Abstract
Perlman syndrome is a rare, autosomal recessive overgrowth disorder. Recently, the deletion of exon 9 and other mutations of the DIS3L2 gene have been reported in patients; however, the mechanism behind this deletion is still unknown. We report the homozygous deletion of exon 9 of DIS3L2 in a Japanese patient with Perlman syndrome. We identified the deletion junction, and implicate a non-allelic homologous recombination (NAHR) between two LINE-1 (L1) elements as the causative mechanism. Furthermore, the deletion junctions were different between the paternal and maternal mutant alleles, suggesting the occurrence of two independent NAHR events in the ancestors of each parent. The data suggest that the region around exon 9 might be a hot spot of L1-mediated NAHR.
Keywords: LINE-1, non-allelic homologous recombination, Perlman syndrome, exon deletion
Introduction
Perlman syndrome (OMIM #267000) is a rare, autosomal recessive overgrowth disorder characterized by polyhydramnios with neonatal macrosomia, nephromegaly, distinctive facies, renal dysplasia, nephroblastomatosis and a predisposition to Wilms tumor. The clinical features are reminiscent of Beckwith–Wiedemann syndrome; however, genetic and epigenetic alterations at 11p15.5 have been excluded from the etiology.1 Recently, DIS3L2 at 2q37.1 was reported as a causative gene, showing homozygous deletions of exon 6 or exon 9 (82.8 and ∼22 kb, respectively) and compound heterozygous mutations in such patients.2 However, the mechanisms behind these deletions are still unknown. In this report, we explore a parentally transmitted homozygous deletion of exon 9 in DIS3L2 responsible for Perlman syndrome in a Japanese patient. We detected the sequence of the deletion junction and found that a rare, non-allelic homologous recombination (NAHR) between two collinear LINE-1 (L1) elements was the causative mechanism of the deletion. To our knowledge, this is the fourth NAHR event to be documented as causing a human disease. Furthermore, the deletion junctions were different between the paternal and maternal mutant alleles, suggesting the occurrence of two independent NAHR events in the ancestors of each parent. Our data suggest that the region around exon 9 of DIS3L2 is a hot spot of L1-mediated NAHR.
Materials and methods
Patient
The male infant was the first child of non-consanguineous, healthy, Japanese parents. Prenatal ultrasound examination showed polyhydramnios and bilateral nephromegaly. He was delivered at 29 weeks and 4 days of gestation. He weighed 2267 g (+6.4 SD) and measured 45.5 cm (+4.3 SD) in length. Low-set ears, large fontanels, micrognathia, a depressed nasal bridge, an everted upper lip, prominent forehead, flexed digits, a micropenis and cryptorchidism were observed. He suffered from cholestasis with coagulation disorder and recurrent adrenal crisis, and died at 175 days of life due to a sepsis. Autopsy revealed visceromegaly and nephroblastomatosis, and he was diagnosed with Perlman syndrome. His karyotype was normal (46,XY). Causative alterations of Beckwith–Wiedemann syndrome, such as loss of methylation at KvDMR1, gain of methylation at H19DMR, paternal uniparental disomy of chromosome 11 and CDKN1C mutations, were ruled out (data not shown). This study was approved by the ethics committee for Human Genome and Gene Analyses of the Faculty of Medicine, Saga University, Japan.
Polymerase chain reaction and sequencing
Genomic DNA was extracted from cord blood, placenta and amniotic fluid of the patient and peripheral blood of his parents. All coding exons, from exon 2 to exon 21, of DIS3L2 were amplified by PCR using primer pairs described previously.2 The copy number of DIS3L2 exon 9 was analyzed by quantitative real-time PCR (qPCR) based on SYBR-Green I. Normalization was performed against GAPDH and TAT.3, 4
L1-A and L1-B, which were located collinearly upstream and downstream of exon 9, were amplified with primer pairs 1a/2, and 3/4 and 5/6, respectively (Figure 1d). The paternal and maternal mutant alleles containing the deletion junctions were amplified with primer pair 1b/6. All PCR products were directly sequenced.
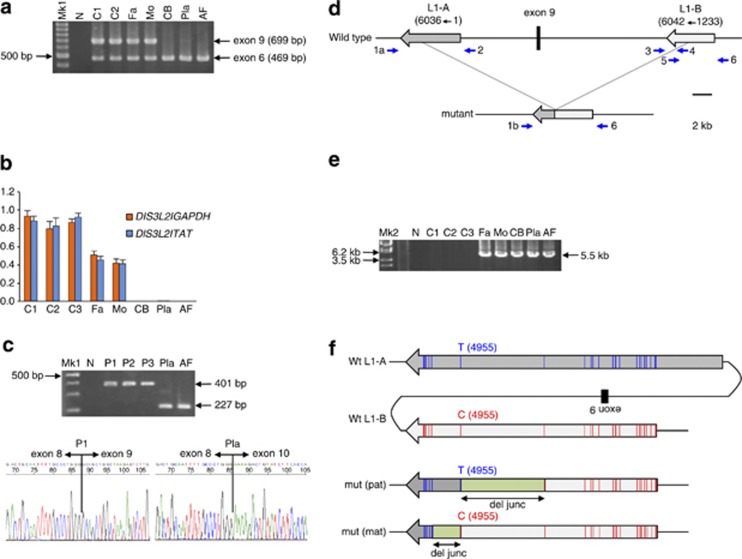
Figure 1.
Homozygous deletion of DIS3L2 exon 9 in a patient with Perlman syndrome. (a) Homozygous deletion of DIS3L2 exon 9. Duplex PCR of exons 6 and 9 showed no amplification of exon 9 in any patient sample, whereas both exons were amplified in the parents and normal controls. Mk1, 100-bp ladders; N, negative control; C1, unrelated normal control #1; C2, unrelated normal control #2; Fa, father of the patient; Mo, mother of the patient; CB, cord blood of the patient; Pla, placenta of the patient; AF, amniotic fluid of the patient. (b) Copy number analysis of exon 9. qPCR of DIS3L2 exon 9 normalized with GAPDH or TAT showed that the copy number of exon 9 in the parents was approximately half that of controls. No PCR product was amplified in any patient sample. The y axis displays arbitrary units. (c) RT-PCR of DIS3L2. RT-PCR was performed with a forward primer in exon 8 and a reverse primer in exon 11. The exon 9 deleted products were seen in the patient samples. Sequencing of the PCR products showed a lack of the exon 9 sequence, which was supposed to be 174 bp in length, in the patient's placenta, whereas the exon 9 sequence existed in normal placentas. P1, normal placenta #1; P2, normal placenta #2; P3, normal placenta #3. (d) Map of exon 9. Two L1 sequences, L1-A and L1-B, were located collinearly upstream and downstream of exon 9. The L1s were in the opposite direction to the DIS3L2 gene. L1-A was full-length, corresponding to nucleotides 1 to 6036 of the reference sequence of L1Hs from Repbase on the GIRI website.5 L1-B was a 5′ truncated form, corresponding to nucleotides 1233 to 6042 of the reference. Primers for PCR amplification are depicted as blue arrows. (e) PCR products containing the deletion junction. Approximately 5.5-kb products were amplified by PCR with primer pair 1b/6 in the patient and the parents, whereas no product was seen in normal controls. Mk2, lambda DNA digested with StyI. (f) The deletion junction. Comparing the sequences among L1-A, L1-B and the PCR products revealed that the deletion junctions of each parental allele were different. Vertical blue or red bars showed the positions of nucleotide differences between L1-A and L1-B. As the patient had two parental mutant alleles, a nucleotide at position 4955 was heterozygous (T/C) in the patient. The deletion junction is shown as a green box.
Total RNA was also extracted from placenta and amniotic fluid and cDNA was synthesized with random primers. RT-PCR was performed with a forward primer in exon 8 and a reverse primer in exon 11. The RT-PCR product was sequenced directly. The novel primers used in this study are shown in Table 1.
Table 1. Original primers used in this study.
| Analyzed region | 5′-Forward primer-3′ | 5′-Reverse primer-3′ | |
|---|---|---|---|
| qPCR for copy number analysis | DIS3L2 exon 9 | GGCGTGGATTTCTCTGATTT | AAGCCTAGCCCCTAGGAAAG |
| RT-PCR | Between exons 8 and 11 | TTTATGTGCCTCTCAAGGAC | AGCAATGTGAACTCCCACTT |
| Identification of deletion junction | Deletion junction | 1a: ACTGATTGAAGCAGCCAACT | 2: AGGACAAAAGGAAGCAAGTG |
| 1b: TGAAGCAGCCAACTCCAAAT | |||
| 3: CCTCTTACCTCAGCCTACCA | 4: GAAGTCAGTGTGGCGATTCC | ||
| 5: TATTCCCCTTCCTGTGTCCA | 6: GGTGACATGATGAAACCTCACTT |
Results
We first examined whether all coding exons of DIS3L2 were amplified by PCR for the patient. In all patient samples, exon 9 could not be amplified whereas all exons were amplified in the patient's parents and normal control individuals (Figure 1a, data not shown). This result indicated a homozygous deletion of exon 9 in the patient, one that has been previously reported.2 qPCR showed that the copy number of exon 9 in the parents was approximately half that of controls, indicating heterozygosity for the deletion (Figure 1b). No PCR amplification was observed in the patient's samples, supporting homozygosity for the deletion. Therefore, the parents were carriers of the deletion and one deleted allele was transmitted to the patient by each parent.
Next, the expression of the mutant allele was investigated by RT-PCR using primers on exons 8 and 11 (Figure 1c). In normal placentas, RT-PCR products matched the estimated normal size and contained the 174-bp exon 9 sequence. In contrast, the product size of the patient's placenta and amniotic fluid was smaller than normal. Sequencing revealed a missing exon 9 sequence and the existence of a junction at exons 8 and 10, indicating the expression of the mutant allele (Figure 1c). The expressed mutant allele would be translated to a mutant protein harboring an in-frame deletion of 58 amino acids, resulting in an abolished RNA-binding domain. Wild-type DIS3L2 has ribonuclease activity, but the mutant lacking exon 9 loses it. Accordingly, it has been speculated that alterations in mRNA turnover might be responsible for the phenotypes of Perlman syndrome.2
Finally, we tried to identify the deletion junction to clarify the deletion mechanism. Two L1 sequences, L1-A and L1-B, were located directly upstream and downstream of exon 9. The directions of the two L1 sequences were opposite to DIS3L2 (Figure 1d). L1-A and L1-B in the parents were amplified and sequenced directly. L1-A sequences were full-length and identical between father and mother with 99.2% similarity to the L1Hs reference sequence obtained from Repbase on the Genetic Information Research Institute (GIRI) website.5 L1-B sequences, which produced a 5′ truncated form with 98.6% similarity to the reference, were also identical between father and mother. The sequence similarity was 99.0% between L1-A and L1-B; however, nucleotide differences were found at 45 positions (Figures 1d and f). In addition, the mutant alleles in both father and mother were successfully amplified by PCR from the parents and the patient (Figure 1e). A sequence comparison among L1-A, L1-B and the mutant alleles revealed that the deletion junctions of each parental allele were different. The paternal deletion junction lay within an interval of 1578 nt corresponding to nucleotides 3377 to 4954 of the reference, whereas the maternal junction lay within an interval of 565 nt corresponding to nucleotides 4956 to 5520 of the reference (Figure 1f, Supplementary Figure S1). Furthermore, a nucleotide difference at position 4955 was heterozygous (T/C) in the patient, supporting the existence of both mutant alleles in the patient (Figure 1f). The results indicated that the deletion was caused by NAHR between the two L1 elements and strongly suggested that the two NAHR events occurred independently in the ancestors of each parent.
Discussion
In this study, we found NAHR between the two L1 elements as the causative mechanism of DIS3L2 exon 9 deletion. We also found that the deletion junctions of each parental allele were different, suggesting the occurrence of two independent NAHRs in the ancestors of each parent.
L1s account for 17% of the human genome.6 A full-length L1 is ∼6 kb and encodes two ORFs (ORF1 and ORF2), which are required for retrotransposition. Mobilization of L1s created several hundred species-specific insertions in humans and chimpanzees, and L1s are still actively expanding in humans, resulting in polymorphisms of L1 elements among individuals.7, 8 L1s are mutagenic agents capable of causing human disease as a result of insertion mutations or insertion-mediated deletions by retrotransposition and NAHR between L1 elements. Twenty-five L1 retrotransposition events have been reported to result in single-gene diseases to date.6 Although Alu-mediated NAHR contributes to a large variety of genetic disorders, L1-mediated NAHR and human endogenous retrovirus-mediated NAHR are very rare causes of human diseases.9, 10, 11, 12 Only three human diseases – glycogen storage disease type IXb, Alport syndrome-diffuse leiomyomatosis, and Ellis-van Creveld syndrome – have been reported to be caused by L1-mediated NAHR.13, 14, 15 To our knowledge, this is the fourth NAHR event to cause human disease, in this case Perlman syndrome. Several possible explanations for the rareness of L1-mediated NAHR have been posed: (1) L1s locate in gene-poor regions, such that recombination events are clinically silent; (2) frequent and extensive mutations over evolutionary time have limited the homology among elements; (3) L1s occur at longer intervals, rendering recombinations involving collinear elements unlikely.13 The NAHR found in this study occurred in a gene, DIS3L2. The similarity between L1-A and L1-B was high (99.0%), and the interval was shorter than that of the human lineage-specific L1 recombination-associated deletion (∼450 kb).16 These conditions might enable the L1-mediated NAHR to cause disease, although the possibility of microhomology-mediated replication-dependent recombination models, such as fork stalling and template switching, microhomology-mediated break-induced replication and serial replication slippage, could not be ruled out.17 The deletion size of exon 9 in the patients reported by Astuti et al2, found in two Dutch pedigrees and one cell line established from a Caucasian patient, strongly suggests the same mechanism at work, although this was not mentioned. In our study, we suggest that two independent NAHRs in ancestors of a Japanese patient occurred. Taken together, this suggests that the region including exon 9 of DIS3L2 might be a hot spot of L1-mediated NAHR. Other disease-causing L1-mediated NAHRs should be studied and analyzed to clarify the precise mechanism.
Perlman syndrome predisposes to Wilms tumor, the most common childhood malignancy, whereas the other three diseases caused by L1-mediated NAHR are not associated with malignancy. The difference in a predisposition to malignancy would depend on the function of the causative genes, not on the genomic instability because of NAHR, because unlike the other genes, DIS3L2 shows tumor-suppressor activity.2
Acknowledgments
This study was supported, in part, by a Grant for Research on Intractable Diseases from the Ministry of Health, Labor and Welfare; a Grant for Child Health and Development from the National Center for Child Health and Development; a Grant-in-Aid for Challenging Exploratory Research; and, a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Alessandri JL, Cuillier F, Ramful D, et al. Perlman syndrome: report, prenatal findings and review. Am J Med Genet A. 2008;146A:2532–2537. doi: 10.1002/ajmg.a.32391. [DOI] [PubMed] [Google Scholar]
- Astuti D, Morris MR, Cooper WN, et al. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet. 2012;44:277–284. doi: 10.1038/ng.1071. [DOI] [PubMed] [Google Scholar]
- Ponchel F, Toomes C, Bransfield K, et al. Real-time PCR based on SYBR-Green I fluorescence: an alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003;3:18. doi: 10.1186/1472-6750-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop R, Zaragoza MV, Gaudette M, Dohrmann U, Scherer G. A homozygous nonsense mutation in SOX9 in the dominant disorder campomelic dysplasia: a case of mitotic gene conversion. Hum Genet. 2005;117:43–53. doi: 10.1007/s00439-005-1295-y. [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH. Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, et al. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet. 2006;78:671–679. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Song L, Grover D, Azrak S, Batzer MA, Liang P. dbRIP: a highly integrated database of retrotransposon insertion polymorphisms in humans. Hum Mutat. 2006;27:323–329. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Deininger PL, Roy-Engel AM. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009;1:97. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Skaletsky H, Rozen S, et al. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum Mol Genet. 2000;9:2291–2296. doi: 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- Blanco P, Shlumukova M, Sargent CA, Jobling MA, Affara N, Hurles ME. Divergent outcomes of intrachromosomal recombination on the human Y chromosome: male infertility and recurrent polymorphism. J Med Genet. 2000;37:752–758. doi: 10.1136/jmg.37.10.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwinkel B, Kilimann MW. Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J Mol Biol. 1998;277:513–517. doi: 10.1006/jmbi.1998.1641. [DOI] [PubMed] [Google Scholar]
- Segal Y, Peissel B, Renieri A, et al. LINE-1 elements at the sites of molecular rearrangements in Alport syndrome-diffuse leiomyomatosis. Am J Hum Genet. 1999;64:62–69. doi: 10.1086/302213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temtamy SA, Aglan MS, Valencia M, et al. Long interspersed nuclear element-1 (LINE1)-mediated deletion of EVC, EVC2, C4orf6, and STK32B in Ellis-van Creveld syndrome with borderline intelligence. Hum Mutat. 2008;29:931–938. doi: 10.1002/humu.20778. [DOI] [PubMed] [Google Scholar]
- Han K, Lee J, Meyer TJ, Remedios P, Goodwin L, Batzer MA. L1 recombination-associated deletions generate human genomic variation. Proc Natl Acad Sci USA. 2008;105:19366–19371. doi: 10.1073/pnas.0807866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.