Abstract
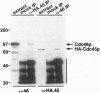
We report the isolation and characterization of CDC45, which encodes a polypeptide of 650 amino acids that is essential for the initiation of chromosomal DNA replication in the budding yeast, Saccharomyces cerevisiae. CDC45 genetically interacts with at least two members of the MCM (minichromosome maintenance) family of replication genes, CDC46 and CDC47, which are proposed to perform a role in restricting initiation of DNA replication to once per cell cycle. Like mutants in several MCM genes, alleles of CDC45 also show a severe minichromosome maintenance defect. Together, these observations imply that Cdc45p performs a role in the control of initiation events at chromosomal replication origins. We investigated this possibility further and present evidence demonstrating that Cdc45p is assembled into complexes with one MCM family member, Cdc46p/Mcm5p. These observations point to a role for Cdc45p in controlling the early steps of chromosomal DNA replication in conjunction with MCM polypeptide complexes. Unlike the MCMs, however, the subcellular localization of Cdc45p does not vary with the cell cycle, making it likely that Cdc45p interacts with MCMs only during the nuclear phase of MCM localization in G1.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Block D. E., Eitzman P. D., Wangensteen J. D., Srienc F. Slit scanning of Saccharomyces cerevisiae cells: quantification of asymmetric cell division and cell cycle progression in asynchronous culture. Biotechnol Prog. 1990 Nov-Dec;6(6):504–512. doi: 10.1021/bp00006a015. [DOI] [PubMed] [Google Scholar]
- Blow J. J., Laskey R. A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988 Apr 7;332(6164):546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Chong J. P., Mahbubani H. M., Khoo C. Y., Blow J. J. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995 Jun 1;375(6530):418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Coué M., Kearsey S. E., Méchali M. Chromotin binding, nuclear localization and phosphorylation of Xenopus cdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO J. 1996 Mar 1;15(5):1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Coxon A., Maundrell K., Kearsey S. E. Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res. 1992 Nov 11;20(21):5571–5577. doi: 10.1093/nar/20.21.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S., Whitbread L. Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2514–2518. doi: 10.1073/pnas.92.7.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H., Dowell S. J., Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994 Jul 29;78(2):303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Laskey R. A. Nuclear targeting sequences--a consensus? Trends Biochem Sci. 1991 Dec;16(12):478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Dohmen R. J., Wu P., Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994 Mar 4;263(5151):1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Gibson S. I., Surosky R. T., Tye B. K. The phenotype of the minichromosome maintenance mutant mcm3 is characteristic of mutants defective in DNA replication. Mol Cell Biol. 1990 Nov;10(11):5707–5720. doi: 10.1128/mcb.10.11.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy K. M., Clark C. D., Botstein D. Subcellular localization of yeast CDC46 varies with the cell cycle. Genes Dev. 1990 Dec;4(12B):2252–2263. doi: 10.1101/gad.4.12b.2252. [DOI] [PubMed] [Google Scholar]
- Hennessy K. M., Lee A., Chen E., Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991 Jun;5(6):958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- Hu B., Burkhart R., Schulte D., Musahl C., Knippers R. The P1 family: a new class of nuclear mammalian proteins related to the yeast Mcm replication proteins. Nucleic Acids Res. 1993 Nov 25;21(23):5289–5293. doi: 10.1093/nar/21.23.5289-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Nozaki N., Sugimoto K. DNA polymerase alpha associated protein P1, a murine homolog of yeast MCM3, changes its intranuclear distribution during the DNA synthetic period. EMBO J. 1994 Sep 15;13(18):4311–4320. doi: 10.1002/j.1460-2075.1994.tb06751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Takizawa N., Nozaki N., Sugimoto K. Molecular cloning of cDNA encoding mouse Cdc21 and CDC46 homologs and characterization of the products: physical interaction between P1(MCM3) and CDC46 proteins. Nucleic Acids Res. 1995 Jun 25;23(12):2097–2104. doi: 10.1093/nar/23.12.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T., Musahl C., Laskey R. A., Knippers R. Human replication proteins hCdc21, hCdc46 and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. J Cell Sci. 1996 Feb;109(Pt 2):309–318. doi: 10.1242/jcs.109.2.309. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Mimura S., Nishimoto S., Takisawa H., Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995 May 19;81(4):601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Madine M. A., Khoo C. Y., Mills A. D., Laskey R. A. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995 Jun 1;375(6530):421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- Miyake S., Okishio N., Samejima I., Hiraoka Y., Toda T., Saitoh I., Yanagida M. Fission yeast genes nda1+ and nda4+, mutations of which lead to S-phase block, chromatin alteration and Ca2+ suppression, are members of the CDC46/MCM2 family. Mol Biol Cell. 1993 Oct;4(10):1003–1015. doi: 10.1091/mbc.4.10.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova J. M., Mikesell G. E., Breeden L. L. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol Biol Cell. 1995 Dec;6(12):1641–1658. doi: 10.1091/mbc.6.12.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thömmes P., Fett R., Schray B., Burkhart R., Barnes M., Kennedy C., Brown N. C., Knippers R. Properties of the nuclear P1 protein, a mammalian homologue of the yeast Mcm3 replication protein. Nucleic Acids Res. 1992 Mar 11;20(5):1069–1074. doi: 10.1093/nar/20.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov I. T., Attaran A., Kearsey S. E. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995 Jun;129(6):1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov I. T., Pepperkok R., Philipova R. N., Kearsey S. E., Ansorge W., Werner D. A human nuclear protein with sequence homology to a family of early S phase proteins is required for entry into S phase and for cell division. J Cell Sci. 1994 Jan;107(Pt 1):253–265. doi: 10.1242/jcs.107.1.253. [DOI] [PubMed] [Google Scholar]
- Treisman J. E., Follette P. J., O'Farrell P. H., Rubin G. M. Cell proliferation and DNA replication defects in a Drosophila MCM2 mutant. Genes Dev. 1995 Jul 15;9(14):1709–1715. doi: 10.1101/gad.9.14.1709. [DOI] [PubMed] [Google Scholar]
- Yan H., Gibson S., Tye B. K. Mcm2 and Mcm3, two proteins important for ARS activity, are related in structure and function. Genes Dev. 1991 Jun;5(6):944–957. doi: 10.1101/gad.5.6.944. [DOI] [PubMed] [Google Scholar]
- Yan H., Merchant A. M., Tye B. K. Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev. 1993 Nov;7(11):2149–2160. doi: 10.1101/gad.7.11.2149. [DOI] [PubMed] [Google Scholar]