Abstract
Uterine leiomyomas, or fibroids, are the most common human tumors. Based on histopathology, they can be divided into common leiomyomas and various relatively rare subtypes that mimic malignancy in one or more aspects. Recently, we showed that exon 2 of mediator complex subunit 12 (MED12) is mutated in up to 70% of common fibroids. To investigate the frequency of MED12 exon 2 mutations in histopathological uterine leiomyoma variants, we screened altogether 206 lesions, including 69 histopathologically common leiomyomas, 59 cellular (23 cellular and 36 highly cellular), 18 atypical and 26 mitotically active leiomyomas, as well as 34 uterine fibroid samples from 14 hereditary leiomyomatosis and renal cell cancer patients with a heterozygous germ line mutation in fumarate hydratase (FH). The uterine leiomyoma variants harbored MED12 exon 2 mutations significantly less frequently than common leiomyomas (P=2.93 × 10−8). In all, 6 mutations were detected among cellular fibroids (6/67; 8.96%), 3 among atypical fibroids (3/18; 16.67%) and 10 among mitotically active fibroids (10/26; 38.46%). Only mitotically active fibroids displayed a mutation frequency that was not statistically different from common leiomyomas (P=0.11). Three MED12 exon 2 mutations were detected among 34 tumors with a heterozygous germ line FH mutation (P=5.28 × 10−7). None of these tumors displayed biallelic inactivation of FH. Our results suggest that MED12 mutation positivity is a key characteristic of common leiomyomas. Cellular and atypical fibroids, in particular, may arise through different molecular mechanisms. The results also propose that MED12 and biallelic FH mutations may be mutually exclusive.
Keywords: uterine fibroid, uterine leiomyoma variants, MED12, FH
Introduction
Uterine leiomyomas, also known as fibroids, are benign tumors that occur in nearly 70% of women by the age of 50 years.1 Regardless of their benign nature, fibroids can cause a variety of health problems, such as abnormal menstrual bleeding and infertility.2 Uterine leiomyomas are the most common cause for hysterectomy and pose a considerable socioeconomic burden.3, 4 Hormonal factors, such as estrogen and progesterone, are known to have a role in the growth of leiomyomas and the lesions may regress when hormone levels decrease.5 Based on histopathology, fibroids can be classified in common leiomyomas and several relatively rare subtypes, such as cellular, atypical, mitotically active, epitheloid and myxoid fibroids. These histopathological variants account for <5% of all uterine leiomyomas. Although these variants mimic malignancy in one or more aspects,6 their behavior is benign.
Fibroids also occur in the context of hereditary leiomyomatosis and renal cell cancer (HLRCC, OMIM # 150800). HLRCC is a tumor susceptibility syndrome predisposing to cutaneous and uterine leiomyomatosis, and in some families also to renal cell cancer.7, 8 It is caused by heterozygous germ line mutations in fumarate hydratase (FH), which encodes the citric acid cycle enzyme fumarase.9 Uterine leiomyomas associated with this syndrome display loss of the normal FH allele, occur at young age and require treatment more often than sporadic uterine leiomyomas.10 The role of FH in sporadic uterine leiomyoma tumorigenesis has previously been investigated and biallelic inactivation of FH, the hallmark of HLRCC leiomyomatosis, has been detected in only 1.3% of the tumors.11, 12
Recently, we discovered specific mutations in exon 2 of mediator complex subunit 12 (MED12) in as many as 70% of the 225 unselected uterine leiomyomas studied.13 The majority represented missense mutations affecting a single codon glutamine 44. MED12 is part of a multiprotein complex called Mediator, which participates in regulation of global as well as gene-specific transcription.14 MED12 forms together with MED13, CDK8 and cyclin C a Mediator subcomplex known as the CDK8 module. This module interacts with the main Mediator complex, but functions outside the complex have also been proposed.15 MED12 is a key regulator of the kinase activity of CDK8 module, and the protein directly interacts with multiple transcription factors, such as β-catenin.14, 15, 16
These recent advances may provide tools for molecular classification of uterine fibroids. To examine the prevalence of MED12 exon 2 mutations in various uterine leiomyoma subtypes, we first made a significant effort to derive an appropriate sample set to allow addressing this question. We then screened altogether 206 uterine leiomyoma samples representing common leiomyomas, the most common histopathological variants of leiomyoma, and fibroids from HLRCC patients for the MED12 mutations.
Materials and methods
Research permits
All the samples used in this study were derived according to Finnish laws and regulations, either after informed consent or, if anonymized, after authorization from the director of the respective health-care unit. The study has been approved by the appropriate ethics review board of Helsinki University Central Hospital, Finland.
Archival tissue samples in MED12 exon 2 mutation analysis
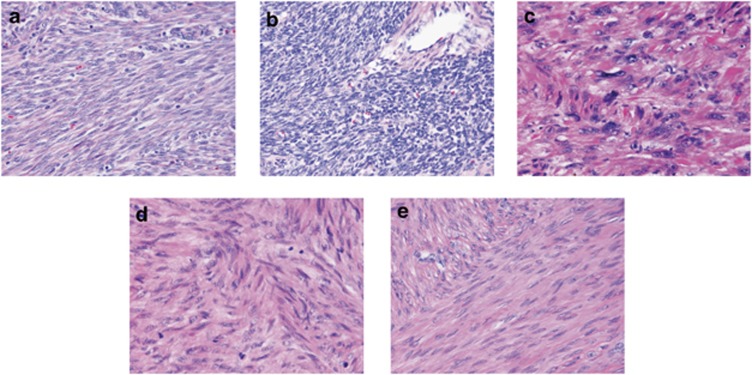
A series of 172 anonymous archival formalin-fixed paraffin-embedded (FFPE) uterine leiomyoma samples was collected at the Department of Pathology, Hospital District of Helsinki and Uusimaa, Finland. The series consisted of 69 common leiomyomas, as well as 59 cellular (23 cellular and 36 highly cellular), 18 atypical and 26 mitotically active leiomyomas (8 lesions showing simultaneously increased cellularity). The representatives of each subtype are shown in Figure 1. Eight mitotically active leiomyomas with increased cellularity were included to both mitotically active and cellular leiomyoma groups for statistical testing. Also a set of 34 uterine fibroid samples from 14 HLRCC patients with known FH mutation status17 was available for MED12 exon 2 mutation screening. None of the samples have been previously published in respect of MED12 mutations.
Figure 1.
Histopathological variants of uterine leiomyoma. (a) Cellular leiomyoma. (b) Highly cellular leiomyoma. (c) Atypical leiomyoma. (d) Mitotically active leiomyoma. (e) Common leiomyoma. HE-stainings are shown with × 40 magnification.
Histopathological evaluation of tissue samples
Histopathological assessment of uterine leiomyoma samples was performed by a pathologist (RB). FFPE blocks and ice blocks from fresh frozen tissue samples harboring primarily tumor tissue were selected, sectioned on a microtome/cryotome to 5 μm in thickness and stained with hematoxylin–eosin (HE) according to standard procedures. HE-stained sections from each specimen were reviewed and the tumors were classified into common, cellular, atypical and mitotically active leiomyomas according to the WHO criteria.6 The number of mitotic figures per 10 high-power fields, the degree of cellularity (normal, cellular and highly cellular) and severity of nuclear atypia (0–3) were recorded for each uterine leiomyoma. See Supplementary Tables S1 and S2 for detailed histopathological data.
MED12 exon 2 mutation screening
MED12 mutation screening was carried out by direct sequencing. Genomic DNA from the FFPE samples was extracted either with NucleoSpin FFPE DNA Kit or with NucleoSpin FFPE RNA/DNA Kit (Macherey-Nagel, Düren, Germany). Oligonucleotide primers were designed with Primer318 using Ensembl release 62 as the reference. The primer sequences in the 5′ to 3′ direction are GCCCTTTCACCTTGTTCCTT (forward) and AAGCTGACGTTCTTGGCACT (reverse) covering all the observed mutation hotspots in MED12 exon 2 and intron 1. PCR analyses were performed in duplicate to ensure accuracy, with two different amounts of DNA: 15 and 25 ng. PCR products were sequenced utilizing Big Dye Terminator v.3.1 Kit (Applied Biosystems, Foster City, CA, USA) on an ABI3730 Automatic DNA Sequencer according to the manufacturer's instructions. The sequence graphs were analyzed both manually and with Mutation Surveyor-software (Softgenetics, State College, PA, USA).
Detection of FH loss of heterozygosity (LOH)
To assess LOH, tumor DNA from patients with a germ line FH mutation was sequenced, as described above. Three different sets of primers were utilized, one for each mutation: 5′-TTTGTTTTTGTTGCCTCTGATTT-3′ and 5′-GGATTTTGCATCAAGAGCATC-3′ (c.587A>G, p.H196R), 5′-TGCTGCTGCAATAGAAGTTCA-3′ and 5′-CCTGCCCAAGAGTAAGTGGA-3′ (c.671_672delAG, p.E224fs), and 5′-GCTCTGGTTGAGCTCAGTGG-3′ and 5′-GGACCTAGTCAAGTTTTAGCTCCA-3′ (c.1027C>T, p.R343X). For each sample, five parallel PCR reactions were carried out to ensure accuracy. LOH was determined manually by comparing the heights of wild-type and mutant allele peaks. When the height of wild-type allele peak was repeatedly significantly lower than the height of mutant allele peak, LOH was scored.
Statistical analyses
Statistical analyses were performed using R software, version 2.14.0 (R Foundation for Statistical Computing, Vienna, Austria). Differences between the frequencies of MED12 exon 2 mutations in clinical uterine leiomyoma subtypes and common uterine leiomyomas were calculated with Fisher's exact test. All P-values are two-sided and P-value <0.05 was considered statistically significant.
Results
MED12 exon 2 mutation statuses were determined for 103 uterine leiomyoma variants and 69 common leiomyomas. Altogether 18 out of 103 histopathological variants (17.48%) harbored a mutation in MED12 exon 2 (Table 1). Ten mutations were observed among mitotically active fibroids (10/26; 38.46%), three among atypical fibroids (3/18; 16.67%) and six among cellular fibroids (6/67; 8.96%). Of the 42 highly cellular fibroids, only two carried a MED12 mutation (2/42; 4.76%). In contrast, the majority of common leiomyomas (41/69; 59.42%) displayed a MED12 mutation. We observed altogether 59 MED12 exon 2 mutations: 39 missense mutations in codon 44, three in codon 36 and one in codon 43, as well as 15 insertion–deletion type mutations (see Supplementary Tables S1 and S3 for more detailed information of MED12 statuses). The majority of these mutations have previously been reported.13 In addition, one novel missense mutation c.122T>A, p.V41E was detected in a common leiomyoma.
Table 1. MED12 exon 2 mutations in clinical uterine leiomyoma subtypes.
| Leiomyoma subtype | Total | Mutation positive lesions | P-value | OR (95% CI) | |
|---|---|---|---|---|---|
| Common leiomyoma | 69 | 41 | 59.42% | ||
| Leiomyoma variants | 103 | 18 | 17.48% | 2.93 × 10−8 | 6.82 (3.26–14.83) |
| Cellular leiomyoma | 67 | 6 | 8.96% | 2.49 × 10−10 | 14.55 (5.34–46.90) |
| Cellular (+) | 25 | 4 | 16.00% | 1.82 × 10−4 | 7.52 (2.21–33.42) |
| Highly cellular (++) | 42 | 2 | 4.76% | 1.51 × 10−9 | 28.45 (6.49–260.56) |
| Atypical leiomyoma | 18 | 3 | 16.67% | 1.40 × 10−3 | 7.16 (1.79–42.15) |
| Mitotically active leiomyoma | 26 | 10 | 38.46% | 1.05 × 10−1 | 2.32 (0.85–6.64) |
| Leiomyomas from HLRCC-patients | 34 | 3 | 8.82% | 5.28 × 10−7 | 14.74 (4.01–82.59) |
Eight tumors were included in both mitotically active and cellular leiomyomas.
Overall, the histopathological variants of uterine leiomyoma harbored MED12 exon 2 mutations significantly less frequently than common leiomyomas (P=2.93 × 10−8; OR, 6.82; 95% CI, 3.26–14.83; Table 1). Only mitotically active fibroids displayed a MED12 mutation frequency that was not statistically different from the common leiomyomas (P=0.11; OR, 2.32; 95% CI, 0.85–6.64). The lowest mutation frequency was observed in highly cellular fibroids (P=1.51 × 10−9; OR, 28.45; 95% CI, 6.49–260.56).
A total of 34 fibroids from 14 HLRCC patients with a known germ line FH mutation were screened for MED12 exon 2. Only three cases (8.82%) displayed a mutation in MED12, which was significantly less frequently compared with common leiomyomas (P=5.28 × 10−7) (Table 1, see Supplementary Table S2 for more detailed information). In our previous studies, biallelic inactivation of FH has been shown in 14 out of these 34 HLRCC fibroids. The remaining lesions were assessed for LOH, as a marker of FH driven tumorigenesis and altogether 21 fibroids (61.76%) exhibited clear biallelic FH inactivation. LOH was not detected in any of the MED12 mutation positive lesions (see Supplementary Table S2 for more detailed information).
Discussion
The discovery of MED12 as an important target for driver mutations in 50–70% of unselected uterine leiomyomas13, 19, 20, 21, 22 has been an important step toward understanding the tumorigenesis of these extremely common lesions. Recently, MED12 exon 2 mutations have also been detected in uterine leiomyosarcomas and colorectal cancer.22, 23, 24 Although these mutations are not restricted to fibroids, the majority seem to be characteristic to uterine malignancies. So far, the relevance of the mutations for molecular classification of fibroids has remained poorly understood. To investigate the association between MED12 mutation positivity and different histopathological variants of uterine leiomyoma, as well as tumors with a germ line FH mutation, we screened MED12 exon 2 in altogether 206 uterine fibroid samples.
MED12 mutations were significantly less frequent among unusual histopathological variants of uterine leiomyoma as compared with common leiomyomas (P=2.93 × 10−8). Only 16.7% of atypical leiomyomas harbored a MED12 exon 2 mutation. Atypical lesions exhibit nuclear pleomorphism, but lack tumor necrosis and mitotic figures, which are other markers of malignancy. These tumors are often also cellular and like common leiomyomas, their course is benign. Cellular leiomyomas are defined as fibroids with cellularity that is greater than in the normal myometrium. In the utmost form, the cells lack spindle-shaped form and fascicular arrangement typical to leiomyomas, and these tumors have been designated as highly cellular leiomyomas. MED12 exon 2 mutations were uncommon in cellular leiomyomas, and particularly rare in highly cellular leiomyomas (P=1.51 × 10−9). The rare occurrence of MED12 exon 2 mutations in this histopathological subtype suggests a distinct molecular pathogenesis. The difference between the frequencies of MED12 mutation positive lesions in mitotically active and common leiomyomas did not reach significance (P=0.11). This is in keeping with the concept that hormonal and other external factors, not directly related to the tumors themselves, are important for this phenotype.25
Uterine leiomyomas associated with HLRCC have been suggested to display increased cellularity, atypia and occasional mitoses, the same histopathological features that characterize uterine leiomyoma variants.26 In addition, they have been proposed to exhibit other distinct features, such as orangeophilic nucleoli surrounded by a perinucleolar halo.27 Previous literature has suggested that FH inactivation associates with larger, more numerous, and symptomatic tumors, although malignant transformation is rare.26, 28 Only three MED12 exon 2 mutations were observed among 34 fibroids from HLRCC patients (P=5.28 × 10−7). Unlike in typical HLRCC lesions8, 11 and while biallelic inactivation of FH was observed in the majority of the studied HLRCC fibroids, loss of the remaining normal FH allele was not detected in the three MED12 mutant tumors. These three cases may represent incidental lesions not related to the germ line FH mutation. Thus, we did not find any tumors that had both a MED12 mutation as well as biallelic FH inactivation, suggesting that MED12 and FH mutations may represent two different pathways for leiomyomagenesis. Recent study by Markowski et al. showed that MED12 mutations were also absent in tumors harboring the most common cytogenetic rearrangement, a translocation involving chromosome bands 12q15 and 14q24.20, 29 This rearrangement is known to upregulate high mobility group AT-hook 2 (HMGA2) gene at 12q15,30, 31 and thus might provide a third pathway for leiomyomagenesis.
In conclusion, our results indicate that MED12 exon 2 mutations are characteristic to common leiomyomas. We also suggest that MED12 and FH mutations might be mutually exclusive. Before significant progress toward targeted therapies on fibroids can be made, the events leading to genesis of these frequent lesions need to be understood in great detail. The emerging molecular classification should provide valuable tools for accurate stratification of fibroids, as well as clues to the exact molecular mechanisms underlying this tumor type.
Acknowledgments
We thank Sini Nieminen, Inga-Lill Svedberg and Mairi Kuris for excellent technical assistance. This study was supported by the Academy of Finland (Center of Excellence in Cancer Genetics Research, grant 250345 and personal grant 260370 to PV), the Sigrid Jusélius Foundation, and the Cancer Society of Finland, as well as Emil Aaltonen Foundation, Orion-Farmos Research Foundation and K Albin Johansson Foundation (personal grants to NM).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Day Baird D, Dunson DB, Hill MC, Cousins D, Schectman JM. High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. Am J Obstet Gynecol. 2003;188:100–107. doi: 10.1067/mob.2003.99. [DOI] [PubMed] [Google Scholar]
- Stewart EA. Uterine fibroids. Lancet. 2001;357:293–298. doi: 10.1016/S0140-6736(00)03622-9. [DOI] [PubMed] [Google Scholar]
- Flynn M, Jamison M, Datta S, Myers E. Health care resource use for uterine fibroid tumors in the United States. Am J Obstet Gynecol. 2006;195:955–964. doi: 10.1016/j.ajog.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Hartmann KE, Birnbaum H, Ben-Hamadi R, et al. Annual costs associated with diagnosis of uterine leiomyomata. Obstet Gynecol. 2006;108:930–937. doi: 10.1097/01.AOG.0000234651.41000.58. [DOI] [PubMed] [Google Scholar]
- Parker WH. Etiology, symptomatology, and diagnosis of uterine myomas. Fertil Steril. 2007;87:725–736. doi: 10.1016/j.fertnstert.2007.01.093. [DOI] [PubMed] [Google Scholar]
- Hendrickson MR, Tavassoli FA, Kempson RL, McCluggage WG, Haller U, Kubik-Huch RA.Mesenchymal tumours and related lesionsin Tavassoli FA, Devilee P, (eds).: World Health Organization Classification of Tumours: Tumours of the Breast and Female Genital Organs Lyon: IARC Press; 2003233–244. [Google Scholar]
- Reed WB, Walker R, Horowitz R. Cutaneous leiomyomata with uterine leiomyomata. Acta Derm Venereol. 1973;53:409–416. [PubMed] [Google Scholar]
- Launonen V, Vierimaa O, Kiuru M, et al. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci USA. 2001;98:3387–3392. doi: 10.1073/pnas.051633798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Stewart L, Glenn GM, Stratton P, et al. Association of germline mutations in the fumarate hydratase gene and uterine fibroids in women with hereditary leiomyomatosis and renal cell cancer. Arch Dermatol. 2008;144:1584–1592. doi: 10.1001/archdermatol.2008.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuru M, Lehtonen R, Arola J, et al. Few FH mutations in sporadic counterparts of tumor types observed in hereditary leiomyomatosis and renal cell cancer families. Cancer Res. 2002;62:4554–4557. [PubMed] [Google Scholar]
- Lehtonen R, Kiuru M, Vanharanta S, et al. Biallelic inactivation of fumarate hydratase (FH) occurs in nonsyndromic uterine leiomyomas but is rare in other tumors. Am J Pathol. 2004;164:17–22. doi: 10.1016/S0002-9440(10)63091-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen N, Mehine M, Tolvanen J, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Xu X, Hecht A, Boyer TG. Mediator is a transducer of Wnt/beta-catenin signaling. J Biol Chem. 2006;281:14066–14075. doi: 10.1074/jbc.M602696200. [DOI] [PubMed] [Google Scholar]
- Lehtonen HJ, Kiuru M, Ylisaukko-Oja SK, et al. Increased risk of cancer in patients with fumarate hydratase germline mutation. J Med Genet. 2006;43:523–526. doi: 10.1136/jmg.2005.036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Mäkinen N, Heinonen HR, Moore S, Tomlinson IP, van der Spuy ZM, Aaltonen LA. MED12 exon 2 mutations are common in uterine leiomyomas from South African patients. Oncotarget. 2011;2:966–969. doi: 10.18632/oncotarget.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowski DN, Bartnitzke S, Loning T, Drieschner N, Helmke BM, Bullerdiek J. MED12 mutations in uterine fibroids-their relationship to cytogenetic subgroups. Int J Cancer. 2012;131:1528–1536. doi: 10.1002/ijc.27424. [DOI] [PubMed] [Google Scholar]
- McGuire MM, Yatsenko A, Hoffner L, Jones M, Surti U, Rajkovic A. Whole exome sequencing in a random sample of North American women with leiomyomas identifies MED12 mutations in majority of uterine leiomyomas. PLoS One. 2012;7:e33251. doi: 10.1371/journal.pone.0033251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je EM, Kim MR, Min KO, Yoo NJ, Lee SH. Mutational analysis of MED12 exon 2 in uterine leiomyoma and other common tumors. Int J Cancer. 2012;131:E1044–E1047. doi: 10.1002/ijc.27610. [DOI] [PubMed] [Google Scholar]
- Perot G, Croce S, Ribeiro A, et al. MED12 alterations in both human benign and malignant uterine soft tissue tumors. PLoS One. 2012;7:e40015. doi: 10.1371/journal.pone.0040015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämpjärvi K, Mäkinen N, Kilpivaara O, et al. Somatic MED12 mutations in uterine leiomyosarcoma and colorectal cancer. Br J Cancer. 2012;107:1761–1765. doi: 10.1038/bjc.2012.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prayson RA, Hart WR. Mitotically active leiomyomas of the uterus. Am J Clin Pathol. 1992;97:14–20. doi: 10.1093/ajcp/97.1.14. [DOI] [PubMed] [Google Scholar]
- Lehtonen HJ. Hereditary leiomyomatosis and renal cell cancer: update on clinical and molecular characteristics. Fam Cancer. 2011;10:397–411. doi: 10.1007/s10689-011-9428-z. [DOI] [PubMed] [Google Scholar]
- Sanz-Ortega J, Vocke C, Stratton P, Linehan WM, Merino MJ. Morphologic and molecular characteristics of uterine leiomyomas in hereditary leiomyomatosis and renal cancer (HLRCC) syndrome. Am J Surg Pathol. 2013;37:74–80. doi: 10.1097/PAS.0b013e31825ec16f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MH, Toure O, Glenn GM, et al. Novel mutations in FH and expansion of the spectrum of phenotypes expressed in families with hereditary leiomyomatosis and renal cell cancer. J Med Genet. 2006;43:18–27. doi: 10.1136/jmg.2005.033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg AA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: leiomyoma. Cancer Genet Cytogenet. 2005;158:1–26. doi: 10.1016/j.cancergencyto.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Tallini G, Vanni R, Manfioletti G, et al. HMGI-C and HMGI(Y) immunoreactivity correlates with cytogenetic abnormalities in lipomas, pulmonary chondroid hamartomas, endometrial polyps, and uterine leiomyomas and is compatible with rearrangement of the HMGI-C and HMGI(Y) genes. Lab Invest. 2000;80:359–369. doi: 10.1038/labinvest.3780040. [DOI] [PubMed] [Google Scholar]
- Gross KL, Neskey DM, Manchanda N, et al. HMGA2 expression in uterine leiomyomata and myometrium: quantitative analysis and tissue culture studies. Genes Chromosomes Cancer. 2003;38:68–79. doi: 10.1002/gcc.10240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.