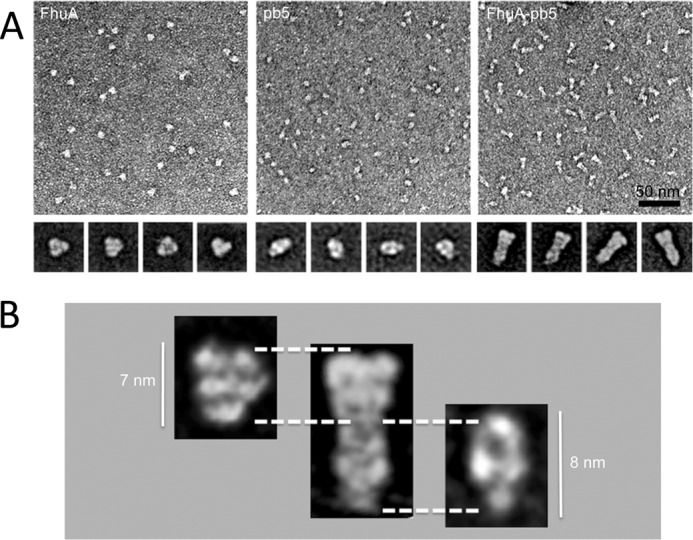
Negative stain electron microscopy. A, negative stain electron microscopy of purified FhuA (left), pb5 (middle), and FhuA-pb5 complex (right). The insets are their corresponding averages of the major classes. Scale bar, 50 nm; inset base line, 20 nm. B, FhuA and pb5 proteins are identified on the FhuA-pb5 complex.
FIGURE 6.
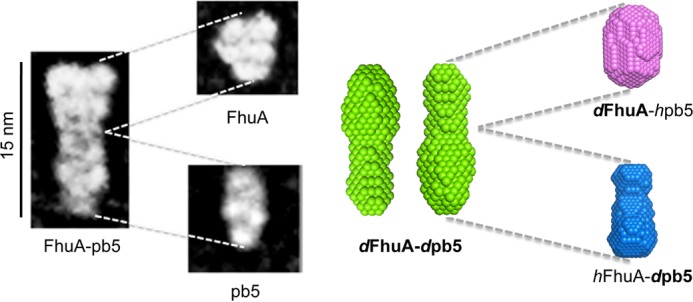
Comparison of the EM and SANS projections. Left, EM; right, SANS. The overall dimensions of the SANS and EM molecules coincide. Two SANS projections are presented, rotated by 180° because the relative position of FhuA and pb5 is not determined by the resolution. Projections are drawn to scale.
DISCUSSION
In this study, we validate the use of F6-DigluM for low resolution studies of individual protein partners within a membrane protein complex using SANS. Conformational changes, if occurring, can be monitored. For all practical purposes, F6-DigluM (34) can be considered as homogeneous, matched on the whole Q range. Therefore, sophisticated modeling of detergent residuals and very careful control of detergent concentration in the sample can be avoided. Usually, hydrophobic tails and hydrophilic heads have different CMPs (due to their different chemical composition) and will contribute to the scattered signal (see Ref. 11 and references therein). Only a very limited number of other detergents, such as Foscholine12 (35), can be considered as homogeneously matched in practice. However, F6-DigluM has the particular property of very similar scattering length densities, for both head and tail on the whole D2O % range, similar to hydrogenated proteins (11). This allows specific investigation of a protein subunit, if deuterated, within a membrane protein complex. A similar achievement was obtained by combining partial protein deuteration and appropriate mixing of deuterated and hydrogenated SDS (36). The advantage of F6-DigluM is that it can be used with a wide range of membrane proteins and that it is potentially less denaturating than most commonly used detergents (15). Importantly, in our study, F6-DigluM provided samples of required homogeneity for SANS.
In this study, we have determined for the first time, using SANS and EM, the low resolution structure of the receptor-binding protein of a Siphoviridae infecting Gram-negative bacteria. From both techniques, pb5 appears to be elongated and compact with a maximum dimension of ∼80–90 Å. This is in agreement with the limited proteolysis results suggesting that the protein folds as a unique and compact domain (10).
SANS, combined with the use of F6-DigluM and specific deuteration of one of the partners, shows no large structural rearrangements of either partner upon formation of the complex (Figs. 3 and 4). Note that even at low resolution, rather minor conformational changes can be distinguished by small angle scattering. For example, in the two multidomain proteins, adenylate cyclase (23.5 kDa) and lactoferrin (78.2 kDa), a small domain (17 and 22% of the protein mass) undergoes ∼60–70° rotation with respect to the rest of the protein upon ligand binding. These conformational changes are reflected by very distinguishable scattering curves (36). The absence of structural rearrangements in FhuA is consistent with its crystal structure alone or bound to different ligands showing only little, rigid body, conformational changes of extracellular loops (see Ref. 9 for a review). A dynamic behavior of the plug has recently been reported from destabilization by urea (37). However, our results definitely rule out the possibility of the plug domain of FhuA or part of it being removed from the barrel upon pb5 binding (e.g. to allow the DNA through the barrel of FhuA, a mechanism already eliminated from electron microscopy studies) (38). Concerning pb5, the invariance of its shape upon binding to FhuA from SANS and EM analysis was instead unexpected because binding information needs to be transmitted to the rest of the phage (see below).
The overall envelope and projection of the FhuA-pb5 complex determined by SANS and EM are similar (Fig. 6), even if the SANS envelope of the complex presents a slight curved shape that is not apparent in the EM projection. We should keep in mind, however, that both techniques sample different objects; the EM projections result from stained, possibly oriented and dried particles that are picked individually and averaged. We cannot rule out that staining induces slight distortions of the particle. SANS, on the other hand, is a mean of all particles present in the solution sample, including aggregates (if present) and averaging conformations in dynamic equilibrium. Significant differences in EM and small angle x-ray scattering envelopes were, for example, observed in subcomplexes of the type 6 secretion system (39). Furthermore, in the EM structure of our membrane complex, the detergent could contribute to the signal, whereas it does not for the SANS data. Thus, the two envelopes do not represent exactly the same object. However, using both techniques, the complex appears as an elongated particle, with a maximum dimension of 150 Å, corresponding to juxtaposition of the two partners along their longer axis. Although pb5 can be clearly recognized within the complex in the EM projection, it is not possible to fit the isolated proteins into the FhuA-pb5 SANS envelope at this resolution without further restraints. In agreement with our data, pb5 was recently unambiguously localized by immunolocalization at the tip of the straight fiber, and EM images show a similar structure of pb5 whether isolated, bound to FhuA (Fig. 5), or within the phage.5 The comparison of EM images further suggests that monomeric pb5 has its long axis parallel to the straight fiber and that one extremity binds to the straight fiber protein, whereas the interaction with FhuA occurs via the other extremity.
Although this is the event that commits the phage to infection, little is known about the irreversible interaction between phage RBPs and their receptors at the surface of the host and subsequent signaling for cell wall puncturing and DNA ejection. An increasing number of structures of caudovirales RBPs (also named tail spikes) that bind cell wall polysaccharides and that are present in multicopy within the phage are now available (for a review, see Ref. 40). A more complete picture of the molecular mechanisms that signal binding to the rest of the phage comes from the resolution of larger baseplate assemblies. The structures of these host recognition apparatus are available for the Myoviridae coliphage T4 (41) and the Siphoviridae lactococcal phage p2 (42) and TP901.1 (43) at high resolution and for the Bacillus siphophage SPP1 at lower resolution (44, 45). T4 and p2 baseplates, whose structure has been solved before and after DNA ejection, experience large conformational changes after irreversible binding of the phage to the host, resulting from low affinity but multiple attachments of receptor binding domains (18 copies in p2) to polysaccharides. For TP901-1, the irreversible and “avid” binding mediated by 54 flexible RBPs is thought to induce minor mechanical deformations or reorientations of the baseplate components. These large or minor conformational changes within the baseplate will trigger DNA ejection. Other structural information comes from the study of the short-tailed Podoviridae ϵ15 (46), BPP-1 (47), and P-SSP7 (48). It is worth noting that for all of these phages, the structures of the isolated RBPs, when available, fit into the density of the low resolution phage structures, indicating that these proteins have an autonomous fold, irrespective of whether they are isolated or integrated in the phage structure. This is also what we observe for pb5. However, T5 differs from the phages for which structural data are available. It does not have a baseplate with multiple copies of the RBP but bears a single copy of pb5 that is located at the distal end of a central straight fiber. This RBP folds as a unique domain and binds with extremely high affinity a protein receptor (8, 10, 49).
T5 shares some structural features of its tail tip with the other well described siphophages λ and SPP1, which bear a central spike-like fiber formed by a trimer of the proteins gpJ and gp21, respectively. gpJ binds the E. coli porin LambB (50), and gp21 binds the Bacillus subtilis protein YueB (44, 45, 51), and in both cases, it is the C-terminal part of the protein that forms the RBP. In all three phages, the host recognition and binding strategy is different from phages bearing a baseplate whose structure is known. Their sophisticated adsorption mechanism relies on two types of receptors: low affinity receptors for saccharides of the LPS (T5), techtoic acids (SPP1), or the porin OmpC (λ) (52) that accelerate adsorption, and high affinity RBP, present in low copy number, that binds a protein receptor. As for T5, the mere interaction of λ (53) and SPP1 (51) with their respective receptor is enough to trigger DNA ejection. The structures of the SPP1 tail before and after DNA release show very nicely how the conformational switch is propagated as a domino-like cascade along the tail. At the level of the spike, gp21N-ter opens to allow DNA transit out of the tail. The rest of the spike is not visible on EM pictures, probably due to the disordering of the receptor binding domain (54). In the case of T5, tomography of empty phages bound to FhuA-containing liposomes indicates a change in the geometry of the straight fiber, which perforates the liposome membrane (38). In the unbound phage, the straight fiber and its tip appear flexible,5 possibly to optimize the interaction with the host. Transmission of the binding information to the rest of the phage requires conformational changes within the single copy of pb5. Because structural rearrangements induced upon binding of pb5 to its receptor FhuA are not detected by either SANS or EM, we assume that they are of small amplitude. We have shown that upon binding to FhuA, pb5 undergoes secondary structure conformational changes (10). These changes could be sufficient to initiate the cascade of signaling events that lead to DNA release. This mechanism would be specific to siphophages that bare a straight fiber/spike and that bind a protein receptor.
Acknowledgments
We are grateful to H. Stahlberg (Center for Cellular Imaging and NanoAnalytics) for support. We thank A. Le Roy (IBS) for performing AUC experiments and S. Raynal (University of Avignon) for the synthesis of F6-DigluM. We also thank the Laue-Langevin Institute for BAG beamtime on D22, P. Callow and G. Zaccai for local contacting, and the European Synchrotron Radiation Facility for BAG beamtime on ID14-3. This work used the Analytical Ultracentrifugation, Surface Plasmon Resonance, Mass Spectrometry, and Membrane Protein Purification platforms of the Grenoble Instruct Centre (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB).
This work was supported by the European Community's Seventh Framework Programme (FP7/2007–2013) under Grant Agreements 211800 (SBMP) and 226507 (NMI3), by French Ministry of Research Grant ANR-07-PCVI-0010-02, and by the Swiss Initiative for Systems Biology, SystemsX.ch.
Y. Zivanovic, L. Confalonieri, L. Ponchon, R. Lurz, M. Chami, A. Flayhan, M. Renouard, A. Huet, P. Decottignies, A. R. Davidson, C. Breyton, and P. Boulanger, submitted for publication.
- RBP
- receptor-binding protein
- SANS
- small angle neutron scattering
- AUC
- analytical ultracentrifugation
- CMP
- contrast match point
- LDAO
- N,N-dimethyldodecylamine N-oxide
- Rg
- radius of gyration
- SEC
- size exclusion chromatography
- hFhuA and hpb5
- hydrogenated FhuA and pb5, respectively
- dFhuA and dpb5
- deuterated FhuA and pb5, respectively.
REFERENCES
- 1. Stern A., Sorek R. (2011) The phage-host arms race. Shaping the evolution of microbes. BioEssays 33, 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Soto C. M., Ratna B. R. (2010) Virus hybrids as nanomaterials for biotechnology. Curr. Opin. Biotechnol. 21, 426–438 [DOI] [PubMed] [Google Scholar]
- 3. Maura D., Debarbieux L. (2011) Bacteriophages as twenty-first century antibacterial tools for food and medicine. Appl. Microbiol. Biotechnol. 90, 851–859 [DOI] [PubMed] [Google Scholar]
- 4. Stone R. (2002) Bacteriophage therapy. Food and agriculture. Testing grounds for phage therapy. Science 298, 730. [DOI] [PubMed] [Google Scholar]
- 5. Cascales E., Cambillau C. (2012) Structural biology of type VI secretion systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heller K. J., Bryniok D. (1984) O antigen-dependent mutant of bacteriophage T5. J. Virol. 49, 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boulanger P., le Maire M., Bonhivers M., Dubois S., Desmadril M., Letellier L. (1996) Purification and structural and functional characterization of FhuA, a transporter of the Escherichia coli outer membrane. Biochemistry 35, 14216–14224 [DOI] [PubMed] [Google Scholar]
- 8. Plançon L., Janmot C., le Maire M., Desmadril M., Bonhivers M., Letellier L., Boulanger P. (2002) Characterization of a high-affinity complex between the bacterial outer membrane protein FhuA and the phage T5 protein pb5. J. Mol. Biol. 318, 557–569 [DOI] [PubMed] [Google Scholar]
- 9. Braun V. (2009) FhuA (TonA), the career of a protein. J. Bacteriol. 191, 3431–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flayhan A., Wien F., Paternostre M., Boulanger P., Breyton C. (2012) New insights into pb5, the receptor binding protein of bacteriophage T5, and its interaction with its Escherichia coli receptor FhuA. Biochimie 94, 1982–1989 [DOI] [PubMed] [Google Scholar]
- 11. Breyton C., Gabel F., Lethier M., Flayhan A., Durand G., Jault J.-M., Juillan-Binard C., Imbert L., Moulin M., Ravaud S., Härtlein M., Ebel C. (2013) Small angle neutron scattering for the study of solubilised membrane proteins. Eur. Phys. J. E Soft Matter 36, 71. [DOI] [PubMed] [Google Scholar]
- 12. Jacrot B. (1976) The study of biological structures by neutron scattering from solution. Rep. Prog. Phys. 39, 911 [Google Scholar]
- 13. Timmins P. A., Zaccai G. (1988) Low resolution structures of biological complexes studied by neutron scattering. Eur. Biophys. J. 15, 257–268 [DOI] [PubMed] [Google Scholar]
- 14. Blesneac I., Ravaud S., Juillan-Binard C., Barret L.-A., Zoonens M., Polidori A., Miroux B., Pucci B., Pebay-Peyroula E. (2012) Production of UCP1 a membrane protein from the inner mitochondrial membrane using the cell free expression system in the presence of a fluorinated surfactant. Biochim. Biophys. Acta 1818, 798–805 [DOI] [PubMed] [Google Scholar]
- 15. Breyton C., Pucci B., Popot J.-L. (2010) Amphipols and fluorinated surfactants. Two alternatives to detergents for studying membrane proteins in vitro. Methods Mol. Biol. 601, 219–245 [DOI] [PubMed] [Google Scholar]
- 16. Cho K. H., Byrne B., Chae P. S. (2013) Hemifluorinated maltose-neopentyl glycol (HF-MNG) amphiphiles for membrane protein stabilisation. Chembiochem 14, 452–455 [DOI] [PubMed] [Google Scholar]
- 17. Nehmé R., Joubert O., Bidet M., Lacombe B., Polidori A., Pucci B., Mus-Veteau I. (2010) Stability study of the human G-protein coupled receptor, Smoothened. Biochim. Biophys. Acta 1798, 1100–1110 [DOI] [PubMed] [Google Scholar]
- 18. Talbot J.-C., Dautant A., Polidori A., Pucci B., Cohen-Bouhacina T., Maali A., Salin B., Brèthes D., Velours J., Giraud M.-F. (2009) Hydrogenated and fluorinated surfactants derived from tris(hydroxymethyl)-acrylamidomethane allow the purification of a highly active yeast F1-F0 ATP-synthase with an enhanced stability. J. Bioenerg. Biomembr. 41, 349–360 [DOI] [PubMed] [Google Scholar]
- 19. Abla M., Durand G., Pucci B. (2008) Glucose-based surfactants with hydrogenated, fluorinated, or hemifluorinated tails. Synthesis and comparative physical-chemical characterization. J. Org. Chem. 73, 8142–8153 [DOI] [PubMed] [Google Scholar]
- 20. Breyton C., Gabel F., Abla M., Pierre Y., Lebaupain F., Durand G., Popot J.-L., Ebel C., Pucci B. (2009) Micellar and biochemical properties of (hemi)fluorinated surfactants are controlled by the size of the polar head. Biophys. J. 97, 1077–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abla M., Durand G., Breyton C., Raynal S., Ebel C., Pucci B. (2012) A diglucosylated fluorinated surfactant to handle integral membrane proteins in aqueous solution. J. Fluor. Chem. 134, 63–71 [Google Scholar]
- 22. Ferguson A. D., Breed J., Diederichs K., Welte W., Coulton J. W. (1998) An internal affinity-tag for purification and crystallization of the siderophore receptor FhuA, integral outer membrane protein from Escherichia coli K-12. Protein Sci. 7, 1636–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ebel C. (2011) Sedimentation velocity to characterize surfactants and solubilized membrane proteins. Methods 54, 56–66 [DOI] [PubMed] [Google Scholar]
- 24. Salvay A. G., Santamaria M., le Maire M., Ebel C. (2007) Analytical ultracentrifugation sedimentation velocity for the characterization of detergent-solubilized membrane proteins Ca2+-ATPase and ExbB. J. Biol. Phys. 33, 399–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghosh R., Egelhaalf S., Rennie A. (2006) A Computing Guide for Small-angle Scattering Experiments, pp.35–100, Institute Laue-Langevin, Grenoble [Google Scholar]
- 26. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. (2003) PRIMUS. A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 27. Jacrot B., Zaccai G. (1981) Determination of molecular weight by neutron scattering. Biopolymers 20, 2413–2426 [Google Scholar]
- 28. Franke D., Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 30. Volkov V. V., Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Svergun D. I., Richard S., Koch M. H., Sayers Z., Kuprin S., Zaccai G. (1998) Protein hydration in solution. Experimental observation by x-ray and neutron scattering. Proc. Natl. Acad. Sci. U.S.A. 95, 2267–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN. Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 33. Mertens H. D., Svergun D. I. (2010) Structural characterization of proteins and complexes using small-angle x-ray solution scattering. J. Struct. Biol. 172, 128–141 [DOI] [PubMed] [Google Scholar]
- 34. Compton E. L., Karinou E., Naismith J. H., Gabel F., Javelle A. (2011) Low resolution structure of a bacterial SLC26 transporter reveals dimeric stoichiometry and mobile intracellular domains. J. Biol. Chem. 286, 27058–27067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clifton L. A., Johnson C. L., Solovyova A. S., Callow P., Weiss K. L., Ridley H., Le Brun A. P., Kinane C. J., Webster J. R., Holt S. A., Lakey J. H. (2012) Low resolution structure and dynamics of a colicin-receptor complex determined by neutron scattering. J. Biol. Chem. 287, 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rambo R. P., Tainer J. A. (2013) Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annu. Rev. Biophys. 42, 415–441 [DOI] [PubMed] [Google Scholar]
- 37. Udho E., Jakes K. S., Finkelstein A. (2012) TonB-dependent transporter FhuA in planar lipid bilayers. Partial exit of its plug from the barrel. Biochemistry 51, 6753–6759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Böhm J., Lambert O., Frangakis A. S., Letellier L., Baumeister W., Rigaud J. L. (2001) FhuA-mediated phage genome transfer into liposomes. A cryo-electron tomography study. Curr. Biol. 11, 1168–1175 [DOI] [PubMed] [Google Scholar]
- 39. Zoued A., Durand E., Bebeacua C., Brunet Y. R., Douzi B., Cambillau C., Cascales E., Journet L. (2013) TssK is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the Type VI secretion system. J. Biol. Chem. 288, 27031–27041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Veesler D., Cambillau C. (2011) A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 75, 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leiman P. G., Arisaka F., van Raaij M. J., Kostyuchenko V. A., Aksyuk A. A., Kanamaru S., Rossmann M. G. (2010) Morphogenesis of the T4 tail and tail fibers. Virol. J. 7, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sciara G., Bebeacua C., Bron P., Tremblay D., Ortiz-Lombardia M., Lichière J., van Heel M., Campanacci V., Moineau S., Cambillau C. (2010) Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl. Acad. Sci. U.S.A. 107, 6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Veesler D., Spinelli S., Mahony J., Lichière J., Blangy S., Bricogne G., Legrand P., Ortiz-Lombardia M., Campanacci V., van Sinderen D., Cambillau C. (2012) Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc. Natl. Acad. Sci. U.S.A. 109, 8954–8958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goulet A., Lai-Kee-Him J., Veesler D., Auzat I., Robin G., Shepherd D. A., Ashcroft A. E., Richard E., Lichière J., Tavares P., Cambillau C., Bron P. (2011) The opening of the SPP1 bacteriophage tail, a prevalent mechanism in Gram-positive-infecting siphophages. J. Biol. Chem. 286, 25397–25405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vinga I., Baptista C., Auzat I., Petipas I., Lurz R., Tavares P., Santos M. A., São-José C. (2012) Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol. Microbiol. 83, 289–303 [DOI] [PubMed] [Google Scholar]
- 46. Chang J. T., Schmid M. F., Haase-Pettingell C., Weigele P. R., King J. A., Chiu W. (2010) Visualizing the structural changes of bacteriophage ϵ15 and its Salmonella host during infection. J. Mol. Biol. 402, 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dai W., Hodes A., Hui W. H., Gingery M., Miller J. F., Zhou Z. H. (2010) Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 107, 4347–4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu X., Zhang Q., Murata K., Baker M. L., Sullivan M. B., Fu C., Dougherty M. T., Schmid M. F., Osburne M. S., Chisholm S. W., Chiu W. (2010) Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat. Struct. Mol. Biol. 17, 830–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Basit H., Sharma K. S., Van der Heyden A., Gondran C., Breyton C., Dumy P., Winnik F. M., Labbé P. (2012) Amphipol mediated surface immobilization of FhuA. A platform for label-free detection of the bacteriophage protein pb5. Chem. Commun. (Camb.) 48, 6037–6039 [DOI] [PubMed] [Google Scholar]
- 50. Wang J., Hofnung M., Charbit A. (2000) The C-terminal portion of the tail fiber protein of bacteriophage λ is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J. Bacteriol. 182, 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. São-José C., Lhuillier S., Lurz R., Melki R., Lepault J., Santos M. A., Tavares P. (2006) The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J. Biol. Chem. 281, 11464–11470 [DOI] [PubMed] [Google Scholar]
- 52. Montag D., Schwarz H., Henning U. (1989) A component of the side tail fiber of Escherichia coli bacteriophage λ can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. J. Bacteriol. 171, 4378–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roessner C. A., Struck D. K., Ihler G. M. (1983) Injection of DNA into liposomes by bacteriophage λ. J. Biol. Chem. 258, 643–648 [PubMed] [Google Scholar]
- 54. Plisson C., White H. E., Auzat I., Zafarani A., São-José C., Lhuillier S., Tavares P., Orlova E. V. (2007) Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 26, 3720–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]