Abstract
Fluorescence-activated cell sorting (FACS) is an essential tool for studies requiring isolation of distinct intestinal epithelial cell populations. Inconsistent or lack of reporting of the critical parameters associated with FACS methodologies has complicated interpretation, comparison, and reproduction of important findings. To address this problem a comprehensive multicenter study was designed to develop guidelines that limit experimental and data reporting variability and provide a foundation for accurate comparison of data between studies. Common methodologies and data reporting protocols for tissue dissociation, cell yield, cell viability, FACS, and postsort purity were established. Seven centers tested the standardized methods by FACS-isolating a specific crypt-based epithelial population (EpCAM+/CD44+) from murine small intestine. Genetic biomarkers for stem/progenitor (Lgr5 and Atoh 1) and differentiated cell lineages (lysozyme, mucin2, chromogranin A, and sucrase isomaltase) were interrogated in target and control populations to assess intra- and intercenter variability. Wilcoxon's rank sum test on gene expression levels showed limited intracenter variability between biological replicates. Principal component analysis demonstrated significant intercenter reproducibility among four centers. Analysis of data collected by standardized cell isolation methods and data reporting requirements readily identified methodological problems, indicating that standard reporting parameters facilitate post hoc error identification. These results indicate that the complexity of FACS isolation of target intestinal epithelial populations can be highly reproducible between biological replicates and different institutions by adherence to common cell isolation methods and FACS gating strategies. This study can be considered a foundation for continued method development and a starting point for investigators that are developing cell isolation expertise to study physiology and pathophysiology of the intestinal epithelium.
Keywords: intestinal epithelium, intestinal stem cell, FACS, epithelial dissociation
the recent and rapid discoveries made in the intestinal epithelial stem cell (IESC) field have heavily relied on separating target IESC populations by fluorescence-activated cell sorting (FACS). A handful of validated biomarkers (e.g., Lgr5, Sox9, Bmi1, Hopx, mTert) have served as proxies for cellular phenotypes defined by varying degrees of “stemness” (2, 7, 13, 15, 19). Transgenic mice expressing enhanced green fluorescent protein as a function of the regulatory regions of these IESC biomarkers have enabled FACS isolation of live cells for downstream applications. This ability to FACS-isolate cell populations from dissociated intestinal epithelium has transformed the IESC field by facilitating detailed mechanistic studies on distinct intestinal cell populations.
Inherent to rapid progress in a field, protocol development for isolating intestinal stem cells continues to evolve, complicating the ability of the field to endorse a single standardized protocol. An emerging problem is that the variability of published tissue dissociation protocols and the lack of data reporting guidelines for FACS-isolated intestinal epithelial cell populations makes it nearly impossible to reproducibly execute, interpret, and accurately compare published findings (7, 16, 17). As new studies focus away from basic IESC biomarker discovery to studies that focus on understanding stem/progenitor cell behavior during homeostasis, injury, and disease, developing common standards for reporting FACS methodology, parameters, and outcomes is necessary and timely.
In response to the challenge of establishing reproducible approaches that facilitate comparison of data across institutions, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH)-funded Intestinal Stem Cell Consortium (ISCC) undertook a comprehensive, multicenter study to evaluate reproducibility of epithelial dissociation from the small intestine, viability of cells pre- and post-FACS isolation, and purity of FACS-isolated target cell populations. The overall goal was to determine whether standardized tissue procurement and manipulation and FACS parameter and data reporting could be reproducibly conducted between ISCC centers. As recommended by Alexander and colleagues (1) in a recent commentary on reducing variability in solid tissue FACS isolation, multiple key parameters must be reported to facilitate interpretation and reproducibility of FACS data. These parameters include 1) antibody binding conditions; 2) FACS instrumentation; 3) display of manual FACS gates; and 4) validation of sorting. To complement these general parameters, this study tests additional parameters that are specific to the intestinal epithelium including 5) dissociation methods to maximize epithelial cell viability, 6) FACS antibodies that enable enrichment of crypt-based epithelial cells, 7) established quality control and functional assays to assess purity of isolated populations, and 8) well-controlled FACS isolation procedures and data reporting. On the basis of the collective expertise of the ISCC, a protocol for intestinal epithelial isolation and guidelines for FACS isolation reporting and validation was established. The caveats for protocols relying on mechanical digesting (yielding cell aggregates) and enzymatic digestion (resulting in differential cleavage of cell surface antigens) was carefully considered for optimal viability of the purified cells. Centers that participated in this study utilized the ISCC standardized methods to FACS-isolate a crypt-based epithelial population (EpCAM+/CD44+) and determine whether there was statistically significant reproducibility in purity of the population within individual centers and between centers.
MATERIALS AND METHODS
Participating laboratories, FACS capabilities.
Eight independent institutions comprising the ISCC participated in intestinal epithelial isolation and FACS isolation comparative analyses. Characteristics of flow cytometers, sorting parameters, and analysis software are presented in Table 1.
Table 1.
Characteristics of flow cytometers, sorting parameters, and analysis software
| Center | Model (Make) | Nozzle Diameter, μm | Sheath Pressure, psi | Sheath Fluid | Flow Rate, events/s | Software |
|---|---|---|---|---|---|---|
| 1 | MoFloXDP (Beckman) | 100 | 29.9 | PBS | 5,000 ± 0 | Summit 5.1 |
| 2 | FACS Vantage SE (BD) | 130 | 10 | PBS | 5,050 ± 1291 | BD Diva 5.0.1 |
| 3 | Aria II (BD) | 100 | 20 | Saline | 3,667 ± 289 | BD Diva 5 |
| 4 | FACS Vantage SE (BD) | 100 | 15 | Coulter Fluid | 6,938 ± 515 | BD Diva 5 |
| 5 | Aria II (BD) | 100 | 20 | PBS | 2,500 ± 0 | BD Diva 5.0.1 |
| 6 | Moflo Legacy (BD) | 150 | 12 | PBS | 5,000 ± 0 | Summit 4.3 |
| 7 | Influx V (BD) | 80 | 16.5 | PBS | 4,133 ± 115 | Spigot 6.3 |
Animals.
Male C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) between 6 and 8 wk old were used for all studies. Mice were housed in facilities approved by the American Association for Accreditation of Laboratory Animal Care and kept on a 12-h light-dark cycle, in a specific pathogen-free environment, fed a standard rodent laboratory chow, and provided water ad libitum. Animal use for this study was approved by each institution's Animal Care and Use Committee.
Logistical considerations: protocol development, training, sample handling.
Detailed protocols, log sheets, and protocol-associated information (Supplemental Movie S1; supplemental material for this article is available online at the Journal website), were prepared to minimize errors and to ensure that sample isolation, collection, and handling were consistent. To control for as many variables as possible, the ISCC established experimental parameters, reporting criteria, and threshold values to determine protocol compliance and subsequent inclusion of data in the analysis. Optimization of antibody saturation and gating strategy was established at Center 1. The standardized protocol was tested for consistency at Centers 1 and 2 prior to the study. An online training session, facilitated by the ISCC Coordinating Center, was conducted with laboratory staff from all eight institutions to clarify critical points and establish consistency in protocol execution. A single operator performed all cell isolations at each center to help ensure intracenter consistency. All centers used antibodies that originated from the same vendor and lot number. Each center received prelabeled vials containing cell lysis buffer for RNA isolation to minimize reagent variation and labeling errors. One center did not participate in FACS analysis because of incompatibility of FACS instrumentation.
All centers were required to meet established experimental thresholds for inclusion in the study. These parameters included 1) ≥75% presort cell viability; 2) ≥90% postsort viability; 3) ≥90% postsort cell purity; and 4) minimum RNA integrity number (RIN) value of 6.5. During FACS isolation, samples were sorted directly into the lysis buffer to minimize RNA degradation, and then FACS-isolated samples were shipped overnight on dry ice and processed at Center 1 and analyzed at Centers 1 and 2. Note that Center 5 submitted two rounds of samples: the first set was analyzed in the intercenter comparison and the second analyzed in the intracenter comparison.
Epithelial isolation and dissociation.
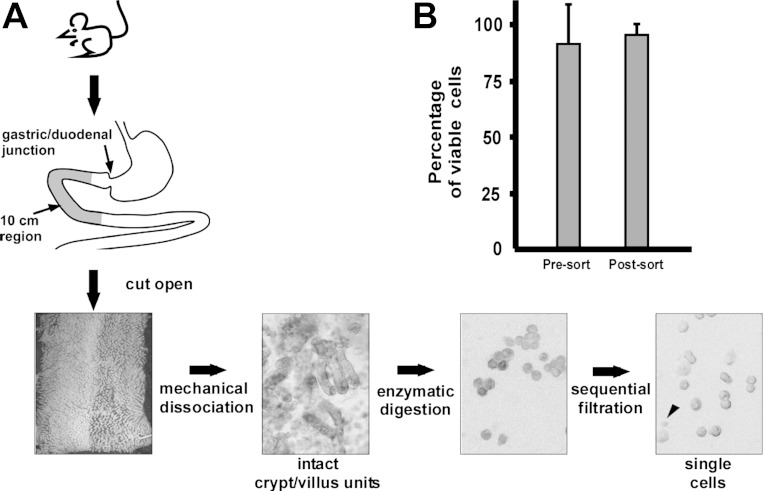
A segment of the proximal intestine representing a 10-cm region from 2 to 12 cm distal to the pyloric sphincter was used for cell isolation (Fig. 1, n = 3 mice). The intestinal segment was flushed with cold PBS, longitudinally cut open and lightly rinsed to remove residual luminal contents. Tissue was incubated in PBS containing 30 mmol/l EDTA and 1.5 mmol/l dithiothreitol on ice for 20 min. Intestinal tissue was transferred to a 15-ml conical tube containing 5 ml of 30 mmol/l EDTA made in PBS, incubated at 37°C for 8 min, then shaken by hand along the tube's long axis. A force of two to three times gravity along the long axis of the tube was used, as measured by the accelerometer in the iPhone (Supplemental Movie S1). Shaking frequency and duration were standardized to 2.5–3 shake cycles per second for 30 s.
Fig. 1.
Epithelial isolation protocol results in reproducibly, high-viability cells. A: combination of mechanical perturbation, enzymatic digestion, and sequential filtration yields single murine intestinal epithelial cells of high viability, reported as means ± SE (B). Arrowhead in the single-cell panel denotes lymphocyte contamination among larger epithelial cells.
After shaking, the remnant muscle layer was removed and the dissociated cells were pelleted at 1,000 g, resuspended in PBS + 10% FBS, and repelleted. The cells were then incubated in 10 ml of modified HBSS containing 0.3 U/ml of dispase (Roche, San Francisco, CA) at 37°C for 10 min with intermittent shaking (3–3.5 cycles per s for 15 s) every 2 min for 15 s to dissociate epithelial sheets to single cells. After 10 min, a 10-μl sample of the cell solution was collected and viewed under light microscopy to monitor dissociation to single cells. If less than 50% of the tissue was single cells, the tissue underwent a second round of 30 s shaking and 2-min incubation at 37°C. To enhance cell viability and decrease cell clumping, 1 ml of FBS and 1,000 U DNase I (Roche) were added to cell suspension, which was sequentially passed through 70-μm then 40-μm filters (BD Falcon, Bedford, MA). Cells were pelleted, then resuspended in 10 ml of HBSS to inactivate the dispase. Cells were repelleted and resuspended at a concentration of ∼2 × 107 cells/ml in IESC media [Advanced DMEM/F12 (GIBCO, Grand Island, NY) supplemented with 1× N2 (Invitrogen), 1× B27 without vitamin A (Invitrogen), 10 mM HEPES (GIBCO), 10 μM Y27632 (Sigma), 100 μg/ml penicillin-streptomycin, and 2 mM l-glutamine]. Cell viability was assessed by Trypan blue (Invitrogen) staining; a 75% cell viability was the threshold to proceed forward.
Antibody staining of isolated cells.
Conjugated antibodies were carefully selected to accommodate laser and detector configurations at each institution and to avoid the need to troubleshoot color compensation for antibody combinations. Approximately 1 × 107 cells were incubated in 1 ml of IESC medium on ice for 30 min with mixing every 10 min with the following antibodies: 10 μl α-CD31-PE/Cy7, cat. no. 102418 Biolegend; 10 μl α-CD45-PE/Cy7, cat. no. 103114 Biolegend; 15 μl α-EpCAM-eFluor 450, cat. no. 48-5791-82 eBioscience; 10 μl α-CD44-APC, cat. no. 103011, Biolegend. A total of 2.5 × 105 cells in 100 μl of IESC media were individually stained with one of the four isotype control antibodies (1 μl rat IgG2a-PE/Cy7, cat. no. 400522 Biolegend; 1 μl rat IgG2b-PE/Cy7, cat. no. 400618, Biolegend; 1.5 μl rat IgG2a-eFluor 450, cat. no. 48-4321-80, eBioscience; 1 μl rat IgG2b-APC, cat. no. 400611 Biolegend), as described above. Cells were then washed with 10 ml of DMEM/F12 containing 0.5% FBS, pelleted, repeated once, then resuspended in 2 ml of IESC media for the experimental samples and 1 ml for the controls. Cells were stored on ice. Ten minutes before sorting, 2 μl of 1 mg/ml propidium iodide solution was added.
FACS isolation of target population.
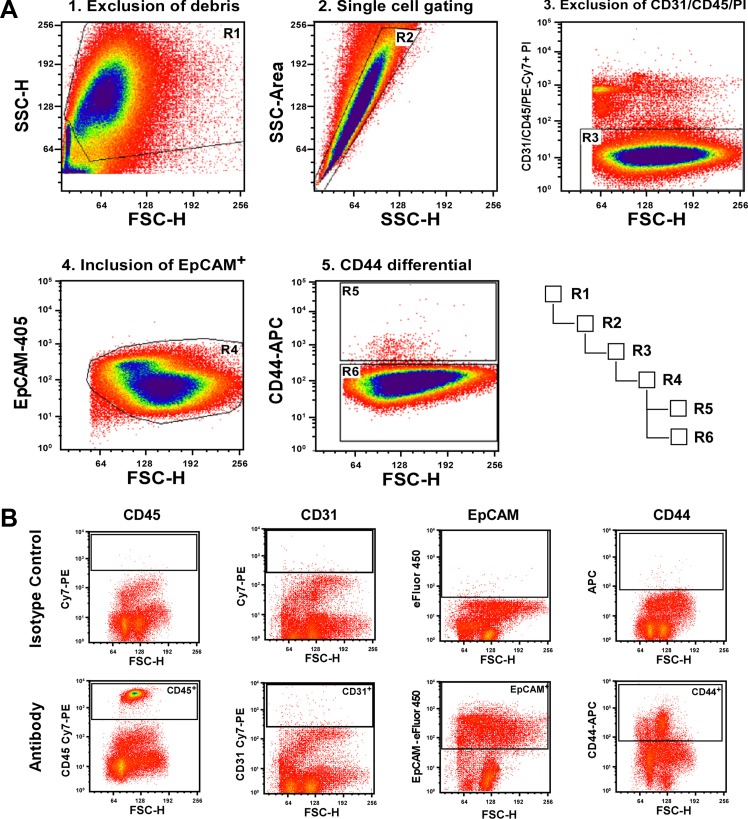
Cells were kept on ice until FACS. Five thousand events were collected from isotype- and antigen-specific stained samples to set the voltages and gates (Fig. 2B). The gating scheme was established by Centers 1 and 2 on the basis of the events depicted in isotype control histograms. Crude live/dead gates were set on the forward scatter (FSC)/side scatter (SSC) histograms, and these cells were then gated for singlets by using SSC height and SSC area. More rigorous dead-cells exclusion was conducted by gate-excluding propidium iodide (PI)-positive cells from the singlet gated cells. CD31 and CD45-positive cells were then excluded from the live PI-negative cells to gate out endothelial cells and lymphocytes, respectively. Note that all cells excluded from the target population were detected on the same channel (PE-Cy7) to minimize the number of instrument detectors required by each center. An additional positive selection for epithelial cells (EpCAM+) was conducted on the PI− CD31− CD45− population to rigorously gate excluding any nonepithelial (EpCAM-negative) populations. From the highly purified EpCAM+ population, CD44+ and CD44− cells were gated and collected. Event files were saved for post hoc analysis. A total of 10,000 cells from three different populations [total epithelium (EpCAM+; gate R4), CD44− (EpCAM+/CD44−; gate R6), and CD44+ (EpCAM+/CD44+; gate R5)] were sorted directly into 500 μl of RNA lysis buffer on ice (RNAqueous Micro Kit, Ambion, Invitrogen; Fig. 2A). Following the sort, collection tubes were vortexed briefly to ensure that all cells had come into contact with the lysis buffer. The lysates were transferred to prelabeled collection tubes, stored at −80°C, and shipped to and processed by Centers 1 and 2 for gene expression analysis. To validate whether the gating scheme isolated the appropriate target populations, 5,000 cells from each population were sorted into 500 μl of 1% paraformaldehyde and fixed for 30 min on ice, washed with PBS containing 1% BSA, and pelleted at 10,000 g for 5 min. Cells were resuspended in 10 μl of PBS with 1% BSA, placed on a glass slide, and assessed for EpCAM and CD44 cell surface staining in 1 μm optical sections using a Zeiss 710 confocal microscope.
Fig. 2.
Gating parameters for fluorescence-activated cell sorting. A series of proposed gates for isolation of a targeted population of intestinal epithelial cells (CD44+) involves exclusion of cellular debris, exclusion of cell doublets, exclusion of nonepithelial cells, inclusion of epithelial cells, gating differential on the target population, and a hierarchical gating relationship (A). B: incubation with isotype control to define the positive cellular population. Gates to identify specific cell populations for inclusion or exclusion (top) were established by staining with isotype control antibodies (bottom). Shown here are all of the antibody and isotype control combinations used for this study.
Reverse transcription and quantitative real-time PCR.
Total RNA was isolated using the RNAqueous Micro Kit (Ambion) according to recommended protocols, except that RNA was eluted using two 12.5-μl elution washes. RNA was DNAse treated directly in the PCR tube at 37°C for 30 min, then DNaseI inactivated prior to RNA quantification by use of a Nanodrop spectrophotometer. A total of 5 ng of RNA was analyzed on a 2100 Agilent Bioanalyzer (Aligent Technologies, Santa Clara, CA) by using the Eukaryotic Pico series II Assay (Agilent). Samples that had RIN values lower than 6.5 were considered degraded and therefore unsuitable for the study (5, 6). cDNA was generated by using the iScript Reverse Transcription Supermix for RT-qPCR cDNA kit (Bio-Rad, Hercules, CA). Quantitative RT-PCR (qRT-PCR) was conducted using SsoFast Probes Supermix with ROX (Bio-Rad) according to manufacturer's protocols. Gene-specific TaqMan probes (18S, internal control: HS99999901_s1; Atoh1, stem/progenitor: Mm00476035_s1; chromogranin A, enteroendocrine: Mm00514341_m1; Lgr5, stem/progenitor: Mm00438890_m1; lysozyme, Paneth: Mm00727183_s1; Mucin2-goblet: Mm00458299_m1; sucrase isomaltase, enterocyte: Mm01210305_m1; Applied Biosystems, Life Technologies) and SYBR probes [CD45, lymphocyte: 5′ CCT GCA GAC ACA GCC TTT CCT (5′ CCC AGA GTG GAT GGT GTA AGA GT)] were used to assess relative gene expression levels when normalized to 18S rRNA. For intracenter comparison each qRT-PCR sample was evaluated in triplicate. For intercenter comparisons, equivalent masses of RNA were pooled from samples represented by n = 2 mice then evaluated in triplicate. The expression of selected genes, relative to 18S rRNA expression, was determined using the 2(−ΔΔCt) method (11, 18). ΔΔCt values were calculated to obtain fold changes for sample comparison.
Comparative analysis.
Data distributions were reported as means ± 1 SD or medians (min, max) for all continuous end points. RNA was preserved individually for all mice in this study. For intracenter comparisons at three centers, variation within each center was determined by comparing the between-mouse vs. between-replicate ΔΔCT SD values in three mice (Centers 1 and 2) or two mice (Center 5b) via a Wilcoxon's rank sum test, in two different sample populations, i.e., CD44− and CD44+. For intercenter comparison across all seven laboratories, RNA from n = 2 mice were pooled and ΔΔCT values determined for all genes, in each sample population, from every center. Next, a principal component (PC) analysis (PCA) was performed using all ΔΔCT values from each pooled sample to evaluate the relative proximity of each center to the population mean via a permutation procedure (10). To evaluate statistical significance of the multicenter PC values, the following three steps were taken: 1) a PCA was repeated in a large number of 1,000+ permuted data sets, where each data set was generated by randomly permuting the ΔΔCT data across all centers within each gene and sample population; 2) the joint effect of all tested genes in both sample populations was quantified by using a summary statistic for the sum of absolute values in the top three PCs weighted by their explained variances; and 3) the statistical significance for the closeness to population mean of each center was evaluated on the basis of the proportion of permutations for which the sum of top three PCs exceeded that of the original data. Statistical significance was indicated if two-sided P < 0.05. Data visualizations were generated in the R programming environment (freely available at http://www.r-project.org/). All other statistical analyses were performed with SAS software, version 9.2 (SAS Institute, Cary, NC).
Functional analysis of CD44+/− population: enteroid forming capacity.
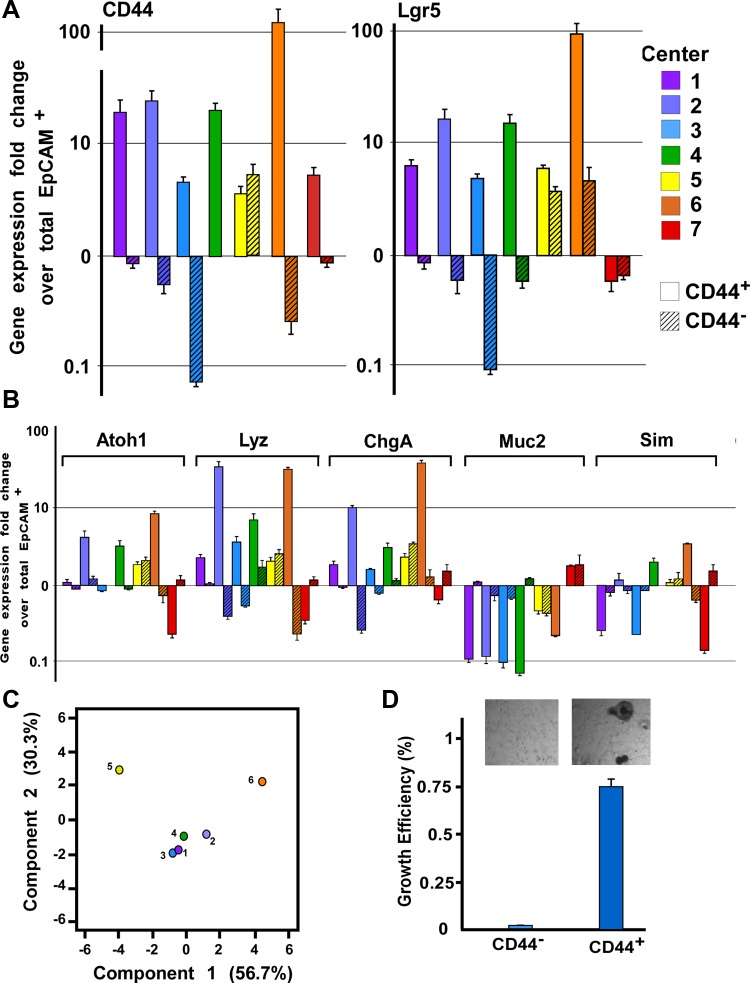
CD44+ cells were collected in compliance with the standardized protocol. A total of 12,000 CD44+ cells were cultured in 50 μl of Matrigel Matrix (BD Biosciences) as previously described (9). Cultures were quantified for enteroid development after 10 days in culture. Enteroid forming capacity assay work was performed by three centers (Fig. 5D).
Fig. 5.
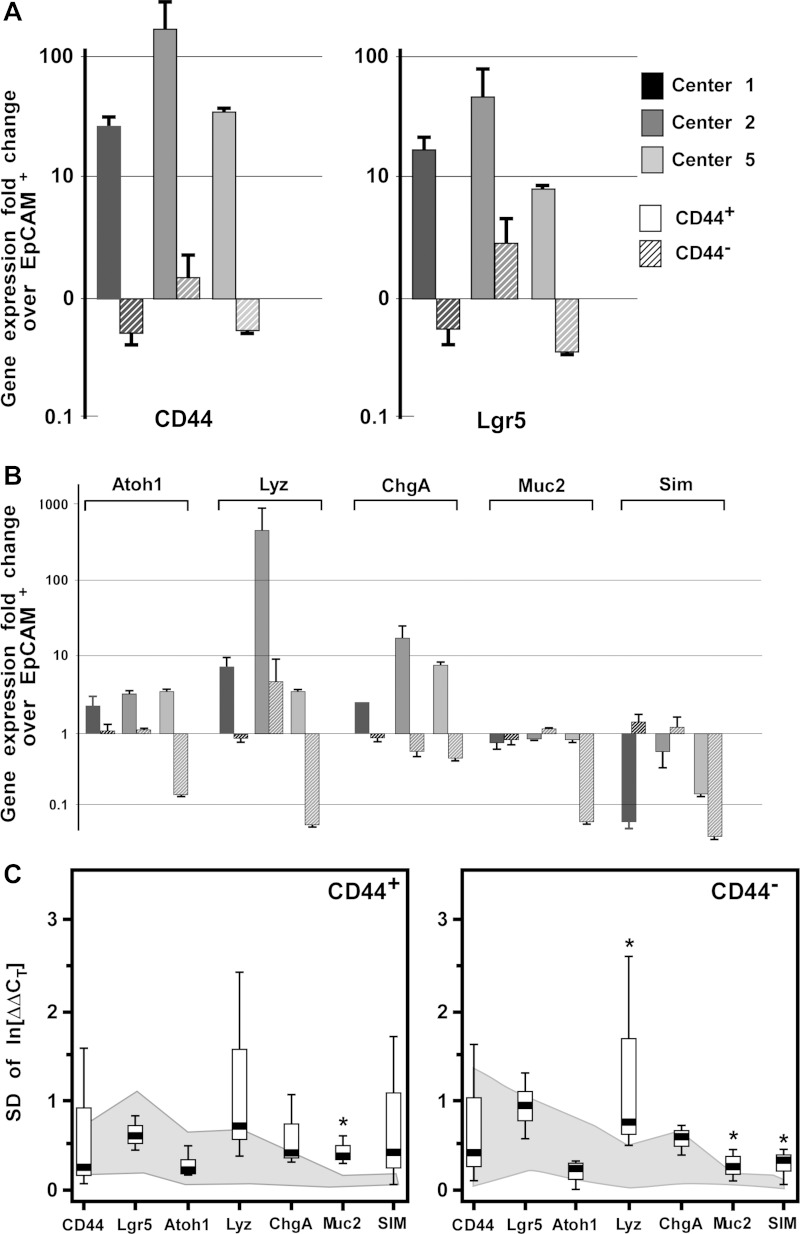
Intergene expression variation within CD44+ and CD44− populations. A: CD44 and Lgr5 gene expression in CD44+ (open bars) and CD44− (hatched bars) cellular populations as determined by quantitative RT-PCR. Each center (1-7) is designated by a different color. B: genes expressed in differentiated subpopulations were evaluated (atonal homology 1: secretory cells; lysozyme: Paneth cells; chromagranin A: enteroendocrine cells; Mucin2: goblet cells; and sucrose isomaltase: enterocytes). As determined by gene expression patterns and principal component analysis (PCA; C), Centers 1–4 display convergent results whereas Centers 5a and 6 do not. Center 7 was not evaluated in the PCA because of contamination with lymphocyte lineages. D: CD44+ and CD44− FACS-isolated cells were evaluated for enteroid growth efficiency.
RESULTS
Standardized intestinal epithelial isolation and dissociation methods results in high cell viability and population purity.
It is well accepted that variability in epithelial isolation approaches can impact cell viability and the ability to isolate comparable epithelial subpopulations (3). Factors such as mouse strain, age, and different cellular composition and gene expression along the proximal-to-distal axis the gastrointestinal tract (15) can increase variability of outcomes confounding data interpretation. The dissociation of contact between the intestinal epithelial monolayer and the underlying mesenchyme followed by disruption of cell-cell contacts (i.e., tight and adherens junctions) also presents a unique challenge to cell isolation and dissociation for FACS. Dissociation methods, which include mechanical force, use of chelating agents, and enzymatic digestion, are especially challenging for isolation of intestinal epithelia, where cell viability and integrity is defined and regulated by physiological association to each other and the supportive basement membrane (4, 12, 14). The significant negative impact that these factors can have on FACS reproducibility and enrichment of target cell populations thereby necessitates rigorous disclosure of dissociation parameters to facilitate comparison and interpretation of reported data.
To minimize sources of variability, standards for protocol training, compliance, and absolute thresholds for experimental progression were established by the ISCC (Table 2). In alignment with suggested reporting parameters (1), the make/model, nozzle size, flow rate, and analysis software for FACS machines used by each center were reported (Table 1). In addition, isolation time, total cells isolated, pre- and postsort viability, purity, and percentages of target populations were carefully documented on a standardized data collection form to establish compliance to parameter thresholds (Table 3). If defined thresholds were not met, ISCC Centers were required to abort the preparation and repeat until the minimal thresholds were met.
Table 2.
ISCC recommended reporting criteria
| Criteria | Threshold |
|---|---|
| Preservation of cell viability | Prep time duration |
| Presort cell viability | |
| Postsort cell viability | |
| Target cell purity | Postsort reanalysis |
| Gene expression of negative marker selection | |
| Gene expression of positive marker selection | |
| Cell surface antibody staining |
ISCC, Intestinal Stem Cell Consortium.
Table 3.
Isolation time, total cells isolated, pre- and postsort viability, purity, and percentages of target populations
| Center | Isolation Time, min | Sort Time, min | Total Cells Isolated, 107 | Presort Viability, % | Postsort Viability, % | Postsort Purity, % | Gate 4 EpCAM+, % | Gate 5 CD44−, % | Gate 6 CD44+, % |
|---|---|---|---|---|---|---|---|---|---|
| Recommendation or threshold | 300 | 60 | N/A | 75 | refer to RIN | ||||
| 1 | 296 | 30 | 3.1 | 75 | 100 | 100 | 99 | 0.3 | 99 |
| 2 | 230 | 30 | 3.5 | 79 | 96 | 96 | 96 | 0.4 | 99 |
| 3 | 254 | 15 | NR | 75 | 96 | 98 | 97 | 6.0 | 86 |
| 4 | 240 | 35 | 3.1 | 92 | 97 | 99 | 99 | 1.2 | 100 |
| 5 | 230 | 6 | 2.5 | 83 | 97 | 100 | 100 | 3.3 | 95 |
| 6 | 206 | 26 | 2.6 | ND | 90 | ND | 98 | 1.2 | 98 |
| 7 | 139 | 50 | ND | 79 | 80 | 98 | 43 | 0.3 | 85 |
Boldface indicates threshold.
N/A, not applicable; RIN, RNA integrity number; NR, not reported, ND, not determined.
A single methodology for removal of the intestinal epithelium from the basement membrane and subsequent dissociation into single cells was established and approved by all ISCC centers. Removal of the epithelial sheets from the underlying mesenchyme was accomplished by using a modified Evans method and dissociation of the intact epithelium to single cells was accomplished by a brief enzymatic digestion using dispase (4). To minimize variability due to age, sex, weight, strain, and regions of the intestine, C57BL/6 male mice between 6–8 wk of age were used for the study, and cells were isolated from a defined region of jejunum (Fig. 1A).
Across eight centers, the average procedural time, defined as the amount of time from euthanasia to the initiation of FACS, was 231 min. Each 10-cm jejunal segment yielded an average of 3.2 × 107 cells (Table 3). Cell viability was quantified by PI exclusion using flow cytometry and determined to be an average of 80.3% (Table 3). To assess the extent of cell death due to FACS, PI-negative epithelial cells were sorted into tubes containing PI and then reanalyzed by flow cytometry. Postsort viabilities averaged 94.3%, indicating limited cell death by the FACS procedure (Table 3). Importantly, differences in FACS instrumentation resulted in a negligible impact on cell viability. These data demonstrate that the standardized protocol resulted in high cell viability, supporting rationale for the next study phase comparing cross-institution FACS isolation of epithelial populations.
A standardized approach results in efficient and reproducible FACS isolation of intestinal epithelial cell populations.
By nature of the technology, FACS has numerous subjective parameters including cell gating, doublet discrimination, and voltage thresholds used to define different populations. Variations in any one of these parameters can result in significant changes in the purity of cells collected. To decrease the impact of subjective FACS parameters on reproducible isolation of target cell populations, the ISCC defined a gating strategy that was used by each center (see materials and methods for complete description of gating logic). Our approach used 1) exclusion of dead cells/debris (Fig. 2A; FSC vs. SSC), 2) exclusion of doublets/multimers (Fig. 2A; SSC height vs. SSC area), 3) exclusion of dead/dying cells (Fig. 2A; PI+); 4) exclusion of nonepithelial cell populations (Fig. 2A; CD31−, CD45−); and 5) inclusion of epithelial populations (Fig. 2A; EpCAM+). Although it is possible to exclude smaller cells or cells with more or less complexity based on FSC and SSC parameters, our study focused on discriminating cells on the basis of their antigenic expression. With the goal of assessing intra- and intercenter variability of sorting target populations by FACS, standardized gating strategies were employed to collect three epithelial populations: 1) total epithelium (EpCAM+), 2) crypt-based cells (CD44+), and 3) villus cells (CD44−).
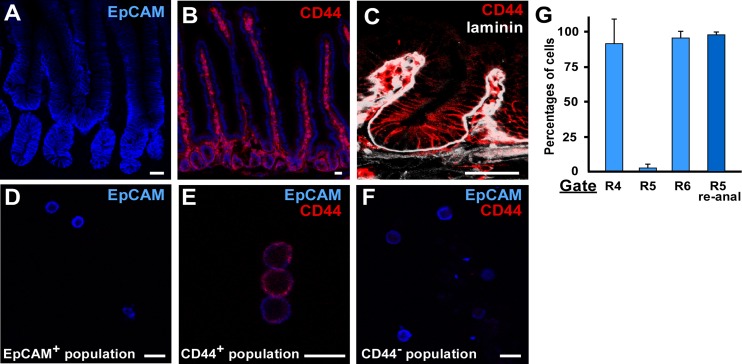
The three FACS-isolated target populations (EpCAM+, CD44+, and CD44−) from each center for n = 3 mice were evaluated for purity and gene expression signatures by qualitative and quantitative measures. Confocal images of intestinal sections demonstrated EpCAM staining in all target populations, and CD44 expression was restricted to the cell surface of the CD44+ FACS-isolated epithelial subpopulation (Fig. 3, A–C). The CD44−/EpCAM+ cells (Gate R6; x = 93.7%) represented a majority of the total epithelial cells population whereas CD44+/EpCAM+ cells (Gate R5; x = 1.7%) represented a minority of the cells, consistent with CD44 immunostaining data (Fig. 3B, Table 3). The purity of the CD44+ population was quantitatively assessed by reanalysis of the FACS-isolated CD44+ population by flow cytometry. The data demonstrate only 2.1% of the cells on average fell outside of the CD44+ gate (Table 3).
Fig. 3.
CD44 antibody staining of intestinal epithelium. A: EpCAM antibody staining (blue) of the proximal intestine is confined to the epithelium. B: CD44 antibody staining (red) is restricted to the epithelium of the base of the crypt but is also expressed on blood-derived cells located in the crypt and villus mesenchyme. Section is also stained with Hoechst (blue). White dashed line in A and B demarcates the boundary between the epithelium and mesenchyme. C: higher magnification of CD44 staining (red) in the crypt region by confocal microscopy. Laminin (white) is expressed in the mesenchyme. D: EpCAM-expressing (blue) cytospun cells isolated from the epithelial inclusion sorting gate. EpCAM-expressing cells were divided into CD44-expressing (red; E) or CD44-nonexpressing (F). G: there was little variation in the percentages of within the EpCAM+ (gate R4), CD44+ (gate R5), and CD44− (gate R6) populations across centers. Purity of the CD44+ cells was high. Data reported as means ± SE.
A standardized approach results in minimal variation of technical and biological replicates.
To determine intracenter variability, three ISCC centers (Centers 1, 2, and 5) conducted the standardized ISCC protocol on separate mice and compared gene expression profiles in EpCAM+, CD44−, and CD44+ populations from the biological replicates. It was predicted that the CD44+ population would be specifically enriched for genes that mark crypt-based cells (i.e., IESCs) and the CD44− population would be enriched for genes that are expressed in cells that are primarily localized to the villus (i.e., fully differentiated absorptive enterocytes). The CD44+ population was highly enriched for CD44 expression while the CD44− population was deenriched for CD44 validating the FACS isolation (Fig. 4A). Furthermore, as predicted, the CD44+ population was also enriched for Lgr5 expression and exhibited enteroid-forming capacity of 0.1% at 10 days in culture (Fig. 5D) consistent with the stem cell capacity of CD44+/Lgr5+ cells (2, 8, 20). Analysis of other lineage-specific markers indicated that the CD44+ population was enriched for secretory progenitors (Atoh1+) and Paneth cells (Lyz1+), which are anatomically restricted to the crypts (Fig. 4B). The data also demonstrated deenrichment of Muc2 and SIM, which is consistent with the villus localization of goblet cells and absorptive enterocytes (Fig. 4B).
Fig. 4.
Intracenter gene expression variation within CD44+ and CD44− populations. A: CD44 and Lgr5 gene expression data in CD44+ and CD44− epithelial populations between n = 3 samples from 3 centers (Centers 1, 2, 5), as determined by quantitative RT-PCR. CD44+ solid boxes, CD44− hatched boxes. B: genes expressed in differentiated subpopulations were evaluated [atonal homology 1 (Atoh1): secretory cells; lysozyme (Lyz): Paneth cells; chromagranin A (ChgA): enteroendocrine cells; Mucin2 (Muc2): goblet cells; and sucrose isomaltase (Sim): enterocytes]. C: comparison of the standard deviations of “between-replicate” (gray shading) and “between-sample” (box plot) indicated marginal intracenter differences, denoted by an asterisk (all P < 0.05).
For intracenter comparison of biological replicates, variation within each center was determined by comparing the ΔΔCT standard deviation (SD) of biological replicate (between-mouse) and technical replicate (between-replicate) values in three mice by using a Wilcoxon's rank sum test in two different sample populations (CD44− and CD44+; Fig. 4C). In the CD44− population, there was a statistically significant difference in the median SD of technical vs. biological replicates for Lyz (P = 0.04), Muc2 (P = 0.04), and SIM (P = 0.04). In the CD44+ population, there was a statistically significant difference in the median SD of technical vs. biological replicates only for Muc2 (P = 0.04). Comparisons for all remaining genes were not statistically significant. Together, these data indicate that following a standardized approach for FACS of target intestinal epithelial cell populations results in minimal technical and biological replicate variability.
Standardized reporting of FACS results performed by different institutions results in consistent FACS enrichment of crypt-based CD44+/− cells and detection of technical error.
The final phase of this multicenter study was to determine whether standardized epithelial and FACS isolation methods could be used at different centers and yield consistent results. The same gene expression analysis that was conducted to assess intracenter variability was also used by seven ISCC Centers to compare gene expression signatures for the three target cell populations (EpCAM+, CD44−, CD44+). At each of the seven centers RNA was isolated from these three populations from three biological replicates. RIN threshold values were limited to 6.5 for inclusion in this phase of the study. RIN analysis on all nine samples from each of the centers indicated that not all of the nine samples from three centers met the RIN threshold of 6.5. However, the analysis showed that two biological replicates from all seven centers did meet the RIN threshold of 6.5; thus pooling of samples within each center was conducted.
Gene expression analysis on the CD44+ population from Centers 1–4 and 6 demonstrated enrichment for the crypt-based biomarker, Lgr5 (Fig. 5A), indicating that the CD44+ population was enriched for actively cycling intestinal epithelial stem cells. Analysis of progenitor and lineage-restricted biomarkers demonstrated consistent enrichment of Atoh1 and Lysozyme1 in CD44+ populations from Centers 1–4 and 6. Together these data are consistent with the immunostaining data demonstrating restricted CD44 expression in crypt-based epithelial cells (Figs. 3 and 5B). Significant enrichment of Muc2 in the CD44− population (Centers 1–4 and 6) and SIM (Centers 1, 3, and 7) further support the interpretation that CD44 can be used to FACS-enrich for crypt-based and villus-localized cells and then differentiate between the two populations.
To assess the consistency of the FACS isolation of populations conducted at each center, PCA was performed using all ΔΔCT values from each sample to evaluate the relative proximity of each center to the population mean (Fig. 5C). This analysis demonstrated close clustering between Centers 1–4 (each P ≥ 0.54), with Centers 5 and 6 falling outside the cluster (each P < 1.2 × 10−5). Center 7 was excluded from the PCA because of verifiable contamination of CD45+ lymphocytes in the CD44+ epithelial population (Fig. 6).
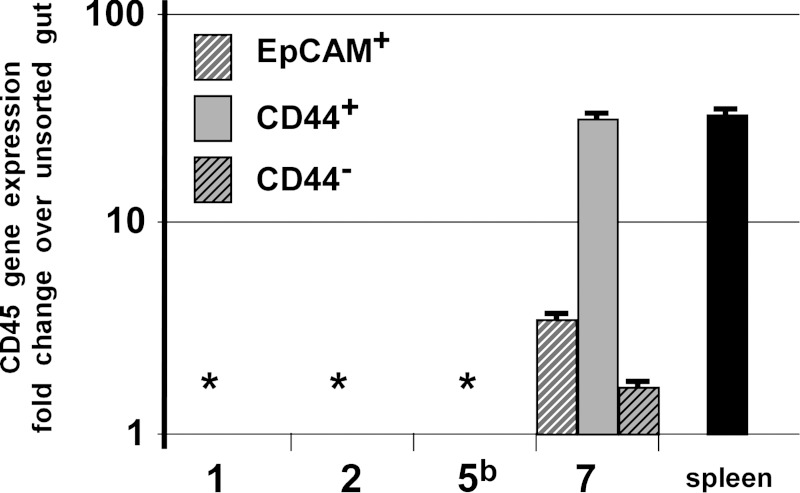
Fig. 6.
Detection of contaminating CD45-expression in isolated epithelial populations. Epcam+ (white hatched), CD44+ (solid bars), and CD44− (black hatched) isolated populations were surveyed for contaminating lymphocyte gene expression. Samples from only 1 center exhibited high levels of CD45 gene expression relative to an unsorted intestinal preparation. Asterisk represents “no detectible signal.”
DISCUSSION
Reproducible FACS of target intestinal epithelial cell populations is challenging. This multicenter ISCC study addresses an urgent and unmet need in the field by standardizing critical elements of epithelial isolation and FACS methods, establishing critical controls, experimental thresholds, and data reporting guidelines. The results indicate that compliance to standardized isolation and data reporting methods lead to reproducible, and therefore comparable, results within an institution and across different institutions. Importantly, this study highlights likely sources of experimental error that can adversely impact reproducibility of target cell isolation and offers recommendations for studies using FACS-based approaches to epithelial cell isolation.
Although the standardized procedures established in this study demonstrated efficient and reproducible FACS isolation of a crypt-based epithelial populations, three centers fell outside the PCA clustering of Centers 1–4. The new data reporting guidelines outlined in this study were used to conduct a post hoc error analysis to identify likely experimental flaws that resulted in the aberrant results from Centers 5–7.
Center 5 collected cell populations that exhibited gene enrichment patterns discordant from other centers. Data from Center 5 showed similar CD44 expression in both the CD44+ and CD44− populations, indicating no population enrichment (Fig. 5). Additionally, biomarker expression for other lineage-specific genes was enriched to the same extent in both the CD44+ and CD44− populations (Fig. 5). Careful review of tissue isolation, cell dissociation, and FACS histogram data indicated that Center 5 had followed all standardized methodologies and had met the minimum requirements for entry into the FACS phase of the study. Histogram data also indicated acceptable FACS gating parameter compliance. Since protocol compliance appeared to be consistent with other centers and gene enrichment patterns were not statistically different between CD44+ and CD44− populations, the aberrant results are consistent with the interpretation that there was an error in FACS gate logic that resulted in inadvertent sorting of CD44+ cells into the CD44+ and CD44− collection tubes. The recommendation to avoid this type of error is to conduct a postsort analysis on all target cell populations. This additional level of quality control will ensure that the correct populations are being sorted prior to collecting cells into the RNA lysis buffer.
Gene expression patterns in the CD44− and CD44+ populations from Center 6 were consistent with Centers 1–4, but the extent of enrichment fell outside the average, resulting in data from Center 6 falling outside the PCA cluster. Analysis of FACS histograms from Center 6 revealed that gating to exclude PI+, CD31+, and CD45+ cells was less stringent than the recommended standard that was used by Centers 1–4 (data not shown). The impact of more rigorous gating is that any contaminating CD44− cells would be excluded from the CD44+ population. This exclusion would result in a purer population of CD44+ cells and gene enrichment profiles that would lie in concordance with Centers 1–4. This interpretation highlights the need for consistent and rigorous gating strategies to minimize contaminating populations, which can have significant and negative impact on downstream applications.
Although the CD44+ populations from Center 7 demonstrated CD44 mRNA enrichment, gene expression data from the CD44+ population indicated Lgr5 mRNA deenrichment. This result is inconsistent with results from other centers that clustered tightly in the PCA. Additionally, gene expression patterns for all other lineage-restricted biomarkers were opposite from values reported from other centers the cluster in PCA (Fig. 5C). Evaluation of the reported data to identify probable causes for these inconsistencies suggested that the CD44+ epithelial population was contaminated with a CD44+ nonepithelial population. Contamination of the EpCAM+/CD45−/CD44+ population with CD45+/CD44+ cells would result in a deenrichment of Lgr5 expression and other epithelial lineage-restricted genes compared with total epithelium. To explore this possibility, gene expression analysis for CD45 was conducted on samples from Center 7 and compared with samples from Centers 1, 2, and 5 as controls. The data demonstrate that all three cell populations from Center 7 possessed CD45 gene expression, consistent with the interpretation of immune cell contamination (Fig. 6).
These three examples of post hoc error analyses were facilitated by our standardized data reporting requirements included in the ISCC epithelial cell isolation protocol and guidelines. The post hoc error analysis revealed that discrepant data was likely introduced during FACS of target cell populations and not the initial epithelial isolation and dissociation phases. These results further highlight the importance of rigorous application of FACS methodology and transparent reporting of supporting FACS data (including FACS gating parameters and post hoc population purity analyses) to mitigate systematic error.
On the basis of this study, we recommend that the purity of the sorted populations be reported to allow investigators the ability to evaluate the extent of potential contamination of nontarget cell populations. Although there are numerous hallmarks for cell purity, it is recommended that FACS-isolated populations be assessed for purity by 1) reanalysis by FACS to show that the original population resorts into the original target FACS gate; 2) verification of cell surface staining to confirm antibody fidelity; and 3) gene expression analysis using positive and negative markers employed during FACS isolation. It is highly recommend that operators of FACS instrumentation be highly trained and experienced and that investigators evaluate all FACS parameters with the FACS operator prior to cell collection. Although a gating strategy guideline is useful in navigating the first-time experience into flow cytometry, it is essential that the operator clearly understand the biological rationale underlying the inclusion and/or exclusion of subpopulations in establishing the gating logic. Clear and rigorous controls should be reported when using multiple antibodies and multiple fluorophores. Although our study was designed to avoid color compensation, each antibody/fluorophore combination should be evaluated for filter compatibility in the initial control experiments. Machine and sorting parameters including all gate logic used in the study should be published as supplemental methods to provide the ability to assess population purity and enable comparison between studies.
The ISCC acknowledges that the rapid evolution of experimental method development for the dissociation and FACS of the intestinal epithelium precludes the endorsement of a single epithelial isolation protocol. However, the methodologies established in this study can be considered a foundation for continued method development and improvement, as well as an initial starting point for laboratories setting up experiments requiring FACS isolation of intestinal epithelial cells. FACS isolation and data reporting recommendations tested in this study will also be useful for human IESC studies that require FACS.
GRANTS
This work was funded by the cooperative efforts of the NIDDK and the NIAID, components of the US NIH. ISCC members include M. H. Wong: U01DK85525; C. J. Kuo: U01DK85527; L. Li: U01DK85507; M. G. Martin: U01DK85535; S. J. Henning: U01DK85547; C. Houchen: U01DK85508; J. Y: U01DK85570; J. Lynch: U01DK85551; J. C. Niland: U01DK085532; and S. T. Magness: R01DK091427.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
S.T.M., B.J.P., J.C.D., S.J.H., C.W.H., J.S.K., C.J.K., L.L., J.P.L., M.G.M., M.S., J.Y., and M.H.W. conception and design of research; S.T.M., B.J.P., M.A.C., R.J.M., J.R.S., F.W., J.W., X.W., K.Y., and M.H.W. performed experiments; S.T.M., M.A.C., J.C.D., S.J.H., C.W.H., J.S.K., C.J.K., L.L., J.P.L., M.G.M., J.C.N., B.O., D.Q., M.S., F.W., X.W., K.Y., and M.H.W. analyzed data; S.T.M., J.C.D., S.J.H., C.W.H., J.S.K., C.J.K., L.L., J.P.L., M.G.M., J.C.N., B.O., D.Q., M.S., F.W., J.Y., and M.H.W. interpreted results of experiments; S.T.M. and M.H.W. prepared figures; S.T.M. and M.H.W. drafted manuscript; S.T.M., S.J.H., C.W.H., J.S.K., C.J.K., L.L., B.O., and M.H.W. edited and revised manuscript; S.T.M., B.J.P., M.A.C., J.C.D., S.J.H., C.W.H., J.S.K., C.J.K., L.L., J.P.L., M.G.M., R.J.M., J.C.N., B.O., D.Q., M.S., F.W., J.W., X.W., K.Y., J.Y., and M.H.W. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Jessica Girard in the coordination and organization of this consortium activity, Richard von Furstenberg for method feasibility studies, Adam D. Gracz for method development, Dr. Nicholas Smith for enteroid growth assays, and Dr. Paige Davies for supplying confocal images of the stained intestine. Finally, we are grateful for assistance from personnel from each Center's FACS Facility.
REFERENCES
- 1.Alexander CM, Puchalski J, Klos KS, Badders N, Ailles L, Kim CF, Dirks P, Smalley MJ. Separating stem cells by flow cytometry: reducing variability for solid tissues. Cell Stem Cell 5: 579–583, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007, 2007. [DOI] [PubMed] [Google Scholar]
- 3.Dufour G, Demers MJ, Gagne D, Dydensborg AB, Teller IC, Bouchard V, Degongre I, Beaulieu JF, Cheng JQ, Fujita N, Tsuruo T, Vallee K, Vachon PH. Human intestinal epithelial cell survival and anoikis. Differentiation state-distinct regulation and roles of protein kinase B/Akt isoforms. J Biol Chem 279: 44113–44122, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Evans GS, Flint N, Somers AS, Eyden B, Potten CS. The development of a method for the preparation of rat intestinal epithelial cell primary cultures. J Cell Sci 101: 219–231, 1992. [DOI] [PubMed] [Google Scholar]
- 5.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med 27: 126–139, 2006. [DOI] [PubMed] [Google Scholar]
- 6.Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol Lett 28: 1601–1613, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol 296: G1108–G1118, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gracz AD, Fuller MK, Wang F, Li L, Stelzner M, Dunn JC, Martin MG, Magness ST. CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells 2013. April 4. 10.1002/stem.1391 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gracz AD, Ramalingam S, Magness ST. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol 298: G590–G600, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landgrebe J, Wurst W, Welzl G. Permutation-validated principal components analysis of microarray data. Genome Biol 3: RESEARCH0019, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 12.Lugo-Martinez VH, Petit CS, Fouquet S, Le Beyec J, Chambaz J, Pincon-Raymond M, Cardot P, Thenet S. Epidermal growth factor receptor is involved in enterocyte anoikis through the dismantling of E-cadherin-mediated junctions. Am J Physiol Gastrointest Liver Physiol 296: G235–G244, 2009. [DOI] [PubMed] [Google Scholar]
- 13.Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S, Breault DT. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci USA 108: 179–184, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pallavicini MG, Levin J, Summers L, Levin F. Multivariate flow cytometric characterization and enrichment of murine megakaryocyte progenitors (CFU-Meg). Exp Hematol 15: 704–709, 1987. [PubMed] [Google Scholar]
- 15.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40: 915–920, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469: 415–418, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265, 2009. [DOI] [PubMed] [Google Scholar]
- 18.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108, 2008. [DOI] [PubMed] [Google Scholar]
- 19.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science 334: 1420–1424, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, Scoville D, He XC, Mahe M, Box A, Perry J, Smith NR, Lei Nanye N, Davies PS, Fuller MK, Haug JS, McClain M, Gracz AD, Ding S, Stelzner M, Dunn JC, Magness ST, Wong MH, Martin M, Helmrath M, Li L. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology 145: 383–395.e21, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.