Abstract
Human and animal studies show that suboptimal intrauterine environments lead to fetal programming, predisposing offspring to disease in later life. Maternal obesity has been shown to program offspring for cardiovascular disease (CVD), diabetes, and obesity. MicroRNAs (miRNAs) are small, noncoding RNA molecules that act as key regulators of numerous cellular processes. Compelling evidence links miRNAs to the control of cardiac development and etiology of cardiac pathology; however, little is known about their role in the fetal cardiac response to maternal obesity. Our aim was to sequence and profile the cardiac miRNAs that are dysregulated in the hearts of baboon fetuses born to high fat/high fructose-diet (HFD) fed mothers for comparison with fetal hearts from mothers eating a regular diet. Eighty miRNAs were differentially expressed. Of those, 55 miRNAs were upregulated and 25 downregulated with HFD. Twenty-two miRNAs were mapped to human; 14 of these miRNAs were previously reported to be dysregulated in experimental or human CVD. We used an Ingenuity Pathway Analysis to integrate miRNA profiling and bioinformatics predictions to determine miRNA-regulated processes and genes potentially involved in fetal programming. We found a correlation between miRNA expression and putative gene targets involved in developmental disorders and CVD. Cellular death, growth, and proliferation were the most affected cellular functions in response to maternal obesity. Thus, the current study reveals significant alterations in cardiac miRNA expression in the fetus of obese baboons. The epigenetic modifications caused by adverse prenatal environment may represent one of the mechanisms underlying fetal programming of CVD.
Keywords: maternal obesity, miRNA, cardiac, fibrosis
early life exposure to either an excess or a deficit in maternal nutrition has been shown to cause modulations in the offspring's body composition and cardiovascular and metabolic function as a result of developmental programming (45). The incidence of maternal overnutrition and obesity is rising rapidly worldwide, and the number of pregnant women who are overweight and obese has increased (83). Children exposed to maternal obesity and gestational diabetes during fetal life have a higher risk of insulin resistance (10), myocardial hypertrophy (117), congenital heart defects (74), and cardiovascular disease (61). Consistent with these human epidemiological data, animal models provide strong evidence that being born to an obese mother increases offspring risk of myocardial dysfunction including ventricular hypertrophy (37) and myocardial fibrosis (51). Although the effects of maternal nutrition on the offspring's epigenetic status are well documented (77), the nature of the fetal molecular pathways modified by maternal obesity remains unknown. There is currently much interest in the use of transcriptional profiling of RNA patterns from potentially affected tissues to identify key regulators involved in initiation and progression of fetal programming.
The recent discovery of microRNAs (miRNAs) has revealed a crucial layer of posttranscriptional gene regulation (73). MiRNAs are a class of small (18–25 nucleotides long) noncoding RNA molecules that posttranscriptionally regulate protein-coding mRNA in both plants and animals. More than 1,000 miRNAs have been identified, many of which are tissue specific and temporally regulated in their expression (38). MiRNAs act as governors of gene expression networks, thereby modifying complex cellular phenotypes in development and pathophysiology. MiRNAs mediate gene silencing by binding to specific target sites within the 3′-untranslated regions (UTR) of mRNA, which will either block the translation or bring about degradation of the transcripts. Many miRNAs are dysregulated in response to cellular stress and can modify essential cellular functions of proliferation, differentiation, and cell death (93). However, the role of miRNAs in fetal programming remains largely unstudied.
The baboon (Papio hamadryas) is a well-characterized nonhuman primate model for biological studies (21, 107). Nonetheless, very little is known about baboon miRNAs. Two recent studies have identified and profiled baboon liver miRNAs that are responsive to dietary fat and cholesterol (56). We and others have studied maternal and fetal baboon physiology in both normal pregnancy and following perturbations that led to developmental programming (65). We have shown that maternal overnutrition in sheep leads to impaired fetal cardiac function and altered insulin signaling (104). Our aim in the present study was to undertake a comprehensive sequencing and profiling analysis of miRNA expression in the hearts of baboon fetuses exposed to maternal overnutrition. We hypothesized that maternal overnutrition combined with high-fat diet and maternal obesity will affect the expression of cardiac miRNA in offspring, potentially changing the expression of key proteins in the heart.
MATERIALS AND METHODS
Animal care and maintenance.
All procedures were approved by the Texas Biomedical Research Institute (Texas Biomed) Institutional Animal Care and Use Committee and conducted in Association for Assessment and Accreditation of Laboratory Animal Care-approved facilities.
System for controlling and recording individual feeding.
The animals were fed between either 0700 and 0900 or 1100 and 1300 as described in detail elsewhere (89). At feeding time, all baboons exited their group cage and passed along a chute and into individual feeding cages. The weight of each baboon was obtained as she crossed an electronic scale system (GSE 665, GSE Scale Systems). The weight recorded was the mean of 50 individual measurements over 3 s. If the standard deviation of the weight measurement was >0.01 of the mean weight, the weight was automatically discarded, and the weighing procedure begun again. Water was continuously available in the feeding cages via individual waterers (Lixit, Napa, CA).
Dietary intervention.
Animals were fed Purina Monkey Diet and Monkey Diet Jumbo (Purina LabDiets, St. Louis, MO). Four months prior to pregnancy, healthy female baboons of similar age and weight were randomly assigned to regular (RD) or high-fat/high-fructose diet (HFD) regimen. The HFD contained 45% energy from fat, 4.62% from glucose, 5.64% from fructose, and 2.32% from sucrose with an energy content of 4.03 kcal/g and free access to a variety of high-fructose sodas. The RD contained 12% energy from fat, 0.29% from glucose, 0.32% from fructose with an energy content of 3.07 kcal/g. The amount of protein and all of the essential minerals and vitamins required for baboon were equalized for the RD and HFD regimens. The dietary intervention was carried through the pregnancy. Pregnant baboons underwent Cesarean sections at 165 days of gestation (0.9 gestation, term 184 days).
Cesarean sections.
Surgical procedures were performed by a fully certified MD or DVM, and postsurgical care was prescribed and monitored by a veterinarian. Cesarean sections were performed at 165 days gestation (0.9 gestation) by standard techniques that have been previously described in detail (22). All baboons were premedicated with ketamine hydrochloride (10 mg/kg im). After intubation, isoflurane (2%) was used to maintain a surgical plane of anesthesia throughout surgery. Following hysterotomy, the umbilical cord was identified and used for fetal exsanguination while the baboons were under general anesthesia as approved by the American Veterinary Medical Association Panel on Euthanasia. The placenta and fetus were removed from the uterus and immediately submitted for morphometric analyses and tissue sampling. Fetal morphometrics were obtained at necropsy. Postoperative maternal analgesia was by buprenorphine hydrochloride (Buprenorphine HCl injection; Hospira, Lake Forest, IL) 0.015 mg/kg/day split as two doses for 3 days. After recovery in individual cages, mothers were returned to their group housing.
Histology.
The excised hearts were washed in PBS, fixed overnight in 4% paraformaldehyde, and embedded in paraffin. After serial sectioning of hearts (apex to base), 7-μm sections were stained with Masson trichrome. Fibrosis areas within sections were measured by visualizing blue-stained areas, exclusive of staining that colocalized with perivascular or intramural vascular structures. Using ImageJ software (http://rsbweb.nih.gov/ij/), we used color-based thresholding to determine blue-stained areas and nonstained myocyte areas from each section. The percentage of total fibrotic area was calculated as the summed blue-stained areas divided by total ventricular area, as described previously (99).
Cardiomyocyte proliferation was assessed in 5 μm paraffin sections with a Ki-67 antibody (Leica Biosystems, Newcastle, UK). Sections were deparaffinized in xylenes, rehydrated through ethanol gradient solutions to PBS, and permeabilized in 0.1% Tween in PBS. Antigen retrieval was achieved using 0.01 M citrate buffer (with 0.1% Triton-X100, Sigma-Aldrich) in a microwave oven for 15 min. Endogenous peroxidase activity was inhibited with 1.5% H2O2/methanol for 10 min. The primary Ki-67 antibody was diluted 1:100 prior to use, and the tissue was incubated with the primary antibody overnight at 4°C in a humidified chamber. Detection was performed with the ABC kit (Vector Labs). The sections were counterstained by hematoxylin for 30 s. Negative controls were tissues that were not incubated with the primary antibody. The percentage of proliferating cells was calculated by dividing the number of Ki-67-positive nuclei by total nuclei and multiplying by 100.
RNA isolation.
Overall, this study used 11 fetal hearts: six from baboons born to RD-fed mothers (n = 3 males and 3 females) and five from baboons born to HFD-fed mothers (n = 3 females and 2 males). Total RNA was isolated with the use of RNAeasy kit (Qiagen). The integrity of RNA was tested by spectroscopic analysis and by resolving on denaturing formaldehyde gel.
miRNA sequencing and profiling.
RNA quality control, microRNA sequencing, profiling, and data analysis were performed by LC Sciences (Houston, TX). We sequenced and profiled baboon fetal cardiac miRNAs from RD and HFD groups (Gene Expression Omnibus accession number for the microarray data: GSE43323). Initially, RNA samples pooled from two RD and two HFD hearts (males and females) were processed to generate a cDNA library, which was then used for deep sequencing. The purified cDNA library was used for cluster generation on Illumina's Cluster Station and then sequenced on Illumina GAIIx following vendor's instruction for running the instrument. Raw sequencing reads were obtained using Illumina's Pipeline v1.5 software following sequencing image analysis by Pipeline Firecrest Module and base calling by Pipeline Bustard Module. The extracted sequencing reads were then used in the standard data analysis. Custom-made microarrays were used to profile the expression of fetal cardiac miRNAs in all 11 RNA samples (6 RD and 5 HFD).
RT-PCR.
For validation of the microarray data RT-PCR was performed. We reverse transcribed 5 ng of total RNA using the TaqMan MicroRNA Reverse Transcription Kit. Taqman reactions were conducted with commercially available validated primer/probe sets (Applied Biosystems) and normalized to U18 as internal control. The changes in the threshold cycle (CT) values were calculated by the equation CT = CT target − CT input. The fold differences were calculated by 2−(dCT) method.
Western blot analysis.
Potential miRNA targets were determined by Western blot analysis. The whole cell homogenates were isolated and resolved on 4–20% gradient SDS-polyacrylamide gels. After electrophoresis, the proteins were transferred to nitrocellulose membranes, the membranes were blocked with 5% skim milk in Tween-20/Tris-buffered saline and probed with mouse monoclonal anti-HIF-1α (BD Biosciences), anti-thrombospondin (TSP)-1 (R&D Systems), anti-connective tissue growth factor (CTGF) (Gentex), and anti-p53 (Novus). The blots were then incubated with horseradish peroxidase-conjugated secondary antibodies. Finally, the enhanced chemiluminescence (ECL) reaction was performed and the blots were visualized by G-Box (Syngene). The intensity of each signal spot was transformed into digital data with autobackground subtraction during spot density analysis with the Syngene GeneTools software.
Ingenuity Pathways Analysis.
MiRNA Target Filter Analysis and Core Analysis of miRNA Target Genes were performed by Ingenuity Pathways Analysis (IPA, Ingenuity Systems). This software analysis lists genes in the context of known biological response and regulatory networks as well as other higher-order response pathways. MiRNA targets that were associated with biological functions in the Ingenuity Pathways Knowledge Base were used in the analysis.
Statistics.
All data are expressed as means ± SE. The statistical significance of differences between experimental groups was determined by ANOVA and unpaired two-tailed Student's t-test. P values of <0.05 were considered statistically significant.
RESULTS
General characteristics of experimental animals.
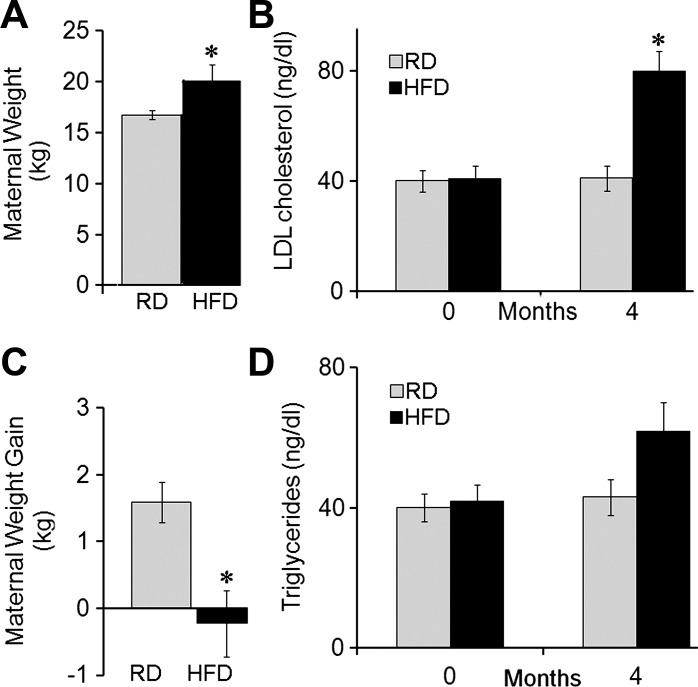
Four months after the beginning of the dietary regimen and prior to conception, dams fed the HFD exhibited increased body weight (Fig. 1A) and LDL-cholesterol (Fig. 1B) and showed a trend for a rise in blood triglycerides compared with RD (Fig. 1D). During pregnancy, dams fed the HFD lost nearly 0.2 kg in body weight, while RD dams gained 1.8 kg (Fig. 1C).
Fig. 1.
Physiological changes in regular diet (RD) and high-fat/high-fructose diet (HFD) mothers prior and during the pregnancy. The HFD increased maternal body weight (A) and blood LDL-cholesterol (B), showing a trend for a rise in blood triglycerides (D) prior to breeding. C: during pregnancy, HFD-fed mothers lost nearly 0.5 kg body wt, while RD-fed mothers gained 1.8 kg. *P < 0.05.
Fetal morphometrics at 165 days of gestation.
Table 1 presents morphometric measures on the placenta and fetuses of RD and HFD dams at cesarean section on day 165. Although there was no difference in placental weight or volume, placental efficiency, expressed as the fetal mass supported per unit placental mass, was reduced in HFD pregnancies compared with control RD (P < 0.02)(Table 1). Fetal body weight was reduced by 16% in HFD pregnancies. Brain and thymus weights were significantly increased in HFD fetuses compared with RD. No change in heart weight was detected (Table 1).
Table 1.
Effect of maternal overnutrition on fetal morphometric parameters at cesarean section (0.9 gestation)
| Groups | RD (n = 22) | HFD (n = 5) | P Value |
|---|---|---|---|
| Fetal measures | mean ± SE | mean ± SE | |
| Body weight, g | 806 ± 24 | 675 ± 37 | < 0.02* |
| Fetal measures as percent fetal weight | |||
| Placenta | 26.0 ± 0.80 | 32.1 ± 2.50 | < 0.01* |
| Brain | 10.4 ± 0.30 | 11.9 ± 0.60 | < 0.02* |
| Heart | 0.61 ± 0.02 | 0.63 ± 0.04 | 0.6 |
| Kidneys | 0.56 ± 0.02 | 0.59 ± 0.03 | 0.5 |
| Liver | 2.97 ± 0.06 | 2.97 ± 0.10 | 1.0 |
| Lungs | 2.16 ± 0.07 | 2.43 ± 0.10 | < 0.09 |
| Pancreas | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.9 |
| Thymus | 0.42 ± 0.02 | 0.52 ± 0.05 | 0.048* |
RD, regular diet; HFD, high-fat diet.
P < 0.05.
Heart histology.
Hematoxylin-and-eosin staining did not reveal any significant differences in myofiber orientation in fetal ventricular tissue from RD and HFD groups (not shown). However, Masson trichrome-stained cardiac sections (Fig. 2) showed increased myocardial fibrosis in HFD fetal hearts (22.05 ± 3.8%, n = 5) compared with RD hearts (2.1 ± 0.76%, n = 6; P = 0.003).
Fig. 2.
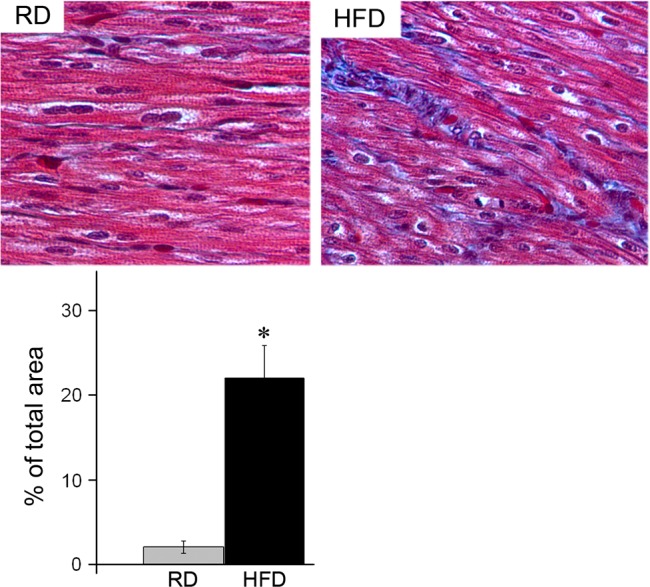
Phenotypic characterization of fetal hearts from RD and HFD baboons at the day 165 of gestation. Representative histology (top) and quantification of fibrosis (bottom) in heart sections from RD and HFD baboon fetuses. Masson trichrome staining; n = 5 for each group, *P < 0.05 vs. RD.
MiRNA sequencing in fetal baboon hearts.
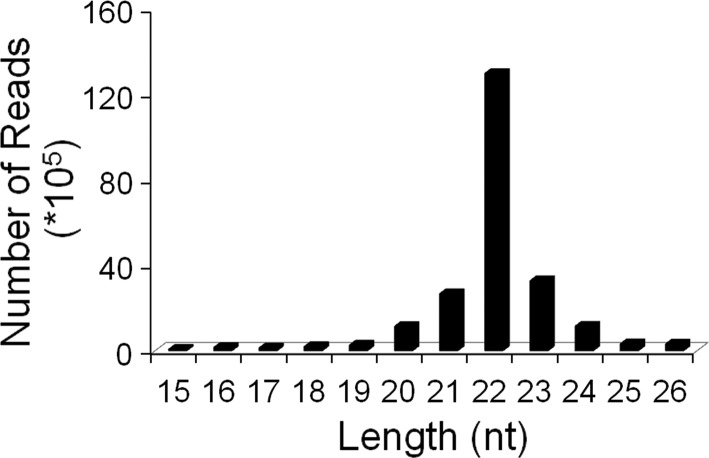
We isolated total RNA from the hearts of baboon fetuses. The RNA samples were checked for RNA quality control and processed to generate a cDNA library on which deep sequencing was performed. Overall, 27,857,063 sequence reads were obtained (Table 2). A total of 2,369,584 unique sequences were annotated to annotated small RNA sequences. The small RNA libraries exhibited a diverse size distribution of sequence reads that aligned to the human genome (Fig. 3). miRNAs were the most abundantly expressed small RNAs with 22,611,923 or 81% of total sequences. Other small noncoding RNAs such as small interfering RNAs, small nucleolar RNAs, small nuclear RNAs, and transfer RNAs comprised only 1% (318,109) of the total sequences. On average 19,708,547 sequences (or 87.2%) were mapped to miRBase. We discovered 961 miRNAs in the hearts from baboon fetuses: 601 (68%) were identical to human miRNAs; 23 were mapped to other mammalian miRNAs. Of these mapped miRNAs, 157 showed new genome location, and 180 baboon miRNAs were novel; of these 86 were unmapped to known miRNAs (Table 3).
Table 2.
Summary of standard data analysis results
| SequSeq, n | Mappable SequSeq, % | Unique Seq, n | Mappable Unique Seq, % | |
|---|---|---|---|---|
| Raw | 27,857,063 | 2,369,584 | ||
| Mapped to mRNA | 749,824 | 14,425 | ||
| Mapped to other RNAs: rRNA, tRNA, snRNA, snoRNA, | 318,109 | 9,810 | ||
| Mapped to RepBase | 5,537 | 304 | ||
| Total mappable for miRNA | 22,611,923 | 100 | 118,674 | 100 |
| Mapped to miRBase | 19,274,534 | 85.2 | 18,955 | 16 |
| Unmapped to miRBase | 2,912,223 | 12.9 | 96,136 | 81.0 |
| Mapped total | 19,274,534 | 85.2 | 18,955 | 16.0 |
| No hit | 3,337,389 | 14.8 | 90,723 | 84 |
rRNA, ribosomal RNA; tRNA, transfer RNA; snRNA, small nuclear RNA, snoRNA, small nucleolar RNA. RepBase is a database of prototypic sequences representing repetitive DNA from different eukaryotic species. MirBase is a searchable database of published miRNA sequences and annotation.
Fig. 3.
Length distribution of microRNA (miRNA)-mappable reads.
Table 3.
Known and predicted miRNAs
| Known miRNAs | Group | Unique miRs, n |
|---|---|---|
| Of Homo sapiens | group 1a | 601 |
| Of mammals but novel to Homo sapiens | group 1b | 23 |
| Of Homo sapiens and mammals, but with new genome locations | group 1c | 157 |
| Predicted miRs | ||
| Mapped to known mammal miRs and genome; within hairpins | group 2 | 17 |
| Mapped to known mammal miRs but unmapped to genome | group 3 | 77 |
| Unmapped to known miRs but mapped to genome and within hairpin | group 4 | 86 |
| Total (Unique miRs) | 961 |
microRNA: miRNA or miR.
Dysregulation of fetal cardiac miRNA expression in response to maternal obesity.
MiRNA expression profiling using microarrays is a powerful high-throughput tool capable of monitoring the regulatory networks of the entire genome. To identify regulatory networks involved into the fetal response to maternal obesity, miRNA microarray analysis was performed by LC Sciences. Comprehensive miRNA profiles were generated for 11 fetal hearts from baboons born to RD (n = 3 males and 3 females) or HFD (n = 3 females and 2 males)-fed mothers. The expression pattern of differentially expressed miRNAs is presented as a clustered heat map (Fig. 4). Overall, 80 miRNAs were altered in response to maternal obesity (P < 0.05). Of those, 55 miRNAs were upregulated and 25 downregulated. In total, 22 miRNAs (27.5%) mapped exclusively to human, nine miRNAs mapped to other mammalian species: three miRNAs to Bos taurus (bta-miRs-2436-3p, -1296, -2889), three to Canis familiaris (cfa-miRs-143, -135a-2, -127), one miRNA to Monodelphis domestica (mdo-miR-139), one to Mus musculus (mmu-miR-326), and one to Pongo pygmaeus (ppy-miR-638). The remaining 49 miRNAs were unmapped at the time of analysis. The greatest expression change, >16-fold upregulation, was found for hsa-miR-1296 in HFD hearts compared with RD. Other miRNAs demonstrating notable (>4-fold) nutrient sensitivity included overexpression of hsa-mir-30a, hsa-mir-1-2, hsa-mir-223, hsa-mir-197, hsa-mir-145, hsa-mir-21, and hsa-mir-133a-1. Of the 80 significantly differentially expressed miRNAs, a group of 14 (10 upregulated and 4 downregulated) has previously been reported to be dysregulated in experimental or human cardiovascular diseases. This group includes miR-133a, -197, miR-30a, miR-499, and miR-451 (Table 4). A second group includes differentially expressed miRNA that have not been previously linked to cardiac development and function but have been shown to be involved in other human diseases (Table 5), such as cancer (miR-326, miR-181a, miR-377, miR-584, miR-593, miR-101-1, miR-193, miR-342, miR-99b), diabetes (miR-223, miR-181a), multiple sclerosis (miR-326), and Alzheimer's disease (miR-146).
Fig. 4.
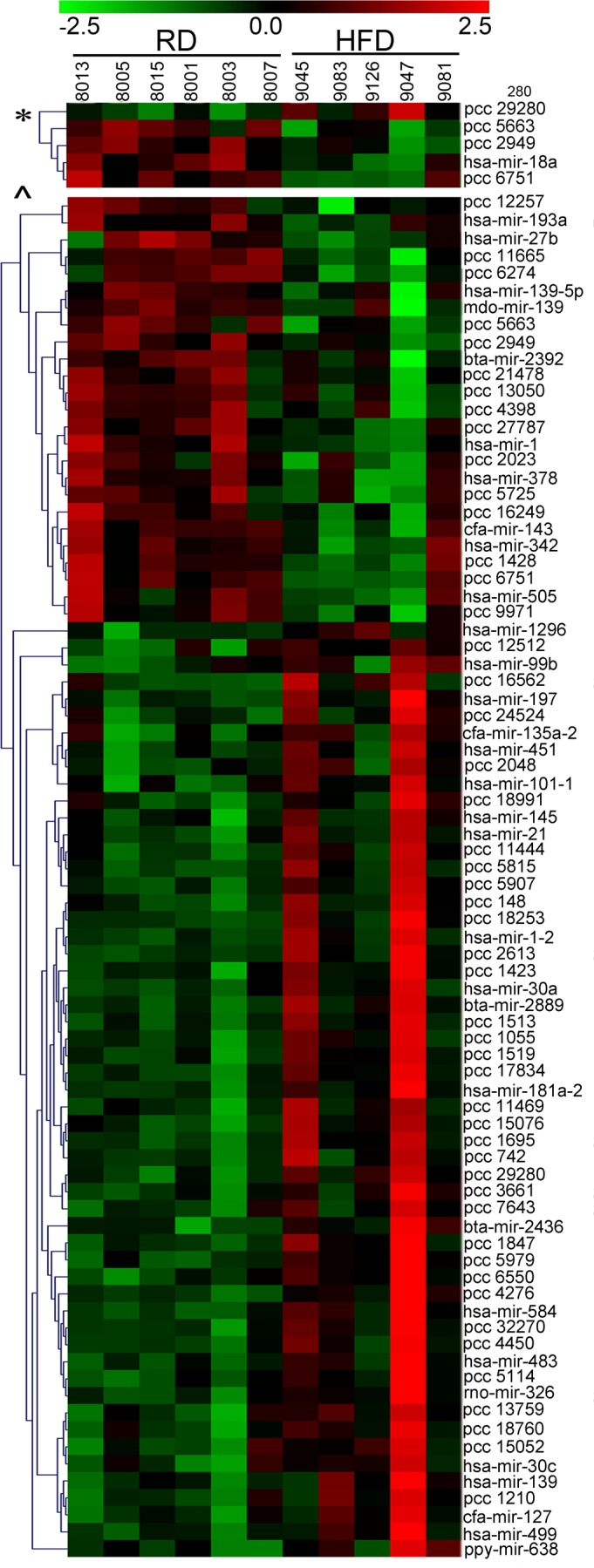
Hierarchical clustering of differentially expressed miRNAs. Heat maps of the known and novel (PCC) miRNA microarray data showing the normalized log2 miRNA expression level for each miRNA (rows) in each sample (columns) for P < 0.001 (top) and P < 0.05 (bottom), according to the indicated color scale. Red indicates overexpression; green represents downregulation.
Table 4.
Differentially expressed fetal cardiac miRNAs previously described as having a functional role in CVDs
| miRNA | Role in CVD |
|---|---|
| Upregulated | |
| hsa-miR-30a | regulates myocardial matrix remodeling (31) and atrial fibrillation (66) |
| hsa-miR-30c | myocardial infarction (31) |
| hsa-miR-145 | regulates smooth muscle cell plasticity (22) |
| hsa-miR-451 | myocardial infarction (11) |
| hsa-miR-499 | regulates mitochondrial dynamics (105) and myosin isoforms (29) |
| hsa-miR-223 | regulates glut4 expression and cardiomyocyte glucose metabolism (69) |
| hsa-miR-133a | involved in cardiac hypertrophy (35) |
| hsa-miR-21 | involved in heart failure (100), myocardial infarction (28), and cardiac hypertrophy (97) |
| hsa-miR-197 | involved in cardiac hypertrophy (17) |
| hsa-miR-139 | involved in myocardial infarction (86) |
| Downregulated | |
| hsa-miR-1-2 | involved in cardiomyopathy (113) |
| hsa-miR-27b | involved in cardiac hypertrophy (113) |
| hsa-mir-378 | targets IGF1R (59), inhibits apoptosis in cardiomyocytes (112) |
| hsa-miR-18a | regulates myocardial matrix remodeling in age related heart failure (102) |
CVD, cardiovascular disease; IGFR1, insulin-like growth factor 1 receptor. The numbers in parentheses are reference list numbers.
Table 5.
Differentially expressed fetal cardiac miRNAs not previously described in the heart
| miRNA | Experimentally Observed Function |
|---|---|
| Upregulated | |
| hsa-miR-326 | cancer (26); multiple sclerosis (30) |
| hsa-miR-483 | pancreatitis (9) |
| hsa-miR-181a | cancer (80); diabetes (58) |
| hsa-miR-377 | nephropathy (106); cancer (72) |
| hsa-miR-99b | myopathy (32), endometriosis (79) |
| hsa-miR-584 | cancer (49); multiple sclerosis (57) |
| hsa-miR-146 | Alzheimer disease (67) |
| hsa-miR-593 | cancer (54) |
| hsa-miR-101-1 | cancer (53) |
| Downregulated | |
| hsa-miR-582-3p | osteoarthritis (25) |
| hsa-miR-505 | tumor suppression (110) |
| hsa-miR-193a | cancer (60) |
| hsa-miR-342 | cancer (48) |
The numbers in parentheses refer to reference list numbers.
Quantitative RT-PCR analysis of selected miRNAs.
To confirm the accuracy of the results in the microarray study, we performed real-time PCR on 11 differentially expressed miRNAs. Four criteria were used to select candidate miRNAs: 1) the miRNA must be highly expressed in the heart; 2) the miRNA has to be previously linked to cardiac disease; 3) only one representative from a given miRNA family should be considered; and 4) the miRNA must be a target of a commercially available RT-PCR assay at the time of the work. Table 6 summarizes the data and illustrates the differences in expression between the HFD and RD RNA populations found by RT-PCR. U18 (Applied Biosystems) was used to normalize the RT-PCR data set. The normalized RT-PCR data yielded a correlation of 0.68 (P < 0.02) with microarrays. We found that compared with microarray: 1) the changes in expression of seven miRNAs (63%) were consistent with those determined by microarrays: hsa-mir-30a, hsa-mir-1-2, hsa-mir-223, hsa-mir-197, hsa-mir-18a, hsa-mir-584, hsa-mir-499; 2) changes in the opposite direction to those shown by microarrays were found for two miRNAs: hsa-mir-451, hsa-mir-30c-1, and 3) two miRNAs remained unchanged in contrast to our microarray data: hsa-mir-145, hsa-mir-21.
Table 6.
Summary of differences in miRNA expression between RD and HFD groups for 11 dysregulated miRNAs
| miRNAs | Fold Change by Microarray | Fold Change by RT-PCR |
|---|---|---|
| Validated | ||
| hsa-mir-30a | 7.0 | 3.9* |
| hsa-mir-1-2 | 7.0 | 3.4* |
| hsa-mir-223 | 6.0 | 1.4* |
| hsa-mir-197 | 6.0 | 1.5* |
| hsa-mir-18a | 0.8 | 0.76* |
| hsa-mir-584 | 4 | 1.4* |
| hsa-mir-499 | 1.7 | 1.3* |
| Validated with opposite direction | ||
| hsa-mir-451 | 2 | 0.7* |
| hsa-mir-30c-1 | 2 | 0.8* |
| Not validated | ||
| hsa-mir-145 | 6.0 | 1 |
| hsa-mir-21 | 4.8 | 1 |
The microarray data were converted to fold change to directly compare with RT-PCR values, n = 6 RD and 5 HFD,
P < 0.05.
Identification of miRNA predicted targets.
miRNAs can regulate a large number of target genes and several databases based on various algorithms are available for predicting the targets of selected miRNAs. Target Scan 5.0, PicTar, and DIANA LAB were used to predict gene targets of the dysregulated miRNAs identified in this study. Overall >1,700 predicted and experimentally observed targets were identified. Using Ingenuity Pathways Analysis IPA), we utilized an miRNA target filter to limit the search to targets expressed in the heart. As a result, >1,500 target genes were identified, 133 of which were experimentally observed and the others classified as “highly predicted.”
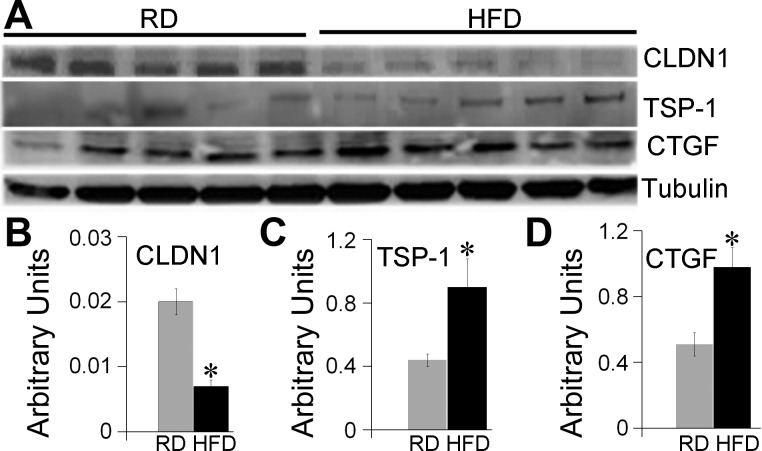
A subset of validated and predicted target genes was selected to confirm the expression changes by Western blot analysis. Two extracellular matrix proteins and mediators of cardiac fibrosis in humans and rodents (95, 96), CTGF, and TSP-1 were among the validated targets. TSP-1 appeared to be a target of upregulated miR-1(4) and two downregulated miRNAs: miR-27b (115) and miR-18a (102). We found significantly increased cardiac protein levels of TSP-1 (P < 0.05) in HFD hearts compared with RD hearts (Fig. 5, A and C). Similarly, the protein level of CTGF, a target gene of three upregulated miRNAs, miR-145 (63), miR-133a, and miR-30c (31) and one downregulated miR-18a (102), was significantly higher in HFD vs. RD hearts (P = 0.01) (Fig. 5, A and D). Among the predicted targets, Claudin1 (CLDN1), a tight junction component, was identified as the potential target for four upregulated miRNAs: miR-139, miR-145, miR-584, miR-30a, and Western blot analysis showed 50% reduction in CLDN1 levels in HFD hearts compared with RD (Fig. 5, A and B). Novel miRNAs differentially expressed in HFD hearts compared with RD also had a great number of predicted target genes. Table 7 summarizes the information regarding cardiac-related potential target genes of five upregulated and five downregulated novel miRNAs.
Fig. 5.
Protein expression of potential and validated genes identified by Ingenuity Pathways Analysis (IPA). Western blots were performed on the hearts from baboon fetuses born to RD and HFD mothers, n = 5 for each group. Representative images (A) and quantification bars (B–D) for CLDN1, TSP-1, and CTGF. Images were analyzed using Syngene GeneTools software, data are presented as means ± SE; *P < 0.005. Statistical analysis was done by 1-way ANOVA.
Table 7.
Novel baboon miRNAs differentially expressed in HFD hearts compared with RD
| PC# | Fold Change | Seed Region | Targets, n | Sample Target Genes |
|---|---|---|---|---|
| Upregulated | ||||
| 32270T4 | 6.3 | ACGGGGA | 4 | PAX2, paired box gene 2; RAP2B, member of RAS oncogene family; PAX2, paired box gene2 |
| 2048T4 | 4.5 | AUGAGAG | 83 | caspase-3; SLC5A6, solute carrier family 5 (sodium-dependent vitamin transporter), member 6; MRPL43, mitochondrial ribosomal protein L43 |
| 11444T3 | 4.3 | GGUCCCC | 242 | CALM1, calmodulin 1; MYH9, myosin, heavy chain 9; ACTN4, actinin, alpha 4, HSPB8, heat shock protein 8 |
| 24524T4 | 2.6 | CAGAAUU | 289 | TP53INP2, tumor protein p53 inducible nuclear protein 2; MAP2, microtubule-associated protein 2, CTGF, connective tissue growth factor |
| 18991T4 | 2.3 | AAGGAUU | 127 | TXNDC13, thioredoxin domain containing 13; ANK2; ankyrin 2; TNPO1, transportin 1 |
| Downregulated | ||||
| 9971T4 | 0.8 | AAUACAU | 656 | KLF3, Kruppel-like factor 3; ADAM10, ADAM metallopeptidase domain 10; APP, amyloid beta (A4) precursor protein |
| 2949T3 | 0.5 | AUCCACG | 48 | ARNTL, aryl hydrocarbon receptor nuclear translocator-like; IGF2BP1, insulin-like growth factor 2 mRNA binding protein 1; HAND2, heart and neural crest derivatives expressed 2 |
| 4398T3 | 0.5 | AGCAGCC | 432 | EIF4E, eukaryotic translation initiation factor 4E; TRAK1, trafficking protein, kinesin binding 1; USP32, ubiquitin specific peptidase 32; CLDN1, claudin1 |
| 5663T3 | 0.4 | CAGTCGG | 6 | AHDC1, AT hook, DNA binding motif, containing 1; FOXJ3, forkhead box J3; GTPBP5, GTP binding protein 5 |
| 6274T3 | 0.4 | AACUGGU | 145 | TPM4, tropomyosin 4; CREB5 cAMP responsive element binding protein 5; GJA1, gap junction protein, alpha 1 |
The fold change, the 2–8 mer seed region, numbers of predicted target genes according to Targetscan and sample target genes, for each novel miRNA are shown. PC, potential candidate.
IPA of predicted targets.
Using the entire list of identified predicted targets as a starting point, we utilized IPA to reveal potential diseases, molecular functions, physiological systems, and canonical pathways associated with differentially expressed miRNAs. Not surprisingly, the analysis identified developmental disorder and cardiovascular disease as the main diseases associated with maternal overnutrition (324 and 251 molecules, respectively; P < 0.05). Cellular death and survival, growth, and proliferation and cellular development were the most affected molecular and cellular functions in response to maternal overnutrition. We next evaluated changes in cell death and proliferation in the RD and HFD hearts. TUNEL assay showed extremely low levels of cardiomyocyte cell death in ventricular tissue of both RD and HFD fetuses (not shown). In contrast, the proliferation rates measured by Ki-67 staining were significantly higher in HFD hearts compared with RD (P < 0.05, Fig. 6).
Fig. 6.
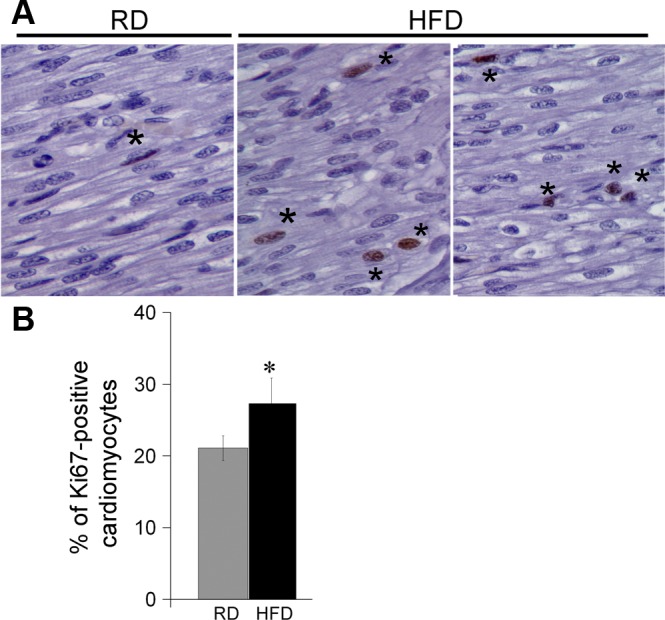
Increase in cell proliferation in HFD hearts compared with RD. Paraffin tissue sections were stained with anti-Ki-67, a marker of cell proliferation, and counterstained with DAPI. The percentage of proliferating cells was calculated by dividing the number of Ki-67-positive nuclei (*) by total nuclei and multiplying by 100. Representative images from RD and 2 HFD fetuses (A) and quantification bars (B); n = 6 for RD and 5 for HFD group. Images were analyzed using Syngene GeneTools software. Data are presented as means ± SE. *P < 0.05 (1-way ANOVA).
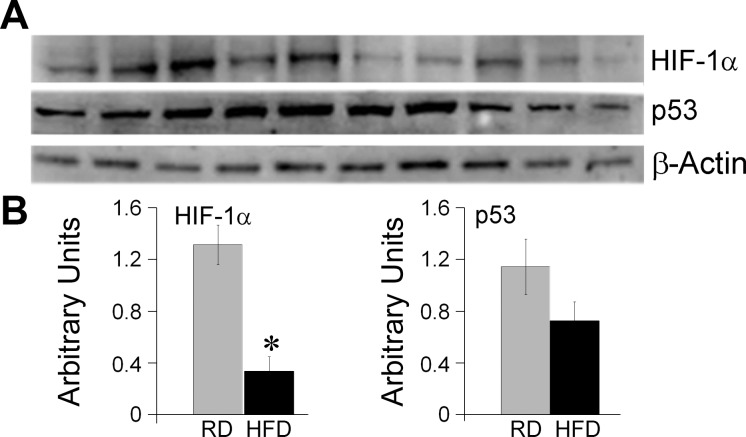
IPA also identified 383 potential transcriptional regulators (not shown). The transcription factors with the highest degree of probability and target molecules were: 1) tumor protein p53 (TP53), 2) peroxisome proliferator-activated receptor gamma (PPAR-γ), and 3) hypoxia-inducible factor 1 alpha (HIF-1α). Western blot analysis showed fourfold decrease in the levels of HIF-1α in HFD hearts compared with RD hearts (P < 0.05, Fig. 7, A and B). The expression of p53 showed a trend toward a decrease, which, however, did not reach a statistical significance (P = 0.1). No differences in PPAR-γ were detected (not shown).
Fig. 7.
Effect of maternal obesity on fetal cardiac expression of transcription factors p53 and HIF-1α. Western blot analysis was performed on the hearts from baboon fetuses born to RD and HFD mothers, n = 5 for each group. Representative images (A) and quantification bars (B). Images were analyzed with Syngene GeneTools software. Data are presented as means ± SE. *P < 0.05 (1-way ANOVA).
DISCUSSION
The incidence of obesity has risen sharply over the past 20 yr and has now reached epidemic proportions, with >1.5 billion adults overweight and 500 million clinically obese adults worldwide (68). In addition to the short-term complications of obesity for both mother and fetus during pregnancy, emerging evidence suggests that maternal obesity has long-term consequences for the health of the offspring (13, 94).
Prenatal and early-life nutrition and stress are among the best documented examples of adverse conditions that predispose the offspring to metabolic and cardiovascular diseases in later life (5). These effects have been confirmed in sheep and rat models of maternal overnutrition (3, 47, 51, 85, 98, 104, 111). The in utero environment can substantially modify how the fetal genome is expressed, thereby exerting stimulatory or inhibitory effects on fetal growth and adiposity.
In this study, we focused on the effect of HFD. We showed a significant increase in maternal body weight and blood LDL-cholesterol in dams fed an obesogenic diet. Despite the fact that HFD-fed mothers did not gain weight during the pregnancy, we found significant physiological changes potentially caused by pregravid maternal obesity. We showed a significant decrease in fetal weight, as well as increase in the weights of brain and thymus in the fetuses of HFD mothers. A reduction in fetal body weights has been shown in nonhuman primate model of maternal obesity (71). In human data, a prospective study of pregnancy outcome in obese women showed that only 13.4% of infants were large for gestational age, while 18.8% of infants were small for gestational age (82). Our recent observations (Maloyan A, Myatt L, unpublished observations) have shown a strongly correlation between birth weight and weight gain during pregnancy. This correlation has been shown before (1). Thus, a 16% reduction in birth weight in our baboon cohort can be at least partially explained by the fact that the HFD mothers lost nearly half a kilogram in body weight during pregnancy. This suggests that cardiac abnormalities seen in the offspring of HFD mothers are due to pre-existing maternal adiposity and cannot be reversed by low weight gain during pregnancy. In contrast to other animal models of maternal obesity (34, 37), the cardiac mass did not change in HFD fetuses. However, we found a significant accumulation of fibrotic tissue in the myocardium of HFD fetuses. Fibrosis is usually a hallmark of aging in various organs including kidney (41), liver (42), pancreas (44), lung (12), and heart (8). Accumulation of myocardial collagen at this early stage of development could lead to progressive increase in ventricular stiffness and impaired cardiac function in offspring. We have previously shown that cardiac function is already impaired by late gestation in a sheep model of maternal obesity (104).
A major role in cardiac fibrosis has been historically attributed to various growth factors, proteolytic enzymes, angiogenic factors, and fibrogenic cytokines (109). However, miRNAs have recently come into focus as regulators of cardiac fibrosis (6). A number of miRNAs have been identified to induce cardiac fibrosis; however, cardiac miR-21 is among the most strongly upregulated in response to variety of cardiac and physiological stresses (62) including the 4.8-fold increase in its levels seen in this study. Further studies are needed to define the role of miR-21 in the cardiac fibrotic remodeling in response to maternal obesity.
Changes in fetal gene expression as result of maternal undernutrition have been previously shown in rat (108), sheep (43), and baboon (77) models. Recent reports from our group have shown that both maternal nutrient reduction and overfeeding in the sheep alters gene transcription in the fetal heart (27, 56) and downregulates fetal skeletal-muscle protein synthesis (116). The mechanisms whereby maternal obesity and nutrient excess in utero increase risk for future metabolic disease are poorly understood but likely include qualitative and quantitative changes in fetal nutrient supply in combination with genetic and epigenetic mechanisms. The in utero environment can substantially modify how the fetal genome is expressed, thereby exerting stimulatory or inhibitory effects on fetal growth and adiposity.
In this study, we have sequenced baboon fetal cardiac miRNA and identified miRNAs that were differentially expressed in response to maternal HFD for the first time. Our initial search has been for differentially expressed miRNAs that have been linked to cardiac development. We show that, for example, downregulation in miR-17-92 cluster has been reported to produce septal defects in mice (103), and miR-181a is involved into cardiac neural crest migration (16). At this point we can speculate that dysregulation of developmentally important miRNAs may explain epidemiological studies that have shown that offspring of obese women are at significantly increased risk for a range of congenital heart defects (14, 74).
Some miRNAs whose expression was affected by maternal HFD were similar to those that are changed in adult cardiac diseases, such cardiac hypertrophy (miR-143, miR-499, and miR-21) (24), heart failure (miR-21 and miR-223) (50), and myocardial infarction (miR-30c, miR-451, and miR-139) (84). There is upregulation of miRNAs involved in enhancing fibrosis (miR-21, miR-499, miRs-133 family, and miRs-30 family) (6) and enhancing intracellular trafficking, and cell adhesion (miR-30 family) (31). Postnatal cardiac function studies are required to determine whether abnormal expression of these miRNAs results into the functional abnormalities in the hearts of HFD offspring later in life. We speculate that the changes we have observed in cardiac miRNA expression will prove detrimental for cardiac function in HFD offspring.
A question facing any researcher with a microarray data set is how much quantitative confidence can be placed in them? RT-PCR is becoming the method of choice in follow-up validation (19) although microarray and RT-PCR data often result in disagreement. In our study, we validated the differential expression of 11 miRNAs with correlation coefficient of 0.68. A survey of the literature reveals widely ranging correlations between microarrays and RT-PCR data of 0.48 to 0.94, illustrating the differences between these technologies (7). Discrepancy could arise from: 1) a distance between the location of the PCR primers and microarray probes; 2) an intensity of array spot with low intensities having considerably lower correlations with RT-PCR data than high intensity spots (7); 3) the difficulties for conventional microarrays to differentiate between members of miRNA families, which often differ by as little as one nucleotide but might exhibit differential expression patterns.
The pathways and biological processes affected by the differentially expressed fetal cardiac miRNAs have yet to be experimentally ascertained. Linking an miRNA to its downstream gene targets is a major challenge in miRNA research. A variety of bioinformatic processes have been developed that predict potential binding sites within the sequence of gene 3′-UTRs in an effort to identify genes regulated by miRNAs. In this study, three algorithms (Target Scan 5.0, PicTar, and DIANA LAB) were used. These are sensitive algorithms with substantial overlap in their predicted targets (92).
Both CTGF and TSP-1 are expressed in the heart during development, whereas their expression is low during normal postnatal life (20, 87). Maternal obesity-mediated upregulation of fetal cardiac CTGF and TSP-1, which regulate extracellular matrix remodeling, suggests that changes in miRNA expression may contribute toward fine tuning of the extracellular matrix proteins potentially leading to fibrosis (20, 88). Claudins regulate many critical developmental processes in vertebrates, with their loss leading to developmental abnormalities or even death (101). We found significant downregulation in CLDN1 protein level in HFD fetal hearts compared with RD. We hypothesize that reduction in CLDN1 might play a role in abnormal fetal cardiac development in response to maternal obesity.
Using IPA, we found that numerous transcriptional regulators, cell signaling molecules, and genes involved in cell death, proliferation, and cardiac development are strongly represented among the potential targets. Interestingly, our data showed a significant increase in cardiomyocyte proliferation in HFD hearts vs. RD. An increase in cell proliferation in the offspring of maternal obesity has been shown before. In rats, maternal HFD increased proliferation of fetal neuroepithelial and neuronal precursor cells (15). In a sheep model of maternal obesity, Ford et al. (39) found an increase in the proliferation of fetal pancreatic β-cell. In a recent study, Eulalio et al. (33) identified a number of miRNAs that had in vitro and in vivo capability to increase cell proliferation in neonatal cardiomyocytes as well as to promote cell cycle re-entry of adult cardiomyocytes. MiRNA(s) responsible for the activation of cardiomyocyte proliferation in our model remains to be elucidated.
Interestingly, some of the largest groups of dysregulated genes in obesity, for example, inflammatory response-related genes, were not represented by the in silico analysis of potential targets. Potential explanations for this observation include that miRNAs may be indirect regulators of inflammatory response genes at the level of transcription control and that our model of maternal HFD in pregnancy does not induce fetal inflammation at 165 days gestation.
It is well known that an evolutionarily conserved orchestra of transcription factors controls cardiac development and function. More recently, the contribution of transcription factors to miRNA expression in the heart has been identified (23). For example, transcription of miR-1, miR-21, miR-206, and miR-133 is directly regulated by serum response factor (SRF) (114); transcription of miR-210 is activated by HIF-1α (46); and transcription of profibrotic miR-21 is regulated by AP-1 (40) and SRF (78). Tight cooperation of three transcription factors, p53, STAT3, and NF-κB, has been shown to regulate miR-21 expression during heart failure (18). In this study, we found significant reduction in the levels of HIF-1α in HFD hearts compared with RD. The involvement of HIF-1α in fetal programming is mostly limited to in utero adaptation to hypoxic conditions (2, 81). It is generally accepted that HIF-1α is stabilized under hypoxia (90). However, prolonged chronic hypoxia, such as seen in obesogenic intrauterine environment (75, 76), has been reported to promote HIF-1α degradation and inhibition of its downstream signaling (70). It has been previously shown that HIF-1α is required for the proper development of the cardiovascular system (55, 91), and reduction in HIF-1α as seen in HFD fetuses might lead to cardiac malformations and abnormal cardiac function during fetal and postnatal life.
In conclusion, maternal HFD before and during pregnancy induces fetal cardiac fibrosis and differential expression of cardiac miRNAs that may contribute to the programming of heart development observed in several animal models. Our study has some limitations. First, because of the sample size of previously archived tissue (three males and two females in the HFD group), we were not able to identify sex-specific changes in gene and miRNA expression. Early studies identified marked sexual dimorphism in the baboon response to high caloric diet with female baboons, overfed as infants, having significantly greater body fat mass, percent of body mass that was fat, and mean fat cell volume compared with females that were underfed or normally fed as infants, while no significant changes were observed in male baboons (64). Second, alteration in the expression of miRNA alone is only a first step in understanding their role in programming and does not prove a functional role. Further experiments using in vitro and in vivo models with genetic manipulation of differentially expressed miRNAs will provide a more complete understanding of their role in fetal heart development in the setting of maternal overnutrition.
GRANTS
This work was supported by grants from Clinical and Translational Science Awards (UL1RR-025767) from the Institute for Integration of Medicine and Science at the University of Texas Health Science Center at San Antonio (to A. Maloyan) and was in part funded by a pilot grant to P. W. Nathanielsz from the Southwest National Primate Research Center base grant (National Institutes of Health P51 RR-33986) and HD-21350.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: A.M., L.A.C., P.W.N., L.M., and M.J.N. conception and design of research; A.M., S.M., P.W.N., and M.J.N. performed experiments; A.M., S.M., S.H., L.A.C., P.W.N., L.M., and M.J.N. analyzed data; A.M., L.A.C., P.W.N., L.M., and M.J.N. interpreted results of experiments; A.M. and S.M. prepared figures; A.M. drafted manuscript; A.M., L.A.C., P.W.N., L.M., and M.J.N. edited and revised manuscript; A.M., L.A.C., P.W.N., L.M., and M.J.N. approved final version of manuscript.
REFERENCES
- 1.Abrams BF, Laros RK., Jr Prepregnancy weight, weight gain, and birth weight. Am J Obstet Gynecol 154: 503–509, 1986 [DOI] [PubMed] [Google Scholar]
- 2.Adams JM, Difazio LT, Rolandelli RH, Lujan JJ, Hasko G, Csoka B, Selmeczy Z, Nemeth ZH. HIF-1: a key mediator in hypoxia. Acta Physiol Hung 96: 19–28, 2009 [DOI] [PubMed] [Google Scholar]
- 3.Armitage JA, Lakasing L, Taylor PD, Balachandran AA, Jensen RI, Dekou V, Ashton N, Nyengaard JR, Poston L. Developmental programming of aortic and renal structure in offspring of rats fed fat-rich diets in pregnancy. J Physiol 565: 171–184, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagnall RD, Tsoutsman T, Shephard RE, Ritchie W, Semsarian C. Global microRNA profiling of the mouse ventricles during development of severe hypertrophic cardiomyopathy and heart failure. PLoS One 7: e44744, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker DJ. Fetal origins of coronary heart disease. BMJ 311: 171–174, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauersachs J. Regulation of myocardial fibrosis by microRNAs. J Cardiovasc Pharmacol 56: 454–459, 2010 [DOI] [PubMed] [Google Scholar]
- 7.Beckman KB, Lee KY, Golden T, Melov S. Gene expression profiling in mitochondrial disease: assessment of microarray accuracy by high-throughput Q-PCR. Mitochondrion 4: 453–470, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Biernacka A, Frangogiannis NG. Aging and cardiac fibrosis. Aging Dis 2: 158–173, 2011 [PMC free article] [PubMed] [Google Scholar]
- 9.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 297: 1901–1908, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Boerschmann H, Pfluger M, Henneberger L, Ziegler AG, Hummel S. Prevalence and predictors of overweight and insulin resistance in offspring of mothers with gestational diabetes mellitus. Diabetes Care 33: 1845–1849, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bostjancic E, Zidar N, Glavac D. MicroRNA microarray expression profiling in human myocardial infarction. Dis Markers 27: 255–268, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calabresi C, Arosio B, Galimberti L, Scanziani E, Bergottini R, Annoni G, Vergani C. Natural aging, expression of fibrosis-related genes and collagen deposition in rat lung. Exp Gerontol 42: 1003–1011, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Catalano PM, Ehrenberg HM. The short- and long-term implications of maternal obesity on the mother and her offspring. BJOG 113: 1126–1133, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Cedergren MI, Kallen BA. Maternal obesity and infant heart defects. Obes Res 11: 1065–1071, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Chang GQ, Gaysinskaya V, Karatayev O, Leibowitz SF. Maternal high-fat diet and fetal programming: increased proliferation of hypothalamic peptide-producing neurons that increase risk for overeating and obesity. J Neurosci 28: 12107–12119, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Wang DZ. microRNAs in cardiovascular development. J Mol Cell Cardiol 52: 949–957, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol 170: 1831–1840, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choy MK, Movassagh M, Siggens L, Vujic A, Goddard M, Sanchez A, Perkins N, Figg N, Bennett M, Carroll J, Foo R. High-throughput sequencing identifies STAT3 as the DNA-associated factor for p53-NF-kappaB-complex-dependent gene expression in human heart failure. Genome Med 2: 37, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuaqui RF, Bonner RF, Best CJ, Gillespie JW, Flaig MJ, Hewitt SM, Phillips JL, Krizman DB, Tangrea MA, Ahram M, Linehan WM, Knezevic V, Emmert-Buck MR. Post-analysis follow-up and validation of microarray experiments. Nat Genet 32, Suppl: 509–514, 2002 [DOI] [PubMed] [Google Scholar]
- 20.Chuva de Sousa Lopes SM, Feijen A, Korving J, Korchynskyi O, Larsson J, Karlsson S, ten Dijke P, Lyons KM, Goldschmeding R, Doevendans P, Mummery CL. Connective tissue growth factor expression and Smad signaling during mouse heart development and myocardial infarction. Dev Dyn 231: 542–550, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Comuzzie AG, Cole SA, Martin L, Carey KD, Mahaney MC, Blangero J, VandeBerg JL. The baboon as a nonhuman primate model for the study of the genetics of obesity. Obes Res 11: 75–80, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460: 705–710, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res 104: 724–732, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Da Costa Martins PA, De Windt LJ. MicroRNAs in control of cardiac hypertrophy. Cardiovasc Res 93: 563–572, 2012 [DOI] [PubMed] [Google Scholar]
- 25.Diaz-Prado S, Cicione C, Muinos-Lopez E, Hermida-Gomez T, Oreiro N, Fernandez-Lopez C, Blanco FJ. Characterization of microRNA expression profiles in normal and osteoarthritic human chondrocytes. BMC Musculoskelet Disord 13: 144, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, Andrew Lister T, Young BD, Debernardi S. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One 3: e2141, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong F, Ford SP, Nijland MJ, Nathanielsz PW, Ren J. Influence of maternal undernutrition and overfeeding on cardiac ciliary neurotrophic factor receptor and ventricular size in fetal sheep. J Nutr Biochem 19: 409–414, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, Wang D, Krall TJ, Delphin ES, Zhang C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem 284: 29514–29525, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorn GW, 2nd, Matkovich SJ, Eschenbacher WH, Zhang Y. A human 3′ miR-499 mutation alters cardiac mRNA targeting and function. Circ Res 110: 958–967, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol 10: 1252–1259, 2009 [DOI] [PubMed] [Google Scholar]
- 31.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res 104: 170–178, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, Lidov HG, Kang PB, North KN, Mitrani-Rosenbaum S, Flanigan KM, Neely LA, Whitney D, Beggs AH, Kohane IS, Kunkel LM. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci USA 104: 17016–17021, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492: 376–381, 2012 [DOI] [PubMed] [Google Scholar]
- 34.Fan X, Turdi S, Ford SP, Hua Y, Nijland MJ, Zhu M, Nathanielsz PW, Ren J. Influence of gestational overfeeding on cardiac morphometry and hypertrophic protein markers in fetal sheep. J Nutr Biochem 22: 30–37, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng B, Chen S, George B, Feng Q, Chakrabarti S. miR133a regulates cardiomyocyte hypertrophy in diabetes. Diabetes Metab Res Rev 26: 40–49, 2010 [DOI] [PubMed] [Google Scholar]
- 37.Fernandez-Twinn DS, Blackmore HL, Siggens L, Giussani DA, Cross CM, Foo R, Ozanne SE. The programming of cardiac hypertrophy in the offspring by maternal obesity is associated with hyperinsulinemia, AKT, ERK, and mTOR activation. Endocrinology 153: 5961–5971, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114, 2008 [DOI] [PubMed] [Google Scholar]
- 39.Ford SP, Zhang L, Zhu M, Miller MM, Smith DT, Hess BW, Moss GE, Nathanielsz PW, Nijland MJ. Maternal obesity accelerates fetal pancreatic beta-cell but not alpha-cell development in sheep: prenatal consequences. Am J Physiol Regul Integr Comp Physiol 297: R835–R843, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol 378: 492–504, 2008 [DOI] [PubMed] [Google Scholar]
- 41.Gagliano N, Arosio B, Santambrogio D, Balestrieri MR, Padoani G, Tagliabue J, Masson S, Vergani C, Annoni G. Age-dependent expression of fibrosis-related genes and collagen deposition in rat kidney cortex. J Gerontol A Biol Sci Med Sci 55: B365–B372, 2000 [DOI] [PubMed] [Google Scholar]
- 42.Gagliano N, Grizzi F, Annoni G. Mechanisms of aging and liver functions. Dig Dis 25: 118–123, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Gilbert JS, Ford SP, Lang AL, Pahl LR, Drumhiller MC, Babcock SA, Nathanielsz PW, Nijland MJ. Nutrient restriction impairs nephrogenesis in a gender-specific manner in the ovine fetus. Pediatr Res 61: 42–47, 2007 [DOI] [PubMed] [Google Scholar]
- 44.Glaser J, Stienecker K. Pancreas and aging: a study using ultrasonography. Gerontology 46: 93–96, 2000 [DOI] [PubMed] [Google Scholar]
- 45.Gluckman PD, Hanson MA, Low FM. The role of developmental plasticity and epigenetics in human health. Birth Defects Res C Embryo Today 93: 12–18, 2011 [DOI] [PubMed] [Google Scholar]
- 46.Goda N, Kanai M. Hypoxia-inducible factors and their roles in energy metabolism. Int J Hematol 95: 457–463, 2012 [DOI] [PubMed] [Google Scholar]
- 47.Gopalakrishnan GS, Gardner DS, Dandrea J, Langley-Evans SC, Pearce S, Kurlak LO, Walker RM, Seetho IW, Keisler DH, Ramsay MM, Stephenson T, Symonds ME. Influence of maternal pre-pregnancy body composition and diet during early-mid pregnancy on cardiovascular function and nephron number in juvenile sheep. Br J Nutr 94: 938–947, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grady WM, Parkin RK, Mitchell PS, Lee JH, Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz AM, Kroh EM, Allen A, Fritz BR, Markowitz SD, Tewari M. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene 27: 3880–3888, 2008 [DOI] [PubMed] [Google Scholar]
- 49.Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, Knuutila S. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma-A miRNA microarray analysis. Genes Chromosomes Cancer 48: 615–623, 2009 [DOI] [PubMed] [Google Scholar]
- 50.Han M, Toli J, Abdellatif M. MicroRNAs in the cardiovascular system. Curr Opin Cardiol 26: 181–189, 2011 [DOI] [PubMed] [Google Scholar]
- 51.Huang Y, Yan X, Zhao JX, Zhu MJ, McCormick RJ, Ford SP, Nathanielsz PW, Ren J, Du M. Maternal obesity induces fibrosis in fetal myocardium of sheep. Am J Physiol Endocrinol Metab 299: E968–E975, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microRNA expression in human heart disease. Physiol Genomics 31: 367–373, 2007 [DOI] [PubMed] [Google Scholar]
- 53.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65: 7065–7070, 2005 [DOI] [PubMed] [Google Scholar]
- 54.Ito T, Sato F, Kan T, Cheng Y, David S, Agarwal R, Paun BC, Jin Z, Olaru AV, Hamilton JP, Selaru FM, Yang J, Matsumura N, Shimizu K, Abraham JM, Shimada Y, Mori Y, Meltzer SJ. Polo-like kinase 1 regulates cell proliferation and is targeted by miR-593* in esophageal cancer. Int J Cancer 129: 2134–2146, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev 12: 149–162, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karere GM, Glenn JP, Vandeberg JL, Cox LA. Differential microRNA response to a high-cholesterol, high-fat diet in livers of low and high LDL-C baboons. BMC Genomics 13: 320, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M, Lenhof HP, Ruprecht K, Meese E. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One 4: e7440, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kloting N, Berthold S, Kovacs P, Schon MR, Fasshauer M, Ruschke K, Stumvoll M, Bluher M. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS One 4: e4699, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knezevic I, Patel A, Sundaresan NR, Gupta MP, Solaro RJ, Nagalingam RS, Gupta M. A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor: implications in postnatal cardiac remodeling and cell survival. J Biol Chem 287: 12913–12926, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res 68: 2094–2105, 2008 [DOI] [PubMed] [Google Scholar]
- 61.Krishnaveni GV, Veena SR, Hill JC, Kehoe S, Karat SC, Fall CH. Intrauterine exposure to maternal diabetes is associated with higher adiposity and insulin resistance and clustering of cardiovascular risk markers in Indian children. Diabetes Care 33: 402–404, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol 8: 706–713, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee HK, Bier A, Cazacu S, Finniss S, Xiang C, Twito H, Poisson LM, Mikkelsen T, Slavin S, Jacoby E, Yalon M, Toren A, Rempel SA, Brodie C. MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS One 8: e54652, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lewis DS, Bertrand HA, Masoro EJ, McGill HC, Jr, Carey KD, McMahan CA. Effect of interaction of gender and energy intake on lean body mass and fat mass gain in infant baboons. J Nutr 114: 2021–2026, 1984 [DOI] [PubMed] [Google Scholar]
- 65.Li C, Schlabritz-Loutsevitch NE, Hubbard GB, Han V, Nygard K, Cox LA, McDonald TJ, Nathanielsz PW. Effects of maternal global nutrient restriction on fetal baboon hepatic insulin-like growth factor system genes and gene products. Endocrinology 150: 4634–4642, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Li S, Yu B, Liu S. Expression of miR-133 and miR-30 in chronic atrial fibrillation in canines. Mol Med Rep 5: 1457–1460, 2012 [DOI] [PubMed] [Google Scholar]
- 67.Li YY, Cui JG, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. Increased expression of miRNA-146a in Alzheimer's disease transgenic mouse models. Neurosci Lett 487: 94–98, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lillycrop KA, Burdge GC. Epigenetic changes in early life and future risk of obesity. Int J Obes (Lond) 35: 72–83, 2011 [DOI] [PubMed] [Google Scholar]
- 69.Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res 86: 410–420, 2010 [DOI] [PubMed] [Google Scholar]
- 70.Marxsen JH, Stengel P, Doege K, Heikkinen P, Jokilehto T, Wagner T, Jelkmann W, Jaakkola P, Metzen E. Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-alpha-prolyl-4-hydroxylases. Biochem J 381: 761–767, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE, Grove KL. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest 119: 323–335, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Melkamu T, Zhang X, Tan J, Zeng Y, Kassie F. Alteration of microRNA expression in vinyl carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis 31: 252–258, 2010 [DOI] [PubMed] [Google Scholar]
- 73.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 148: 1172–1187, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mills JL, Troendle J, Conley MR, Carter T, Druschel CM. Maternal obesity and congenital heart defects: a population-based study. Am J Clin Nutr 91: 1543–1549, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Myatt L. Review: Reactive oxygen and nitrogen species and functional adaptation of the placenta. Placenta 31, Suppl: S66–S69, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Myatt L, Cui X. Oxidative stress in the placenta. Histochem Cell Biol 122: 369–382, 2004 [DOI] [PubMed] [Google Scholar]
- 77.Nijland MJ, Mitsuya K, Li C, Ford S, McDonald TJ, Nathanielsz PW, Cox LA. Epigenetic modification of fetal baboon hepatic phosphoenolpyruvate carboxykinase following exposure to moderately reduced nutrient availability. J Physiol 588: 1349–1359, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol 19: 618–627, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, Print CG, Hull LM. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol 23: 265–275, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pallasch CP, Patz M, Park YJ, Hagist S, Eggle D, Claus R, Debey-Pascher S, Schulz A, Frenzel LP, Claasen J, Kutsch N, Krause G, Mayr C, Rosenwald A, Plass C, Schultze JL, Hallek M, Wendtner CM. miRNA deregulation by epigenetic silencing disrupts suppression of the oncogene PLAG1 in chronic lymphocytic leukemia. Blood 114: 3255–3264, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patterson AJ, Zhang L. Hypoxia and fetal heart development. Curr Mol Med 10: 653–666, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rajasingam D, Seed PT, Briley AL, Shennan AH, Poston L. A prospective study of pregnancy outcome and biomarkers of oxidative stress in nulliparous obese women. Am J Obstet Gynecol 200: 395 e391–e399, 2009 [DOI] [PubMed] [Google Scholar]
- 83.Reece EA. Obesity, diabetes, and links to congenital defects: a review of the evidence and recommendations for intervention. J Matern Fetal Neonat Med 21: 173–180, 2008 [DOI] [PubMed] [Google Scholar]
- 84.Salic K, De Windt LJ. MicroRNAs as biomarkers for myocardial infarction. Curr Atheroscler Rep 14: 193–200, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Samuelsson AM, Matthews PA, Argenton M, Christie MR, McConnell JM, Jansen EH, Piersma AH, Ozanne SE, Twinn DF, Remacle C, Rowlerson A, Poston L, Taylor PD. Diet-induced obesity in female mice leads to offspring hyperphagia, adiposity, hypertension, and insulin resistance: a novel murine model of developmental programming. Hypertension 51: 383–392, 2008 [DOI] [PubMed] [Google Scholar]
- 86.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 100: 416–424, 2007 [DOI] [PubMed] [Google Scholar]
- 87.Schellings MW, Pinto YM, Heymans S. Matricellular proteins in the heart: possible role during stress and remodeling. Cardiovasc Res 64: 24–31, 2004 [DOI] [PubMed] [Google Scholar]
- 88.Schellings MW, van Almen GC, Sage EH, Heymans S. Thrombospondins in the heart: potential functions in cardiac remodeling. J Cell Commun Signal 3: 201–213, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schlabritz-Loutsevitch NE, Howell K, Rice K, Glover EJ, Nevill CH, Jenkins SL, Bill Cummins L, Frost PA, McDonald TJ, Nathanielsz PW. Development of a system for individual feeding of baboons maintained in an outdoor group social environment. J Med Primatol 33: 117–126, 2004 [DOI] [PubMed] [Google Scholar]
- 90.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE 2007: cm8, 2007 [DOI] [PubMed] [Google Scholar]
- 91.Semenza GL, Agani F, Iyer N, Kotch L, Laughner E, Leung S, Yu A. Regulation of cardiovascular development and physiology by hypoxia-inducible factor 1. Ann NY Acad Sci 874: 262–268, 1999 [DOI] [PubMed] [Google Scholar]
- 92.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Meth 3: 881–886, 2006 [DOI] [PubMed] [Google Scholar]
- 93.Seto AG. The road toward microRNA therapeutics. Int J Biochem Cell Biol 42: 1298–1305, 2010 [DOI] [PubMed] [Google Scholar]
- 94.Sewell MF, Huston-Presley L, Super DM, Catalano P. Increased neonatal fat mass, not lean body mass, is associated with maternal obesity. Am J Obstet Gynecol 195: 1100–1103, 2006 [DOI] [PubMed] [Google Scholar]
- 95.Shi-Wen X, Leask A, Abraham D. Regulation and function of connective tissue growth factor/CCN2 in tissue repair, scarring and fibrosis. Cytokine Growth Factor Rev 19: 133–144, 2008 [DOI] [PubMed] [Google Scholar]
- 96.Sweetwyne MT, Murphy-Ullrich JE. Thrombospondin1 in tissue repair and fibrosis: TGF-beta-dependent and independent mechanisms. Matrix Biol 31: 178–186, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol 42: 1137–1141, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taylor PD, McConnell J, Khan IY, Holemans K, Lawrence KM, Asare-Anane H, Persaud SJ, Jones PM, Petrie L, Hanson MA, Poston L. Impaired glucose homeostasis and mitochondrial abnormalities in offspring of rats fed a fat-rich diet in pregnancy. Am J Physiol Regul Integr Comp Physiol 288: R134–R139, 2005 [DOI] [PubMed] [Google Scholar]
- 99.Teekakirikul P, Eminaga S, Toka O, Alcalai R, Wang L, Wakimoto H, Nayor M, Konno T, Gorham JM, Wolf CM, Kim JB, Schmitt JP, Molkentin JD, Norris RA, Tager AM, Hoffman SR, Markwald RR, Seidman CE, Seidman JG. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-beta. J Clin Invest 120: 3520–3529, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456: 980–984, 2008 [DOI] [PubMed] [Google Scholar]
- 101.Tsukita S, Furuse M, Itoh M. Molecular dissection of tight junctions. Cell Struct Funct 21: 381–385, 1996 [DOI] [PubMed] [Google Scholar]
- 102.van Almen GC, Verhesen W, van Leeuwen RE, van de Vrie M, Eurlings C, Schellings MW, Swinnen M, Cleutjens JP, van Zandvoort MA, Heymans S, Schroen B. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell 10: 769–779, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132: 875–886, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang J, Ma H, Tong C, Zhang H, Lawlis GB, Li Y, Zang M, Ren J, Nijland MJ, Ford SP, Nathanielsz PW, Li J. Overnutrition and maternal obesity in sheep pregnancy alter the JNK-IRS-1 signaling cascades and cardiac function in the fetal heart. FASEB J 24: 2066–2076, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, Li YR, Li PF. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med 17: 71–78, 2011 [DOI] [PubMed] [Google Scholar]
- 106.Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X, Quigg RJ. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J 22: 4126–4135, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang XL, Wang J, Shi Q, Carey KD, VandeBerg JL. Arterial wall-determined risk factors to vascular diseases: a nonhuman primate model. Cell Biochem Biophys 40: 371–388, 2004 [DOI] [PubMed] [Google Scholar]
- 108.Welham SJ, Riley PR, Wade A, Hubank M, Woolf AS. Maternal diet programs embryonic kidney gene expression. Physiol Genomics 22: 48–56, 2005 [DOI] [PubMed] [Google Scholar]
- 109.Wynn TA. Common and unique mechanisms regulate fibrosis in various fibroproliferative diseases. J Clin Invest 117: 524–529, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yamamoto Y, Yoshioka Y, Minoura K, Takahashi RU, Takeshita F, Taya T, Horii R, Fukuoka Y, Kato T, Kosaka N, Ochiya T. An integrative genomic analysis revealed the relevance of microRNA and gene expression for drug-resistance in human breast cancer cells. Mol Cancer 10: 135, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yan X, Huang Y, Zhao JX, Rogers CJ, Zhu MJ, Ford SP, Nathanielsz PW, Du M. Maternal obesity downregulates microRNA let-7g expression, a possible mechanism for enhanced adipogenesis during ovine fetal skeletal muscle development. Int J Obes (Lond) 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhao X, Park J, Ho D, Gao S, Yan L, Ge H, Iismaa S, Lin L, Tian B, Vatner DE, Graham RM, Vatner SF. Cardiomyocyte overexpression of the alpha1A-adrenergic receptor in the rat phenocopies second but not first window preconditioning. Am J Physiol Heart Circ Physiol 302: H1614–H1624, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell 129: 303–317, 2007 [DOI] [PubMed] [Google Scholar]
- 114.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436: 214–220, 2005 [DOI] [PubMed] [Google Scholar]
- 115.Zhou Q, Gallagher R, Ufret-Vincenty R, Li X, Olson EN, Wang S. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23∼27∼24 clusters. Proc Natl Acad Sci USA 108: 8287–8292, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhu MJ, Ford SP, Nathanielsz PW, Du M. Effect of maternal nutrient restriction in sheep on the development of fetal skeletal muscle. Biol Reprod 71: 1968–1973, 2004 [DOI] [PubMed] [Google Scholar]
- 117.Zielinsky P, Piccoli AL., Jr Myocardial hypertrophy and dysfunction in maternal diabetes. Early Hum Dev 88: 273–278, 2012 [DOI] [PubMed] [Google Scholar]