Abstract
Spinocerebellar ataxia type 10 (SCA10) is an autosomal dominant neurodegenerative disorder caused by a noncoding ATTCT pentanucleotide expansion. An inverse correlation between SCA10 expansion size and age at onset has been reported, and genetic anticipation has been documented. Interruptions in the ATTCT expansion are known to occur within the expansion. In order to determine the effect of repeat interruptions in SCA10 expansions, we designed a PCR assay to easily identify ATCCT repeat interruptions in the 5′-end of the expansion. We screened a cohort of 31 SCA10 families of Mexican, Brazilian and Argentinean ancestry to identify those with ATCCT repeat interruptions within their SCA10 expansions. We then studied the effects of ATCCT interruptions on intergenerational repeat instability, anticipation and age at onset. We find that the SCA10 expansion size is larger in SCA10 patients with an interrupted allele, but there is no difference in the age at onset compared with those expansions without detectable interruptions. An inverse correlation between the expansion size and the age at onset was found only with SCA10 alleles without interruptions. Interrupted expansion alleles show anticipation but are accompanied by a paradoxical contraction in intergenerational repeat size. In conclusion, we find that SCA10 expansions with ATCCT interruptions dramatically differ from SCA10 expansions without detectable ATCCT interruptions in repeat-size-instability dynamics and pathogenicity.
Keywords: repeat instability, repeat interruptions, anticipation
Introduction
Spinocerebellar ataxia type 10 (SCA10 (MIM ID #603516)), an autosomal dominant, neurodegenerative disorder, typically presents with progressive ataxia, which may be accompanied by ethnicity-dependent occurrences of epileptic seizures1, 2, 3, 4, 5 and variable extracerebellar symptoms such as hyperreflexia, polyneuropathy, low IQ and neuropsychiatric symptoms.1 Genetic anticipation is a noted disease feature.4, 6
SCA10 results from a noncoding pentanucleotide repeat (ATTCT) expansion within intron 9 of Ataxin 10.7 Normally ranging from 9 to 32 repeats, the ATTCT repeat expands up to 4500 repeats in SCA10 patients.7, 8, 9 Past analysis on patients of one SCA10 family revealed that the 5′-end of the expansion contained ATATTCT and ATTTTCT heptanucleotide interruptions, disrupting the long ATTCT repeat, whereas the SCA10 expansion in members of a second family lacked these interruptions.8 Further investigations of the effect of these repeat interruptions have been hampered by laborious identification of repeat interruptions by end sequencing of the SCA10 expansion.
To examine the effects of the repeat interruption in SCA10 expansion alleles, we developed a novel PCR assay to quickly identify the repeat interruptions. We then applied this assay to a large cohort of SCA10 families to study the effect of repeat interruptions on expansion size, repeat instability, age at onset and anticipation.
Materials and methods
DNA samples
Blood samples were obtained from 298 members of 31 SCA10 families, including 18 Mexican families, 9 Brazilian families and 1 Argentinean family, after obtaining informed consent under protocols approved by local institutional review boards. Some of these families have been previously described.1, 2, 3, 4, 10 Genomic DNA was extracted from peripheral blood leukocytes using conventional methods.
3′ end sequence from the SCA10 expansion
Genomic DNA isolated from SCA10 somatic cell hybrid lines11 was used as template in a long-range PCR using HotStarTaq Master Mix (Qiagen, Germantown, MD, USA) with forward (5′- GTCATCAGGTTGTTGCTGTGGAAAGGGGCGGTA-3′) and reverse (5′- GCCACTGCACTCCAGCCTGGGCAACATAGAGAG-3′) primers flanking the repeat. PCR products were cloned into the TOPO-TA vector (Invitrogen, Grand Island, NY, USA) and propagated in Escherichia coli grown at 25 °C. Sixteen transformants were selected and sequenced on an ABI Prism 377 Genetic Analyzer (Applied Biosystems, Grand Island, NY, USA) using primers specific to unique, non-repeat sequences flanking the SCA10 expansion.
Identification of SCA10-positive individuals
Normal SCA10 alleles were identified by PCR as described.2, 12 Samples containing a single normal allele were further analyzed by repeat-primed PCR (RP-PCR) as described.13 RP-PCR-positive samples were subjected to Southern blot analysis to determine the expansion size as previously described.7
ATCCT repeat interruption PCR (ATCCT-PCR)
ATCCT reverse primer (L2RT: 5′- TACGCATCCCAGTTTGAGACGG(AGGAT)6-3′) and forward primer (LP-L: 5′-GGAATTCGGCTTAAATATCCAACTAAAAGACTACTAGAATGG-3′) were paired in a PCR reaction using KOD Hot Start DNA Polymerase (EMD Millipore, Billerica, MA, USA). The 1.2-kb PCR products were gel purified (Qiagen) and used as template for sequence reactions on an ABI 3130 Genetic Analyzer (Applied Biosystems).
Statistical analysis
Prism 5 (Synergy Software, La Jolla, CA, USA) was used to generate graphs and perform statistical analyses. For comparisons of two groups, Student's t-test was performed. For comparisons of four groups, one-way analysis of variance with post-hoc Bonferroni's multiple comparison test was performed. Pearson's correlation coefficient was calculated for correlations of age at onset and expansion size. Two-tailed P-values were calculated. Standard deviation of the mean is expressed throughout.
Results
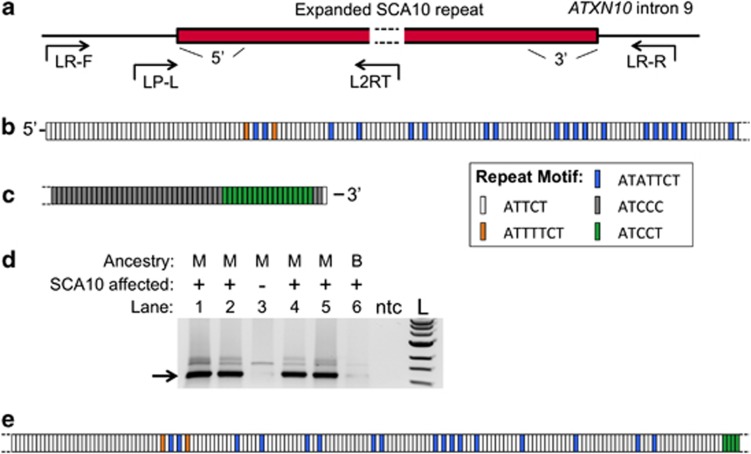
Long-range PCR products across the SCA10 expansion (Figure 1a, primer combination LR-F and LR-R) were generated from the genomic DNA of somatic cell hybrid lines derived from SCA10 patients,11 subcloned in E. coli and then sequenced at the ends. We identified heptanucleotide (ATATTCT and ATTTTTCT) repeat interruptions from the 5′-end of the repeat, matching those previously identified8 (Figure 1b). In addition, we identified a novel ATCCT interruption located at the 3′-end of the expansion (Figure 1c).
Figure 1.
Configuration of ATCCT repeat interruptions within the SCA10 pentanucleotide expansion. (a) Schematic of the SCA10 repeat expansion (red rectangle) in intron 9 of the Ataxin 10 gene, depicting the 5′- and 3′-ends of the repeat. Primers used to amplify full-length (LR-F and LR-R) and ATCCT-PCR (LP-L and L2RT) products are indicated by arrows relative to the repeat. (b) Schematic of the motif structure of the 5′-end of the expansion obtained from SCA10 hybrid cell lines. Sequence obtained is as follows: 5′-ATTCT40–41 ATTTTCT ATTCT (ATATTCT ATTCT)2 ATTTTCT ATTCT11 ATATTCT ATTCT5 ATATTCT ATTCT10 ATATTCT ATTCT2 ATATTCT ATTCT12 (ATATTCT ATTCT)2 ATTCT11 (ATATTCT ATTCT)4 ATTCT2 ATATTCT ATTCT8–10 (ATATTCT ATTCT)5 ATTCT7–8 ATATTCT ATTCTn-3′ and matches previously identified interruption sequences.8 (c) Schematic of the motif structure of the 3′-end of the expansion obtained from SCA10 hybrid lines. Sequence obtained is as follows: 5′-(ATCCT)34+ (ATCCC)4–28 (ATCCT)2 ATTCT1-3′. (d) Typical ATCCT-PCR results from affected (+) or unaffected (−) samples of Mexican (M) or Brazilian (B) ancestry. Lanes containing no template control (ntc) or DNA size standard (L) are indicated. The arrow indicates the 1.2-kb ATCCT-PCR product size. The faint large (∼2.2 kb) band may be a product that resulted from annealing of the reverse primer to interrupted sequence further downstream, whereas the faint middle band is a constant background band found even in unaffected individuals. (e) An example of sequence obtained by direct sequencing of 1.2-kb band of the ATCCT-positive PCR product (from lane 1 above; 5′-ATTCT30+ ATTTTCT ATTCT (ATATTCT ATTCT)2 ATTTTCT ATTCT11 ATATTCT ATTCT5 ATATTCT ATTCT10 ATATTCT ATTCT2 ATATTCT ATTCT12 ATATTCT ATTCT ATATTCT ATTCT12 (ATATTCT ATTCT)4 ATTCT2 ATATTCT ATTCT10 (ATATTCT ATTCT)5 ATTCT7 ATATTCT ATTCT14 ATATTCT ATTCT3 ATATTCT ATTCT16 (ATCCT)2+-3′). An exact count of ATTCT repeats at the 5′-end cannot be made as the sequencing primer anneals near the beginning of the repeat and sequence data began with ATTCT sequences. However, at least 30–35 repeats were observed in various samples. In panels b, c and e: white rectangle, ATTCT motif; orange rectangle, ATTTTCT interruption motif; blue rectangle, ATATTCT interruption motif; gray rectangle, ATCCT interruption motif; green rectangle, ATCCC interruption motif as indicated.
We then designed an ATCCT repeat annealing primer for use in a PCR assay and paired it with an upstream primer that annealed to unique sequence outside the expansion (Figure 1a, primer combination LP-L and L2RT). With these primers, we obtained a 1.2-kb PCR product from some but not all SCA10-positive patients (Figure 1d). Direct sequencing of the purified PCR product revealed stretches of pure repeats frequently interrupted by a combination of heptanucleotide ATTTTCT and ATATTCT interruptions similar to what was seen at the 5′-end, in addition to the more distally located pentanucleotide ATCCT motifs (Figure 1e). As the reverse primer specifically anneals to the ATCCT motif, we refer to this PCR protocol as well as positive SCA10 patients as ‘ATCCT-PCR' and ‘ATCCT-positive', respectively. Consequently, ‘ATCCT-positive' expansions are defined as containing an ATCCT repeat 1.2-kb downstream of the forward primer annealing site. With this novel ATCCT-PCR assay, we can easily and quickly screen for the presence of ATCCT repeat interruptions within SCA10 expansions.
After screening individuals from SCA10 families for the presence of the expanded SCA10 allele by RP-PCR and determining the size of the expansion by Southern blotting, we then checked for the presence of ATCCT repeat interruptions by our new ATCCT-PCR protocol. We then, using family pedigrees, identified whether the allele was maternally or paternally transmitted. In our analysis, we also included published data from an additional 15 SCA10-positive individuals from the Mexican–American family6 that was originally identified as not carrying ATATTCT and ATTTTCT heptanucleotide interruptions within their SCA10 expansions.8
Individuals originally identified with heptanucleotide repeat interruptions8 are positive for the ATCCT-PCR reaction. The ATCCT interruption was seen in all SCA10-positive family members of five families of Mexican origin. Furthermore, all SCA10-positive members of the nine Brazilian, the single Argentinean and the remaining thirteen Mexican families tested ATCCT negative. As negative controls, seven Brazilian, three Argentinean and 77 Mexican SCA10-negative samples were tested and determined to be ATCCT-negative as expected. In addition, in families where multiple members were tested, ATCCT-positive or -negative status was maintained within the family, including 16 parent–child pairs. The faithful heritability of these repeat interruptions was previously noted.8
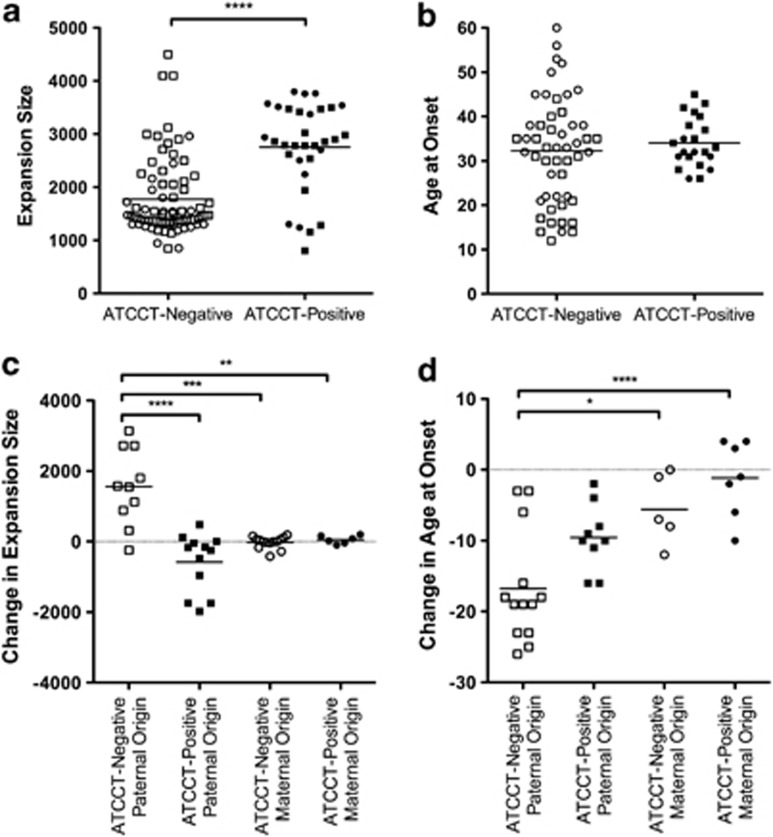
We first examined the difference in expansion size between alleles with ATCCT interruption and those without (Figure 2a). We found that ATCCT-positive SCA10 alleles (2757±835 repeats, n=32) are significantly longer (t(108)=6.132, P<0.0001) than ATCCT-negative SCA10 alleles (1773±734 repeats, n=78).
Figure 2.
Effects of ATCCT interruptions on SCA10 expansions and age at onset. (a) Mean SCA10 expansion size in ATCCT-negative alleles (white squares, paternally transmitted, and circles, maternally transmitted) and ATCCT-positive alleles (black squares, paternally transmitted, and circles, maternally transmitted). (b) Mean age at onset, symbols as in panel a. (c) Mean intergenerational change in expansion size as calculated by subtracting the parents' expansion size from that of their childs' in paternally transmitted, ATCCT-negative alleles (white squares), paternally transmitted, ATCCT-positive alleles (black squares), maternally transmitted, ATCCT-negative alleles (white circles) and maternally transmitted, ATCCT-positive alleles (black circles). (d) Mean intergenerational change in age at onset as calculated by subtracting the parent's age at onset from that of their child, symbols as in c. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
We next compared the age at onset in patients with ATCCT-positive versus ATCCT-negative SCA10 expansions (Figure 2b). We found no significant difference (Welch-corrected t(70)=0.8693, P=0.3877) in the mean age at onset in SCA10 patients with ATCCT-positive (34.0±5.5 years, n=22) and ATCCT-negative (32.3±11.9 years, n=51) SCA10 expansions. However, the variance between the two groups differed significantly (F(50,21)=4.595, P=0.0003).
We then identified 43 parent–child pairs (Supplementary Table 1), where we had obtained the size of the SCA10 expansion in our pedigrees. We calculated the intergenerational change in expansion size by subtracting the size of the parents' expansion from the size of the child's expansion (Figure 2c) and found a significant effect (F(3,38)=17.97, P<0.0001). Here, differences were noted in the gender of the parent. Maternally transmitted expansions behaved similarly, whether ATCCT-negative (−22±167 repeats, n=14) or ATCCT-positive expansions (54±116 repeats, n=6), and were relatively stable. However, intergenerational instability in paternally transmitted alleles was dramatically different: ATCCT-positive expansions contracted (−577±827 repeats, n=12), whereas ATCCT-negative expansions enlarged (1558±1091 repeats, n=10). Significant differences were seen between paternally transmitted, ATCCT-negative and paternally transmitted, ATCCT-positive expansions (P<0.0001). We also found a statistical difference between paternally transmitted, ATCCT-negative and maternally transmitted, ATCCT-negative alleles (P<0.001), as well as between paternally transmitted, ATCCT-negative and maternally transmitted, ATCCT-positive (P<0.01) alleles.
We next identified 36 parent–child pairs in our pedigrees where we had obtained the age at onset (Supplementary Table 1). We calculated the change in age at onset by subtracting the age at onset of the parent from the age at onset of the child (Figure 2d) and found a statistically significant effect on the change in age at onset (F(3,30)=10.41, P=0.0002). All four groups demonstrated anticipation with an earlier average age at onset in subsequent generations although the magnitude of the change differed between groups. Maternally transmitted alleles demonstrated the least amount of change in the age at onset between generations (−1.1±5.4 years in ATTCT-positive alleles, n=9, versus −5.6±5.0 years in ATTCT-negative alleles, n=5). Paternally transmitted alleles demonstrated marked anticipation. ATCCT-positive alleles had a mean change in age at onset of −9.6±4.7 years (n=9), whereas ATCCT-negative alleles had a mean change in age at onset of −16.8±7.9 years (n=13). Between these groups a significant difference in the change in age at onset was found between paternally and maternally transmitted ATCCT-negative alleles (P<0.05) as well as between paternally transmitted, ATCCT-negative alleles and maternally transmitted ATCCT-positive alleles (P<0.001).
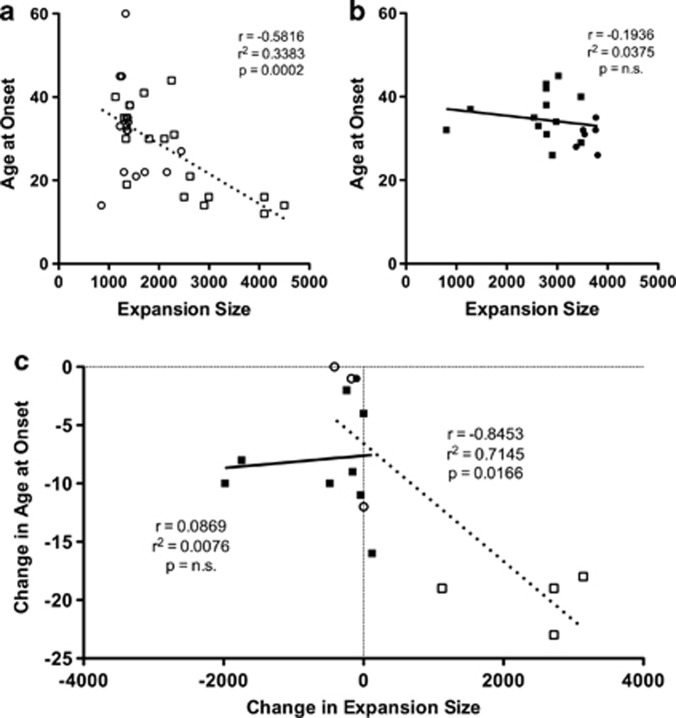
We then examined the effects of the ATCCT repeat interruption on the correlation between age at onset and expansion size (Figure 3). We found a moderate, inverse correlation between age at onset and expansion size in ATCCT-negative individuals (Figure 3a; r=−0.5816, r2=0.3383, P=0.002, n=35), and a non-significant inverse correlation in ATCCT-positive individuals (Figure 3b; r=−0.1936, r2=0.03749, P=0.4271, n=19).
Figure 3.
Effects of ATCCT interruptions on the relationship between expansion size and age at onset and genetic anticipation. (a) Age at onset plotted against expansion size for ATCCT-negative alleles (white circles, maternally transmitted alleles; white squares, paternally transmitted alleles, dashed line). (b) Age at onset plotted against expansion size for ATCCT-positive alleles (black circles, maternally transmitted alleles; black squares, paternally transmitted alleles, solid line). (c) Intergenerational changes in age at onset plotted against intergenerational changes in expansion size, symbols and lines as in panels a and b.
To further examine the effects of ATCCT interruptions on the relationship between anticipation and intergenerational changes in expansion size, we plotted the change in age at onset against the change in expansion size in 16 parent–child pairs (Figure 3c). In ATCCT-negative transmissions, we find a strong, inverse correlation (r=−0.8453, r2=0.7145, P=0.0166, n=7), indicating that as the expansion size increases in subsequent generations, the age at onset decreases. Conversely, for ATCCT-positive transmissions, we find a negligible, positive correlation (r=0.08691, r2=0.0076, P=0.8241, n=9).
Discussion
In this study, we used a new assay to identify SCA10 expansions with ATCCT-interrupted motifs, and report on their effect on SCA10 expansion size, correlations between age at onset and expansion size as well as anticipation and intergenerational instability of the expansion. The ATCCT-PCR assay can rapidly determine the presence of ATCCT repeat interruptions within the 5′-end of SCA10 expansions. However, negative results from this assay need to be interpreted with caution as this may not necessarily indicate that an ATCCT-negative SCA10 expansion is a ‘pure' or non-interrupted expansion. The SCA10 expansion may contain repeat interruptions, which yet give a negative result for a number of reasons including: (1) ATCCT-interrupted repeats are present but are farther interior, preventing amplification of a larger PCR product, (2) interrupted repeats contain heptanucleotide interruptions, but lack a sufficient length of ATCCT repeats for primer annealing, or (3) interrupted repeats of an entirely different motif are present as seen in a SCA10 patient with an unusually small, 280 repeat expansion allele and ATGCT interruptions.8
ATCCT interruptions, age at onset and anticipation
Many coding and noncoding repeat expansion disorders share the common genetic feature of anticipation, the phenomenon whereby symptoms appear at an earlier age than previous generations. Anticipation is explained by increases in repeat expansion size and a strong inverse correlation between repeat length and age at onset. In SCA10, anticipation as well as an inverse correlation between repeat length and age at onset was noted in Brazilian and some Mexican families.1, 4, 6 Yet, a separate Mexican–American family, where repeat interruptions were later identified, demonstrated anticipation but lacked the correlation between age at onset and repeat length.6, 8, 14 Our current study has confirmed this lack of correlation and has also demonstrated it in additional families. This study highlights the presence of ATCCT repeat interruptions as a contributing factor in these disparate results. For ATCCT-positive expansions, there is a statistically insignificant correlation between the age at onset and expansion size. Notably, for similarly sized alleles, ATCCT-positive expansions generally confer a later age at onset than those that are ATCCT negative. How can the presence of ATCCT interruptions counterbalance the effects of age at onset?
The majority of parent–child pairs we examined demonstrated anticipation, but to varying degrees (paternal, ATCCT-negative>paternal, ATCCT-positive>maternal, ATCCT-negative>maternal, ATCCT-positive). However, only paternally transmitted, ATCCT-negative alleles demonstrated large gains in expansion size. This highlights a seemingly paradoxical effect of repeat interruptions on the relationship between anticipation and changes in expansion size between generations. Typically, decreasing the expansion size would be expected to have a beneficial effect by conferring a later age at onset in subsequent generations. However, in every ATCCT-positive transmission, but one, there is a decrease in expansion size with a concomitant decrease in the age at onset. Notably, in these ATCCT-positive transmissions, the child inherits the ATCCT interruption from the parent and is ATCCT positive. The molecular mechanisms of this paradox remain unknown. However, a small minority of CTG expanded alleles in myotonic dystrophy type 1 undergo a similar, paradoxical contraction process.15, 16 How do ATCCT interruptions counteract the beneficial effects on the age at onset from decreasing the expansion size in subsequent generations?
ATCCT interruptions and repeat instability
The substantial difference on intergenerational repeat instability seen between paternally transmitted ATCCT-positive and ATCCT-negative expansions indicates possible differences in the underlying mechanism of repeat instability of these different alleles. A model might be proposed whereby strand slippage of complimentary strands of tandem repeats during DNA replication could generate instability of the SCA10 expansion. In such a model, ATATTCT, ATTTTCT and ATCCT repeat interruptions are then hypothesized to counteract strand slippage and act to stabilize the SCA10 expansions. This proposed mechanisms is similar to what was demonstrated for the CAT interruptions in SCA1 and AGG interruptions in FRAXA repeat expansions.17 Accordingly, the presence of AGG interruptions in maternal premutation alleles of the fragile X syndrome decreases the chance of expansion to a full mutation allele in their children.18 Such a mechanism may be in place for maternally transmitted, ATCCT-positive expansions. Although the sizable contractions (ATCCT-positive) and expansions (ATCCT-negative) of paternally transmitted expansions could still involve misaligned strands due to slippage, a different mechanism may also be considered. Such large changes in expansion size could come about via unequal crossing-over during genetic recombination as is proposed in the CCTG expansions from myotonic dystrophy type 2. In this case, the ‘crossover hotspot' may be a result of the low thermodynamic stability of the CCTG repeats.19 Along these lines, long normal ATTCT repeats appear to act as an in vitro DNA-unwinding element,20 which are thermodynamically unstable genomic regions that can support initiation of replication.21
ATCCT interruptions and SCA10 expansion size
SCA10 expansions containing ATCCT repeat interruptions tend to be longer than those without the interruptions. As paternal transmission of ATCCT-positive expansions results in large contractions in size, whereas maternal transmissions are relatively stable, this begs the questions of the origin of large SCA10 expansions with ATCCT interruptions. The common disease haplotype that exists between SCA10 families, including those that whose expansions do as well as do not carry ATCCT interruptions, supports the idea of a single origin of the expansion.22 Did the uninterrupted expansion occur first and then gather interruptions? Or did interruptions arise first and were later lost from large expansions? Large normal SCA10 alleles (≥17 repeats) typically contain interrupted ATTGT or TTTCT motifs next to the most distal ATTCT motif, whereas smaller, normal alleles (≤16 repeats) are uninterrupted.8 Such interruptions in large, normal alleles are thought to act to stabilize these alleles from further expansion, as is proposed for SCA1 and SCA2, where interruptions are also found in long, normal alleles but are absent from pathogenically expanded alleles.23, 24 However, the smallest disease-causing SCA10 allele (280 repeats) with reduced penetrance has multiple ATGCT interruptions, suggesting that the expansion origin of SCA10 repeat is not straightforward.8 Further studies will be needed to decipher the origin of SCA10 expansions.
Other effects of the repeat interruption on disease duration, age at death and other disease phenotypes remains unexplored. Why the ATCCT interruption can counterbalance the beneficial effect of a smaller expansion size on the age at onset is a question that may be explored by testing the hypothesis that interrupting sequences modify the pathogenic potency of the expanded alleles. As SCA10 is considered to act through an RNA mediated gain-of-function,11, 25, 26 further study on RNA toxicity of AUCCU-positive and -negative alleles may shed light on this question. Finally, it will be interesting to determine whether there are additional repeat interruptions in the expansions of other SCA10 patients.
Acknowledgments
The study was supported by NS041547 (TA).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Rasmussen A, Matsuura T, Ruano L, et al. Clinical and genetic analysis of four Mexican families with spinocerebellar ataxia type 10. Ann Neurol. 2001;50:234–239. doi: 10.1002/ana.1081. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Achari M, Khajavi M, Bachinski LL, Zoghbi HY, Ashizawa T. Mapping of the gene for a novel spinocerebellar ataxia with pure cerebellar signs and epilepsy. Ann Neurol. 1999;45:407–411. doi: 10.1002/1531-8249(199903)45:3<407::aid-ana21>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Gatto EM, Gao R, White MC, et al. Ethnic origin and extrapyramidal signs in an Argentinean spinocerebellar ataxia type 10 family. Neurology. 2007;69:216–218. doi: 10.1212/01.wnl.0000265596.72492.89. [DOI] [PubMed] [Google Scholar]
- Teive HA, Roa BB, Raskin S, et al. Clinical phenotype of Brazilian families with spinocerebellar ataxia 10. Neurology. 2004;63:1509–1512. doi: 10.1212/01.wnl.0000142109.62056.57. [DOI] [PubMed] [Google Scholar]
- Teive HA, Munhoz RP, Raskin S, et al. Spinocerebellar ataxia type 10: Frequency of epilepsy in a large sample of Brazilian patients. Mov Disord. 2010;25:2875–2878. doi: 10.1002/mds.23324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal RP, Achari M, Matsuura T, et al. Clinical features and ATTCT repeat expansion in spinocerebellar ataxia type 10. Arch Neurol. 2002;59:1285–1290. doi: 10.1001/archneur.59.8.1285. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Yamagata T, Burgess DL, et al. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat Genet. 2000;26:191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Fang P, Pearson CE, et al. Interruptions in the expanded ATTCT repeat of spinocerebellar ataxia type 10: repeat purity as a disease modifier. Am J Hum Genet. 2006;78:125–129. doi: 10.1086/498654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL, Wu YQ, Lei LF, et al. Polynucleotide repeat expansion of nine spinocerebellar ataxia subtypes and dentatorubral-pallidoluysian atrophy in healthy Chinese Han population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010;27:501–505. doi: 10.3760/cma.j.issn.1003-9406.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Raskin S, Ashizawa T, Teive HA, et al. Reduced penetrance in a Brazilian family with spinocerebellar ataxia type 10. Arch Neurol. 2007;64:591–594. doi: 10.1001/archneur.64.4.591. [DOI] [PubMed] [Google Scholar]
- Wakamiya M, Matsuura T, Liu Y, et al. The role of ataxin 10 in the pathogenesis of spinocerebellar ataxia type 10. Neurology. 2006;67:607–613. doi: 10.1212/01.wnl.0000231140.26253.eb. [DOI] [PubMed] [Google Scholar]
- Cagnoli C, Michielotto C, Matsuura T, et al. Detection of large pathogenic expansions in FRDA1, SCA10, and SCA12 genes using a simple fluorescent repeat-primed PCR assay. J Mol Diagn. 2004;6:96–100. doi: 10.1016/S1525-1578(10)60496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Ashizawa T. Polymerase chain reaction amplification of expanded ATTCT repeat in spinocerebellar ataxia type 10. Ann Neurol. 2002;51:271–272. doi: 10.1002/ana.10049. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Fang P, Lin X, et al. Somatic and germline instability of the ATTCT repeat in spinocerebellar ataxia type 10. Am J Hum Genet. 2004;74:1216–1224. doi: 10.1086/421526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashizawa T, Anvret M, Baiget M, et al. Characteristics of intergenerational contractions of the CTG repeat in myotonic dystrophy. Am J Hum Genet. 1994;54:414–423. [PMC free article] [PubMed] [Google Scholar]
- Monckton DG, Wong LJ, Ashizawa T, Caskey CT. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum Mol Genet. 1995;4:1–8. doi: 10.1093/hmg/4.1.1. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Eichler EE, Lorenzetti D, et al. Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37:2701–2708. doi: 10.1021/bi972546c. [DOI] [PubMed] [Google Scholar]
- Yrigollen CM, Durbin-Johnson B, Gane L, et al. AGG interruptions within the maternal FMR1 gene reduce the risk of offspring with fragile X syndrome. Genet Med. 2012;14:729–736. doi: 10.1038/gim.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere R, Wells RD. DM2 CCTG*CAGG repeats are crossover hotspots that are more prone to expansions than the DM1 CTG*CAG repeats in Escherichia coli. J Mol Biol. 2006;360:21–36. doi: 10.1016/j.jmb.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Liu G, Bissler JJ, Sinden RR, Leffak M. Unstable spinocerebellar ataxia type 10 (ATTCT*(AGAAT) repeats are associated with aberrant replication at the ATX10 locus and replication origin-dependent expansion at an ectopic site in human cells. Mol Cell Biol. 2007;27:7828–7838. doi: 10.1128/MCB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D, Eddy MJ. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989;8:4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida T, Alonso I, Martins S, et al. Ancestral origin of the ATTCT repeat expansion in spinocerebellar ataxia type 10 (SCA10) PLoS One. 2009;4:e4553. doi: 10.1371/journal.pone.0004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MY, Ranum LP, Duvick LA, Servadio A, Zoghbi HY, Orr HT. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- Pulst SM, Nechiporuk A, Nechiporuk T, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- White M, Xia G, Gao R, et al. Transgenic mice with SCA10 pentanucleotide repeats show motor phenotype and susceptibility to seizure: A toxic RNA gain-of-function model. J Neurosci Res. 2012;90:706–714. doi: 10.1002/jnr.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MC, Gao R, Xu W, et al. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010;6:e1000984. doi: 10.1371/journal.pgen.1000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.