Abstract
Focal segmental glomerulosclerosis (FSGS) and collapsing glomerulopathy are common causes of nephrotic syndrome. Variants in >20 genes, including genes critical for mitochondrial function, have been associated with these podocyte diseases. One such gene, PDSS2, is required for synthesis of the decaprenyl tail of coenzyme Q10 (Q10) in humans. The mouse gene Pdss2 is mutated in the kd/kd mouse model of collapsing glomerulopathy. We examined the hypothesis that human PDSS2 polymorphisms are associated with podocyte diseases. We genotyped 377 patients with primary FSGS or collapsing glomerulopathy, together with 900 controls, for 9 single-nucleotide polymorphisms in the PDSS2 gene in a case-control study. Subjects included 247 African American (AA) and 130 European American (EA) patients and 641 AA and 259 EA controls. Among EAs, a pair of proxy SNPs was significantly associated with podocyte disease, and patients homozygous for one PDSS2 haplotype had a strongly increased risk for podocyte disease. By contrast, the distribution of PDSS2 genotypes and haplotypes was similar in AA patients and controls. Thus a PDSS2 haplotype, which has a frequency of 13% in the EA control population and a homozygote frequency of 1.2%, is associated with a significantly increased risk for FSGS and collapsing glomerulopathy in EAs. Lymphoblastoid cell lines from FSGS patients had significantly less Q10 than cell lines from controls; contrary to expectation, this finding was independent of PDSS2 haplotype. These results suggest that FSGS patients have Q10 deficiency and that this deficiency is manifested in patient-derived lymphoblastoid cell lines.
Keywords: nephrotic syndrome, collapsing glomerulopathy, focal segmental glomerulosclerosis, mitochondria, ubiquinone
focal segmental glomerulosclerosis (FSGS) comprises several distinct pathological entities that start with injury to the glomerular podocytes and progress to loss of these normally postmitotic cells and, eventually, to focal and segmental glomerular scarring. FSGS has become the leading cause of idiopathic nephrotic syndrome in adults and is responsible for ∼6% of cases of end-stage kidney disease in children (24).
Approximately 50% of patients with FSGS reach end-stage kidney disease within 10 yr of diagnosis, which represents the worst prognosis among common glomerular diseases. One recent classification recognizes three major variants: genetic FSGS, primary FSGS, and adaptive FSGS (3). Genetic mutations that have been associated with familial FSGS have been identified in the genes encoding nephrin (NPHS1), podocin (NPHS2), α-actinin-4 (ACTN4), CD2-associated protein (CD2AP), and the TRPC6 cation channel (41, 63), as well as numerous other genes. Common variants in the adjacent WT1 and WIT1 genes, encoding a transcription factor that, when mutated, can cause Wilms' tumor, were associated with idiopathic FSGS in African Americans (AAs). Coding variants in the APOL1 gene are strongly associated with risk for FSGS, collapsing glomerulopathy associated with HIV infection, and nondiabetic chronic kidney disease in AAs (16, 25). Collapsing glomerulopathy accounts for ∼11% of FSGS cases and is characterized by podocyte proliferation and dedifferentiation, which eventually lead to glomerular scarring (55).
There is reason to believe that additional genes that affect susceptibility to FSGS and collapsing glomerulopathy remain to be identified. The kd/kd mouse, which originated as a spontaneous mutant and was initially thought to be similar to nephronophthisis (29), is a promising animal model for these types of syndromes. Mutant homozygotes develop a severe tubulointerstitial nephritis (36), but the same phenotype also develops in Rag-1−/−kd/kd mice in the absence of an adaptive immune response (21). By linkage analysis and positional cloning, the kd allele was shown to have a missense mutation in a prenyltransferase-like mitochondrial protein that corresponds to a human gene product originally identified as “candidate tumor suppressor protein” (9, 40). This gene is now known to be an ortholog of polyprenyl diphosphate synthase in Schizosaccharomyces pombe; it consists of two subunits (Dlp1 and Dps1) that form a heterotetramer with decaprenyl diphosphate synthase activity (46). This heterotetramer catalyzes the synthesis of a polyisoprenoid chain that forms the tail of ubiquinone, which in mice consists predominantly of coenzyme Q9 (Q9) and in humans consists predominantly of coenzyme Q10 (Q10) (45).
The mutant kd/kd mice have immunohistochemical, ultrastructural, and biochemical features of defects that are similar to those seen in collapsing glomerulopathy (2). When associated with certain other mutations, heterozygotes, as well as homozygotes, may manifest glomerular disease (30), and environmental conditions such as caloric restriction and a germ-free environment affect disease expression (20). A child with encephalomyopathy and nephrotic syndrome who was a compound heterozygote for mutations at the PDSS2 locus was recently reported (28), and we subsequently demonstrated that kd/kd mice have neuromuscular defects (66). We have therefore examined DNA samples from individuals with or without FSGS with regard to single-nucleotide polymorphisms (SNPs) in the human gene that corresponds to kd, designated prenyl (decaprenyl) diphosphate synthase subunit 2 (PDSS2) by the Human Gene Nomenclature Committee. We have conducted a case-control study to address the hypothesis that particular haplotypes of the PDSS2 gene are associated with FSGS. B cell lines derived from FSGS patients and controls were prepared and assayed for Q10 content. Surprisingly, FSGS was associated with a decreased content of Q10, irrespective of PDSS2 haplotype.
MATERIALS AND METHODS
Clinical ascertainment.
Study inclusion criteria included diagnosis of primary FSGS or collapsing glomerulopathy on the basis of a biopsy of the native kidney or within the 1st yr following renal transplant. Since no approach has been proven to ensure consensus in diagnosis of these two podocyte disorders among pathologists, the distinction between primary FSGS and collapsing glomerulopathy cannot be made reliably in this study population: for simplicity, the diagnostic term FSGS is used for all patients. Study exclusion criteria include a diagnosis of adaptive FSGS or medication-associated FSGS. All patients except two had undergone at least one renal biopsy documenting FSGS. Two patients did not have a native kidney biopsy but had a presentation and course compatible with FSGS and then developed FSGS in the renal transplant.
Demographics of the study population.
DNA samples were obtained from FSGS patients identified at 21 US-based academic medical centers as part of the National Institutes of Health (NIH) FSGS Genetic Study (26, 38), which was initiated in 1994 and continues to accrue subjects. The study enrolled 377 patients with FSGS and 900 subjects without known kidney disease (controls) (Table 1). Written informed consent was obtained from all participants. Racial classification was based on self-identification. There was no evidence of population substructure among FSGS patients and controls (38). The NIH FSGS Genetic Study has enrolled relatively few Hispanic subjects, Native American subjects, or subjects of Asian descent; therefore, these population groups were not included.
Table 1.
Racial background and HIV infection status of FSGS patients and controls
| FSGS Patients (n = 377) |
Controls (n = 900) |
|||
|---|---|---|---|---|
| HIV-seronegative | HIV-seropositive | HIV-seronegative | HIV-seropositive | |
| EA | 130* | 0 | 259 | 0 |
| AA | 195 | 52 | 393 | 248 |
| Total | 325 | 52 | 652 | 248 |
FSGS, focal segmental glomerulosclerosis; EA, European American; AA, African American.
94 FSGS patients were HIV-seronegative and 36 were not tested; most or all of the untested patients lacked risk factors for HIV-1 infection.
There were 130 European American (EA) FSGS patients. At the time of renal biopsy, 13 EA patients were children (<18 yr of age) and 117 were adults. EAs rarely, if ever, develop HIV-associated collapsing glomerulopathy (here, termed FSGS for simplicity); 94 FSGS patients were found to be HIV-seronegative and 36 were not tested, most or all of whom lacked risk factors for HIV-1 infection; therefore, we consider this group to be HIV-1-uninfected. A family history of biopsy-documented FSGS was present in three patients. Among these families, only one family member was studied, so all patients were unrelated individuals. All three kindreds were studied for NPHS2, ACTN4, CD2AP, and WT1, and no disease-associated mutations were found.
There were 195 AA HIV-seronegative patients. At the time of renal biopsy, 22 of the AA patients were children (<18 yr of age) and 173 were adults. Among these, 181 were found to be HIV-seronegative and 14, most or all of whom lacked risk factors for HIV-1 infection, were not tested; therefore, we consider this group to be HIV-1-uninfected patients. A family history of biopsy-proven FSGS was present in 10 patients. Only one member was studied among these families, so all patients were unrelated individuals.
There were 52 HIV-seropositive FSGS patients, and all were AAs. The renal biopsies were consistent with HIV-associated collapsing glomerulopathy, with collapse of glomerular capillaries and podocyte abnormalities. No patient reported a family history of FSGS.
The control groups included 900 adults, 652 of whom were HIV-1-uninfected and 248 of whom were HIV-1-seropositive. The HIV-uninfected controls were recruited at the NIH Clinical Center (Bethesda, MD) and at the Frederick National Laboratory for Cancer Research. All controls were in self-defined good health, none were known to have kidney disease, and all were tested at the time of blood donation and found to be HIV-1-seronegative.
The 248 HIV-1-seropositive controls were enrolled in the Johns Hopkins AIDS Link to the Intravenous Drug Experience (ALIVE) cohort in Baltimore, MD (61). This control group represents a hypernormal population, defined as individuals who do not have FSGS, do have a genetic susceptibility to FSGS (AA descent), and have been exposed to an environmental factor (HIV-1) known to cause FSGS. The controls had been HIV-1-infected for ≥8 yr, had a serum creatinine <1.5 mg/dl, and had a random urine protein-to-creatinine ratio <0.5. These normal tests for renal function strongly suggest that HIV-associated FSGS was absent at the time of recruitment. The duration of HIV infection was determined as the time since recruitment into the study (for seroprevalent individuals) or as the date of HIV seroconversion, imputed as the midpoint between last negative test date and first positive test date (for seroconverting individuals). The duration of HIV infection was a mean and median of 10 yr.
Institutional Review Board approval was obtained at each participating institution, and written informed consent was obtained from all subjects.
SNP genotyping.
SNPs were selected from the National Center for Biotechnology Information SNP database within the PDSS2 gene, the human ortholog of the mouse gene that encodes the kd allele, considering their map positions within PDSS2, minor allele frequency >5% in EAs or AAs, and linkage disequilibrium (LD) among SNPs using HapMap data or based on HapMap data for CEU and YRI. The nine selected SNPs were distributed at approximately equal distances across the PDSS2 gene, as shown in Fig. 1A. Genotypes were determined using TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA) according to the manufacturer's protocols. TaqMan genotypes were automatically called and plotted on the basis of a two-parameter plot using fluorescence intensities of FAM and VIC dye-labeled MGB probes. Eight water controls were included on each 386-well plate to monitor potential PCR contamination, and 10% of the DNA samples were duplicated between plates to control for sampling-handling error or genotyping errors. We observed 100% concordance among duplicate DNA samples. All water controls were negative, indicating no PCR contamination.
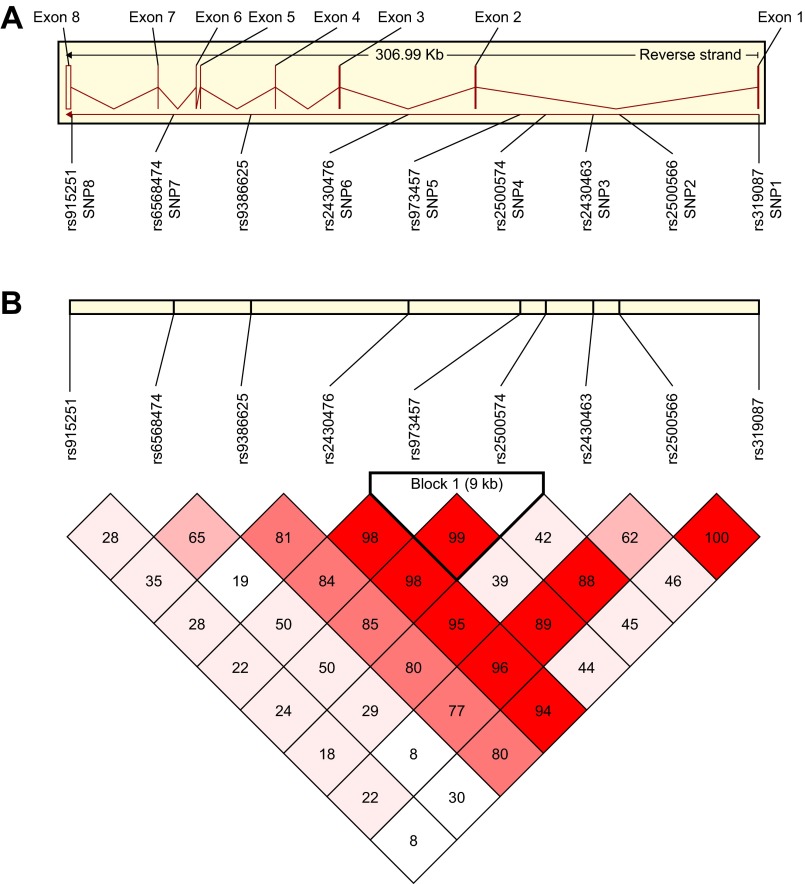
Fig. 1.
Organization of the prenyl (decaprenyl) diphosphate synthase subunit 2 (PDSS2) locus and haplotype block structure. A: exon/intron organization of the PDSS2 locus showing positions of single-nucleotide polymorphisms (SNPs). Eight exons are designated by vertical bars; lines between bars represent introns. Gene image was obtained from the Ensembl Web site (http://www.ensembl.org/Homo_sapiens/geneview?gene=ENSG00000164494). B: haplotype block structure defined by 8 SNPs at the PDSS2 locus. White blocks indicate low D′ [a measure of linkage disequilibrium (LD), where 1.00 = complete LD] and, consequently, high recombination rates, with deepening shades of pink to red indicating increasing D′ values. Numbers in boxes represent actual D′ values multiplied by 100.
Statistical analysis.
Allele frequencies observed in the patients and the racially matched control population were compared by Fisher's exact test; the odds ratios (ORs) with 95% confidence intervals (CIs) were calculated using SAS software version 9.1 (SAS Institute, Cary, NC).
The SNP alleles were arranged into possible haplotypes by a Bayesian statistical method using PHASE version 2.1 (51, 52). Associations between haplotypes and FSGS were assessed by comparison of frequencies between patients and controls by Fisher's exact test or the χ2 test; ORs and CIs were also calculated using SAS version 9.1. LD and correlation coefficients (r2) were assessed by the Haploview program (Whitehead Institute, Cambridge, MA) (4).
Generation and culture of lymphoblastoid cell lines.
Lymphoblastoid cell lines (LCLs) were generated from subjects using Epstein-Barr virus to transform B lymphocytes from peripheral blood mononuclear cells; cells were cultured in 15% fetal calf serum and RPMI culture medium (GIBCO, Life Technologies, Grand Island, NY) (37).
Determination of Q10 content in LCL lipid extracts by HPLC tandem mass spectroscopy.
LCL cell pellets were thawed on ice and then suspended in 3 ml of phosphate-buffered saline (0.14 M NaCl, 1.2 mM NaH2PO4, and 8.1 mM Na2HPO4, pH 7.4) and homogenized for 1.5 min with a Polytron (Kinematica PT2000) at maximum speed on ice. Aliquots of the homogenate (50 μl) were removed to determine the protein content [bicinchoninic acid (BCA) kit, Pierce], with bovine serum albumin as standard. Remaining aliquots (300 μl) were transferred to borosilicate tubes and stored at −20°C. Lipid extracts were prepared by addition of 1.2 ml of methanol followed by 1.8 ml of petroleum ether. Diethoxy-Q10 (12) was used as an internal standard for determination of Q10 in human cell lipid extracts. Samples were vortexed for 45 s. After removal of the organic upper layer, another 1.8 ml of petroleum ether was added, samples were vortexed again, and the combined organic phase was dried under a stream of nitrogen gas and resuspended in 200 μl of ethanol (USP, Aaper Alcohol and Chemical, Shelbyville, KY). A standard curve, composed of tubes containing 3, 15, 72, 204, or 1,021 pmol of Q10 and a constant amount of diethoxy-Q10, was prepared in duplicate. Lipid extraction and resuspension of each standard in 200 μl of ethanol was performed as described above.
HPLC-tandem mass spectrometry (MS/MS) analyses were performed as previously described for determination of Q9 and Q10 in mouse lipid extracts (12). Briefly, a 4000 QTRAP linear MS/MS spectrometer (Applied Biosystems, Foster City, CA) was used. Analyst version 1.4.2 (Applied Biosystems) was used for data acquisition and processing. A binary HPLC solvent delivery system was used with a Luna phenyl-hexyl column (particle size 3 μm, 50 × 2.00 mm; Phenomenex). The mobile phase consisted of solvent A (95:5 methanol-isopropanol with 2.5 mM ammonium formate) and solvent B (isopropanol and 2.5 mM ammonium formate). The percentage of solvent B for the first 1.5 min was 0% and increased linearly to 15% by 2 min. The percentage of solvent B remained unchanged for the next minute and decreased linearly to 0% by 4 min. A constant flow rate of 600 μl/min was used. All samples and standards (10 μl injected) were analyzed in multiple reaction monitoring mode (MRM); MRM transitions were as follows: mass-to-charge ratio (m/z) 880.7/197.0 (Q10 with ammonium adduct), m/z 882.7/197.0 (Q10H2 with ammonium adduct), m/z 908.7/225.1 (diethoxy-Q10 with ammonium adduct), and m/z 910.7/225.1 (diethoxy-Q10H2 with ammonium adduct).
The standard curve was a simple linear fit forced through the origin. The limit of detection (LOD) and the limit of quantitation (LOQ) of Q10 analysis using the 4000 QTRAP were defined as the concentrations that give a peak with signal-to-noise ratio of 3 and 10, respectively. For the assays presented, we determined that the LOD and LOQ of Q10 using 4000 QTRAP were 0.02 and 0.05 pmol, respectively. Thus the first point of the standard curve (0.15 pmol of Q10) was well above the LOD and LOQ. Each of the samples analyzed (lipid extracts from FSGS patients or controls) contained >0.15 pmol of Q10 and, thus, was also well above the established LOQ.
Comparisons of Q10 content were restricted to samples that were prepared and treated in the same way and analyzed at the same time. Q10 in control and haplotype H2-positive (H2+) FSGS patient LCLs were measured first, and Q10 in a new set of control and H2− LCLs was measured ∼2 yr later. During the intervening period, the mass spectrometer went through numerous maintenance procedures and adjustments, and cells cultured during the different times may have been subjected to slight differences in culture conditions. These variables preclude the direct comparison of the two control groups or the H2+ and H2− patients.
RESULTS
Association of SNPs with FSGS.
PDSS2 spans ∼307 kb on chromosome 6 and has seven introns and eight exons encoding a 399-amino acid protein (Fig. 1A). All the SNPs that were genotyped occurred within noncoding regions of the gene: SNP1 (rs319087), SNP2 (rs2500566), SNP3 (rs2430463), SNP4 (rs2500574), and SNP5 (rs973457) were in intron 1; SNP6 (rs2430476) was in intron 2; rs9386625 was in intron 4; SNP7 (rs6568474) was in intron 6; and SNP8 (rs915251) was near the 3′-untranslated region. None of the SNPs are in known splice sites. Genotypic frequencies in rs9386625 did not conform to Hardy-Weinberg equilibrium (HWE) in the control group (P = 0.01) because of a deficiency in the observed number of heterozygotes, so the results for rs9386625 were not included in further calculations regarding differences between patients and controls. All exons at ∼3 kb upstream were resequenced in eight control cell lines and eight cell lines from FSGS patients. Although additional SNPs were identified and were also resequenced in patients and controls, none could explain the susceptibility differences that were observed (M. Peng, unpublished observations).
The extent of LD and correlation (r2) among the remaining SNPs is shown in Fig. 1B and Table 2 for EAs. There was complete LD between the two SNPs near the COOH terminus of the molecule: the SNP1 A allele always occurred with the SNP2 A allele (D′ = 1.00, r2 = 0.507). SNP4 and SNP5 in a 9-kb block were highly correlated (r2 = 0.951) and showed near-absolute LD (D′= 0.99) and are, therefore, proxies (i.e., each SNP conveys equivalent information).
Table 2.
Linkage disequilibrium and correlation among eight PDSS2 SNPs in EAs
| SNP7 rs6568474 | SNP6 rs2430476 | SNP5 rs973457 | SNP4 rs2500574 | SNP3 rs2430463 | SNP2 rs2500566 | SNP1 rs319087 | |
|---|---|---|---|---|---|---|---|
| SNP8 rs915251 | 0.29 (0.03) | 0.29 (0.03) | 0.23 (0.04) | 0.24 (0.05) | 0.19 (0.02) | 0.22 (0.04) | 0.08 (0.00) |
| SNP7 rs6568474 | 0.19 (0.01) | 0.50 (0.07) | 0.50 (0.07) | 0.29 (0.03) | 0.08 (0.00) | 0.30 (0.02) | |
| SNP6 rs2430476 | 0.98 (0.28) | 0.99 (0.29) | 0.96 (0.50) | 0.96 (0.45) | 0.95 (0.22) | ||
| SNP5 rs973457 | 0.99 (0.95) | 0.39 (0.08) | 0.89 (0.47) | 0.44 (0.17) | |||
| SNP4 rs2500574 | 0.43 (0.10) | 0.89 (0.48) | 0.46 (0.17) | ||||
| SNP3 rs2430463 | 0.63 (0.35) | 0.46 (0.10) | |||||
| SNP2 rs2500566 | 1.00 (0.51) |
Values represent linkage disequilibrium (D′), with correlation coefficient (r2) in parentheses. SNP, single-nucleotide polymorphism; PDSS2, prenyl (decaprenyl) diphosphate synthase subunit 2.
No significant difference in allele frequencies was observed between the affected and unaffected AAs (Table 3; P ≥ 0.06), but there were significant differences in allele frequencies between the EA patients and controls for the proxy SNPs, SNP4 (P = 0.001) and SNP5 (P = 0.008; Table 4). We next tested the null hypothesis that genotype frequencies within the patient groups would be in conformance with HWE for a binomial distribution. The observed genotypic frequencies for the minor allele showed significant distortions from expectation for SNP5, SNP4, and SNP1 (P = 0.03, 0.004, and 0.04, respectively) and a trend for SNP2 (P = 0.061), with heterozygotes being underrepresented and homozygotes overrepresented in FSGS patients, but not in controls (P ≥ 0.13; Table 4). No distortions in HWE were observed in AAs (P ≥ 0.05).
Table 3.
PDSS2 minor allele frequencies for FSGS patients and controls
| AA |
EA |
||||
|---|---|---|---|---|---|
| SNP | FSGS patients | Controls | FSGS patients | Controls | |
| rs319087 | 1 | 0.457 | 0.459 | 0.349 | 0.324 |
| rs2500566 | 2 | 0.155 | 0.140 | 0.462 | 0.479 |
| rs2430463 | 3 | 0.160 | 0.130 | 0.445 | 0.484 |
| rs2500574 | 4 | 0.277 | 0.241 | 0.417* | 0.355 |
| rs973457 | 5 | 0.383 | 0.362 | 0.406† | 0.351 |
| rs2430476 | 6 | 0.084 | 0.082 | 0.319 | 0.336 |
| rs6568474 | 7 | 0.412 | 0.401 | 0.341 | 0.310 |
| rs915251 | 8 | 0.393 | 0.383 | 0.481 | 0.425 |
P = 0.001, †P = 0.008 (by Fisher's exact test).
Table 4.
PDSS2 SNP associations with FSGS in EAs for the recessive genetic model
| Number (Frequency) |
||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Group | TT | TA | AA | OR | 95% CI | P Value | HWE |
| SNP1 rs319087 | FSGS (n = 130) | 60 (0.465) | 48 (0.372) | 21 (0.163) | 1.9 | 1.0–3.5 | 0.063 | 0.041 |
| Control (n = 258) | 115 (0.446) | 119 (0.461) | 24 (0.093) | 0.387 | ||||
| GG | GT | TT | ||||||
| SNP2 rs2500566 | FSGS (n = 130) | 33 (0.254) | 54 (0.415) | 43 (0.331) | 1.7 | 1.0–2.6 | 0.031 | 0.061 |
| Control (n = 258) | 70 (0.271) | 129 (0.500) | 59 (0.229) | 0.977 | ||||
| GG | GA | AA | ||||||
| SNP4 rs2500574 | FSGS (n = 127) | 51 (0.402) | 46 (0.362) | 30 (0.236) | 2.6 | 1.5–4.7 | 0.001 | 0.004 |
| Control (n = 258) | 102 (0.395) | 129 (0.500) | 27 (0.105) | 0.135 | ||||
| AA | AG | GG | ||||||
| SNP5 rs973457 | FSGS (n = 128) | 51 (0.398) | 50 (0.391) | 27 (0.211) | 2.2 | 1.2–3.9 | 0.008 | 0.03 |
| Control (n = 258) | 105 (0.407) | 125 (0.484) | 28 (0.108) | 0.304 | ||||
Recessive genetic model compares homozygotes for the minor allele with homozygotes and heterozygotes for the major allele. OR, odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium. In the FSGS group, SNP4 and SNP5 are in strong linkage disequilibrium; therefore, their associations are not independent. TT, TA, AA, GG, GT, GA, and AG refer to the SNP genotypes.
Disease-associated haplotype.
On the basis of the distortions in HWE for the four SNPs, we next tested the hypothesis that homozygosity for SNP5, SNP4, SNP2, or SNP1 minor alleles predicts risk of FSGS in EAs. For each SNP, we tested the recessive genetic model, comparing homozygotes for the minor allele with heterozygotes and homozygotes for the major allele in patients and controls. The strongest associations were observed for the proxy SNP4 and SNP5 (OR = 2.6, 2.2, P = 0.001 and 0.008), with weaker associations observed for SNP1 and SNP2 (Table 4). Since risk factors for HIV were self-reported and there remains a possibility that some individuals within the HIV-1-uninfected control group might actually be HIV-1-infected, we reanalyzed the data excluding the 36 FSGS patients who were not tested for HIV-1 sero status. The results of these analyses corroborated the results described above (data not shown).
Six of the eight SNPs were in strong LD, and these were used to infer haplotypes (Fig. 1B). The six most frequent (>5%) haplotypes and their frequencies are arranged in descending order of frequency in Table 5. Also, in Table 5, using a recessive genetic model comparing homozygotes for each haplotype with all others, we show the frequencies of the three haplotype combinations and the ORs. Haplotype H2 homozygotes were more frequent in the FSGS patients than controls (8 of 130 vs. 3 of 259) and were significantly associated with FSGS (OR = 5.6, 95% CI = 1.5–21.5, P = 0.008). This haplotype contained three of the four SNP alleles that were associated with FSGS patients (Table 5). In the eight patients with the H2/H2 genotype, onset age ranged from 14 to 59 yr, with a median of 40 yr. The ages of the three controls were 47, 54, and 61 yr. The renal biopsies showed primary FSGS without unusual features, and electron microscopy did not demonstrate unusual features of mitochondrial structure. No significant differences in haplotype frequencies were seen in AAs (Table 5).
Table 5.
PDSS2 haplotype frequencies for AAs and EAs and associations in EAs for the six most common haplotypes (frequency >5%) in FSGS patients and controls
| SNP |
Haplotype Frequency |
EA Haplotype Distribution |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | Group | AA | EA | Number (frequency) |
OR | 95% CI | P value | |||
| Haplotype 1 | T | G | A | G | A | C | Hx/Hx | Hx/H1 | H1/H1 | ||||||
| FSGS | 0.08 | 0.31 | 65 (0.500) | 48 (0.369) | 17 (0.131) | 1.7 | 0.9–3.4 | 0.15 | |||||||
| Control | 0.07 | 0.31 | 117 (0.452) | 121 (0.467) | 21 (0.081) | ||||||||||
| Haplotype 2 | T | T | G | A | G | T | Hx/Hx | Hx/H2 | H2/H2 | ||||||
| FSGS | 0.25 | 0.16 | 96 (0.738) | 26 (0.200) | 8 (0.061) | 5.6 | 1.5–21.4 | 0.008 | |||||||
| Control | 0.26 | 0.13 | 191 (0.737) | 65 (0.251) | 3 (0.012) | ||||||||||
| Haplotype 3 | T | G | G | G | A | T | Hx/Hx | Hx/H3 | H3/H3 | ||||||
| FSGS | 0.03 | 0.11 | 103 (0.792) | 26 (0.200) | 1 (0.077) | 0.7 | 0.1–6.4 | 1 | |||||||
| Control | 0.02 | 0.13 | 195 (0.753) | 61 (0.235) | 3 (0.012) | ||||||||||
| Haplotype 4 | A | T | G | G | A | T | Hx/Hx | Hx/H4 | H4/H4 | ||||||
| FSGS | 0.08 | 0.11 | 130 (0.792) | 26 (0.200) | 1 (0.077) | 0.5 | 0.1–1.5 | 0.67 | |||||||
| Control | 0.09 | 0.12 | 199 (0.768) | 56 (0.216) | 4 (0.015) | ||||||||||
| Haplotype 5 | A | T | G | A | G | T | Hx/Hx | Hx/H5 | H5/H5 | ||||||
| FSGS | 0.30 | 0.14 | 98 (0.754) | 27 (0.201) | 5 (0.038) | 2 | 0.6–7.1 | 0.31 | |||||||
| Control | 0.29 | 0.09 | 218 (0.842) | 36 (0.139) | 5 (0.019) | ||||||||||
| Haplotype 6 | A | T | A | A | G | T | Hx/Hx | Hx/H6 | H6/H6 | ||||||
| FSGS | 0.03 | 0.08 | 108 (0.831) | 22 (0.169) | 0 | 0.31 | |||||||||
| Control | 0.04 | 0.10 | 213 (0.822) | 42 (0.162) | 4 (0.015) | ||||||||||
Recessive model compares homozygotes for the haplotype with all others. Hx represents all other haplotypes. n = 130 FSGS patients and 259 controls.
Measurement of Q10 in LCLs.
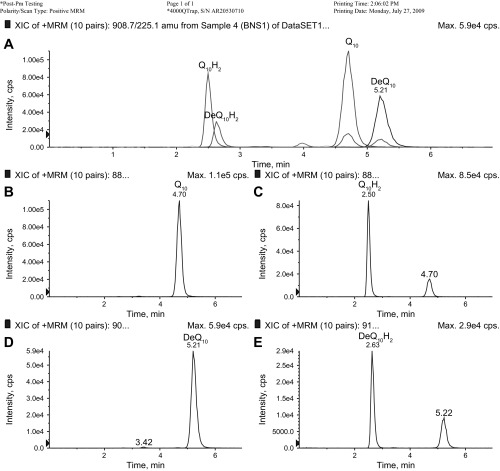
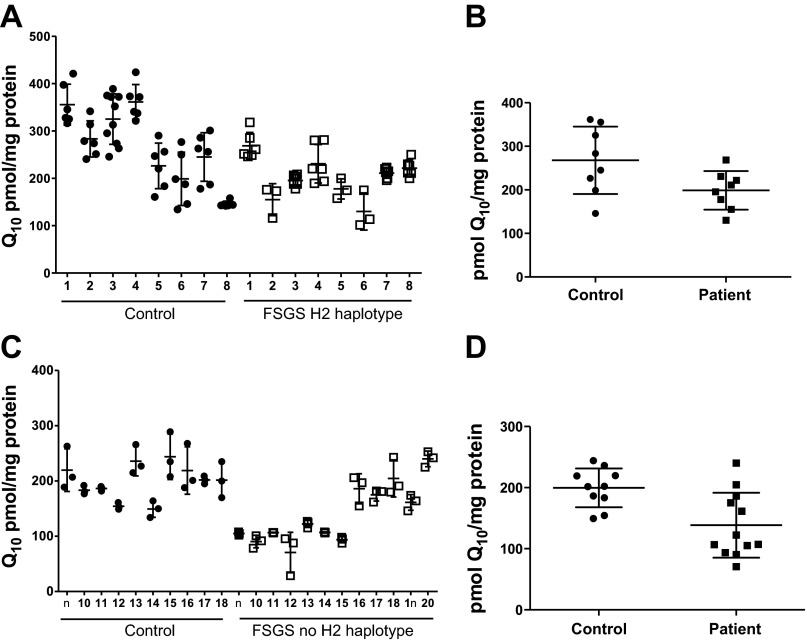
We utilized HPLC-MS/MS and MRM to detect and quantify Q10 in lipid extracts of LCLs (Fig. 2). Diethoxy-Q10 was used as an internal standard in lipid extracts of samples and standards, and a typical standard curve is shown in Fig. 3. As PDSS2 affects the synthesis of coenzyme Q, we examined the Q10 content of LCLs that were derived from the FSGS patients with the H2/H2 homozygous genotype and control individuals. The content of Q10 was significantly decreased in the cell lines of the H2/H2 homozygotes (198.8 ± 44.2 pmol Q10/mg protein) compared with controls analyzed at the same time (267.7 ± 77.3 pmol Q10/mg protein; Fig. 4, A and B). Cell lines from FSGS patients without the H2/H2 haplotype might be expected to have normal Q10 content. However, the content of Q10 was also significantly decreased in the FSGS patient non-H2 haplotype cell lines (138.6 ± 53.2 pmol Q10/mg protein) compared with control LCLs analyzed at the same time (199.6 ± 31.8 pmol Q10/mg protein; Fig. 4, C and D). FSGS patient-derived LCLs manifest a significant Q10 deficiency when analyzed as one group (n = 20, 162.7 ± 57.2 pmol Q10/mg protein) compared with controls (n = 18, 229.9 ± 64.9 pmol Q10/mg protein, P = 0.0017).
Fig. 2.
Sample chromatogram showing detection of reduced and oxidized coenzyme Q10 (Q10) and diethoxy-Q10 (DeQ10). Lipid extract from a lymphoblastoid cell line (LCL) pellet was subjected to HPLC-tandem mass spectrometry (MS/MS) multiple-reaction monitoring. A: precursor-to-product transitions for ammonium adducts of reduced and oxidized Q10 and DeQ10. x-Axis, retention time (min); y-axis, arbitrary units of counts per second (cps). Scale is the same for overlaid traces within A. B–E: individual transitions. B: Q10 with ammonium adduct (880.7/197.0). C: Q10H2 with ammonium adduct (882.7/197.0). D: DeQ10 with ammonium adduct (908.7/225.1). E: DeQ10H2 with ammonium adduct (910.7/225.1). The sum of peak areas corresponding to Q10 and Q10H2 was divided by the sum of peak areas corresponding to DeQ10 and DeQ10H2, and the ratio was used to derive total Q10 content from a standard curve (see Fig. 3).
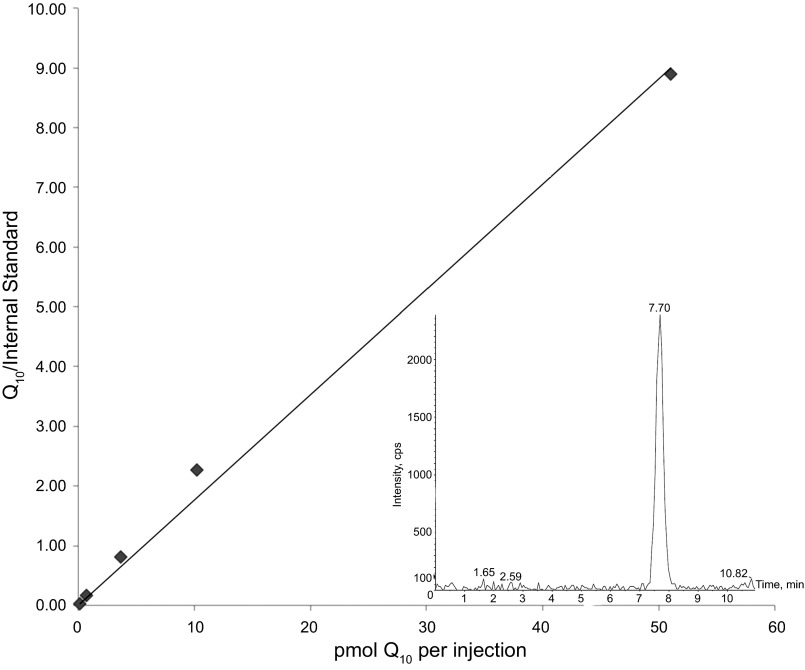
Fig. 3.
Determination of Q10 content with a standard curve. x-Axis, amount of coenzyme Q10 contained in each injection (10 μl); y-axis, peak area of Q10 divided by peak area of the internal standard (DeQ10). A line forced through the origin was plotted to generate the standard curve (slope = 0.176, r2 = 0.995). Inset: a representative peak for the lowest amount of the Q10 standard injected for preparation of the standard curve (0.15 pmol, signal-to-noise ratio = 20:1).
Fig. 4.
Q10 content in LCLs from focal segmental glomerulosclerosis (FSGS) patients is decreased relative to that of controls. A: total Q10 content for each LCL derived from control subjects or patients. FSGS patients are the 8 individuals who were homozygous for the H2/H2 haplotype. Q10 content is presented as a scatter plot, with average ± SD denoted by 3 horizontal lines (n = 6, except n = 10 for control sample 3 and n = 3 for FSGS samples 2, 5, and 6). B: Q10 content of FSGS patients' cell lines (n = 8, 198.8 ± 44.2 pmol/mg protein) is significantly decreased compared with Q10 content of control cell lines (n = 8, 267.7 ± 77.3 pmol/mg protein). Each of the average values from A is represented as a scatter plot, with average ± SD denoted by 3 horizontal lines. P < 0.05 was determined by Student's 2-tailed t-test. C: total Q10 content for each LCL derived from 10 additional control subjects and 12 FSGS patients. FSGS patients did not have the H2/H2 genotype. Q10 content is presented as described in A (n = 3). D: Q10 content of FSGS patients' cell lines (n = 10, 138.6 ± 53.2 pmol/mg protein) is significantly decreased compared with Q10 content of control cell lines (n = 12, 199.6 ± 31.8 pmol/mg protein). Each of the average values from C is represented in a scatter plot, with average ± SD denoted by 3 horizontal lines. P < 0.005 was determined by Student's 2-tailed t-test.
DISCUSSION
This project began as an attempt to determine whether a genetic locus associated with FSGS in mice would also be associated with the corresponding disease in humans. We demonstrated that homozygous genotypes for variant alleles for the PDSS2 gene were indeed more common in FSGS patients than controls and that a single haplotype containing three of these SNPs was more common in EA, but not AA, FSGS patients, suggesting that PDSS2 may be an FSGS susceptibility gene. Completely unexpected, however, was our observation that the Q10 content of LCLs from FSGS patients was significantly lower than that of LCLs from ethnically matched controls.
With regard to the mode of inheritance, the recessive model is consistent with observations in kd/kd mice. None of the results for individual SNPs (OR = 1.7–2.6) were as predictive as haplotype 2 (OR = 5.6), comprising all six SNPs. It should be noted that the SNP associations are not independent, since SNP4 and SNP5 track each other, and the SNP1 A allele always occurs on a haplotype also carrying the SNP2 A allele. The results for SNP5 and haplotype 2 remain significant after correction for multiple comparisons (P = 0.008 and 0.048, respectively) suggesting that these associations are not due to type 1 error. These data suggest that polymorphism in PDSS2 may contribute to increased risk of disease and that people of European ancestry homozygous for haplotype 2 may be at increased risk for developing FSGS. The frequency of haplotype 2 homozygotes was 6.1% among EA FSGS patients and 1.2% in the control EA population (Table 5). The lack of association in AAs could have occurred because an independent effect of PDSS2 is masked by APOL1 renal risk alleles, which explain a large fraction of FSGS in the AA population (25). Alternatively, the causal variants tracked by these SNPs in EAs may be less frequent or on different haplotypes in AAs.
Mutations in the mitochondrial genome that are associated with tubulointerstitial nephritis (53, 58, 67), steroid-resistant FSGS (49), and MELAS (mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes) syndrome have been reported (18, 31). Some patients with MELAS syndrome also have renal lesions such as FSGS or tubulointerstitial nephropathy (18), and other families predominantly express diabetes and deafness (8, 59). The FSGS patients described in this report are also associated with a defective mitochondrial function, but one that is encoded by a nuclear gene. Our findings support the hypothesis recently proposed by Yan and colleagues (65) that emphasizes the importance of energy metabolism for normal podocyte function.
The biochemical function of the gene product suggests that levels of Q10 will be affected (45), and this is supported by the data shown in Fig. 4. LCLs from FSGS patients who were homozygous for the H2 haplotype had significantly less Q10 than control LCLs (Fig. 4B). When 12 additional LCLs from FSGS patients were compared with 10 additional controls, the patients' cell lines again had significantly less Q10 than the control cells (Fig. 4D). This finding was unexpected and suggests that FSGS patients on average have lower Q10 levels than controls, although 12 of the 20 FSGS cell lines examined had an average Q10 content within the normal range reported for human white blood cells (159–369 pmol/mg protein) (19). As described in materials and methods, we have restricted our comparisons of Q10 content to samples that were analyzed at the same time. Nevertheless, a significant Q10 deficiency was manifested when FSGS patient-derived LCLs were compared as one group (n = 20, 162.7 ± 57.2 pmol Q10/mg protein) with the pooled controls (n = 18, 229.9 ± 64.9 pmol Q10/mg protein, P = 0.0017).
It may seem counterintuitive that valid information about Q10 content relevant to FSGS could be obtained from LCLs. There are several interpretations regarding the finding of decreased Q10 content in all cases of FSGS. One possible interpretation is that Q10 content is decreased in all cases of FSGS nonspecifically. Perhaps the azotemia in FSGS patients leads to modifications of protein function or gene expression, including epigenetic modifications with the potential to persist through the process of immortalization. If this were the case, the Q10 content might be decreased in patients with chronic kidney disease of other causes. In this scenario, the Q10 content would be closer to control values in FSGS patients with very early-onset disease than in those with late-stage disease. A second interpretation is that decreased Q10 content may be a common pathway of predisposition to FSGS. Previous studies have established the precedence of LCLs for the genetic analysis of gene expression. Cheung and colleagues (6) used cDNA microarrays to measure gene expression among monozygotic twins, siblings, and unrelated individuals and found evidence for a genetic contribution to the polymorphic variation in the level of gene expression. More recently, polymorphic trans-regulators were identified by RNA-sequencing analyses of human B cell lines from individuals from large families (7). Ding and colleagues (10) developed a statistical method for measuring the overlap between skin cells and lymphoblastoid cells in levels of gene expression and found that ∼70% of the quantitative trait loci detected in lymphoblastoid cells were also observed in skin. Our findings are consistent with the idea that Q10 content in cells reflects not only expression of ≥11 genes required for de novo synthesis, but also the assembly of a mitochondrial multisubunit protein-lipid complex known to be modulated by phosphorylation (54, 56, 64). Such multisubunit complexes may be particularly sensitive to polymorphisms. Indeed, patients and diploid yeast with only one copy of the COQ4 gene show haploinsufficiency of coenzyme Q content (48).
At least 11 genes are known to be involved in the biosynthesis of coenzyme Q in yeast (5, 23, 32, 39, 56). Patients with deficiencies in Q10 have been identified with defects in homologs of at least six yeast genes (22, 43). The first to be associated with a human disease was infantile encephalomyopathy and FSGS, which occurred in two patients with Q10 deficiency caused by a mutation in COQ2 (42). The second report was that of Lopez et al. (28), which described severe encephalomyopathy with nephrotic syndrome in a child with mutations in the PDSS2 gene. Subsequent studies identified mutations in PDSS1 and COQ9 in patients with infantile-onset multisystem disorder (11, 34). More recently, mutations in the COQ6 gene were associated with nephrotic syndrome and sensorineural deafness (22). Our results suggest that a substantial number of FSGS patients have less Q10, which could be caused by any one of a number of genes, and this deficiency can be detected in patient-derived LCLs. If it were possible to measure the Q10 content of the podocytes, the data might be even more informative.
Q10 deficiency has been identified in many other patients with neurological, muscular, and/or renal defects in whom the responsible genes have not been identified, but many of these conditions have proven to be responsive to Q10 supplementation (17, 27, 35, 43, 47, 60). This is encouraging with regard to the possibility of treating some FSGS patients with oral supplementation of Q10, but there are tissue differences with regard to uptake of Q10 from the diet (57), so it remains to be determined whether this approach will be beneficial. In at least one patient, oral Q10 improved the neurological symptoms, but not the renal dysfunction. This could have been because renal damage had already occurred before therapy was initiated, which demonstrates the importance of early diagnosis (57). Thus, PDSS2 joins the list of mitochondrial genes, present in the nuclear or mitochondrial genome, in which mutations or tagging SNPs are associated with FSGS. Podocytes, the target cell in FSGS, are particularly reliant on mitochondrial function for energy generation (1), and thus podocytes may be particularly susceptible to genetic disorders leading to mitochondrial dysfunction.
An association between the PDSS2 gene and FSGS could be demonstrated in EAs but not AAs in this study. The causal variation in or near the PDSS2 gene responsible for FSGS has not been identified. The data suggest that such an allele is in significant LD with one or more of the SNP alleles that have been tested and likely to occur on haplotype 2 in EAs, but extensive sequencing studies may be required before the causal allele or alleles associated with FSGS can be identified precisely. We also have no mechanistic information about how allelic differences in the PDSS2 gene could lead to FSGS. However, the intronic location of the most closely associated SNPs is consistent with previous genome-wide association studies. In the majority (∼93%) of such studies, the disease-associated variant lies within a noncoding sequence (33).
Furthermore, it is not known to what extent Q10 levels might affect disease susceptibility. Although a severely defective allele could possibly behave as a Mendelian factor in the transmission of FSGS, alleles with more moderate effects may only act as contributing factors and only slightly increase the risk of FSGS within the context of other genetic and environmental factors. These findings suggest that PDSS2 variation and Q10 levels may affect respiratory capacity; this pathway warrants further study for its potential role in podocyte injury leading to FSGS and related syndromes. As with any genetic study, the results of this investigation require independent confirmation.
If it is indeed correct that many cases of FSGS involve Q10 deficiencies, animal models of these conditions will be useful for studying possible therapies. The kd/kd mouse phenotype can be rescued to some extent by Q10 supplementation (44) and to an even greater extent by probucol, which has the effect of increasing endogenous Q9 content (12). Diabetic nephropathy in db/db mice can also be significantly ameliorated by Q10 supplementation (50). It is also well established that mitochondrial deficits are associated with several animal models of hypoxia and acute kidney injury (13–15, 62). Our results suggest that further study of coenzyme Q deficiency in FSGS and its possible treatment may have significant therapeutic benefits.
GRANTS
This project has been funded in whole or in part with federal funds from the National Cancer Institute under Contract HHSN26120080001E. Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and National Institute of Diabetes and Digestive and Kidney Diseases Intramural Research Programs and was supported by NIH Grants R01 DK-55852 (D. L. Gasser), R01 GM-45952 (C. F. Clarke), and National Cancer Institute Division of Cancer Treatment and Diagnosis under contract N01 CO-12400. L. X. Xie was supported by the Ruth L. Kirschstein National Service Award GM-007185. The HPLC-MS/MS determination of quinones was supported in part by National Center for Research Resources Grant S10 RR-024605. Recruitment of controls through the ALIVE study was supported by National Institute on Drug Abuse Grant R01-DA-04334.
DISCLAIMER
The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
D.L.G., C.A.W., L.X.X., C.F.C., and J.B.K. are responsible for conception and design of the research; D.L.G., C.A.W., M.P., P.A., L.A.M., G.D.K., Y.S., L.X.X., B.N.M., C.F.C., and J.B.K. analyzed the data; D.L.G., C.A.W., M.P., P.A., L.A.M., G.D.K., L.X.X., B.N.M., C.F.C., and J.B.K. interpreted the results of the experiments; D.L.G. drafted the manuscript; D.L.G., C.A.W., C.F.C., and J.B.K. edited and revised the manuscript; D.L.G., C.A.W., M.P., P.A., L.A.M., G.D.K., Y.S., L.X.X., B.N.M., C.F.C., and J.B.K. approved the final version of the manuscript; C.A.W., P.A., Y.S., L.X.X., and C.F.C. prepared the figures; M.P., P.A., L.A.M., Y.S., L.X.X., B.N.M., and C.F.C. performed the experiments.
ACKNOWLEDGMENTS
We thank Dr. Kathy Ewens, Carl McIntosh, and Randall Johnson for valuable help and advice. We are extremely grateful for the contributions of the following clinical collaborators: Drs. Tejinder S. Ahuja (University of Texas Medical Branch), Diego Aviles (Louisiana State University), Jeffrey S. Berns (University of Pennsylvania), William A. Briggs (Johns Hopkins University School of Medicine), Stephen M. Korbet (Rush University Medical Center), Richard A. Dart (Marshfield Clinic), Dollie Green (University of Miami), Florence Hutchison (University of South Carolina), Paul L. Kimmel (George Washington University Medical Center), Oliver Lenz (University of Miami), Roslyn Mannon (Duke University Medical Center), Donna Michel (West Virgina University), Michele Mokrzycki (Albert Einstein College of Medicine), Patrick Nachman (University of North Carolina), T. K. S. Rao (State University of New York), Elizabeth Ripley (Medical College of Virginia), Jeffrey R. Schelling (Case Western Reserve University), Eric E. Simon (Tulane University School of Medicine), Michael C. Smith (University Hospitals, Cleveland), and Howard Trachtman (Schneider Children's Hospital).
REFERENCES
- 1.Abe Y, Sakairi T, Kajiyama H, Shrivastav S, Beeson C, Kopp JB. Bioenergetic characterization of mouse podocytes. Am J Physiol Cell Physiol 299: C464–C476, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barisoni L, Madaio MP, Eraso M, Gasser DL, Nelson PJ. The kd/kd mouse is a model of collapsing glomerulopathy. J Am Soc Nephrol 16: 2847–2851, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barisoni L, Schnaper HW, Kopp JB. A proposed taxonomy for the podocytopathies: a reassessment of the primary nephrotic diseases. Clin J Am Soc Nephrol 2: 529–542, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Belogrudov GI, Lee PT, Jonassen T, Hsu AY, Gin P, Clarke CF. Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. Arch Biochem Biophys 392: 48–58, 2001 [DOI] [PubMed] [Google Scholar]
- 6.Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet 33: 422–425, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Cheung VG, Nayak RR, Wang IX, Elwyn S, Cousins SM, Morley M, Spielman RS. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biol 8: 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinnery PF, Turnbull DM. Mitochondrial DNA and disease. Lancet 354 Suppl 1: SI17–SI21, 1999 [DOI] [PubMed] [Google Scholar]
- 9.Dell KM, Li YX, Peng M, Neilson EG, Gasser DL. Localization of the mouse kidney disease (kd) gene to a YAC/BAC contig on chromosome 10. Mamm Genome 11: 967–971, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, Chen W, Weichenthal M, Ellinghaus E, Franke A, Cookson W, Nair RP, Elder JT, Abecasis GR. Gene expression in skin and lymphoblastoid cells: refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet 87: 779–789, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan AJ, Bitner-Glindzicz M, Meunier B, Costello H, Hargreaves IP, Lopez LC, Hirano M, Quinzii CM, Sadowski MI, Hardy J, Singleton A, Clayton PT, Rahman S. A nonsense mutation in COQ9 causes autosomal-recessive neonatal-onset primary coenzyme Q10 deficiency: a potentially treatable form of mitochondrial disease. Am J Hum Genet 84: 558–566, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falk MJ, Polyak E, Zhang Z, Peng M, King R, Maltzman JS, Okwuego E, Horyn O, Nakamaru-Ogiso E, Ostrovsky J, Xie LX, Chen JY, Marbois B, Nissim I, Clarke CF, Gasser DL. Probucol ameliorates renal and metabolic sequelae of primary CoQ deficiency in Pdss2 mutant mice. EMBO Mol Med 3: 410–427, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldkamp T, Kribben A, Roeser NF, Senter RA, Kemner S, Venkatachalam MA, Nissim I, Weinberg JM. Preservation of complex I function during hypoxia-reoxygenation-induced mitochondrial injury in proximal tubules. Am J Physiol Renal Physiol 286: F749–F759, 2004 [DOI] [PubMed] [Google Scholar]
- 14.Feldkamp T, Kribben A, Weinberg JM. Assessment of mitochondrial membrane potential in proximal tubules after hypoxia-reoxygenation. Am J Physiol Renal Physiol 288: F1092–F1102, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Funk JA, Schnellmann RG. Persistent disruption of mitochondrial homeostasis after acute kidney injury. Am J Physiol Renal Physiol 302: F853–F864, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gironi M, Lamperti C, Nemni R, Moggio M, Comi G, Guerini FR, Ferrante P, Canal N, Naini A, Bresolin N, DiMauro S. Late-onset cerebellar ataxia with hypogonadism and muscle coenzyme Q10 deficiency. Neurology 62: 818–820, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Guery B, Choukroun G, Noel LH, Clavel P, Rotig A, Lebon S, Rustin P, Bellane-Chantelot C, Mougenot B, Grunfeld JP, Chauveau D. The spectrum of systemic involvement in adults presenting with renal lesion and mitochondrial tRNA(Leu) gene mutation. J Am Soc Nephrol 14: 2099–2108, 2003 [DOI] [PubMed] [Google Scholar]
- 19.Hahn SH, Kerfoot S, Vasta V. Assay to measure oxidized and reduced forms of CoQ by LC-MS/MS. Methods Mol Biol 837: 169–179, 2012 [DOI] [PubMed] [Google Scholar]
- 20.Hallman TM, Peng M, Meade R, Hancock WW, Madaio MP, Gasser DL. The mitochondrial and kidney disease phenotypes of kd/kd mice under germfree conditions. J Autoimmun 26: 1–6, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock WW, Tsai TL, Madaio MP, Gasser DL. Multiple autoimmune pathways in kd/kd mice. J Immunol 171: 2778–2781, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Heeringa SF, Chernin G, Chaki M, Zhou W, Sloan AJ, Ji Z, Xie LX, Salviati L, Hurd TW, Vega-Warner V, Killen PD, Raphael Y, Ashraf S, Ovunc B, Schoeb DS, McLaughlin HM, Airik R, Vlangos CN, Gbadegesin R, Hinkes B, Saisawat P, Trevisson E, Doimo M, Casarin A, Pertegato V, Giorgi G, Prokisch H, Rotig A, Nurnberg G, Becker C, Wang S, Ozaltin F, Topaloglu R, Bakkaloglu A, Bakkaloglu SA, Muller D, Beissert A, Mir S, Berdeli A, Varpizen S, Zenker M, Matejas V, Santos-Ocana C, Navas P, Kusakabe T, Kispert A, Akman S, Soliman NA, Krick S, Mundel P, Reiser J, Nurnberg P, Clarke CF, Wiggins RC, Faul C, Hildebrandt F. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest 121: 2013–2024, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson A, Gin P, Marbois BN, Hsieh EJ, Wu M, Barros MH, Clarke CF, Tzagoloff A. COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J Biol Chem 280: 31397–31404, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Kitiyakara C, Kopp JB, Eggers P. Trends in the epidemiology of focal segmental glomerulosclerosis. Semin Nephrol 23: 172–182, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, Friedman D, Briggs W, Dart R, Korbet S, Mokrzycki MH, Kimmel PL, Limou S, Ahuja TS, Berns JS, Fryc J, Simon EE, Smith MC, Trachtman H, Michel DM, Schelling JR, Vlahov D, Pollak M, Winkler CA. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 22: 2129–2137, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopp JB, Winkler C. HIV-associated nephropathy in African Americans. Kidney Int Suppl S43–S49, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Lalani SR, Vladutiu GD, Plunkett K, Lotze TE, Adesina AM, Scaglia F. Isolated mitochondrial myopathy associated with muscle coenzyme Q10 deficiency. Arch Neurol 62: 317–320, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Lopez LC, Schuelke M, Quinzii CM, Kanki T, Rodenburg RJ, Naini A, Dimauro S, Hirano M. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am J Hum Genet 79: 1125–1129, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyon MF, Hulse EV. An inherited kidney disease of mice resembling human nephronophthisis. J Med Genet 8: 41–48, 1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madaio MP, Ahima RS, Meade R, Rader DJ, Mendoza A, Peng M, Tomaszewski JE, Hancock WW, Gasser DL. Glomerular and tubular epithelial defects in kd/kd mice lead to progressive renal failure. Am J Nephrol 25: 604–610, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majamaa K, Moilanen JS, Uimonen S, Remes AM, Salmela PI, Karppa M, Majamaa-Voltti KA, Rusanen H, Sorri M, Peuhkurinen KJ, Hassinen IE. Epidemiology of A3243G, the mutation for mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes: prevalence of the mutation in an adult population. Am J Hum Genet 63: 447–454, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marbois B, Gin P, Faull KF, Poon WW, Lee PT, Strahan J, Shepherd JN, Clarke CF. Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J Biol Chem 280: 20231–20238, 2005 [DOI] [PubMed] [Google Scholar]
- 33.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mollet J, Giurgea I, Schlemmer D, Dallner G, Chretien D, Delahodde A, Bacq D, de Lonlay P, Munnich A, Rotig A. Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J Clin Invest 117: 765–772, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musumeci O, Naini A, Slonim AE, Skavin N, Hadjigeorgiou GL, Krawiecki N, Weissman BM, Tsao CY, Mendell JR, Shanske S, De Vivo DC, Hirano M, DiMauro S. Familial cerebellar ataxia with muscle coenzyme Q10 deficiency. Neurology 56: 849–855, 2001 [DOI] [PubMed] [Google Scholar]
- 36.Neilson EG, McCafferty E, Feldman A, Clayman MD, Zakheim B, Korngold R. Spontaneous interstitial nephritis in kdkd mice. I. An experimental model of autoimmune renal disease. J Immunol 133: 2560–2565, 1984 [PubMed] [Google Scholar]
- 37.Neitzel H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet 73: 320–326, 1986 [DOI] [PubMed] [Google Scholar]
- 38.Orloff MS, Iyengar SK, Winkler CA, Goddard KA, Dart RA, Ahuja TS, Mokrzycki M, Briggs WA, Korbet SM, Kimmel PL, Simon EE, Trachtman H, Vlahov D, Michel DM, Berns JS, Smith MC, Schelling JR, Sedor JR, Kopp JB. Variants in the Wilms' tumor gene are associated with focal segmental glomerulosclerosis in the African American population. Physiol Genomics 21: 212–221, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Ozeir M, Muhlenhoff U, Webert H, Lill R, Fontecave M, Pierrel F. Coenzyme Q biosynthesis: Coq6 is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem Biol 18: 1134–1142, 2011 [DOI] [PubMed] [Google Scholar]
- 40.Peng M, Jarett L, Meade R, Madaio MP, Hancock WW, George AL, Jr, Neilson EG, Gasser DL. Mutant prenyltransferase-like mitochondrial protein (PLMP) and mitochondrial abnormalities in kd/kd mice. Kidney Int 66: 20–28, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollak MR. Inherited podocytopathies: FSGS and nephrotic syndrome from a genetic viewpoint. J Am Soc Nephrol 13: 3016–3023, 2002 [DOI] [PubMed] [Google Scholar]
- 42.Quinzii C, Naini A, Salviati L, Trevisson E, Navas P, Dimauro S, Hirano M. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am J Hum Genet 78: 345–349, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahman S, Clarke CF, Hirano M. Diagnosis and treatment of coenzyme Q10 deficiency. Neuromuscul Disord 23: 506–515, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saiki R, Lunceford AL, Shi Y, Marbois B, King R, Pachuski J, Kawamukai M, Gasser DL, Clarke CF. Coenzyme Q10 supplementation rescues renal disease in Pdss2kd/kd mice with mutations in prenyl diphosphate synthase subunit 2. Am J Physiol Renal Physiol 295: F1535–F1544, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saiki R, Nagata A, Kainou T, Matsuda H, Kawamukai M. Characterization of solanesyl and decaprenyl diphosphate synthases in mice and humans. FEBS J 272: 5606–5622, 2005 [DOI] [PubMed] [Google Scholar]
- 46.Saiki R, Nagata A, Uchida N, Kainou T, Matsuda H, Kawamukai M. Fission yeast decaprenyl diphosphate synthase consists of Dps1 and the newly characterized Dlp1 protein in a novel heterotetrameric structure. Eur J Biochem 270: 4113–4121, 2003 [DOI] [PubMed] [Google Scholar]
- 47.Salviati L, Sacconi S, Murer L, Zacchello G, Franceschini L, Laverda AM, Basso G, Quinzii C, Angelini C, Hirano M, Naini AB, Navas P, DiMauro S, Montini G. Infantile encephalomyopathy and nephropathy with CoQ10 deficiency: a CoQ10-responsive condition. Neurology 65: 606–608, 2005 [DOI] [PubMed] [Google Scholar]
- 48.Salviati L, Trevisson E, Rodriguez Hernandez MA, Casarin A, Pertegato V, Doimo M, Cassina M, Agosto C, Desbats MA, Sartori G, Sacconi S, Memo L, Zuffardi O, Artuch R, Quinzii C, Dimauro S, Hirano M, Santos-Ocana C, Navas P. Haploinsufficiency of COQ4 causes coenzyme Q10 deficiency. J Med Genet 49: 187–191, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scaglia F, Vogel H, Hawkins EP, Vladutiu GD, Liu LL, Wong LJ. Novel homoplasmic mutation in the mitochondrial tRNATyr gene associated with atypical mitochondrial cytopathy presenting with focal segmental glomerulosclerosis. Am J Med Genet A 123A: 172–178, 2003 [DOI] [PubMed] [Google Scholar]
- 50.Sourris KC, Harcourt BE, Tang PH, Morley AL, Huynh K, Penfold SA, Coughlan MT, Cooper ME, Nguyen TV, Ritchie RH, Forbes JM. Ubiquinone (coenzyme Q10) prevents renal mitochondrial dysfunction in an experimental model of type 2 diabetes. Free Radic Biol Med 52: 716–723, 2012 [DOI] [PubMed] [Google Scholar]
- 51.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73: 1162–1169, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68: 978–989, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szabolcs MJ, Seigle R, Shanske S, Bonilla E, DiMauro S, D'Agati V. Mitochondrial DNA deletion: a cause of chronic tubulointerstitial nephropathy. Kidney Int 45: 1388–1396, 1994 [DOI] [PubMed] [Google Scholar]
- 54.Tauche A, Krause-Buchholz U, Rodel G. Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res 8: 1263–1275, 2008 [DOI] [PubMed] [Google Scholar]
- 55.Thomas DB, Franceschini N, Hogan SL, Ten Holder S, Jennette CE, Falk RJ, Jennette JC. Clinical and pathologic characteristics of focal segmental glomerulosclerosis pathologic variants. Kidney Int 69: 920–926, 2006 [DOI] [PubMed] [Google Scholar]
- 56.Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7 Suppl: S62–S71, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim Biophys Acta 1660: 171–199, 2004 [DOI] [PubMed] [Google Scholar]
- 58.Tzen CY, Tsai JD, Wu TY, Chen BF, Chen ML, Lin SP, Chen SC. Tubulointerstitial nephritis associated with a novel mitochondrial point mutation. Kidney Int 59: 846–854, 2001 [DOI] [PubMed] [Google Scholar]
- 59.van den Ouweland JM, Lemkes HH, Ruitenbeek W, Sandkuijl LA, de Vijlder MF, Struyvenberg PA, van de Kamp JJ, Maassen JA. Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat Genet 1: 368–371, 1992 [DOI] [PubMed] [Google Scholar]
- 60.Van Maldergem L, Trijbels F, DiMauro S, Sindelar PJ, Musumeci O, Janssen A, Delberghe X, Martin JJ, Gillerot Y. Coenzyme Q-responsive Leigh's encephalopathy in two sisters. Ann Neurol 52: 750–754, 2002 [DOI] [PubMed] [Google Scholar]
- 61.Vlahov D, Graham N, Hoover D, Flynn C, Bartlett JG, Margolick JB, Lyles CM, Nelson KE, Smith D, Holmberg S, Farzadegan H. Prognostic indicators for AIDS and infectious disease death in HIV-infected injection drug users: plasma viral load and CD4+ cell count. JAMA 279: 35–40, 1998 [DOI] [PubMed] [Google Scholar]
- 62.Weinberg JM, Venkatachalam MA, Roeser NF, Saikumar P, Dong Z, Senter RA, Nissim I. Anaerobic and aerobic pathways for salvage of proximal tubules from hypoxia-induced mitochondrial injury. Am J Physiol Renal Physiol 279: F927–F943, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winn MP, Conlon PJ, Lynn KL, Farrington MK, Creazzo T, Hawkins AF, Daskalakis N, Kwan SY, Ebersviller S, Burchette JL, Pericak-Vance MA, Howell DN, Vance JM, Rosenberg PB. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science 308: 1801–1804, 2005 [DOI] [PubMed] [Google Scholar]
- 64.Xie LX, Hsieh EJ, Watanabe S, Allan CM, Chen JY, Tran UC, Clarke CF. Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim Biophys Acta 1811: 348–360, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan K, Ito N, Nakajo A, Kurayama R, Fukuhara D, Nishibori Y, Kudo A, Akimoto Y, Takenaka H. The struggle for energy in podocytes leads to nephrotic syndrome. Cell Cycle 11: 1504–1511, 2012 [DOI] [PubMed] [Google Scholar]
- 66.Ziegler CG, Peng M, Falk MJ, Polyak E, Tsika E, Ischiropoulos H, Bakalar D, Blendy JA, Gasser DL. Parkinson's disease-like neuromuscular defects occur in prenyl diphosphate synthase subunit 2 (Pdss2) mutant mice. Mitochondrion 12: 248–257, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zsurka G, Ormos J, Ivanyi B, Turi S, Endreffy E, Magyari M, Sonkodi S, Venetianer P. Mitochondrial mutation as a probable causative factor in familial progressive tubulointerstitial nephritis. Hum Genet 99: 484–487, 1997 [DOI] [PubMed] [Google Scholar]