Abstract
Background:
Osteosarcoma is the most common primary malignant bone tumour, predominantly affecting children and adolescents. Cancer cell line models are required to understand the underlying mechanisms of tumour progression and for preclinical investigations.
Methods:
To identify cell lines that are well suited for studies of critical cancer-related phenotypes, such as tumour initiation, growth and metastasis, we have evaluated 22 osteosarcoma cell lines for in vivo tumorigenicity, in vitro colony-forming ability, invasive/migratory potential and proliferation capacity. Importantly, we have also identified mRNA and microRNA (miRNA) gene expression patterns associated with these phenotypes by expression profiling.
Results:
The cell lines exhibited a wide range of cancer-related phenotypes, from rather indolent to very aggressive. Several mRNAs were differentially expressed in highly aggressive osteosarcoma cell lines compared with non-aggressive cell lines, including RUNX2, several S100 genes, collagen genes and genes encoding proteins involved in growth factor binding, cell adhesion and extracellular matrix remodelling. Most notably, four genes—COL1A2, KYNU, ACTG2 and NPPB—were differentially expressed in high and non-aggressive cell lines for all the cancer-related phenotypes investigated, suggesting that they might have important roles in the process of osteosarcoma tumorigenesis. At the miRNA level, miR-199b-5p and mir-100-3p were downregulated in the highly aggressive cell lines, whereas miR-155-5p, miR-135b-5p and miR-146a-5p were upregulated. miR-135b-5p and miR-146a-5p were further predicted to be linked to the metastatic capacity of the disease.
Interpretation:
The detailed characterisation of cell line phenotypes will support the selection of models to use for specific preclinical investigations. The differentially expressed mRNAs and miRNAs identified in this study may represent good candidates for future therapeutic targets. To our knowledge, this is the first time that expression profiles are associated with functional characteristics of osteosarcoma cell lines.
Keywords: osteosarcoma, tumorigenicity, functional characterisation, gene expression, biomarkers
Osteosarcoma is the most common primary malignant bone tumour, and it has a strong tendency to metastasize early. While overall incidence is low, accounting for <1% of all cancers (Mirabello et al, 2009), osteosarcoma predominantly affects children and adolescents, representing 5–10% of malignancies in this group (van den Berg et al, 2008). The prognosis of osteosarcoma is very poor, and despite advances in surgery and multiagent chemotherapy, the 5-year survival rate remains at 60–70% for patients with localised disease, and as low as 20–30% for patients with metastasis (reviewed in PosthumaDeBoer et al, 2011). This makes osteosarcoma the second most important cause of cancer-related death in children and adolescents. The poor prognosis of this disease is mainly due to lack of response to drug and radiation therapies. It is therefore important to better understand the underlying mechanisms of tumour progression and metastasis to identify or develop more efficient therapies. Cancer cell lines and xenograft models are important tools for studying these mechanisms, as well as for evaluation of novel targeted therapies.
Mesenchymal stem cells are highly motile, and can migrate through the body to repair tissue damage. This mesenchymal phenotype can be reactivated in cancer cells, making also well-organised epithelial cells motile and invasive through the epithelial–mesenchymal transition (EMT), which is shown to increase the tumour aggressiveness (Mani et al, 2008). Properties associated with cancer development and aggressiveness include the capability to sustain proliferative signalling, evade growth suppressors, resist cell death, enable replicative immortality, induce angiogenesis and activate invasion and metastasis (reviewed in Hanahan and Weinberg, 2011).
MicroRNAs (miRNAs) are small non-coding RNA molecules that regulate gene expression by inducing mRNA degradation or by translational repression. Aberrant miRNA expression has been observed in several diseases, including cancer (reviewed in Kong et al, 2012). miRNAs can target multiple genes simultaneously, and this property increases their potential as therapeutics, preventing tumours to escape the inhibition of a single pathway by using compensatory mechanisms. Several deregulated miRNAs have recently been identified for osteosarcoma (reviewed in Drury et al, 2012).
We have previously characterised a collection of osteosarcoma cell lines and xenografts at the genomic, epigenomic and phenotypic level as part of EuroBoNet, an European Network of Excellence for research on bone tumours (Ottaviano et al, 2010; Kuijjer et al, 2011; Mohseny et al, 2011; Kresse et al, 2012; Namløs et al, 2012). In this study, we have extended this panel, and evaluated 22 osteosarcoma cell lines for properties important for cancer development; that is, in vivo tumorigenicity, in vitro colony-forming ability, invasive/migratory potential and proliferation capacity. The mRNA and miRNA expression profiles associated with these phenotypes were also identified. The differentially expressed mRNAs and miRNAs identified in this study may be relevant to osteosarcoma tumorigenesis and metastasis, and they may represent good candidates for future therapeutic targets.
Materials and methods
Cell lines and culturing
Twenty-two osteosarcoma cell lines derived from patient material were analysed. Nineteen of the cell lines were collected within the EuroBoNeT (Ottaviano et al, 2010). The cell lines HAL, KPD, MHM and OHS were established at the Norwegian Radium Hospital. Seven of the cell lines were established at the Instituto Ortopedico Rizzoli; IOR/MOS, IOR/OS9, IOR/OS10, IOR/OS14, IOR/OS15, IOR/OS18 and IOR/SARG. The cell line ZK-58 was kindly provided by Dr Karl-Ludwig Schäfer at the Heinrich-Heine University, Düsseldorf, Germany. The remaining cell lines were obtained from ATCC (www.lgcstandards-atcc.org); HOS (CRL-1543), HOS-143B (CRL-8303, derived from HOS), HOS-MNNG (CRL-1547, derived from HOS), MG-63 (CRL-1427), OSA (CRL-2098), Saos-2 (HTB-85) and U2OS (HTB-96). We also included in our panel the three cell lines Cal72, kindly provided by Prof. Adrienne Flanagan (University College London, UK), 11T254, a kind gift from Florence Pedeutour (Nice University Hospital, France) and G-292, obtained from ATCC (www.lgcstandards-atcc.org) (CRL-1423).
All cell lines were cultured in RPMI-1640 medium (Lonza, Basel, Switzerland) containing 10% fetal bovine serum (FBS) (PAA Laboratories GmbH, Pashing, Austria), 1 × Glutamax (Life Technologies, Carlsbad, CA, USA) and 1 × Penicillin/streptomycin (Life Technologies) at 37 °C with 5% CO2. Cell line identity was verified by short tandem repeats (STRs) DNA fingerprinting using Powerplex 16 (Promega, Madison, WI, USA), and compared against profiles at the EuroBoNeT cell bank (Ottaviano et al, 2010) and ATCC.
In vivo tumorigenicity
Animal experiments were performed according to protocols approved by the National Animal Research Authority in compliance with the European Convention of the Protection of Vertebrates Used for Scientific Purposes (approval ID1499 and 3275; http://www.fdu.no/). In all, 1 × 106 cells in 100 μl serum-free RPMI-1640 were injected subcutaneously into each flank of locally bred NOD/SCID IL2R-gamma-0 (NSG) mice. The tumour size was measured weekly, and tumour volume was calculated by the formula (length × width × height)/2. When tumours reached a 1000-mm3 size the mice were killed, and for the low and non-tumorigenic cell lines the experiments were stopped after 6 months.
Colony formation assay
In all, 1000 cells (in parallels or triplicates) were plated out in CELLSTAR 12-well suspension plate (VWR International, West Chester, PA, USA) in Methocult H410 (Stem Cell Technology, Grenoble, France) and DMEM/F12, supplemented with final concentrations of 1 × B27 (Life Technologies), 1 × Glutamax, 1 × Penicillin/streptomycin, 20 ng ml−1 epidermal growth factor (PeproTech, Stockholm, Sweden) and 20 ng ml−1 basic fibroblast growth factor (PeproTech), as recommended by Stem Cell Technology. After 2 weeks, the cells were stained with Thiazolyl Blue Tetrazolium Bromide (MTT) (Sigma, St Louis, MO, USA) for 24 h. The number of colonies larger than 50 μm was quantified using the GelCount system (Oxford Optronix, Oxford, England).
Invasion and migration assays
Cell invasion and migration were determined by plating 25 000 cells (in duplicates) in RPMI-1640 containing 1% FBS into 24-well invasion chambers with either Matrigel-coated or uncoated membranes, 8.0 μm pores (BD Biosciences, Franklin Lakes, NJ, USA). RPMI-1640 containing 10% FBS was used as a chemoattractant in the lower compartment. Non-invasive/non-migratory cells on the upper surface of the membrane were removed by wiping with a cotton swab, and the remaining cells on the lower surface of the membrane were fixed and stained with Hemacolor (Merck KGaA, Darmstadt, Germany). Nine pictures (10 × ) of each well/membrane were taken, and the number of cells was manually counted.
Proliferation assay
Cellular proliferation rates were analysed by live-cell imaging using the IncuCyte system (Essens Bioscience, Birmingham, UK). Cells were seeded into 96-well plates (2000 cells per well, quadruplets) and phase-contrast photographs (2 per well) were taken automatically every second hour for 5 days. The proliferation rates are presented as cell confluence (%) over time (h).
Genome-wide expression profile comparison and statistical analyses
Comparisons between aggressive vs non-aggressive cell lines (high/low comparisons) were performed on 17 of the 22 cell lines (marked with an asterisk in the Figures and Tables). Global mRNA and miRNA profiles were already available (Kresse et al, 2012; Namløs et al, 2012) and were done by using the Illumina Human-6 v.2.0 Expression BeadChip (Illumina, San Diego, CA, USA) and Agilent Human miRNA Microarrays v.2.0 (Agilent, Santa Clara, CA, USA), respectively. The cell lines HOS-143B and HOS-MNNG were excluded from the gene expression analysis since they originate from HOS and thereby could influence the analysis. Identification of significantly differentially expressed mRNAs and miRNAs was done using Rank Product analysis in J-Express (Dysvik and Jonassen, 2001). mRNAs and miRNAs with q-value<0.05 and fold change >2-fold were selected. The functional annotation tool of DAVID (Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.ncifcrf.gov/home.jsp) was used for functional enrichment analysis of the gene lists, using the DAVID default population background for Homo sapiens. The mRNA and miRNA expression data sets have been deposited in the GEO data repository (www.ncbi.nlm.nih.gov/geo/, accession number GSE28425).
Results
Analysis of in vivo tumorigenicity and identification of putative genes involved
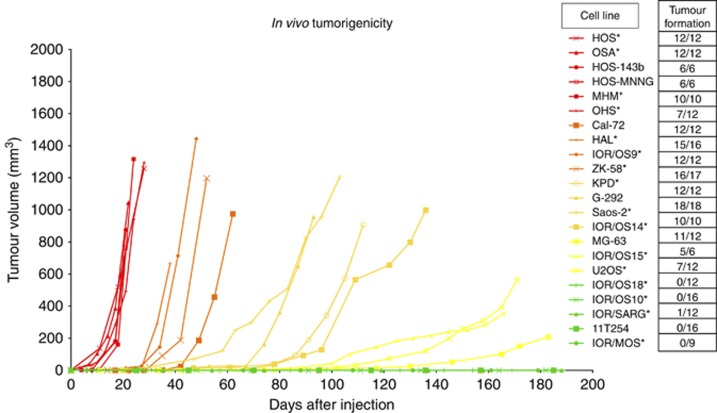
To investigate the tumorigenic capacity of the different cell lines in vivo, we injected 1 × 106 cells subcutaneously into each flank of NSG mice. The majority of the cell lines formed tumours, and the number of injections, number of tumours formed and the growth curves for each cell line are given in Figure 1. The cell lines formed five distinct groups, and were scored from 0 to 4 with regard to speed of tumour formation. The cell lines HOS, HOS-143B, HOS-MNNG, MHM, OHS and OSA formed tumours most rapidly; reaching a volume of ≈1000 mm3 within 10–20 days (Figure 1, red curves), whereas four cell lines (11T254, IOR/MOS, IOR/OS10 and IOR/OS18) did not form tumours at all during the 6 months duration of the experiments. IOR/SARG was scored as non-tumorigenic, although 1 of 12 injections resulted in tumour growth. The remaining cell lines were divided into two groups, cell lines that formed tumours over 600 mm3 between 30 and 60 days post injection (orange curves), and cell lines forming tumours after 60 days (yellow curves).
Figure 1.
In vivo tumorigenicity of 22 osteosarcoma cell lines. The tumorigenic capacity is presented as tumour volume vs time (days after injection). The number of tumours formed/injections is also indicated. The curves show a representative experiment for all the cell lines. The cell lines fell into five groups depending on how fast they gave tumour (shown in Table 3); from highly tumorigenic cell lines (red), to less tumorigenic (orange, light orange and yellow) and non-tumorigenic cell lines (green). *Cell lines included in the high/low expression profile comparison. For clarity, error bars are omitted, but can be seen in Supplementary Figure S1.
To identify genes that were associated with tumorigenicity, we compared the mRNA and miRNA expression profiles (Kresse et al, 2012; Namløs et al, 2012) of highly tumorigenic and non-tumorigenic cell lines. We compared four highly tumorigenic cell lines (HOS, MHM, OHS and OSA) and three non-tumorigenic cell lines (IOR/MOS, IOR/OS10 and IOR/OS18). (The cell lines HOS-143B and HOS-MNNG were excluded from the gene expression analysis since they are derived from the HOS cell line). The analysis revealed several mRNAs and two miRNAs that had significantly different expression levels between the two groups. In all, 354 mRNAs were significantly changed by two-fold or more between the two groups (Supplementary Table S1). A ‘top-ten' list of genes (based on the q-value and fold change) is presented in Table 1. Furthermore, both the miRNAs miR-199b-5p and miR-100-3p were highly expressed in all the non-tumorigenic cell lines, but absent or barely detectable in the highly tumorigenic cell lines (Table 2).
Table 1. Top-ten list of genes that were significantly upregulated or downregulated in the aggressive cell lines for each functional assay (P<0.05, FC; fold change).
|
Tumorigenicity |
Colony-forming ability |
Invasion/Migration |
Proliferation |
|||||
|---|---|---|---|---|---|---|---|---|
| Genes | FC | Genes | FC | Genes | FC | Genes | FC | |
|
Upregulated |
QPCT |
8.5 |
ACTG2 |
10.4 |
IER3 |
15.5 |
LAMA5 |
9.9 |
| |
NPPB |
6.5 |
COL4A1 |
8.7 |
COL4A1 |
10.3 |
COL4A1 |
9.6 |
| |
EPB41L3 |
5.1 |
LAMA5 |
8.1 |
IGFBP7 |
9.3 |
IER3 |
7.5 |
| |
MAOA |
3.6 |
NPPB |
10.5 |
OCIAD2 |
7.9 |
ALDH1A3 |
7.5 |
| |
KRT17 |
6.7 |
C9orf58 |
7.8 |
RBP1 |
7.4 |
KRT18 |
6.4 |
| |
FLJ10154 |
3.3 |
LEPREL1 |
7.2 |
EEF1A2 |
7.0 |
PRDX2 |
5.7 |
| |
LOC647322 |
3.7 |
HRASLS3 |
6.6 |
TAGLN |
6.8 |
S100A16 |
5.4 |
| |
PTPLA |
2.9 |
S100A16 |
6.0 |
SNRPN |
6.6 |
C9orf58 |
5.4 |
| |
GYG2 |
2.9 |
TMEM205 |
6.2 |
SNURF |
6.5 |
ACTG2 |
5.2 |
| |
IL1A |
4.7 |
RASL12 |
5.3 |
UPP1 |
6.2 |
TM4SF1 |
5.2 |
|
Downregulated |
BGN |
11.3 |
COL1A2 |
7.5 |
DCN |
18.4 |
COL1A2 |
10.7 |
| |
MGP |
11.0 |
HAPLN1 |
7.2 |
COL1A2 |
7.8 |
ALPL |
8.4 |
| |
DKK1 |
8.2 |
ALPL |
7.1 |
S100A4 |
6.8 |
MAFB |
6.9 |
| |
LOX |
7.3 |
KYNU |
5.9 |
PDGFRA |
5.2 |
LUM |
6.4 |
| |
TM4SF1 |
6.7 |
MAFB |
5.4 |
MT1X |
4.3 |
CTHRC1 |
4.8 |
| |
MLPH |
6.6 |
BMP4 |
5.2 |
P8 |
3.6 |
TMEM119 |
4.2 |
| |
FAM20C |
6.2 |
IGFBP5 |
4.4 |
BGN |
4.5 |
ENPP2 |
3.9 |
| |
IGFBP4 |
5.8 |
LUM |
4.1 |
GAS1 |
3.9 |
HAPLN1 |
3.8 |
| |
KRT18 |
5.6 |
NINJ2 |
4.1 |
RPS4Y1 |
3.8 |
CDKN2A |
3.2 |
| KIAA1199 | 5.6 | OLFML2B | 4.0 | STEAP1 | 3.6 | ANGPT1 | 3.1 | |
The top-ten lists are based on the q-value, then fold change. See Supplementary Tables S1–S4, for the whole lists of genes significantly changed by a two-fold or more after the different high/low comparisons (q<0.05).
Table 2. miRNAs that were significantly upregulated or downregulated in the aggressive cell lines for each functional assay (q<0.05, FC; fold change).
| High/low comparison | miRNA | Expression (FC) | q-value |
|---|---|---|---|
| Tumorigenicity | hsa-miR-199b-5p | Down (24.1) | |
| |
hsa-mir-100-3p |
Down (12.2) |
0.015 |
|
Colony-forming ability |
hsa-miR-155-5p |
Up (55.6) |
|
| Invasion/Migration | hsa-miR-135b-5p | Up (47.8) | 0.003 |
| |
hsa-miR-146a-5p |
Up (18.4) |
0.031 |
| Proliferation | hsa-miR-155-5p | Up (22.4) | |
| hsa-miR-146a-5p | Up (12.8) | ||
| hsa-miR-135b-5p | Up (11.4) | ||
| hsa-miR-495-3p | Up (9.2) | 0.002 | |
| hsa-miR-136-5p | Up (8.0) | 0.006 | |
| hsa-miR-376c-3p | Up (6.7) | 0.002 | |
| hsa-miR-411-5p | Up (5.6) | 0.045 | |
| hsa-miR-126-3p | Up (5.4) | 0.018 | |
| hsa-miR-758-3p | Up (5.4) | 0.045 | |
| hsa-miR-376a-3p | Up (5.3) | 0.041 | |
| |
hsa-miR-377-3p |
Up (5.4) |
0.045 |
| hsa-miR-886-3p | Down (11.3) | ||
| hsa-miR-143-3p | Down (6.7) | 0.005 | |
| hsa-miR-452-5p | Down (6.1) | 0.003 | |
| hsa-miR-199b-5p | Down (5.9) | 0.001 | |
| hsa-miR-449a | Down (5.4) | 0.012 | |
| hsa-miR-145-5p | Down (5.1) | 0.014 | |
| hsa-miR-34c-5p | Down (5.1) | 0.014 | |
| hsa-miR-145-3p | Down (4.2) | 0.016 | |
| hsa-miR-214-5p | Down (3.7) | 0.016 | |
| hsa-miR-15b-3p | Down (3.3) | 0.033 |
Analysis of colony-forming ability and identification of putative genes involved
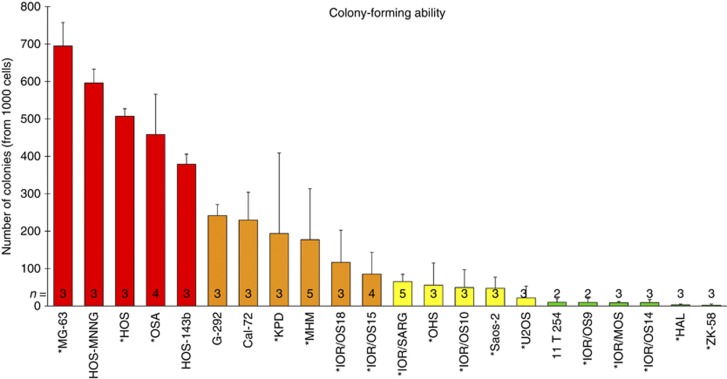
The in vitro colony-forming ability of cells growing under anchorage-independent conditions was further investigated. The number of colonies larger than 50 μm after 2 weeks of growth is presented for each cell line in Figure 2. The cell lines HOS, HOS-143B, HOS-MNNG, MG-63 and OSA showed a very high ability to form colonies (Figure 2, red bars), whereas 11T254, HAL, IOR/MOS, IOR/OS9, IOR/OS14 and ZK-58 had very low colony-forming ability (Figure 2, green bars). The latter cell lines formed only 2–10 colonies per 1000 cells plated, which is ∼100 times lower than the highly clonogenic cell lines. The remaining cell lines showed intermediate colony-forming ability.
Figure 2.
Colony-forming ability of 22 osteosarcoma cell lines. The number of colonies formed (larger than 50 μm) is presented. The error bars show the deviation between 2 and 5 independent experiments. The cell lines are grouped into four groups; from highly clonogenic (red), to gradually less clonogenic (orange, yellow and green, respectively). *Cell lines included in the high/low expression profile comparison.
Comparing the miRNA expression profiles of high (MG-63, HOS and OSA) and low clonogenic cell lines (HAL, IOR/MOS, IOR/OS9, IOR/OS14 and ZK-58), 35 mRNAs that were significantly changed by two-fold or more were identified (Supplementary Table S2). The ‘top-ten' list (based on the q-value and fold change) is presented in Table 1. A similar comparison at the miRNA level revealed one miRNA, miR-155-5p, to be differentially expressed between the two groups (Table 2). This miRNA was highly expressed in all cell lines that formed a high number of colonies, and less expressed or absent in cell lines having a low clonogenic potential.
Analysis of invasive and migratory abilities and identification of putative genes involved
The in vitro migration capacity was determined as the ability of cells to migrate through pores in non-coated membranes, and the in vitro invasion capacity was determined as the ability to degrade and migrate through pores of Matrigel-coated membranes. The cell lines HAL, HOS, HOS-143B, HOS-MNNG, OSA and U2OS showed a high invasion and migration potential (Figure 3, red bars), whereas 11T254, G-292, IOR/MOS, MG-63 and ZK-58 had a poor ability both to invade and to migrate (Figure 3, green bars).
Figure 3.
Invasive and migratory potential of 22 osteosarcoma cell lines. The number of invasive/migratory cells counted from nine pictures (10 × ) of each membrane is presented. The error bars show the deviation between 2 and 4 independent experiments. The cell lines are grouped into four groups; from highly invasive or migratory (red), to gradually less invasive/migratory (orange, yellow and green, respectively). *Cell lines included in the high/low expression profile comparison.
When comparing expression profiles of highly invasive and motile cell lines (HAL, HOS, OSA and U2OS) vs non-invasive and stationary cell lines (IOR/MOS, IOR/SARG, MG-63 and ZK-58), 206 mRNAs were significantly changed by two-fold or more between the two groups (Supplementary Table S3). A ‘top-ten' list of genes (based on the q-value and fold change) is presented in Table 1. At the miRNA level, the miRNAs miR-135b-5p and miR-146a-5p were identified as highly upregulated in the highly invasive group (Table 2).
Analysis of proliferation rate and identification of putative genes involved
The proliferation rates of the cell lines were determined by live-cell imaging, and are presented as increased cell confluence (in %) over time in Figure 4. Most of the cell lines proliferated very fast, reaching 60% confluence within 72 h when seeded at 5% confluence (Figure 4, red and yellow curves). The cell lines G-292, HAL, IOR/MOS, IOR/OS9, IOR/OS14 and KPD proliferated more slowly (Figure 4, green curves). The cell line 11T254 also proliferated slowly but is not included in the figure as we were not able to get satisfying live-cell images of this cell line due to weak cell contrast.
Figure 4.
Proliferation capacity of 22 osteosarcoma cell lines. The proliferation rates are presented as cell confluence (%) over time (h), and are the average of 2–8 independent experiments. See Supplementary Figure S2 for information about n and STDEV for each cell line. The cell lines were grouped depending on their proliferative rate (shown in Table 3); from highly proliferative (red), to less proliferative (orange, yellow and green). *Cell lines included in the high/low expression profile comparison.
The comparison of expression profiles of rapidly (HOS, MHM, OHS and OSA) vs slowly proliferating cell lines (HAL, IOR/MOS, IOR/OS9, IOR/OS14 and KPD) identified 300 mRNAs that were significantly differently expressed, by two-fold or more, when comparing the two groups (Supplementary Table S4). A ‘top-ten' list of genes (based on the q-value and fold change) is presented in Table 1. Furthermore, 11 miRNAs were upregulated in the rapidly proliferating group (miR-126-3p, miR-135b-5p, miR-136-5p, miR-146a-5p, miR-155-5p, miR-376a-3p, miR-376c-3p, miR-377-3p, miR-411-5p, miR-495-3p and miR-758-3p) and 10 miRNAs were downregulated (miR-15b-3p, miR-34c-5p, miR-143-3p, miR-145-3p, miR-145-5p, miR-199b-5p, miR-214-5p, miR-449a, miR-452-5p and miR-886-3p) (Table 2).
Functional enrichment analyses of the differently expressed mRNAs
To evaluate the relevance of the differently expressed mRNAs from the tumorigenic, clonogenic, invasion/migration and proliferation comparisons, the function of the regulated genes was examined by functional enrichment analyses using DAVID (Database for Annotation, Visualization and Integrated Discovery). There was a significant overrepresentation of genes involved in extracellular matrix, growth factor binding, cellular adhesion, cell motility, angiogenesis, bone development and osteoblast differentiation (Supplementary Tables S5–S8), supporting that these mRNAs have relevance for processes related to osteoblast function and to tumorigenesis in osteosarcoma.
Discussion
To improve the understanding of oncogenic mechanisms and how they affect the efficacy of therapies, it is important to have relevant preclinical models. The collection of osteosarcoma cell line models characterised here is to date the most extensive panel of this kind available. As an overture to more focussed mechanistic studies, we have characterised these cell lines for the most relevant malignant phenotypes, and we anticipate that this will also facilitate preclinical studies on this orphan disease.
We determined in vitro abilities to migrate, invade, proliferate and form colonies, as well as the in vivo ability to form tumours for 22 osteosarcoma cell lines. Furthermore, we identified mRNAs and miRNAs that were differently expressed in aggressive vs non-aggressive cell lines, and associated them with specific properties of osteosarcoma tumorigenesis.
Growth in immunodeficient mice is probably the best way to evaluate the tumour-forming ability of patient samples, although false negatives are common. Our data demonstrate that osteosarcoma cell lines in general are highly tumorigenic, as 17 of the 22 cell lines in the panel were able to form tumours in NSG mice within the 6-month duration of the experiments. Out of these 17, 10 of the cell lines formed tumours extremely rapidly. Mohseny et al (2011) have previously investigated in vivo tumorigenicity in nude mice of 19 of the cell lines in our panel. All lines scored as non-tumorigenic here got the same score by Mohseny et al, and the majority of their highly tumorigenic lines fall in the same group here. However, in their study only 8 of 19 cell lines were found to be tumorigenic. Quintana et al (2008) demonstrated how the choice of mouse strain or conditions for implantation greatly affected the efficiency of tumour formation in mice. It is also known that the age and health conditions of the mice may affect these properties. It should therefore not be surprising that some of the lines that were tumorigenic here (HOS, MG-63, Saos2, IOR/OS15, KPD, HAL and ZK58) were scored as non-tumorigenic in nude mice by Mohseny et al, even though those cultures originated from the same cell bank as used here, and the assay was of the same duration.
In this study, we find a strong correlation between invasion, motility and colony formation, especially for the highly aggressive cell lines, such as HOS, HOS-143B and OSA, which were highly aggressive for all phenotypes analysed. 11T254 and IOR/MOS, on the other hand, were in general non-aggressive, as they were non-tumorigenic, non-clonogenic, as well as non-invasive/non-migrative.
Furthermore, we identified mRNAs and miRNAs that were differentially expressed in cell lines with high vs low capacity for these cancer-related phenotypes, which could suggest some mechanistic association. Most notably, four genes—COL1A2, KYNU, ACTG2 and NPPB—were differentially expressed in all the high/low comparisons performed (tumorigenic, clonogenic, invasive/motile and proliferative). COL1A2 and KYNU were downregulated in the highly aggressive cell lines for all properties investigated, and are thereby suggested to be associated with tumour suppression in osteosarcoma. COL1A2 encodes the pro-α2 chain of type I collagen, which is the fibrillar collagen found in most connective tissues, and the main component of the organic part of bone. The gene is mutated in some skeletal diseases (Horton, 1996; Marini et al, 2007) and is epigenetically silenced in several cancer types (Sengupta et al, 2003; Chiba et al, 2005; Mori et al, 2009; Bonazzi et al, 2011; Caren et al, 2011; Misawa et al, 2011). Mori et al (2009) has further presented data indicating that COL1A2 inactivation contributes to increased proliferation and migration activity of bladder cancer. Our findings would be consistent with this, showing reduced expression of COL1A2 in the highly proliferative and motile cell lines.
The other downregulated gene, KYNU, encodes the protein kynureninase, which is involved in the biosynthesis of NAD cofactors from tryptophan. KYNU expression is reduced in invasive ductal carcinoma vs normal fibroadenoma (Perou et al, 2000). In addition, the level of the metabolite kynuerine is 27-fold higher in human embryonic stem cells than in their oncogenic counterpart human embryonal carcinoma (Dawud et al, 2012). Interestingly, in a recent study, gene expression profiles of acute myeloid leukemia patients with IDH mutation were compared with patients with wild-type IDH, revealing a deregulated tryptophan metabolism and a significant downregulation of KYNU expression in the IDH-mutated cases (Damm et al, 2011). This connects KYNU to the rediscovered concept of metabolic switching in cancer development. The switch from oxidative phosphorylation to aerobic glycolysis appears to be a hallmark also for stem-cell function in normal tissue growth, and it is also associated with epigenetic programming (reviewed in Ward and Thompson, 2012). It seems likely that such epigenetic reprogramming could be involved in the downregulation of both KYNU and COL1A2.
Both the genes ACTG2 and NPPB were upregulated in the aggressive group for all the properties analysed, and they were the two most significantly upregulated genes in the clonogenic high/low comparison. Since gamma actins are involved in cellular motility, it makes sense that expression of ACTG2, which encodes actin gamma 2, is increased. Such increased expression in cancer has previously been reported (Edfeldt et al, 2011; Lin and Chuang, 2012). NPPB is a member of the natriuretic peptide family and encodes the secreted protein brain natriuretic peptide (BNP), which functions as a cardiac hormone and has a key role in cardiovascular homeostasis. To our knowledge, this gene has not been linked to the process of tumorigenesis before. However, accumulating evidence suggests that natriuretic peptides also have a role outside the cardiovascular system, and they have been reported to be important regulators of bone and cartilage differentiation and maintenance; that is, overexpression of BNP results in skeletal overgrowth in transgenic mice (Suda et al, 1998), and variations in the promoter of NPPB is linked to rapid bone loss (Kajita et al, 2003). In agreement with a regulative role of NPPB in bone differentiation and growth, dysregulation of this gene might be associated with impaired differentiation in cancer.
Another interesting finding was that RUNX2 expression inversely correlated with proliferation capacity. RUNX2 encodes a master transcription regulator of bone development and osteoblast maturation (reviewed in Komori, 2010), and is shown to be highly elevated in osteosarcoma tumours (reviewed in Martin et al, 2011). In accordance with this, we observe an upregulation of RUNX2 expression in the highly tumorigenic cell lines. Interestingly, our results further show that RUNX2 expression is reduced in the highly proliferative cell lines, which is consistent with a recent study demonstrating that overexpression of RUNX2 suppresses cell growth of several osteosarcoma cell lines (Lucero et al, 2013). Thus, the cancer cells must be able to fine-tune RUNX2 expression to promote tumorigenesis without repressing tumour growth.
miRNAs are interesting as their expression patterns appear to be more tissue and cancer type specific, and their small size make therapeutic applications possible. Here, we found that miR-199b-5p and mir-100-3p were downregulated, or even absent, in the highly tumorigenic cell lines. This is in agreement with recent publications showing that low levels of miR-199b-5p are observed in several cancer types (Garzia et al, 2009; Andolfo et al, 2012), and inversely correlate with survival rates (Wang et al, 2011). Garzia et al (2009) also more directly demonstrated that miR-199b-5p overexpression reduces the cancer stem-cell population in a xenograft mouse model, resulting in impaired medulloblastoma tumour development. Our results further revealed that miR-155-5p was upregulated in the highly clonogenic cell lines. Oncogenic functions of miR-155 have been reported previously (reviewed in Lujambio and Lowe, 2012); in fact, miR-155 was the first example of an miRNA that in a transgenic setting was able to initiate cancer (Costinean et al, 2006; O'Connell et al, 2008). The level of miR-155 is found to be high in mesenchymal stroma cells, and overexpression blocks their differentiation (Skårn et al, 2012). Another oncogenic miRNA described in the literature is miR-135b, which was recently identified as a biomarker for pancreatic ductal adenocarcinoma (Munding et al, 2012). Furthermore, its expression was found to correlate with metastasis and clinical stage in colorectal cancer (Faltejskova et al, 2012; Xu et al, 2012). Interestingly, miR-135b-5p was in our analyses upregulated in the highly motile and invasive cell lines, consistent with a possible function of this miRNA in cancer metastasis. Overexpression of this miRNA in osteosarcoma has also been reported by others (Lulla et al, 2011). Also, miR-146a-5p was upregulated in the highly motile and invasive cell lines, and might also have a role in the process of osteosarcoma metastasis.
A large number of miRNAs were correlated with proliferation. miR-155-5p, miR-135b-5p and miR-146a-5p were highly upregulated in the rapidly proliferating group, whereas miR-199b-5p was downregulated, consistent with findings in the other comparisons. Interestingly, upregulation of miR-155 and miR-135b in rapidly proliferating and clonogenic stem-like cells fits well with the requirement for downregulation for mesenchymal differentiation (Schaap-Oziemlak et al, 2010; Skårn et al, 2012). miR-34c-5p and miR-449a, both members of the miR-34/449 family, were downregulated in the highly proliferating cell lines, in agreement with the role of this family in tumour suppression through induction of cell-cycle arrest, senescence and apoptosis (reviewed in Hermeking, 2010). Inactivation of these miRNAs has been shown for a diverse set of cancer types, including osteosarcoma (He et al, 2009). Interestingly, it has been suggested that miR-34a expression might be a potential predictor of therapy response, and that restoration of miR-34a expression might prevent chemotherapy resistance (Hermeking, 2010). If that is the case also for the miR-34/449 family of miRNAs, which are downregulated in osteosarcoma, this would be of great importance, since the poor prognosis of this disease is mainly due to lack of response to chemotherapy. Interestingly, intravenous administration of an miR-34a mimic has been shown to inhibit tumour growth and metastasis of human prostate and lung cancer cells in mouse models (Liu et al, 2010; Wiggins et al, 2010). On the basis of the current data, it would be interesting to investigate the use of such mimics in our osteosarcoma models. Notably, the miRNAs downregulated in the aggressive groups might be good candidates as therapeutics.
In conclusion, the data obtained from this study demonstrate that osteosarcoma cell lines in general are highly tumorigenic, and points to some candidate biomarkers for aggressive tumours. The panel should be a valuable resource for studies of the various cancer-related phenotypes, of the mechanisms involved and for preclinical therapy studies. Furthermore, we identified a number of candidate genes associated with these phenotypes, the involvement of which in osteosarcoma development should be pursued further. However, further studies are required to evaluate whether any of the candidate genes are actual drivers of the disease.
Depending on the preclinical questions investigated, our classification of the cell line panel can be used to identify relevant models. We encourage the exchange of model systems, both to allow critical evaluation by other scientists, and as common tools to the advantage of the scientific community. An overview of model systems and research groups working on osteosarcomas will be maintained at www.osteosarcomaresearch.org.
Table 3. Overview of the aggressiveness of the cell lines for each functional assay; from high aggressiveness (red) to lower aggressiveness (orange, yellow and green, respectively).

Acknowledgments
We thank Petros Gebregziabher, Alexandr Kristian and Karianne Giller Fleten for assistance with the in vivo tumour assays. We are grateful to Anna Wennerstrøm, Russell Castro and Jeanette Daffinrud for technical assistance. This work was supported by the Norwegian Research Council SFI grant no. 174938/I30 and the Norwegian Cancer Society.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Andolfo I, Liguori L, De Antonellis P, Cusanelli E, Marinaro F, Pistollato F, Garzia L, De Vita G, Petrosino G, Accordi B, Migliorati R, Basso G, Iolascon A, Cinalli G, Zollo M. The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro Oncol. 2012;14:596–612. doi: 10.1093/neuonc/nos002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazzi VF, Nancarrow DJ, Stark MS, Moser RJ, Boyle GM, Aoude LG, Schmidt C, Hayward NK. Cross-platform array screening identifies COL1A2, THBS1, TNFRSF10D and UCHL1 as genes frequently silenced by methylation in melanoma. PLoS One. 2011;6:e26121. doi: 10.1371/journal.pone.0026121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caren H, Djos A, Nethander M, Sjoberg RM, Kogner P, Enstrom C, Nilsson S, Martinsson T. Identification of epigenetically regulated genes that predict patient outcome in neuroblastoma. BMC Cancer. 2011;11:66. doi: 10.1186/1471-2407-11-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Yokosuka O, Fukai K, Hirasawa Y, Tada M, Mikata R, Imazeki F, Taniguchi H, Iwama A, Miyazaki M, Ochiai T, Saisho H. Identification and investigation of methylated genes in hepatoma. Eur J Cancer. 2005;41:1185–1194. doi: 10.1016/j.ejca.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm F, Thol F, Hollink I, Zimmermann M, Reinhardt K, van den Heuvel-Eibrink MM, Zwaan CM, de Haas V, Creutzig U, Klusmann JH, Krauter J, Heuser M, Ganser A, Reinhardt D, Thiede C. Prevalence and prognostic value of IDH1 and IDH2 mutations in childhood AML: a study of the AML-BFM and DCOG study groups. Leukemia. 2011;25:1704–1710. doi: 10.1038/leu.2011.142. [DOI] [PubMed] [Google Scholar]
- Dawud RA, Schreiber K, Schomburg D, Adjaye J. Human embryonic stem cells and embryonal carcinoma cells have overlapping and distinct metabolic signatures. PLoS One. 2012;7:e39896. doi: 10.1371/journal.pone.0039896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury R, Verghese ET, Hughes TA. The roles of microRNAs in sarcomas. J Pathol. 2012;227:385–391. doi: 10.1002/path.4037. [DOI] [PubMed] [Google Scholar]
- Dysvik B, Jonassen I. J-Express: exploring gene expression data using Java. Bioinformatics. 2001;17:369–370. doi: 10.1093/bioinformatics/17.4.369. [DOI] [PubMed] [Google Scholar]
- Edfeldt K, Bjorklund P, Akerstrom G, Westin G, Hellman P, Stalberg P. Different gene expression profiles in metastasizing midgut carcinoid tumors. Endocr Relat Cancer. 2011;18:479–489. doi: 10.1530/ERC-10-0256. [DOI] [PubMed] [Google Scholar]
- Faltejskova P, Svoboda M, Srutova K, Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K, Kiss I, Vyzula R, Slaby O. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med. 2012;16:2655–2666. doi: 10.1111/j.1582-4934.2012.01579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia L, Andolfo I, Cusanelli E, Marino N, Petrosino G, De Martino D, Esposito V, Galeone A, Navas L, Esposito S, Gargiulo S, Fattet S, Donofrio V, Cinalli G, Brunetti A, Vecchio LD, Northcott PA, Delattre O, Taylor MD, Iolascon A, Zollo M. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS One. 2009;4:e4998. doi: 10.1371/journal.pone.0004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- He C, Xiong J, Xu X, Lu W, Liu L, Xiao D, Wang D. Functional elucidation of MiR-34 in osteosarcoma cells and primary tumor samples. Biochem Biophys Res Commun. 2009;388:35–40. doi: 10.1016/j.bbrc.2009.07.101. [DOI] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Horton WA. Molecular genetic basis of the human chondrodysplasias. Endocrinol Metab Clin North Am. 1996;25:683–697. doi: 10.1016/s0889-8529(05)70347-9. [DOI] [PubMed] [Google Scholar]
- Kajita M, Ezura Y, Iwasaki H, Ishida R, Yoshida H, Kodaira M, Suzuki T, Hosoi T, Inoue S, Shiraki M, Orimo H, Emi M. Association of the -381T/C promoter variation of the brain natriuretic peptide gene with low bone-mineral density and rapid postmenopausal bone loss. J Hum Genet. 2003;48:77–81. doi: 10.1007/s100380300010. [DOI] [PubMed] [Google Scholar]
- Komori T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010;339:189–195. doi: 10.1007/s00441-009-0832-8. [DOI] [PubMed] [Google Scholar]
- Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- Kresse SH, Rydbeck H, Skårn M, Namløs HM, Barragan-Polania AH, Cleton-Jansen AM, Serra M, Liestøl K, Hogendoorn PC, Hovig E, Myklebost O, Meza-Zepeda LA. Integrative analysis reveals relationships of epigenetic alterations in osteosarcoma. PLoS One. 2012;7:e48262. doi: 10.1371/journal.pone.0048262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijjer ML, Namløs HM, Hauben EI, Machado I, Kresse SH, Serra M, Llombart-Bosch A, Hogendoorn PC, Meza-Zepeda LA, Myklebost O, Cleton-Jansen AM. mRNA expression profiles of primary high-grade central osteosarcoma are preserved in cell lines and xenografts. BMC Med Genomics. 2011;4:66. doi: 10.1186/1755-8794-4-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZY, Chuang WL. Genes responsible for the characteristics of primary cultured invasive phenotype hepatocellular carcinoma cells. Biomed Pharmacother. 2012;66:454–458. doi: 10.1016/j.biopha.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2010;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucero C, Vega O, Osorio M, Tapia JC, Antonelli M, Stein GS, van Wijnen AJ, Galindo MA. The cancer-related transcription factor Runx2 modulates cell proliferation in human osteosarcoma cell lines. J Cell Physiol. 2013;228:714–723. doi: 10.1002/jcp.24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla RR, Costa FF, Bischof JM, Chou PM, de F Bonaldo M, Vanin EF, Soares MB. Identification of differentially expressed microRNAs in osteosarcoma. Sarcoma. 2011;2011:732690. doi: 10.1155/2011/732690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini JC, Forlino A, Cabral WA, Barnes AM, San Antonio JD, Milgrom S, Hyland JC, Körkkö J, Prockop DJ, De Paepe A, Coucke P, Symoens S, Glorieux FH, Roughley PJ, Lund AM, Kuurila-Svahn K, Hartikka H, Cohn DH, Krakow D, Mottes M, Schwarze U, Chen D, Yang K, Kuslich C, Troendle J, Dalgleish R, Byers PH. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat. 2007;28:209–221. doi: 10.1002/humu.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JW, Zielenska M, Stein GS, van Wijnen AJ, Squire JA. The Role of RUNX2 in osteosarcoma oncogenesis. Sarcoma. 2011;2011:282745. doi: 10.1155/2011/282745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa K, Kanazawa T, Misawa Y, Imai A, Endo S, Hakamada K, Mineta H. Hypermethylation of collagen alpha2 (I) gene (COL1A2) is an independent predictor of survival in head and neck cancer. Cancer Biomark. 2011;10:135–144. doi: 10.3233/CBM-2012-0242. [DOI] [PubMed] [Google Scholar]
- Mohseny AB, Machado I, Cai Y, Schaefer KL, Serra M, Hogendoorn PC, Llombart-Bosch A, Cleton-Jansen AM. Functional characterization of osteosarcoma cell lines provides representative models to study the human disease. Lab Invest. 2011;91:1195–1205. doi: 10.1038/labinvest.2011.72. [DOI] [PubMed] [Google Scholar]
- Mori K, Enokida H, Kagara I, Kawakami K, Chiyomaru T, Tatarano S, Kawahara K, Nishiyama K, Seki N, Nakagawa M. CpG hypermethylation of collagen type I alpha 2 contributes to proliferation and migration activity of human bladder cancer. Int J Oncol. 2009;34:1593–1602. doi: 10.3892/ijo_00000289. [DOI] [PubMed] [Google Scholar]
- Munding JB, Adai AT, Maghnouj A, Urbanik A, Zollner H, Liffers ST, Chromik AM, Uhl W, Szafranska-Schwarzbach AE, Tannapfel A, Hahn SA. Global microRNA expression profiling of microdissected tissues identifies miR-135b as a novel biomarker for pancreatic ductal adenocarcinoma. Int J Cancer. 2012;131:E86–E95. doi: 10.1002/ijc.26466. [DOI] [PubMed] [Google Scholar]
- Namløs HM, Meza-Zepeda LA, Baroy T, Ostensen IH, Kresse SH, Kuijjer M, Serra M, Bürger H, Cleton-Jansen AM, Myklebost O. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7:e48086. doi: 10.1371/journal.pone.0048086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviano L, Schaefer KL, Gajewski M, Huckenbeck W, Baldus S, Rogel U, Mackintosh C, de Alava E, Myklebost O, Kresse SH, Meza-Zepeda LA, Serra M, Cleton-Jansen AM, Hogendoorn PC, Buerger H, Aigner T, Gabbert HE, Poremba C. Molecular characterization of commonly used cell lines for bone tumor research: a trans-European EuroBoNet effort. Genes Chromosomes Cancer. 2010;49:40–51. doi: 10.1002/gcc.20717. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- PosthumaDeBoer J, Witlox MA, Kaspers GJ, van Royen BJ. Molecular alterations as target for therapy in metastatic osteosarcoma: a review of literature. Clin Exp Metastasis. 2011;28:493–503. doi: 10.1007/s10585-011-9384-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–599. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap-Oziemlak AM, Raymakers RA, Bergevoet SM, Gilissen C, Jansen BJ, Adema G, Kögler G, le Sage C, Agami R, van der Reijden BA, Jansen JH. MicroRNA hsa-miR-135b regulates mineralization in osteogenic differentiation of human unrestricted somatic stem cells. Stem Cells Dev. 2010;19:877–885. doi: 10.1089/scd.2009.0112. [DOI] [PubMed] [Google Scholar]
- Sengupta PK, Smith EM, Kim K, Murnane MJ, Smith BD. DNA hypermethylation near the transcription start site of collagen alpha2(I) gene occurs in both cancer cell lines and primary colorectal cancers. Cancer Res. 2003;63:1789–1797. [PubMed] [Google Scholar]
- Skårn M, Namløs HM, Noordhuis P, Wang MY, Meza-Zepeda LA, Myklebost O. Adipocyte differentiation of human bone marrow-derived stromal cells is modulated by microRNA-155, microRNA-221, and microRNA-222. Stem Cells Dev. 2012;21:873–883. doi: 10.1089/scd.2010.0503. [DOI] [PubMed] [Google Scholar]
- Suda M, Ogawa Y, Tanaka K, Tamura N, Yasoda A, Takigawa T, Uehira M, Nishimoto H, Itoh H, Saito Y, Shiota K, Nakao K. Skeletal overgrowth in transgenic mice that overexpress brain natriuretic peptide. Proc Natl Acad Sci USA. 1998;95:2337–2342. doi: 10.1073/pnas.95.5.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg H, Kroon HM, Slaar A, Hogendoorn P. Incidence of biopsy-proven bone tumors in children: a report based on the Dutch pathology registration ‘PALGA'. J Pediatr Orthop. 2008;28:29–35. doi: 10.1097/BPO.0b013e3181558cb5. [DOI] [PubMed] [Google Scholar]
- Wang C, Song B, Song W, Liu J, Sun A, Wu D, Yu H, Lian J, Chen L, Han J. Underexpressed microRNA-199b-5p targets hypoxia-inducible factor-1alpha in hepatocellular carcinoma and predicts prognosis of hepatocellular carcinoma patients. J Gastroenterol Hepatol. 2011;26:1630–1637. doi: 10.1111/j.1440-1746.2011.06758.x. [DOI] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Qian JC, Deng ZL, Cai Z, Tang T, Wang P, Zhang KH, Cai JP. Expression of miR-21, miR-31, miR-96 and miR-135b is correlated with the clinical parameters of colorectal cancer. Oncol Lett. 2012;4:339–345. doi: 10.3892/ol.2012.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.