Abstract
Objectives. HLA-B*27:05 is associated with AS whereas HLA-B*27:09 is not associated. We hypothesized that different interactions with KIR immune receptors could contribute to the difference in disease association between HLA-B*27:05 and HLAB*27:09. Thus, the objective of this study was to compare the formation of β2m-free heavy chain (FHC) including B27 dimers (B272) by HLA-B*27:05 and HLA-B*27:09 and their binding to KIR immunoreceptors.
Methods. We studied the formation of HLA-B*27:05 and HLA-B*27:09 heterotrimers and FHC forms including dimers in vitro and in transfected cells. We investigated HLA-B*27:05 and HLA-B*27:09 binding to KIR3DL1, KIR3DL2 and LILRB2 by FACS staining with class I tetramers and by quantifying interactions with KIR3DL2CD3ε-reporter cells and KIR3DL2-expressing NK cells. We also measured KIR expression on peripheral blood NK and CD4 T cells from 18 HLA-B*27:05 AS patients, 8 HLA-B27 negative and 12 HLA-B*27:05+ and HLA-B*27:09+ healthy controls by FACS staining.
Results. HLA-B*27:09 formed less B272 and FHC than HLA-B*27:05. HLA-B*27:05-expressing cells stimulated KIR3DL2CD3ε-reporter T cells more effectively. Cells expressing HLA-B*27:05 promoted KIR3DL2+ NK cell survival more strongly than HLA-B*27:09. HLA-B*27:05 and HLA-B*27:09 dimer tetramers stained KIR3DL1, KIR3DL2 and LILRB2 equivalently. Increased proportions of NK and CD4 T cells expressed KIR3DL2 in HLA-B*27:05+ AS patients compared with HLA-B*27:05+, HLA-B*27:09+ and HLA-B27− healthy controls.
Conclusion. Differences in the formation of FHC ligands for KIR3DL2 by HLA-B*27:05 and HLA-B*27:09 could contribute to the differential association of these alleles with AS.
Keywords: spondyloarthritis, HLA-B*27:05, HLA-B*27:09, B27 homodimer, KIR3DL1, KIR3DL2
Introduction
HLA-B27 is strongly associated with a group of inflammatory arthritic disorders collectively known as the SpA, typified by AS [1]. Theories for B27 disease involvement include presentation of arthritogenic peptides to CD8 T cells and induction of a proinflammatory misfolded protein response (reviewed in [2]). B27 is expressed at the surface of antigen presenting cells (APCs) as classical β2m-associated class I heterodimers and disulphide-bonded β2m-FHC forms including homodimers (termed B272) [3–5]. We have proposed that disease results from B27 FHC immune receptor interactions promoting arthritis [6, 7]. Such immune receptors include members of the killer cell immunoglobulin-like receptor (KIR) and leukocyte immunoglobulin-like receptor (LILR) families.
HLA-B27 has multiple allelic variants [8], with some alleles such as the prototypic dominant Caucasian HLA-B*27:05 subtype being strongly associated with disease while others such as HLA-B*27:09 are not associated [9]. Different B27 variants differ by one or more amino acid substitutions, frequently resulting in altered residues within their peptide binding clefts [10]. HLA-B*27:09 [11] is not associated with disease and differs from HLA-B*27:05 by one amino acid substitution. Aspartic acid at position 116, in the α1 domain, is substituted to a histidine (D116H).
KIR are expressed by NK cells and minor subsets of T cells [12]. B27 binds to KIR family members with three extracellular immunoglobulin-like domains (DoD1D2) and a long cytoplasmic tail termed KIR3DL1/2. KIR3DL1 recognizes HLA-class I molecules including HLA-B*27:05 that express the Bw4 epitope [13]. While both HLA-B*27:05 FHC and heterodimer bind to KIR3DL1, only FHC binds to KIR3DL2 [3, 14]. Although the sequence of peptide bound to class I heterodimers affects KIR3DL1 and KIR3DL2 recognition, by contrast, recombinant B27 homodimer binding to KIR is independent of complexed peptide [14]. KIR ligation inhibits activation-induced cell death (AICD) [15, 16]. Ligation of KIR3DL2 by B27 FHC promotes the survival of NK and Th17 cells [17, 18]. AS patients have increased proportions of activated Th17 and NK cells expressing KIR3DL2 [17, 18].
The LILR receptors LILRB1 and LILRB2 bind a broad range of β2m-associated classical and non-classical class I [19]. LILRB2 but not LILRB1 binds to β2m-free class I heavy chain forms including HLA-B27 FHC and B27 dimer [20, 21]. While LILRB1 is broadly expressed on B cell, myeloid cells NK and T cells, LILRB2 is expressed by cells of myeloid origin.
We hypothesized that the differential association of HLA-B*27:09 with disease could be due to differences in its interaction with KIR and LILR immune receptors compared with HLA-B*27:05. Thus, we compared the formation of dimer and FHC by HLA-B*27:05 and HLA-B*27:09 and their interaction with KIR3DL1 and KIR3DL2. Our results suggest that differences in formation of FHC ligands for KIR3DL2 by HLA-B*27:05 and HLA-B*27:09 could play a role in the differential association of these alleles with AS.
Materials and methods
HLA-B27 plasmid constructs
Prokaryotic plasmids and PHR-SIN lentiviral cassettes for HLA-B*27:09 expression were generated by mutagenesis of pLM1-HLA-B*27:05 and PHR-SIN HLA-B*27:05, respectively, using the QuickChange site-directed mutagenesis kit (Invitrogen) following the manufacturer’s instructions.
Lentiviral transduction of cell lines and immunoprecipitations
LBL.721.221 cells (hereafter referred to as 221 cells) were transduced with lentivirus expressing HLA-B*27:05 or HLA-B*27:09 as described previously [21]. 20 × 106 parental 221 cells or 221B*27:05 and 221B*27:09 cells were surface biotinylated with NHS-LC-biotin (Thermoscientific UK Ltd) according to the manufacturers instructions. Subsequently cells were stained with HC10 or W632 antibody (10µg), washed with ice-cold PBS and lysed as previously described [14]. Lysates were precipitated with anti-mouse immunoglobulins Dynabeads, resolved by non-reducing or reducing SDS-PAGE and western blots developed with streptavidin-HRP (Sigma) or HRP-conjugated HC10.
KIR3DL2CD3ε reporter cell assay
Jurkat T cell reporter cells transduced with a KIR3DL2CD3ε lentiviral expression cassette were prepared as previously described [22]. All experiments for cytokine assay were set up in RPM1640 medium (Sigma) supplemented with 10% fetal calf serum and antibiotics [14]. 200 000 transduced T cells were incubated with 200 000 parental 221 cells or HLA-B*27:05 or HLA-B*27:09 transduced 221 cells for 24 h in with/without HC10, W632, ME1 or isotype control IgG1/G2a mAbs (MOPC 123 and MG1-45; Biolegend) at 50 µg/ml. Subsequently, supernatants were harvested for IL-2 ELISA according to the manufacturer’s instructions (EBiosciences Ltd UK).
Recombinant protein and tetramer generation
HLA-B27 heterodimer and homodimer were expressed and produced as previously described [3]. Recombinant biotinylated HLA-G dimers were prepared as previously described [20]. Yields of refolded protein were calculated by FPLC peak integration using the Unicorn software. HC10 and HD6 ELISAs were performed as described previously [23].
Shared HLA-B*27:05 and HLA-B*27:09 peptides used in this study: NRIKGIPKL; GRIGPNIRL; ARLQTALLV and RRFFPYYVY [24]; and RRRWRRLTV [25]. HLA-B*27:05 peptides: RRIYDLIEL [26]; KRWIILGLNK (GAG, [27]). SRYWAIRTR (Flu NP [28]) HLA-B*27:09 peptides: TRIPKIQKL [24]. Proteins were biotinylated and tetramers made with extravidin PE (Sigma) as previously described [14].
Tetramer staining
Tetrameric complexes were used to stain Baf3 cells transfected with KIR3DL1, KIR3DL2, LILRB1 or LILRB2 receptor as previously described [14].
Generation of NK cell lines
KIR3DL2+ NK cell lines were FACS-sorted from PBMC using the DX31 mAb (Gift from Jo Phillips, DNAX), and maintained by allogeneic stimulation as previously described [17].
Coculture of NK cell lines with HLA-B27-expressing APCs
NK cell lines were labelled with CFSE according to the manufacturer’s instructions (Invitrogen). 100 000–500 0000 γ-irradiated 221 APCs were incubated with equal numbers of NK cells as previously described [17]. On day 5 cells were first stained for KIR3DL2 expression and then stained with allophycocyanin-conjugated Annexin V (BD Biosciences) and pacific blue Live-Dead stain (Invitrogen) according to the manufacturer’s instructions.
Patients and controls
18 HLA-B*27:05 AS patients (12 males and 6 females, mean age 42.8 ± 8.2 years, mean disease duration 17.1 ± 11 years), 12 HLA-B*27:05 (mean age 35.8 years), 12 HLA-B*27:09 (mean age 37.5 years) and 8 HLA-B27 negative (mean age 36.6 years), sex-matched healthy controls (HCs), were recruited under written informed consent. All patients fulfilled the modified New York criteria for Ankylosing Spondylitis and were not on anti-TNF-α drugs at the time of taking samples. All patients and HCs selected for this study were of Sardinian Caucasian origin dating back at least three generations from both the maternal and paternal side. This study was approved by the Institutional Ethical Committee of the University Hospital, Cagliari, Italy (365/09/CE).
Reagents and antibodies
KIR3DL2 or KIR3DL1 expression was detected by indirect staining with DX31 MAb or DX9 MAbs (Biolegend) then Fluorescein isothiocyanate-conjugated goat F(ab')2 anti-mouse (GAM) IgG (F0479, DAKO). Cells were then stained with allophycocyanin-conjugated anti-CD56 (N901/NKH-1, Beckman Coulter, Fullerton, CA, USA), PerCP-conjugated anti-CD3 (SK7, BD Biosciences), PE-anti-CD4 (SFCI12T4D11, Coulter) or PE-Cy7-anti-CD4 (BD Biosciences).
The specificities of anti-HLA class I antibodies used in this study are summarized in Table 1. HD6 antibody binds to HLA-B27 dimers and multimers [23]. HC10 antibody recognizes a linear epitope in HLA class I A, B and C heavy chains incorporating amino acids 55–64 of the heavy chain [29]. W6/32 recognizes a discontinuous epitope in the α2 and α3 domains of HLA-A, B and C incorporating residue 121 of heavy chain formed by both β2 m-associated and free forms of HLA class I [30, 31]. ME1 binds a conformational β2m-dependent epitope in the α1 domain of HLA-B27, critically dependent on interactions between residues 67–71 [32].
Table 1.
Summary of the anti-HLA class I antibodies used in this study
| Antibody | Isotype | Specificity | References |
|---|---|---|---|
| HD6 | IgG1 | β2m-free HLA-B27 dimers and multimers | [23] |
| HC10 | IgG2a | β2m-free HLA-A, B and C heavy chain. Linear epitope incorporates residues 55-64 of the HLA class I heavy chain | [29] |
| ME1 | IgG1 | β2m-associated HLA-B27 and some other HLA-B alleles. Recognizes a conformational epitope in the a1 helix criticall dependent on interaction between residues 67 and 71 | [32] |
| W6/32 | IgG2a | Recognizes a discontinuous epitope in the α2 and α3 domains of HLA-A, B and C incorporating residue 121 of heavy chain formed by both β2m-associated and free forms of HLA-class I | [30, 31, 34] |
Flow cytometric analysis
PBMCs were isolated by density gradient centrifugation using Lympholyte-H (Cedarlane Laboratories, Hornby, Ontario, Canada) and immediately stained for KIR expression. Stained cells were fixed with 1% paraformaldehyde in PBS and analysed within 24 h. Flow cytometric analyses were performed using a FACSCanto (BD Biosciences, Franklin Lakes, NJ) and data were analysed with FACSDiva 6.1.3 software (BD Biosciences).
Percentages of cells expressing KIR3DL2 and KIR3DL1 and KIR receptor density were evaluated on CD3−/CD56+ NK and CD3+/CD4+ T cells. Briefly, for each sample, the mean fluorescence channel was expressed as Relative Channel Number on a linear scale, compared with five different standards of beads coated with defined mouse antibodies and converted into Antibody Binding Capacity (ABC) (DAKO Denmark). Saturating concentrations of primary and secondary antibodies were used. Fluorescence correlates with the number of bound primary antibody molecules on the cells and on the beads and ABC units correspond to the number of binding sites [33].
Data analysis
Values are expressed as mean percentages ± s.d. positive cells and as mean cell surface antigen density in ABC units. Differences between AS and HCs were analysed by one-way analyses of variance (ANOVA) with a Bonferroni post-test and a two-tailed unpaired t-test with Welch’s correction. Statistical analysis was performed using Prism 5.0 software (GraphPad Inc.).
Results
HLA-B*27:05 forms more cell surface FHC forms than HLA-B*27:09
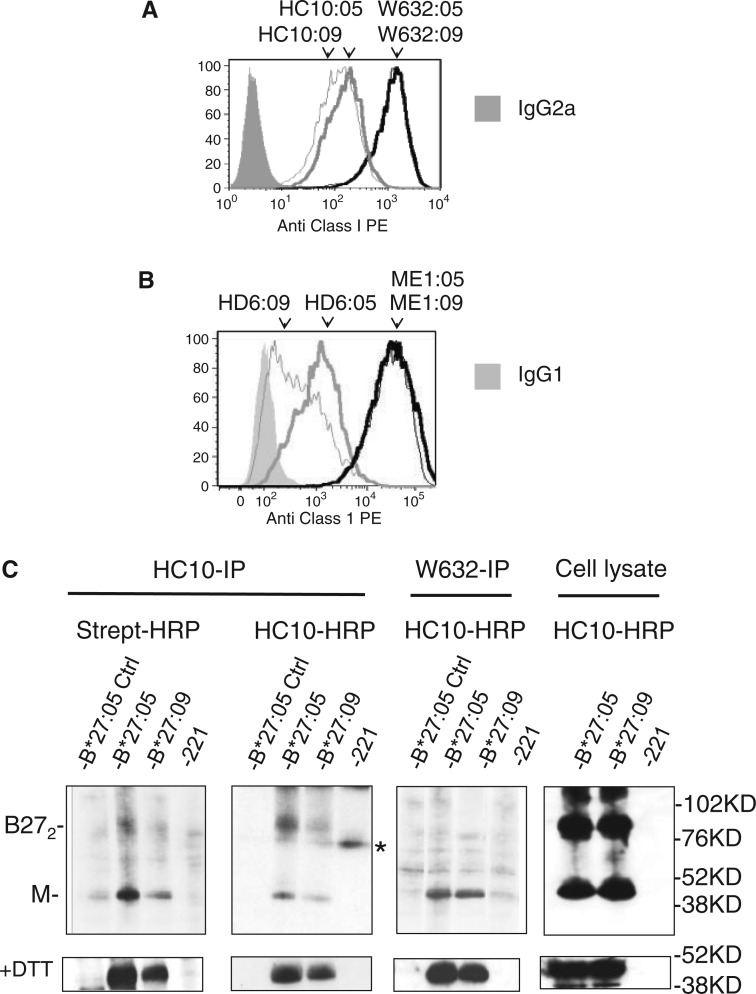
The specificities of anti-HLA-B27 and anti-HLA-class I antibodies used in this study are summarized in Table 1. The ME1 antibody recognizes β2m-associated HLA-B27, whereas W6/32 has been reported to recognize both β2m-associated and β2m-free forms of HLA-B27 and other HLA class I [30, 32, 34]. HC10 antibody binds to HLA-B27 and other β2m-free HLA class I heavy chains and HD6 antibody recognizes dimeric and multimeric forms of HLA-B27 β2m-FHC [23, 29]. We first compared levels of expression of B27 β2m-FHC on 221 cells which expressed comparable levels of HLA-B*27:05 or HLA-B*27:09 of ME1- and W6/32-reactive B27 by FACS analysis (Fig. 1A and B). Both HC10 and HD6 antibodies stained 221B*27:05 more strongly than 221B*27:09 cells (Fig. 1A and B).
Fig. 1.
HLA-B*27:05 forms more cell surface free heavy chain dimer and multimer than HLA-B*27:09.
Representative FACS stains of HLA-B*27:05 and HLA-B*27:09 transduced 221 cells with in (A) W632 and HC10, (B) ME1 and HD6 antibodies. Representative stain from one of four independent stains. MFIs for HLA-B*27:05 transduced 221 cells stained with ME1, W632, HC10 and HD6 were and respectively. MFIs for HLA-B*27:09 transduced 221 cells stained with ME1, W632, HC10 and HD6 were and respectively. (C) Western blot with streptavidin HRP and HC10-HRP of HC10 and W632 immunoprecipitates from cell-surface biotinylated 221B*27:05 and 221B*27:09 cells. Lower panels (+DTT): western blots of reduced samples of HC10 and W632 immunoprecipitates and whole cell lysates from cell-surface biotinylated 221B*27:05 and HLA-B*27:09 cells. Far right hand panels: western blots with HC10-HRP of whole cell lysates from 221B*27:05 and HLA-B*27:09 cells before immunoprecipitation. Representative western blots from one of three independent experiments.
Next we immunoprecipitated HC10-reactive class I heavy chains from surface biotinylated 221B*27:05 and 221B*27:09 cells. Immunoprecipitates were resolved by SDS-PAGE and analysed by western blot with HRP-conjugated streptavidin or HRP-conjugated HC10 (Fig. 1C). Increased quantities of dimers and multimeric FHC forms were detectable in immunoprecipitates from 221B*27:05 cells compared with 221B*27:09 cells (Fig. 1C). In the presence of dithiothreitol, higher molecular weight forms reduced to monomeric heavy chains (Fig. 1C). W6/32 immunoprecipitated monomeric B27 heavy chains from 221B*27:05 and 221B*27:09 cells (Fig. 1C). Western blots of whole cell lysates from 221B*27:05 and 221B*27:09 cells show equivalent levels of expression of HC10-reactive class I heavy chains (Fig 1C).
221B*27:05 cell lines interact more strongly with KIR3DL2 than 221B*27:09
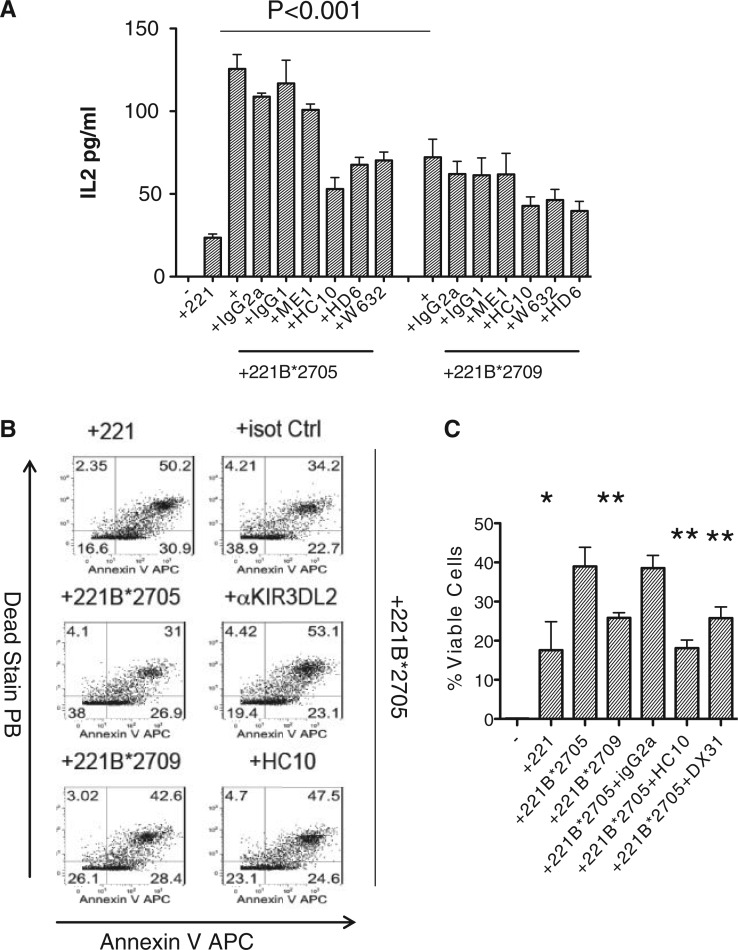
We asked whether increased levels of expression of FHC on 221B*27:05-expressing cells compared with HLA-B*27:09-expressing cell lines would result in enhanced interactions with KIR3DL2. KIR3DL2CD3ε-expressing Jurkat T reporter cells produce more IL-2 when stimulated with B27-expressing B cell lines compared with cells expressing other HLA class I [22]. KIR3DL2CD3ε-expressing Jurkat reporter cells consistently produced more IL-2 when stimulated with HLA-B*27:05-expressing 221 cells compared with stimulation with HLA-B*27:09-expressing cells (Fig. 2A). KIR3DL2CD3ε-reporter cell B27 interactions were inhibited by FHC and dimer reactive antibodies HC10 and HD6, respectively, and by W6/32 but not by isotype control mAbs (Fig. 2A). W6/32 has been reported to bind to some forms of β2m-free HLA class I including HLA-B27. ME1 antibody which binds to β2m-associated HLA-B27 had no effect (Fig. 2A).
Fig. 2.
KIR3DL2 binds more strongly to cell surface HLA-B*27:05 than HLA-B*27:09.
(A) IL-2 secretion by KIR3DL2CD3ε-reporter Jurkat T cells stimulated with (A) HLA-B*27:05 and HLA-B*27:09 transduced 221 cells. Effect of HC10, HD6, ME1 and W632 MAbs on IL-2 secretion by KIR3DL2CD3ε-transduced Jurkat T cells is also shown. Representative of three independent experiments. (B) Left hand panels: proportions of viable (Annexin V, Live-Dead negative) KIR3DL2 + NK cells after 6 day stimulation with parental 221 cells or 221B*27:05 and 221B*27:09 cells. Right hand panels: pproportions of viable (Annexin V, Live-Dead negative) KIR3DL2 + NK cells after 6 day stimulation with 221B*27:05 cells and anti-KIR3DL2 (DX31) or HC10 and W632 antibodies or isotype control antibody (IgG2a). Representative FACS stains from one of three independent experiments with an NK cell line from a healthy B27 control. (C) Proportions of viable KIR3DL2 + NK cells after 6 day stimulation with 221, 221B*27:05 or 221B*27:09 cells or 221B*27:05 cells with anti-KIR3DL2 (DX31), HC10, W632 or isotype control (IgG2a) antibodies. Mean proportions of surviving cells ± 1s.d. from three independent experiments. *P < 0.05 and **P < 0.01 ANOVA.
We studied survival of KIR3DL2-expressing cells stimulated with transduced 221 cells for 5 days. Viable NK cells do not stain with Annexin V and dead stains. 221B*27:05 cells stimulated greater survival of KIR3DL2-expressing NK cells compared with 221B*27:09 cells (Fig. 2B and C). NK cell survival was reduced by stimulation in the presence of HC10, W632 and anti-KIR3DL2 (DX31) mAbs (Fig. 2B and C).
HLA-B*27:05 forms more heavy chain homodimer (B272) in vitro than HLA-B*27:09
In order to determine whether increased dimer formation is an inherent property of HLA-B*27:05, we next asked whether HLA-B*27:05 and HLA-B*27:09 subtypes differed in their ability to form heavy chain homodimers in vitro. Identical quantities of HLA-B*27:05 and HLA-B*27:09 heavy chains were refolded with β2m and B27-binding peptide or without β2m and the yield and purity of resulting B27 heterodimers and dimers assessed biochemically by FPLC and SDS PAGE.
Fig. 3A shows representative FPLC plots of refolded protein from HLA-B*27:05 and HLA-B*27:09, folded in parallel in the presence of β2m and peptide. A panel of HLA-B*27:05- and HLA-B*27:09-specific peptides and shared epitopes (summarized in Materials and methods section) were used. Peaks corresponding to homodimers and heterodimers, defined by SDS PAGE, were quantified by gel exclusion chromatography. Refolds were performed for seven peptides and repeated up to five times. A representative purification is shown in Fig. 4A and the yields of dimeric and heterodimeric protein, expressed as a proportion of total dimeric and heterodimeric protein, are summarized in Fig. 3B. Although we consistently observed heterodimers, in some refolds with HLA-B*27:09 dimer peaks were absent (results not shown). HLA-B*27:05 consistently yielded more B27 dimer compared with HLA-B*27:09.
Fig. 3.
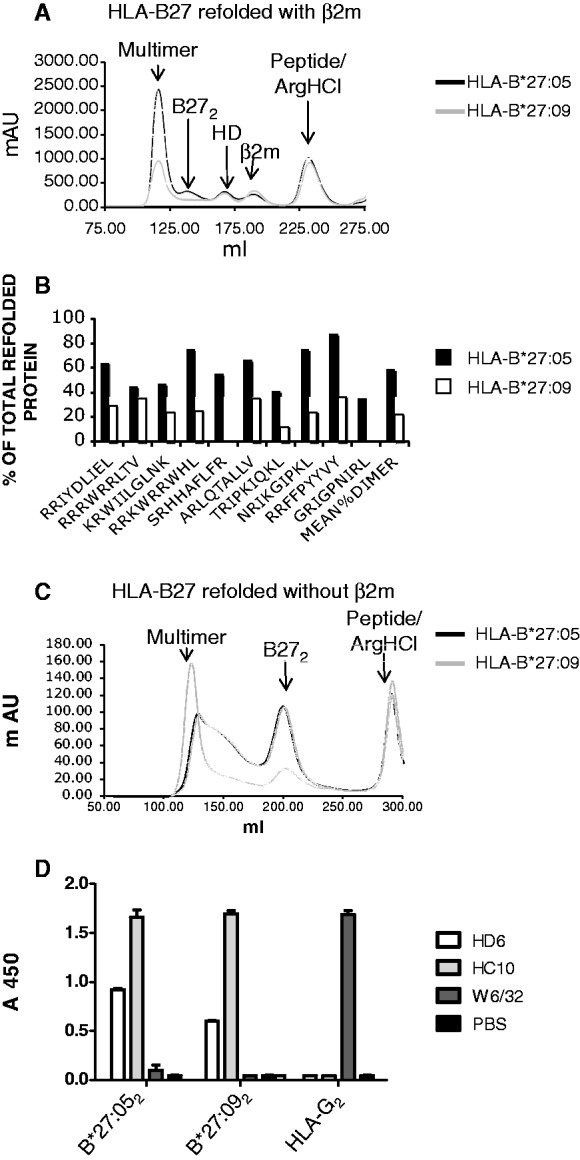
HLA-B*27:05 forms more heavy chain homodimer (B272) in vitro than HLA-B*27:09.
(A) Representative FPLC plot of in vitro refolds of B*27:05 and B*27:09 refolded in the presence of β2m and peptide. Peaks corresponding to heterodimer (HD) and homodimer (B272) are indicated. (B) Ratios of the yield of dimers and heterodimers for HLA-B*27:05 and HLA-B*27:09 refolds with the peptides indicated. FPLC peaks were integrated to obtain yields of the different molecular species. Refolds were performed up to five times. (C) Representative FPLC plot of HLA-B*27:05 and HLA-B*27:09 heavy chain dimer (representative FPLC of six independent refolds). (D) ELISA of HLA-B*27:05 (B*27:052), HLA-B*27:09 (B*27:092) and HLA-G dimers (HLA-G2) with HC10, W632 and HD6 antibodies.
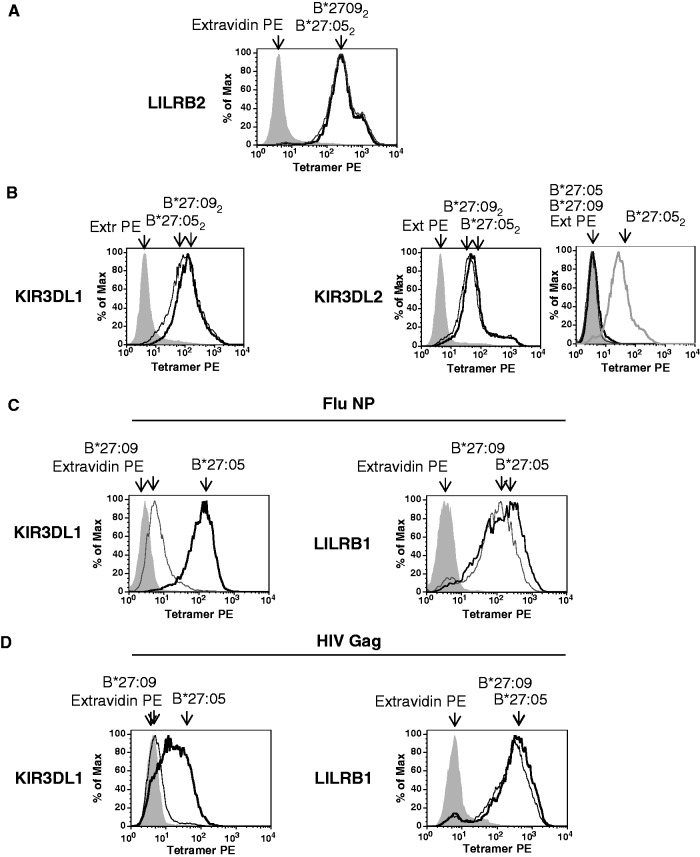
Fig. 4.
Similar binding of HLA-B*27:05 and HLA-B*27:09 dimers to KIR3DL1, KIR3DL2 and LILRB2. HLA-B*27:05 and HLA-B*27:09 heterodimers bind differently to KIR3DL1.
(A) Representative FACS staining of LILRB2-transduced Baf3 cells with HLAB*27:05 (B*27:052) and HLA-B*27:09 (B*27:092) dimer tetramers. Cells were stained with extravidin PE (EX PE) as a negative control stain. (B) Representative FACS staining of KIR3DL1- and KIR3DL2-transduced Baf3 cells with HLA-B*27:05 (B*27:052) and HLA-B*27:09 (B*27:092) heavy chain dimer tetramers. Representative staining of KIR3DL2-transduced Baf3 cells with HLA-B*27:05 (B*27:05) and HLA-B*27:09 heterodimer (B*27:09) tetramers. Staining with HLA-B*27:05 (B*27:052) heavy chain dimer tetramers (B272) is shown for comparison. Cells were stained with extravidin PE (EX PE) as a negative control stain. (C) Representative FACS staining of KIR3DL1- and LILRB1-transduced Baf3 cells with HLA-B*27:05 (B*27:05) and HLA-B*27:09 (B*27:09) heterodimer tetramers formed with the FluNP epitope. Cells were stained with extravidin PE (EX PE) as a negative control stain. (D) Representative FACS staining of KIR3DL1- and LILRB1-transduced Baf3 cells with HLA-B*27:05 (B*27:05) and HLA-B*27:09 (B*27:09) heterodimer tetramers formed with the HIV GAG epitope. Cells were stained with extravidin PE (EX PE) as a negative control stain.
HLA-B*27:05 heavy chains folded in the absence of β2m consistently yielded more B27 dimer compared with HLA-B*27:09 (Fig. 3C). Recombinant HLA-B*27:05 and HLA-B*27:09 dimers bound equivalently strongly to HC10 antibody in ELISA (Fig. 3D). In contrast HLA-B*27:05 dimers bound more strongly to HD6 antibody compared with HLA-B*27:09 dimers in ELISA (Fig. 3D). As previously observed neither HLA-G dimers nor HLA-B27 heterodimers bound to HD6 antibody (Fig. 3D and results not shown).
Recombinant HLA-B*27:05 and HLA-B*27:09 dimer tetramers bind similarly to KIR3DL1, KIR3DL2 and LILRB2; HLA-B*27:05 and HLA-B*27:09 heterodimers bind differently to KIR3DL1
Differences in KIR3DL2 binding to HLA-B*27:05 and HLA-B*27:09 could occur as a consequence of differences in their propensity to form B27 dimers and other FHC species and/or differences in their interaction with KIR receptors. In order to address whether HLA-B*27:05 and HLA-B*27:09 bound differently to immune receptors, we studied the ability of HLA-B*27:05 and HLA-B*27:09 dimer and heterodimer tetramers to stain KIR3DL1/2-transduced cells. In parallel we stained LILRB1- and LILRB2-transduced cells with tetramers to control for tetramer integrity.
HLA-B*27:05 and HLA-B*27:09 dimer tetramers stained LILRB2-transduced Baf3 cells similarly (Fig. 4A). Neither HLA-B*27:05 nor HLA-B*27:09 dimer tetramers bound LILRB1-transduced Baf3 cells (results not shown). HLA-B*27:05 and HLA-B*27:09 dimers bound KIR3DL1 and KIR3DL2 transfectants similarly (Fig. 4B).
HLA-B*27:05 heterodimer tetramers formed with FluNP and Gag epitopes stain KIR3DL1 transfectants. In contrast HLA-B*27:09 heterodimer tetramers complexed with these epitopes did not stain KIR3DL1-transfected cells as strongly as HLA-B*27:05 heterodimers, although these tetramers nevertheless stained LILRB1/ILT2-transfected cells equivalently (Fig. 4C and D).
Increased proportions of peripheral blood NK and CD4 T cells express KIR3DL2 in HLA-B*27:05+ AS patients compared with healthy B*27:05+, B*27:09+ and B27 negative controls
We have previously demonstrated increased proportions of peripheral blood NK and CD4 T cells expressing KIR3DL2 in HLA-B*27:05+ SpA patients compared with B27− RA, ulcerative colitis and HCs [17, 18]. Although KIR3DL2 is also expressed on CD8 T cells, we did not observe significant increases in the proportions of CD8 T cells expressing KIR3DL2 in SpA patients in these studies. Thus, we chose to focus on studying on KIR3DL2 expression by NK and CD4 T cells in this study. Our results indicated a reduced propensity of HLA-B*27:09 to form dimeric and other B27 FHC forms and decreased interactions with KIR3DL2 occurring as a consequence. We predicted that this would translate into differences in the proportions of NK and CD4 T cells expressing KIR3DL2 in HLA-B*27:09+ individuals compared with HLA-B*27:05+ AS patients. Thus we compared the proportions of NK and CD4 T cells expressing KIR3DL2 in the peripheral blood of HLA-B*27:05+ AS patients and HLA-B*27:05, HLA-B*27:09 and HLA-B27− HCs by ex vivo FACS staining. We also studied the proportions of NK and CD4 T cells in each of these groups expressing KIR3DL1 and KIR3DS1 which bind to HLA-B27 and have been reported to be associated with SpA respectively. We observed increased proportions of NK and CD4 T cells expressing KIR3DL2 in the peripheral blood of AS patients with HLA-B*27:05 compared with healthy HLA-B*27:05+, HLA-B*27:09+ and HLA-B27 negative controls (Fig. 5A and B). In contrast we did not see any significant differences in the proportions of NK and CD4 T cells expressing KIR3DL1 and KIR3DS1 between each of these groups. A mean of 20.7 ± 11.9 AS NK cells expressed KIR3DL1 compared with 13.0 ± 8.9 (P = ns) HLA-B*27:05 HCs, 12.8 ± 10.7 HLA-B*27:09 HCs (P = ns) and 23.7 ± 17.7 HLA-B27 negative (P = ns) (results expressed as mean values ± 1s.d.). AS patient NK cells expressed a mean of 9301.2 ± 3175.5 ABC units KIR3DL1 compared with 7429.5 ± 4784.2 units in HLA-B*27:05 HCs (P = ns), 6027.4 ± 4082.6 units in HLA-B*27:09 HCs (P = ns) and 7512 ± 5088 units in HLA-B27 negative HCs (P = ns). 0.5% ± 0.4% AS CD4 T cells expressed KIR3DL1 compared with an aggregated mean of 0.3 ± 0.3 in the HC groups (P = ns). KIR3DS1 staining was only detected in one HLA-B*27:09 HC in this study.
Fig. 5.
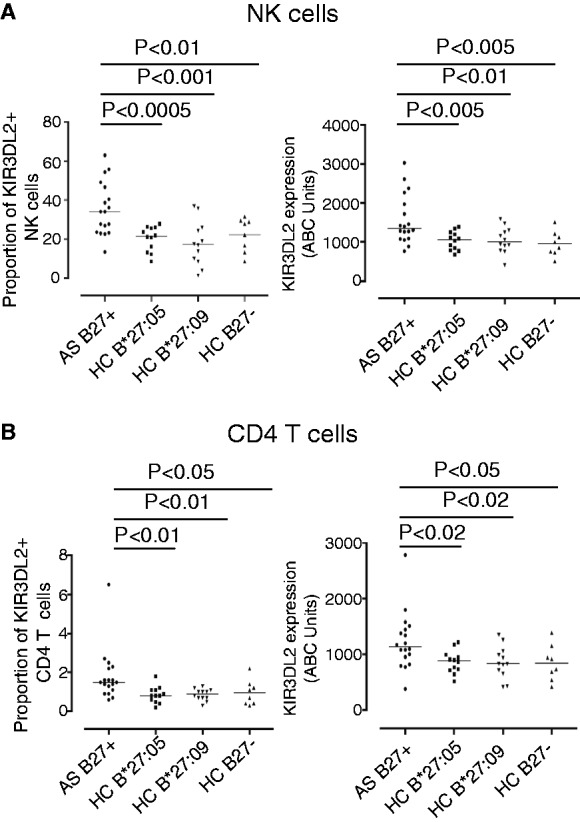
Increased expression of KIR3DL2 by leucocytes from HLA-B*27:05+ patients compared with HLA-B27− and HLA-B*27:05+ and HLA-B*27:09+ healthy controls (HCs).
(A) Left hand panel: percentage of NK cells expressing KIR3DL2 in HLA-B*27:05 + SpA patients, HLA-B*27:05+ HC, HLA-B*27:09 HC and HLA-B27 negative HC. Percentages of NK cells expressing KIR3DL2 were 35.6 ± 13.6 (mean ± 1s.d.), 20.3 ± 6.3 (P = 0.0004), 17.9 ± 11.3 (P = 0.0007) and 22.3 ± 8.5 (P = 0.007) for each of the respective groups. Right hand panel: level of expression of KIR3DL2 (ABC units) by SpA patient NK cells, HLA-B*27:05+, HLA-B*27:09+ and HLA-B27− HC. HLA-B*27:05+ SpA patients expressed 1571 ± 631.9 ABC units compared with 1038 ± 235.7 units (P = 0.003) for HLA-B*27:05 HC, 1049 ± 331.5 (P = 0.007) for HLA-B*27:09+ HC and 961 ± 318 units (P = 0.003) for HLA-B27 negative HC. (B) Left hand panel: percentages of CD4 T cells expressing KIR3DL2 in HLA-B*27:05 + SpA patients, HLA-B*27:05+, HLA-B*27:09 + and HLA-B27 negative HC. A mean of 1.8 ± 1.3 SpA CD4 T cells, 0.8 ± 0.4 HLA-B*27:05 HC (P = 0.01), 0.9 ± 0.3 B*27:09 HC (P = 0.009) and 1.0 ± 0.7 (P = 0.04) B27 negative HC expressed KIR3DL2. Right hand panel: SpA patient CD4 T cells expressed 1232 ± 513.8 ABC units of KIR3DL2, compared with 878.3 ± 199 HLA-B*27:05 HC (P = 0.011), 859.3 ± 288.7 HLA-B*27:09 HC (P = 0.016) and 847 ± 321 B27 negative HC (P = 0.030).
Discussion
Here we show that HLA-B*27:05, which is strongly associated with SpA, forms more B27 FHC ligands for KIR3DL2 compared with HLA-B*27:09 which is not associated with disease.
We detected more cell surface heavy chain dimers and FHC forms on 221B*27:05 compared with 221*27:09 cells biochemically. 221B*27:05 cells also stained more strongly than 221B*27:09 cells with HD6 antibody, which recognizes B27 FHC dimers and other FHCs [23]. Western blots showed that 221B*27:05 and 221B*27:09 cells expressed equivalent amounts of class I heavy chains in cell lysates. Moreover 221B*27:05 and 221B*27:09 cells expressed similar levels of cell surface ME1 and W6/32-reactive B27. These findings support previous preliminary work showing increased proportions of heavy chain dimers on the surface of 221B*27:05 cells compared with 221B*27:09 cells [35].
We have previously shown that B27 FHC is a ligand for KIR3DL2 [3, 22]. Consistent with 221B*27:05 cells expressing more cell surface FHC forms than 221B*27:09, 221B*27:05-expressing cells stimulated KIR3DL2CD3ε reporter cells to a greater extent than 221B*27:09. Moreover KIR3DL2 ligation by HLA-B*27:05 promoted leucocyte survival to a greater degree than HLA-B*27:09.
Reduced interactions with KIR3DL2 could result from decreased cell surface ligand density and/or differences in the strength of ligand binding to these receptors. HLA-B*27:05 and HLA-B*27:09 dimer tetramers bound equivalently to KIR3DL2. Thus, the reduced interaction of HLA-B*27:09 with KIR3DL2 compared with HLA-B*27:05 probably occurs as a consequence of lower levels of B27 FHC forms expressed by this allele.
HLA-B*27:09 heterodimer tetramers bound more weakly or did not bind to KIR3DL1 compared with HLA-B*27:05 heterodimers with the same peptide. The amino acid change of aspartic acid in HLA-B*27:05 to histidine in HLA-B*27:09 at position 116 influences the conformation of bound peptide and recognition by CD8 T cells [36]. KIR3DL1 recognition of HLA-Bw4 is dependent on the sequence of the C-terminal positions 7 and 8 of bound peptide [13]. Differences in the preferred C-terminal anchor and the C-terminal conformations of bound peptide between HLA-B*27:05 and HLA-B*27:09 could thus affect KIR3DL1 recognition.
HLA-B*27:05+ SpA patients had increased proportions of KIR3DL2-expressing peripheral blood NK and CD4 T cells compared with healthy HLA-B*27:05+, HLA-B*27:09+ and B27− controls. Although we have previously shown increased KIR3DL2 expression on leucocytes from B27+SpA patients compared with leucocytes from arthritis and B27 negative HCs, we did not study expression on HLA-B*27:09 controls [17, 18]. KIR3DL2 + CD4 Th17 cells, highly enriched for the expression of Th17 phenotypic markers, are expanded in SpA patients [18]. Increased levels of HLA-B27 FHC dimers and FHC are expressed by monocytes from SpA patients [23, 37–39]. Thus increased expression of B27 FHC ligands for immune receptors such as KIR3DL2 in SpA patients with HLA-B*27:05 could promote the survival of proinflammatory leucocytes.
Our results suggest that the increased binding of HLA-B*27:05 FHC to KIR3DL2 is primarily due to increased formation of FHC compared with HLA-B*27:09. HLA-B*27:05 and HLA-B*27:09 dimer tetramers bound similarly to KIR and LILRB2 receptors. HLA-B*27:05 shows an increased propensity to form dimers compared with HLA-B*27:09. The reactive cysteine 67 is critical for B27 heavy chain dimer formation [5]. Differences in residues at position 116 of the class I heavy chain affect both the reactivity of cysteine 67 and the way in which these two alleles fold and unfold and could affect dimer and FHC formation [5, 40, 41].
B27 FHC forms including B27 dimers form more readily from β2m-associated HLA-B27 heterodimers suboptimally loaded with peptide [5]. It is possible that HLA-B*27:05 is less stringent in the optimization of peptide cargo compared with HLA-B*27:09. Thus, the increased instability of β2m-associated HLA-B*27:05 compared with HLA-B*27:09 could lead to the formation of more B27 FHC forms in disease.
Here we show that disease-associated HLA-B*27:05 forms more B27 heavy chain dimer and FHC in vitro and on transfected cells than HLA-B*27:09, which is not associated with disease. Increased expression of cell surface B27 FHC forms by HLA-B*27:05 correlates with stronger interaction with KIR3DL2 compared with HLAB*27:09. HLA-B*27:05 + AS leucocytes have increased proportions of KIR3DL2. Thus, the increased propensity of HLA-B*27:05 to form FHC ligand compared with HLA-B*27:09 results in enhanced survival of KIR3DL2-expressing leucocytes and could contribute to the differential association of these B27 alleles with AS.
Rheumatology key messages.
The SpA-associated HLA-B*27:05 subtype forms more B27 free heavy chain ligands for KIR3DL2 than HLA-B*27:09.
The AS-associated HLA-B*27:05 subtype promotes greater survival of KIR3DL2-expressing leucocytes than HLA-B*27:09.
Acknowledgements
We thank Jo Phillips, DNAX, and Hidde Ploegh for provision of DX31 and HC10 hybridomas and Kati DiGleria for peptides synthesized for this study. J.S., J.G. O.R. and S.K. were funded by Arthritis Research UK, S.K. by NIHR, H.H. by the Japan Society for the Promotion of Science, K.M. by Furlong, M.A.B.G. and C.L.L. by (FICYT) PC10-70 and P.B. by Oxford NIHR Biomedical Research Centre.
Dr Kollnberger has full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study design: A.C., S.K., P.B., A.M. Acquisition of data: J.G., J.S., A.C., H.H., K.M., O.R., S.F., S.H., A.C., G.D., G.P., E.D., A.V., M.P., V.I., P.G., G.L.N., S.K., C.L.-L., M.A.B.-G. Analysis and interpretation of data: J.G., J.S., A.C., H.H., K.M., O.R., S.F., S.H., A.C., S.P., S.K., P.B. Manuscript preparation: A.C., J.G., J.S., H.H., O.R., P.B., S.K. Statistical analysis: A.C., S.K.
Disclosure statement: The authors have declared no conflicts of interest.
References
- 1.Brown MA, Pile KD, Kennedy LG, et al. HLA class I associations of ankylosing spondylitis in the white population in the United Kingdom. Ann Rheum Dis. 1996;55:268–70. doi: 10.1136/ard.55.4.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melis L, Elewaut D. Progress in spondylarthritis. immunopathogenesis of spondyloarthritis: which cells drive disease? Arthritis Res Ther. 2009;11:233. doi: 10.1186/ar2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kollnberger S, Bird L, Sun MY, et al. Cell-surface expression and immune receptor recognition of HLA-B27 homodimers. Arthritis Rheum. 2002;46:2972–82. doi: 10.1002/art.10605. [DOI] [PubMed] [Google Scholar]
- 4.Kollnberger S, Bird LA, Roddis M, et al. HLA-B27 heavy chain homodimers are expressed in HLA-B27 transgenic rodent models of spondyloarthritis and are ligands for paired Ig-like receptors. J Immunol. 2004;173:1699–710. doi: 10.4049/jimmunol.173.3.1699. [DOI] [PubMed] [Google Scholar]
- 5.Bird LA, Peh CA, Kollnberger S, et al. Lymphoblastoid cells express HLA-B27 homodimers both intracellularly and at the cell surface following endosomal recycling. Eur J Immunol. 2003;33:748–59. doi: 10.1002/eji.200323678. [DOI] [PubMed] [Google Scholar]
- 6.Allen RL, O'Callaghan CA, McMichael AJ, et al. Cutting edge: HLA-B27 can form a novel beta 2-microglobulin-free heavy chain homodimer structure. J Immunol. 1999;162:5045–8. [PubMed] [Google Scholar]
- 7.Kollnberger S, Bowness P. The role of B27 heavy chain dimer immune receptor interactions in spondyloarthritis. Adv Exp Med Biol. 2009;649:277–85. doi: 10.1007/978-1-4419-0298-6_21. [DOI] [PubMed] [Google Scholar]
- 8.Zino E, Di Terlizzi S, Carugo C, et al. Rapid detection of all HLA-B*27 alleles (B*2701-B*2725) by group-specific polymerase chain reaction. Tissue Antigens. 2004;63:88–92. doi: 10.1111/j.1399-0039.2004.00158.x. [DOI] [PubMed] [Google Scholar]
- 9.D'Amato M, Fiorillo MT, Carcassi C, et al. Relevance of residue 116 of HLA-B27 in determining susceptibility to ankylosing spondylitis. Eur J Immunol. 1995;25:3199–201. doi: 10.1002/eji.1830251133. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Larrea C, Gonzalez-Roces S, Alvarez V. HLA-B27 structure, function, and disease association. Curr Opin Rheumatol. 1996;8:296–308. doi: 10.1097/00002281-199607000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Khan MA, Mathieu A, Sorrentino R, et al. The pathogenetic role of HLA-B27 and its subtypes. Autoimmun Rev. 2007;6:183–9. doi: 10.1016/j.autrev.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Lanier LL. Follow the leader: NK cell receptors for classical and nonclassical MHC class I. Cell. 1998;92:705–7. doi: 10.1016/s0092-8674(00)81398-7. [DOI] [PubMed] [Google Scholar]
- 13.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 14.Kollnberger S, Chan A, Sun MY, et al. Interaction of HLA-B27 homodimers with KIR3DL1 and KIR3DL2, unlike HLA-B27 heterotrimers, is independent of the sequence of bound peptide. Eur J Immunol. 2007;37:1313–22. doi: 10.1002/eji.200635997. [DOI] [PubMed] [Google Scholar]
- 15.Young NT, Uhrberg M. KIR expression shapes cytotoxic repertoires: a developmental program of survival. Trends Immunol. 2002;23:71–5. doi: 10.1016/s1471-4906(01)02113-5. [DOI] [PubMed] [Google Scholar]
- 16.Ugolini S, Arpin C, Anfossi N, et al. Involvement of inhibitory NKRs in the survival of a subset of memory-phenotype CD8+ T cells. Nat Immunol. 2001;2:430–5. doi: 10.1038/87740. [DOI] [PubMed] [Google Scholar]
- 17.Chan AT, Kollnberger SD, Wedderburn LR, et al. Expansion and enhanced survival of natural killer cells expressing the killer immunoglobulin-like receptor KIR3DL2 in spondyloarthritis. Arthritis Rheum. 2005;52:3586–95. doi: 10.1002/art.21395. [DOI] [PubMed] [Google Scholar]
- 18.Bowness P, Ridley A, Shaw J, et al. Th17 cells expressing KIR3DL2+ and responsive to HLA-B27 homodimers are increased in ankylosing spondylitis. J Immunol. 2011;186:2672–80. doi: 10.4049/jimmunol.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown D, Trowsdale J, Allen R. The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens. 2004;64:215–25. doi: 10.1111/j.0001-2815.2004.00290.x. [DOI] [PubMed] [Google Scholar]
- 20.Shiroishi M, Kuroki K, Rasubala L, et al. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc Natl Acad Sci USA. 2006;103:16412–7. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giles J, Shaw J, Piper C, et al. HLA-B27 homodimers and free H chains are stronger ligands for leukocyte Ig-like receptor B2 than classical HLA class I. J Immunol. 2012;188:6184–93. doi: 10.4049/jimmunol.1102711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong-Baeza I, Ridley A, Shaw J, et al. KIR3DL2 binds to HLA-B27 dimers and free H chains more strongly than other HLA class I and promotes the expansion of T cells in ankylosing spondylitis. J Immunol. 2013;190:3216–24. doi: 10.4049/jimmunol.1202926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Payeli SK, Kollnberger S, Marroquin Belaunzaran O, et al. Inhibiting HLA-B27 homodimer-driven immune cell inflammation in spondylarthritis. Arthritis Rheum. 2012;64:3139–49. doi: 10.1002/art.34538. [DOI] [PubMed] [Google Scholar]
- 24.Ramos M, Paradela A, Vazquez M, et al. Differential association of HLA-B*2705 and B*2709 to ankylosing spondylitis correlates with limited peptide subsets but not with altered cell surface stability. J Biol Chem. 2002;277:28749–56. doi: 10.1074/jbc.M204155200. [DOI] [PubMed] [Google Scholar]
- 25.Fiorillo MT, Ruckert C, Hulsmeyer M, et al. Allele-dependent similarity between viral and self-peptide presentation by HLA-B27 subtypes. J Biol Chem. 2005;280:2962–71. doi: 10.1074/jbc.M410807200. [DOI] [PubMed] [Google Scholar]
- 26.Brooks JM, Murray RJ, Thomas WA, et al. Different HLA-B27 subtypes present the same immunodominant Epstein-Barr virus peptide. J Exp Med. 1993;178:879–87. doi: 10.1084/jem.178.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nixon DF, Townsend AR, Elvin JG, et al. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature. 1988;336:484–7. doi: 10.1038/336484a0. [DOI] [PubMed] [Google Scholar]
- 28.Kvist S, Hamann U. A nucleoprotein peptide of influenza A virus stimulates assembly of HLA-B27 class I heavy chains and beta 2-microglobulin translated in vitro. Nature. 1990;348:446–8. doi: 10.1038/348446a0. [DOI] [PubMed] [Google Scholar]
- 29.Perosa F, Luccarelli G, Prete M, et al. Beta 2-microglobulin-free HLA class I heavy chain epitope mimicry by monoclonal antibody HC-10-specific peptide. J Immunol. 2003;171:1918–26. doi: 10.4049/jimmunol.171.4.1918. [DOI] [PubMed] [Google Scholar]
- 30.Tanabe M, Sekimata M, Ferrone S, et al. Structural and functional analysis of monomorphic determinants recognized by monoclonal antibodies reacting with the HLA class I alpha 3 domain. J Immunol. 1992;148:3202–9. [PubMed] [Google Scholar]
- 31.Martayan A, Sibilio L, Tremante E, et al. Class I HLA folding and antigen presentation in beta 2-microglobulin-defective Daudi cells. J Immunol. 2009;182:3609–17. doi: 10.4049/jimmunol.0802316. [DOI] [PubMed] [Google Scholar]
- 32.McCutcheon JA, Smith KD, Valenzuela A, et al. HLA-B*0702 antibody epitopes are affected indirectly by distant antigen residues. Hum Immunol. 1993;36:69–75. doi: 10.1016/0198-8859(93)90108-d. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz A, Fernandez-Repollet E. Primary performance parameters as quality control indicators. Flow Cytometry Standardization Forum. 1994;6:9–10. [Google Scholar]
- 34.Dangoria NS, DeLay ML, Kingsbury DJ, et al. HLA-B27 misfolding is associated with aberrant intermolecular disulfide bond formation (dimerization) in the endoplasmic reticulum. J Biol Chem. 2002;277:23459–68. doi: 10.1074/jbc.M110336200. [DOI] [PubMed] [Google Scholar]
- 35.Blanco-Gelaz MA, Suarez-Alvarez B, Diaz-Pena R, et al. HLA-B27 polymorphism at position 116 critically influences the association with TAP/tapasin, intracellular trafficking and conformational homodimers formation. Mol Immunol. 2009;46:1304–11. doi: 10.1016/j.molimm.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 36.Hulsmeyer M, Fiorillo MT, Bettosini F, et al. Dual, HLA-B27 subtype-dependent conformation of a self-peptide. J Exp Med. 2004;199:271–81. doi: 10.1084/jem.20031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai WC, Chen CJ, Yen JH, et al. Free HLA class I heavy chain-carrying monocytes—a potential role in the pathogenesis of spondyloarthropathies. J Rheumatol. 2002;29:966–72. [PubMed] [Google Scholar]
- 38.Raine T, Brown D, Bowness P, et al. Consistent patterns of expression of HLA class I free heavy chains in healthy individuals and raised expression in spondyloarthropathy patients point to physiological and pathological roles. Rheumatology. 2006;45:1338–44. doi: 10.1093/rheumatology/kel305. [DOI] [PubMed] [Google Scholar]
- 39.Cauli A, Dessole G, Nurchis PP, et al. [The role of HLA-B27 molecules in the pathogenesis of ankylosing spondylitis] Reumatismo. 2002;54:266–71. doi: 10.4081/reumatismo.2002.266. [DOI] [PubMed] [Google Scholar]
- 40.Fussell H, Nesbeth D, Lenart I, et al. Novel detection of in vivo HLA-B27 conformations correlates with ankylosing spondylitis association. Arthritis Rheum. 2008;58:3419–24. doi: 10.1002/art.23990. [DOI] [PubMed] [Google Scholar]
- 41.Hillig RC, Hulsmeyer M, Saenger W, et al. Thermodynamic and structural analysis of peptide- and allele-dependent properties of two HLA-B27 subtypes exhibiting differential disease association. J Biol Chem. 2004;279:652–63. doi: 10.1074/jbc.M307457200. [DOI] [PubMed] [Google Scholar]