Abstract
Myocardial interstitial fibrosis is an important contributor to the development of heart failure. Type 3 p90 ribosomal S6 kinase (RSK3) was recently shown to be required for concentric myocyte hypertrophy under in vivo pathological conditions. However, the role of RSK family members in myocardial fibrosis remains uninvestigated. Transgenic expression of α-tropomyosin containing a Glu180Gly mutation (TM180) in mice of a mixed C57BL/6:FVB/N background induces a cardiomyopathy characterized by a small left ventricle, interstitial fibrosis, and diminished systolic and diastolic function. Using this mouse model, we now show that RSK3 is required for the induction of interstitial fibrosis in vivo. TM180 transgenic mice were crossed to RSK3 constitutive knockout (RSK3−/−) mice. Although RSK3 knockout did not affect myocyte growth, the decreased cardiac function and mild pulmonary edema associated with the TM180 transgene were attenuated by RSK3 knockout. The improved cardiac function was consistent with reduced interstitial fibrosis in the TM180;RSK3−/− mice as shown by histology and gene expression analysis, including the decreased expression of collagens. The specific inhibition of RSK3 should be considered as a potential novel therapeutic strategy for improving cardiac function and the prevention of sudden cardiac death in diseases in which interstitial fibrosis contributes to the development of heart failure.
Keywords: ribosomal S6 kinase, α-tropomyosin, fibrosis
hypertrophic cardiomyopathy (HCM) is the most commonly inherited heart defect (1 in 500 individuals) and the leading cause of sudden death in children, accounting for 36% of sudden deaths in young athletes (16). HCM is caused by dominant mutations in sarcomeric proteins that typically are identified due to their induction of cardiac hypertrophy. However, the phenotype and clinical course resulting from HCM mutations is highly variable, and despite the name of the disorder, hypertrophic cardiomyopathy is not always present in those significantly affected. For example, genotype-positive patients without left ventricular hypertrophy can have clinically significant myocardial fibrosis, diastolic dysfunction, and electrocardiographic (ECG) abnormalities (16, 25). Studies using transgenic mice have also demonstrated that the phenotype of HCM mutations depends on genetic background (17, 20). In FVB/N mice, expression of the HCM mutation TM180 (Glu180Gly amino acid substitution of the thin filament protein α-tropomyosin) resulted in concentric left ventricular hypertrophy, extensive fibrosis, atrial enlargement, and death within 6 mo (20). In contrast, expression of the TM180 mutation in C57BL/6 mice resulted in no ventricular hypertrophy or fibrosis and a lower heart weight (17). As shown below, we found that in a mixed C57BL/6:FVB/N background, the TM180 transgene induced another phenotypic variant, including a small left ventricle with small myocytes, interstitial fibrosis, and impaired systolic and diastolic cardiac function.
Present in HCM regardless of myocyte hypertrophy, remodeling of the extracellular matrix and the induction of myocardial interstitial fibrosis are important factors contributing to the development of heart failure in this disease (5, 24). Increased deposition of fibrillar collagen and the disruption of the normal cellular architecture of the myocardium can result in decreased compliance and both diastolic and systolic dysfunction, as well as arrhythmias due to interference with the electrical conduction system. p90 ribosomal S6 kinases (RSK) are pleiotropic protein kinases that are activated in myocytes in response to many stress-related stimuli (1, 2, 13, 15). We recently reported that type 3 RSK (RSK3) is required for the induction of concentric myocyte hypertrophy in mice subjected to pressure overload or catecholamine infusion, even though RSK3 comprised a minority of the total RSK enzyme in cardiac myocytes (14). Consequently, we have been interested in how general a role RSK3 might play in the regulation of pathological remodeling, including whether RSK3 contributes to fibrosis. The four RSK family members are activated by sequential phosphorylation by extracellular signal-regulated kinases (ERKs) and 3′-phosphoinositide-dependent kinase 1 (PDK1) (2). Like ERK1/2 (6), RSKs are activated in TM180 FVB/N mice (Li J, unpublished observations). Data presented below reveal that RSK3 contributes to the development of interstitial fibrosis and cardiac insufficiency in the TM180 C57BL/6:FVB/N mouse.
MATERIALS AND METHODS
Reagents.
Primary antibodies included mouse 1F6 monoclonal anti-RSK3 (catalog no. H00006196-M01; Abnova), which actually detects all RSK family members (14), OR43 rabbit anti-RSK3 (14), and N-16 goat anti-RSK3 (Santa Cruz Biotechnology). Secondary antibodies included horseradish peroxidase-conjugated donkey secondary antibodies (Jackson ImmunoResearch). RSK3 immunoprecipitation was performed as previously described (14).
Mice.
All experiments involving animals were approved by the Institutional Animal Care and Use Committee at the University of Miami. The RSK3−/− C57BL/6 mouse was mated to the TM180 transgenic FVB/N mouse (14, 20). All mice studied were littermates from RSK3−/+ × TM180;RSK3−/+ breedings such that the background strain was 50:50 C57BL/6:FVB/N. All four genotypes were present in typical Mendelian proportion. All experiments were performed with mice that were 16 wk of age. Genotyping was performed at weaning by PCR using tail biopsy samples as previously described (14, 20).
Echocardiography.
A Vevo 770 high-resolution in vivo imaging system (VisualSonics) with an RMVTM 707B “high frame” scan head was used for imaging. Mice were anesthetized with 2.0% isoflurane for M-mode imaging.
Left ventricular catheterization.
Intact heart hemodynamic analysis was performed using miniaturized conductance micromanometry in mice anesthetized with 2.0% isoflurane and then paralyzed by intraperitoneal 2 mg/kg pancuronium. All mice were ventilated via endotracheal intubation. A four-electrode catheter (SPR-839; Millar Instruments) was inserted into the right carotid artery and advanced into the left ventricle for simultaneous measurement of pressure and volume. Ventilation was transiently interrupted during the acquisition of data. Hemodynamic data was acquired closed chested. To measure load-independent parameters, preload was decreased by occluding the inferior vena cava through a limited right anterior thoracotomy. Data were acquired and analyzed using LabChart 7 Pro software (ADInstrument). Three different intervals of at least 10 cardiac cycles were analyzed for both open- and closed-chest conditions for each mouse.
Histochemistry.
Heart tissue was fixed in 3.7% formaldehyde. Deparaffinized 5-μm tissue sections were stained using Masson's trichrome (Richard Allen Scientific), picrosirius red (Polysciences), Alexa Fluor 555 wheat germ agglutinin conjugate (Invitrogen), and terminal deoxynucleotidyl nick-end labeling (TUNEL; in situ cell death detection kit, TMR red; Roche), as recommended by the manufacturers. The cross-sectional areas of >150 myocytes in >3 distinct regions of the left ventricle were measured per heart using the wheat germ agglutinin sections. Collagen content was assayed using the picrosirius red-stained sections and linearly polarized light microscopy of the entire left atrium and three to five ×4 objective fields per left ventricle. Morphometrics and collagen content were measured using IPLab microscope software (BD Biosciences). All analyses were performed by a blinded investigator.
RNA assay.
Total RNA was quantified with a Nanodrop 8000 Spectrophotometer (ThermoScientific) and quality controlled using a Bioanalyzer 2100 and the RNA 6000 Nano kit (Agilent). The NanoString assay is based on direct, multiplexed measurement of gene expression without amplification, utilizing fluorescent molecular barcodes and single-molecule imaging to identify and count multiple transcripts in a single reaction. Briefly, 100 ng of total RNA were hybridized in solution to a target-specific “codeset” overnight at 65°C. The codeset contained dual, adjacently placed 50-bp oligonucleotide probes against the entire panel of genes, one set of probes fluorescently bar-coded and the other biotinylated. The hybridization reactions were loaded onto the NanoString prep station, which removes excess oligonucleotides and binds the hybridized mRNA to the streptavidin-coated cartridge surface. The cartridges were loaded onto the NanoString digital analyzer, and >500 fields of view were fluorescently scanned to count only those individual mRNAs bound to both a biotinylated and fluorescently bar-coded probe. Data sets for each RNA sample were normalized to internal positive controls and background subtracted. Probe sequences are available on request.
Statistics.
For all experiments, n refers to the number of individual mice. All data are means ± SE. P values were calculated using two-tailed Student's t-tests, paired or unpaired as appropriate, and are not corrected for multiple comparisons. Repeated symbols represent P values of different orders of magnitude: *P < 0.05, **P < 0.005, ***P < 0.0005.
RESULTS
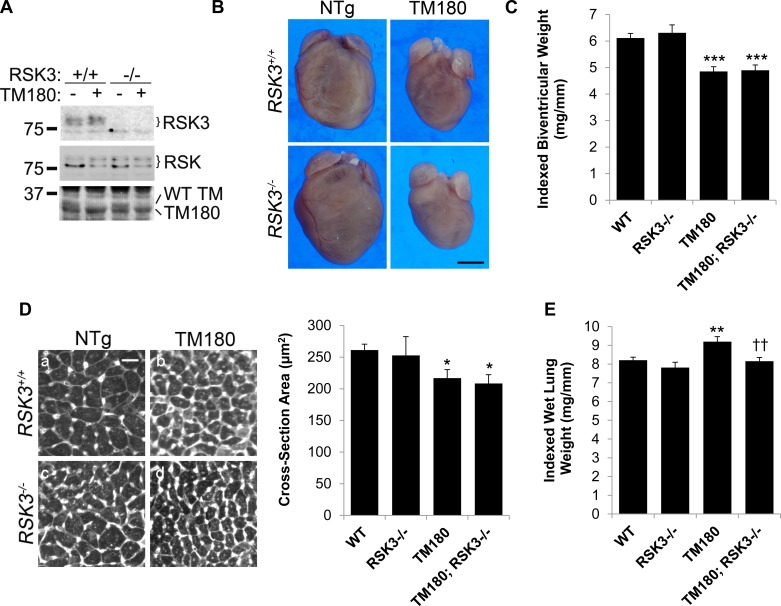
To study the function of RSK3 in TM180 mice, we crossed FVB/N TM180 transgenic mice with C57BL/6 RSK3 knockout mice such that all mice were of a mixed 50:50 background. These mice bred according to Mendelian genetics, and there was no excess mortality for any of the cohorts noted through 4 mo of age. Expression of the TM180 mutant protein that migrates faster in SDS-PAGE and the concomitant downregulation of the endogenous wild-type tropomyosin was evident by the expected change in α-tropomyosin bands detected by total protein stain (Fig. 1A) (20). RSK3 expression was not significantly increased in TM180-expressing mice, while absent in RSK3−/− mice. There was no evidence of compensatory changes in the expression of other RSK family members upon RSK3 knockout. In addition, loss of RSK3 did not change the total level of RSK in the heart, consistent with our previous findings that RSK3 is expressed at a much lower level in the heart than other RSK family members (14).
Fig. 1.
TM180 (α-tropomyosin containing a Glu180Gly mutation) C57BL/6:FVB/N mice have a small heart phenotype. A, top: type 3 p90 ribosomal S6 kinase (RSK3) protein was immunoprecipitated using N-16 antibody and detected using OR43 antibody. Middle, total RSK protein in heart extracts was detected using mouse anti-RSK antibody. Bottom, Ponceau staining for total heart protein shows that the major α-tropomyosin (α-TM) wild-type (WT) band is replaced by a lower TM180 band in transgenic hearts (20). n = 4. B: representative photographs of mouse hearts. NTg, nontransgenic. Bar, 2 mm. C: biventricular weight indexed to tibial length. D: wheat germ agglutinin-strained heart sections (left; bar, 50 μm) and cross-sectional areas (right; n = 6 for each cohort). E: wet lung weight indexed to tibial length. Values are means ± SE. *P < 0.05; **P < 0.005; ***P < 0.0005 compared with WT cohort. ††P < 0.005 compared with TM180 cohort. Cf. Table 1 for C and E.
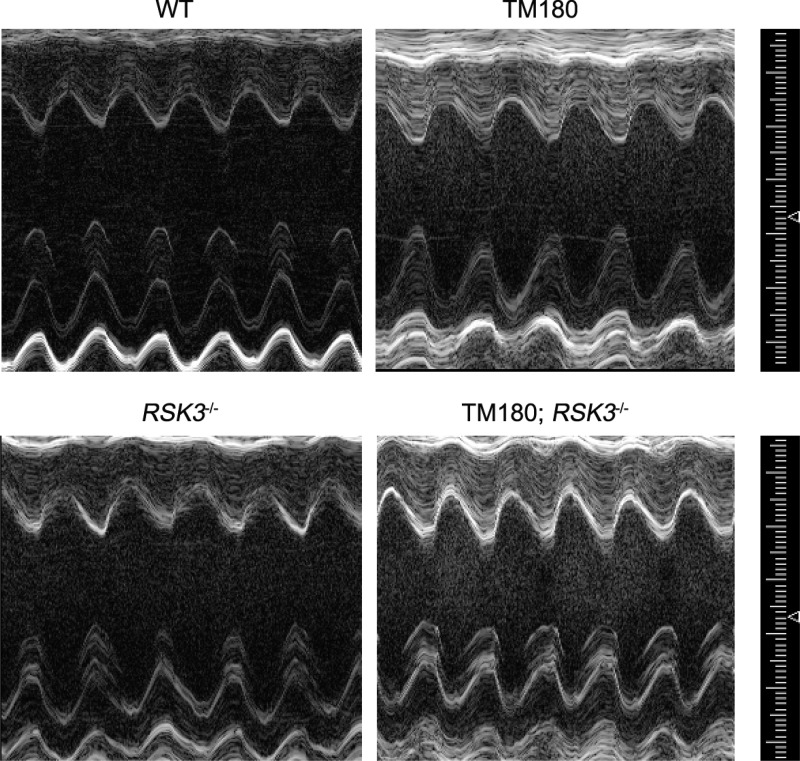
In these mice of mixed lineage, the TM180 transgene induced a small heart phenotype that included a reduced biventricular weight (21%) and left ventricular myocytes with a similarly smaller cross-sectional area (17%; Fig. 1, B–D, and Table 1). As shown by M-mode echocardiography (Fig. 2 and Table 2), the TM180 mice had reduced left ventricular internal dimensions (diastolic left ventricular internal diameter and volume) but similar wall thicknesses (diastolic left ventricular posterior and anterior wall thicknesses) to those for wild-type mice, resulting in a smaller calculated left ventricular mass similar to that gravimetrically measured postmortem (Table 1). The TM180 left ventricle was hyperdynamic with increased fractional shortening (increased by 22%) and ejection fraction (increased by 16%). That the changes in the TM180 left ventricle were pathologically important was implied by both a 39% increased atrial weight and a significant, albeit small (12%), increase in wet lung weight, consistent with the presence of mild pulmonary edema and left-sided cardiac insufficiency (Fig. 1E and Table 1).
Table 1.
Gravimetric data
| WT | RSK3−/− | TM180 | TM180;RSK3−/− | |
|---|---|---|---|---|
| n | 24 | 18 | 14 | 21 |
| Body weight, g | 27.8 ± 0.8 | 28.1 ± 1.4 | 26.7 ± 1.1 | 26.6 ± 1.0 |
| Tibial length, mm | 17.7 ± 0.1 | 17.6 ± 0.2 | 17.6 ± 0.1 | 17.5 ± 0.1 |
| Biventricular weight, mg | 108 ± 3 | 111 ± 6 | 85 ± 3*** | 86 ± 4*** |
| Biatrial weight, ms | 7.75 ± 0.45 | 8.11 ± 0.59 | 10.71 ± 0.68** | 11.33 ± 0.73*** |
| Wet lung weight, mg | 145 ± 3 | 138 ± 6 | 162 ± 5* | 143 ± 4†† |
| Biventricular weight/tibial length, mg/mm | 6.11 ± 0.18 | 6.31 ± 0.30 | 4.85 ± 0.18*** | 4.90 ± 0.20*** |
| Biatrial weight/tibial length, mg/mm | 0.44 ± 0.03 | 0.46 ± 0.03 | 0.61 ± 0.04*** | 0.65 ± 0.04*** |
| Wet lung weight/tibial length, mg/mm | 8.21 ± 0.16 | 7.81 ± 0.29 | 9.19 ± 0.27** | 8.16 ± 0.20†† |
Values are means ± SE. WT, wildtype; RSK3−/−, type 3 p90 ribosomal S6 kinase RSK3 knockout; TM180, α-tropomyosin containing a Glu180Gly mutation.
P < 0.05;
P < 0.005;
P < 0.0005 compared with WT.
P < 0.005 compared with TM180.
Fig. 2.
Echocardiography. Representative M-mode images for 16-wk-old mice. See Table 2 for echocardiographic values.
Table 2.
M-mode echocardiographic data
| WT | RSK3−/− | TM180 | TM180;RSK3−/− | |
|---|---|---|---|---|
| n | 24 | 27 | 26 | 32 |
| LVID;d, mm | 4.04 ± 0.06 | 4.02 ± 0.07 | 3.78 ± 0.06** | 3.60 ± 0.05***† |
| LVPW;d, mm | 0.83 ± 0.03 | 0.80 ± 0.03 | 0.80 ± 0.03 | 0.83 ± 0.02 |
| LVID;s, mm | 2.72 ± 0.08 | 2.85 ± 0.08 | 2.26 ± 0.06*** | 2.13 ± 0.04*** |
| LVPW;s, mm | 1.18 ± 0.03 | 1.10 ± 0.03* | 1.21 ± 0.03 | 1.21 ± 0.03 |
| LVAW;d, mm | 0.90 ± 0.02 | 0.88 ± 0.03 | 0.86 ± 0.03 | 0.94 ± 0.03† |
| LVAW;s, mm | 1.32 ± 0.03 | 1.26 ± 0.04 | 1.33 ± 0.03 | 1.39 ± 0.03 |
| LV Vol;d, μl | 72.4 ± 2.5 | 71.4 ± 2.7 | 61.9 ± 2.1** | 55.0 ± 1.6***† |
| LV Vol;s, μl | 28.3 ± 1.8 | 31.9 ± 2.0 | 18.0 ± 1.2*** | 15.1 ± 0.6***† |
| EF, % | 61.3 ± 1.8 | 56.1 ± 1.5* | 70.9 ± 1.4*** | 72.4 ± 0.8*** |
| FS, % | 32.9 ± 1.3 | 29.2 ± 1.0* | 40.2 ± 1.2*** | 41.0 ± 0.7*** |
| LV mass, mg | 107 ± 4 | 103 ± 5 | 91 ± 4* | 92 ± 3* |
| Heart rate, beats/min | 527 ± 12 | 499 ± 15 | 492 ± 12 | 500 ± 14 |
Values are means ± SE. LVID, left ventricular internal diameter; LVPW, left ventricular posterior wall thickness; LVAW, left ventricular anterior wall thickness; d, diastole; s, systole; LV Vol, left ventricular volume = [7.0/(2.4 + LVID)] × LVID3 for either diastole or systole; EF, ejection fraction = (LV Vol;d − LV Vol;s)/LV Vol;d; FS, fractional shortening = (LVID;d − LVID;s)/(LVID;d); and LV mass = 0.84 [(LVID;d + LVPW;d + LVAW;d)3 − LVID;d3].
P < 0.05;
P < 0.005;
P < 0.0005 compared with WT.
P < 0.05 compared with TM180.
RSK3 knockout had little effect on the heart in the absence of the TM180 transgene. Although RSK3 knockout did not prevent the decrease in size of the TM180 left ventricle and its myocytes or inhibit the atrial enlargement (Fig. 1, B–D, and Tables 1 and 2), an improvement in cardiac function upon RSK3 knockout was suggested by the fact that the wet lung weight of TM180;RSK3−/− mice (8.16 ± 0.20 mg/mm tibial length) was significantly decreased compared with that for mice expressing only the TM180 allele (9.19 ± 0.27 mg/mm tibial length) and, in fact, was comparable to the wet lung weight of wild-type mice (8.21 ± 0.16 mg/mm tibial length).
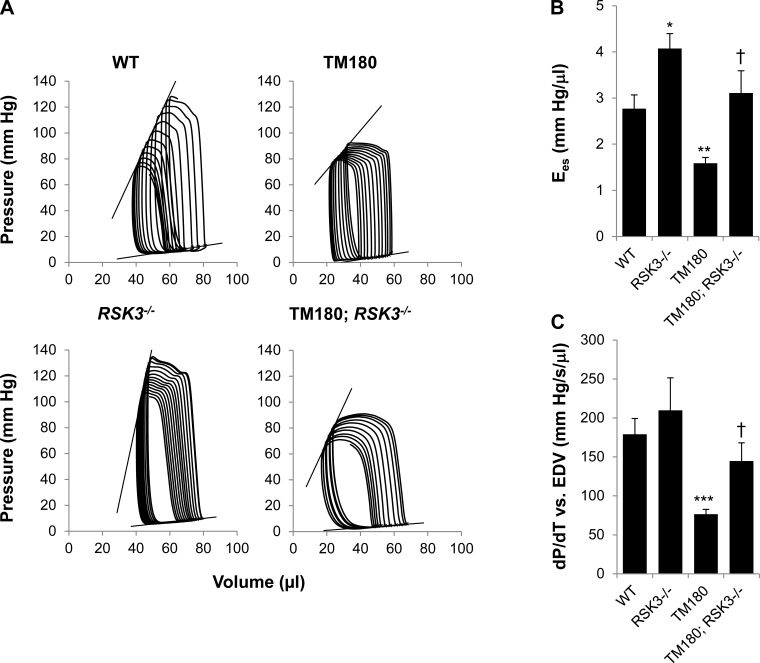
To determine why the cardiac insufficiency of the TM180 mouse was improved by RSK3 knockout, we studied these mice via left ventricular catheterization (Figs. 3 and 4). Hemodynamics measurements revealed that the TM180 allele induced both systolic and diastolic dysfunction that was improved in TM180;RSK3−/− mice (Table 3). TM180 hearts had decreased contractility compared with wild-type mice (Fig. 4), as evident by a significant decrease in end-systolic elastance (the slope of the end-systolic pressure-volume relationship; 1.6 ± 0.1 vs. 2.8 ± 0.3 mmHg/μl for TM180 vs. WT) and dP/dtmax vs. EDV (the slope of the maximum rate of left ventricular pressure rise vs. end-dystolic volume relationship; 76 ± 6 vs. 179 ± 20 mmHg·s−1·μl−1 for TM180 vs. WT). These indexes, as well as preload recruitable stroke work, were significantly increased upon crossing to the RSK3 knockout, becoming comparable to that for wild-type mice (Table 3).
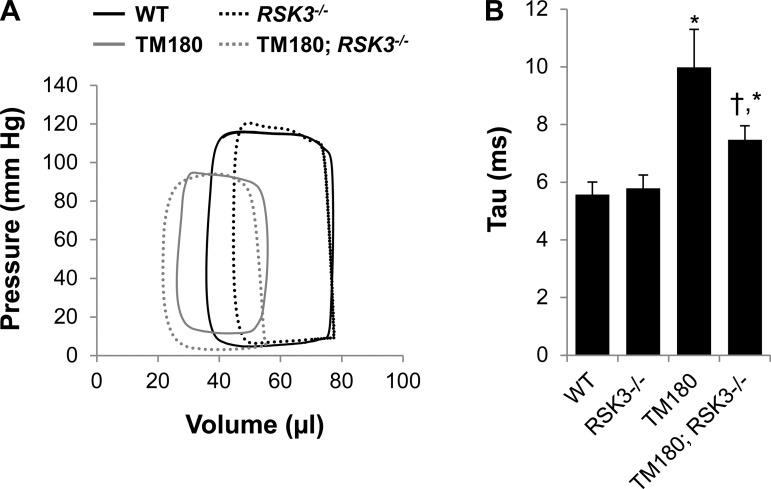
Fig. 3.
Diastolic dysfunction in TM180 mice was improved by RSK3 knockout. A: representative pressure-volume loops for mice studied by left ventricular catheterization with closed chest. B: tau (Weiss's method). Values are means ± SE; n = 5–12. *P < 0.05 compared with WT cohort. †P < 0.05 compared with TM180 cohort. See Table 3 for complete hemodynamic values.
Fig. 4.
Systolic dysfunction in TM180 mice was improved by RSK3 knockout. A: representative pressure-volume loops for mice studied by left ventricular catheterization to acquire preload-independent parameters. Loop series were obtained by inferior vena cava compression following a limited right thoracotomy. B: end-systolic elastance (Ees) is the slope of the end-systolic pressure-volume relationship. C: slope obtained by regression of maximum rate of left ventricular pressure rise (dP/dtmax) vs. end-diastolic volume (EDV). Like Ees, this index for contractility was reduced in TM180 mice but normalized by RSK3 knockout. Values are means ± SE; n = 5–12. *P < 0.05; **P < 0.005; ***P < 0.0005 compared with WT cohort. †P < 0.05 compared with TM180 cohort. See Table 3 for complete hemodynamic values.
Table 3.
Hemodynamics data
| WT | RSK3−/− | TM180 | TM180; RSK3−/− | |
|---|---|---|---|---|
| n | 5 | 5 | 8 | 12 |
| HR, beats/min | 605 ± 24 | 627 ± 17 | 585 ± 9 | 596 ± 11 |
| ESV, μl | 37 ± 4 | 42 ± 5 | 27 ± 4 | 22 ± 4* |
| EDV, μl | 69 ± 3 | 74 ± 5 | 65 ± 4 | 52 ± 3**† |
| SV, μl | 35 ± 3 | 37 ± 2 | 44 ± 3 | 35 ± 3† |
| CO, μl/min | 20,828 ± 1,493 | 23,006 ± 1,613 | 25,810 ± 1,950 | 20,454 ± 1,681 |
| Ea, mmHg/μl | 3.9 ± 0.3 | 3.5 ± 0.5 | 2.2 ± 0.2*** | 2.9 ± 0.3 |
| Systolic function | ||||
| EF, % | 51.1 ± 4.6 | 51.0 ± 3.7 | 67.1 ± 4.1* | 67.7 ± 5.1 |
| dP/dtmax, mmHg/s | 13,697 ± 1,791 | 13,071 ± 1,252 | 10.130 ± 895 | 10,978 ± 515 |
| ESP, mmHg | 130 ± 6 | 125 ± 9 | 94 ± 4** | 92 ± 4*** |
| Ees. mmHg/μl | 2.8 ± 0.3 | 4.1 ± 0.3* | 1.6 ± 0.1** | 3.1 ± 0.5† |
| dP/dtmax vs. EDV, mmHg·s−1·μl−1 | 179 ± 20 | 210 ± 42 | 76 ± 6*** | 145 ± 24† |
| PRSW, mmHg | 61 ± 3 | 95 ± 4*** | 50 ± 5 | 65 ± 3† |
| Diastolic function | ||||
| −dP/dtmin, mmHg/s | 12,689 ± 943 | 10,945 ± 655 | 7,192 ± 715** | 8,012 ± 482*** |
| EDP, mmHg | 10.7 ± 2.4 | 11.0 ± 3.8 | 15.3 ± 3.3 | 8.9 ± 1.9 |
| P@dV/dtmax, mmHg | 5.0 ± 1.2 | 4.8 ± 1.1 | 12.7 ± 2.8 | 6.2 ± 1.5† |
| Tau, ms | 5.6 ± 0.4 | 5.8 ± 0.5 | 10.0 ± 1.3* | 7.5 ± 0.5*† |
| EDPVR, mmHg/μl | 0.14 ± 0.04 | 0.16 ± 0.03 | 0.29 ± 0.12 | 0.21 ± 0.07 |
Values are means ± SE. HR, heart rate; ESV, end-systolic volume; EDV, end-diastolic volume; SV, stroke volume; CO, cardiac output; Ea, arterial elastance; EF, ejection fraction; dP/dtmax, maximum value of the pressure derivative; ESP, end-systolic pressure; Ees, end-systolic elastance; dP/dtmax vs. EDV, slope of the relationship between dP/dtmax and EDV; PRSW, preload-recruitable stroke work; −dP/dtmin, opposite of minimum value of the pressure derivative; EDP, end-diastolic pressure; P@dV/dtmax, pressure at the point where the maximum volume derivative occurs (fastest filling); tau, time constant for isovolumic relaxation (Weiss's method); EDPVR, slope of end-diastolic pressure-volume relationship.
P < 0.05;
P < 0.005;
P < 0.0005 compared with WT.
P < 0.05 compared with TM180.
The TM180 mouse also exhibited diastolic dysfunction as revealed by a significantly increased tau (Weiss's method; Fig. 3B; 10.0 ± 1.3 vs. 5.6 ± 0.4 ms for TM180 vs. WT) and dP/dtmin (Table 3; −7,192 ± 715 vs. −12,689 ± 943 mmHg/s for TM180 vs. WT), two indexes for ventricular relaxation. In addition, diastolic pressures and the end-diastolic pressure-volume relationship tended to be increased in the presence of the mutant tropomyosin. All of these parameters tended to be improved by RSK3 knockout, with both tau (7.5 ± 0.5 ms for TM180;RSK3−/−) and diastolic pressure during rapid ventricular filling (12.7 ± 2.8 vs. 6.2 ± 1.5 mmHg for TM180 vs. TM180;RSK3−/−) significantly different between the TM180 and TM180;RSK3−/− cohorts. Taken together, these data suggest that RSK3 is involved in the remodeling contributing to the cardiac insufficiency present in the TM180 mouse, including both the diminished contractility during systole and the diminished compliance during diastole of the left ventricle.
Although no RSK3 substrates have been validated in the heart, there are many candidate targets for RSK family members that are expressed in myocytes (2). Active RSK3 is thought to regulate transcription because it is enriched in the nucleus (2). This idea is consistent with our previous observations that the changes in gene expression induced by pressure overload were prevented by RSK3 knockout (14). Using NanoString technology, we surveyed the expression in the left ventricle of a panel of genes involved in cardiac function and remodeling (Table 4). As indicated by ANOVA, overall expression was significantly different between the TM180 cohort and the wild-type and TM180;RSK3−/− cohorts, but not between the wild-type cohort and the RSK3−/− and TM180;RSK3−/− cohorts. Post hoc testing revealed that TM180 transgene had mainly an effect on the expression of genes involved in cardiac fibrosis. Profibrotic genes significantly induced by the TM180 transgene included Tgfb2 (transforming growth factor-β2, TGF-β2), Postn (periostin), the collagen genes Col1a1, Col1a2, Col6a1, and Col8a1, Fgl2 (fibrinogen-like protein 2), Mmp2 (matrix metallopeptidase 2), Pcolce (procollagen C-endopeptidase enhancer), and Rtn4 (Nogo). The expression of all of the profibrotic genes was less following RSK3 knockout, although with varying statistical significance.
Table 4.
Gene expression data
| Gene | Protein | WT | RSK3−/− | TM180 | TM180;RSK3−/− |
|---|---|---|---|---|---|
| Related to interstitial fibrosis | |||||
| Col1a1 | Collagen type I α1 | 1.00 ± 0.13 | 1.08 ± 0.08 | 1.42 ± 0.07* | 1.02 ± 0.09† |
| Col1a2 | Collagen type I α2 | 1.00 ± 0.12 | 1.14 ± 0.07 | 1.47 ± 0.05** | 1.22 ± 0.11 |
| Col3a1 | Collagen type III α1 | 1.00 ± 0.17 | 1.11 ± 0.10 | 1.37 ± 0.12 | 1.04 ± 0.08† |
| Col5a1 | Collagen type V α1 | 1.00 ± 0.13 | 1.08 ± 0.09 | 1.11 ± 0.08 | 0.84 ± 0.09 |
| Col6a1 | Collagen type VI α1 | 1.00 ± 0.12 | 1.08 ± 0.04 | 1.33 ± 0.09* | 0.95 ± 0.09† |
| Col8a1 | Collagen type VIII α1 | 1.00 ± 0.24 | 0.95 ± 0.11 | 2.08 ± 0.31* | 1.73 ± 0.13 |
| Tgfb1 | Transforming growth factor β1 | 1.00 ± 0.12 | 1.11 ± 0.08 | 1.11 ± 0.09 | 0.98 ± 0.12 |
| Tgfb2 | Transforming growth factor β2 | 1.00 ± 0.21 | 0.97 ± 0.10 | 2.61 ± 0.56* | 1.96 ± 0.24 |
| Postn | Periostin | 1.00 ± 0.23 | 1.07 ± 0.08 | 2.48 ± 0.30** | 1.76 ± 0.16 |
| Fbn1 | Fibrillin-1 | 1.00 ± 0.19 | 1.10 ± 0.14 | 1.31 ± 0.12 | 0.92 ± 0.13 |
| Fgl2 | Fibrinogen-like protein 2 | 1.00 ± 0.25 | 1.16 ± 0.13 | 1.99 ± 0.25* | 1.44 ± 0.11 |
| Fn1 | Fibronectin 1 | 1.00 ± 0.14 | 1.07 ± 0.04 | 1.33 ± 0.10 | 0.92 ± 0.12† |
| Mfap5 | Microfibrillar associated protein 5 | 1.00 ± 0.21 | 1.07 ± 0.16 | 1.74 ± 0.35 | 1.52 ± 0.17 |
| Mmp2 | Matrix metallopeptidase 2 | 1.00 ± 0.12 | 1.09 ± 0.09 | 1.63 ± 0.15* | 1.23 ± 0.10† |
| Pcolce | Procollagen C-endopeptidase enhancer | 1.00 ± 0.08 | 1.14 ± 0.03 | 1.31 ± 0.09* | 1.04 ± 0.10 |
| Rtn4 | Reticulon 4 (a.k.a Nogo) | 1.00 ± 0.20 | 1.18 ± 0.09 | 1.55 ± 0.11* | 1.19 ± 0.09† |
| Srf | Serum response factor | 1.00 ± 0.16 | 1.17 ± 0.17 | 1.24 ± 0.14 | 0.90 ± 0.09 |
| Sarcomeric proteins | |||||
| Acta1 | Skeletal muscle α-actin | 1.00 ± 0.34 | 0.78 ± 0.17 | 1.16 ± 0.29 | 0.81 ± 0.15 |
| Actc1 | Cardiac muscle α-actin | 1.00 ± 0.11 | 1.05 ± 0.09 | 0.88 ± 0.06 | 0.61 ± 0.09† |
| Myh6 | Cardiac muscle α-myosin heavy chain | 1.00 ± 0.11 | 1.10 ± 0.16 | 3.39 ± 0.50** | 2.26 ± 0.37 |
| Myh7 | Cardiac muscle β-myosin heavy chain | 1.00 ± 0.24 | 0.47 ± 0.10 | 1.95 ± 0.48 | 1.30 ± 0.19 |
| Tnni3 | Cardiac muscle troponin I | 1.00 ± 0.09 | 1.09 ± 0.10 | 0.91 ± 0.13 | 0.65 ± 0.07 |
| Tnnt2 | Cardiac muscle troponin T | 1.00 ± 0.16 | 1.14 ± 0.11 | 0.88 ± 0.11 | 0.66 ± 0.06 |
| Capg | Gelsolin-like capping protein | 1.00 ± 0.10 | 1.08 ± 0.02 | 1.29 ± 0.11 | 1.16 ± 0.10 |
| Related to calcium cycling | |||||
| Atp2a2 | Sarco(endo)plasmic reticulum Ca2+-ATPase 2 | 1.00 ± 0.17 | 1.15 ± 0.10 | 0.88 ± 0.09 | 0.52 ± 0.09† |
| Cacna1c | L-type channel channel subunit α1c | 1.00 ± 0.18 | 1.13 ± 0.15 | 0.98 ± 0.08 | 0.59 ± 0.11† |
| Pln | Phospholamban | 1.00 ± 0.25 | 1.18 ± 0.15 | 1.08 ± 0.16 | 0.86 ± 0.09 |
| Signal transduction | |||||
| Adra1a | α1-Adrenergic receptor | 1.00 ± 0.16 | 1.39 ± 0.26 | 1.09 ± 0.21 | 0.67 ± 0.09 |
| Adrb1 | β1-Adrenergic receptor | 1.00 ± 0.13 | 1.23 ± 0.11 | 0.97 ± 0.15 | 0.66 ± 0.08 |
| Adrb2 | β2-Adrenergic receptor | 1.00 ± 0.16 | 1.52 ± 0.22 | 1.43 ± 0.20 | 1.51 ± 0.13 |
| Akap6 | mAKAP | 1.00 ± 0.21 | 1.04 ± 0.12 | 1.24 ± 0.11 | 0.75 ± 0.09† |
| Dusp4 | Dual-specificity protein phosphatase 4 | 1.00 ± 0.23 | 1.19 ± 0.19 | 1.11 ± 0.19 | 0.80 ± 0.12 |
| Mapk1 | Extracellular signal-regulated kinase 2 | 1.00 ± 0.19 | 1.14 ± 0.09 | 1.13 ± 0.13 | 0.80 ± 0.09 |
| Mapk3 | Extracellular signal-regulated kinase 1 | 1.00 ± 0.09 | 1.17 ± 0.07 | 1.06 ± 0.10 | 0.85 ± 0.07 |
| Mapk7 | Extracellular signal-regulated kinase 5 | 1.00 ± 0.18 | 1.36 ± 0.11 | 1.80 ± 0.14* | 1.33 ± 0.17 |
| Rps6ka1 | Ribosomal S6 kinase 1 | 1.00 ± 0.14 | 1.14 ± 0.05 | 0.81 ± 0.08 | 0.83 ± 0.09 |
| Rps6ka3 | Ribosomal S6 kinase 2 | 1.00 ± 0.25 | 1.14 ± 0.13 | 1.31 ± 0.12 | 1.03 ± 0.07 |
| Fhl1 | Four and a half LIM domains protein 1 | 1.00 ± 0.19 | 1.07 ± 0.08 | 1.19 ± 0.11 | 0.81 ± 0.06† |
| Rcan1 | Regulator of calcineurin 1 | 1.00 ± 0.19 | 1.04 ± 0.08 | 1.13 ± 0.12 | 0.93 ± 0.07 |
| Other | |||||
| Nppa | Atrial natriuretic factor | 1.00 ± 0.32 | 0.95 ± 0.28 | 3.74 ± 1.73 | 4.26 ± 0.97 |
| Nppb | Brain natriuretic factor | 1.00 ± 0.16 | 0.90 ± 0.04 | 1.23 ± 0.29 | 1.55 ± 0.26 |
| Max | MYC associated factor X | 1.00 ± 0.15 | 1.16 ± 0.07 | 1.18 ± 0.10 | 0.81 ± 0.13 |
Total mouse left ventricular RNA was assayed using NanoString technology for the indicated mRNAs. All data (means ± SE) are fold expression normalized to the mean for the WT cohort; n = 6 for all cohorts. ANOVA (2 factor with replication) was significant for TM180 vs. WT (P = 0.005) and TM180;RSK3−/− vs. TM180 (P = 0.0002), but not for WT vs. TM180;RSK3−/− (P = 0.40) and WT vs. RSK3−/− (P = 0.16). Uncorrected post hoc test P values (2-sided, unpaired t-tests) are provided for TM180 vs. WT (
P < 0.05) and TM180;RSK3−/− vs. TM180 (
P < 0.05).
P < 0.005.
Few other genes were affected by either the TM180 transgene and/or RSK3 knockout. Although it is unclear why Myh6 mRNA (α-myosin heavy chain) was upregulated in both the presence and absence of RSK3 by TM180, the sustained increased expression of Nppa (atrial natriuretic factor) was consistent with the incomplete reversal of TM180 diastolic dysfunction by RSK3 knockout, as well as the continued atrial hypertrophy. Mapk7 (ERK5), whose cardiac myocyte-specific knockout blocked the interstitial fibrosis associated with pressure overload (11), was also significantly induced in expression by the TM180 allele. RSK3 knockout had no significant effect, as shown by ANOVA, on the expression of the gene panel in the absence of the TM180 transgene. In addition to the profibrotic genes, the expression levels in the TM180;RSK3−/− left ventricle of Actc1 (cardiac muscle α-actin), Atp2a2 [sarco(endo)plasmic reticulum Ca2+-ATPase 2, SERCA2A], Cacna1c (L-type Ca2+ channel subunit α1c), AKAP6 (muscle A-kinase anchoring protein, mAKAP), and Fhl1 (four and a half LIM domains protein 1) were significantly different from TM180 tissue, albeit with uncertain functional significance. Two of these genes, AKAP6 and Fhl1, do encode proteins that regulate pathological remodeling (12, 23). In fact, RSK3 anchoring by mAKAP is required for concentric myocyte hypertrophy (14).
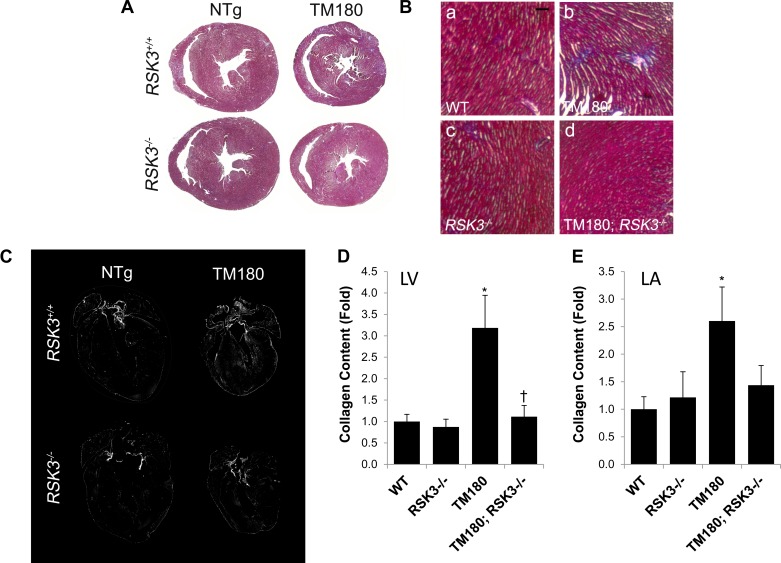
Because of the RSK3-dependent induction of fibrosis-related genes by the TM180 allele, we assayed for additional evidence of pathological remodeling in the mutant hearts. As surveyed by TUNEL assay, there was no detectable increase in cell death associated with the TM180 transgene at 16 wk of age (∼10−4 TUNEL-positive nuclei for all cohorts; data not shown). However, trichrome staining of the TM180 hearts did reveal a patchy interstitial fibrosis in the myocardium not present in wild-type mice that was greatly reduced in the absence of RSK3 (Fig. 5, A and B). Images obtained by linearly polarized light microscopy of picrosirius red-stained sections showed that fibrosis was induced by the TM180 transgene in all four cardiac chambers (Fig. 5, C–E). Quantitation of the picrosirius red images showed that fibrosis was induced 3.2-fold in the left ventricle and 2.6-fold in the left atria. Importantly, TM180; RSK3−/− mice lacked fibrosis in both the ventricle and the atria. Together with the gene expression data, these results suggest that RSK3-dependent interstitial fibrosis contributes to the cardiac insufficiency induced by the TM180 mutation.
Fig. 5.
RSK3 is required for TM180-induced interstitial fibrosis. A: trichrome staining of transverse sections. B: higher magnification of trichrome-stained sections. Bar, 100 μm. C: images of picrosirius red-stained heart sections were acquired by linearly polarized light microscopy at ×40 magnification and assembled as a montage using Photoshop. Each montage represents at least 16 overlapping images. Collagen is bright in these black-and-white images. D: collagen detected by picrosirius red staining within the left ventricle (LV). Values are means ± SE; n = 14–15. E: collagen detected by picrosirius red staining within the left atrium (LA). Values are means ± SE; n = 7–14. *P < 0.05 compared with WT cohort. †P < 0.05 compared with TM180 cohort.
DISCUSSION
Tropomyosin is a 284-amino acid residue protein identical in mice and humans that is found as a coiled-coil dimer bound to the major groove of actin thin filaments and that together with the troponin complex controls actin-myosin interactions during the contractile cycle. Binding of Ca2+ to troponin C results in a conformational shift in the position of tropomyosin and troponin I that permits myosin head binding to filamentous actin. Located within the troponin T binding site on tropomyosin, the TM180 mutation affects the local flexibility of the molecule and the response of the complex to Ca2+ (4). The mutation is thought to induce cardiomyopathy as a result of the increased Ca2+ sensitivity and increased maximum tension generation of TM180 filaments (20).
When expressed in transgenic FVB/N mice under the control of the cardiac myocyte-selective α-myosin heavy chain promoter, TM180 induces a severe phenotype (20). These mice exhibit concentric left ventricular hypertrophy with extensive fibrosis and grossly enlarged atria, obvious diastolic dysfunction, and decreased survival to less than 6 mo of age. When the same mutation was expressed in C57BL/6 mice, a mild phenotype resulted, with no fibrosis, a <10% smaller heart, increased fractional shortening, and diastolic but not systolic dysfunction at 1 yr of age (17). We have found that in a mixed C57BL/6:FVB/N background, a third phenotype intermediate in severity was induced by the TM180 transgene. This phenotype featured a small heart with small myocytes, interstitial fibrosis, and both diastolic and systolic dysfunction, with evidence of cardiac insufficiency provided by the presence of mild pulmonary edema. These strain-dependent differences in phenotype are reminiscent of the phenotypic variability present in humans genotype positive for HCM mutations (16). Of the affected members of the original TM180 family MZ, only 7/15 had left ventricular hypertrophy by echocardiography or at autopsy, with the remainder having only clinical or electrocardiographic findings of heart disease, including various arrhythmias (29).
Other HCM mutant mice have been found to have a similar phenotype to the TM180 C57BL/6:FVB/N mouse. Like the TM180 model, mice bearing a truncated cardiac troponin T lacking the COOH-terminal 28 amino acid residues (cTnT ΔCT) had small left ventricles with ∼15% smaller myocytes and mild fibrosis, as well as atrial hypertrophy (26). Accordingly, cTnT ΔCT mice also displayed diastolic and systolic dysfunction. One difference between the two mutant lines is that we did not observe an increase in apoptosis due to the TM180 transgene, whereas the cTnT ΔCT mouse had a higher rate of cell death and ∼10% fewer myocytes, as well as decreased survival. cTnT transgenic mice with an Arg92Gln mutation (cTnT R92Q) within the cTnT domain that binds tropomyosin also had smaller left ventricles and atrial hypertrophy (27). In addition, cTnT R92Q myocytes were smaller in width, and the ventricles contained patchy fibrosis. The phenotypes of these cTnT mutant mice correlated with the high frequency of sudden cardiac death and lack of significant hypertrophy in human patients bearing cTnT mutations (25). Thus it is clear that sarcomeric mutations that confer a higher thin-filament Ca2+ sensitivity can induce cardiomyopathy with or without myocyte hypertrophy. Although the extent of hypertrophy apparently depends on other genetic factors that vary among individual human family members or mouse strains, this observation suggests that the primary consequence of the change in sarcomere function is an increase in profibrotic signaling and/or myocyte death.
As an important and potentially early feature of HCM, interstitial fibrosis contributes to left ventricular dysfunction, arrhythmias, heart failure, and sudden cardiac death (9). Important factors known to promote interstitial fibrosis include TGF-β and periostin, as shown for HCM transgenic mouse models expressing α-myosin heavy chain Arg403Gln and Arg719Trp mutant proteins (28). We found that interstitial fibrosis was induced in the TM180 mice of mixed C57BL/6:FVB/N background, both as detected histologically and by assay of relevant gene expression, including the genes for TFG-β2 and periostin. This fibrosis presumably contributed to both the diastolic and systolic dysfunction observed by hemodynamics analysis, as well as to the mild pulmonary edema measured postmortem.
We recently reported that RSK3 was increased in expression and activated by α-adrenergic stimulation of neonatal rat ventricular myocytes in vitro (14). RSK3 was required for the concentric hypertrophy of cardiac myocytes in mice stressed by pressure overload or chronic catecholamine infusion. Although interstitial fibrosis was minimal in the mice subjected to transverse aortic constriction for two wk, the significant induction of Tgfb2 and Col6a1 gene expression by pressure overload was attenuated by RSK3 knockout. Although we assumed that the decrease in Tgfb2 and Col6a1 expression was secondary to the lack of hypertrophy in the RSK3 knockout stressed by transverse aortic constriction, the data obtained for the TM180 mouse now suggest that RSK3 contributes to profibrotic signaling independently of myocyte hypertrophy.
RSK family members are activated by ERK MAP kinases, including ERKs 1, 2, and 5 (14). ERK1/2 has been shown to be activated in two HCM models, the TM180 FVB/N mouse (6) and the β-myosin heavy chain Q403 rabbit (18), potentially contributing to pathological remodeling. We found using a phospho-specific antibody that total RSK was activated threefold in the TM180 FVB/N heart (Li J, unpublished observations). Consistent with the milder phenotype, RSK activation was much less when TM180 was present in a mixed background (61% increase as detected by the phospho-specific RSK antibody; data not shown). Accordingly, we were unable to detect either a significant increase in RSK3 total protein (Fig. 1A) or phosphorylation (data not shown) in total heart extracts. Nevertheless, RSK3 knockout inhibited the induction of profibrotic gene expression and interstitial fibrosis by the TM180 transgene. Importantly, RSK3 knockout significantly improved both the diastolic and systolic cardiac function of the TM180 mixed C57BL/6:FVB/N mouse, as well as preventing the development of mild pulmonary edema, a cardinal sign of cardiac insufficiency. These results imply that either baseline RSK3 activity is permissive for the pathologic process or that a subset of total cardiac RSK3 restricted to a particular cell type or intracellular compartment is relevant to the TM180 phenotype.
The present findings complement our previous observation that RSK3 is required for pathological cardiac hypertrophy despite the higher level of expression of other RSK isoenzymes in the heart (14). A potential mechanism permitting RSK3 to play a significant, unique role in intracellular signaling despite its low level of expression is the anchoring of RSK3 through its unique NH2-terminal domain to specifically localized scaffold proteins such as mAKAP (14). Future work is required to identify which substrate(s) mediate RSK3 signaling in cardiac fibrosis. Given the enrichment of active RSK3 in the nucleus (2), it is possible that the primary function of RSK3 is to regulate gene expression.
Regardless of the underlying mutation, there are no definitive treatments available for HCM. Current therapy for HCM includes the use of beta-blockers and Ca2+ and Na+ channel blockers to modulate cardiac function, surgical myectomy or alcohol ablation in severe cases of outflow obstruction, and placement of an implantable cardioverter-defibrillator in those at risk for sudden cardiac death (7). Increasing Ca2+ reuptake during diastole to compensate for the increased Ca2+ sensitivity of the contractile machinery has been proposed as a therapeutic strategy for HCM. Adenovirus-based expression of SERCA2A delivered via intraventricular injection of neonatal mice attenuated the hypertrophy and fibrosis present at 3–4 mo of age in the TM180 FVB/N mouse (19). In addition, genetic knockout of phospholamban, the endogenous inhibitor of SERCA2A and lusitropy, conferred long-term improvement in fibrosis, hypertrophy, and cardiac function (6). That RSK3 knockout both improved the cardiac function of the TM180 mouse and prevented the cardiac hypertrophy associated with pressure overload and catecholamine infusion suggests that RSK3 should be considered as a new therapeutic target in both fibrotic and hypertrophic cardiac diseases. RSK3 is an attractive therapeutic target, since global, constitutive RSK3 knockout is benign in wild-type mice (14). In particular, we suggest that the development of RSK3-specific inhibitors may provide an opportunity to prevent the fibrosis and cardiac dysfunction associated with HCM, potentially decreasing the risk of sudden death for these patients.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grant HL075398 (to M. S. Kapiloff) and an American Heart Association Scientific Development Grant (to J. Li).
DISCLOSURES
Catherine L. Passariello, Michael D. Kritzer, Jinliang Li, and Michael S. Kapiloff hold a provisional patent related to the use of RSK3 inhibitors to treat heart failure.
AUTHOR CONTRIBUTIONS
C.L.P., M.G., and M.S.K. conception and design of research; C.L.P., M.G., M.D.K., H.T., Z.C., and F.R. performed experiments; C.L.P., M.G., J.L., and M.S.K. analyzed data; C.L.P., M.G., M.S., J.L., and M.S.K. interpreted results of experiments; C.L.P., M.G., and M.S.K. prepared figures; C.L.P. and M.S.K. drafted manuscript; C.L.P., D.F.W., and M.S.K. edited and revised manuscript; C.L.P., M.G., D.F.W., M.S., J.L., and M.S.K. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Toumy Guettouche, Loida Navarro, and Yoslayma Cardentey of the Oncogenomics Core Facility, Sylvester Cancer Center, University of Miami School of Medicine, for their assistance in mRNA analyses.
REFERENCES
- 1.Amirak E, Fuller SJ, Sugden PH, Clerk A. p90 Ribosomal S6 kinases play a significant role in early gene regulation in the cardiomyocyte response to Gq-protein-coupled receptor stimuli, endothelin-1 and alpha1-adrenergic receptor agonists. Biochem J 450: 351–353, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol 9: 747–758, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Balza RO, Jr, Misra RP. Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J Biol Chem 281: 6498–6510, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Bing W, Knott A, Redwood C, Esposito G, Purcell I, Watkins H, Marston S. Effect of hypertrophic cardiomyopathy mutations in human cardiac muscle alpha-tropomyosin (Asp175Asn and Glu180Gly) on the regulatory properties of human cardiac troponin determined by in vitro motility assay. J Mol Cell Cardiol 32: 1489–1498, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Edgley AJ, Krum H, Kelly DJ. Targeting fibrosis for the treatment of heart failure: a role for transforming growth factor-beta. Cardiovasc Ther 30: e30–e40, 2012 [DOI] [PubMed] [Google Scholar]
- 6.Gaffin RD, Pena JR, Alves MS, Dias FA, Chowdhury SA, Heinrich LS, Goldspink PH, Kranias EG, Wieczorek DF, Wolska BM. Long-term rescue of a familial hypertrophic cardiomyopathy caused by a mutation in the thin filament protein, tropomyosin, via modulation of a calcium cycling protein. J Mol Cell Cardiol 51: 812–820, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, Naidu SS, Nishimura RA, Ommen SR, Rakowski H, Seidman CE, Towbin JA, Udelson JE, Yancy CW, and American College of Cardiology Foundation/American Heart Association Task Force on Practice G. 2011 ACCF/AHA Guideline for the Diagnosis and Treatment of Hypertrophic Cardiomyopathy: a Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines Developed in Collaboration with the American Association for Thoracic Surgery, American Society of Echocardiography, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J Am Coll Cardiol 58: e212–e260, 2011 [DOI] [PubMed] [Google Scholar]
- 8.Hanlon M, Sturgill TW, Sealy L. ERK2- and p90(Rsk2)-dependent pathways regulate the CCAAT/enhancer-binding protein-beta interaction with serum response factor. J Biol Chem 276: 38449–38456, 2001 [DOI] [PubMed] [Google Scholar]
- 9.Ho CY, Lopez B, Coelho-Filho OR, Lakdawala NK, Cirino AL, Jarolim P, Kwong R, Gonzalez A, Colan SD, Seidman JG, Diez J, Seidman CE. Myocardial fibrosis as an early manifestation of hypertrophic cardiomyopathy. N Engl J Med 363: 552–563, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janknecht R, Hipskind RA, Houthaeve T, Nordheim A, Stunnenberg HG. Identification of multiple SRF N-terminal phosphorylation sites affecting DNA binding properties. EMBO J 11: 1045–1054, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura TE, Jin J, Zi M, Prehar S, Liu W, Oceandy D, Abe JI, Neyses L, Weston AH, Cartwright EJ, Wang X. Targeted deletion of the extracellular signal-regulated protein kinase 5 attenuates hypertrophic response and promotes pressure overload-induced apoptosis in the heart. Circ Res 106: 961–970, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kritzer MD, Li J, Dodge-Kafka K, Kapiloff MS. AKAPs: the architectural underpinnings of local cAMP signaling. J Mol Cell Cardiol 52: 351–358, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le NT, Takei Y, Shishido T, Woo CH, Chang E, Heo KS, Lee H, Lu Y, Morrell C, Oikawa M, McClain C, Wang X, Tournier C, Molina CA, Taunton J, Yan C, Fujiwara K, Patterson C, Yang J, Abe J. p90RSK targets the ERK5-CHIP ubiquitin E3 ligase activity in diabetic hearts and promotes cardiac apoptosis and dysfunction. Circ Res 110: 536–550, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Kritzer MD, Michel JJ, Le A, Thakur H, Gayanilo M, Passariello CL, Negro A, Danial JB, Oskouei B, Sanders M, Hare JM, Hanauer A, Dodge-Kafka K, Kapiloff MS. Anchored p90 ribosomal S6 kinase 3 is required for cardiac myocyte hypertrophy. Circ Res 112: 128–139, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Liu Z, Hu X, Ma K, Zhou C. Involvement of ERK-RSK cascade in phenylephrine-induced phosphorylation of GATA4. Biochim Biophys Acta 1823: 582–592, 2012 [DOI] [PubMed] [Google Scholar]
- 16.Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet 381: 242–255, 2013 [DOI] [PubMed] [Google Scholar]
- 17.Michele DE, Gomez CA, Hong KE, Westfall MV, Metzger JM. Cardiac dysfunction in hypertrophic cardiomyopathy mutant tropomyosin mice is transgene-dependent, hypertrophy-independent, and improved by beta-blockade. Circ Res 91: 255–262, 2002 [DOI] [PubMed] [Google Scholar]
- 18.Patel R, Nagueh SF, Tsybouleva N, Abdellatif M, Lutucuta S, Kopelen HA, Quinones MA, Zoghbi WA, Entman ML, Roberts R, Marian AJ. Simvastatin induces regression of cardiac hypertrophy and fibrosis and improves cardiac function in a transgenic rabbit model of human hypertrophic cardiomyopathy. Circulation 104: 317–324, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pena JR, Szkudlarek AC, Warren CM, Heinrich LS, Gaffin RD, Jagatheesan G, del Monte F, Hajjar RJ, Goldspink PH, Solaro RJ, Wieczorek DF, Wolska BM. Neonatal gene transfer of Serca2a delays onset of hypertrophic remodeling and improves function in familial hypertrophic cardiomyopathy. J Mol Cell Cardiol 49: 993–1002, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prabhakar R, Boivin GP, Grupp IL, Hoit B, Arteaga G, Solaro JR, Wieczorek DF. A familial hypertrophic cardiomyopathy alpha-tropomyosin mutation causes severe cardiac hypertrophy and death in mice. J Mol Cell Cardiol 33: 1815–1828, 2001 [DOI] [PubMed] [Google Scholar]
- 21.Rivera VM, Miranti CK, Misra RP, Ginty DD, Chen RH, Blenis J, Greenberg ME. A growth factor-induced kinase phosphorylates the serum response factor at a site that regulates its DNA-binding activity. Mol Cell Biol 13: 6260–6273, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tonjes M, Dunkel I, Sperling SR. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet 7: e1001313, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheikh F, Raskin A, Chu PH, Lange S, Domenighetti AA, Zheng M, Liang X, Zhang T, Yajima T, Gu Y, Dalton ND, Mahata SK, Dorn GW, 2nd, Brown JH, Peterson KL, Omens JH, McCulloch AD, Chen J. An FHL1-containing complex within the cardiomyocyte sarcomere mediates hypertrophic biomechanical stress responses in mice. J Clin Invest 118: 3870–3880, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spinale FG, Janicki JS, Zile MR. Membrane-associated matrix proteolysis and heart failure. Circ Res 112: 195–208, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tardiff JC. Thin filament mutations: developing an integrative approach to a complex disorder. Circ Res 108: 765–782, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tardiff JC, Factor SM, Tompkins BD, Hewett TE, Palmer BM, Moore RL, Schwartz S, Robbins J, Leinwand LA. A truncated cardiac troponin T molecule in transgenic mice suggests multiple cellular mechanisms for familial hypertrophic cardiomyopathy. J Clin Invest 101: 2800–2811, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tardiff JC, Hewett TE, Palmer BM, Olsson C, Factor SM, Moore RL, Robbins J, Leinwand LA. Cardiac troponin T mutations result in allele-specific phenotypes in a mouse model for hypertrophic cardiomyopathy. J Clin Invest 104: 469–481, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teekakirikul P, Eminaga S, Toka O, Alcalai R, Wang L, Wakimoto H, Nayor M, Konno T, Gorham JM, Wolf CM, Kim JB, Schmitt JP, Molkentin JD, Norris RA, Tager AM, Hoffman SR, Markwald RR, Seidman CE, Seidman JG. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-beta. J Clin Invest 120: 3520–3529, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thierfelder L, MacRae C, Watkins H, Tomfohrde J, Williams M, McKenna W, Bohm K, Noeske G, Schlepper M, Bowcock A, Vosberg HP, Seidman JG, Seidman C. A familial hypertrophic cardiomyopathy locus maps to chromosome 15q2. Proc Natl Acad Sci USA 90: 6270–6274, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]