Abstract
Both innate and adaptive immunity in birds are different from their mammalian counterparts. Understanding bird immunity is important because of the enormous potential impact of avian infectious diseases, both in their role as food animals and as potential carriers of zoonotic diseases in man. The anti-inflammatory protein tristetraprolin (TTP) is an important component of the mammalian innate immune response, in that it binds to and destabilizes key cytokine mRNAs. TTP knockout mice exhibit a severe systemic inflammatory syndrome, and they are abnormally sensitive to innate immune stimuli such as LPS. TTP orthologs have been found in most vertebrates studied, including frogs. Here, we attempted to identify TTP orthologs in chicken and other birds, using database searches and deep mRNA sequencing. Although sequences encoding the two other widely expressed TTP family members, ZFP36L1 and ZFP36L2, were identified, we did not find sequences corresponding to TTP in any bird species. Sequences corresponding to TTP were identified in both lizards and alligators, close evolutionary relatives of birds. The induction kinetics of Zfp36l1 and Zfp36l2 mRNAs in LPS-stimulated chicken macrophages or serum-stimulated chick embryo fibroblasts did not resemble the normal mammalian TTP response to these stimuli, suggesting that the other two family members might not compensate for the TTP deficiency in regulating rapidly induced mRNA targets. Several mammalian TTP target transcripts have chicken counterparts that contain one or more potential TTP binding sites, raising the possibility that birds express other proteins that subsume TTP's function as a rapidly inducible regulator of AU-rich element (ARE)-dependent mRNA turnover.
Keywords: tristetraprolin, zinc finger proteins, innate immunity, AU-rich elements, mRNA turnover
chickens and other birds have immune systems that are different from those of mammals in many respects (17). Differences include responding cell types, the cytokines and chemokines produced by those cells, and others (18). The innate immune system in chickens and other birds recognizes many of the same pathogen products as their mammalian counterparts and is thought to perform similar functions in protecting birds from biologically derived environmental hazards. As summarized recently by Wu and Kaiser, “The chicken has a different repertoire of tissues, cells, and genes of the immune response compared with mammals, yet generally survives infection with viral, bacterial, protozoal, and fungal pathogens, and also worms and ectoparasites, just like mammals” (32).
One important component of the mammalian innate immune response is the CCCH tandem zinc finger protein tristetraprolin, or TTP (7). TTP biosynthesis in macrophages is rapidly induced by innate immune agonists, such as LPS. The TTP protein, thus produced, can bind to the AU-rich regions of certain mRNAs, and then promote their deadenylation and rapid destruction. The best known target for TTP is the Tnf-α mRNA, coding for the prototypical proinflammatory cytokine. TTP knockout (KO) mice exhibit a severe, chronic, systemic inflammatory syndrome that can largely be attributed to excess TNF (31). The importance of TTP in the acute response to innate immune stimuli can also be seen in mice with myeloid cell-specific TTP deficiency (19, 26). These animals appear normal until challenged with low doses of LPS. They then exhibit extreme and lethal sensitivity to LPS concentrations that have little effect on control mice, associated with massive increases in blood TNF concentrations. From these and other observations, we and others have proposed that TTP is a natural, fundamental physiological regulator of TNF biosynthesis and secretion, and a critical component of innate immunity in vertebrates.
The TTP family of CCCH tandem zinc finger proteins has three members in humans and most other mammals, and all three are conserved among vertebrates from humans through amphibians. A fourth family member appears to be specific to rodents and is expressed only in the yolk sac and placenta (6, 15). In the current study, we have attempted to identify TTP-encoding sequences in chicken and other bird genomes. Although a negative conclusion is impossible to prove, we found no evidence for TTP's existence in the chicken or other birds, despite its presence in mammals, amphibians, and reptiles. We propose that the chicken, and presumably other birds, can regulate mRNAs that are normally TTP targets in other species by other means, possibly by using one of the other two TTP family members expressed in birds, but also possibly through a hypothetical, unrelated, inducible protein that could bind to AU-rich elements and promote mRNA decay.
MATERIALS AND METHODS
Vectors and cells.
Subcloning of the mouse, human, or bovine genomic TTP into plasmid vectors; isolation and culturing of primary chicken embryonic fibroblasts (CEF); and transient transfection of CEF with plasmid DNA were performed as described previously (22). Fetal liver-derived mouse macrophages were isolated from E14.5 embryos and cultured as described previously (11). The mouse monocyte-macrophage cell line Raw264.7 was maintained in MEM with 10% FBS. The virus-transformed chicken macrophage cell line HD11 (3) was a generous gift from Dr. Uma Babu [Immunobiology Branch, U.S. Food and Drug Administration (FDA)], and it was maintained in RPMI-1640 medium with 10% FBS.
RNA purification and Northern blotting.
RNA isolation from CEF was performed as described previously (22), and RNA was purified from mouse fetal liver-derived macrophages, Raw264.7 cells and HD11 cells using the illustra RNAspin isolation kit (GE Healthcare), according to the manufacturer's protocol. Northern blotting was performed as described previously (11, 22). The chicken cDNAs used as probes for Northern blotting were inserts in plasmids containing the chicken Zfp36l1 cDNA (∼1.6 kb, clone pgf1n.pk002.d18), or one containing the partial chicken Zfp36l2 cDNA (∼0.6 kb, clone pgf1n.pk009.l15). These were purchased from Delaware Biotechnology Institute and verified by sequencing. A probe that comprised the chicken Zfp36l1 mRNA 3′-UTR was also used for some Northern blots. It was obtained by PCR using the Zfp36l1 vector backbone pgf1n.pk002.d18 as a template, with a 5′ primer (TCCATCTCCGACGACTAAGCG) that began 15 b before the stop codon (underlined), and the SP6 primer that recognized the 3′ cloning site. Northern blots hybridized with the TTP probes were washed at 60°C. Unless otherwise indicated, blots hybridized with the rat or the chicken Zfp36l1 probes, or the mouse or chicken Zfp36l2 probes, were washed at 70°C. The RNA samples from the CEF and HD11 cells that were used for deep sequencing were also used for Northern blotting.
Chicken cell RNA samples for illumina RNA sequencing.
After serum deprivation for 24 h, CEF or HD11 cells at ∼90% confluence were treated for 60 min with FBS (10%) or LPS (1 μg/ml), respectively. Total RNA was prepared using the GE Healthcare illustra RNAspin kit, and RNA concentrations were quantitated using A260. Initial RNA quality for both samples was assessed using denaturing formaldehyde/agarose gels and Northern blotting, and was confirmed using an Agilent 2100 bioanalyzer, on which both samples had RNA quality scores of at least 10.
mRNA library construction for sequencing.
mRNA from 10 μg total RNA was polyA selected using Dynal oligo DT beads (Invitrogen) and sheared to ∼400 b using the Covaris S2 system. The sample was concentrated to 25 μl using an Amicon Ultra 20K filter. cDNA synthesis was performed using random hexamers and the SuperScript double-stranded cDNA synthesis kit (Invitrogen). A library was constructed from the resulting cDNA, according to the Illumina protocol in the paired-end DNA sample prep oligo only kit. All additional enzymes were purchased from New England Biolabs, with the exception of Platinum Pfx from Invitrogen. After paired-end adapter ligation, the sample was size selected on a 2% agarose gel, stained with SYBRGold, and visualized on a Dark Reader. To ensure that the library was not overamplified, a test amplification was performed in which PCR reaction aliquots were removed every two cycles from 4 to 16. These aliquots were evaluated on a 2% agarose gel, and an optimal cycle number of 12 was selected for the subsequent large-scale amplification. Amplification reactions were cleaned up using two rounds of Agencourt AMPure Beads. Libraries were quantitated by qPCR using Applied Biosystems Power SYBRGreen PCR master mix.
mRNASeq.
Total cellular RNA samples from CEF (sample ID: ES90) and HD11 cells (sample ID: ES91) were sequenced at the National Institutes of Health Intramural Sequencing Center using Illumina GAIIx technology with version 4 chemistry, running the standard CASAVA/ELAND pipeline. Data were processed using RTA1.6.32.0 and GERALD 1.15. Sequencing of RNA fragments generated paired-end 51mers, including 33,635,637 pairs from ES90 and 34,451,770 pairs from ES91. Sequence quality was assessed using FastQC v0.8.0. Reads were not trimmed. However, low-quality pairs (either end with mean Phred score <20) were removed from downstream analyses, retaining 81% and 83% of CEF and HD11 pairs, respectively. Alignment to the reference genome assembly (November 2011 chicken Gallus gallus v4.0, including the canonical chromosomes and 15,900 unordered genomic sequences) was performed with TopHat v2.0.4 using bowtie 0.12.8 as the underlying aligner, and resulted in the successful mapping of 25,989,012 (95%) and 27,697,482 (96%) of CEF and HD11 quality filtered fragments, respectively. Up to 10 alignments per pair were allowed, and empirically determined fragment lengths and standard deviations were passed to the aligner. Otherwise, default parameters were used. Transcript abundances were estimated using cufflinks v2.0.2, based on current (June 2013) Gallus gallus v4.0 RefSeq Gene models (downloaded using University of California Santa Cruz table tools), under default parameters.
Anole and alligator sequence searches.
We searched GenBank for anole sequences using TTP family member protein sequences from Xenopus laevis, using tblastn to search the nr/nt and EST databases for Anolis caroliniensis. We identified expressed cDNA sequences from anole lung that encoded full-length anole TTP and ZFP36L1. These are GenBank numbers gi 463551648 and 463552106, respectively. The encoded full-length proteins were used for both the alignments and the evolutionary tree construction. We also detected a nearly full-length expressed Zfp36l2 cDNA from adult lung (gi 463552097), which was combined with a genomic trace (AAWZ02020961.1) to assemble a full-length predicted protein sequence. The full-length predicted anole TTP protein sequence was also used to search the GenBank “bird” genomes, RefSeq mRNA, ESTs, and whole genome shotgun contigs on 6/13/2013, using tblastn.
For the alligator, we searched a genomic scaffolds database from Alligator mississippiensis downloaded from ftp://ftp.crocgenomes.org/pub/. These were searched with tblastn using the predicted, full-length anole protein sequences. Scaffolds identified as containing likely matches were analyzed manually, and probable protein coding mRNA sequences were assembled, on the basis of the orthologous anole sequence. Alligator TTP was from a conceptual translation within the aMiss:scaffold-4061 from this database. Alligator ZFP36L1 was within the aMiss:scaffold-5190, and alligator ZFP36L2 was within aMiss:scaffold-5785.
Other bird genomes.
Predicted polypeptide sequences were obtained from the Avian Phylogenomic Project (http://phybirds.genomics.org.cn/index.jsp), subject to their data usage agreement, for 47 avian species and two reptilian species (alligator and green turtle). For each species, between 13,500 and 19,000 predicted gene products were available for analysis, with a median length of 303 amino acids per predicted protein. Sequence searches were performed using the alligator-predicted TTP protein sequence, using blastp from BLAST+ version 2.2.27, available from http://blast.ncbi.nlm.nih.gov. Results were filtered to 1e-10, and all results were inspected manually.
We also searched these sequences using a protein profile hidden Markov model (HMM) search, to improve sensitivity over standard blastp alignments. Profiles were built using 64 amino acid tandem zinc finger (TZF) domain peptide sequences from 10 or 11 peptide sequences from TTP, ZFP36L1, and ZFP36L2 from mammals, reptiles, and amphibians, and, for ZFP36L1 and ZFP36L2, from chicken as well. The HMMER tool suite version 3.1b1 (May 2013) was used to build the profile and then run subsequent searches vs. the all-avian peptide database. As positive controls, searches were performed on full-length ZFP36 family member protein sequences from multiple species, using their respective family profile HMM, to confirm that the alignments correctly discriminated among the different proteins.
Alignments and phylogenetic comparisons.
Full-length protein alignments were conducted with ClustalW, using the default parameters for protein multiple comparisons, on either the MacVector 11.1.2 or MEGA5.05 platforms. We used the Gonnet matrix for multiple comparisons, with an open gap penalty of 10 and an extended gap penalty of 0.2 (default options in this version of ClustalW). Phylogenetic comparisons were performed using MEGA5.05 (30) on the ClustalW alignment, and we used the maximum parsimony bootstrap method to infer the evolutionary history of the full length proteins.
RESULTS
Database searches.
On August 9, 2012, the tandem zinc finger domain (TZF domain) from mouse TTP (NP_035886.1; residues 95–158) was used to search the most recent reference genomes for chicken (Build 3.1), turkey (Build 1.1), and zebra finch (Build 1.1), using the tblastn function. In all three cases, there was only a single high scoring hit, which turned out to be ZFP36L2 (see below). The mouse sequence was then used to search all bird ESTs in the database on that date. The same ZFP36L2 sequence was identified in a turkey EST. In addition, a second related sequence was identified in several chicken ESTs (see below), which turned out to be the chicken ortholog of ZFP36L1. No third sequence corresponding to TTP was identified in any species by this search.
As described in the materials and methods, we also identified expressed cDNAs from Anolis carolinensis that encoded orthologs of TTP, ZFP36L1, and almost full-length ZFP36L2 (see below). The TTP protein sequence was used to search the GenBank “bird” genomes, RefSeq mRNA, ESTs, and whole genome shotgun contigs on 6/13/2013, using tblastn. No sequences corresponding to any domains of a bird version of TTP were identified.
We also conducted a hidden Markov model (HMM) search, using the combined TTP TZF domain sequences from 10 mammals, reptiles, and amphibians. The results of this search showed the presence of ZFP36L1 and ZFP36L2 in numerous avian species, but not TTP. On the other hand, searching the mixed mammalian/reptile TTP/ZFP36L1/ZFP36L2 protein set using the TTP profile HMM correctly identified all TTP orthologs at much greater significance than either ZFP36L1 or ZFP36L2.
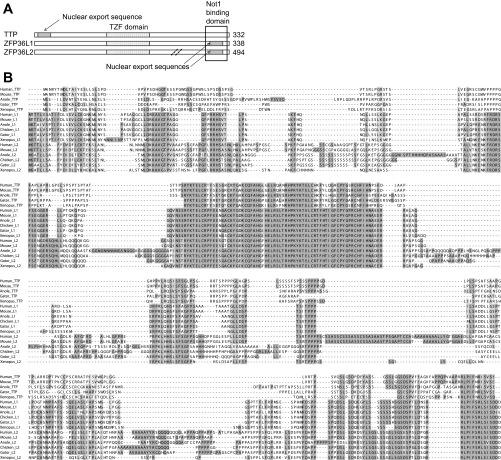
In the search of chicken ESTs, we identified cDNA clones that clearly represented two different TTP family members, based on the differences in their translated protein sequences. We obtained two plasmids representing these ESTs from the Delaware Biotechnology Institute, i.e., clones pgf1n.pk002.d18 (GenBank accession number BI067709.1) and pgf1n.pk009.l15 (BI066858.1), and sequenced their inserts. The first contained a 1.6-kb insert that appeared to contain the complete open reading frame of a chicken protein that we will refer to as chicken ZFP36L1 (see below), based on its orthology with the mammalian and Xenopus proteins of this name. The complete insert sequence has been deposited in GenBank (accession number KC242625), and the predicted translated protein sequence is shown in Fig. 1.
Fig. 1.
Alignments of tristetraprolin (TTP) and its family members from mammals, birds, reptiles, and amphibians. A: schematic representation of the three human TTP family members, with important sequence elements that are conserved in the proteins displayed in B. The approximate locations of the nuclear export sequences are shown in gray shading; the TZF domains are stippled; the conserved NOT1 binding domain is indicated by a box; and the diagonal dashed arrows near the ZFP36L2 COOH-terminus indicate sequences not present in either TTP or ZFP36L1 that account for its greater length. B: alignment of the full-length protein sequences discussed in the text, performed using ClustalW. The highly conserved tandem zinc finger domains are in the middle of the second line of the alignment, and the highly conserved COOH-termini are also highlighted. Dark gray represents amino acid identity at a position, and lighter grey represents amino acid similarity at that position. Sources of these sequences are as follows, with sequence names representing GenBank accession numbers unless otherwise stated. For TTP: Human, NP_003398.2; mouse, NP_035886.1; Xenopus laevis, NP_001081884.1. For ZFP36L1: Human, NP_001231627.1; mouse, NP_031590.1; Xenopus laevis, NP_001084214.1. For ZFP36L2, human, NP_008818.3; mouse, NP_001001806.1; Xenopus laevis, NP_001080610.1. Sequences for the anole, alligator and chicken proteins are described in the text.
The second EST contained a ∼0.6 kb insert that corresponded to ∼120 amino acids containing the TZF domain within the proposed chicken ZFP36L2. A full-length cDNA was prepared as follows. EST clone pgf1n.pk009.l15 was sequenced and covered the eventual mRNA sequence from b530 to b1167. Using this sequence and various other EST sequences from GenBank, we added additional sequences from a cDNA library prepared from the HD11 macrophage mRNA, or genomic DNA from CEF, as templates, and obtained a 732-bp fragment that covered the eventual mRNA sequence from b763 to b1494. The primers used to obtain this fragment were forward primer, 5′-CTCACCCGCCACCCCAAGTACAAG-3′, and reverse primer, 5′-CAGGTAGGAAAGGCGGCGCAG-3′. The EST sequences used to fill in the sequence gaps were GenBank accession numbers BI390306.1 and CD216054.1, which formed bases 1–572 of the eventual sequence: BI066858.1, which matched eventual sequence 528–897; CD218942.1, which matched 902–1058 of the eventual sequence; and DR412026.1, which filled in from 1228 to the 3′ end of the eventual mRNA sequence. The final mRNA sequence has been deposited in GenBank (accession no. KC242626), and the predicted protein sequence is shown in Fig. 1.
Because of the close evolutionary relationship between chickens and lizards, we used similar strategies to identify or assemble TTP, ZFP36L1, and ZFP36L2 sequences from the Carolina anole, Anolis carolinensis, and the American alligator, Alligator mississippiensis. As described in materials and methods, we were able to identify expressed cDNA sequences encoding full-length anole TTP, ZFP36L1, and nearly full-length ZFP36L2; the latter sequence was completed with anole genomic sequence at the extreme 3′-end. Anole ZFP36L2 was predicted by NCBI on the basis of genomic sequences to have the protein sequence contained in XP_003224094.1. However, there were two introns predicted in that sequence that were not present in the expressed cDNA sequence we used, or in several other ESTs from anole lung. The sequence shown in Fig. 1 is the one described above rather than XP_003224094.1.
We were also able to assemble predicted complete protein sequences for all three family members for the alligator (Fig. 1), although in all three cases, these were based on genomic sequences, and expression has not been demonstrated.
To determine whether the predicted anole or alligator ZFP36L1 and ZFP36L2 sequences were closest to the corresponding chicken sequences, we compared the predicted full-length proteins encoded by ZFP36L1 and ZFP36L2. In the ZFP36L1 comparison, the chicken and anole proteins were 90% identical using blastp, whereas the chicken and alligator proteins were 92% identical. In the ZFP36L2 comparison, the chicken and anole full-length proteins were 62% identical, whereas the chicken and alligator proteins were 73% identical. Both alligator and anole sequences for all three family members were used in the phylogenetic comparisons described below.
We also searched the recently completed genome for the Western painted turtle, Chrysemys picta bellii (1), using tblastn with the anole full-length protein sequences. Although a sequence corresponding to nearly full-length TTP was present in the genome for this species (see GenBank accession number AHGY01118920.1), as well as sequences corresponding to ZFP36L1 and ZFP36L2, we were not able to assemble a complete protein sequence for TTP, or determine that it was expressed on the basis of ESTs. However, the nearly complete open reading frame suggests that this turtle also expresses a version of TTP.
Similarly, we found whole genome shotgun sequence reads in GenBank that corresponded to all or part of the TZF domain for all three family members from the Indian Python (Python molurus). These included TTP (AEQU010784169.1), ZFP36L1 (AEQU010065815), and ZFP36L2 (AEQU010255586). We did not find evidence of expression of TTP in the GenBank EST collection for this species.
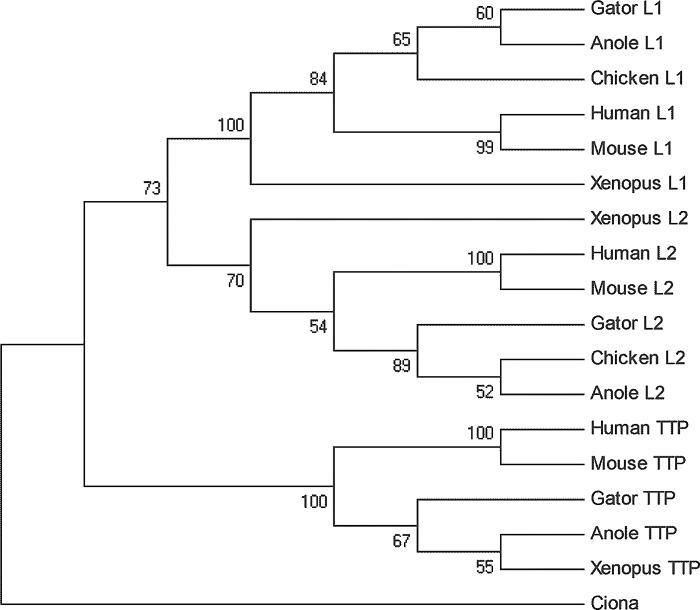
A ClustalW alignment of the mammalian, reptile, and chicken protein sequences is shown in Fig. 1, which highlights the highly conserved RNA binding TZF domain (5), as well as the recently described, highly conserved COOH-terminal NOT1 binding domain (14). It also emphasizes the presence of amino acids within the TZF domain of TTP that are not present in the other two family members. A phylogenetic comparison of the full-length sequences, along with those from human, mouse, and Xenopus laevis, showed that the chicken ZFP36L1 and ZFP36L2 protein sequences were closely related to those of the alligator and anole (Fig. 2). This is in keeping with previous conclusions concerning the evolution of birds and reptiles (2). We used as an outgroup comparison the predicted protein sequence from the single TTP family member identified to date in Ciona intestinalis (GenBank accession number NP_001071879.1). To date, therefore, TTP, ZFP36L1, and ZFP36L2 have been documented in mammals, amphibians, and reptiles. Within the reptiles, the expression of all three mRNAs has been documented in lizards (this paper), and sequences encoding at least the TZF domains of all three proteins have been found in the genomes of turtles, alligators, and snakes.
Fig. 2.
Phylogenetic relationships among full-length TTP family members from mammals, birds, reptiles and amphibians. Shown is a phylogenetic tree of the full-length protein sequences shown in Fig. 1, illustrating the evolutionary relatedness of each protein in amphibians through mammals, and the relative closeness of the alligator and anole to chicken sequences. Sources of each sequence are described in materials and methods and in the legend to Fig. 1, except for the single TTP family member found to date in Ciona intestinalis (NP_001071879.1). The tree was constructed using the maximum parsimony bootstrap method, for which full documentation can be obtained from MEGA5.05 (30). The numbers next to each node refer to the percentage of trees (from 500 replicates), in which the associated sequences are clustered together in the bootstrap test. See the text and MEGA5.05 documentation for further details.
We used the predicted alligator TTP protein sequences to research the genomic and EST databases for chicken (Build 3.1), zebra finch (Build 1.1), and turkey (Build 1.1), using tblastn. As in previous searches, there were no “hits” that would correspond to a bird TTP. We also used the alligator TTP protein sequence to search the predicted peptide sequences of 47 birds represented in the Avian Phylogenomic Project (http://phybirds.genomics.org.cn/index.jsp), using blastp. ZFP36L1 sequences were identified in many birds, and a few ZFP36L2 sequences were also identified, but no sequences corresponding to TTP were found.
We concluded from these database searches that chicken and other birds express ZFP36L1 and ZFP36L2, but not TTP, whereas the anole, alligator, and Xenopus also have TTP orthologs, as well as ZFP36L1 and ZFP36L2. We then did further experiments to search for the “missing” TTP transcript in chicken cellular RNA and also examined the expression patterns of the other two TTP family members in cultured chicken cells.
TTP family member expression in cultured chicken cells.
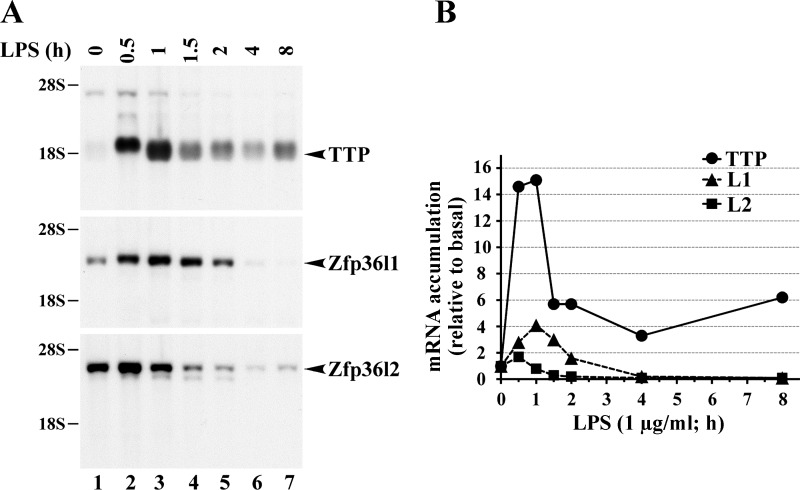
In mammalian cells, a characteristic feature of TTP expression is its extreme, rapid, and transient inducibility in serum-deprived fibroblasts stimulated with serum or growth factors (21), and in macrophages stimulated with LPS (13). Such an experiment is shown in Fig. 3, in which the expression patterns of all three major mouse family members are shown in primary cultured mouse macrophages isolated from fetal liver and stimulated with the Toll-like receptor 4 agonist LPS (4). RNA samples from this experiment were then probed with cDNAs for TTP, Zfp36l1, and Zfp36l2. Fig. 3A shows the characteristic pattern of rapid and dramatic TTP mRNA induction under these conditions, and the quantitatively much lower expression, as well as minimal inducibility, of the Zfp36l1 and Zfp36l2 transcripts. Although probe affinity may vary in this type of experiment, the probes were labeled to the same specific activity, and the exposures of the autoradiographs were the same, allowing us to compare the expression patterns after PhosphorImager mRNA quantitation (Fig. 3B).
Fig. 3.
Expression of TTP family members in LPS-stimulated, fetal liver-derived mouse macrophages. Cells were serum-deprived for 24 h and then treated with LPS (1 μg/ml) for the indicated times. A: each lane was loaded with 8 μg of total cellular RNA, and the resulting Northern blot was hybridized with the 32P-labeled mouse TTP cDNA probe (top), the rat Zfp36l1 cDNA probe (middle), or the mouse Zfp36l2 cDNA probe (bottom). Note that the mouse Zfp36l2 probe also cross-hybridized to a lower-molecular-weight species (the fainter band) that corresponds to the Zfp36l1 mRNA. B: plot of these data, showing the relative expression of TTP, Zfp36l1, and Zfp36l2 mRNAs after phosphorimager quantitation of the northern blot shown in A.
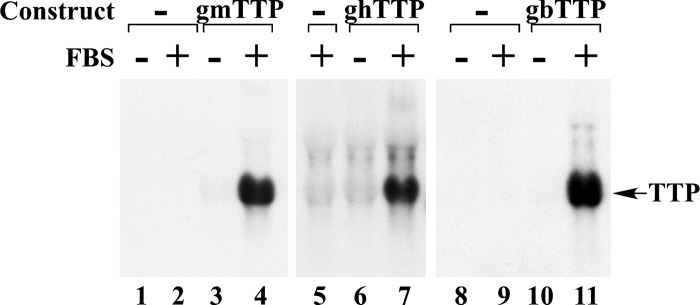
To test whether mammalian TTP cDNAs would cross hybridize to a potential endogenous TTP-like molecule in chicken embryo fibroblasts (CEF), we transfected CEF with inducible genomic TTP constructs from mouse, human, and bovine TTP, or vector alone, and then induced the cells with 10% FCS for 60 min. We then hybridized a Northern blot containing RNA extracted from these cells with the respective cDNAs. As shown in Fig. 4, serum treatment strikingly induced the exogenous mammalian TTP mRNA expression, based on three different genomic constructs containing endogenous promoters, but in no case was there a detectable band corresponding to the hypothetical TTP mRNA in CEF.
Fig. 4.
Expression of mammalian TTP mRNA in chicken embryonic fibroblasts (CEF). CEF were either mock-transfected (Construct -), or transfected with plasmids containing the genomic mouse TTP (gmTTP, lanes 3 and 4), human genomic TTP (ghTTP, lanes 6 and 7), or genomic bovine TTP (gbTTP, lanes 10 and 11) DNA. Twenty four hours later, the cells were serum-deprived for a further 24 h, and then 10% FBS was added to groups of dishes (FBS +) for 60 min. Each lane was loaded with 20 μg of total cellular RNA, and the Northern blot was hybridized with the 32P-labeled mouse TTP cDNA probe (lanes 1–4), the human TTP cDNA probe (lanes 5–7), or the bovine TTP cDNA probe (lanes 8–11).
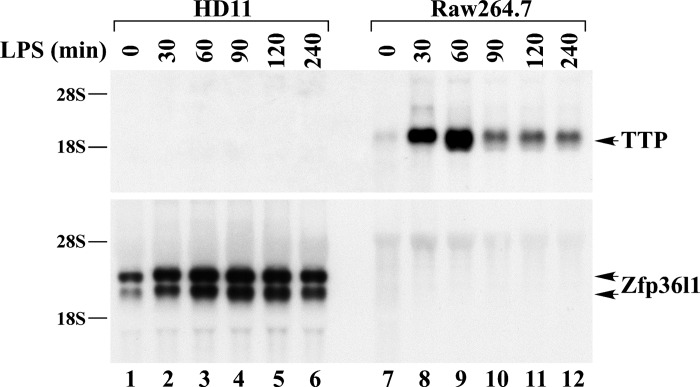
In addition to the CEF experiments, we also studied the cultured chicken macrophage cell line HD11 (3) (a generous gift from Uma Babu, Immunobiology Branch, FDA), before and after LPS stimulation (1 μg/ml). For comparison, we used the mouse macrophage cell line Raw 264.7, in which LPS is known to be a potent TTP inducer (8). A Northern blot using samples taken at intervals from these cells was hybridized with the 32P-labeled mouse TTP cDNA probe (Fig. 5, upper panel), or the chicken Zfp36l1 cDNA probe (lower panel). Mouse Zfp36l1 mRNA was expressed at very low levels in Raw 264.7 cells and was barely detectable after hybridization with a mouse Zfp36l1 cDNA probe (not shown). As shown in Fig. 5 (upper panel, right side), the LPS addition to mouse Raw 264.7 cells resulted in a rapid and dramatic TTP mRNA induction. Under the same conditions, LPS did not induce detectable expression of an mRNA in the chicken HD11 cells that cross-hybridized to the mouse TTP cDNA probe (Fig. 5, upper panel, left side). When the same RNA samples were probed with a chicken Zfp36l1 probe, there was no detectable hybridization to an mRNA in the mouse cells (Fig. 5, lower panel, right side). However, there was clear hybridization to two bands, ∼2.3 and 2.8 kb in size, that were modestly induced by LPS in the HD11 cells. The upper band was predominant in this experiment. We concluded that there was no evidence for chicken TTP existence in these cells, and that the chicken Zfp36l1 probe appeared to be hybridizing to two mRNA species.
Fig. 5.
TTP and Zfp36l1 mRNA expression in mouse and chicken macrophages. Chicken HD11 and mouse Raw264.7 cells were serum-deprived for 24 h and then treated with LPS (1 μg/ml) for the indicated times. Each lane was loaded with 15 μg (lanes 1–6) or 5 μg (lanes 7–12) of total cellular RNA, and the Northern blot was hybridized with the 32P-labeled mouse TTP cDNA probe (top), or the chicken Zfp36l1 cDNA probe (bottom). Zfp36l1 mRNA expresses at low levels in Raw264.7 cells and is barely detectable, even using the rat Zfp36l1 cDNA probe (not shown).
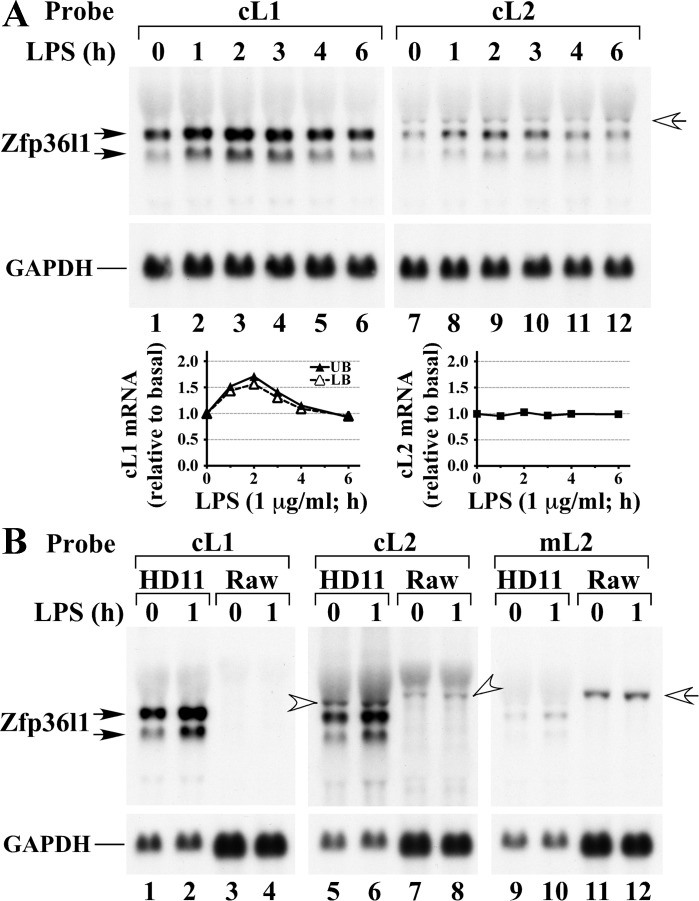
Preliminary experiments indicated that the full-length chicken Zfp36l2 probe cross-hybridized strongly to the two bands of Zfp36l1, so the blots were hybridized at higher stringency. When RNA from the LPS-stimulated HD11 cells was probed with chicken Zfp36l1 3′UTR and Zfp36l2 probes, the same two chicken mRNA species were labeled with the Zfp36l1 probe (Fig. 6A, upper left panel) and, to a lesser extent, the Zfp36l2 probe. In this experiment, the expression of both species of Zfp36l1 mRNA expression was quantitated after normalization for Gapdh mRNA expression (Fig. 6A, lower left panels), and showed a modest degree of induction by LPS. The Zfp36l2 probe also identified a faint, larger species that corresponded to the predicted Zfp36l2 mRNA size (Fig. 6A, upper right panel, open arrow). This band, which we believe represents Zfp36l2 mRNA, did not change appreciably upon cell stimulation with LPS (Fig. 6A, upper right panel). This expression was quantitated using a phosphorimager with Gapdh mRNA normalization (Fig. 6A, lower right panels), and demonstrates the lack of LPS induction.
Fig. 6.
Zfp36l1 and Zfp36l2 mRNA expression in chicken and mouse macrophages. Chicken HD11 and mouse Raw 264.7 cells were serum-deprived for 24 h and then treated with LPS (1 μg/ml) for the indicated times. Each lane was loaded with 15 μg of HD11 cell RNA (lanes 1–12 in A; lanes 1 and 2, 5 and 6, and 9 and 10 in B), or 10 μg of Raw264.7 cell RNA (lanes 3 and 4, lanes 7 and 8, and lanes 11 and 12 in B). The blots hybridized to the chicken Zfp36l1 3′UTR probe were washed at 75°C, and those hybridized to the chicken or mouse Zfp36l2 probes were washed at 70°C. A: Northern blot of HD11 cell RNA that was hybridized with the 32P-labeled chicken Zfp36l1 3′UTR cDNA probe (lanes 1–6), or the chicken Zfp36l2 cDNA probe (lanes 7–12). The open arrow in the right panel indicates putative Zfp36l2 mRNA, whereas the lower bands represent cross-hybridization to Zfp36l1 mRNA. Under each panel is shown the abundance of Gapdh mRNA in the same Northern blot as a loading control. In both cases, the Zfp36l1 and Zfp36l2 mRNA bands were quantitated by a phosphorimager and normalized to Gapdh mRNA expression, and these values were plotted below their respective Northern blots. UB, upper band; LB, lower band. B: Northern blot was hybridized with the 32P-labeled chicken Zfp36l1 3′UTR cDNA probe (lanes 1–4), or with the chicken Zfp36l2 cDNA probe (lanes 5–8), or with the mouse Zfp36l2 cDNA probe (lanes 9–12). The bands in the HD11 cells that hybridized to both the chicken Zfp36l1 and Zfp36l2 probes are indicated as Zfp36l1 by the double-arrows to the left of the blots. The bands in the HD11 cells that hybridized to the chicken Zfp36l2 probe (lanes 5 and 6 in the middle panel of B), or the bands in the Raw264.7 cells that hybridized to both the chicken (lanes 7 and 8) and the mouse Zfp36l2 probes (lanes 11 and 12, right) are indicated by the open arrows and arrowheads. The hybridization of the Gapdh mRNA in each case is shown below the relevant Northern blots.
We used the same stringent conditions to compare hybridization patterns in LPS-stimulated chicken HD11 cells and mouse Raw 264.7 cells (Fig. 6B), using Gapdh mRNA as an internal control. As before, the chicken Zfp36l1 3′UTR probe hybridized strongly to two mRNA bands in the HD11 cells but did not cross-hybridize with anything in the mouse cells (Fig. 6B, left panel). The chicken Zfp36l2 probe hybridized to both bands of Zfp36l1 mRNA and a fainter, larger band that we have tentatively identified as Zfp36l2 mRNA (Fig. 6B, middle panel, left open arrow). The probe also hybridized to a band corresponding to mouse Zfp36l2 in RAW 264.7 cell RNA (Fig. 6B, middle panel, right open arrow). A mouse Zfp36l2 probe hybridized weakly to all three chicken mRNA bands (Fig. 6B, right panel), as well as the mouse Zfp36l2 mRNA (Fig. 6B, right panel, open arrow). Both the chicken and mouse Zfp36l2 bands showed no appreciable induction with LPS in their respective cell types.
We predicted the mature transcript sizes for chicken Zfp36l1 and Zfp36l2 mRNA, based on the predicted length of the protein-coding domains and 3′ and 5′UTRs, and on GenBank genomic and EST sequences and predicted polyadenylation signals with EST confirmation of 3′ ends. For Zfp36l1 mRNA, there were two predicted sizes, 1681 b and 2629 b (not including the poly A tail), based on two optimal polyadenylation signals (AATAAA) that were perfectly conserved in the corresponding sequences from turkey and zebra finch. However, histograms from the mRNASeq experiments (see below) demonstrated that only the 3′-most polyadenylation signal was used in the two cell types studied, suggesting that the longer transcript was the mature Zfp36l1 mRNA. The same histograms showed that there was no appreciable expression of a pre-mRNA, i.e., an RNA species with the single intron retained, in either cell type. The exact nature of the two mRNA species for Zfp36l1 on Northern blots remains unclear. For Zfp36l2 mRNA, the predicted size was 3377 bp (not including the poly A tail), based on a single optimal polyadenylation signal sequence that was conserved with turkey and zebra finch. These predicted sizes correlate well with the respective mRNA bands on Northern blots.
We concluded from these experiments that chicken Zfp36l1 and Zfp36l2 mRNAs were detectable in both CEFs and HD11 macrophages, but that cell stimulation with FBS and LPS, respectively, did not result in detectable expression of an mRNA species that cross-hybridized with the mouse TTP cDNA probe. A further conclusion is that neither family member transcript behaved like the TTP mRNA in mammalian cells in terms of rapid, large-magnitude, and transient induction.
Deep sequencing of mRNAs from cultured and induced chicken cells.
In mouse embryonic fibroblasts and primary macrophages or cultured Raw 264.7 macrophages, stimulation with FCS or LPS, respectively, causes a rapid and transient TTP mRNA induction, which peaks at 30–60 min in both cell types. To determine whether we could detect a chicken ortholog in the CEFs or HD11 macrophages under these conditions by deep mRNA sequencing, we subjected RNA samples from the two cell types after FCS or LPS stimulation for 60 min, respectively, to deep sequencing (mRNASeq). The most conserved region within these proteins is the tandem zinc finger domain (TZF domain), and in most species, including mammals, reptiles, and Xenopus, there are characteristic amino acid differences among the three family members from a given animal species (see Fig. 1). Therefore, our sequence analysis focused on the TZF domain sequences.
The RNA samples used in this sequencing experiment were subjected to Northern blotting with chicken probes to Zfp36l1 and Zfp36l2, as shown in Fig. 7. These confirmed that the RNA used for mRNASeq was high quality, as did analysis on an Agilent Bioanalyzer 2100, and that both samples expressed hybridizing mRNA. We examined sequence reads spanning the TZF domains between the two samples; since the same sequence length was examined, the results should be proportional to the relative expression of the two genes in the same sample. In the RNA samples from cell line 91, 394 reads corresponded exactly to the chicken Zfp36l1 TZF domain. There were 47 additional reads that were less than 100% in the initial alignment, but also aligned with Zfp36l1 upon manual assignment, with a few possible sequencing errors or polymorphisms. This represents 441 Zfp36l1 representations in this cell type. When the L2 TZF domain was used in a similar search, there were 195 total reads that were 100% matched. Another 199 matched to Zfp36l1 (not Zfp36l2) but with occasional sequencing mistakes that were nonsystematic. In cell line 90 (CEF), 56 reads aligned with the Zfp36l1 TZF domain exactly, and 12 others aligned but were less than 100%. These 12 were all Zfp36l1 reads with occasional sequence mistakes or polymorphisms. There were 32 reads that aligned exactly with the ZFP36L2 TZF domain, and 25 additional reads that aligned with the Zfp36l2 mRNA but not at 100%. All of these aligned with the Zfp36l1 mRNA with occasional sequence mistakes and one apparent polymorphism.
Fig. 7.
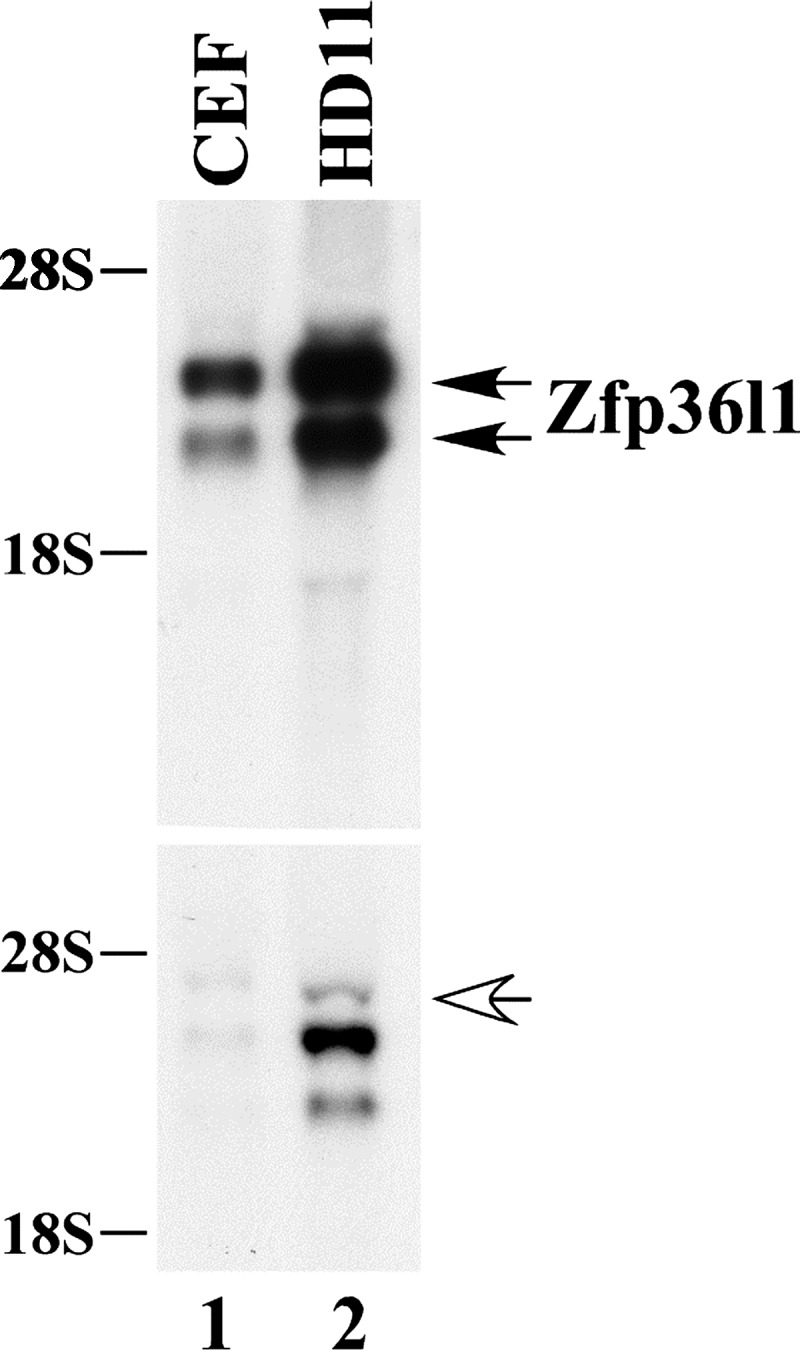
Chicken cell RNA samples for mRNASeq. After serum deprivation for 24 h, CEF or HD11 cells were treated for 60 min with FBS (10%) or LPS (1 μg/ml), respectively. Each gel lane was loaded with 10 μg of total cellular RNA, and the Northern blot was hybridized with the 32P-labeled chicken Zfp36l1 cDNA probe (top) or the chicken Zfp36l2 cDNA probe (bottom). The double arrows in the upper panel indicate the two bands of Zfp36l1 mRNA, and the single arrow in the lower panel indicates the single species of Zfp36l2 mRNA. The lower bands apparent in the HD11 cells represent cross-hybridization of the Zfp36l2 probe with Zfp36l1 mRNA.
The sequence reads from both libraries have been deposited in the Array Express (http://www.ebi.ac.uk/cgi-bin/microarray/magetab.cgi; accession numbers pending).
For comparison, in cell type 90 (CEF) there were 2,145 sequences corresponding to the entire mRNA of NADH dehydrogenase subunit 4 (NADH4) (GenBank RefSeq NP_006924.2). In cell type 91 (HD11), there were 7,255 reads corresponding to NADH4. We also determined the transcript abundance, in FPKM (fragments per kilobase of exon, per million mapped fragments), of Zfp36l1 and Zfp36l2 mRNAs, as well as all RefSeq annotated transcripts, based on the genomic alignments of each sample. In CEF, these values were determined to be 174 and 25 for the Zfp36l1 and Zfp36l2 transcripts, respectively, placing them in the 95th and 78th percentiles of expression. In HD11 cells, the values for the Zfp36l1 and Zfp36l2 transcripts were 279 (96th percentile) and 12 (58th percentile). The chicken Zfp36l1 and Zfp36l2 transcripts were defined as the regions of the reference assembly with continuous RNAseq read coverage, overlapping regions of annotated homology to mammalian gene models (chr5:27618992-27621373 and chr3:24329114-24331061 for Zfp36l1 and Zfp36l2 mRNAs, respectively). Because of the similarity of these transcripts and the alignment strategy of allowing reads to map to multiple locations, the abundance of the Zfp36l2 transcript may be overestimated in these statistics.
In neither case was there evidence for a third sequence that corresponded to a TZF domain from a different species like TTP. As one additional check, we attempted to align the chicken reads from both cell types to the anole TTP sequence. These alignments yielded “hits” to both chicken Zfp36l1 and Zfp36l2 mRNAs, but not to a putative TTP sequence.
We also identified a single genomic trace in GenBank that, when translated, contained a TZF domain-like sequence, very near to a sequence resembling the extreme TTP COOH-terminus (ti 250984745). We examined the deep sequencing results to determine whether this sequence was transcribed to an appreciable extent in the two chicken cell types under consideration. Averaging both sequencing runs, there were an average of 9 reads corresponding to this sequence in sample 90, and an average of 22 reads in sample 91. Examination of the putative TZF domain from this sequence revealed that only one of the five TTP-specific residues was present in the new sequence, arguing against its identity as a TTP ortholog. In addition, there is a stop codon immediately 5′ of the putative TZF domain in the genomic trace, and there are no encoded methionines between this stop codon and the TZF domain-encoding sequence. Our tentative conclusion is that this is a poorly transcribed segment of noncoding RNA, but we cannot rule out a poorly expressed polypeptide. We did not find analogous sequences in either completed genomes or genomic traces from turkey or zebra finch.
Genomic considerations.
Since a TTP ortholog is expressed in mammals, anoles, and frogs, and appears to be present in alligators, turtles, and snakes, but is apparently not present in birds, it seems likely that the gene has been effectively deleted in birds. To examine this question, we searched genomic databases and other information for genes surrounding the gene encoding TTP in humans, ZFP36, on chromosome 19q13.1. As described by Smith et al. (27, 28), the chicken genome contains 8 macrochromosomes and 29 microchromosomes, as well as the sex chromosomes Z and W. These authors described several genes from this region of human chromosome 19q13.1 as members of linkage group 25, which corresponds to chicken microchromosome 31. Specifically, Smith et al. (27) mentioned a series of loci on E25, in the following order: CAPN4, RYR1, TGFB1. These are all syntenic in the same order to a region on human chromosome 19q13.1. According to the human genome sequence, ZFP36 should be very close to RYR1, and between it and TGFB1. However, to our knowledge, there are no existing data mapping any of these four genes to an actual chicken sequence contig, presumably on microchromosome 31, so the state of the sequences in this chicken genomic region is unclear.
We rechecked the chicken and zebra finch genomes for sequences corresponding to human RYR1 and TGFB1 on November 19, 2012. There were no sequences in either genomic database that appeared to be orthologous to these two mammalian genes in either bird species.
TTP target mRNAs in birds.
If TTP is absent in birds, then an important question is, what happens to the mRNAs whose stability and perhaps translation are affected by TTP in mammalian cells? To begin to answer this question, we analyzed some of the best known TTP target sequences in mammals for orthologs in birds. If the target orthologs were identified, we determined whether they contained potential TTP binding target sequences in their 3′UTRs. The TTP target mRNAs investigated included tumor necrosis factor alpha (Tnf), Ier3, Cxcl1, Cxcl2, Csf2 (GM-CSF), Il10, and Il23a.
Tumor necrosis factor-α mRNA is the major physiological target for TTP in mammals. TNF orthologs have been identified in the anole (XP_003218070) and in Xenopus (NP_001108250), but not to date in chicken or other birds (18). When the TNF ortholog from the anole was used to search the chicken genome, the top hit (at 6e-11) was a TNF superfamily member 15 (NP_001019749). When this was, in turn, blasted against human proteins, the top hit was the apparent ortholog, TNF superfamily member 15 (NP_005109), but not TNF itself.
We next searched the deep sequence reads from the LPS-stimulated chicken macrophages for TNF-like sequences, using the Xenopus and anole TNF protein sequence. We did not identify any DNA sequences that, when translated, yielded a high degree of similarity to the Xenopus or mammalian TNF sequence. From these studies, it does not appear that birds express a clear TNF ortholog.
A search of the chicken reference genome also failed to identify IER3, CXCL1 or CXCL2 orthologs. However, we did identify sequences corresponding to several known TTP target mRNAs, including Csf2 (GM-CSF), Il10, Il23a, Lif, and Plk3. In all cases, the 3′UTRs of the encoded mRNAs contained at least one 7-mer potential TTP binding site, and often contained more than one (Fig. 8). These data suggest that, even in the chicken, these mRNAs could be subject to ARE-mediated mRNA turnover.
Fig. 8.
Selected potential TTP family member binding sites within chicken orthologs of known mammalian TTP target transcripts. Shown are the 3′UTR sequences from several apparent chicken orthologs of known TTP target transcripts in the mouse. The minimum TTP binding heptamers (UAUUUAU) are highlighted in gray. The stop codons and polyadenylation signals are underlined. There was no obvious polyadenylation signal in the case of the IL10 transcript, so the sequence was cut off after the potential TTP family member protein binding sites. Initial studies demonstrating that these transcripts were direct TTP targets in the mouse were Csf2 (12), Il23a (25), Il10 (29), Plk3 (16), and Lif (20).
DISCUSSION
Understanding avian immunity is important for many reasons. These include the enormous economic impact of avian farm animals as meat and egg producers, and the great importance of avian species as intermediates in zoonotic diseases in humans (17). Major differences exist between avian and mammalian immune systems in both innate and adaptive immunity (18). Within mammalian innate immunity, the CCCH tandem zinc finger protein TTP is an important regulator of cytokine mRNA stability (7). Of the cytokine mRNAs thought to be destabilized by TTP, Tnf mRNA is the best known, and its resulting excess contributes to virtually all aspects of the TTP deficiency syndrome in KO mice (10, 31). Although TTP orthologs have been identified in many vertebrate species, ranging from mammals to amphibians, it has not been identified in any avian species.
The major finding of this paper is that chickens, and apparently other birds, do not express an ortholog of mammalian TTP. This is despite the fact, as we show here, that orthologous mRNAs are expressed in reptiles, such as Anolis caroliniensis; amphibians, such as Xenopus laevis and Silurana tropicalis; and genomic sequences corresponding to TTP are present in Alligator mississipiensis, Chrysemys picta bellii, and Python molurus. Expression of this mRNA in these amphibians and reptiles, as well as in mammals, indicates that the gene encoding TTP, ZFP36 in humans, is an ancient one whose expression predated the separation of reptiles and amphibians from birds. The mechanism of this apparent loss in birds is not clear, but it may have occurred during the generation of the microchromosomes, which are components of avian genomes, presumably with fragmentation of some expressed genes. Against this argument is the presence of TTP-encoding sequences in reptiles, which also have microchromosomes (2).
It is not possible to prove this negative result unequivocally, but we used several lines of evidence to arrive at this conclusion. First, a sequence-based search of the available avian genomic and EST resources, with protein sequences from mammalian, amphibian, and reptile TTP, did not yield any orthologs. We readily identified fragments of the sequences coding for the other two major TTP family members, ZFP36L1 and ZFP36L2, in chicken and other birds, and completed these sequences in the chicken. As expected, these are highly conserved with their corresponding sequences in mammals, reptiles, and amphibians. We also performed Northern blotting on RNA from serum-stimulated chicken embryo fibroblasts and LPS-stimulated chicken macrophages, both situations in which TTP expression is greatly induced in mammalian cells. In neither case was there evidence for an mRNA species that hybridized to any of three mammalian TTP cDNA probes, or a Xenopus TTP cDNA probe, even though mammalian probes for ZFP36L1 and ZFP36L2 hybridized to their apparent orthologs in the chicken. Finally, we also performed deep sequencing (mRNASeq) on polyA-selected RNA from the serum-stimulated fibroblasts and LPS-stimulated macrophages. In both cases, numerous sequences were identified that corresponded to ZFP36L1 and ZFP36L2, but there were no sequences that were assignable to a putative chicken TTP mRNA. Our tentative conclusion from these experiments is that TTP is not expressed in birds.
TTP is important in mammalian physiology, as exemplified by the TTP KO mouse phenotype (31). These animals appear normal at birth, but soon fail to gain weight like their wild-type (WT) littermates, and develop severe arthritis, dermatitis, conjunctivitis, body fat loss, splenomegaly, lymphadenopathy, myeloid hyperplasia, and autoimmunity. Most of these phenomena can be prevented by injecting the newborn pups with antibodies to TNF-α, generally regarded as the most important proinflammatory cytokine. The TTP deficiency syndrome can also be largely prevented by interbreeding the TTP KO mice with mice deficient in both TNF receptors, again stressing the importance of TNF in the pathogenesis of this syndrome (9). The excess TNF that is characteristic of these mice is due to the lack of TTP's regulatory control on the Tnf mRNA stability (11, 13). Abnormal Tnf mRNA and protein accumulation occur constitutively in these mice, with the resulting severe chronic inflammatory syndrome.
In addition to the chronic TTP deficiency syndrome, TTP's importance in the acute innate immune response can be demonstrated in several ways. One striking recent example is in mice deficient in TTP only in myeloid lineage cells. These mice exhibit a normal external phenotype at ages when the original TTP-deficient mice are quite sick. When these mice are challenged with a low-dose LPS injection, one that does not cause significant illness in WT mice, the myeloid TTP-deficient mice develop lethal endotoxin shock (19, 26). Their basal TNF levels are elevated compared with control, but the most striking difference is a nearly 100-fold increase in serum TNF compared with control animals after the low-dose LPS injection. These data are a dramatic demonstration of TTP's importance in mice, and presumably other mammals, in preventing the conversion of a mild infection into lethal septic or endotoxin shock.
In chicken cells, we examined two situations in which TTP should be rapidly induced, serum-stimulated fibroblasts and LPS-stimulated macrophages, to determine whether either or both TTP family members expressed in these cells were induced, as expected for TTP. In both cases, there was readily detectable constitutive mRNA expression, and a modest increase in expression after stimulation in the case of Zfp36l1 mRNA, but in neither case did the expression pattern resemble TTP in terms of rapid induction, many-fold increase, and rapid return to baseline. The role of these two proteins in avian physiology is not known, but, on the basis of these expression patterns, it seems unlikely that they fill in for TTP in the acute regulation of innate immunity.
What other factors, then, might control the mRNA stability of ARE-containing orthologs of mammalian TTP target mRNAs in birds? There are several other well-known families of AU-rich element binding proteins, and many of these have been identified in the chicken, including the AUF1 proteins heterogeneous nuclear ribonucleoprotein D0 (NP_001026314.1) and heterogeneous nuclear ribonucleoprotein D-like (NP_001026313.1); the ELAV family proteins ELAV-like protein 1 (NP_990164), ELAV-like protein 4 (NP_990161), and RNA-binding protein HuC (NP_990163); the probable E3 ubiquitin-protein ligase roquin (XP_001234605); far upstream element-binding protein 2 (KSRP;NP_989608); and nucleolysin TIAR isoforms 1 and 2 (NP_989687 and NP_001244132). These could all contribute to changes in stability of AU-rich element-containing mRNAs, and it will be interesting to determine whether one or more of them behaves kinetically like mammalian TTP in terms of rapid, large-scale induction by LPS in macrophages.
Several well-known TTP target transcripts do not seem to be expressed in chicken, including that encoding TNF, the prototypical proinflammatory cytokine in mammals. In a recent review of avian cytokine genomics, Kaiser et al. (18) speculated that the apparent absence of TNF expressed by the chicken genome is due to extensive rearrangement of the major histocompatibility complex (MHC), which is severely contracted and rearranged compared with the mammalian MHC. However, because of reports of TNF-like activity in the chicken, and the presence of at least one TNF receptor-like protein in the chicken genome (18), the possibility remains that a TNF ortholog is present but as yet undiscovered. In the present study, we were unable to find sequences corresponding to TNF in the LPS-stimulated chicken macrophages, a situation in mammals in which Tnf mRNA expression should be maximal.
In addition to the Tnf mRNA, TTP has been shown to be important for the stability and regulation of several other mRNAs. Several of these do have chicken orthologs, and most transcripts contain AREs with potential TTP family member binding sites in their 3′UTRs. Among these potential targets, transcripts in the chicken were mRNAs encoding CSF2, IL-10, IL-23A, LIF, and PLK3. In future work, it will be important to determine what factors are required for the regulation of the ARE-mediated decay of these transcripts in the avian innate immune response.
Does TTP absence in birds result in a defect in innate immunity, or in any other aspect of physiology? One possible way to address this question would be to use the recently developed methods for chicken transgenesis (24) to add back a regulated TTP genomic construct from another species, perhaps using a reptile sequence. The feasibility of this approach was recently demonstrated by a transgenic experiment that resulted in decreased ability to spread the H5N1 influenza virus (23). Although the outcome of this hypothetical experiment is difficult to predict, the possibility of developing a chicken that has an improved innate immune response to environmental exposures cannot be ruled out.
Perspectives and Significance
Our finding that modern birds appear to lack TTP is interesting from both evolutionary and immunological perspectives. From an evolutionary point of view, it is clear from its presence in numerous reptiles and amphibians that a gene encoding TTP was present before the evolutionary divergence of modern birds. It is conceivable that it is still present in one or more existing bird species, and it will be interesting to monitor this possibility as more avian genomes are sequenced. It also may be possible to determine its existence in ancient DNA from progenitor species, but this seems unlikely at present. From an immunological standpoint, it will be both interesting and important to determine how birds cope differently with the environmental and microbiological assaults that stimulate the acute innate immune response in mammals. This will be important to understand, both to protect birds from infections, and to protect man from bird-transmitted zoonoses.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: W.S.L., J.M.W., D.L.F., and P.J.B. conception and design of research; W.S.L., D.J.S., E.A.K., A.B.B., and J.M.W. performed experiments; W.S.L., D.J.S., E.A.K., A.B.B., J.M.W., D.L.F., and P.J.B. analyzed data; W.S.L., D.J.S., E.A.K., A.B.B., J.M.W., D.L.F., and P.J.B. interpreted results of experiments; W.S.L. and P.J.B. prepared figures; W.S.L. drafted manuscript; W.S.L., D.J.S., E.A.K., A.B.B., J.M.W., D.L.F., and P.J.B. approved final version of manuscript; D.L.F. and P.J.B. edited and revised manuscript.
ACKNOWLEDGMENTS
We are grateful to the members of the National Institutes of Health (NIH) Intramural Sequencing Center for their help with mRNASeq. We thank Drs. Jacqueline Smith and David Burt for helpful discussions, Dr. Uma Babu of the FDA for the HD11 cells, and Dr. Tom Randall for help with the phylogenetic analysis. We thank Drs. Dori Germolec and Bill Schrader for critical review of the manuscript. This study was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
REFERENCES
- 1.Abramyan J, Badenhorst D, Biggar KK, Borchert GM, Botka CW, Bowden RM, Braun EL, Bronikowski AM, Bruneau BG, Buck LT, Capel B, Castoe TA, Czerwinski M, Delehaunty KD, Edwards SV, Fronick CC, Fujita MK, Fulton L, Graves TA, Green RE, Haerty W, Hariharan R, Hillier LH, Holloway AK, Janes D, Janzen FJ, Kandoth C, Kong L, de Koning J, Li Y, Literman R, Mardis ER, McGaugh SE, Minx P, Mork L, O'Laughlin M, Paitz RT, Pollock DD, Ponting CP, Radhakrishnan S, Raney BJ, Richman JM, St John J, Schwartz T, Sethuraman A, Shaffer B, Shedlock AM, Spinks PQ, Storey KB, Thane N, Thomson RC, Valenzuela N, Vinar T, Warren DE, Warren WC, Wilson RK, Zimmerman LM, Hernandez O, Amemiya CT. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol 14: R28, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfoldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, Ray DA, Boissinot S, Shedlock AM, Botka C, Castoe TA, Colbourne JK, Fujita MK, Moreno RG, ten Hallers BF, Haussler D, Heger A, Heiman D, Janes DE, Johnson J, de Jong PJ, Koriabine MY, Lara M, Novick PA, Organ CL, Peach SE, Poe S, Pollock DD, de Queiroz K, Sanger T, Searle S, Smith JD, Smith Z, Swofford R, Turner-Maier J, Wade J, Young S, Zadissa A, Edwards SV, Glenn TC, Schneider CJ, Losos JB, Lander ES, Breen M, Ponting CP, Lindblad-Toh K. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477: 587–591, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beug H, von Kirchbach A, Doderlein G, Conscience JF, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18: 375–390, 1979 [DOI] [PubMed] [Google Scholar]
- 4.Beutler B, Moresco EM. The forward genetic dissection of afferent innate immunity. Curr Top Microbiol Immunol 321: 3–26, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Blackshear PJ, Lai WS, Kennington EA, Brewer G, Wilson GM, Guan X, Zhou P. Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. J Biol Chem 278: 19947–19955, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Blackshear PJ, Phillips RS, Ghosh S, Ramos SB, Richfield EK, Lai WS. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the Tristetraprolin family of CCCH tandem zinc finger proteins. Biol Reprod 73: 297–307, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Brooks SA, Blackshear PJ. Tristetraprolin (TTP): Interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim Biophys Acta 1829: 666–679, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao H, Tuttle JS, Blackshear PJ. Immunological characterization of tristetraprolin as a low abundance, inducible, stable cytosolic protein. J Biol Chem 279: 21489–21499, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carballo E, Blackshear PJ. Roles of tumor necrosis factor-α receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood 98: 2389–2395, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Carballo E, Cao H, Lai WS, Kennington EA, Campbell D, Blackshear PJ. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J Biol Chem 276: 42580–42587, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carballo E, Gilkeson GS, Blackshear PJ. Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (−/−) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNF-α overproduction. J Clin Invest 100: 986–995, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95: 1891–1899, 2000 [PubMed] [Google Scholar]
- 13.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281: 1001–1005, 1998 [DOI] [PubMed] [Google Scholar]
- 14.Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N. Structural basis for the recruitment of the CCR4-NOT complex by tristetraprolin. Nat Struct Mol Biol 20: 735–739, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frederick ED, Ramos SB, Blackshear PJ. A unique C-terminal repeat domain maintains the cytosolic localization of the placenta-specific tristetraprolin family member ZFP36L3. J Biol Chem 283: 14792–14800, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horner TJ, Lai WS, Stumpo DJ, Blackshear PJ. Stimulation of polo-like kinase 3 mRNA decay by tristetraprolin. Mol Cell Biol 29: 1999–2010, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser P. Advances in avian immunology—prospects for disease control: a review. Avian Pathol 39: 309–324, 2010 [DOI] [PubMed] [Google Scholar]
- 18.Kaiser P, Poh TY, Rothwell L, Avery S, Balu S, Pathania US, Hughes S, Goodchild M, Morrell S, Watson M, Bumstead N, Kaufman J, Young JR. A genomic analysis of chicken cytokines and chemokines. J Interferon Cytokine Res 25: 467–484, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Kratochvill F, Machacek C, Vogl C, Ebner F, Sedlyarov V, Gruber AR, Hartweger H, Vielnascher R, Karaghiosoff M, Rulicke T, Muller M, Hofacker I, Lang R, Kovarik P. Tristetraprolin-driven regulatory circuit controls quality and timing of mRNA decay in inflammation. Mol Syst Biol 7: 560, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol 26: 9196–9208, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem 265: 16556–16563, 1990 [PubMed] [Google Scholar]
- 22.Lai WS, Thompson MJ, Taylor GA, Liu Y, Blackshear PJ. Promoter analysis of Zfp-36, the mitogen-inducible gene encoding the zinc finger protein tristetraprolin. J Biol Chem 270: 25266–25272, 1995 [DOI] [PubMed] [Google Scholar]
- 23.Lyall J, Irvine RM, Sherman A, McKinley TJ, Nunez A, Purdie A, Outtrim L, Brown IH, Rolleston-Smith G, Sang H, Tiley L. Suppression of avian influenza transmission in genetically modified chickens. Science 331: 223–226, 2011 [DOI] [PubMed] [Google Scholar]
- 24.Mozdziak PE, Petitte JN. Status of transgenic chicken models for developmental biology. Dev Dyn 229: 414–421, 2004 [DOI] [PubMed] [Google Scholar]
- 25.Qian X, Ning H, Zhang J, Hoft DF, Stumpo DJ, Blackshear PJ, Liu J. Posttranscriptional regulation of IL-23 expression by IFN-γ through tristetraprolin. J Immunol 186: 6454–6464, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu LQ, Stumpo DJ, Blackshear PJ. Myeloid-specific tristetraprolin deficiency in mice results in extreme lipopolysaccharide sensitivity in an otherwise minimal phenotype. J Immunol 188: 5150–5159, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith J, Paton IR, Bruley CK, Windsor D, Burke D, Ponce de Leon FA, Burt DW. Integration of the genetic and physical maps of the chicken macrochromosomes. Anim Genet 31: 20–27, 2000 [DOI] [PubMed] [Google Scholar]
- 28.Smith J, Paton IR, Murray F, Crooijmans RP, Groenen MA, Burt DW. Comparative mapping of human chromosome 19 with the chicken shows conserved synteny and gives an insight into chromosomal evolution. Mamm Genome 13: 310–315, 2002 [DOI] [PubMed] [Google Scholar]
- 29.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem 283: 11689–11699, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, Blackshear PJ. A pathogenetic role for TNF-α in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4: 445–454, 1996 [DOI] [PubMed] [Google Scholar]
- 32.Wu Z, Kaiser P. Antigen presenting cells in a non-mammalian model system, the chicken. Immunobiology 216: 1177–1183, 2011 [DOI] [PubMed] [Google Scholar]