Abstract
Incorporation of aza-β3-amino acids into endogenous neuropeptide from mollusks (ALSGDAFLRF-NH2) with weak antimicrobial activities allows us to design new AMPs sequences. We find that, depending on the nature of the substitution, these could result either in inactive pseudopeptides or in a drastic enhancement of the antimicrobial activity without high cytotoxicity resulted. Structural studies perform by NMR and circular dichroism on the pseudopeptides show the impact of aza-β3-amino acids on the peptide structures. We obtain the first three-dimensional structures of pseudopeptides containing aza-β3-amino acids in aqueous micellar SDS and demonstrate that hydrazino turn can be formed in aqueous solution. Overall, these results demonstrate the ability to modulate AMPs activities through structural modifications induced by the nature and the position of these amino acid analogs in the peptide sequences.
Keywords: Amino Acids; chemical synthesis; chemistry; pharmacology; Animals; Anti-Bacterial Agents; chemical synthesis; chemistry; pharmacology; Antimicrobial Cationic Peptides; chemistry; Aza Compounds; chemical synthesis; chemistry; pharmacology; CHO Cells; Circular Dichroism; Cricetinae; Cricetulus; Gram-Negative Bacteria; drug effects; Gram-Positive Bacteria; drug effects; Hemolysis; Magnetic Resonance Spectroscopy; Microbial Sensitivity Tests; Models, Molecular; Molecular Conformation; Neuropeptides; chemical synthesis; chemistry; pharmacology; Oligopeptides; chemical synthesis; chemistry; pharmacology; Rabbits; Stereoisomerism; Structure-Activity Relationship
Introduction
Antimicrobial α-peptides (AMPs) are natural occurring peptides that play essential roles in the host innate defense of wide array of organisms and represent one of the major anti-infectious agents against bacterial pathogens.[1–5]At this time, databases report over 1000 sequences for natural AMPs (http://www.bbcm.units.it/~tossi/amsdb.html), whereas several thousand others have been designed de novo and produced synthetically.[6]Although they differ widely in sequence, AMPs are usually short (12–50 residues) with hydrophobic and cationic residues, spatially segregated in amphipathic structures. Many AMPs kill microorganisms by permeabilization of the cytoplasmic membrane whereas others penetrate into the cell and target additional anionic intracytoplasmic constituents (e.g., DNA, RNA, proteins, or cell wall components).[7]These mechanisms of action require an interaction with the cell membrane and numerous interaction models have been proposed, including the barrel-stave pore,[2]the toroidal pore,[8]the carpet like,[2]the aggregate [9]and the detergent like models.[10,11]
Due to the low degree of selectivity between microbial and host cells, and the vulnerability of peptides to the rapid in vivo degradation, AMPs have yet to see widespread clinical use.[3]Therefore, a strategy to improve the activity, selectivity and bioavailability of natural peptides is to design pseudopeptides. Recently, numerous peptidomimetics have been developed in this field of biological application including β-peptides,[12–14]peptoids,[15–17]β-peptoids,[18,19]oligoureas [20]and in this work antimicrobial properties of aza-β3-amino acids-containing pseudopeptides are investigated.
We try to improve the biological activity of a natural occurring AMP using aza-β3-amino acids which are known to enhance the bioavailability of biological active peptides, notably for immunological applications.[21]These monomers are aza analogs of β3-aminoacids in which the CHβ is replaced by a nitrogen stereocenter atom. This confers a better flexibility to the pseudopeptide due to the side chain attached to a chiral nitrogen atom with non-fixed configuration.[22]The nonnatural oligomers have an extended conformational space and are supposed to adopt non-canonical secondary structures. To date, no structures of peptides containing aza-β3-amino acids in solution have been determined. The X-ray crystal structures of linear and macrocyclic aza-β3-peptides have been solved and exhibit an internal hydrogen-bond network leading to bifidic eight-membered ring pseudocycles, called N-N turn or hydrazino turn.[23–25]Recently, it was demonstrated that the nitrogen configuration inversion could be fixed, by incorporating chiral monomers among heteromacrocyles.[26]
Seawater contain many invading microorganisms (up to 106 bacteria/mL and 109 virus/mL) and marine invertebrates have a large number of AMPs. [27]. It was reported that some neuropeptides or peptidic hormones display structural similarities with antimicrobial peptides and exhibit antimicrobial activities.[28]First, we identified a short neuropeptide, H-ALSGDAFLRF-NH2 (AD) of the cuttlefish Sepia officinalis, belonging to the FMRF-amide-related peptides (FaRPs), involved in control of chromatophores and reproductive functions in cuttlefish.[27] The decapeptide AD has a weak antimicrobial activity and a very low cytotoxicity on mammalian cells and constitutes a suitable template to study the impact of the incorporation of aza-β3-amino acids on the antimicrobial activity of an AMP. Then, a large range of Nβ-Fmoc-aza-β3-amino acids with proteinogenic and non-proteinogenic side chains have been synthesized to be incorporated into peptide sequences via solid-phase peptide synthesis (SPPS).[29–32]Finally, all synthesized peptides and pseudopeptides are tested on various Gram-positive and Gram-negative bacteria including marine bacteria classically encountered by the cuttlefish and typical human pathogens and their structures are determined by NMR.
Substitutions of α-amino acids by aza-β3-amino acids induce a strong improvement or, in contrast a complete loss of the antimicrobial activity of this marine neuropeptide. We found that the AD or AK (H-ALSGKAFLRF-NH2) C-terminal amphiphilic helical moiety in SDS micelles is destabilized by the aza-β3 residues incorporation and can tentatively rationalize the structure-activity relationship of the studied peptides and pseudopeptides.
Results and Discussion
Design of antimicrobial analogs
The neuropeptide AD possesses a net charge of +1 at physiological pH. Antimicrobial activity can often be improved upon increasing the peptide positive net charge as a result of enhancing the electrostatic interactions with the negatively charged bacterial membranes. Consequently, we chose first to replace the aspartic acid residue in position 5 by a lysine residue. This AD analog, noted AK, keeps the same hydrophobic ratio of 50% but possesses a net charge of +3, at physiological pH, and an isoelectric point (pI) of 14, instead of 11 for AD. Subsequently, based on the AK sequence, four pseudopeptides containing standard α-amino acids and aza-β3-residues are synthesized. Substitution of α-amino acids by aza-β3-amino acids prevents stereochemical constraints and may contribute to enhance proteolytic resistance.[21]
The first pseudopeptide Aβ3K (H-ALSG-aza-β3-K-AFLRF-NH2) is designed based on the AK, replacing a lysine in position 5 by an aza-β3-lysine. Then, one phenylalanine residue of the AK peptidic sequence is substituted by an aza-β3-2-naphthylalanine (K-2Nal7: H-ALSGKA-aza-β3-2-Nal-LRF-NH2), which displays good antimicrobial activities (Table 1). As a result, this led us to synthesize two new pseudopeptides (K-1Nal: H-ALSGKA-aza-β3-1-Nal-LR-azaβ3-1-Nal-NH2 and K-2Nal: H-ALSGKA-aza-β3-2-Nal-LR-aza-β3-2-Nal-NH2) based on the template where both phenylalanine residues are replaced by aza-β3-naphthylalanine. Substitution of phenylalanine by aza-β3-naphthylalanine residues has already been reported to improve antimicrobial activities with a therapeutic potential.[33]Nevertheless, this improvement is very often coupled with a decrease in the cell selectivity [34,35]since synthetic peptides containing naphthylalanine amino acid are classically more hemolytic with regard to the parent peptides. Indeed, this residue is already known to enhance the interaction of antimicrobial peptides with the lipid bilayers,[36]as their aromatic nucleus structurally close to the hydrophobic indole nucleus, can promote the AMPs burying within the hydrophobic core of the lipid bilayer.
Table 1.
Antibacterial Activities of peptides and pseudopeptides minimal inhibitory concentration (MIC) expressed in μg.mL−1.
| Cuttlefish peptide | Analogs of cuttlefish peptide | Antibiotics | |||||
|---|---|---|---|---|---|---|---|
| AD | AK | Aβ3K | K-2Nal7 | K-1Nal | K-2Nal | Ampicillin | |
|
| |||||||
| Gram positive | |||||||
|
| |||||||
| Bacillus megaterium | 640 | 80 | na | 80 | 20 | 80 | 20 |
| Enterococcus faecalis | naa | na | na | >640 | >640 | >640 | 5 |
| Listeria monocytogenes | na | na | na | 80 | 80 | 160 | 5 |
| Staphylococcus aureus | na | 320 | na | 80 | 20 | 40 | 5 |
|
| |||||||
| Gram negative | |||||||
|
| |||||||
| Escherichia coli | 640 | 80 | na | 320 | 40 | 160 | 5 |
| Salmonella typhimurium | na | 320 | na | 80 | 80 | 160 | 5 |
| Pseudomonas aeruginosa | na | 640 | >640 | 160 | 160 | 320 | 320 |
| Klebsiella pneumoniae | na | >640 | na | na | na | >640 | 5 |
|
| |||||||
| Vibrio harveyi | >640 | 320 | na | 40 | 640 | 640 | >320 |
| Vibrio alginolyticus | 320 | 320 | na | 80 | >640 | >640 | >320 |
| Vibrio aestuarianus | 640 | 80 | na | 40 | 80 | 80 | 5 |
| Vibrio splendidus | 320 | 40 | 320 | 20 | 320 | 640 | 10 |
na : not active
Antibacterial activities
In screening experiment, antibacterial activity was assayed against a representative set of Gram-positive and Gram-negative bacteria (Table 1). The results allow us to compare the antibacterial activities of these analogs with the endogenous peptide AD. Ampicillin, a common antibiotic, serves as a positive control. The neuropeptide AD possesses a narrow-spectrum antibacterial activity and acts more particularly on marine bacteria of the genus Vibrio with relatively high MICs (> 320 μg.mL−1). Indeed, its MICs are widely superior to other marine antimicrobial peptides so it is unlikely that this marine neuropeptide is responsible for the cuttlefish’s innate immunity. For example, the defensin MGD-1 of Mytilus galloprovincialis exhibit MICs that are inferior by 25 to 100 times.[37]The AD antibacterial properties may result from the close structural resemblance between short neuropeptides and AMPs.
As expected, the simple D5>K substitution considerably improves the antibacterial activity of the natural peptide. The peptide AK possesses a larger antibacterial spectrum than native AD. AK is active on all Gram-negative bacteria tested and becomes active on S. aureus. AK’s MICs on bacterial strains are significantly lowered compared to AD’s MICs (MICs ≤ 80 μg.mL−1 on B. megaterium, E. coli, on V. splendidus and aestuarianus). For these bacteria MICs of AK are globally 8 times smaller compared to the MICs of AD and marine bacteria are more affected by this peptide since a concentration of 40 μg.mL−1 inhibits the growth of V. splendidus.
Our first attempt to design AMPs with aza-β3-amino acids is unsuccessful since the pseudopeptide Aβ3K is inactive on all bacteria except on V. splendidus. These results demonstrate that the incorporation of aza-β3-amino acid can switch off the biological activity. Interestingly, the side chains being similar for AK and Aβ3K, the loss of activity is exclusively related with the backbone atom variation inducing a global fold modification.
Fortunately, in contrast to Aβ3K, the three other synthesized pseudopeptides, K-2Nal7, K-1Nal and K-2Nal, exhibit very broad antimicrobial spectra. First, considering the Gram-positive bacteria, they become active against E. Faecalis and L. Monocytogenes, and are globally more than four times more active than AK against S. aureus. Then, on the Gram-negative non-marine bacteria, we can notice an increase of the activities when compared to AK, especially on S. typhimurium and K. pneumoniae. Considering these first eight bacterial strains, we observe that K-1Nal globally possesses the lowest MICs with, for example, MICs of 20 μg.mL−1 against B. megaterium and S. aureus, and 40 μg.mL−1 against E. coli. Surprisingly, K-2Nal, which contains also two aza-β3-naphthylalanine residues in position 7 and 10, but aza-β3-2-naphthylalanine instead of aza-β3-1-naphthylalanine, exhibits large variations in antimicrobial activities when compared to K1-Nal, as the MICs of K-2Nal are always superior or equal to that of K-1Nal when considering the same bacterial strain. This result points out the possible fine tuning of the activity related to the macrocycle interaction with the bacterial walls. Concerning K-2Nal7, MICs are close to those of the K-1Nal and are better than those of K-2Nal. Finally, considering the four studied marine bacteria, we observe this time that K-1Nal and K2-Nal antimicrobial activities are globally similar but not better than AD and AK while K-2Nal7 exhibits the lowest MICs for all the tested marine bacteria. Interestingly, whereas no antimicrobial activities improvement has been measured for K-1Nal and K2-Nal when compared to AK, K-2Nal7 has MICs much lower than AK. These good activities are encountered for all the four marine bacteria and are even better than those of ampicilline against V. harveyi and V. alginolyticus.
Hemolytic activity and in vitro cytotoxicity of analogs
The hemolytic activities are measured on rabbit erythrocytes (Figure 1) and the peptide cytotoxicities are determined on Chinese Hamster Ovary (CHO-K1) cell line (Figure 2). No significant hemolytic activity (<1,5%) of AK is observed for concentrations up to 320 μg.mL−1 and very low cytotoxicity is measured up to 640 μg.mL−1 with 87% of viability indicating a good selectivity for bacterial cells over mammalian cells. The pseudopeptide K-1Nal is hemolytic with a 50% hemolytic dose (HD50) close to 200 μg.mL−1. This concentration is lower than some MICs but is ten times higher than MIC observed on the pathogen S. aureus. K-2Nal is less hemolytic than K-1Nal with only 14% of hemolysis at 320 μg.mL−1. Concerning the cytotoxicity, at 640 μg.mL−1 of K-2Nal, we find 47% of cell viability while the K-1Nal appears to be highly toxic. For K-1Nal, the smallest doses (up to 40 μg.mL−1) are not cytotoxic to CHO-K1 cells, slightly cytotoxic (78% of viability) at 160 μg.mL−1 and toxic at 640 μg.mL−1, but this later concentration is 8 times higher than its MICs against human pathogens L. monocytogenes and S. typhimurium and 32 times higher than its MICs against S. aureus. As often observed, the improvement of the antimicrobial activity is associated with a decline of the selectivity between prokaryotic and eukaryotic cells. The presence of non proteinogenic naphthylalanine side chains seems to decrease the peptide selectivity[35]and notable differences appear between K-1Nal and K-2Nal demonstrating that physico-chemical parameters such as the orientation of the aromatic naphthylalanine moieties have a great influence on the biological activities and selectivity. It has also been pointed out that K-2Nal7 which contains one naphthylalanine side chain displays only very weak hemolytic (5% at 320 μg.mL−1) or cytotoxic (87% of viability at 640 μg.mL−1). These behaviors, very similar to those of the a-peptide AK, support the idea that naphthylalanine residue do not induced a systematic cell selectivity decrease.
Figure 1.
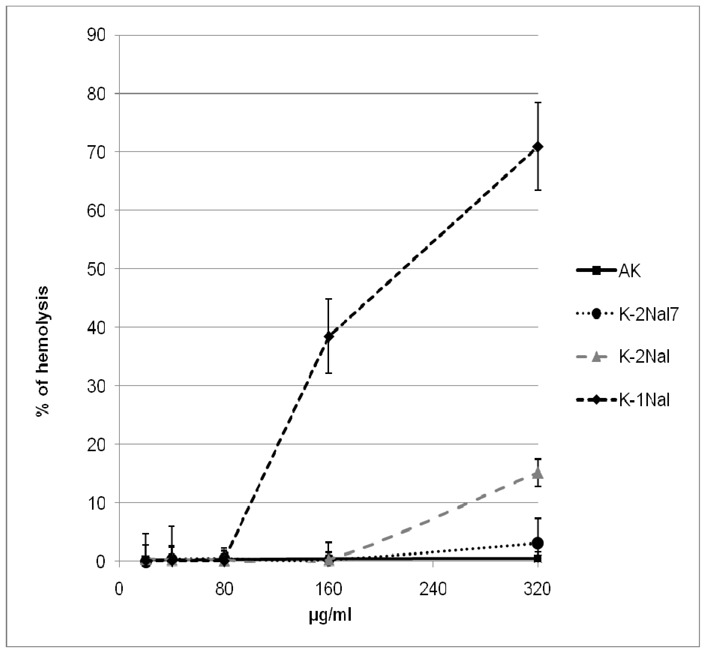
Hemolytic activity of analogs on rabbit erythrocytes.
Figure 2.
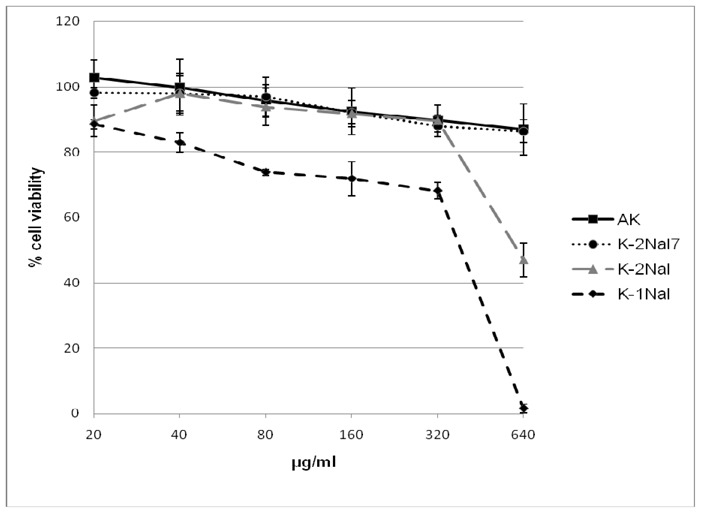
Cytotoxic activity of analogs on Chinese Hamster Ovarian cells (CHO-K1 cell line).
Circular dichroism analysis
The CD spectra of the endogenous peptide AD and its analog AK are similar, in agreement with an identical structural behavior in several media (Figure 3A). They present a random coil CD profile in phosphate buffer (10 mM KP, pH 7.4) and a typical helical signature in hydrophobic environments. Indeed, in 50% TFE or in the presence of SDS micelles, the CD spectra exhibit a maximum at 190 nm and two minima towards 208 and 222 nm showing that AD and AK adopt to some extent a well-defined helical character in these two lipid membranes mimicking media. Nevertheless, we notice that these short peptides (10 residues) cannot form more than one to two helix turns (3.6 residues per turn).
Figure 3.
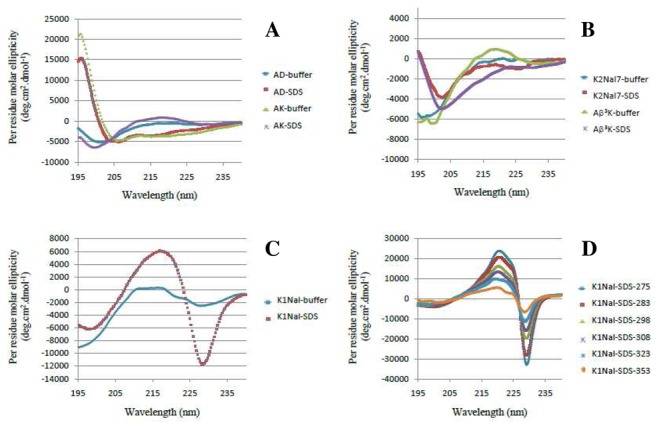
CD spectra of peptides AD and AK (A), and pseudopeptides Aβ3K, K2-Nal7 (B), K-1Nal (C), temperature dependence of K-2Nal (D). Buffer: 10 mM Phosphate buffer, SDS: 30 mM SDS in buffer
Interestingly the Aβ3K displays a random profile in phosphate buffer, but an atypical CD profile in the presence of micelles of SDS with a thin minimum at 202 nm (Figure 3B), which may be attributed to the presence of turns within the peptidic backbone. The extinction of the activity of Aβ3K may be caused by the loss of helicity in membrane mimicking media due to the incorporation of the aza-β3-K in position 5. K-2Nal7 exhibits a similar profile to that of Aβ3K and no helical conformation can be detected (Figure 3B). In the case of pseudopeptides K-1Nal and K-2Nal (Figures 3C and 3D), interpretation of the CD spectra is difficult due to the strong absorbance of the aromatic nuclei of aza-β3-naphthylalanine residues that perturbs their CD profiles. Nevertheless, the pseudopeptides K-1Nal and K-2Nal are clearly in random coil conformation in phosphate buffer. In contrast, dramatic changes are observed in 50% TFE and especially in the presence of SDS micelles. This K-1Nal specific profile is characterized by a couplet centered at 223 nm with a maximum at 217 nm and a minimum at 228 nm. K-2Nal spectrum exhibits the same pattern with a couplet centered at 226 nm with a minimum at 221 nm and a maximum at 229 nm. Based on previous work realized on poly(L-1-naphthyalanine), poly(L-2-naphthyl alanine) peptides,[38,39]the center wavelength around 225 nm is related with the 1Bb transition of a naphthalene group in presence of SDS micelles. Two other residues along its long axis separate the aromatic nuclei of the aza-β3-naphthylalanine residues. This CD spectrum suggests a distance between the aromatic residues that is short enough to allow dipole-dipole interaction. This interpretation is further supported by a similar spectrum when studying the K-2Nal pseudopeptide in the presence of SDS micelles. Dathe and coworkers obtained the same type of CD profiles with cyclic antimicrobial hexapeptide RNal (cyclo[-R-R-1Nal-1Nal-R-F-]) in contact with SDS micelles or POPG Small Unilamellar Vesicles (SUVs).[34]This cyclopeptide contains two α-L-naphthylalanine residues and exhibits, in membrane like environment (micelles or SUVs), a CD spectrum displaying a couplet centered at 227 nm with a maximum of positive ellipticity at 222 nm and a maximum of negative ellipticity at 232 nm.[34]It is important to notice that this type of couplet is inverted in the case of an antimicrobial D-peptide containing β-D-naphthylalanine residues. D-Nal-Pac-525 (Ac-k-nal-r-r-nal-v-r-nal-i-NH2) in the presence of phospholipids or micelles displays the same type of couplet centered towards 225 nm but with opposite sign for the maxima (a positive ellipticity maximum at 229 nm and a negative maximum at 220 nm).[35]In the case of our pseudopeptides, the aromatic side chains are carried by chiral nitrogen atoms with a non-fixed configuration. Thus, CD spectra of pseudopeptides in SDS micelles seem to indicate that the side chains are preferentially positioned in an orientation similar to α-naphthylalanine with L configuration (absolute configuration S). Moreover, K-2Nal CD spectra, in the presence of SDS micelles (reported in Figure 3D), show a strong temperature dependence. An increase in temperature results in a strong decrease in the intensities of the CD ellipticity maxima, possibly related to a fluctuation of the distance between the aromatic rings due to the thermal motion or rapid inversion of the nitrogen stereochemistry.
NMR experiments and structure calculations
Since the CD spectra revealed that the five peptides AD, AK, Aβ3K, K-2Nal7 and K-2Nal could adopt particular conformations in presence of SDS micelles, NMR structural studies are performed in aqueous solution with SDS detergent. In the case of Aβ3K, K-2Nal7 and K-2Nal, as the aza-β3-amino acids side chain attached to an unprotonated nitrogen atom no classical spin systems through magnetization transfer from the amidic proton can be seen in the TOCSY spectra. However, starting from the typical amide proton singlet near 9 ppm of the aza-β3-residue, we can easily identify the side chain proton resonance of the aza-β3 residues on the NOESY spectra. Then the strong NOEs between Hα of the aza-β3 residues and the amide proton of the previous amino acid allow us to perform the sequential assignment. The peptide resonances are reported in the supplementary Table S1.
Where possible, NOEs are assigned based on unambiguous chemical shift assignments, but due to chemical shift overlap, ambiguous NOEs are also incorporated and used by ARIA. In the final ARIA run, the AD structure is calculated using 147 distance restraints and 5 dihedral restraints for the Leu2, Asp5, Ala6, Leu8 and Arg9 residues. At the last iteration, 60 structures are refined in a shell of water, the 20 lowest energy conformers with no distance violations > 0.3 Å and a good stereochemical quality are represented in Figure 4. The global fold of AD in SDS micelles displays a disordered N-terminal domain and a well-defined helical encompassing residues 5 to10. Indeed, the backbone RMSD on the whole structure is 1.312 Å and fall to 0.163 Å if the poorly defined N-terminal moiety is ignored (see supplementary Table S2). This peculiarity is similar to structural features of other short neuropeptides belonging to the tachykinin family.[40]In addition, the AD structure is amphipathic, displayed in Figure 4A, which shows the polar residues Asp5 and Arg9 aligned on the same side of the helix turn and at the opposite side, a large hydrophobic surface composed of the Ala6, Phe7, Leu8 and Phe10. Then, to solve the AK structure, 170 distance restraints and 5 dihedral restraints (Lys5, Leu6, Phe7, Leu8, Arg9) are used in the final iteration of ARIA. The proton chemical shifts of AK are very close to those of AD and, as expected, the global fold of AK (Figure 4B) is similar to the AD structure with a disordered N-terminal moiety and a well-defined helical turn from Lys5 to Phe10 (Figure 5). Consequently, the effect of the mutation D5K on the neuropeptide structure is very slight and do not alter its amphipathic property.
Figure 4.
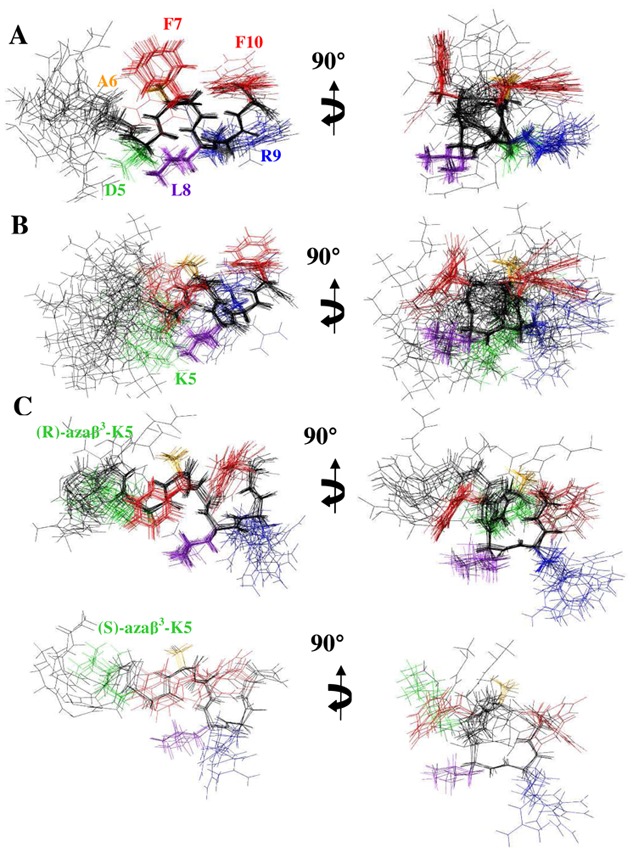
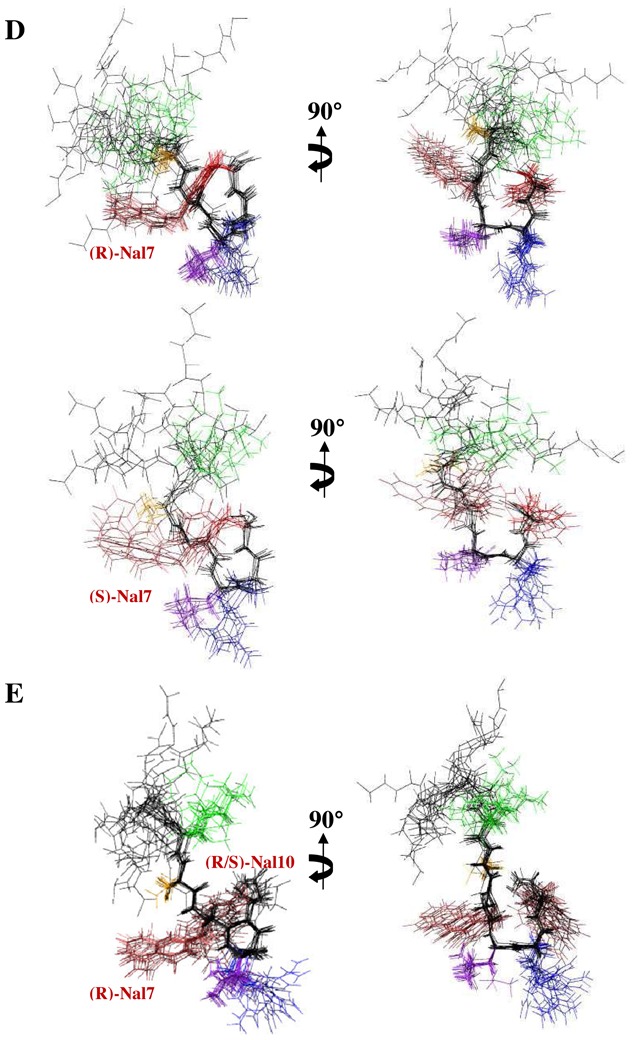
The 20 lowest energy structures with no violations > 0.3 Å from the 60 structures calculated in the final iteration of ARIA for neuropeptide analogs in SDS micelles. (A) AD, (B) AK, the two set of structures of (C) Aβ3K, (D) K-2nal7 and (E) K-2Nal for the two possible configurations of the aza-β3-lysine and the aza-β3-naphtylalanine, respectively. The superposition was performed using the backbone atoms of residues 5–10. For clarity, the side chains of residues 1–4 are not drawn. The backbone of all structures is shown in black while the side chains of the Phe residues are displayed in red, the Asp or Lys residues in green, the Ala residues in orange, the Leu residues in purple, and the Arg residues in blue.
Figure 5.
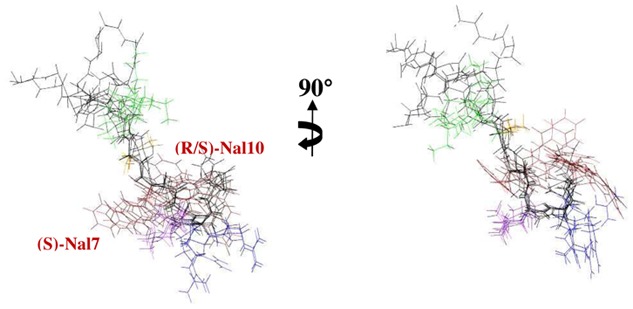
The best structures (with the lowest energies) of AD (green), AK (cyan), Aβ3K (yellow), K-2Nal7 (pink) and K-2Nal (grey). The side chains of the polar and hydrophobic residues are respectively in blue and orange. Aza-β3 residues snapshots are available for the Aβ3K and K-2Nal7 peptides (see also Table S2).
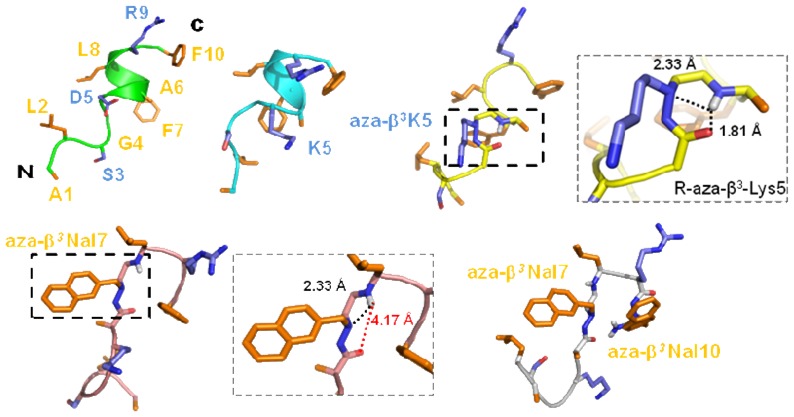
NMR solution structure of Aβ3K is clearly different than AD and AK peptides. The stereochemistry of the nitrogen bearing the side chain of the aza-β3 residue is not fixed, thus the configuration of the stereocenter of the aza-β3-Lys5 is not fixed in our dedicated CNS topology and parameter files. The final restraint file of Aβ3K comprises a set of 213 distance restraints and 3 dihedral restraints. Despite a unique set of signals, we notice in the 20 lowest energy selected structures, two conformations according to the configuration R or S of the Nα stereocenter of the aza-β3-Lys5. Respectively, 15 and 5 structures are composed with a R- and a S-aza-β3-Lys5. The Aβ3K NOESY spectrum exhibits 20% more NOE peaks and as opposed to the AD and AK structures, the backbone of the Aβ3K peptide is quite well-defined all along the sequence (Figures 4 and 5). Moreover, its global fold is very different and does not exhibit a helical character in contrast to the two a-peptides AD and AK. We observe, in Figure 6, that the introduction of the aza-β3-Lys induces a hydrazino turn or N-N turn which has been previously shown in CDCl3 and described in aza-β3-peptide crystal structures.[23-25]The amide proton of Leu6 is H-bonded with both the lone pair of the sp3 nitrogen atom of the aza-β3-Lys5 (6.HN-5.Nα = 2.33 Å) and the carbonyl of Gly4 (6.HN-4.CO = 1.81 Å). We have also measured four characteristic torsional angles ω, ϕ, θ and ψ for the R- and the S- hydrazino turn (Supplementary material, Table S3). Interestingly, this particular turn is surrounded by two β-turns composed of the residues 2-5 and 5-8 on each side of the hydrazino turn (Figure 6). They are stabilized by hydrogen bonds between the carbonyl of Leu2 and amide protons of Gly4 (2.O-4.HN = 1.83 Å) and aza-β3-Lys5 (2.O-5.HN = 2.31 Å), and the carbonyl of aza-β3-Lys5 and amide protons of Phe7 (5.O-7.HN = 1.82 Å) and Leu8 (5.O-8.HN = 2.33 Å). This specific folding feature is particularly stable and strong NOEs peaks can be detected on the NOESY spectrum between the amide protons 2.HN-3.HN, 4.HN-5.HN, 7.HN-8.HN, 9.HN-10.HN.
Figure 6.
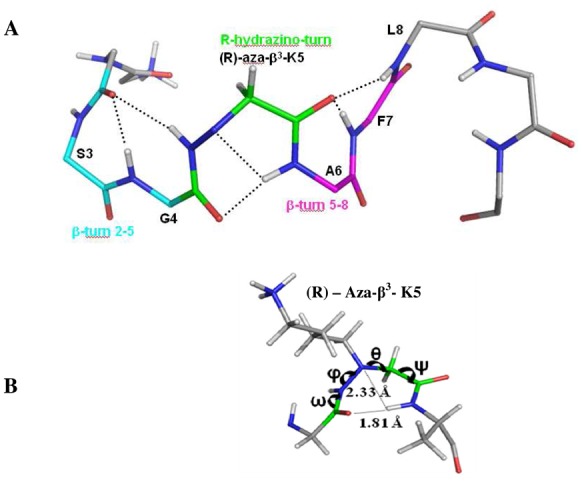
(A) The structural features of the Aβ3K. The carbon atoms of the R-hydrazino turn are displayed in green and the two β-turn carbon atoms involving the residue 2-5 and 5-8, surrounding this N-N turn, are respectively shown in cyan and in magenta, side chains have been removed for clarity. Hydrogen bonds are represented with dashed lines. (B) Distances and torsion angles in the hydrazino turn.
To our knowledge, this work describes the first NMR three-dimensional structure of pseudopeptides including aza-β3-residue in an aqueous medium. The presence of two sets of structure with the R and S configuration of the aza-β3-lysine does not necessary mean that the two populations can be found in solution but reflect the fact that the distance restraints used in the NMR structure calculations can be satisfied by the R and S configuration.
The AD and AK well-ordered helix moieties are dramatically modified by the incorporation of only one aza-β3-residue which induces a loss of the amphipathic properties. This amphipathic character can be associated with the antimicrobial peptide activities of AD and AK. For the peptides K-2Nal7 and K-2Nal, only their C-terminal moieties are ordered forming β-turn in contrast with the helical structure of the α-peptides AD and AK (Figures 4D, 4E). They share a similar turn, which slightly approaches the two hydrophobic aromatic residues in position 7 and 10. Nevertheless, the aromatic rings are too far to affect each other (~9 Å) in agreement with the absence of NOEs between the residues 7 and 10 aromatic protons. NMR 2D-NOESY spectra of the pseudopeptides K-2Nal7 and K-2Nal in SDS micelles display less NOEs comparing to the three other peptides pointing out the relative flexibility of these pseudopeptides. Surprisingly, the hydrazino turn NOE markers previously noticed on the Aβ3K NOESY spectrum cannot be detected for these pseudopeptides and the K-2Nal7 and K-2Nal structures do not exhibit a hydrazino turn.
Overall, structural characterization of the α-peptides and the pseudopeptides permits us to study the influence of the aza-β3 residue. The D5K substitution does not cause any loss of the helix encountered with the native neuropeptide, for both peptides the amphipathic character is conserved. All the introductions of aza-β3 residue induce drastic structural modification, especially in the case of Aβ3K with a well-defined structure along all the sequence. This rigidity has to be related with the disappearance of any antibacterial activity. In the case of aza-β3-naphthylalanine derivatives only the C-terminal moieties are well-defined adopting a β-turn but the remaining of the peptides display large flexibility. This flexibility, along with the properties of the naphthylalanine side chains, is related to the increase in the antimicrobial activities. Such an observation is consistent with the hypothesis that peptide structural flexibility improves ntimicrobial activity.[41]
Conclusion
This article reports the first observation of antibacterial activity for a modified marine invertebrate neuropeptide. CD and NMR data show that this peptide adopts an amphiphilic α-helical conformation in a hydrophobic medium as seen for the majority of natural antimicrobial peptides. The importance of the cationic net charge is demonstrated by the improvement of the antibacterial activity due to a simple substitution of aspartic acid to lysine without affecting the secondary structure. On the other hand, the incorporation of only one aza-β3-amino acid can lead to a drastic rearrangement of the peptidic backbone resulting in a rigid structure and the loss of antibacterial activity. However the first three-dimensional structure of a pseudopeptide with a heterogeneous backbone containing an aza-β3-amino acid was solved in aqueous micellar environment and this structure provided essential information on the impact of aza-β3-residues incorporation in peptidic sequences. Interestingly, the two more biologically active pseudopeptides do not display helical character in contrast to AD/AK peptides and appear to be less structurally defined than the Aβ3K pseudopeptide.
Finally, as it was recently reported for heterogenous helical urea/amide backbones,[42]“mixed” pseudopeptides containing α- and aza-β3-amino acids appear as a promising therapeutic tool to fight multiresistant bacteria. The present work demonstrates that aza-β3-amino acid can modify pseudopeptide structures and modulate the biological activities depending on the number and the position of the substitutions.
Experimental Section
Peptides and Pseudopeptides Synthesis
Peptides (AD and AK) and the pseudopeptide Aβ3K were synthesized on an Advanced Chem Tech 440 Mos synthesizer®. Pseudopeptides K-2Nal7, K-1Nal and K-2Nal were synthesized on a Pioneer Peptide Synthesis System®. Synthesis was accomplished using commercially available N -Fmoc-amino acid, Nβ-Fmoc-aza-β3-amino acid [29–32]and a Rink Amide resin. Peptides were synthesized via Fmoc solid phase synthesis methods using a 4-fold excess of amino acid, TBTU, and HOBt in the presence of a 8-fold excess of DIPEA for 1 h for standard residues and 2 h for aza-β3 residues. The Fmoc group was removed with 20% piperidine in DMF for 10 min. At the end of the synthesis, the resin was washed with CH2Cl2 and dried. Side chain deprotection and cleavage of peptides from the resin were performed simultaneously by treatment with TFA/ H2O/TIS (95/2.5/2.5, v/v/v) for 3h. After filtration of the resin, the TFA solutions were concentrated in vacuo and peptides were precipitated by addition of cold diethyl ether. Peptides were purified by RP-HPLC on a C18 XTerra® semi-preparative column (10 μm, 19 mm × 300 mm, Waters) with a linear gradient of water, 0.08% TFA (A)/acetonitrile, 1% TFA (B) (5–60% B in 40 min and 60–100% B in 20 min, 8 ml/min, 215nm) to a final purity of ≥95% and lyophilized. Characterization of purified peptides by RP-HPLC analyses were performed on a C18 XTerra® (4.6x250 mm, 5 μm) column using water 0.08% TFA (A)/acetonitrile, 1% TFA (B) linear gradient (5–60% B in 20 min, 1 ml/min, 30°C, 215nm). Peptide concentrations for all experiments were calculated as the TFA salt (assuming association of one molecule of TFA per cationic residue, determined by 13C NMR).
H-ALSGDAFLRF-NH2 (AD): Yield after purification 26 %; white powder; RP-HPLC tR : 17.30 min; MWt C51H79N14O13.TFA 1208.6 g/mol. LC-ESI-MS/MS mass: found m/z 1095.27 [M+H+], C51H79N14O13 (without TFA) requires.1095.59.
H-ALSGKAFLRF-NH2 (AK): Yield after purification 40 %; white powder; RP-HPLC tR : 16.93 min; MWt C53H85N15O11.TFA 1222.0 g/mol. LC-ESI-MS/MS mass: found m/z 1108.60 [M+H+], C53H85N15O11 (without TFA) requires 1108.66.
H-ALSG-aza-β3-K-AFLRF-NH2 (Aβ3K): Yield after purification 18 %; white powder; RP-HPLC tR : 15.17 min; MWt C53H86N16O11.TFA 1237.0g/mol. LC-ESI-MS/MS mass: found m/z 1123.65 [M+H+]; C53H86N16O11 (without TFA) requires 1123.67.
H-ALSGKA-aza-β3-2Nal-LRF-NH2 (K2-Nal7): Yield after purification 58 %; white powder; RP-HPLC tR : 14.85 min; MWt C57H88N16O11.TFA 1286.0 g/mol. LC-ESI-MS/MS mass: found m/z 1173.70 [M+H+]; C57H88N16O11.(without TFA) requires 1173.69.
H-ALSGKA-aza-β3-1Nal-LR-aza-β3-1Nal-NH2 (K1-Nal): Yield after purification 21 %; white powder; RP-HPLC tR : 19.13 min; MWt C61H91N17O11.TFA 1352.1 g/mol. LC-ESI-MS/MS mass: found m/z 1238.65 [M+H+]; C61H91N17O11 (without TFA) requires 1238.71.
H-ALSGKA-aza-β3-2Nal-LR-aza-β3-2Nal-NH2 (K2-Nal): Yield after purification 55 %; white powder; RP-HPLC tR : 20.56 min; MWt C61H91N17O11.TFA 1352.1 g/mol. LC-ESI-MS/MS mass: found m/z 1238.65 [M+H+]; C61H91N17O11 (without TFA) requires 1238.71.
Antibacterial assays
The antibacterial activity of peptides was measured by liquid growth inhibition assay, performed in 96-wells microtiter plates.[43]Microbial growth was assessed by measurement of the optical density at D595 after the 20 h incubation at 30°C or 18°C for Vibrios. The minimal inhibitory concentration (MIC) is defined as the lowest concentration required to inhibite growth (100% inhibition). The bacteria species tested were the Gram-positive Bacillus megaterium, Enterococcus faecalis, Listeria monocytogenes, Staphylococcus aureus; and the Gram-negative Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Salmonella typhimurium and marine Vibrio (V. aestuarianus, V. alginolyticus, V. harveyi and V. splendidus). For antibacterial tests, distilled water is used to dissolve peptides. Briefly, 10 μl of water or 10μl of peptide solution at a final concentration ranging from 20 to 640 μg.mL-1, were incubated with 100 μl of a mid-logarithmic growth phase culture of bacteria at a starting optical density of D600 = 0.001. Poor-broth nutrient medium (PB: 1% peptone, 0.5% NaCl, w/v, pH 7.5) was used for standard bacterial culture. Pathogenic bacteria were grown in PB with 0.3% of beef extract (BD) or in brain heart infusion for Enterococcus and Listeria. Vibrios were grown in Saline PB (SPB: 1% peptone, 1.5% NaCl, w/v, pH 7.2).
Cytotoxicity assay
Cytotoxicity of peptides on CHO-K1 cells were determined by the colorimetric MTT (3-[4,5-dimethylthiozol-2-yl]-2,5-diphenyltetrazolium bromide) dye reduction assay. Briefly, 1.34×104 cells/well in Ham F12 supplemented with L-glutamine (Gibco) and 10% foetal calf serum (Eurobio) were placed into 96-well plates. After incubation for 6h under a fully humidified atmosphere of 95% room air and 5% CO2 at 37 °C, analogs dissolved in phosphate buffered saline (PBS) were added to cell cultures at a final concentration ranging from 20 to 640 μg.mL−1. After 36-hours incubation, toxicity was evaluated by measuring the optical density of the culture at 570 nm using the cell growth determination kit (Sigma) based on conversion of the yellow tetrazolium salt MTT into purple formazan crystals by metabolically active cells. 100% of Viability and 0% of viability were determined in PBS buffer and 1% Triton X-100, respectively. The experiments were set in triplicate.
Hemolytic activity
The hemolytic activity of analogs was determined with freshly isolated rabbit erythrocytes as already describe.[44]Erythrocytes solution was incubated for 1 hour at 37°C with peptide dissolved in PBS to reach 20 at 320 μg.mL−1. Hemolysis was determined by measuring the optical density of the supernatant at 415 nm. Zero hemolysis (blank) and 100% hemolysis were determined in PBS buffer and 1% Triton X-100, respectively. For each concentration and control, the experiments were set in triplicate.
Circular dichroism
CD experiments were carried out using a JASCO J-815 spectropolarimeter (Easton, USA) equipped with Peltier devices for temperature control. All of the spectra were obtained with a quartz cell of 0.2 mm path length. Spectra were recorded at a peptide concentration of 100 mM in different environments: water, phosphate buffer 10 mM, in 50% TFE, in presence of SDS detergent (30 mM). The baseline-corrected spectra were smoothed, ellipticities were converted to mean residue molar ellipticities in degree cm2.d.mol−1, and the helical content was estimated as previously described [45]
NMR experiments
All spectra were recorded on a Bruker Avance 500 spectrometer equipped with a 5 mm triple-resonance cryoprobe (PRISM, Rennes). 1D spectra were first recorded at concentration around 1 mM for each peptide, dissolved in aqueous (90% H2O, 10% D2O) increasing SDS d-25 (Euriso-top) concentrations, the pH was adjusted to 5.0 (until 200 mM SDS). Up to 50 mM SDS, variations in chemical shifts and line width narrowing could be observed, and then no change occurred at higher concentrations for all the peptides. The detergent concentrations were subsequently set at 50 mM for the further experiments. Homonuclear 2-D spectra DQF-COSY, TOCSY (MLEV) and NOESY were recorded in the phase-sensitive mode using the States-TPPI method as data matrices of 256 real (t1) × 2048 (t2) complex data points; 64 scans per t1 increment with 1.5 s recovery delay and spectral width of 5341 Hz in both dimensions were used. The mixing times were 100 ms for TOCSY and 200 ms for the NOESY experiments. In addition, 2D heteronuclear spectra 13C-HSQC and 13C-HMBC were acquired to help to fully assign the naphthylalanyl side chains. Spectra were processed with Topspin (Bruker Biospin) or the NMRpipe/NMRdraw software package[46]and visualized with Topspin or NMRview[47]on a Linux station. The matrices were zero-filled to 1024 (t1) × 2048 (t2) points after apodization by shifted sine-square multiplication and linear prediction in the F1 domain. Chemical shifts were referenced to the solvent chemical shifts.
Structure calculations
1H chemical shifts were assigned according to classical procedures.[48]NOE cross-peaks were integrated and assigned within the NMRView software.[47]Structure calculations were performed with ARIA 2.2.[49]Representation and quantitative analysis of the calculated structures were performed using MOLMOL[50]and PyMOL (Delano Scientific). The PROCHECK program[51]was used to assess the stereochemical quality of the structures. The calculations were initiated using the default parameters of ARIA and a nearly complete set of manually assigned NOE. The CNS[52]and ARIA topology and parameter files were modified to define the aza-β3 residues based on the characteristic angles and distances in the previously published crystalline structures.[23–25]According to the Nα pyramidal inversion, the stereochemistry of the nitrogen bearing the side chain of the aza-β3 residues is not fixed. The torsion angle ϕ was restrained to −60 ± 40° for 3JHN-HA < 6 Hz. At the end of each run, violations and assignment proposed by ARIA were checked before starting a new run. This process was repeated until all the NOE were correctly assigned and no restraints were rejected. A last run of 60 structures was performed and refined in water, a set of 20 structures of lowest energies with no violation > 0.3 Å were considered as representative of the peptide structure.
Acknowledgments
We thank SERB Laboratories and Region Bretagne for their financial support and IFR 140 and Biogenouest for access to PRISM and Spectroscopies facilities. We are grateful to Pr. Françoise Vovelle for her helpful participation in the aza-β3 residues description in CNS/ARIA parameter files. We thank Dr Helena Wee for post-editing the English style.
Abbreviations
- AMPs
Antimicrobial Peptides
- SDS
sodiumdodecyl sulphate
- NMR
Nuclear Magnetic Resonance
- pI
isoelectric point
- na
not active
- SPPS
Solid Phase Peptide Synthesis
- MIC
Minimal Inhibitory Concentration
- MD
Molecular Dynamics
- NOESY
Nuclear Overhauser Effect Spectroscopy
- ROESY
Rotating Frame Overhauser Spectroscopy
- MS
Mass Spectroscopy
- HPLC
High Performance Liquid Cromatography
Footnotes
Supporting Information Available: Chemical shift of AD, AK, A K and K-Nal in presence of 50 mM SDS (Table S1); Structural statistics for the 20 models of AD, AK, Aβ3K, K-2Nal7 and K-2Nal bound to SDS micelles (Table S2) and (A) Average torsion angles of the (R)- and (S)-Hydrazino turn and (B) of the β-turn 2-5 and 5-8 for the Aβ3K peptide (Table S3) are available free of charge.
References
- 1.Andreu D, Rivas L. Animal antimicrobial peptides: an overview. Biopolymers. 1998;47:415–433. doi: 10.1002/(SICI)1097-0282(1998)47:6<415::AID-BIP2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 2.Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochim Biophys Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 4.Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 5.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 6.Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 7.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 8.Matsuzaki K. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim Biophys Acta. 1998;1376:391–400. doi: 10.1016/s0304-4157(98)00014-8. [DOI] [PubMed] [Google Scholar]
- 9.Wu M, Maier E, Benz R, Hancock RE. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry. 1999;38:7235–7242. doi: 10.1021/bi9826299. [DOI] [PubMed] [Google Scholar]
- 10.Bechinger B, Lohner K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim Biophys Acta. 2006;1758:1529–1539. doi: 10.1016/j.bbamem.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Legrand B, Laurencin M, Sarkis J, Duval E, Mouret L, Hubert JF, Collen M, Vie V, Zatylny-Gaudin C, Henry J, Baudy-Floc’h M, Bondon A. Structure and mechanism of action of a de novo antimicrobial detergent-like peptide. Biochim Biophys Acta. 2011;1808:106–116. doi: 10.1016/j.bbamem.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 12.Arvidsson PI, Ryder NS, Weiss HM, Gross G, Kretz O, Woessner R, Seebach D. Antibiotic and hemolytic activity of a beta2/beta3 peptide capable of folding into a 12/10-helical secondary structure. Chembiochem. 2003;4:1345–1347. doi: 10.1002/cbic.200300698. [DOI] [PubMed] [Google Scholar]
- 13.Porter EA, Weisblum B, Gellman SH. Mimicry of host-defense peptides by unnatural oligomers: antimicrobial beta -peptides. J Am Chem Soc. 2002;124:7324–7330. doi: 10.1021/ja0260871. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt MA, Weisblum B, Gellman SH. Unexpected relationships between structure and function in alpha, beta -peptides: antimicrobial foldamers with heterogeneous backbones. J Am Chem Soc. 2004;126:6848–6849. doi: 10.1021/ja048546z. [DOI] [PubMed] [Google Scholar]
- 15.Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, Zuckermann RN, Barron AE. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc Natl Acad Sci U S A. 2008;105:2794–2799. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song YM, Park Y, Lim SS, Yang ST, Woo ER, Park IS, Lee JS, Kim JI, Hahm KS, Kim Y, Shin SY. Cell selectivity and mechanism of action of antimicrobial model peptides containing peptoid residues. Biochemistry. 2005;44:12094–12106. doi: 10.1021/bi050765p. [DOI] [PubMed] [Google Scholar]
- 17.Zhu WL, Song YM, Park Y, Park KH, Yang ST, Kim JI, Park IS, Hahm KS, Shin SY. Substitution of the leucine zipper sequence in melittin with peptoid residues affects self-association, cell selectivity, and mode of action. Biochim Biophys Acta. 2007;1768:1506–1517. doi: 10.1016/j.bbamem.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Olsen CA, Bonke G, Vedel L, Adsersen A, Witt M, Franzyk H, Jaroszewski JW. alpha -Peptide/beta -peptoid chimeras. Org Lett. 2007;9:1549–1552. doi: 10.1021/ol070316c. [DOI] [PubMed] [Google Scholar]
- 19.Shuey SW, Delaney WJ, Shah MC, Scialdone MA. Antimicrobial beta-peptoids by a block synthesis approach. Bioorg Med Chem Lett. 2006;16:1245–1248. doi: 10.1016/j.bmcl.2005.11.075. [DOI] [PubMed] [Google Scholar]
- 20.Violette A, Fournel S, Lamour K, Chaloin O, Frisch B, Briand JP, Monteil H, Guichard G. Mimicking helical antibacterial peptides with nonpeptidic folding oligomers. Chem Biol. 2006;13:531–538. doi: 10.1016/j.chembiol.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 21.Dali H, Busnel O, Hoebeke J, Bi L, Decker P, Briand JP, Baudy-Floc’h M, Muller S. Heteroclitic properties of mixed alpha- and aza-beta3-peptides mimicking a supradominant CD4 T cell epitope presented by nucleosome. Mol Immunol. 2007;44:3024–3036. doi: 10.1016/j.molimm.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 22.Cheguillaume A, Salaün A, Sinbandhit S, Potel M, Gall P, Baudy-Floc’h M, Le Grel P. Solution synthesis and characterization of hydrazinopeptoidic oligomers. J Org Chem. 2001;66:4923–4929. doi: 10.1021/jo001500d. [DOI] [PubMed] [Google Scholar]
- 23.Le Grel P, Salaun A, Potel M, Le Grel B, Lassagne F. Aza-beta3-cyclohexapeptides: pseudopeptidic macrocycles with interesting conformational and configurational properties slow pyramidal nitrogen inversion in 24-membered rings! J Org Chem. 2006;71:5638–5645. doi: 10.1021/jo0608467. [DOI] [PubMed] [Google Scholar]
- 24.Salaun A, Potel M, Roisnel T, Gall P, Le Grel P. Crystal structures of aza-beta3-peptides, a new class of foldamers relying on a framework of hydrazinoturns. J Org Chem. 2005;70:6499–6502. doi: 10.1021/jo050938g. [DOI] [PubMed] [Google Scholar]
- 25.Salaun A, Mocquet C, Perochon R, Lecorgne A, Le Grel B, Potel M, Le Grel P. Aza-beta3-cyclotetrapeptides. J Org Chem. 2008;73:8579–8582. doi: 10.1021/jo8013963. [DOI] [PubMed] [Google Scholar]
- 26.Mocquet C, Salaun A, Claudon P, Le Grel B, Potel M, Guichard G, Jamart-Gregoire B, Le Grel P. Aza-beta3-cyclopeptides: a new way of controlling nitrogen chirality. J Am Chem Soc. 2009;131:14521–14525. doi: 10.1021/ja9058074. [DOI] [PubMed] [Google Scholar]
- 27.Henry J, Zatylny C, Boucaud-Camou E. Peptidergic control of egg-laying in the cephalopod Sepia officinalis: involvement of FMRFamide and FMRFamide-related peptides. Peptides. 1999;20:1061–1070. doi: 10.1016/s0196-9781(99)00102-3. [DOI] [PubMed] [Google Scholar]
- 28.Brogden KA, Guthmiller JM, Salzet M, Zasloff M. The nervous system and innate immunity: the neuropeptide connection. Nat Immunol. 2005;6:558–564. doi: 10.1038/ni1209. [DOI] [PubMed] [Google Scholar]
- 29.Busnel O, Baudy-Floc’h M. Preparation of new monomers aza-beta 3-amino acids for solid-phase syntheses of aza-beta 3-peptides. Tetrahedron Lett. 2007;48:5767–5770. [Google Scholar]
- 30.Busnel O, Bi L, Baudy-Floc’h M. Synthesis of Fmoc-protected aza-beta 3-amino acids via reductive amination of glyoxylic acid. Tetrahedron Lett. 2005;46:7073–7075. [Google Scholar]
- 31.Busnel O, Bi L, Dali H, Cheguillaume A, Chevance S, Bondon A, Muller S, Baudy-Floc’h M. Solid-phase synthesis of “mixed” peptidomimetics using Fmoc-protected aza-beta3-amino acids and alpha-amino acids. J Org Chem. 2005;70:10701–10708. doi: 10.1021/jo051585o. [DOI] [PubMed] [Google Scholar]
- 32.Laurencin M, Bauchat P, Baudy-Floc’h M. Preparation of Nbeta-Fmoc-protected aza-beta 3-amino acids with nonproteinogenic hydrophobic side chains for solid-phase syntheses of pseudopeptides. Synthesis. 2009:1007–1013. [Google Scholar]
- 33.Baudy-Floc’h M, Zatylny-Gaudin C, Henry J, Duval E, Laurencin M. N° FR 07 09054 1808. SERB Patent. 2007:106–116.
- 34.Dathe M, Nikolenko H, Klose J, Bienert M. Cyclization increases the antimicrobial activity and selectivity of arginine- and tryptophan-containing hexapeptides. Biochemistry. 2004;43:9140–9150. doi: 10.1021/bi035948v. [DOI] [PubMed] [Google Scholar]
- 35.Wu JM, Wei SY, Chen HL, Weng KY, Cheng HT, Cheng JW. Solution structure of a novel D-ss-naphthylalanine substituted peptide with potential antibacterial and antifungal activities. Biopolymers. 2007;88:738–745. doi: 10.1002/bip.20736. [DOI] [PubMed] [Google Scholar]
- 36.Haug BE, Svendsen JS. The role of tryptophan in the antibacterial activity of a 15-residue bovine lactoferricin peptide. J Pept Sci. 2001;7:190–196. doi: 10.1002/psc.318. [DOI] [PubMed] [Google Scholar]
- 37.Hubert F, Noel T, Roch P. A member of the arthropod defensin family from edible Mediterranean mussels (Mytilus galloprovincialis) Eur J Biochem. 1996;240:302–306. doi: 10.1111/j.1432-1033.1996.0302h.x. [DOI] [PubMed] [Google Scholar]
- 38.Sisido M, Egusa S, Imanishi Y. One-Dimensional Aromatic Crystals in Solution .2. Synthesis, Conformation, and Spectroscopic Properties of Poly(L-2-Naphthylalanine) J Am Chem Soc. 1983;105:4077–4082. [Google Scholar]
- 39.Sisido M, Egusa S, Imanishi Y. One-Dimensional Aromatic Crystal in Solution .1. Synthesis, Conformation, and Spectroscopic Properties of Poly(L-1-Naphthylalanine) J Am Chem Soc. 1983;105:1041–1049. [Google Scholar]
- 40.Dike A, Cowsik SM. Solution structure of amphibian tachykinin Uperolein bound to DPC micelles. J Struct Biol. 2006;156:442–452. doi: 10.1016/j.jsb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Ivankin A, Livne L, Mor A, Caputo GA, DeGrado WF, Meron M, Lin B, Gidalevitz D. Role of the Conformational Rigidity in the Design of Biomimetic Antimicrobial Compounds. Angew Chem Int Ed Engl. 2010;49:8462–8465. doi: 10.1002/anie.201003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Claudon P, Violette A, Lamour K, Decossas M, Fournel S, Heurtault B, Godet J, Mely Y, Jamart-Gregoire B, Averlant-Petit MC, Briand JP, Duportail G, Monteil H, Guichard G. Consequences of Isostructural Main-Chain Modifications for the Design of Antimicrobial Foldamers: Helical Mimics of Host-Defense Peptides Based on a Heterogeneous Amide/Urea Backbone. Angew Chem Int Ed Engl. 2010;49:333–336. doi: 10.1002/anie.200905591. [DOI] [PubMed] [Google Scholar]
- 43.Hetru C, Bulet P. Strategies for the isolation and characterization of antimicrobial peptides of invertebrates. Methods Molecular Biology. 1997;78:35–49. doi: 10.1385/0-89603-408-9:35. [DOI] [PubMed] [Google Scholar]
- 44.Duval E, Zatylny C, Laurencin M, Baudy-Floc’h M, Henry J. KKKKPLFGLFFGLF: a cationic peptide designed to exert antibacterial activity. Peptides. 2009;30:1608–1612. doi: 10.1016/j.peptides.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 45.Javadpour MM, Juban MM, Lo WC, Bishop SM, Alberty JB, Cowell SM, Becker CL, McLaughlin ML. De novo antimicrobial peptides with low mammalian cell toxicity. J Med Chem. 1996;39:3107–3113. doi: 10.1021/jm9509410. [DOI] [PubMed] [Google Scholar]
- 46.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 47.Johnson BA, Blevins R. NMRVIEW: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 48.Wüthrich K. NMR of Proteins and Nucleic acids. Wiley-Interscience; New York: 1986. [Google Scholar]
- 49.Linge JP, O’Donoghue SI, Nilges M. Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 2001;339:71–90. doi: 10.1016/s0076-6879(01)39310-2. [DOI] [PubMed] [Google Scholar]
- 50.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 51.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 52.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]


