Abstract
Opiates are among the oldest medications available to manage a number of medical problems. Although pain is the current focus, early use initially focused upon the treatment of dysentery. Opium contains high concentrations of both morphine and codeine, along with thebaine, which is used in the synthesis of a number of semisynthetic opioid analgesics. Thus, it is not surprising that new agents were initially based upon the morphine scaffold. The concept of multiple opioid receptors was first suggested almost 50 years ago (Martin, 1967), opening the possibility of new classes of drugs, but the morphine-like agents have remained the mainstay in the medical management of pain. Termed mu, our understanding of these morphine-like agents and their receptors has undergone an evolution in thinking over the past 35 years. Early pharmacological studies identified three major classes of receptors, helped by the discovery of endogenous opioid peptides and receptor subtypes—primarily through the synthesis of novel agents. These chemical biologic approaches were then eclipsed by the molecular biology revolution, which now reveals a complexity of the morphine-like agents and their receptors that had not been previously appreciated.
I. Historical Overview
“If the entire materia medica at our disposal were limited to the choice and use of only one drug, I am sure that a great many, if not the majority, of us would choose opium; and I am convinced that if we were to select, say half a dozen of the most important drugs in the Pharmacopeia, we should all place opium in the first rank. If we were to inquire, however, into how much the great majority of the medical men know about the history of this wonderful product of plant life, which, when judiciously employed, has proved such a boon to suffering humanity, if we were to ask about the origin of some of our most familiar remedies–laudanum or paregoric, for instance—I fear the information gleaned would be meager.”
Opium has been used for thousands of years, and its clinical value cannot be overstated. Pain transcends the boundaries of all medical specialties and impacts almost everyone at some stage of their life. There are many classes of drugs used to relieve pain. Mild to moderate pain is typically treated with acetaminophen or aspirin or other nonsteroidal anti-inflammatory drugs (NSAID), but the mainstay of pain management for severe pain remains the opiates. Their effects on pain are quite intriguing. Unlike local anesthetics that relieve pain by blocking all sensory transmission, opiates selectively modulate the perception of pain without interfering with basic sensations, such as light touch, temperature, position sense, and discrimination of sharp and dull. The opioids target the subjective component of pain, an integrated sensation. It is not uncommon for a patient to remark after taking an opiate that “the pain is still there, but it does not hurt.”
Medically, opium was first used by Arabic physicians, with its use spreading outward from the Middle East to India and China, where it was used to treat dysentery. Indeed, the use of opium in China was relatively late, being fostered first by the Portugese in the mid- to late-1700s and then through the British East India Company. Its trade increased dramatically and eventually became a major political issue, leading to the opium wars between China and England (Macht, 1915).
The early preparations of opium were oral and were used primarily for relief of diarrhea associated with dysentery, a common problem. Indeed, oral opium preparations to treat diarrhea, such as laudanum (tincture of opium), are still available. As the use of opium and opiates spread and increased, their euphoric and addictive properties became more apparent, along with significant abuse (Macht, 1915; Terry and Pellens, 1928). This had become prominent in the United States in the mid-1800s, around the time of the Civil War when the development of the hypodermic syringe permitted morphine to be administered parenterally, which greatly enhanced its euphoric activity. Opioid abuse became so problematic in the early 1900s that international treaties limiting its trafficking were instituted.
The importance of opioids in the treatment of pain has never been contested. However, the desire to develop analgesics dissociating pain and abuse potential drove massive synthetic efforts over the years that generated hundreds, if not thousands, of analogs and eventually provided clinicians with dozens of opiate drugs. Although the vast majority of these agents did not separate analgesia from abuse potential and/or many of problematic side effects seen with traditional opiates, the clinical use of these synthesized drugs has given many insights into opiate action. Indeed, opiates are in the rare position where their clinical pharmacology preceded the development of corresponding animal models and molecular mechanisms of action. Thus, the clinical pharmacology of opiates has driven much of the basic preclinical research into their mechanism of action.
The initial pharmacologic studies of opiates focused on the general effects of morphine in humans (reviewed by Martin, 1963, 1967; Reisine and Pasternak, 1996). Analgesia is very difficult to study, primarily because of its extreme subjectivity. Painful stimuli, their thresholds, and neuronal pathways have been well characterized both neurophysiologically and neuroanatomically. However, the clinical perception of pain cannot be defined as concretely. It is very dependent on the emotional makeup of the individual, as well as the emotional state and expectations and desires of the individual at the time. In his classic study comparing wounded soldiers to civilians with postoperative pain, Beecher (1946) found that 80% of the civilians asked for pain relief, whereas only 25% of the soldiers made the same request. This difference was made even more dramatic by the fact that the nociceptive stimuli were thought to be comparable or more severe in the soldiers. Clearly, the stress of combat altered the perception of the nociceptive stimuli. Clinically, opiates act upon the subjective aspects of pain (Beecher, 1946, 1960; Lasagna, 1964). Patients receiving narcotics are able to discern noxious stimuli but report no pain. Thus, the study of analgesia in humans is extremely difficult, especially its quantification. The subtle nuances and the importance of context further illustrate the complexity of pain measurements and must be taken into consideration when comparing experimental and clinical pain models. These factors also demonstrate the inadequacies and limitations of preclinical studies of analgesics.
Despite their limitations, animal models were needed to evaluate new compounds and explore basic questions in mechanism. The mouse hot-plate test (Woolfe and MacDonald, 1943) and the tail flick (D'Amour and Smith, 1941) were the first two widely used models. Both are reproducible and applicable over wide dose ranges (Janssen and Jageneau, 1957) and highly predictive of analgesic activity in people. The tail-flick assay is dependent upon a spinal reflex, with descending supraspinal influences (Irwin et al., 1951), whereas the hot plate relies upon a more integrated escape response. However, in most paradigms, the nociceptive input in these thermal assays is sufficiently severe that only potent analgesics show activity, with partial agonists or mixed opioid agonist/antagonist drugs and most nonopioids, such as the NSAIDs, often having little effect. Over time, a wide range of additional assays have been developed and used to explore different types of pain (Le Bars et al., 2001), including mechanical stimuli, inflammation, and neuropathic pain associated with nerve injury to name a few. The activity, or “efficacy,” of a drug commonly varies among the different pain models. When assessing these assays, it is important to consider both the intensity of the nociceptive stimulus and its nature. Weaker drugs may show activity only against lower intensities of nociceptive stimuli and may be inactive against more severe ones. Efficacy also may differ depending upon the nature of the pain involved. One example is the difference between the radiant heat tail-flick assay, which focuses upon a small region of the tail, and the tail immersion assay. Both are thermal assays. However, the radiant heat assay might be like a drop of hot water on a finger, whereas the immersion assay would be equivalent of putting the whole finger into the hot water bath. Both are widely used and have been validated, but they are different. Another would be the traditional abdominal constriction (writhing) assay, where the ED50 values for opioids are typically approximately 10-fold lower than in the thermal assays and where drugs such as the tricyclic antidepressants are analgesic despite their inactivity in thermal assays (Spiegel et al., 1983). A full discussion of pain models is beyond the scope of this review, and the reader is referred to another review from this journal (Le Bars et al., 2001).
Opioids were first used for their actions on gastrointestinal motility, as noted above. Opiates decrease propulsive peristaltic contractions, while increasing circular muscle tone and intraluminal pressure (Reisine and Pasternak, 1996). Early investigators found it easier to examine opiate mechanisms in isolated organ systems, such as the guinea pig ileum (Trendelenburg, 1917; Schaumann, 1955; Kosterlitz and Robinson, 1957a; Paton, 1957), and found that opiates inhibited the release of acetylcholine (Paton, 1957; Schaumann, 1957; Trendelenburg, 1957; Cox and Weinstock, 1966). The activity of guinea pig ileum assays rested with the opioid receptors located in the myenteric plexus that surrounds the muscle layers of the gut in these organ preparations. Similar assays using alternative organ systems have also been used, such as the mouse vas deferens assay that Kosterlitz used to follow the isolation and purification of the enkephalins (Hughes et al., 1975).
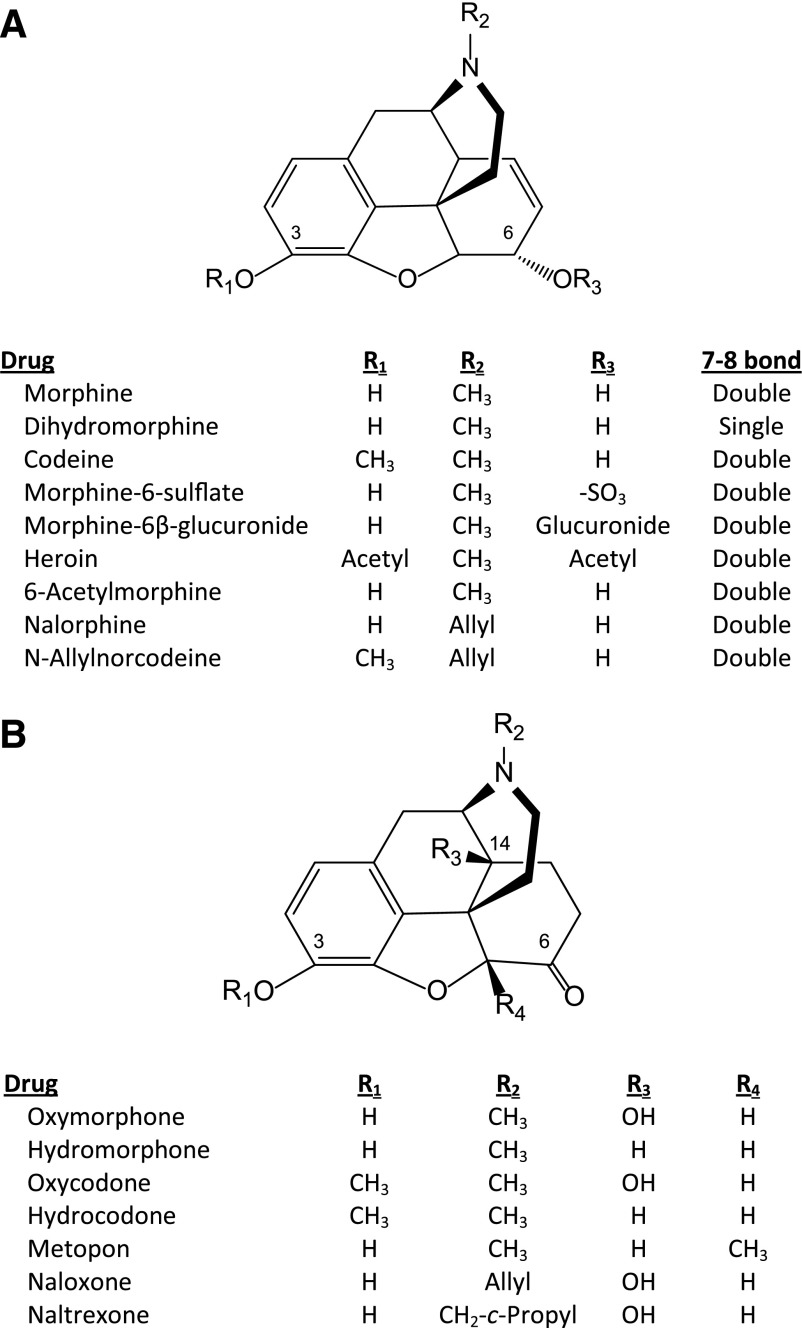
Morphine (Fig. 1A) was isolated from opium in 1805 (Serturner, 1805; Macht, 1915) and first sold by Merck in 1827, with its popularity increasing with the development of the hypodermic needle in 1857. Its synthesis was delayed by its complex ring structure until 1956 (Gates and Tschudi, 1956). However, modifications of its structure were made much earlier. Indeed, diacetylmorphine (heroin) (Fig. 1A) was synthesized in 1874 and marketed as a nonaddictive cough suppressant by Bayer in the late 1800s—a claim we now find amazing and clearly wrong.
Fig. 1.
Selected 4,5α-epoxymorphinan compounds.
In an attempt to eliminate respiratory depression, Von Braun (1916) synthesized a new opiate derivative, N-allylnorcodeine (Fig. 1A), a compound that stimulated respiration when given alone and reversed morphine-induced respiratory depression (Pohl, 1915). This discovery of the first antagonist was buried in the literature for 25 years. The next attempt to use an allyl nitrogen substitution to make an opiate free from respiratory depression came in the 1940s with the synthesis and studies of N-allyl-normorphine (nalorphine) (McCawley et al., 1941; Weijlard and Erickson, 1942; Unna, 1943). The N-allyl replacement of the N-methyl group created a new class of drugs that enabled the reversal of respiratory depression seen with morphine, but which also precipitated withdrawal in dependent subjects.
The modern era of opioid research came with the demonstration of opioid receptors in 1973 (Pert et al., 1973; Simon et al., 1973; Terenius, 1973) using binding assays based upon stereoselectivity (Goldstein et al., 1971). The concept of opioid receptors has a long history, with selective recognition sites being proposed much earlier based upon the rigid structural requirements for activity (Beckett and Casy, 1965; Portoghese, 1965, 1970). This was followed by extensive studies on opioid receptor binding, leading to the cloning and expression of the family of opioid receptors. This review will focus upon the mu drugs, their receptors, and their actions.
II. Opioids
A. Alkaloids
The original opiates, morphine and codeine (Fig. 1A), were isolated from opium. Their structures provided the scaffolds upon which many of the current mu opiates are based. Thebaine, another major component of opium, is a valuable precursor in the synthesis of many of these derivatives. Most opiates fall under six chemical classes: 4,5α-epoxymorphinans, morphinans, benzomorphans, phenylpiperidines, acylic analgesics, and oripavine. These chemical classes conceptually can be visualized by a systematic dismantling of the morphine structure.
1. Morphine Analogs.
The two major natural opiates are morphine and codeine (Fig. 1A), which are present at high levels in opium. As noted above, it took 120 years to establish morphine’s structure (Gulland and Robinson, 1925) after its initial isolation (Serturner, 1805) and another 30 for its total synthesis (Gates and Tschudi, 1956). Morphine and codeine analogs were made before the structure was determined, one of the earliest being heroin (diacetylmorphine or diamorphine).
Morphine belongs to the 4,5α-epoxymorphinan opiate class. The structure-activity relationships for morphine activity have been well established. Although early studies based upon traditional pharmacological assays provided many insights into activity, more recent receptor binding studies have further refined the criteria influencing the selectivity/affinity of the ligands. Only the natural (−)isomers are active. A free 3-hydroxyl group is essential for activity. Blockade of this position dramatically lowers affinity for mu receptors (Pert et al., 1973). Thus, the activity of codeine is thought to be attributed to its demethylation to morphine, which helps explain the varying sensitivity of patients because of differences in CYP2D6 enzymatic activity. Indeed, ultrarapid metabolizers may even overdose or encounter respiratory depression. This is well described in several studies looking at the use of codeine after surgery in children (Kelly et al., 2012; Voelker, 2012; Kuehn, 2013). It is presumed that similar situations exist for oxycodone and hydrocodone.
In contrast, changes at the 6-position have proven very useful, as several morphine metabolites have shown. Morphine-6β-glucuronide (M6G) is an excellent example. A morphine metabolite in humans, M6G has long been known to have activity (Shimomura et al., 1971), but its extraordinary potency was not appreciated until more than a decade later (Pasternak et al., 1987; Paul et al., 1989b). The sugar moiety impedes the passage of M6G through the blood-brain barrier, but when that is avoided by direct injection into the brain, M6G is over 50-fold more potent than morphine. [Unlike humans, rodents do not glucuronidate morphine at the 6-position (Inturrisi et al., 1996), which has greatly simplified its study in rodent models.] Clinically, M6G is now recognized as a contributor to the overall analgesic activity of morphine, particularly with chronic dosing (Tiseo et al., 1995). Indeed, M6G blood levels can accumulate and exceed those of morphine itself, particularly in the presence of renal insufficiency or failure. A second morphine derivative, morphine-6-sulfate (Fig. 1A) has been isolated from brain (Donnerer et al., 1987). Similar to M6G, this modification at the 6-position markedly enhances potency (Zuckerman et al., 1999).
Heroin (Fig. 1A) has acetyl groups at both the 3- and 6-positions, yet it is more potent than morphine. There are likely several reasons. Acetylation enhances the ability of the drug to pass the blood-brain barrier, resulting in greater penetration into the brain from an equivalent dose. Second, the 3-position is rapidly deacetylated enzymatically in the blood to form 6-acetylmorphine (Fig. 1A), a potent metabolite thought to be responsible for its actions (Inturrisi et al., 1983,1984). Thus, heroin provides another example of how modifications of the 6-position can influence activity.
Conversion of the 6-hydroxyl group to a ketone, along with reduction of the 7–8 double bond, has given us potent analgesics, including hydromorphone, hydrocodone, and metopon, and a number of 14-hydroxy analogs, such as the analgesics oxymorphone and oxycodone and the antagonists naloxone and naltrexone (Fig. 1B). One particularly interesting derivative is 14-hydroxymetopon (Schmidhammer et al., 1990; Fürst et al., 1993; Freye et al., 2000; King et al., 2003). This derivative is an exceedingly potent analgesic with limited respiratory depression or inhibition of gastrointestinal transit. Peripherally acting agents, such as 3-methylnaltrexone, which is used clinically to counter opioid-induced constipation, are generated by quaternizing the nitrogen, greatly diminishing their ability to transverse the blood-brain barrier.
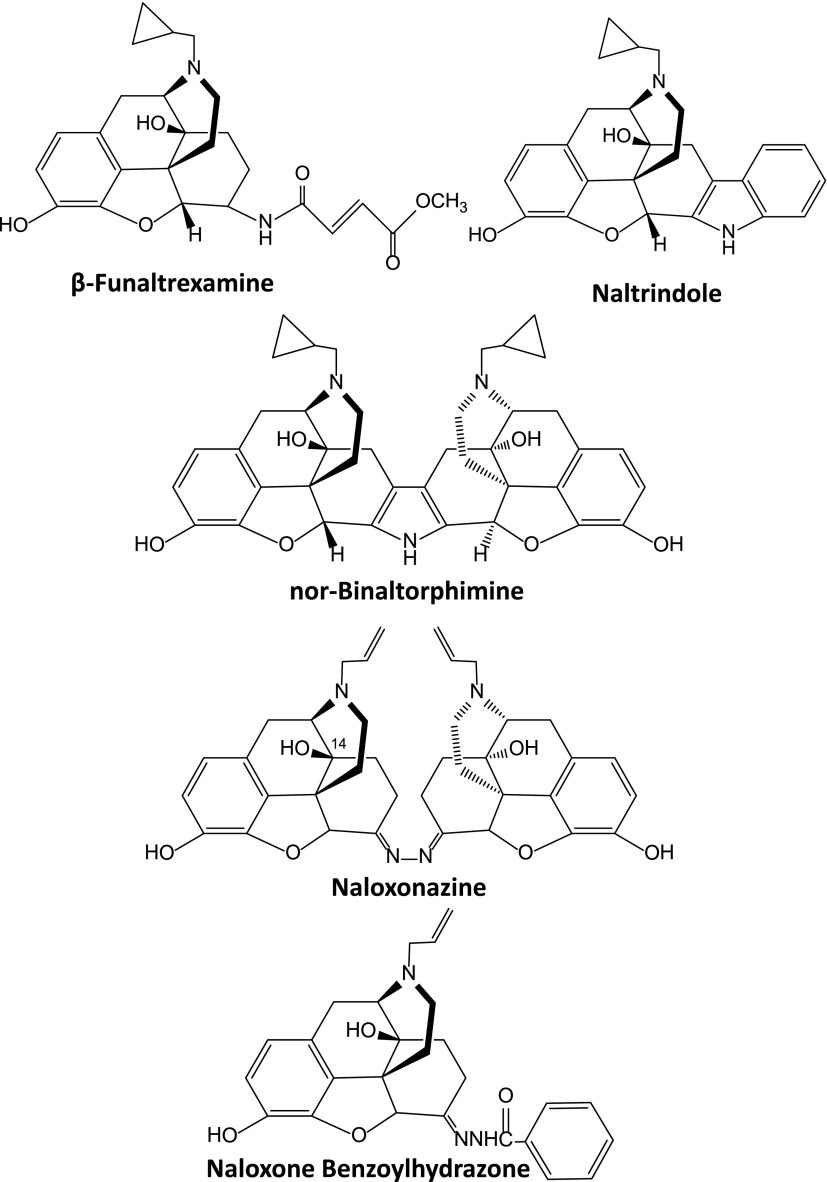
6-Position modifications have also generated a large number of unique agents that have proven valuable in the laboratory in investigating opioid mechanisms (Fig. 2), including β-funaltrexamine, naltrindole, and nor-binaltorphimine, that selectively inhibit mu, delta, and kappa1 receptors, respectively (Portoghese et al., 1980, 1987, 1988); naloxonazine and naloxazone, which block a mu-receptor subtype (Pasternak et al., 1980a,b; Childers and Pasternak, 1982; Hahn et al., 1982; Hahn and Pasternak, 1982); and naloxone benzoylhydrazone, a mu-antagonist/kappa3 agonist (Hahn et al., 1985; Luke et al., 1988; Clark et al., 1989). Indeed, these have been extensively used to define receptor classes responsible for drug actions.
Fig. 2.
Selective opioid antagonists.
A number of other modifications have been examined. Reduction of the C7–C8 double bond in morphine generates dihydromorphine (Fig. 1A), which is more potent than morphine. Substitutions on the nitrogen greatly influence the pharmacology of the compound, with replacement of the N-methyl group of the 4,5α-epoxymorphinans with either an N-allyl or N-methylcyclopropyl group typically converting the compound from an agonist to antagonist, as first demonstrated with N-allylnorcodeine (Von Braun, 1916) and N-allylnormorphine (nalorphine) (McCawley et al., 1941; Weijlard and Erickson, 1942; Unna, 1943) (Fig. 1A). Nalorphine proved to be a very unusual compound. It readily reversed morphine actions, including analgesia and respiratory depression. However, at higher doses it was analgesic (Lasagna and Beecher, 1954; Houde and Wallenstein, 1956), actions that led Martin (1967) to propose that the drug was an antagonist at the M (morphine; μ) receptor and an agonist at the N (nalorphine) receptor, a concept he termed receptor dualism. The N receptor probably corresponds to the current kappa1 receptor. Thus, nalorphine is a mixed agonist/antagonist and led to the first proposal of opioid receptor subtypes. Subsequently, several mixed agonist/antagonists have been developed for clinical use, including nalbuphine and nalmephene.
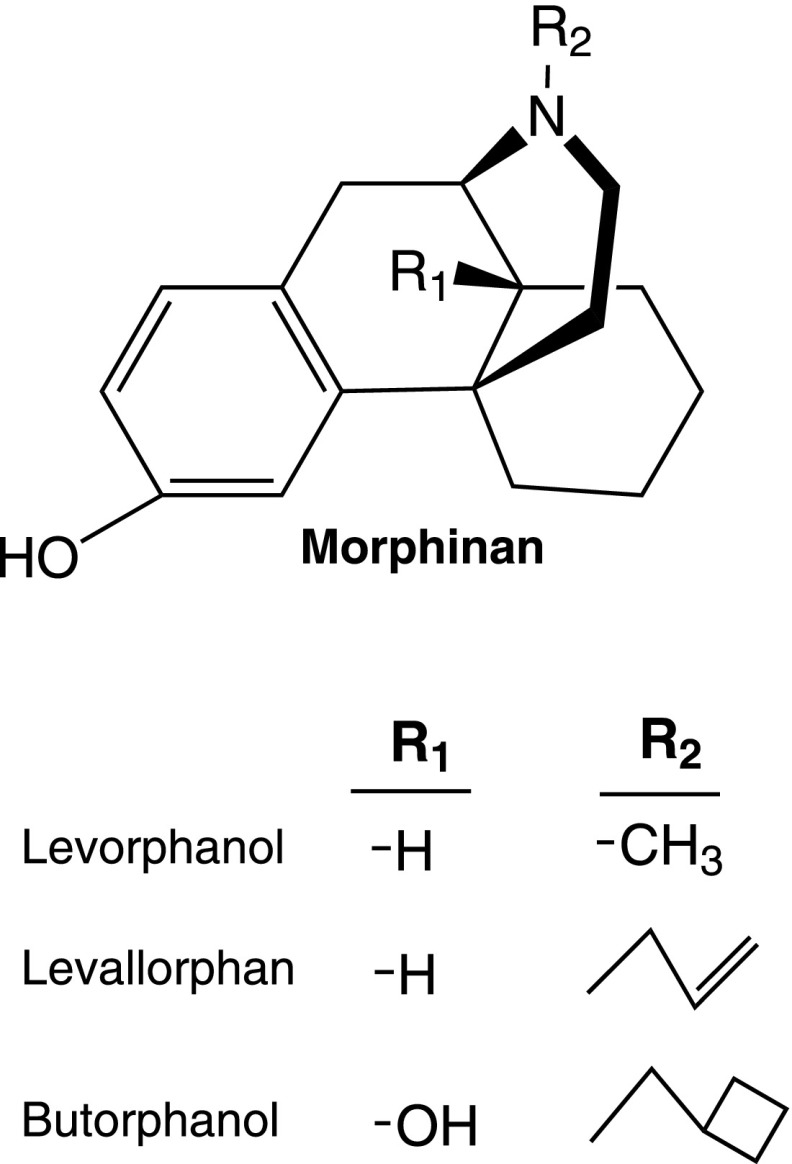
2. Morphinans.
These analogs lack the 4,5-epoxy bridge seen in morphine and lack both the C7–C8 double bond and the 6-hydroxyl group (Fig. 3). As with morphine, the morphinans show rigid stereoselectivity, with the levo (−)isomers possessing all the opioid activity. There are three significant morphinans as follows: the agonists levorphanol and butorphanol and the antagonist levallorphan. Levorphanol has an N-methyl group and the antagonist levallorphan an N-allyl group. Butorphanol is closely related, the difference being an N-methylcyclobutyl group and a 14-OH. They all display high affinity for mu receptors in binding assays, but these changes also lead to a loss of selectivity, with the compounds displaying high affinity for other receptor classes (Moulin et al., 1988; Tive et al., 1992; Inturrisi, 2002; Rowbotham et al., 2003; Majumdar et al., 2011). Their lack of selectivity for mu receptors may help explain their complex pharmacology, especially with evidence now showing a role for a truncated MOR-1 splice variant in their actions (see section VIII.C.3.b). Both levorphanol and butorphanol have been widely used clinically and are effective analgesics in people.
Fig. 3.
Morphinan.
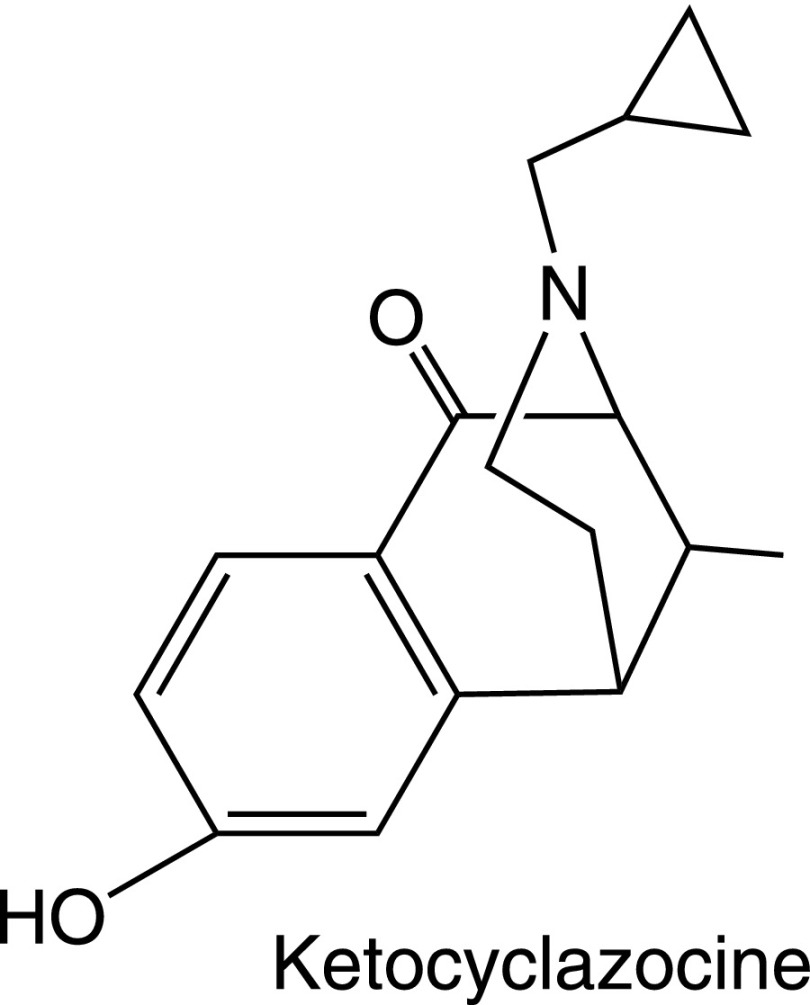
3. Benzomorphans.
Removal of the “C ring” of morphine led to the benzomorphans (Archer et al., 1962, 1996; Fraser and Harris, 1967). The prototypic benzomorphan is ketocyclazocine (Fig. 4), which Martin used to formally define kappa receptors over 35 years ago (Martin et al., 1976). Similar to most of the benzomorphans, ketocyclazocine has high affinity for kappa receptors but shows little selectivity among the other opioid receptor classes. Ethylketocyclazocine is another benzomorphan that has proven valuable in preclinical studies. However, it also is relatively nonselective, as is cyclazocine. These structures typically have prominent psychotomimetic properties.
Fig. 4.
Ketocyclazocine.
Of the vast array of benzomorphans that have been synthesized, only pentazocine is used clinically. Similar to nalorphine, pentazocine is a mixed kappa agonist/mu antagonist, with its analgesic actions residing primarily in its kappa interactions. Its actions are highly stereoselective. Its opioid activity is restricted to the (−)isomer. However, the (+)isomer is a potent sigma-receptor agonist, which is interesting because sigma agonists lower opioid analgesic activity (Chien and Pasternak, 1993, 1994, 1995a). Because the clinical formulation of pentazocine in the United States is racemic, containing both stereoisomers, the presence of the (+)isomer may actually lower the analgesic actions of the (−)isomer.
4. Oripavines.
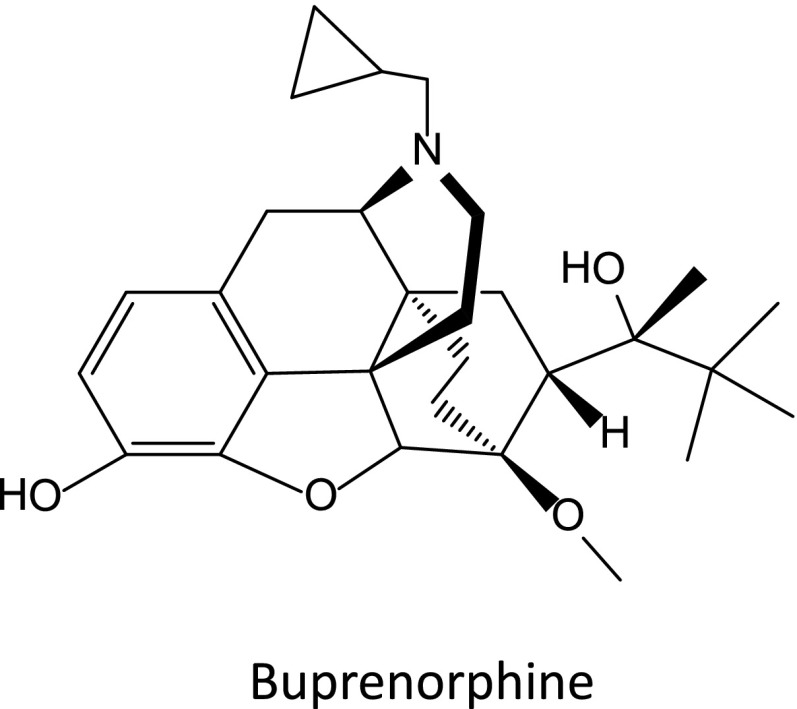
Etorphine, diprenorphine, and buprenorphine are the best known oripavine derivatives (Bentley and Hardy, 1967). All three oripavines display little selectivity among the various opioid receptor classes. Etorphine is about 1000 times more potent an analgesic than morphine, whereas diprenorphine is a potent antagonist. Buprenorphine (Fig. 5) is clinically useful both as an analgesic and in addiction treatment programs. It is ~20-fold more potent than morphine, but with a complex selectivity profile (Cowan et al., 1977; Dum and Herz, 1981; Lewis, 1985; Leander, 1987; Kamei et al., 1995, 1997; Pick et al., 1997; Lutfy and Cowan, 2004; Ding and Raffa, 2009; Pergolizzi et al., 2010; Davis, 2012).
Fig. 5.
Buprenorphine.
5. Other.
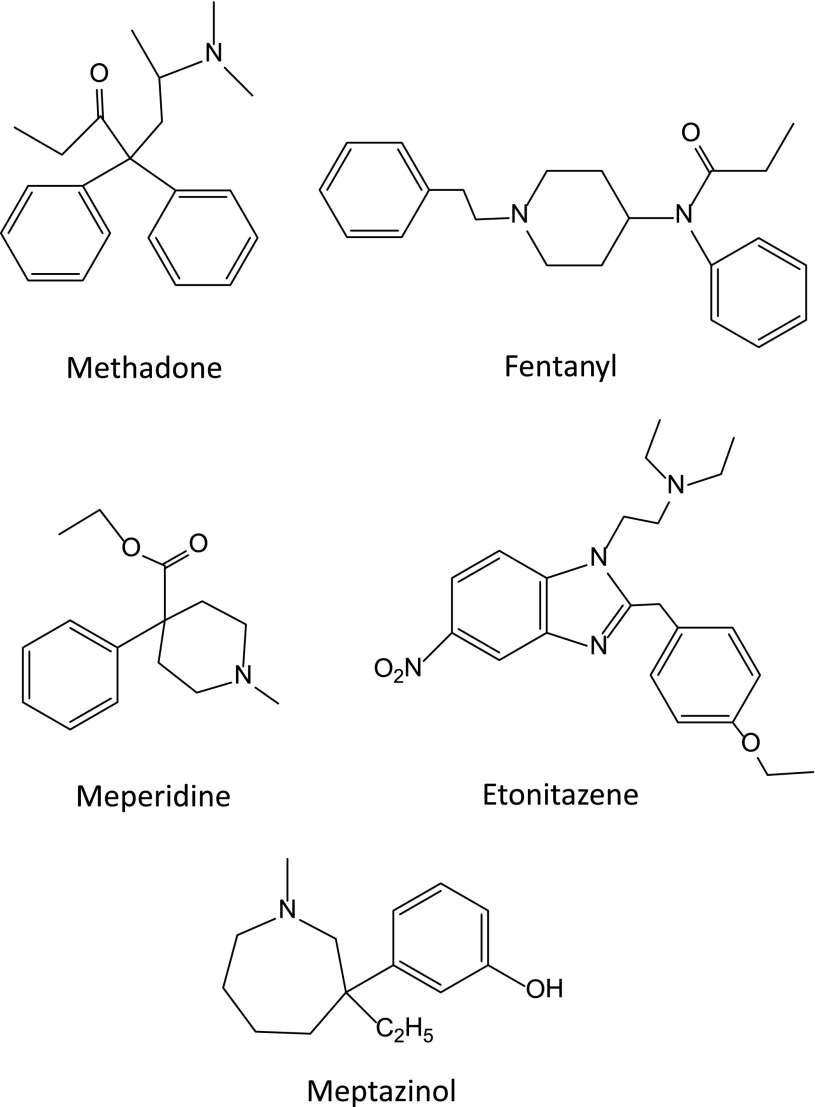
A variety of totally synthetic opiates avoiding the need for opium have been synthesized over the years. Meperidine, the prototypic 4-phenylpiperidine, is a widely used clinical analgesic, although its utility is limited by the toxicity and epileptic actions of its N-demethylated metabolite normeperidine (Kaiko et al., 1983; Inturrisi, 2002). Methadone is another mu-selective analgesic, and is the major drug in the acyclic analgesic class. Synthesized in 1937 in Germany to avoid interruption of their supplies of opium in case of war (Bockmuhl, 1948), it was first marketed in the United States in 1947 by Eli Lilly. A potent analgesic, for many decades it was used in the United States primarily in addiction treatment programs, based upon the work of Dole et al. (1966). More recently, its use has extended to pain management as well. However, the clinical use of methadone as an analgesic is complex because of its prolonged (typically ∼24 hours) and widely variable half-life. A lack of appreciation of the prolonged time needed to reach steady-state drug levels, typically from 3 to 5 days, has led to inadvertent overdosing and even death. Its analgesic activity is limited to the (−)isomer, which is marketed in Europe. In the United States, it is marketed as the racemic mixture, which may be important because the (+)isomer has been reported to have NMDA antagonist actions (Gorman et al., 1997; Davis and Inturrisi, 1999). Recent studies have also observed that methadone can prolong the QT interval of the electrocardiogram, an issue of potential clinical significance (Raffa et al., 2012).
Among the most widely used of the synthetic opiates are the 4-anilidopiperidines, originally developed by Janssen et al. (1959). This class includes fentanyl (Fig. 6), sufentanil, remifentanil, alfentanil, carfentanil, lofentanil, and ohmefentanil, which are mu selective and 500- to more than 10,000-fold more potent than morphine. Another agent is loperimide, which is poorly absorbed from the gastrointestinal tract and is widely used as an antidiarrheal. The benzimazone opioids also are potent mu analgesics (Hunger et al., 1957), particularly etonitazene (Fig. 6), which is over 1000-fold more potent than morphine in animal models. It is not used clinically.
Fig. 6.
Other opioid structures.
Meptazinol (Fig. 6) is a unique opioid. It has been used clinically to treat moderate to severe pain in Europe. It produces its analgesic actions through mu receptors, as clearly shown by its sensitivity to the antagonist naloxonazine (Robson, 1983; Spiegel and Pasternak, 1984). Yet, it has little respiratory depression or constipation and is said to have a diminished abuse potential.
Agents selective for other opioid receptor classes, such as the kappa1-selective arylacetamides such as U50,488H [2-(3,4-dichlorophenyl)-N-methyl-N-[(1R,2R)-2-pyrrolidin-1-ylcyclohexyl]acetamide], spiradoline, and endaloline, have been synthesized, but these compounds have not proven useful clinically because of psychomimetic and dysphoric actions, as well as a pronounced diuresis. Other classes of agents selective for delta receptors are also under investigation.
B. Opioid Peptides
1. Endogenous Opioids.
Soon after the discovery of the opioid receptors, investigators identified materials within the brain with opioid-like activity and affinity for the receptors (Table 1). Kosterlitz and Hughes were the first to sequence the pentapeptide enkephalins (Hughes et al., 1975), which soon expanded into the following three families of peptides, each with its own precursor peptide: preproenkephalin, preprodynorphin, and β-lipotropin (Berezniuk and Fricker, 2011). Kosterlitz named the two pentapeptides the enkephalins, referring to their presence within the brain, whereas Avram Goldstein coined the term dynorphin for the 17mer he isolated based upon its very high potency. The enkephalins have been associated with the delta receptors, dynorphin A with the kappa1 receptors, and β-endorphin with mu receptors, although it also retains a similar high affinity for delta receptors. An early article reported the existence of a high-affinity β-endorphin binding site on lymphocytes (KD 3 nM) (Hazum et al., 1979). What makes this high-affinity β-endorphin site so interesting is that it is insensitive to a number of traditional opioids at concentrations as high as 1 µM, including naloxone, cyclazocine, morphine, and the enkephalins. Together, all three families of opioid peptides are referred to as the endorphins at the suggestion of Eric Simon, reflecting a contraction of “endogenous morphine.”
TABLE 1.
Structures of selected endogenous opioid peptides
| [Leu5]enkephalin | Tyr-Gly-Gly-Phe-Leu |
| [Met5]enkephalin | Tyr-Gly-Gly-Phe-Met |
| Dynorphin A | Tyr-Gly-Gly-Phe-Leu-Arg-Arg-Ile-Arg–Pro-Lys–Leu-Lys-Trp-Asp-Asn-Gln |
| Dynorphin B | Tyr-Gly-Gly-Phe-Leu-Arg-Arg-Gln-Phe-Lys-Val-Val-Thr |
| a-Neoendorphin | Tyr-Gly-Gly-Phe-Leu-Arg-Lys-Tyr-Pro-Lys |
| β-Neoendorphin | Tyr-Gly-Gly-Phe-Leu-Arg-Lys-Tyr-Pro |
| βh-Endorphin | Tyr-Gly-Gly-Phe-Met-Thr-Ser-Glu-Lys-Ser-Gln-Thr-Pro-Leu-Val-Thr-Leu-Phe-Lys-Asn-Ala-Ile-Ile-Lys-Asn-Ala-Tyr-Lys-Lys-Gly-Glu |
| Endomorphin-1 | Tyr-Pro-Trp-Phe-NH2 |
| Endomorphin-2 | Tyr-Pro-Phe-Phe-NH2 |
| Orphanin FQ/nociceptin | Phe-Gly-Gly-Phe-Thr-Gly-Ala-Arg-Lys-Ser-Ala-Arg-Lys-Leu-Ala-Asn-Gln |
The first public disclosure of the endogenous opioids came at a meeting of the Neuroscience Research Program in Boston in 1974 sponsored by Massachusetts Institute of Technology (Snyder and Matthysse, 1975). A small conference of less than 50 scientists, it included most of the major investigators in the opioid field (Fig. 7). The disclosures were quite dramatic. Hans Kosterlitz announced that he and John Hughes had isolated, but not fully purified, a material from the brain that was active in the mouse vas deferens bioassay and that was reversed by the opioid specific antagonist naloxone. By using receptor binding techniques, both Lars Terenius and we independently reported an endogenous material in brain that competed opioid binding. Early in the course of our own studies characterizing receptor binding, we observed that incubating brain membranes prior to the binding assay increased binding by 50%, which was the result of the dissociation of a morphine-like factor from the receptor (Table 2) (Pasternak et al., 1975c). [Many groups continue to incubate membrane preparations prior to binding assays to dissociate endogenous opioids and increase binding.] The earliest reports described partially purified extracts (Hughes, 1975; Pasternak and Snyder, 1975a; Pasternak et al., 1975a; Terenius and Wahlstrom, 1975), but these were rapidly followed by determination of the structure of the enkephalins by Kosterlitz (Hughes et al., 1975).
Fig. 7.
Neuroscience Research Program Meeting. Photograph of the attendees at the Neuroscience Research Program meeting in Boston, MA, May 19–21, 1974. Seated (left to right): Gavril W. Pasternak, William Bunney, John Hughes, Hans Kosterlitz, Steven Matthysse, Francis O. Schmitt, Solomon H. Snyder, Avram Goldstein, E. Leong Way, Vincent P. Dole, and Aki Takemori. Middle row (left to right): L. Everett Johnson, Frederic G. Worden, Robert D. Hall, Candace D. Pert, Yvonne M. Homsy, Parvati Dev, Huda Akil, Floyd E. Bloom, Agu Pert, Peter A. Mansky, William H. Sweet, Albert Herz, William R. Martin, and Harriet Schwenk. Top row (left to right): Ian Creese, David J. Mayer, Eric J. Simon, Leslie Iversen, Diana Schneider, Pedro Cuatrecasas, Horace Loh, Arnold J. Mandell, Arthur E. Jacobson, Jose M. Musacchio, and Lars Terenius. From Snyder and Matthysse (1975).
TABLE 2.
Release of opioid factor with incubation of brain tissue
Rat brain homogenates were centrifuged, resuspended, and incubated at the indicated temperature/time. The soluble extracts were then tested in a [3H]naloxone binding assay and the inhibition determined. From Pasternak et al. (1975c).
| Addition | [3H]Naloxone Binding |
|
|---|---|---|
| cpm | Inhibition | |
| % | ||
| None | 4142 | |
| 0° Supernatant, 40 min | 3154 | 24 |
| 25° Supernatant, 40 min | 1905 | 54 |
| 37° Supernatant, 40 min | 1493 | 64 |
| 37o Supernatant, 120 min | 1166 | 72 |
The isolation and structural determination of the enkephalins was quickly followed by the identification of dynorphin A and β-endorphin (Cox et al., 1975; Teschemacher et al., 1975; Birdsall and Hulme, 1976; Goldstein, 1976; Li and Chung, 1976; Li et al., 1976; Goldstein et al., 1979). The extended peptides all contain an enkephalin sequence at the N terminus (Table 1) and all are generated by processing their respective larger precursors (for review, see Berezniuk and Fricker, 2011). Both preproenkephalin and preprodynorphin yield a number of opioid peptides, many of which remain uncharacterized pharmacologically. In contrast, β-endorphin is the only opioid generated from a β-lipotropin, which also produces adrenocorticotropin and α-melanocyte-stimulating hormone.
All of the endogenous opioid peptides share the enkephalin sequence (Tyr-Gly-Gly-Phe-Leu or Tyr-Gly-Gly-Phe-Met) at the N terminus, with differing extensions at the C terminus. Dynorphin A is a 17mer containing the TGGFL sequence at its N terminus. The physiologic roles and the importance of dynorphin A have been well established, but there was controversy in the early days about whether dynorphin was simply a precursor of an enkephalin or whether the enkephalins were simply breakdown products of dynorphin. It is said that after a single malt whisky or two Kosterlitz referred to dynorphin as an extended enkephalin, whereas Goldstein considered the enkephalins to be truncated dynorphins. The identification of their different precursors put that question to rest. It is notable that the sequences of both precursors predict a number of additional opioid ligands. Although their pharmacological significance is not clear, it is possible that each of these additional peptides might have its own receptor and functions. This is well illustrated by BAM22, a 22-amino acid peptide containing the traditional N-terminal enkephalin motif (TGGFM). In competition studies, it displays high affinity for all the traditional opioid receptors. Yet, it also labels a family of sensory neuron-specific G protein-coupled receptors, which are quite distinct from traditional cloned opioid receptors (Baird et al., 1982; Lembo et al., 2002; Grazzini et al., 2004; Hong et al., 2004).
The opioid peptides have been extensively studied and reviewed (Hook et al., 2008; Berezniuk and Fricker, 2011). The enkephalins are the endogenous ligands for the delta-opioid receptor (DOR-1), whereas dynorphin A is the endogenous ligand for the kappa1-opioid receptor (KOR-1). β-Endorphin has high affinity for both mu and delta sites, and some investigators have suggested that it is an endogenous mu peptide. However, this classification may be an oversimplification. For example, dynorphin A potently inhibits all kappa1-selective radioligand [3H]U50,488H binding in brain membranes monophasically, consistent with a single site. Yet, the peptides dynorphin B and α-neoendorphin, two other opioid peptides containing the [Leu5]enkephalin sequence that are generated by the dynorphin A precursor, both compete [3H]U69,593 [[3H]methyl-2-phenyl-N-[(5R,7S,8S)-7-(pyrrolidin-1-yl)-1-oxaspiro[4.5]dec-8-yl]acetamide] binding in a biphasic manner. These findings imply that [3H]U50,488H labels two classes of kappa1 binding sites (kappa1A and kappa1B), with dynorphin A having similar high affinities for both (Clark et al., 1989). These additional classes of kappa receptors have not been fully characterized at the molecular level. With all the potential opioid peptides that can be generated from the precursors, it is likely that many more classes of opioid-associated receptors will be identified in the future.
Another set of endogenous opioid peptides have been reported, the endomorphins, endomorphin 1 (Tyr-Pro-Trp-Phe-NH2) and endomorphin 2 (Tyr-Pro-Phe-Phe-NH2) (Zadina et al., 1997). These are quite distinct from the other opioid peptides in that they are highly selective for mu receptors and do not contain the common enkephalin sequence located at the N terminus of all the other opioid peptides. These peptides have been extensively studied (Fichna et al., 2007), but many questions remain. Foremost is our lack of understanding of how these peptides are generated because no precursor peptide has been identified (Terskiy et al., 2007).
2. Synthetic Mu Peptides.
Early studies with the endogenous opioids were complicated by their rapid enzymatic degradation. However, substitution of a d-amino acid in the 2-position, typically d-Ala, markedly enhanced their stability (Pert et al., 1976b). Since then, a vast array of opioid peptides have been synthesized with striking differences in their receptor binding selectivity and pharmacology (Hawkins et al., 1989; Schiller et al., 1989, 1992; Dooley et al., 1995; Schiller, 1999, 2005). One of the most important is [d-Ala2,MePhe4,Gly(ol)5]enkephalin (DAMGO), which is used extensively to label mu-opioid receptors in binding assays and as a mu opioid pharmacologically. Conversely, [d-Pen2,d-Pen5]enkephalin (DPDPE) is a highly selective and enzymatically stable delta ligand. Over the years thousands of compounds have been generated, many of which are highly selective for mu or delta receptors.
Although a comprehensive overview of opioid peptides is beyond the scope of this review, two other compounds deserve mentioning. The tetrapeptide TAPS (Tyr-d-Arg-Phe-Sar) is a mu-selective ligand in binding assays, as well as a potent analgesic (Paakkari et al., 1992, 1993; Vonhof et al., 2001). Unlike the other mu opioids, TAPS is a respiratory stimulant. This dissociation between analgesia and respiratory depression is quite unique and potentially useful. Early studies indicated that morphine analgesia and respiratory depression could be distinguished by naloxonazine, which antagonized morphine analgesia but not its respiratory depressant effects (Ling et al., 1983, 1985). Although TAPS was not developed as a drug, it illustrates the potential of analgesics lacking respiratory depression.
The other peptide is metkephamid (Frederickson et al., 1981; Burkhardt et al., 1982). One of the early enkephalin derivatives, it was initially thought to be selective for delta receptors but actually labels both delta and mu receptors. What sets metkephamid apart is that it underwent clinical trials (Calimlim et al., 1982; Bloomfield et al., 1983; Pasanisi et al., 1985) and was the first to show enkephalin effects in humans.
C. Mu Antagonists
A major step forward in opioid pharmacology came with the synthesis of naloxone, the first pure antagonist (Blumberg et al., 1961; Lewenstein and Fishman, 1966; Garfield, 1983). As noted on the awarding of the 1982 John Scott Award to Fishman and Blumberg, in 1960, Jack Fishman was studying steroid chemistry at Sloan-Kettering Institute in New York while he also was working in the private laboratory of Mozes Lewenstein where he synthesized naloxone, N-allylnoroxymorphone. At the time that he synthesized the compound, Fishman did not know Harold Blumberg, but both Blumberg and Lewenstein worked at Endo Laboratories. When Lewenstein licensed the drug to Endo Laboratories, Blumberg immediately evaluated it, which lead to the first report of the antagonist properties of the compound the following year (Blumberg et al., 1961). The importance of this compound cannot be overstated. Naloxone is one of the most widely used opioids and is the primary treatment of overdose and opioid-induced respiratory depression. It also has been instrumental in assessing and defining opioid actions in preclinical studies. As Hans Kosterlitz would always ask after a presentation, “Is it naloxone reversible?”
Over the years, a number of selective mu-opiate antagonists have been synthesized based upon the pure antagonist naltrexone/naloxone scaffold that has proven invaluable in defining the pharmacology of opioids. The work of Portoghese et al. (1980, 1987, 1988) truly stands out. They synthesized the prototypic mu-antagonist β-funaltrexamine (β-FNA), the delta antagonist naltrindole, and the kappa1 antagonist nor-binaltorphimine (norBNI) (Fig. 2), which are still used to define the pharmacology of the different receptor classes. However, there are many caveats with their use. For example, β-FNA is a selective, irreversible mu antagonist, lasting for several days after its administration. However, it has reversible kappa actions, requiring that it be given 24 hours before testing to enable free drug (and thus the kappa activity) to be eliminated. Although naltrindole and norBNI can both be used to selectively block delta and kappa1 actions, it is also important to remember that their selectivity is limited and higher doses may block alternative receptor sites. Finally, norBNI is intriguing in that its antagonist actions last for weeks or longer, despite the elimination of the drug itself. Thus, its actions are not those of a simple competitive antagonist (Melief et al., 2011).
Several very useful mu-selective peptide antagonists were developed based upon the somatostatin sequence (Maurer et al., 1982; Pelton et al., 1985a,b; Gulya et al., 1986). H-d-Phe-Cys-Tyr-d-Trp-Orn-Thr-Pen-Thr-NH2 (CTOP) and d-Phe-Cys-Tyr-d-Trp-Arg-Thr-Pen-Thr-NH2 (CTAP) are both are highly selective mu-opioid antagonists and have been extensively used to define the pharmacology of mu receptors. [3H]CTAP has been used in receptor binding assays, providing a more selective alternative to either [3H]naloxone or [3H]naltrexone to examine muantagonist binding sites.
β-FNA, CTAP, and CTOP all are highly selective mu antagonists and block all mu actions. However, there also are analogs that can distinguish among mu activities, raising the question of multiple subtypes of mu receptors (Wolozin and Pasternak, 1981). Naloxazone was the first described, followed by naloxonazine, which supplanted the former’s use (Fig. 2). Although β-FNA, CTAP, and CTOP block all mu actions, naloxonazine and naloxazone are more selective (Pasternak et al., 1980a,b, 1983; Wolozin and Pasternak, 1981; Childers and Pasternak, 1982; Spiegel et al., 1982b; Holaday et al., 1983; Ling et al., 1983, 1984, 1985, 1986; Wood and Pasternak, 1983; Heyman et al., 1988; Paul and Pasternak, 1988). In these studies, naloxazone and/or naloxonazine block morphine analgesia without blocking respiratory depression, the inhibition of gastrointestinal transit, and most signs of physical dependence, all actions that were readily reversed by β-FNA. As with β-FNA, naloxazone and naloxonazine have both reversible and irreversible actions, with the irreversibly blocked sites defined as mu1, whereas the reversibly blocked ones are designated as mu2. Given short term, naloxonazine and naloxazone antagonize all mu actions and compete with all mu receptors. To achieve selectivity in vivo, these compounds, like β-FNA, are given 24 hours prior to testing to permit the elimination of free drug and the reversible blockade of mu2 receptors. Given immediately prior to testing, the compounds lose their selectivity. The naloxonazine-sensitive receptors have an unusual binding profile as well as pharmacology, which is discussed below (section V.A).
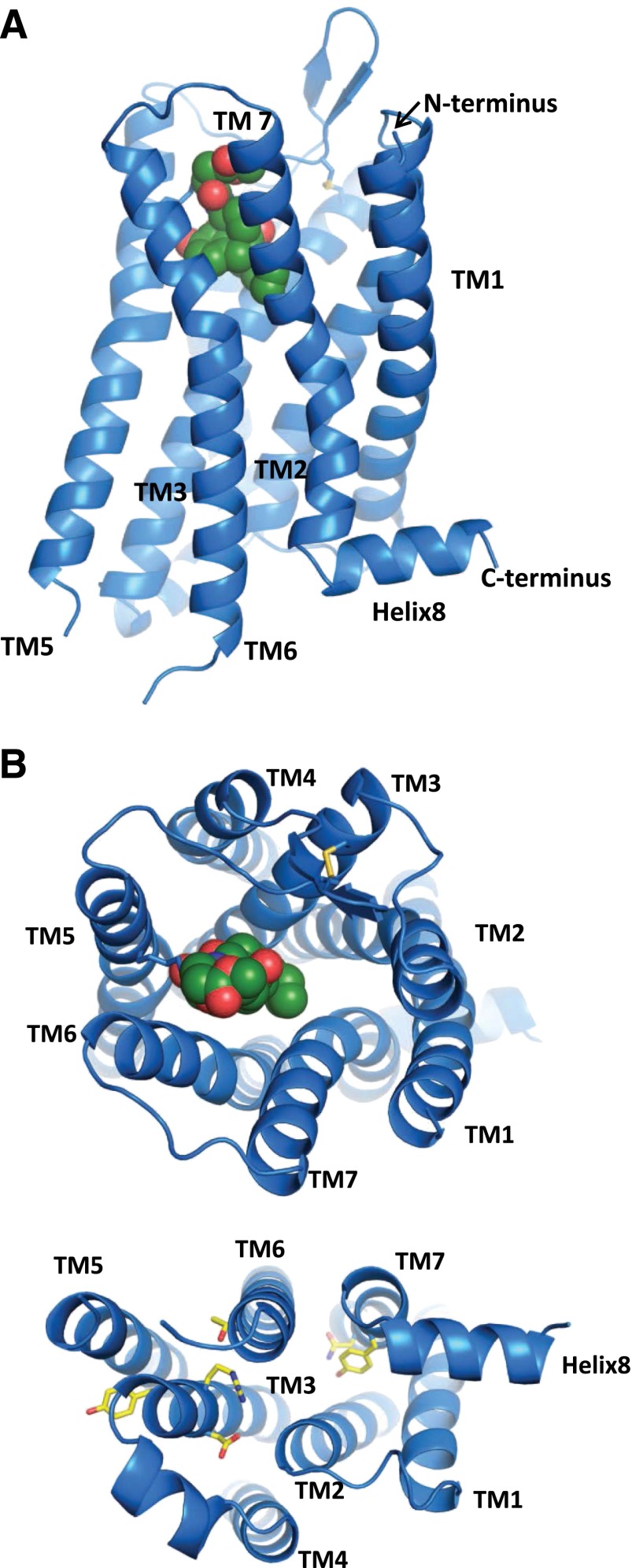
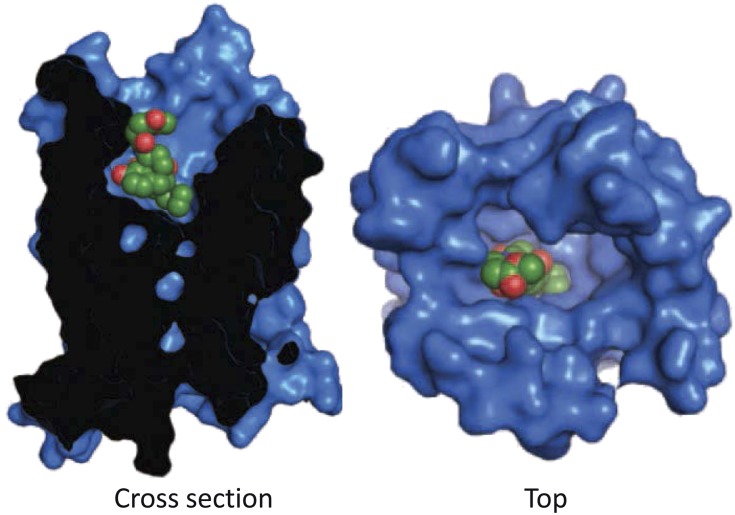
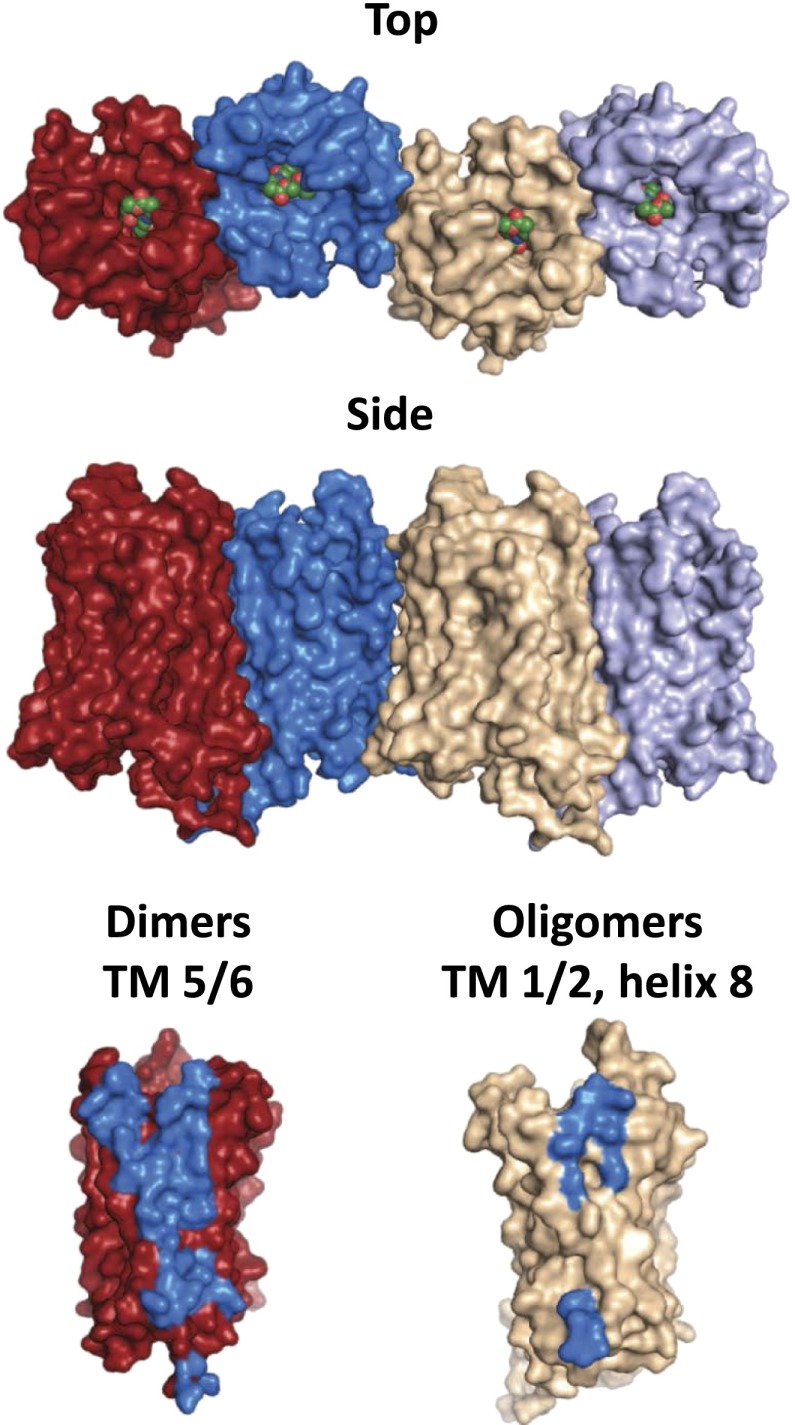
Naloxonazine is a dimer, the equivalent of two molecules of naloxone with a hydrazine bridge. Naltrexonazine and oxymorphonazine, the naltrexone and oxymorphone analogs of naloxonazine, also display intriguing pharmacologies, as do analogs containing dihydrazide bridges in place of the hydrazine (Hahn et al., 1982). As with the antagonists, the agonist oxymorphazone has a prolonged activity, producing analgesia lasting over 24 hours that correlates with an irreversible blockade of the receptor. Yet, the analgesic response is readily reversed by naloxone (Galetta et al., 1982). Inasmuch as the blockade of the receptor by oxymorphazone is irreversible and is not lost when naloxone is given, naloxone cannot act by simply displacing the oxymorphazone, implying the presence of interacting sites in which naloxone binds to one site and reverses activity of the other despite the continued occupation by the agonist. This suggestion of receptor dimerization came long before the cloning of the receptors and 30 years before it was confirmed by the crystal structure studies of the mu receptor (Manglik et al., 2012).
Portoghese suggested that the long duration of action of his bifunctional compounds reflect the simultaneous binding to two receptor pockets (Erez et al., 1982). However, this is not the case for naloxonazine and its analogs. The irreversible inhibition also is seen with asymmetrical azines in which one of the opiates does not bind to the receptors. For example, methylation of the 3-position eliminates the affinity of naloxone for mu receptors. Yet, the asymmetric azine composed of naloxone and 3-methylnaloxone still elicits the same long-lasting activity seen with naloxonazine. Similar results are seen with the hydrazide series as well. Thus, these agents do not simultaneously label two binding pockets, and their mechanism(s) of prolonged activity differ from that proposed by Portoghese for his bivalent ligands (Erez et al., 1982; Portoghese et al., 1986; Bolognesi et al., 1996).
Over the years, a vast array of additional opioids has been synthesized, with widely varying structures and selectivities. However, a full overview of all opiates and their structure-activity relationships is beyond this review.
D. Endogenous Mu Alkaloids
There is strong evidence for the presence of endogenous morphine and codeine within the brain. First proposed by Gintzler and Spector using immunologic techniques (Gintzler et al., 1978; Shorr et al., 1978; Donnerer et al., 1986, 1987; Grobe et al., 2010), these observations are supported by several other laboratories (Hazum et al., 1981b; Killian et al., 1981; Goldstein et al., 1985; Weitz et al., 1986). The question arose whether the endogenous morphine is synthesized in the brain or whether it is absorbed from foodstuffs, particularly with its isolation from milk and plants other than the poppy (Hazum et al., 1981b). Although ingestion remains one potential mechanism for accumulation of endogenous morphine, there is evidence it can be synthesized de novo by mammals (Goldstein et al., 1985; Weitz et al., 1987; Poeaknapo et al., 2004; Boettcher et al., 2005; Grobe et al., 2010) and even in isolated cells (Poeaknapo et al., 2004; Boettcher et al., 2005; Han et al., 2010). In addition to morphine, other metabolites have been isolated from brain, including morphine-6β-glucuronide and morphine-6-sulfate (Fig. 1A). These two analogs are notable because of their far greater analgesic potency than morphine (Pasternak et al., 1987; Paul et al., 1989b; Zuckerman et al., 1999).
III. Mu Opiate Pharmacology
A. Historical Overview
The opiates have been used for millennia, but the modern history of opiate analgesics starts with the isolation of morphine in 1805 (Serturner, 1805). The full addictive potential of morphine was not fully realized until the availability of both the hypodermic needle and pure morphine enabled parenteral administration of very high doses. The realization of the profound addictive properties of morphine initiated the quest for nonaddictive alternatives. In the early 20th century the United States, the National Academy of Sciences/National Research Council Committee on Drug Addiction and Narcotics established a program to identify nonaddictive morphine substitutes (Isbell, 1977; Lasagna, 1984). Initially funded by the Rockefeller Foundation and the City of New York Bureau of Social Hygiene, the effort soon received support from the pharmaceutical industry. This effort had a number of important aspects. As noted by Lasagna (1984), “First, it provided an identifiable motivating force and research support umbrella. Second, it was able not only to achieve a ‘marriage’ between investigators (both in industry and academia) and drugs deserving investigation, but to arrange for the addiction liability testing of promising drugs both at the primate facility at the University of Michigan and at the clinical testing facility at the Addiction Research Center in Lexington, Kentucky. Third, the program supported methodologic research, which was badly needed at that time.” The National Research Council Committee evolved to become the Committee on Problems of Drug Dependence.
Although efforts were made to develop nonaddictive drugs, few inroads were made. The partial agonist/antagonists came closest to minimizing the addictive potential but suffered from limited analgesic activity and/or the precipitation of withdrawal in patients dependent upon other opiates. Many, including nalorphine, also were associated with severe psychotomimetic side effects. However, the research yielded much information, particularly the development by Houde and Wallenstein of methodology and statistical analysis permitting the formalized establishment of the relative potency of the different opiate drugs (Wallenstein, 1984). The availability of these tables giving relative doses of the various opiates provided valuable information for physicians when prescribing the drugs. Indeed, it is still common to see physicians carrying cards with these drug ratios.
With the availability of an array of morphine-like (i.e., mu) opiates, clinicians made a number of important observations. First, it was not uncommon for patients to respond differently to each opiate. This was not a simple matter of sensitivity to opioids in general. Some patients would respond well to one mu opiate and not another, whereas the reverse might be seen in a different patient. Second, physicians found that although the equianalgesic dosing tables were helpful in naive patients, the relative potencies of the drugs in tolerant patients differed markedly. All the mu opiates show cross-tolerance, but this is often not complete (section III.G). This is best illustrated when switching a morphine-tolerant patient from morphine to methadone, when the equianalgesic methadone dose determined from the tables needs to be reduced by 50–75% (Cherny et al., 2001). It is clear that incomplete cross-tolerance among mu opiates can be quite profound and has major clinical implications, including being the basis for opioid rotation. Third, the side effect profile of different opiates within a single patient can differ. It is not unusual for a patient incapable of tolerating morphine because of nausea and/or vomiting to take methadone without a problem. Together, these observations raised major questions regarding the receptor mechanism(s) of action of these drugs. They also illustrate the value of clinical insights and experience in focusing preclinical opiate research.
For those interested in the early pharmacology of opioids, Krueger et al. (1941) prepared an extensive and comprehensive two volume overview of opiate action prior to World War II. What is impressive about this treatise is how much was known about opioid pharmacology before the current “molecular era.” Indeed, it is humbling to see how much we forgot and what we have “rediscovered.”
Within the opioid field, addiction and analgesia are intimately intertwined. Many approaches have been used to explore addiction, but one of the major sources of our clinical understanding of these drugs came from the Addiction Research Center, which was originally the Research Division of the U.S. Narcotics Farm, a 1200-bed facility established in 1935 in Lexington, Kentucky, to treat addicts. This facility explored the actions of opioids in established addicts, obtaining detailed descriptions and comparisons of the opioids. It was unique in its ability to provide clinical insights into the pharmacological actions and perceptions of a range of drugs. The Lexington facility was moved to Baltimore as part of the intramural program of the National Institute on Drug Abuse in 1979. The history of the Center has been well reviewed (Campbell, 2006, 2010; Campbell et al., 2008).
B. Bioassays
Although the hallmark of opiate pharmacology is its in vivo pharmacology, bioassays have played a major role in defining these drugs (Cox, 2011). The guinea pig ileum contraction assay was first described over 50 years ago (Schaumann, 1955; Kosterlitz and Robinson, 1957a; Paton, 1957). This assay involves measuring the strength of a contraction of the muscle, induced by the electrically induced release of acetylcholine. Opiates such as morphine inhibit this release (Schaumann, 1957; Cox and Weinstock, 1966), leading to a decreased contraction. These assays provide a means of screening a vast array of drugs, both agonists and antagonists, quickly and inexpensively, requiring very small amounts of material, and were the standard for the evaluation of novel agents before the availability of opioid binding assays. Preparations incubated with opioids for extended times provide a model of tolerance, with the subsequent removal of the opiate or the addition of an antagonist yielding a model of withdrawal. The identification of the opioid binding sites quickly replaced the use of bioassays as screening, with the “sodium shift” (section IV.D.1) even providing an estimate of the agonist/antagonist nature of the compound (Pert et al., 1973; Snyder et al., 1975), but bioassays still were quite useful. In the mid-1970s several groups attempted to identify endogenous opioid materials from the brain. Kosterlitz used the mouse vas deferens bioassay to identify the material and monitor its purification. They termed the material “enkephalin” to indicate it was isolated from the brain and subsequently labeled the enkephalin receptors “delta,” based upon the “d” in the vas deferens assay that was used in its isolation.
C. Analgesia
The actions of morphine have been well studied, and analgesia remains the major focus of mu opioid action. Studies of morphine analgesia even go back to Darwin, who tried to assess whether morphine was active in plants “but with no certain result” (Darwin, 1875; Krueger et al., 1941). Early studies also examined analgesia in a host of species ranging from the frog to mammals (Krueger et al., 1941). It is extraordinary how much was known before the “modern era.” The Straub tail is still considered by many to be a hallmark of mu opiate action, but most investigators are not aware that it was first reported over a century ago (Straub, 1911). Early investigators also emphasized differences in responses among species and even among strains of mice—supporting a physiologic basis of the clinical observations. Many of these very early studies, which have not been digitized and are not listed in many search engines, are difficult to retrieve, particularly as libraries have scaled down their collections, essentially losing a vast repository of opioid pharmacology.
The study of analgesia requires the presence of pain. A large number of assays have been developed and reviewed (Le Bars et al., 2001). Detailed descriptions and analysis of the various assays is beyond the scope of this review. Several factors must be considered whenever analyzing opioid analgesia. The first is the intensity of the nociceptive stimulus. Clinically, lower doses of drug are required to relieve mild pain than severe pain. Drugs with ceiling effects may be effective for mild pain and not for more severe pain. The second is more difficult to assess and involves the quality/type/nature of the pain. Pain encompasses many different sensations. Three major types of pain have been proposed: somatic, visceral, and neuropathic. However, they rarely exist alone, and most clinical situations are combinations of them. The various types of pain differ in their sensitivities to therapeutic drugs and are described by terms such as sharp, dull, aching, burning, throbbing, shooting, and cramping. Finally, in the clinical situation, the meaning of the pain is crucial. The perception of pain does not always correlate with nociceptive intensity. Context is important. In Beecher’s (1946) reports with soldiers during World War II, he noted that wounded soldiers requested less morphine than patients undergoing elective surgery back in the United States despite the greater severity of the soldiers’ wounds. Anecdotal evidence for the role of context in the perception of pain abounds, as shown by athletes being unaware of a painful injury until the game is over. The brain “filters” pain depending upon its context. In addition to its minimization in stressful situations or with distraction, the perception of pain may be enhanced in other situations. Clinically, this is can be seen when a patient equates pain with a serious medical problem, such as progression of cancer. The importance of context led many investigators to believe that experimental pain models in humans, or in animals, are not accurate models of the clinical situation (Raymond Houde, personal communication). Although the animal models are valuable and provide important information and insights, they are limited in that they do not take into consideration the subtleties of human pain perception.
Morphine is a potent analgesic given systemically. However, the doses needed vary enormously among species. For example, most mice and rats show an ED50 for morphine of approximately 5 mg/kg s.c. in thermal assays such as tail flick and hot plate, whereas a total morphine dose of 10 mg is adequate in most people (corresponding to ∼0.15 mg/kg s.c.). Likewise, the analgesic/respiratory depression therapeutic index is far smaller in humans than in rodents. Even within a species, different strains can vary markedly, as can sex (Mogil, 1999; Mogil et al., 2000a; Mogil and Bailey, 2010). These differences in sensitivity recapitulate clinical observations.
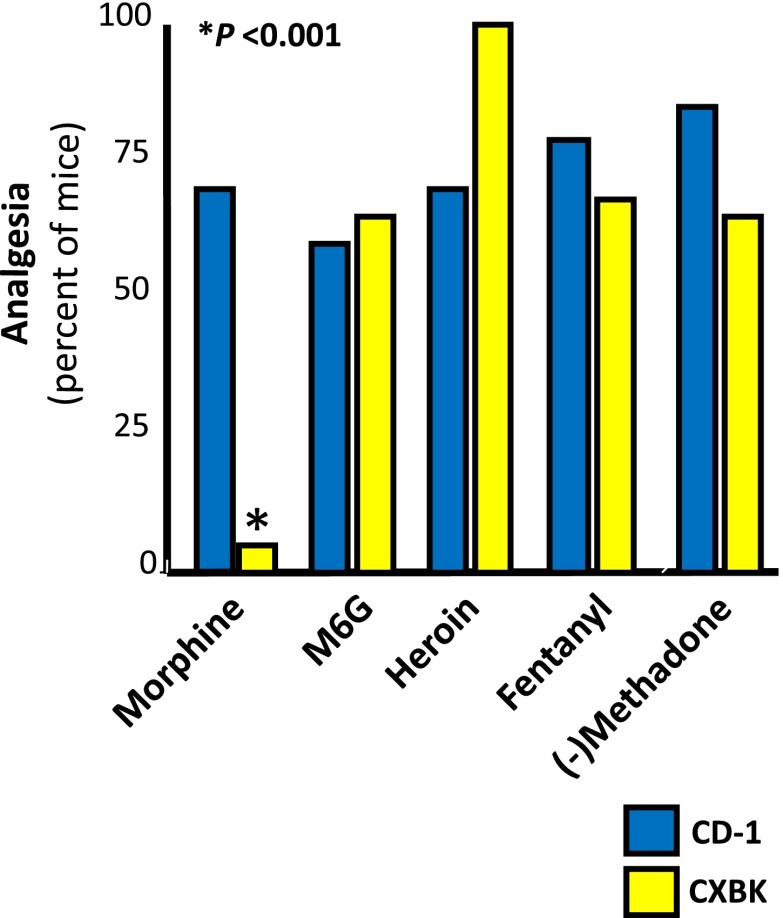
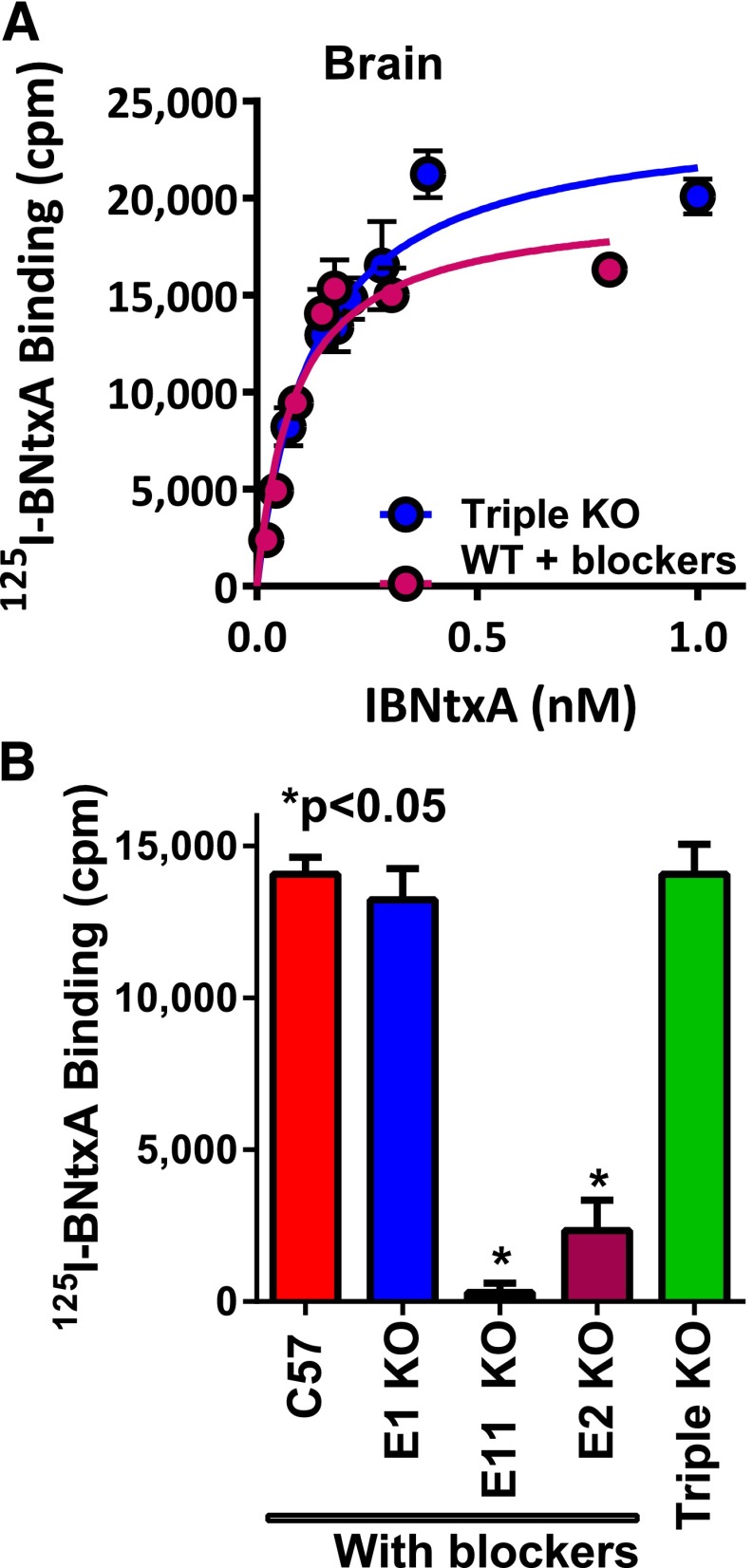
One of the most dramatic examples of strain differences is the CXBK mouse, which is relatively insensitive to morphine (Baron et al., 1975; Reith et al., 1981; Kest et al., 1998a). However, CXBK mice retain their sensitivity toward many other mu opioids (Fig. 7), including methadone, heroin, fentanyl, and morphine-6β-glucuronide (Rossi et al., 1996; Chang et al., 1998; Pasternak, 2004, 2010). Additional studies have explored the roles of genetic backgrounds and sex issues in more detail (Kepler et al., 1991; Gear et al., 1999; Kest et al., 1999; Lariviere et al., 2002; Wilson et al., 2003a,b; Gintzler et al., 2008; Chakrabarti et al., 2010; Mogil and Bailey, 2010). Thus, these preclinical studies suggest a genetic basis for differing sensitivities of individuals to each opiate and to different relative potencies of mu opiates from patient to patient.
1. Sites of Action.
The site of action of opiate action has been explored since the early 1900s (reviewed in Krueger et al., 1941). Regions of the central nervous system sensitive to morphine were initially defined by administering morphine into discrete areas using microinjection techniques (Pert and Yaksh, 1974). The most prominent sites in the brain stem include the periaqueductal gray, n. raphe magnus, n. reticularis gigantocellularis, and the locus coeruleus, whereas at the spinal level the dorsal horn is most important. Much effort has been devoted to defining the circuits and interactions among these regions using both pharmacological and electrophysicological approaches (Bodnar et al., 1991; Morgan et al., 1992; Kiefel et al., 1993; Rossi et al., 1993, 1994b; Gutstein et al., 1998; Mitchell et al., 1998; Fields and Martin, 2001; Julius and Basbaum, 2001; Basbaum and Julius, 2006; Basbaum et al., 2009). Evidence also supports a peripheral opioid component of pain relief (Stein and Yassouridis, 1997; Stein et al., 2003). Opioid receptors are present on dorsal root ganglion cells and on peripheral nerves. Administered topically in a model that lacks any appreciable systemic absorption, morphine and other opioids are potent analgesics, indicating a peripheral effect (Kolesnikov et al., 1996a,b, 2000, 2004; Kolesnikov and Pasternak, 1999).
2. Synergy.
Synergy is an important component of mu analgesia. There are many aspects of synergy that need to be considered when evaluating both clinical and preclinical studies. First is the interaction between opioids and other classes of drugs. Opioids have long been marketed as combination products, initially with acetaminophen and aspirin and then with NSAIDs such as ibuprofen. Clinicians appreciate the advantages of both classes of drugs together, and there is preclinical evidence suggesting the possibility of synergy between these agents (Kolesnikov et al., 2003b; Zelcer et al., 2005). Opiates also are used extensively with local anesthetics, particularly when administered epidurally or intradurally (Nordberg, 1984; Payne, 1987; Arner et al., 1988), an approach validated in animal models (Durant and Yaksh, 1986). Synergy also exists between opiates and local anesthetics when they are administered topically (Kolesnikov et al., 2000, 2003a). Synergy is seen with opioids and clonidine. Reports of interactions between mu and delta drugs go back to the 1980s, with a more recent report demonstrating synergy between mu opioids (Porreca et al., 1987; Sutters et al., 1990; Horan et al., 1992; Ossipov et al., 1997; He and Lee, 1998; Fairbanks and Wilcox, 1999; Grabow et al., 1999; Kovelowski et al., 1999; Bolan et al., 2002). In the mu synergy study, the combination of morphine and methadone elicits the most robust synergy, whereas most other combinations are additive.
Regional synergy is important in mu opioid actions. Yeung and Rudy (1980) first demonstrated regional interactions between spinal and supraspinal morphine in rats. In this classic study, they reported profound synergy when morphine was given both spinally and supraspinally simultaneously (Table 3). It has been suggested that morphine tolerance may reflect, in part, the loss of this synergy (Roerig et al., 1984). However, regional interactions are not limited to spinal/supraspinal sites. A number of studies have documented other synergistic regional interactions (Rossi et al., 1993, 1994b; Kolesnikov et al., 1996b; Pavlovic and Bodnar, 1998; Bodnar, 2000). Within the brain stem, microinjection studies reveal multiplicative interactions among several brain stem nuclei, including the periaqueductal gray, rostral ventral medulla, locus coeruleus, and amygdala.
TABLE 3.
Morphine spinal/supraspinal synergy in rats
Morphine analgesia was assessed with either administration supraspinally, spinally, or simultaneously in both regions in a 1:1 ratio. Adapted from Yeung and Rudy (1980).
| Morphine ED50 |
||
|---|---|---|
| Supraspinal | Spinal | |
| μg | ||
| Each site alone | 10 | 4.2 |
| Both sites simultaneously | 0.35 | 0.35 |
Clinically, synergy probably plays a major role in the efficacy of epidural opioid analgesia. Administration of an opioid into the epidural space leads to high concentrations of drug at the spinal level because of its diffusion into the cerebrospinal fluid within the thecal sac. However, the epidural space is highly vascular and Batson’s venous plexus provides an efficient means for absorbing the drug directly into the blood. Thus, in this route of administration, the patient gains the advantages of both local (i.e., spinal) and systemic drug, a combination that produces profound synergy in a mouse model (Kolesnikov et al., 1996b). In a mouse model, intrathecal administration of a morphine dose that gives less than a 20% analgesic response alone shifts the systemic analgesic ED50 value over 10-fold to the left while a lower dose shifts the curve approximately 6-fold (Table 4). This intrathecal/systemic interaction may help explain the prolonged, potent analgesic responses seen clinically with epidural morphine analgesia.
TABLE 4.
Intrathecal/systemic morphine synergy in mice
Morphine intrathecal ED50 is 305 ng (153, 501). Morphine given intrathecally at a dose less than 10% of its intrathecal ED50 shifted the systemic analgesic dose-response curve over 6-fold. From Kolesnikov et al. (1996b).
| Systemic Morphine ED50 | Systemic Shift | |
|---|---|---|
| mg/kg s.c. | -fold | |
| Systemic alone | 3.1 (1.6, 4.4) | |
| + 25 ng of Intrathecal morphine | 0.5 (0.4, 0.8) | >6 |
Synergy between peripheral and central sites also is important (Kolesnikov et al., 1996b; King et al., 2001a) because systemic drugs activate both at the same time. Peripheral/central synergy even complicates interpretation of drugs administered intracerebroventricularly. Morphine and most opiates are substrates for transporters important in maintaining the blood-brain barrier, such as P-glycoprotein and multidrug resistant protein. These transporters secrete opiates that diffuse from the blood into brain back into the systemic circulation, thereby providing a “barrier” to their entry into the brain. It is not as widely appreciated that this same system secretes opioids given intracerebroventricularly into the systemic circulation where they can contribute to the overall pharmacological actions (King et al., 2001a) as well as other agents such as tumor necrosis factor (Bodnar et al., 1989).
D. Opioid/Sigma1 Interactions
Although originally proposed by Martin, sigma receptors now refer to a class of “receptor” without affinity for active opiates. The sigma1 receptors have been cloned in a number of species (Hanner et al., 1996; Kekuda et al., 1996; Seth et al., 1997; Pan et al., 1998a; Mei and Pasternak, 2001). Structurally, the proteins are highly conserved among species and do not fall within any known receptor class, with two transmembrane domains and intracellular C and N termini. Furthermore, there is no known transduction mechanism for the receptors. They do not couple to G proteins, and there is no indication of any kinase activity. However, they physically associate with a range of G protein-coupled receptors, including the opioid receptors, as well as potassium and NMDA calcium channels (Yamamoto et al., 1995; Wilke et al., 1999; Lupardus et al., 2000; Aydar et al., 2002; Kim et al., 2010). Although active opioids do not label sigma receptors, many of the inactive (+)benzomorphans, such as (+)pentazocine will potently label the site.
What makes sigma1 receptors relevant is their ability to modulate opioid action. This can be seen biochemically, where sigma antagonists enhance the intrinsic activity of opioid drugs (Kim et al., 2010). This is illustrated by the ability of sigma1 antagonists to shift leftward the opioid dose-response curves for stimulation of [35S]GTPγS binding without altering either the maximal stimulation by the opioid or the binding affinity of the opioid for its receptor. In other words, in the presence of sigma1 antagonists opioids attain their maximal stimulation of [35S]GTPγS binding at lower receptor occupancy—i.e., increasing intrinsic activity.
Sigma1/opioid interactions also have been documented in vivo with opioid analgesia. Administration of a sigma1 antagonist or downregulation of the sigma1 receptor with antisense potentiates morphine and kappa analgesia (Chien and Pasternak, 1994, 1995b; Pasternak, 1994; King et al., 1997a,b; Mei and Pasternak, 2007; Cobos et al., 2008; Entrena et al., 2009a,b; Kim et al., 2010; Sanchez-Fernandez et al., 2013). Sigma1 receptors also play a role in differences in sensitivity among several strains of mice. For example, the sigma1 antagonist haloperidol enhances the potency of the kappa1 analgesic U50,488H and eliminates the difference in sensitivity between CD-1 and BALB-c mice (Chien and Pasternak, 1994). These haloperidol effects are not mediated thorough dopamine receptors because the D2-selective drug sulpiride is ineffective and the haloperidol effect persists in a D2 knockout mouse (King et al., 2001b).
E. Other Actions
Opiates have a variety of additional actions, including depression of respiration; inhibition of gastrointestinal transit, nausea, and vomiting; and a host of endocrinological effects. Full descriptions of these are beyond the scope of this review, and readers are referred to a number of reviews (Cicero, 1980; Reisine and Pasternak, 1996; Pattinson, 2008; Diego et al., 2009, 2011; Cox, 2011; Elliott et al., 2011; Wald, 2012). However, it is worthwhile to highlight several aspects of these additional functions.
Constipation and respiratory depression remain among the most troublesome of all opioid side effects. Respiratory depression is always a clinical concern, although it is rarely a problem in the outpatient setting in the absence of underlying pulmonary disease. On the other hand, constipation is almost universal, requiring laxatives in a high percentage of patients on opioids for more than a couple of days. This problem has now been addressed pharmacologically with two of the following U.S. Food and Drug Administration-approved peripherally acting antagonists: methylnaltrexone and alvimopam. Methylnaltrexone achieves its peripheral activity by quaternizing the nitrogen, limiting its ability to traverse the blood-brain barrier. Alvimopam, on the other hand, is administered orally and its actions are restricted to the gastrointestinal tract because, in large part, of its very poor water solubility and limited systemic absorption.
Pruritus, or itching, is a common problem with morphine and many of the other analgesics, especially with epidural administration. Although many had assumed that that this was attributed to histamine release mediated through the same mu receptors as analgesia, several reports suggest that this may not entirely explain the symptom. According to Andoh et al. (2008), intracisternal morphine and morphine-6β-glucuronide are both analgesic, but only morphine produces pruritus. Furthermore, the facial scratching seen with morphine is insensitive to naloxonazine while analgesia is blocked, leading them to conclude that morphine analgesia and pruritus are mediated through different receptor subtypes. More recently, a study has suggested that pruritus is produced through a heterodimer of the MOR-1D splice variant with the gastrin-releasing peptide receptor (Liu et al., 2011).
Although the endocrine effects of opioids have been well described, there has been little attention to them clinically. Morphine impacts prolactin and growth hormone release (Spiegel et al., 1982a) as well as altering a wide range of other hormones (Cicero, 1980). Decreased testosterone levels have garnered most of the attention over the past few years. The loss of muscle tone, sexual dysfunction, and psychological effects can be a significant problem with chronic administration of opiates, particularly in the setting of opioid maintenance programs.
F. Tolerance/Dependence/Withdrawal
Chronic administration of opioids leads to a reduction in the response—tolerance. Alternatively, tolerance can be defined as the need to increase the dose of a drug to maintain a response. Clinically, this often is seen over a period of days to weeks, but in animal models tolerance can be seen in rats in hours (Cox et al., 1968). A wide range of different and unrelated mechanisms impact tolerance, indicating that tolerance involves the convergence of many pathways to a common behavioral response. Interference with any one is sufficient to impact tolerance, much like a tug of war with many different people pulling on the same rope. Each is contributing to the final effort and the loss of any one of them leads to a similar effect.
It is also important to recognize that there is a wide range of tolerance. Many paradigms used in preclinical models are associated with modest 2-fold changes in tolerance. In the clinic, patients have developed far more extensive tolerance. Indeed, in our experience at Memorial Sloan-Kettering, we treated a patient who required and tolerated intravenous morphine doses ~1000-fold higher than those used in naive patients. Despite the extraordinary dose, the patient remained awake and communicative (Richard Payne, personal communication). It may well be that the mechanisms responsible for producing tolerance may be dependent upon the dose/duration of drug treatment. If so, treatments active against modest levels of tolerance may not be as effective as the degree of tolerance increases, which may need to be considered in clinical trials.
Although tolerance develops to all mu-opioid actions, the rate of tolerance development may vary. For example, tolerance develops slowly, if at all, to opioid-induced miosis and its characteristic pinpoint pupils. Clinically, many feel that tolerance to the respiratory depressant and constipating actions develop more slowly than to analgesia, leading to a decreased therapeutic index with chronic administration and increasing analgesic tolerance and thereby increasing problems with side effects. When side effects become intolerable, clinicians often use “Opioid Rotation,” in which the patient is switched or rotated to a different mu opioid, often regaining analgesic sensitivity because of incomplete cross-tolerance (Cherny et al., 2001; Pasternak, 2001; Inturrisi, 2002; Chou et al., 2009). Thus, understanding tolerance and finding ways to avoid it would be advantageous.
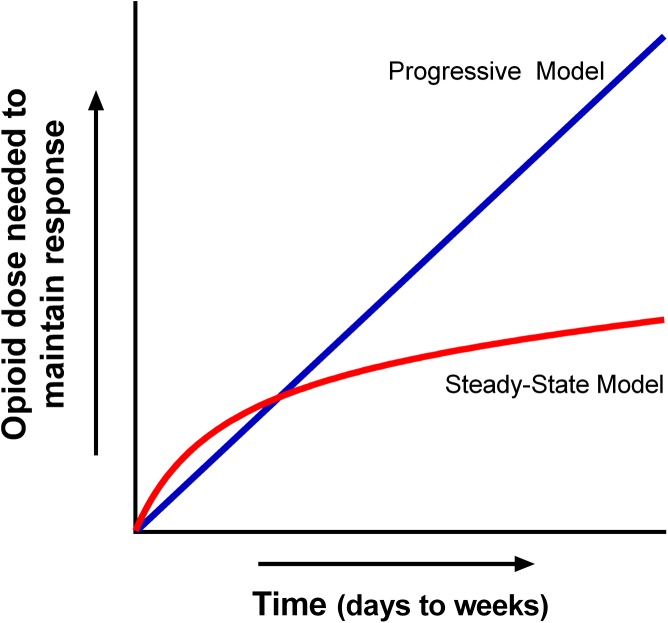
A basic question clinically is whether tolerance is a continuously progressive response to opiate use or whether tolerance eventually achieves a “steady state” with a constant drug dose/regimen (Fig. 8) (Pasternak, 2007). Clinical experience suggests that a steady state does develop (Foley, 1993). Once a cancer patient has been titrated to an effective dose, it is not unusual to manage them with a stable dose of opiate for prolonged periods of time (weeks to months or longer). Indeed, in cancer patients, the need for dose escalation in these stabilized patients is usually an indication of an increase in the severity of the pain attributed to the progression of disease rather than increased tolerance. However, stable dosing does not mean that the patient is not tolerant. Because the doses of analgesics in these patients are often far greater than those needed in naive patients, they are likely tolerant, but their “tolerance” can be stable for extended periods of time.
Fig. 8.
Two hypothetical models of opioid tolerance. In the Progressive Model tolerance continues to increase over the full duration of the drug administration. In the Steady-state Model, tolerance increases, but reaches a steady state that can be maintained over long periods of time. Adapted from Pasternak (2007).
Opioid tolerance encompasses a diverse range of mechanisms, making it difficult to integrate them into a unified theory. The ability to modulate morphine tolerance was first reported with the cholecystokinin antagonist proglumide based upon the observation that cholecystokinin is a functional “antagonist” of opiate analgesia and the suggestion that the cholecystokinin antagonist proglumide reversed morphine tolerance (Faris et al., 1983; Watkins et al., 1984). Although proglumide potentiates morphine analgesia in tolerant animals, it has similar actions in naive animals (Bodnar et al., 1990). Thus, simply enhancing morphine analgesia in tolerant animals is not sufficient to imply a role in tolerance. To assess the role of a drug in tolerance, it is necessary to show that it lowers the shift in the opioid dose-response curve between naive and chronically treated subjects. Testing the drug only in tolerant animals cannot differentiate between simple potentiation and reversal/prevention of tolerance. A number of systems can modulate opioid tolerance.
1. Adenyl Cyclase.
The first suggestion of a biochemical basis for opioid tolerance was put forward by Way and then Cox in the 1960s when they showed that morphine tolerance is dependent upon protein synthesis (Way et al., 1968; Cox and Osman, 1969). Collier extended this concept of a biochemical mechanism for tolerance development to “hypertrophy of the cyclic AMP system” (Collier and Roy, 1974; Collier, 1980). Opiates, similar to many drugs acting through Gi and/or Go, inhibit the stimulation of adenyl cylcase. Collier showed that growing and then testing neuroblastoma NG108 cells in the presence of morphine had little effect upon basal cAMP production or on its stimulation by either prostaglandin E1 or adenosine. However, addition of naloxone increased the stimulation of cAMP levels 3- to 5-fold in the morphine-exposed cells but not in control cells. On the basis of these observations, he proposed that tolerance involves a compensatory increase in the adenyl cyclase enzyme so that in the continued presence of morphine cAMP levels return to relatively “normal” levels despite the continued inhibition by the opiate. Removing the opioid inhibition by the administration of the antagonist naloxone then uncovers this compensatory upregulation of adenyl cyclase activity. This model is consistent with the dependence of tolerance on protein synthesis (Way et al., 1968; Cox and Osman, 1969). Although not appreciated at the time, it should be noted that the NG108 cell line used expresses delta receptors but not mu receptors. However, similar results have been observed in cells containing mu-opioid receptors.
2. N-Methyl-d-Aspartate Antagonists and Nitric-Oxide Synthase Inhibitors.
The NMDA antagonist MK-801 [(5S,10R)-(+)-5-methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine maleate or dizocilpine] was the first compound that convincingly prevented/reversed opioid tolerance (Trujillo and Akil, 1991; Ben-Eliyahu et al., 1992), followed by similar reports for a range of NMDA antagonists (Gutstein and Trujillo, 1993; Kolesnikov et al., 1993a, 1994; Tiseo and Inturrisi, 1993; Elliott et al., 1994, 1995; Trujillo and Akil, 1994). Nitric oxide is closely linked to NMDA receptor functions in a variety of systems. Thus, it is not surprising that a series of nitric-oxide synthase (NOS) inhibitors also prevent/reverse morphine tolerance (Kolesnikov et al., 1992, 1993b; Babey et al., 1994), implicating the involvement of the full NMDA-nitric oxide cascade.
Further evidence for the role of NMDA receptors comes from the 129/SvEv mouse (Kolesnikov et al., 1998), results that were subsequently confirmed (Crain and Shen, 2000a). Unlike traditional mice, 129/SvEv mice do not develop tolerance to morphine because of a defect at the level of the NMDA receptor that can be by bypassed by stimulating nitric-oxide synthase directly (Kolesnikov et al., 1998).
Crain and Shen have long proposed that tolerance may involve the enhanced function of an opioid excitatory system that is blocked by ultralow doses of antagonists, perhaps explaining the ability of ultralow antagonist doses to potentiate morphine analgesia as well as the “hyperalgesia” seen in experimental models (Crain et al., 1988; Crain and Shen, 1990; Shen and Crain, 1992, 1994). This excitatory opioid system is reportedly inactive in the SvEv mice, consistent with the lack of tolerance development and consistent with the earlier work localizing the problem to the level of the NMDA receptor. Attempts to use NMDA antagonists clinically have not been successful, possibly because of the inability to escalate doses sufficiently due to the psychotomimetic side effects associated with their use.
3. Enkephalin Systems.
The enkephalin system has been implicated in morphine tolerance, starting with the observation by Takemori and colleagues that morphine tolerance is prevented by delta-receptor antagonists (Abdelhamid et al., 1991), followed by the demonstration that an antisense targeting the delta-opioid receptor has a similar effect (Kest et al., 1996). The absence of morphine tolerance in both a DOR-1 and in an enkephalin knockout mouse further validated the role of the delta-receptor/enkephalin system (Zhu et al., 1999; Nitsche et al., 2002).
4. P-Glycoprotein.
Evidence also suggests a distributional aspect of morphine tolerance (Aquilante et al., 2000; Lötsch et al., 2000; Zong and Pollack, 2000; King et al., 2001a). Morphine is a substrate for the transporter P-glycoprotein (Pgp) (Callaghan and Riordan, 1993), an important component of the blood-brain barrier (Schinkel et al., 1994). Its disruption increases morphine’s analgesic potency. Long-term morphine administration upregulates Pgp expression in the brain, raising the possibility that these increased Pgp levels may contribute to morphine tolerance by lowering its penetration into the brain. This was confirmed with studies showing that removing Pgp in a knockout model or downregulating it through antisense approaches prevents the development of morphine tolerance (Lötsch et al., 2000; Zong and Pollack, 2000; King et al., 2001a). Similar findings have been reported with a second ATP transporter, multidrug-resistant protein (Su and Pasternak, 2013).
The importance of distributional factors is consistent with observations that mice tolerant to systemic morphine show no change in morphine’s analgesic potency when the drug is given supraspinally or spinally, thereby bypassing the blood-brain barrier (Roerig et al., 1984). These findings were replicated, showing no difference in the spinal and supraspinal morphine ED50 values despite over a 2-fold shift in the systemic ED50 value (Kolesnikov et al., 1996b). The later study went one step further and shows a dramatic 19-fold shift in the analgesic activity of peripheral morphine in these same animals, raising the possibility of an important peripheral component of morphine analgesia and tolerance (King et al., 2001a; Su and Pasternak, 2013).
5. Trafficking.
Similar to other membrane proteins and G protein-coupled receptors, mu-opioid receptors undergo extensive trafficking (Keith et al., 1998; Whistler et al., 1999; Von Zastrow et al., 2003; Von Zastrow, 2011). The intriguing aspect of these studies is the observation that in some paradigms morphine does not internalize MOR-1, whereas other mu agonists such as DAMGO do (Keith et al., 1998). It has been suggested that failure to internalize MOR-1 by morphine might be responsible for the development of tolerance (Whistler et al., 1999). The trafficking aspects of these studies are quite intriguing and offer sophisticated insights into the cell biology of opioid receptors. However, several issues argue against internalization being a prime factor in morphine tolerance in vivo. Inherent in this concept is that drugs that induce internalization, such as methadone, would not be expected to produce tolerance. However, methadone tolerance is a widely recognized entity. Second, several lines of evidence indicate that these trafficking patterns do not hold for all the MOR-1 splice variants. MOR-1C internalizes in vivo after morphine or DAMGO administration, whereas MOR-1 internalized only with DAMGO (Abbadie and Pasternak, 2001). In other studies, Von Zastrow and colleagues found marked differences in trafficking among a series of MOR-1 splice variants, with MOR-1B, MOR-1D, and MOR-1E showing far more degradation than MOR-1 itself and emphasizing that alternative splicing of the C terminus of the receptor affects trafficking (Tanowitz et al., 2008).
It also has been suggested that mu/delta heterodimers play a role in morphine tolerance (Gupta et al., 2010; He et al., 2011). He and colleagues proposed that activation of delta receptors in a mu/delta heterodimer leads to a diminished response due to degradation of the mu receptor. Although this hypothesis is intriguing and may help explain the earlier observations linking enkephalin systems to morphine tolerance, it has not yet been fully validated.
A particularly intriguing aspect of their study was their result with an engineered protein, MORTM1-TAT. This construct corresponds to the first transmembrane domain of MOR-1 coupled to TAT (transactivator of transcription), which enables its insertion into the membrane. They report that the construct disrupts the mu/delta dimer, implicating TM1 in the association of DOR-1 with MOR-1, and show that the construct enhances morphine analgesia and blocks tolerance. What makes the MORTM1-TAT construct particularly interesting is the fact that a number of single-domain splice variants of MOR-1 corresponding to TM1 have been reported (Du et al., 1997; Pan and Pasternak, 2011; Xu et al., 2013), raising the question as to whether these endogenous single TM MOR-1 variants play a physiologic role in tolerance.
6. Other.
Several other mechanisms also have been linked to opioid tolerance. Protein kinase C has been implicated by several groups (Mayer et al., 1995; Granados-Soto et al., 2000), as has β-arrestin2 (Bohn et al., 1999, 2000, 2002, 2003; Raehal and Bohn, 2011; Groer et al., 2011). In these studies, knocking out β-arrestin2 increases the sensitivity of the mice to morphine and delays the production of tolerance. Structural changes associated with membrane sialic acid at the spinal level are seen after long-term administration of morphine (El Maarouf et al., 2011). Another intriguing report implicates platelet-derived growth factor receptor-β (PDGFβ) signaling in morphine tolerance in rats (Wang et al., 2012). This is particularly interesting because this pathway can be blocked by imatinib, which is currently approved for clinical use. The question, however, will be whether the side effects of imatinib, which is used as a chemotherapeutic drug, will preclude its use in blocking opioid tolerance.
G. Incomplete Cross-Tolerance
Cross-tolerance among opiates is an accepted and important clinical concept. Patients highly tolerant to one mu opiate display tolerance to them all. However, clinicians also appreciate that cross-tolerance among drugs is not always complete. Indeed, the presence of incomplete cross-tolerance is the foundation for the practice of Opioid Rotation (Cherny et al., 2001; Chou et al., 2009). As the dose of an opioid is increased, it is common for side effects to become limiting, preventing additional dose escalation. If the pain is not under good control despite dose-limiting side effects, clinicians typically will switch patients to a different opiate, often regaining analgesic activity at doses of the second drug far lower than expected based upon the level of tolerance to the first agent. As noted earlier, these differences can be quite dramatic when switching from morphine to methadone. Incomplete tolerance can be demonstrated in animal models. Whereas complete tolerance is seen between morphine and codeine, incomplete cross-tolerance is observed between morphine and several other mu drugs, including morphine-6β-glucuronide, heroin, and methadone (Lange et al., 1980; Rossi et al., 1996). Unidirectional cross-tolerance, an extreme example of incomplete cross-tolerance, exists between morphine and levorphanol (Moulin et al., 1988). Rats tolerant to morphine remain sensitive to levorphanol, whereas animals tolerant to levorphanol are also tolerant to morphine. In this situation, levorphanol acts through multiple opioid receptors (section VIII.C.3.b), sharing only the mu receptor with morphine. Long-term administration of levorphanol produces tolerance at all of the receptors, including mu, resulting in cross-tolerance to morphine. Long-term dosing with morphine, on the other hand, produces tolerance only at the mu receptor, leaving levorphanol’s activity at the other receptor classes intact.
IV. Opioid Binding Sites: Early Studies
A. Identification of Opioid Binding Sites
The concept of receptors has had an integral role in pharmacology for over a hundred years (Langley, 1909; Ehrlich, 1913; Clark, 1933; Gaddum, 1937). The proposal of opioid receptors goes back many decades, first being proposed based upon the rigid structure-activity relationships of the opiates (Beckett and Casy, 1954, 1965; Beckett, 1956; Beckett et al., 1956a,b; Janssen et al., 1960, 1966; de Stevens, 1965; Portoghese, 1966, 1970; Jacobson et al., 1970). However, they were not demonstrated experimentally until 1973 (Pert and Snyder, 1973; Simon et al., 1973; Terenius, 1973), after many unsuccessful attempts (Chernov and Woods, 1965; Ingoglia and Dole, 1970; Navon and Lajtha, 1970; Berkowitz and Way, 1971; Goldstein et al., 1971; Hug and Oka, 1971; Seeman et al., 1972; Clouet and Williams, 1973; Van Praag and Simon, 1966). Most used the concept of stereospecificity, looking at the difference in binding of active and inactive stereoisomers, as described by Goldstein et al. (1971). In hindsight, these early attempts failed because of technical problems, in particular, the limited sensitivity of radioligands of low specific activity. On the basis of estimations that the concentration of morphine in the brain needed for activity was approximately 0.1 µM or higher, investigators assumed micromolar affinities of opiates for their receptors, whereas the actual affinities were in the low nanomolar range, concentrations not feasible with low specific activity 14C radioligands. Second, the low abundance of the receptors further complicates their detection with 14C radioligands.
Three laboratories reported the biochemical demonstration of opioid binding sites using different radioligands as follows: [3H]naloxone (Pert et al., 1973), [3H]dihydromorphine (Terenius, 1973), or [3H]etorphine (Simon et al., 1973). The binding was validated in a variety of approaches. First, the binding is stereospecific and highly selective for opiates (Table 5), fulfilling the criteria needed for a receptor. Second, the receptor is present in nervous tissues where opiates are assumed to act. Prior to receptor binding, many investigators used bioassays to assess opiate actions, particularly the guinea pig ileum contraction bioassay (Schaumann, 1955; Kosterlitz and Robinson, 1957a,b; Paton, 1957). The demonstration of opioid binding sites in the myenteric plexus of the guinea pig ileum provides additional strong support for the relevance of the binding sites (Table 5) (Pert and Snyder, 1973; Creese and Snyder, 1975).
TABLE 5.
Opioid-receptor binding affinities
The ability of drugs to compete [3H]naloxone binding was determined using either brain homogenates or the intestine. It is given as an ED50—the concentration able to compete half the specific binding. From Pert and Snyder (1973).
| Drug | ED50 |
|---|---|
| M | |
| Brain homogenate | |
| (−)-Naloxone | 1 × 10−8 |
| (−)-3-Hydroxy-N-allylmor-phinan (levallorphan) | 1 × 10−9 |
| Levorphanol | 2 × 10−9 |
| (−)-Nalorphine | 2 × 10−9 |
| (−)-Morphine | 6 × 10−9 |
| (−)-Methadone | 2 × 10−8 |
| (±)-Pentazocine | 5 × 10−8 |
| (+)-Methadone | 2 × 10−7 |
| (±)-Propoxyphene | 1 × 10−6 |
| (+)-3-Hydroxy-N-allylmorphinan | 5 × 10−6 |
| Dextrorphan | 8 × 10−6 |
| (−)-Codeine | 2 × 10−5 |
| Phenobarbital | * |
| Serotonin | * |
| Norepinephrine | * |
| Carbamylcholine | * |
| Choline | * |
| Atropine | * |
| Histamine | * |
| Colchicine | * |
| Intestine, minced preparation | |
| Levorphanol | 8 × 10−8 |
| (−)-Morphine | 3 × 10−6 |
| Dextrorphan | 4 × 10−5 |
| Codeine | 1 × 10−4 |
No effect at 10−4 M.
At the time, few distinctions were appreciated between the three radioligands used to describe the binding. Of the three, the agonist [3H]dihydromorphine is clearly the most mu selective followed by the antagonist [3H]naloxone, which at the concentrations used is still relatively selective for mu sites. However, the agonist [3H]etorphine is nonselective, labeling additional receptor classes that were not appreciated at the time, including delta and kappa1 (Martin et al., 1976; Lord et al., 1977) and kappa2 and kappa3 receptors (Zukin et al., 1988; Clark et al., 1989; Rothman et al., 1992).
Despite the limited available structural information on the receptors, a number of important characteristics of the receptors were described, both biochemical and pharmacological. The protein nature of the receptor was established by its sensitivity to the proteases trypsin and chymotrypsin and insensitivity toward DNase, RNase, neuraminidase, and phospholipase C (Simon et al., 1973; Pasternak and Snyder, 1974a, 1975c) and by the sensitivity of opioid binding to protein modifying reagents, particularly those targeting sulfhydryl groups (Pasternak et al., 1975c; Wilson et al., 1975). However, these early studies also reveal a marked sensitivity of opioid binding to detergents, possibly explaining the difficulties many laboratories encountered when trying to isolate and purify the receptors. For example, low Triton X-100 concentrations potently inhibit mu opioid binding without solubilizing the receptor, as shown by the restoration of binding when the detergent is washed away from the membranes. This sensitivity to detergent may explain the sensitivity of opioid binding to phospholipase A (Pasternak and Snyder, 1974b; Abood et al., 1978), which liberates fatty acids. Additionally, cerebroside sulfate was implicated in opioid binding (Law et al., 1978, 1979).
B. Localization of Binding Sites
The validation of the binding sites was further established with the localization of the binding sites, both at the cellular level and regionally.
1. Cellular Localization of Opioid Receptors.
The initial studies confirmed the association of the binding sites with synaptosomal membranes (Pert and Snyder, 1973; Pert et al., 1974b; Mule et al., 1975), consistent with a neurotransmitter receptor. The binding sites are associated with both pre- and postsynaptic membranes, as revealed by lesioning the dorsal roots of the spinal cord, which lowers spinal binding by approximately 50% (Lamotte et al., 1976). Receptors also are observed on peripheral nerves (Young et al., 1980). In these studies, the investigators autoradiographically labeled receptors on the vagus nerve, documenting their presence and demonstrating axonal flow.
2. Regional Localization of Opioid Receptors.
Homogenate binding quickly established marked regional binding differences among brain structures (Table 6) (Hiller et al., 1973; Kuhar et al., 1973; Wong and Horng, 1973; Lee et al., 1975). These homogenate studies established a relationship between binding and the limbic system, regions important in opioid action (Herz et al., 1970; Pert and Yaksh, 1974), whereas the absence of binding to white matter emphasizes the neuronal association of the receptors.
TABLE 6.
Opioid-receptor binding in regions of monkey brain
The indicated regions of monkey brain were dissected, homogenized, and receptor binding performed with the agonist [3H]dihydromorphine. Adapted from Kuhar et al. (1973).
| [3H]Dihydromorphine Binding | |||
|---|---|---|---|
| fmol/mg protein | fmol/mg protein | ||
| Cerebral hemispheres | Extrapyramidal areas | ||
| Frontal pole | 11.9 | Caudate nucleus (head) | 19.4 |
| Superior temporal gyrus | 10.1 | Caudate nucleus (body) | 9.0 |
| Middle temporal gyrus | 7.1 | Caudate nucleus (tail) | 8.9 |
| Inferior temporal gyrus | 6.0 | Putamen | 11.7 |
| Precentral gyrus | 3.4 | Globus | 7.7 |
| Postcentral gyrus | 2.8 | Internal capsule | 5.4 |
| Occipital pole | 2.3 | Midbrain | |
| White matter areas | Superior colliculi | 10.6 | |
| Corpus callosum | <2 | Inferior colliculi | 6.7 |
| Corona radiata | <2 | Interpeduncular nucleus | 13.7 |
| Anterior commissure | 5.4 | Raphe area | 8.2 |
| Fornix | <2 | Periaqueductal gray | 31.1 |
| Optic chiasm | <2 | Cerebellum-lower brain stem | |
| Limbic cortex | Pons (ventral) | 1.4 | |
| Anterior amygdala | 65.1 | Cerebellar cortex | <2 |
| Posterior amygdala | 34.1 | Dentate nucleus | 1.9 |
| Hippocampus | 12.5 | Floor of fourth ventricle | 6.3 |
| Hypothalamus | Pyramidal tract | 3.0 | |
| Medial hypothalamus | 24.2 | Lower medulla | 5.8 |
| Anterior hypothalamus | 24.3 | Spinal cord (thoracic) | |
| Posterior hypothalamus | 24.7 | Dorsal column (white) | 3.1 |
| Hypothalamus | 32.2 | Lateral cord (white) | 3.3 |
| Mammillary bodies | 5.0 | Gray matter | 8.8 |
| Thalamus | |||
| Medial thalamus | 24.6 | ||
| Lateral thalamus | 7.8 | ||
Homogenate binding studies have some advantages, including the ability to quantify results and characterize the binding more readily. However, they are limited by the size of the dissected region and the amount of tissue needed for the assays. Autoradiography provides detailed distributions of the binding sites not achievable with homogenate binding. Opiate receptor autoradiographic approaches were among the very first demonstrating the distribution of neurotransmitter receptors (Pert et al., 1975, 1976a; Schubert et al., 1975; Atweh and Kuhar, 1977a,b,c; Young and Kuhar, 1979). The most detailed early studies correspond most closely to the distribution of mu receptors despite the use of [3H]diprenorphine. The later availability of selective agents enabled the mapping of mu, kappa, and delta sites (Goodman et al., 1980; Duka et al., 1981; Foote and Maurer, 1982; Wamsley et al., 1982; Maurer et al., 1983; Goodman and Snyder, 1982a,b; Mansour et al., 1987). Opioid receptor autoradiography also was one of the first to use digital subtraction (Goodman and Pasternak, 1985). Digital subtraction autoradiography permits the virtual visualization of competed (i.e., “lost”) sites. This approach was used to map the mu1 receptors in the brain (Goodman and Pasternak, 1985), showing a distribution quite similar to the mapping predicted by homogenate binding and from studies in the CXBK mouse (Moskowitz and Goodman, 1985a,b).
The cloning of the receptors has enabled more selective and comprehensive mapping at both the mRNA level with in situ hybridization and at the protein level using imunohistochemical approaches (Mansour et al., 1994a,b; Arvidsson et al., 1995a,b; Ji et al., 1995). However, there remains some debate on immunohistochemical approaches because of issues of antibody selectivity. Thus, autoradiographic approaches that are dependent upon “function” (i.e., the ability to bind a ligand) still provide a valuable resource.
C. Development
Developmental studies quickly established the caudal to rostral appearance of binding sites in the rat, with profound increases in binding levels in the first several weeks postnatally (Clendeninn et al., 1976; Coyle and Pert, 1976; Garcin and Coyle, 1977). Pharmacologically, the high sensitivity of young rats to the respiratory effects of morphine (Kupferberg and Way, 1963) contrasts with the later development of analgesic sensitivity (Zhang and Pasternak, 1981). Young 2-day-old rats are more sensitive to morphine’s respiratory depressant effect (ED50 2.5 mg/kg s.c.) than its analgesic one (ED50 56 mg/kg s.c.), whereas their relative potency is reversed at day 14 when the effects on respiratory depression are less (ED50 9.3 mg/kg s.c.) and those on analgesia are greater (ED50 1.4 mg/kg s.c.). The increased sensitivity of the older rats to morphine analgesia is even more surprising in view of the increasing blood-brain barrier over this same period. The appearance of morphine’s increasing analgesic sensitivity corresponds to the appearance of the mu1 binding site, whereas respiratory depression correlates with the traditional mu2 site, a dissociation also observed with the antagonists naloxazone and naloxonazine in which blockade of the mu1 sites prevents morphine analgesia but not respiratory depression (Pasternak and Hahn, 1980; Pasternak et al., 1980a,b; Pasternak, 1981; Wolozin and Pasternak, 1981).
D. Discrimination of Agonist and Antagonist Binding
One of the most intriguing aspects of the early studies of opioid binding is the ability to distinguish between agonist and antagonist binding states of the receptor, first shown with sodium ions (Table 7) (Pert et al., 1973; Snyder et al., 1974). A major advantage of the opioid system is the availability of many pairs of structurally close opioid agonists and antagonists, a number of which were available in radiolabeled form, thus permitting a direct comparison of their binding. A wide range of biochemical approaches subsequently distinguished agonist and antagonist binding, including monovalent and divalent cations, enzymes, reagents, and even assay incubation temperatures (Pert et al., 1973; Creese et al., 1975; Pasternak et al., 1975b,c; Pasternak and Snyder, 1975c; Wilson et al., 1975; Blume, 1978a,b; Childers and Snyder, 1978; Blume et al., 1979). It is presumed that the treatments affect the stabilization of different, interconverting receptor states, with the antagonist conformation being the most stable.
TABLE 7.
Discrimination of opioid agonist and antagonist binding
The effects of many of the treatments that diminish agonist binding are more pronounced in the presence of sodium chloride. Although EGTA preferentially chelates Ca2+ over Mg2+ or Mn2+, EDTA binds them all. Both EDTA and EGTA will chelate the heavy metals. Note that binding is extremely sensitive to trypsin, which at only 1 µg/ml lowers agonist binding by approximately 90% and antagonist binding by approximately a third. From the literature (Pert et al., 1973; Creese et al., 1975; Pasternak and Snyder, 1975c; Pasternak et al., 1975b,c; Wilson et al., 1975).
| Treatment | Effect on Receptor Binding |
|
|---|---|---|
| Agonist | Antagonist | |
| Ions | ||
| Na1+ | ↓ | —a |
| Mg2+ and Mn2+ | ↑ | − |
| Cu2+ and Ni2+ and Fe2+ | ↓↓ | ↓ |
| Anions | — | — |
| Chelators | ||
| EDTA | ↓ | —a |
| EGTA | — | |
| Enzymes | ||
| Trypsin | ↓↓ | ↓ |
| Chymotrypsin | ↓ | ↓ |
| Phospholipase A | ↓ | ↓ |
| Reagents | ||
| N-Ethylmaleimide | ↓ | — |
| Iodoacetate | ↓ | — |
| Carbodiimide | ↓ | — |
| Temperature (0° compared with 25°C) | ↓ | ↑ |
Binding may increase due to the dissociation of endogenous agonists in brain tissue.
The concept of agonist and antagonist receptor conformations is important in binding assays. It is presumed that antagonists label both agonist and antagonist states with similar high affinity, whereas high-affinity agonist binding is restricted to only the agonist conformation. This concept is supported by the observation that the Bmax values for radiolabeled agonists and antagonists in brain or cell lines are not the same, with antagonist binding typically 2- to 3-fold greater than agonist binding. When first observed in brain, the question arose of whether antagonists might simply label additional classes of receptors. However, similar differences have also been observed in cells expressing the cloned mu receptor.
The concept that antagonists label both conformations while agonists only label one becomes important when interpreting binding data, particularly competition approaches. Many of the early studies used the agonist [3H]dihydromorphine, whereas [3H]DAMGO is commonly used today. Both are agonists at mu receptors and therefore will yield Bmax values that typically are only a fraction of those that would be labeled by an antagonist such as [3H]naloxone or the more mu-selective ligand, [3H]CTAP. Thus, [3H]agonist binding does not provide a full estimate of the expression levels of the receptor. On the other hand, competitions against [3H]agonists may provide more insightful estimates for Ki values for the active receptor conformation.
Radiolabeled antagonists have their own issues, particularly with competition studies. Because agonists have poor affinity for the antagonist conformation of the receptor, which represents the majority of the labeled sites, the apparent IC50 value of an agonist in a competition study actually corresponds to a composite value obtained by competing both the high-affinity agonist and the lower affinity antagonist conformations. Thus, the Ki value may appear significantly greater (i.e., a lower affinity) than when using a radiolabeled agonist. This can be even more pronounced if conditions include sodium ions. An argument can be made to use a radiolabeled antagonist when looking at changes in total receptor number (i.e., Bmax), whereas radiolabeled agonists might be more suitable for determining relative affinities (i.e., Ki) in competition studies. Thus, conditions of a binding assay are crucial when interpreting affinities and binding studies.
1. Sodium Effect.
Sodium ions have the ability to stabilize the antagonist conformation of G protein-coupled receptors (GPCR), as first demonstrated for opioid receptors in 1973 (Pert et al., 1973) and subsequently extended to a wide range of other GPCRs. This discovery resulted from a disagreement between Adele Snowman and me (G.W.P.). In December of 1972, I observed that ethylenediaminotetracetic acid (Na2EDTA) dramatically enhanced [3H]naloxone binding and had suggested that she perform the experiment. Her findings contradicted mine—her binding dramatically decreased. She used the agonist [3H]dihydromorphine, whereas I used the antagonist [3H]naloxone, providing the first evidence of the sodium effect and of general differences in binding between GPCR agonists and antagonists. The demonstration that sodium ions enhance [3H]antagonist binding and lower [3H]agonist binding was then generalized to a series of paired [3H]labeled agonists and antagonists (Table 8). The increased antagonist binding resulted from the “uncovering” of additional binding sites attributed to the temperature- and time-dependent dissociation of an endogenous inhibitor in the tissue—the opioid peptides (Table 2) (Pasternak et al., 1975c). The effect is limited to sodium ions and is not seen with other monovalent cations, such as potassium or ammonium. Likewise, the binding is not sensitive to a range of anions, including chloride, sulfate, fluoride, bromide, iodide, thiocyanate, perchlorate, and formate.
TABLE 8.
Effects of sodium on binding of 3H-labeled opiate agonists and antagonists to rat brain homogenates
Each radioligand was labeled with tritium and stereospecific binding assessed in rat brain membranes. Binding in the presence of sodium chloride (100 mM) was determined and is presented as a percentage of the control binding. Modified from Pert et al. (1973).
| Radiolabel | Control Binding | Binding in NaCl (100 mM) |
|---|---|---|
| cpm | % control | |
| Agonists | ||
| [3H]Dihydromorphine | 2256 ± 113 | 30 |
| [3H]Oxymorphone | 669 ± 31 | 56 |
| [3H]Levorphanol | 1292 ± 61 | 72 |
| Antagonists | ||
| [3H]Nalorphine | 408 ± 22 | 145 |
| [3H]Naloxone | 1582 ± 80 | 241 |
| [3H]Levallorphan | 2861 ± 123 | 129 |
The ability of sodium to lower agonist, but not antagonist, affinity was then used to establish the “sodium shift,” which enabled the assessment of the agonist/antagonist character of a ligand in a simple competition binding assay (Table 9) (Pert et al., 1973). The shift is defined as the ratio of the IC50 values of the compound in question against the antagonist [3H]naloxone in the presence and absence of sodium chloride (Table 9). The affinity of agonists in the presence of sodium ions is decreased, leading to a ratio greater than 1, whereas antagonist affinity is unaffected, leading to a ratio of approximately 1. The ratio of partial agonists is intermediate. This approach has proven helpful in assessing new opioids, and its use has been extended to other GPCR systems. The effect of sodium ions is particularly dramatic on a very high-affinity binding site (Ki < 1 nM) common to both [3H]naloxone and [3H]dihydromorphine (Pasternak and Snyder, 1975b). Sodium ions increase the high-affinity [3H]naloxone site while eliminating the high-affinity [3H]dihydromorphine one, consistent with an interconverting single binding site.
TABLE 9.
Effects of sodium on inhibition of opioid agonists and antagonists of stereospecific [3H]naloxone binding in rat brain homogenates
The IC50 values for the designated competitor against [3H]naloxone binding in the absence (Control) and presence of sodium chloride (100 mM) were determined. The sodium shift was calculated as the ratio of the IC50 values in the presence and absence of sodium chloride (IC50 sodium chloride/IC50 control). Modified from Pert et al. (1973).
| Competitor | Control | NaCl (100 mM) | Sodium Shift |
|---|---|---|---|
| Naloxone | 1.5 | 1.5 | 1 |
| Naltrexone | 0.5 | 0.5 | 1 |
| Diprenorphine | 0.g | 0.g | 1 |
| Cyclazocine | 0.9 | 1.5 | 1.7 |
| Levallorphan | 1 | 2 | 2 |
| Nalorphine | 1.5 | 4 | 2.7 |
| Pentazocine | 15 | 50 | 3.3 |
| Etorphine | 0.5 | 6 | 12 |
| Meperidine | 3,000 | 50,000 | 17 |
| Levorphanol | 1 | 15 | 15 |
| Oxymorphone | 1 | 30 | 30 |
| Dihydromorphine | 3 | 140 | 47 |
| Propoxyphene | 200 | 12,000 | 60 |
| Phenazocine | 0.6 | 80 | 133 |
Although the opioid receptor was the first to demonstrate the sodium effect, it generalizes to a wide range of G protein-coupled receptors. Recently, crystallographic analysis of the adenosine A2A receptor identified the binding pocket for sodium (Liu et al., 2012). It is located deep in within the transmembrane portion of the receptor, similar to a hypothetical location suggested in 1976 for both sodium and divalent (i.e., magnesium/manganese) ions (Pasternak et al., 1976).
2. Divalent Cations and GTP.
In contrast to sodium ions, certain divalent cations enhance agonist binding, effectively counteracting the effects of sodium ions (Table 7) (Pasternak et al., 1975b). Although they are effective alone, the relative increases in agonist binding are far more pronounced when examined in the presence of sodium ions. Mn2+ is most effective followed by Mg2+. Interestingly, Ca2+ ions are without activity. Many laboratories now routinely include MgCl2 in their binding buffers. These divalent cation actions are concentration dependent, with much higher concentrations lowering both agonist and antagonist binding. A reduction in both agonist and antagonist binding is seen with heavy metal divalent cations, including Ni2+, Cu2+, and Fe2+. Much like sodium shifts, manganese and/or magnesium shifts can be used to assess the agonist/antagonist character of a ligand and there is an excellent correlation between the two, but divalent shifts are not often used.
The role of endogenous divalent cations is illustrated by the ability of the chelator EDTA to lower agonist binding and render it more sensitive to sodium ions. EGTA, which is selective for calcium ions over either magnesium or manganese, had little effect upon opioid agonist binding, implicating a role for endogenous magnesium and/or manganese in receptor function. Inasmuch as many proteolytic enzymes are calcium dependent, EDTA is often used in the buffers along with protease inhibitors to minimize receptor degradation when preparing tissues. Because EDTA also chelates both Mg2+ and Mn2+, investigators should consider adding back MgCl2 during the binding assay. GTP and its stabilized nonhydrolyzable analogs GppNHp (guanosine 5′-[β,γ-imido]triphosphate) and GTPγS also lower opioid agonist, but not antagonist, binding (Childers and Snyder, 1978), results similar to those of the β-adrenergic receptor (Lefkowitz et al., 1976, 1982, 1984).
3. Protein Modifying Reagents, Enzymes, and Temperature.
Agonist and antagonist binding also are differentiated by a number of other treatments (Creese et al., 1975; Pasternak and Snyder, 1975c; Pasternak et al., 1975b,c; Wilson et al., 1975). These include a wide range of protein modifying reagents, particularly those targeting sulfhydryl groups, as well as the proteolytic enzymes trypsin and chymotrypsin and even the temperature of the binding assay. Again, their ability to selectively lower agonist binding is more pronounced in the presence of sodium ions. These observations clearly impact the design and approach to receptor binding assays and further imply a greater stability of the antagonist conformation of the receptor.
V. Pharmacological Evidence for Multiple Mu Receptor Subtypes
Clinicians have had extensive experience with a wide range of opiate analgesics, most of them mu, and have compiled general principles in their use (Payne and Pasternak, 1992; Jacox et al., 1994; Foley, 1996; Chou et al., 2009). Foremost is the need to individualize therapy, both with regards to the choice of opioid and its dose. Insensitivity to one mu opioid or its association with severe side effects, such as nausea and vomiting, may not extend to a different mu opioid. Clinical experience also tells us that there is no way to predict the optimal drug or its dose. Whereas one mu opioid may work well in one patient, a different patient may respond better to a different mu opioid.
Many of these observations have been recapitulated in preclinical models. The importance of genetic backgrounds in the sensitivity of mouse strains to opiates has long been known (Baran et al., 1975; Reith et al., 1981; Pick et al., 1991; Brodsky et al., 1994; Mogil et al., 1994, 1995, 1996; Woolfolk and Holtzman, 1995; Kamei et al., 1997; Chang et al., 1998; Elmer et al., 1998; Hoffmann et al., 1998; Kest et al., 1998b; Lacroix-Fralish and Mogil, 2009), starting with the early studies on the insensitivity of CXBK mice to morphine. However, perhaps the most intriguing aspect of the CXBK mice is their ability to discriminate between morphine and other mu opioids (Fig. 9) (Rossi et al., 1996; Chang et al., 1998; Neilan et al., 2003). Although the potency of morphine in CXBK mice is markedly decreased relative to a standard mouse (CD-1), the mu opiates methadone, heroin and 6-acetylmorphine (the active component of heroin), morphine-6β-glucuronide, and fentanyl retain equivalent potencies in both animal strains. Thus, genetic backgrounds clearly impact the sensitivity of the mouse strains to opioids and, in some situations, can differentiate among different mu opioids. This is hard to reconcile with the concept that all mu opioids share a single mechanism through a single mu-opioid receptor.
Fig. 9.
Opioid analgesia in CXBK mice. With use of the radiant heat tailflick assay, the indicated opiates were given at equianalgesic doses to either CD-1 or CXBK mice, and the responses were determined. Adapted from the literature (Rossi et al., 1996; Chang et al., 1998).
A. Receptor Binding
The concept of multiple mu receptors was first formally proposed in 1981 (Wolozin and Pasternak, 1981) based in large part upon earlier binding studies (Pasternak and Snyder, 1975b). The earlier study reported the presence of a very high-affinity binding site for both opioid agonists and antagonists in addition to the previously described sites. The later paper extended this to enkephalin sites and demonstrated that both mu and delta ligands labeled this new site with very high affinity, clearly distinguishing it from mu-selective and enkephalin-selective (delta) sites. The common site for mu opiates and enkephalins was termed mu1, whereas the mu-selective site was labeled mu2. The enkephalin-selective site corresponded to the delta receptor (Lord et al., 1977; Chang and Cuatrecasas, 1979). The binding paradigms underlying this proposal were subsequently validated by Munson and Rodbard and colleagues using a computer modeling approach (Lutz et al., 1984; Munson et al., 1984). The regional distribution of these sites has been established autoradiographically (Moskowitz and Goodman, 1985a,b), including through the use of digital subtraction approaches (Goodman and Pasternak, 1985).
Many of the studies looking at multiple mu receptors were facilitated by two antagonists, naloxazone and naloxonazine (Fig. 2) (Pasternak et al., 1980a,b; Hazum et al., 1981a; Childers and Pasternak, 1982; Hahn and Pasternak, 1982; Hahn et al., 1982). Similar in structure, these two antagonists are capable of selectively and irreversibly blocking the mu1 binding sites (Table 10). However, to use these drugs, it is important to recognize that they also will label mu2 sites reversibly. In vivo, sufficient time must be given to permit the elimination of free drug before a selective antagonism of mu1 sites can be seen. Likewise, in vitro, the free drug must be washed away to achieve a selective blockade of the mu1 sites. Their ability to selectively block mu1 sites provided important validation of the proposal of multiple mu receptors, as shown by the loss of the mu1 binding site in brain tissue after treatment with either naloxazone or naloxonazine, leaving the mu-selective (mu2) and enkephalin-selective (delta) receptors. In competition studies, this treatment eliminates the morphine-sensitive component of [3H]enkephalin binding and the enkephalin-sensitive component of [3H]dihydromorphine binding.
TABLE 10.
Opioid binding to multiple mu and delta sites in brain
As shown, mu1 sites have very high affinity for both mu and most delta ligands, whereas mu2 and delta sites are selective. Adapted from Wolozin and Pasternak (1981).
| Approximate KD Values |
|||
|---|---|---|---|
| Mu1 | Mu2 | Delta | |
| nM | |||
| Morphine | |||
| Saturation studies | 0.4 | 11 | — |
| Competition studies | <1 | 8 | 71 |
| [d-Ala2,d-Leu5]Enkephalin | |||
| Saturation studies | 0.5 | — | 5 |
| Competition studies | <1 | 50 | 8 |
Soon after our proposal of multiple mu receptors, Rothman and Westfall (1982a,b, 1983) suggested three classes of mu and delta receptors. In his model, Rothman proposed a physical association (i.e., “complex”) between the mu and delta receptors, but conceptually it is similar to the mu1/mu2/delta model in that it proposed the two selective sites as well as a “complexed” site with very high affinity for both mu and delta ligands. At that time, there was no experimental evidence to support a physical association of the mu and delta receptors because their structures were unknown. However, subsequent studies have confirmed that mu and delta receptors can dimerize (Cvejic and Devi, 1997; George et al., 2000; Gomes et al., 2000, 2002, 2004; Law et al., 2005; Rutherford et al., 2008; He et al., 2011; Pasternak and Pan, 2011). Most recently, Rothman and co-workers suggested that the mu/delta heterodimer may correspond to the “complexed” site (i.e., mu1) site (Rutherford et al., 2008). Others also report the appearance of novel binding sites with the coexpressed MOR-1 and DOR-1 receptors, consistent with the possibility that the heterodimer may represent the mu1 site (George et al., 2000; Gomes et al., 2004; Law et al., 2005; Kabli et al., 2010). Although strongly suggestive, the complexity of the mu receptor MOR-1 gene and its splice variants (section VIII) raises a number of issues. With dozens of splice variants identified in mice, rats, and humans (Bare et al., 1994; Du et al., 1997; Pan et al., 1998b, 1999, 2000, 2001, 2003, 2005a,b; Pasternak et al., 2004; Doyle et al., 2007b; Xu et al., 2009, 2011; Gris et al., 2010), it is not yet clear which one(s) dimerize with delta receptors and how this affects their pharmacology.
B. Pharmacology
1. Analgesia.
As with the binding studies, both naloxazone and naloxonazine associate the mu1 and mu2 receptors with various pharmacological actions (Table 11). Administered the day before pharmacological testing, the drugs provide a valuable model to assess the role of mu1 sites. The first studies associate mu1 receptors with systemic morphine analgesia (Pasternak et al., 1980a,b). Subsequent studies explored the role of mu1 sites in supraspinal and spinal morphine analgesia, with interesting results (Paul et al., 1989a). Naloxonazine blocks supraspinal morphine analgesia, but not spinal analgesia, suggesting that different mu receptor mechanisms act in these two regions. CXBK mice further support different mu analgesic mechanisms (Pick et al., 1993). CXBK mice are insensitive to systemic morphine, requiring doses 5- to 10-fold higher than in traditional mouse models, such as CD-1 mice. Likewise, intracerebroventricular morphine given into CXBK mice is ineffective at doses 10-fold greater than the ED50 in control CD-1 mice. In contrast, at the spinal level, morphine given intrathecally retains full analgesic activity and is equipotent in the CXBK and the CD-1 mice. These results are consistent with a supraspinal mu1 system that is deficient in the CXBK mouse and a spinal mu2 analgesic system that is intact. The lack of mu1 receptors in CXBK mice was demonstrated autoradiographically, supporting this proposal (Moskowitz and Goodman, 1985a,b). Differences between the supraspinal mu receptor mechanisms for supraspinal analgesia and spinal/supraspinal synergy also exist. Yeung and Rudy (1980) report profound synergy between spinal and supraspinal sites in rats, a synergy also present in both CXBK mice and CD-1 controls. In contrast to supraspinal administration in CXBK where morphine is relatively inactive, supraspinal morphine synergizes with spinal morphine in CXBK mice as effectively as in control CD-1 mice. This implies a mu1 analgesic mechanism for supraspinal morphine alone and a mu2 mechanism for its contributions to spinal/supraspinal synergy.
TABLE 11.
Naloxonazine-sensitive and -insensitive actions
Compiled from the literature (Pasternak et al., 1980b, 1983; Hahn and Pasternak, 1982; Ling et al., 1983, 1984, 1985, 1986; Munson, 1983; Wood and Pasternak, 1983; Bodnar et al., 1988; Heyman et al., 1988; Mann et al., 1988a,b; Paul and Pasternak, 1988; Paul et al., 1989a; Janik et al., 1992; Pick et al., 1993; Honkanen et al., 1996).
| Morphine Effect | Sensitivity |
|
|---|---|---|
| β-Funaltrexamine | Naloxonazine | |
| Analgesia | ||
| Systemic | Yes | Yes |
| Supraspinal | Yes | Yes |
| Spinal | Yes | No |
| Supraspinal/spinal synergy | Yes | No |
| Respiratory depression | Yes | No |
| Inhibition of gastrointestinal transit | Yes | No |
| Prolactin release | Yes | Yes |
| Growth hormone release | Yes | No |
| Physical dependence | Yes | Mixed |
| Feeding | Yes | Yes |
| Nigrostriatal dopamine release | No | |
| Alcohol intake | Yes | |
| Pruritus | No | |
2. Other Opioid Actions.
Constipation and respiratory depression accompany all the traditional mu opioids. Although the peripheral antagonists alvimopan and N-methylnaltrexone are helpful clinically, the optimal approach remains the development of analgesics lacking gastrointestinal transit and respiratory side effects. The feasibility of this concept rests upon the ability of several drugs to dissociate mu analgesia from these side effects. For example, naloxonazine blocks morphine analgesia without antagonizing morphine-induced inhibition of gastrointestinal transit (Heyman et al., 1988; Paul and Pasternak, 1988) or respiratory depression (Ling et al., 1983, 1985). Morphine analgesia developmentally also is dissociable from respiratory depression (Zhang and Pasternak, 1981) (section IV.C); neonatal rats are far more sensitive to the respiratory depressant effects of morphine than analgesia, whereas adolescent and adult animals show a reversed sensitivity.
An early approach to minimize side effects involved the development of mixed agonists/antagonists and partial mu agonists. Many of the mixed agonists/antagonists are potent antagonists at mu receptors and elicit their analgesic actions through alternative sites, such as traditional kappa1 receptor or kappa3 receptors (section VIII.C.3.b). However, several other agents have unique actions. Meptazinol is an effective mu analgesic lacking significant respiratory depression or constipation (Robson, 1983; Spiegel and Pasternak, 1984). Likewise, the peptide TAPS is a potent mu analgesic with respiratory stimulant activity (Paakkari et al., 1993; Vonhof et al., 2001). These pharmacological profiles might be explained if agents were mu1 agonists/mu2 antagonists. However, the most promising approach may involve new receptor targets as described below.
VI. Molecular Biology of Mu Receptors
The breakthrough in our understanding of the molecular aspects of opioid receptors came with the cloning of the delta receptor DOR-1 in 1992 (Evans et al., 1992; Kieffer et al., 1992). This was followed closely by the cloning of the mu (MOR-1) (Chen et al., 1993a; Eppler et al., 1993; Thompson et al., 1993; Wang et al., 1993) and kappa1 (KOR-1) opiate receptors (Chen et al., 1993b; Li et al., 1993; Meng et al., 1993) and the closely related ORL1 (also known as KOR-3) receptors (Bunzow et al., 1994; Chen et al., 1994; Fukuda et al., 1994; Keith et al., 1994; Mollereau et al., 1994; Nishi et al., 1994; Pan et al., 1994, 1995; Homberg et al., 2009). Structurally, they fall into the family of G protein-coupled receptors. Only a single gene was identified for each of the three classes of opioid receptors, raising the question of the relationship of the gene with the pharmacological evidence for subtypes of mu (Pasternak and Snyder, 1975b; Pasternak et al., 1980b; Wolozin and Pasternak, 1981), delta (Jiang et al., 1991), and kappa receptors (Zukin et al., 1988; Clark et al., 1989; Rothman et al., 1990; Reisine and Pasternak, 1996). Correlating the pharmacologically defined subtypes and the gene has proven difficult. One of the kappa subtypes, kappa2, appears to be a heterodimer between DOR-1 and KOR-1 (Jordan and Devi, 1999), but the molecular basis for the two U50,488H-sensitive sites (Clark et al., 1989; Rothman et al., 1990) or delta-receptor subtypes (Cowan et al., 1985; Portoghese et al., 1990, 1993; Jiang et al., 1991; Mattia et al., 1991; Chakrabarti et al., 1993; Sofuoglu et al., 1993; Buzas et al., 1994; Miyamoto et al., 1994; Ohkawa et al., 1997; Rossi et al., 1997; Zhu et al., 1999) remains unknown. It is interesting, however, that some delta ligands still elicit analgesia in a DOR-1 knockout animal (Nitsche et al., 2002), raising the possibility of a second delta-opioid receptor gene. Finally, evidence suggests that the kappa3 receptor may correspond to a heterodimer of a truncated MOR-1 splice variant (see section VIII.C.3b) (Majumdar et al., 2011).
A. MOR-1
Mu-opioid receptor genes have been identified in over 30 different species ranging from nonmammalian vertebrates such as white suckerfish and frog to a wide range of mammals (Fig. 10). Similar to other members of the GPCR family, computer modeling of the cloned MOR-1 predicts a seven-transmembrane protein that has now been confirmed crystallographically (Figs. 11, 12, 13) (Manglik et al., 2012). GPCRs represent one of the largest gene families in the mammalian genome (Attwood and Findlay, 1994; Kolakowski, 1994; Fredriksson et al., 2003), with seven transmembrane domains, an extracellular N terminus and intracellular C terminus. The structure of GPCRs was first extrapolated from rhodopsin (Palczewski et al., 2000) and the β2 receptor (Rasmussen et al., 2011). All of the full-length opioid receptors contain several conserved motifs of the rhodopsin family, such as the DRY motif at the boundary between TM3 and the second intracellular loop, xBBxxB (where B represents a basic amino acid residue) motif in the third intracellular loop, NSxxNPxxY motif in TM7, and cysteine residues in the carboxyl-terminal tail for potential prenylation with the transmembrane domains organized in a counterclockwise manner.
Fig. 10.
Phylogency of mu-opioid receptors. Overview of the species variations in MOR-1. (A) Schematic of species. (B) Comparison of exons 4, 7, 11 in mammals. From Pan and Pasternak (2011).
Fig. 11.
Crystal structure of MOR-1. The crystal structure of MOR-1 was determined with β-funaltrexamine covalently attached within the binding pocket. (A) Side view of the crystal structure of MOR-1 with β-funaltrexamine docked within the binding pocket. Note that the binding pocket involves TM3, TM5, TM6, and TM7 and that the C and N termini have been truncated. (B) View from extracellular side (top) and form the intracellular side (bottom). Reprinted by permission from Macmillan Publishers Ltd: [Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, and Granier S (2012) Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature 485:321–326].
Fig. 12.
Crystal structure of the binding pocket of MOR-1 with β-funaltrexamine. The crystal structure of MOR-1 was determined with β-funaltrexamine covalently attached within the binding pocket. The left is a side view of the structure with a “transection” of the receptor to show the docking of the ligand. The view on the right is looking at the docking of the ligand from the extracellular surface. Reprinted by permission from Macmillan Publishers Ltd: [Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, and Granier S (2012) Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature 485:321–326].
Fig. 13.
Crystallization of MOR-1 dimers. Schematic showing the dimerization structure of MOR-1. The formation of homodimers involves interactions between TM5 and TM6. Oligomerization forms from interactions of the homodimers through TM1, TM2, and helix 8. Reprinted by permission from Macmillan Publishers Ltd: [Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, and Granier S (2012) Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature 485:321–326].
At the amino acid level, MOR-1 is 60–70% homologous to the other opioid receptor families, including ORL-1, particularly in the transmembrane and intracellular loop regions. The N terminus, C terminus, and the second and third extracellular loops provide most of the diversity (Fig. 14). The opioid binding pocket and the G protein-coupling domain were originally assessed by chimera and mutagenesis approaches (for review, see Minami and Satoh, 1995; Law and Loh, 1999; Chavkin et al., 2001). Chimeras involving mu/delta and mu/kappa receptors implicate the extracellular loops 1 and 3 and TMs 2, 6, and 7 in mu ligand binding (Fukuda et al., 1995; Wang et al., 1995; Watson et al., 1996; Dietrich et al., 1998; Seki et al., 1998). A number of individual residues involved with binding have been identified by site-directed mutagenesis (Xue et al., 1994; Minami et al., 1996; Xu et al., 1999a; Zhang et al., 1999; Bonner et al., 2000; Ulens et al., 2000). The domains and residues involved in G protein coupling, mu agonist-induced receptor phosphorylation, internalization, and desensitization have also been described using similar approaches (Minami and Satoh, 1995; Law and Loh, 1999; Chavkin et al., 2001). Among the more interesting observations was the ability of a D164Q point mutation in the DRY motif to produce a constitutively active mu receptor (Li et al., 2001). However, all of these predictions now need to be reassessed using the recently published crystal structure (Manglik et al., 2012).
Fig. 14.
Amino acid sequence of MOR-1 in mice, rats, and humans. The predicted amino acid sequences of MOR-1 in mice, rats, and humans are shown. The transmembrane domains are shown by the underlined regions. The exon junctions are also indicated. The consensus sequences are shown below the individual species.
The mature MOR-1 receptor on Western blots or with affinity labeling is approximately 70 kDa, far greater than the predicted protein weight of approximately 45 kDa. Western blots also can show additional bands depending upon the tissue and the methods used to isolate and purify the receptor, possibly due to combinations of degradation and ubiquitination (Xu et al., 2013). Glycosylation is important because the 70-kDa band is lowered to the predicted protein molecular mass of 45 kDa after treatment with N-glycanase (Liu-Chen and Phillips, 1987; Liu-Chen et al., 1993). The N terminus has several predicted N-linked glycosylation sites (NxS/T). N-Glycosylation may also have functional relevance because there is evidence suggesting differential N-glycosylation among brain regions (Huang et al., 2008), and the A118G mutation altering glycosylation has been implicated in addiction (Bart et al., 2005; Kreek and LaForge, 2007; Kroslak et al., 2007).
The recent report of the crystal structures of all three classes of opioid receptors has provided important insights (Granier et al., 2012; Manglik et al., 2012; Wu et al., 2012). By use of the T4 lysozyme fusion protein strategy with truncation of the N and C termini to facilitate crystallization, Manglik and co-workers confirm that MOR-1 is composed of seven-transmembrane α-helices with a conserved disulfide bridge between C140 and C217. The structure was solved with the antagonist β-funaltrexamine covalently attached. The ligand makes contact with TM3, TM5, TM6, and TM7, with a covalent attachment to K233, confirming that it is a member of the class A family of G protein-coupled receptors, with a binding pocket within the transmembrane domains. The binding site is quite intriguing. Unlike other GPCRs where the ligands are typically buried within transmembrane domains, the study demonstrates that the binding pocket of MOR-1 is open and exposed to the extracellular surface (Fig. 12). This observation is consistent with numerous studies indicating that large bulky groups can be placed at the 6-position of morphine and morphine-like scaffolds without impairing MOR-1 binding affinity. Finally, they observed that MOR-1 readily dimerizes and forms oligomers. Homodimerization involves an association between TM5 and TM6, whereas the oligomers show an association of the homodimers through an association between TM1, TM2, and helix 8, which is located on the intracellular C terminus (Fig. 13).
The insights into the binding pocket and the transmembrane domain interactions in the formation of dimers and oligomers are valuable. However, several features of the resolved structure of the mu receptor deserve comment. Crystallization of membrane proteins is often quite difficult, leading to the use of the T4 lysozyme and the truncation of the N and C termini to facilitate crystallization of the mu receptor. The binding pockets in all the full-length splice variants are identical and are contained within the crystals solved in the current study. However, the mu-opioid receptor undergoes extensive 5′ and 3′ splicing that involves the N and C termini, regions that have been removed from the constructs used in the current study (section VIII). Thus, the current structure is the first step to a better understanding of mu-opioid receptor function.
B. Phylogeny and Evolution
The initial receptor binding studies reported binding in vertebrates, with low levels in invertebrates (Pert et al., 1974a). However, these early studies were quite limited in their sensitivity compared with current molecular biologic techniques (section VIII) as were the predominately mu radioligands. Subsequent studies reveal binding in insects and in mollusks (Stefano et al., 1980, 1982), although these studies typically do not attempt to address the concept of receptor classes
The opioid receptor genes OPRM1 (MOR-1), OPRD1 (DOR-1), OPRK1 (KOR-1), and OPRL1 (ORL1) are expressed only in vertebrates (Fig. 10), consistent with the early binding studies (Pert et al., 1974a). They have been identified in over 45 vertebrate species directly by molecular cloning or by bioinformatic analysis of available genomic sequence data. Phylogenetic analysis suggests two rounds of genome-wide duplication (paleoploidization) from a single ancestral opioid gene (unireceptor) (Ohno, 1999; Escriva et al., 2002; Lundin et al., 2003), with the first yielding the ancestral DOR-1/MOR-1 and ORL-1/KOR-1 genes. The duplication then led to DOR-1 and MOR-1, as well as KOR-1 and ORL-1 (Dreborg et al., 2008; Larhammar et al., 2009; Stevens, 2009). The predicted MOR-1 protein sequences from 27 species reveals four major clades as follows: 1) fish, 2) amphibians, 3) birds, and 4) mammals, mimicking the evolutionary tree of life (Fig. 10A). Sequence alignments of MOR-1 from multiple species show the regions with the highest homology among the species are in the transmembrane domains and the three intracellular loops, the structures important for mu ligand binding and G protein coupling.
The structure of the OPRM1 gene (Fig. 15) evolved (Herrero-Turrion and Rodríguez, 2008). In the early teleosts, the OPRM1 gene contains five exons, with the first two exons encoding the receptor from the N terminus through TM4. Evolutionarily the two introns between the last three exons were lost, generating a single third exon in zebrafish and mammals that encodes the last three transmembrane domains. Thus, all seven transmembrane domains are encoded by three exons, a structure that is conserved in the other opioid receptor genes. Only the OPRM1 gene further evolved to contain both 3′ and/or 5′ splicing that led to coding sequence differences, starting with the chicken (Fig. 10B).
Fig. 15.
Schematic of human, mouse and rat OPRM1. A schematic representation of the OPRM1 gene in humans, mice, and rats is shown. The exon and intron distances are not drawn to scale. Exons and introns are shown as boxes and horizontal lines, respectively. Intron sizes are indicated as kilobases (kb). The exon and intron distances are not drawn to scale. Promoters are indicated by arrows. Exons are numbered based upon the published data (Pan and Pasternak, 2011).
Alternative splicing of MOR-1 was first observed at the 3′ region, with the replacement of exon 4 and its 12 amino acids by a variety of exon combinations encoding 1 (hMOR-1B4 and rMOR-1D) to 88 amino acids (mMOR-1U) (Figs. 15, 16, 17, 18). The mouse OPRM1 gene contains ten alternatively spliced exons downstream of exon 3, whereas the rat has 9 and humans have 6 (Figs. 15–18). The full length 3′-splice variants differ only at the tip of the C terminus. Thus, they share identical binding pockets and intra- and extracellular loops. Exon 4 is the predominant 3′ exon in the C-terminal tail of MOR-1 in 19 species (Fig. 10B). The 12 amino acid exon 4 sequences are identical in thirteen mammalian species, with one amino acid variation in four species and three amino acid differences in two other species. Exon 7 was first identified in the mouse (Pan et al., 1999) and has been identified in 12 mouse variants (Pan et al., 1999, 2000, 2001, 2005b; Doyle et al., 2007a,b). It is translated in only three variants and is within the 3′-untranslated region (UTR) in the other nine because of upstream termination of translation. Exon 7-associated splice variants have been isolated and mapped to the rat and human OPRM1 genes, with sequence alignments showing its presence in an additional 13 mammalian OPRM1 genes (Fig. 10B). Among species, the predicted amino acid sequences from exon 7 show high homology. Exon 7 has another distinction. The 70 bp of the gene for cytohesin exchange factor 1 (IPCEF1), which is in the opposite orientation from OPRM1, overlaps with the exon 7 sequence in the mouse (Pan et al., 2005a). This overlap is conserved in all the species containing exon 7.
Fig. 16.
Schematic of MOR-1 splicing in the mouse. A composite schematic of the various splice variants of mMOR-1 reported in the literature is shown (Pan and Pasternak, 2011). Variants are grouped as full length, 7TM, 6TM, and 1TM with the predicted structure shown to the right and the exons color coded to match the splicing schematic. Predicted protein sequences for the spliced sequences for the variant are documented in Tables 13–15. The exon composition of mMOR-1Eii, mMOR-1Eiii and mMOR-1Eiv is identical to that of mMOR-1E, except for an insertion of exon 19 between exons 7 and 8. Their predicted protein sequences, however, are identical to mMOR-1E due to termination of translation in exon 6. Only partial sequences were reported for mMOR-1Eii and mMOR-1Vi (Doyle et al., 2007b).
Fig. 17.
Schematic of MOR-1 splicing in the rat. A composite schematic of the various splice variants of rMOR-1 reported in the literature is shown. Variants are grouped as full length, 7TM, 6TM, and 1TM with the predicted structure shown to the right and the exons color coded to match the splicing schematic. Predicted protein sequences for the spliced sequences for the variant are documented in Tables 13–15.
Fig. 18.
Schematic of MOR-1 splicing in humans. A composite schematic of the various splice variants of hMOR-1 reported in the literature is shown. Variants are grouped as full length, 7TM, 6TM, and 1TM with the predicted structure shown to the right and the exons color coded to match the splicing schematic. Predicted protein sequences for the spliced sequences for the variant are documented in Tables 13–15.
Exon 11 is the major exon associated with 5′ splicing. Originally identified in mice and located approximately 30 kb upstream of exon 1 (Pan et al., 2001), exon 11, and its splice variants have been identified in an additional eight mammalian OPRM1 genes by homologous cloning or bioinformatic searches, including rat and human (Pan et al., 2001, 2009; Xu et al., 2006, 2009; Gris et al., 2010) (section VIII). Exon 11 contains its own promoter that is distinct from the promoter responsible for generating the exon 1 variants. In addition to several full-length variants with predicted protein sequences identical to MOR-1 itself (i.e., mMOR-1H, mMOR-1i, mMOR-1J), these exon 11-associated variants include a number of truncated proteins predicting only six transmembrane domains. Initially considered of questionable relevance, recent work using an exon 11 knockout mouse now reveals that they are important in the actions of many established opioid analgesics (section VIII.C.3.b) (Pan et al., 2009; Majumdar et al., 2011, 2012).
The binding characteristics of mu-opioid receptors have also evolved. Studies in white suckerfish, frog, and rough-skinned newt reveal high-affinity [3H]naloxone binding with a mu-like selectivity (Newman et al., 2000, 2002). However, the receptors display lower affinities for mu agonists such as DAMGO and decreased activity in the inhibition of cAMP accumulation and coupling to a G protein-gated inward-rectifying potassium channel (Darlison et al., 1997). Amino acids known to be crucial for either mu agonist or antagonist binding, such as D114 in TM2 (Surratt et al., 1994; Bot et al., 1998; Xu et al., 1999b), D147 in TM3 (Li et al., 1999), and W318 in EL3 (Bonner et al., 2000; Ulens et al., 2000; Xu et al., 2000), are conserved. As MOR-1 evolved, the affinity of DAMGO for the receptor increases.
C. Single Nucleotide Polymorphisms
Scientists have long desired a marker to predict vulnerability to opioid abuse. With an estimated 10 million single nucleotide polymorphisms (SNPs) in the human genome, they are an important source of variability among individuals. Over 4,000 SNPs have been identified in the human mu-opioid receptor gene based upon the International HapMap Project database (International HapMap3 Consortium, 2010), raising questions on their role(s) in contributing to vulnerability of opioid addiction, abuse, and dependence (Uhl et al., 1999; Ikeda et al., 2005; Kreek et al., 2005a,b; Mayer and Hollt, 2006; Mogil et al., 2000b). Several have been correlated with clinical vulnerability to opioid addiction (Hoehe et al., 2000; Tan et al., 2003; Bart et al., 2004, 2005) and substances of abuse, including alcohol (Ray and Hutchison, 2004; Bart et al., 2005; Kim et al., 2004b,c) and nicotine (Lerman et al., 2004; Berrettini and Lerman, 2005). The A118G SNP has been the most extensively studied and proposed to be associated with heroin addiction. In model systems, the A118G SNP reportedly increases the affinity of MOR-1 for β-endorphin (Bond et al., 1998; LaForge et al., 2000; Kreek and LaForge, 2007; Kroslak et al., 2007) and/or lowers MOR-1 expression (Zhang et al., 2005), although some controversy remains. A mouse model carrying an A112G mutation mimicking the human A118G SNP displays a lower level of mMOR-1 mRNA and protein expression and demonstrates a reduced analgesic response to morphine and sex-specific reductions in morphine-induced rewarding properties (Mague et al., 2009). Interpreting the consequences of the A118G SNP in vivo is complicated by the fact that this mutation is present in all the full-length MOR-1 splice variants, making it difficult to determine which one(s) are responsible for these actions (section VIII).
Other SNPs have been examined. R265H and S268P reportedly alter receptor/G protein coupling and calmodulin binding, as well as mu agonist-induced receptor signaling (Koch et al., 2000; Befort et al., 2001b; Wang et al., 2001). Exon 1 promoter activity is influenced by several other SNPs (Hoehe et al., 2000; Kraus et al., 2001; Bayerer et al., 2007). One SNP altered a STAT (signal transducers and activators of transcription type) 6 binding site and thereby lowered promoter activity (Kraus et al., 2001). CXBK mice are insensitive to morphine analgesia (sections III.C and V.B.1). Interestingly, an A to C polymorphism in the exon 1 promoter lowers Sp1 binding, leading to decreased MOR-1 transcripts (Lee et al., 2004). However, CXBK mice also contain an intracisternal A-particle element in the 3′-noncoding region that also influences expression levels (Han et al., 2006)
D. Binding Studies
Although mu-opioid receptor binding had long been studied in brain, we now know that these results represent the labeling of a heterogeneous mixture of mu receptors because of the presence of an array of splice variants in the brain with similar affinities for mu opioids. Working with the cloned receptor avoids these issues. When transiently or stably expressed in mammalian cell lines the cloned rat (Chen et al., 1993a; Thompson et al., 1993; Wang et al., 1993; Bunzow et al., 1995), human (Pan et al., 2003; Wang et al., 1994c; Raynor et al., 1995) and mouse MOR-1 (Kaufman et al., 1995; Pan et al., 1999) receptor display high affinity and selectivity for known mu-selective ligands such as DAMGO and morphine. Indeed, the binding characteristics of the cloned receptor are remarkably similar to those previously seen in brain. Correlating the cloned receptors with the pharmacologically defined subtypes has been difficult, particularly because dozens of MOR-1 splice variants potentially have the ability to form both homodimers and heterodimers with other members of the opioid receptor family as well as unrelated G protein-coupled receptors (Bunzow et al., 1994; Chen et al., 1994; Keith et al., 1994; Pan et al., 1994, 1995, 2002; Uhl et al., 1994; Wang et al., 1994b; Jordan and Devi, 1999; George et al., 2000; Gomes et al., 2000; Befort et al., 2001a; Liu et al., 2011).
E. Regional Expression of MOR-1 in the Central Nervous System
1. mRNA.
Northern blots of MOR-1 mRNA reveal a major band of approximately 11–16 kb in mice (Kaufman et al., 1995; Pan et al., 2001), rats (Thompson et al., 1993; Minami et al., 1994), and humans (Raynor et al., 1995; Pan et al., 2003). Most of the mRNA corresponds to UTR because the coding region of MOR-1 covers only ∼1.2 kb. In both mouse and human transcripts, exon 4 contains a poly(A) site and is expressed as a 10- to 13-kb exon (Ide et al., 2005; Wu et al., 2005). Northern blot analysis also reveals several smaller bands dependent upon the probes used in the mouse (Pan et al., 2001), rat (Thompson et al., 1993), and human brains (Pan et al., 2003; Raynor et al., 1995), raising the possibility of alternative splicing. Ribonuclease protection assays reveal the highest MOR-1 mRNA levels in the thalamus and the lowest in the cerebellum, with moderate levels in the hypothalamus, brain stem, and spinal cord (Wang et al., 1993), results also seen with the superior quantification obtained with solution hybridization analysis (Brodsky et al., 1995a,b).
The regional distribution of MOR-1 mRNA in the rat was examined using in situ hybridization approaches soon after the cloning of the receptor (Delfs et al., 1994a,b; Mansour et al., 1994a; Minami et al., 1994; Kaufman et al., 1995). Overall, there is an excellent correlation between the distribution of the mRNA and previous autoradiographic approaches (section IV.B.2) (Atweh and Kuhar, 1977a,b,c; Goodman and Pasternak, 1985; Waksman et al., 1986; Mansour et al., 1995a, 1987). Regions high in MOR-1 mRNA include the periaqueductal gray, the locus coeruleus, and raphe magnus, all of which have been implicated in supraspinal mu analgesia. Somatosensory regions associated with pain perception, including the dorsal root ganglia, the spinal trigeminal nucleus, the dorsal horn of the spinal cord, and the thalamus, also contain MOR-1 mRNA, as does the nucleus of the solitary tract, the nucleus ambiguus, the parabrachial nucleus, and the bed nucleus.
Although the in situ hybridization mapping corresponds closely to earlier receptor distributions, there are several mismatches. Despite relatively high levels of mu binding in the cortex and dorsal raphe nucleus, little mRNA is observed there. It is presumed that the receptors in these regions are localized presynaptically on neurons targeting these regions from elsewhere.
Studies in human brain are more limited. Overall, the expression patterns of MOR-1 mRNA in most regions of human brain are similar to those in the rat (Peckys and Landwehrmeyer, 1999). However, there is stronger labeling in human cortex and hippocampus than in the rat and weaker labeling of brain stem nuclei.
Expression levels of many of the variants generated by alternative splicing of the OPRM1 gene contain a premature termination codon that may subject them to regulation by nonsense-mediated mRNA degradation (NMD), a process that degrades a mRNA with a stop codon located more than 50 nucleotides upstream of the last exon-exon junction. Among a total of 32 full-length splice variants in the mouse OPRM1 gene, 11 are subjective to be regulated by NMD (Table 12). Two of the 17 splice variants in the rat OPRM1 gene and 4 of the 20 variants in the human ORPM1 gene are potential targets (Table 12). NMD may account for low expression levels of these NMD-targeted variant mRNAs within the brain. It remains to be seen whether the marked differences of expression of the variants targeted by NMD, such as mMOR-1E and mMOR-1V, reflect region-specific NMD or other RNA processing mechanisms.
TABLE 12.
MOR-1 splice variants subject to nonsense-mediated mRNA degradation
| Mouse OPRM1 Variant | Rat OPRM1 Variant | Human OPRM1 Variant |
|---|---|---|
| mMOR-1E | rMOR-1D | SV1 |
| mMOR-1Eiii | rMOR-1Z | SV2 |
| mMOR-1Eiv | hMOR-1Y | |
| mMOR-1F | hMOR-1Z | |
| mMOR-1Q | ||
| mMOR-1R | ||
| mMOR-1T | ||
| mMOR-1T(D2)* | ||
| mMOR-1Vii | ||
| mMOR-1W | ||
| mMOR-1Z |
mMOR-1T(D2) is similar to mMOR-1 except that it contains additional exons 6/7/19/8/9 downstream of exon 4 in the 5′-untranslated region (see Fig 16).
2. Immunohistochemistry.
The anatomic distribution of mu opioid binding sites was established long before the cloning of MOR-1. However, immunohistochemical approaches offer a number of advantages and provide an important validation. Whereas autoradiographic approaches are dependent upon the binding characteristics of the receptor, immunohistochemistry documents the presence of an epitope, a short amino acid sequence specific to the receptor. This epitope may, or may not, correspond to an intact and functional receptor. The immunohistochemical distribution of MOR-1 was evaluated by several groups (Arvidsson et al., 1995b; Mansour et al., 1995b; Ding et al., 1996; Moriwaki et al., 1996). The antisera in these studies were generated against epitopes on the C terminus encoded by exon 4. Attempts to generate antibodies against the N terminus or the extracellular loops have proven difficult, presumably because of the extensive glycosylation of the extracellular surface of the receptor. Overall, the anatomic distribution of the immunoreactivity corresponds quite well with autoradiographic studies, although some discrepancies exist. For example, autoradiographic binding studies indicate high levels of binding in the cerebral cortex, the lateral and basolateral nuclei of the amygdala, and the medial geniculate nucleus of the thalamus, although these regions show little or no immunoreactivity. These differences might simply reflect technical and/or sensitivity issues, but they may also be explained by the multiple MOR-1 splice variants. The full-length MOR-1 splice variants bind opiates with affinities and selectivities similar to MOR-1 and would be labeled autoradiographically. Yet they do not contain exon 4 and would not be recognized by the antibodies used in these early studies. Differences in the regional distribution of some these splice variants have been reported (Abbadie et al., 2000a,b,c, 2004; Zhang et al., 2006).
The opposite also is seen, with several regions showing immunoreactivity but no mu opioid binding, including the dentate gyrus of the hippocampus, stratum oriens and radiatum of Ammon’s horn, some pretectal nuclei, and the accessory facial nucleus (Mansour et al., 1995b; Ding et al., 1996). Again, these variations may be due to technical differences, but it is important to note that there are splice variants containing exon 4 (MOR-1G) that, when expressed alone, do not bind traditional mu opiates (Majumdar et al., 2011). Thus, autoradiographic and immunohistochemical approaches are complementary.
The in situ hybridization distributions correlate well with the immunohistochemistry (Arvidsson et al., 1995b; Mansour et al., 1995a; Ding et al., 1996; Moriwaki et al., 1996). Yet several mismatches exist, such as the dorsal horn of the spinal cord. Here, the superficial layers I and II are labeled autoradiographically and immunohistochemically but contain little MOR-1 mRNA (Arvidsson et al., 1995b; Mansour et al., 1995b; Ding et al., 1996). This mismatch is most easily explained by the presence of presynaptic receptors on nerve terminals from neurons projecting from the dorsal root ganglion and from rostral sites (Gamse et al., 1979; Fields et al., 1980; Besse et al., 1990).
3. Developmental Expression of MOR-1.
Early studies examining the developmental appearance of mu receptors used receptor binding and autoradiographic approaches (Gintzler et al., 1980; Bardo et al., 1981; Kent et al., 1981; Zhang and Pasternak, 1981). Although helpful, most of these studies were limited by technical constraints that were solved with the cloning of MOR-1. With more sensitive probes it was possible to establish the spatial and temporal expression of MOR-1. For example, in situ hybridization studies reveal the expression of MOR-1 mRNA as early as embryonic day E10.5 in the facial-vestibulocochlear preganglion complex of the mouse, 1 day later than the expression of KOR-1 mRNA in the gut epithelium (E9.5) and 2 days earlier than the expression of DOR-1 mRNA in peripheral tissues (E12.5) (Zhu et al., 1998). MOR-1 mRNA labeling in most regions resembles that in adult brain by E17.5 (Zhu et al., 1998). In these studies, in situ hybridization detects MOR- 1 mRNA earlier (E10.5) than receptor binding (E12.5), illustrating the importance of the sensitivity of the approach. This is further supported by the ability of reverse-transcription polymerase chain reaction (RT-PCR) to detect MOR-1 mRNA as early as E8.5 or E9.5 (Ko et al., 2002; Xu et al., 2006). The MOR-1 mRNA in striatal anlage is homogeneously labeled at E14–E19, but starts showing the characteristics patches seen autoradiographically in adults by E20–E21 (Georges et al., 1998).
VII. The OPRM1 Gene
Initially, the mu-receptor gene OPRM1 was defined by MOR-1 and was considered to contain only four coding exons and three introns. Subsequent studies have isolated a number of splice variants, extending the gene at both the 5′ and 3′ regions (Fig. 15). Currently, the mouse OPRM1 gene contains at least 18 exons spanning over 270 kb, whereas the human one is composed of at least 12 exons over 210 kb. It remains to be seen whether additional exons will be uncovered. The gene is complex, with two independent promoters, one associated with exon 1 and the other with exon 11. This is important in understanding some of the pharmacology seen in the knockout animals, because there are models that selectively delete exon 1-associated variants or exon 11-associated variants.
A. Chromosomal Mapping of OPRM1
A single copy of the mu-opioid receptor gene (OPRM1) is present in a wide range of species. The human OPRM1 initially was mapped to 6q24–25 using in situ hybridization (Wang et al., 1994a), whereas the mouse gene is on proximal chromosome 10, a region corresponding to the human chromosome 6q by linkage analysis using different interspecific backcrosses (Kozak et al., 1994; Giros et al., 1995; Kaufman et al., 1995). Morphine analgesia and hypothermia is mapped to the same location in proximal chromosome 10 in the mouse, supporting their association with the OPRM1 gene (Berrettini et al., 1994; Belknap et al., 1995). Their precise chromosomal localization was then determined by sequence analysis of the genome databases, mapping the gene in over 30 different species. The mouse OPRM1 gene is located at 3.27–3.61 Mb of chromosome 10, and the human OPRM1 gene is located at 154.3–154.7 Mb of chromosome 6.
Despite its localization to different chromosomes among species, adjacent gene loci are relatively conserved among mammalian species. For example, analysis of the mammalian genes reveal that the OPRM1 gene locus is flanked by a regulator of G protein signaling 17 (RGS17) gene and subunit 5 of the splicing factor 3b (SF3b5) gene with similar distances at the 5′ and 3′ regions, respectively (Herrero-Turrion and Rodríguez, 2008). The IPCEF1 gene overlaps with the OPRM1 gene at the 3′ ends with opposite orientations in several mammalian species (Pan, 2005), raising questions about their respective regulation during chromatin remodeling and transcription.
B. Promoters
The regulation of the OPRM1 gene is complex. Although initial studies identified and characterized the promoter associated with exon 1, the subsequent identification of exon 11 and its associated variants led to the identification of a second upstream promoter associated with exon 11 (Fig. 19). Thus, the gene has two independent promoters that modulate the expression of two sets of MOR-1 splice variants.
Fig. 19.
Schematic of the OPRM1 gene promoters in mice. Exons 11 and 1 are indicated by black boxes. The 5′-flanking regions and defined promoter regions are shown by lines. Transcription start points (tsp) and translation start codon (ATG) are indicated by arrows. Cis-acting elements that bind to corresponding trans-acting factors are shown by boxes of various shapes/colors along the promoter regions. TATA, TATA-binding protein; AP-1, activator protein 1; AP-2, activator protein 2; CCAAT, CCAAT box binding factors; C/EBP, CCAAT/enhancer-binding proteins; cMyc/Max, cMyc and Max factors; NRSE, neurorestrictive silencer element; Oct-1, octamer-1; Sp1, specificity protein 1; Sp3, specificity protein 3; CRE, cyclic adenosine monophosphate (cAMP) response element binding protein; 34 bp, 34-bp element; Sox, Sry-like high-mobility-group box gene; PARP1, poly(ADP-ribose polymerase 1; PCBP, polyC-binding protein; STAT1, signal transducers and activators of transcription type 1.
1. Exon 1 Promoter.
The exon 1-associated (E1) promoter is located to a ∼1.5-kb region immediately upstream of exon 1 and has been the focus of many investigations in mouse, rat, and humans (Min et al., 1994; Kraus et al., 1995; Liang et al., 1995; Wendel and Hoehe, 1998; Mayer et al., 1996; Wei and Loh, 2011) and has been reviewed (Wei and Loh, 2011). The E1 promoter conforms to a dual-promoter model with a separation of approximately 500 bp between the two active regions and is conserved at the nucleotide level in both mouse and human (Ko et al., 1997; Liang and Carr, 1997; Xu and Carr, 2001). The activity of the proximal promoter is stronger than the distal promoter (Liang and Carr, 1997; Xu and Carr, 2001; Ko et al., 2002). Ribonuclease protection assays and RT-PCR reveal multiple transcriptional start points (Min et al., 1994; Kraus et al., 1995; Mayer et al., 1996). The E1 promoter exhibits many characteristics of a “housekeeping gene,” with cis-acting elements with high GC content such as Sp1 and Ap1 but lacking a TATA box.
The mouse E1 promoter contains proximal and distal regions with a number of cis-acting elements regulating promoter activity such as the Sp binding element, NF-κB, cyclic adenosine monophosphate response element-binding protein, Oct-1, interleukin (IL)-4, Sox, STAT6, and neurorestrictive suppressor element (NRSE) (Liang and Carr, 1996; Ko et al., 1998; Andria and Simon, 2001; Kraus et al., 2001, 2003; Lee et al., 2003; Kim et al., 2004a). For example, expression in nonneuronal cells is suppressed by binding of the neuron-restrictive silencer factor (NRSF) to a 21-bp cis-acting NRSE element in the proximal promoter region (Kim et al., 2004a). The binding of Sp1 and Sp3 to a 10-bp Sp binding element in the proximal promoter stimulated promoter activity while the same Sp binding element interacted with the M1 and M2 isoforms of Sp3 to suppress the promoter activity (Choi et al., 2005). A number of transacting factors also have been identified. By use of DNA affinity purification coupled with matrix-assisted laser desorption ionization/time of flight mass spectrometry, two repressors, αCP3 and poly(ADP-ribose) polymerase-1 (PARP-1), were found (Choi et al., 2008a,b).
Cytokines, including the interleukins, tumor necrosis factor, and interferon-γ, also modulate MOR-1 expression through the induction of transacting factors that act through their corresponding cis-acting elements (Kraus, 2009). For example, IL-4-induced de novo synthesis of MOR-1 transcripts is induced in a number of immune cells by IL-4 acting through a STAT6 cis-acting element with the induced STAT6 factor (Borner et al., 2004). Likewise, MOR-1 transcripts are increased by TNF, which induces NF-κB, which then interacts with three NF-κB binding sites (Kraus et al., 2003).
The expression of MOR-1 is also under epigenetic control, with DNA methylation and histone modification in the E1 promoter region accounting for silencing and activating the gene in undifferentiated and differentiated P19 cells, respectively, and for the unique regional expression of MOR-1 mRNA in some brain regions (Hwang et al., 2007, 2009), with methyl-CpG-binding protein 2 and the chromatin remodeling factors, including Brg1 and Dnmt1, playing a role.
2. Exon 11 Promoter.
After the identification of the exon 11-associated splice variants (section VIII.B.1), a second promoter associated with exon 11 (E11) was located approximately 30 kb upstream of exon 1 and characterized (Pan, 2002). The activity of the E11 promoter correlates well with the expression of the exon 11-associated splice variants, as seen by its greater activity in neuronal-like cells, such as the neuroblastoma cell lines NIE-115 and SHSY-5Y, than in Chinese hamster ovary (CHO) cells. The promoter has a basal core region with a putative TATA box, a negative region, and a positive region. The TATA box appears to be crucial for the E11 promoter activity, as revealed by mutational analysis. There are several other cis-acting elements near the TATA box that modulate activity, including NF-1 and cMyc/Max. It has been proposed that a preinitiation complex is assembled through binding TBP to the TATA box and the subsequent recruitment of other general transcription factors to the basal core region, whereas the assembling of the transcription complex and initiation is further regulated by cMyc-Max-like and NF-1-like proteins (Pan, 2002).
There are clear differences between the E1 and the E11 promoters. First, the exon 11 promoter contains a TATA box, whereas the exon 1 promoter does not. Second, the exon 11 promoter contains a single major transcription start point, contrasting with the multiple transcription start points in the exon 1 promoter. Third, the exon 1 promoter has several GC-rich cis-acting elements like Sp1 and AP-2 that are involved in transcriptional regulation in TATA-less promoters that are absent in the exon 11 promoter. Thus, the E11 promoter appears to be a eukaryote class II promoter associated with RNA polymerase II, whereas the E1 promoter seems to be similar to a housekeeping gene. Finally, each promoter controls the expression of a different set of splice variants.
Further evidence for differences are seen in the temporal expression of exon 11- and exon 1-associated mRNA transcripts in C57BL/6J mice during ontogeny and in a transgenic mouse model (Xu et al., 2006). The expression of the exon 11 transcripts (E13.5) can be detected embryonically 4 days later than exon 1 promoter-driven transcripts (E9.5). A transgenic model incorporating a construct, in which a 3.7-kb exon 11 promoter and a 8.9-kb exon 1 promoter drove expression of tau/LacZ and tau/green fluorescent protein reporters, respectively, further documents differences in activity in some brain regions such as the hippocampus and substantia nigra.
VIII. Alternative Splicing of the OPRM1 Genes
Pharmacological studies suggesting mu-opioid receptor subtypes are hard to reconcile with a single copy of the OPRM1 gene. Thus, it is not surprising to find that the OPRM1 gene undergoes extensive alternative splicing (Figs. 16–18). The similar splicing patterns in rodents and humans imply that alternative splicing is important evolutionarily and functionally. Of the three opioid receptor genes, only the mu receptor undergoes extensive alternative splicing of coding exons. Indeed, the complexity of the splicing far exceeds the pharmacologically implied subtypes. Three sets of variants have been identified in both rodents and in humans. One comprises the full-length 7TM variants that all contain exons 1, 2, and 3 that encode the N terminus and all transmembrane domains with extensive 3′ splicing at the tip of the C terminus in which exon 4 is replaced by a series of alternative exons. The second set involves exon 11-associated variants with their 5′ splicing, many of which generate truncated 6TM proteins because of the absence of exon 1 due to exon skipping. The last group comprises truncated variants with a single transmembrane domain encoded by exon 1, also a result of exon skipping. The first evidence of alternative splicing of MOR-1 came from the human variant hMOR-1A (Bare et al., 1994) and the rat variant rMOR-1B (Zimprich et al., 1995), with similar variants in mice (Pan et al., 1999, 2005b; Pan, 2005).
A. Full-length Variants and 3′ Splicing
1. Rodent.
The mouse OPRM1 gene undergoes extensive 3′ splicing, in which exon 4 is replaced by a series of alternative exons (Fig. 16) (Y.-X. Pan, J. Xu, M. Xu, R. Yu, E. Bolan, A.K. Gilbert, and G.W. Pasternak, submitted manuscript; Pan et al., 1999, 2000, 2005b; Kvam et al., 2004; Doyle et al., 2007a,b; Pan and Pasternak, 2011). All of the full-length variants contain exons 1, 2, and 3, which encode the entire MOR-1 receptor except for the terminal 12 amino acids. Thus, all of the full-length variants contain identical binding pockets, consistent with the high affinity of all of the variants for mu opioids. Although many of the variants share 3′ exons, the amino acid sequences downstream of those encoded by exon 3 are all unique for each variant due to reading-frame shifts and/or early termination (Table 13). It is interesting that the length of the C terminus encoded by these alternatively spliced exons varies extensively, from only two amino acids in mMOR-1B5 to 88 in mMOR-1U with some containing putative phosphorylation sites for protein kinase C, cAMP- and cGMP-dependent protein kinase, and casein kinase II. A variety of splice patterns exist with the full-length variants, including intron-retention, exon inclusion/skipping, and alternative 3′ splicing.
TABLE 13.
Amino acid sequences downstream of exon 3 resulting from 3′ splicing of OPRM1
The amino acid sequences resulting from 3′ splicing are presented. Although some of the variants share the same downstream exons, the translated amino acid sequences for all sets of exons are unique because of frame shifts and the early termination. Note that 5′ splicing results in several variants with identical exon compositions downstream of exon 3 and these all share the same amino acid sequences.
| Species | Variant | Exons | Amino Acid Sequence Downstream of Exon 3 |
|---|---|---|---|
| Mouse | mMOR-1 | 4 | LENLEAETAPLP |
| mMOR-1A | 3b | VCAF | |
| mMOR-1B1 | 5a | KIDLF | |
| mMOR-1B2 | 5ab | KLLMWRAMPTFKRHLAIMLSLDN | |
| mMOR-1B3 | 5abc | TSLTLQ | |
| mMOR-1B4 | 5abcd | AHQKPQECLKCRCLSLTILVICLHFQHQQFFIMIKKNVS | |
| mMOR-1B5 | 5abcde | CV | |
| mMOR-1C | 7/8/9 | PTLAVSVAQIFTGYPSPTHVEKPCKSCMDRGMRNLLPDDGPRQESGEGQLGR | |
| mMOR-1D | 8/9 | RNEEPSS | |
| mMOR-1E/Eii/Eiiii/Eiv | 6/7/8/9 | KKKLDSQRGCVQHPV | |
| mMOR-1F | 10/6/7/8/9 | APCACVPGANRGQTKASDLLDLELETVGSHQADAETNPGPYEGSKCAEPLAISLVPLY | |
| mMOR-1H/-1i/-1J | 4 | LENLEAETAPLP | |
| mMOR-1O | 7ab | PTLAVSVAQIFTGYPSPTHVEKPCKSCMDR | |
| mMOR-1P | 15 | IMKFEAIYPKLSFKSWALKYFTFIREKKRNTKAGALPPLPTCHAGSPSQAHRGVAAWLLPLRHMGPSYPS | |
| mMOR-1V/-1Vii | 18/6/7/8/9 | KQEKTKTKSAWEIWEQKEHTLLLGETHLTIQHLS | |
| mMOR-1U | 7/19/8/9 | PTLAVSVAQIFTGYPSPTHVEKPCKSCMDSVDCYNRKQQTGSLRKNKKKKKRRKNKQNILEAGISRGMRNLLPDDGPRQESGEGQLGR | |
| mMOR-1W(D2) | 18/6/7/8/9 | LAFGCCNEHHDQR | |
| mMOR-1T(D2) | 4 | LENLEAETAPLP | |
| Human | hMOR-1 | 4 | LENLEAETAPLP |
| hMOR-1A | 3b | VRSL | |
| hMOR-1B1 | 5a | KIDLFQKSSLLNCE | |
| hMOR-1B2 | 5ab | RERRQKSDW | |
| hMOR-1B3 | 5abc | GPPAKFVADQLAG | |
| hMOR-1B4 | 5abcd | S | |
| hMOR-1B5 | 5abcde | VELNLDCHCENAKPWPLSYNAG | |
| hMOR-1O | O | PPLAVSMAQIFTRYPPPTHREKTCNDYMKR | |
| hMOR-1X | X | CLPIPSLSCWALEHGCLVVYPGPLQGPLVRYDLPAILHSSCLRGNTAPSPSGGAFLLS | |
| hMOR-1Y | Y/5abc | IRDPISNLPRVSVF | |
| hMOR-1i | 4 | LENLEAETAPLP | |
| Rat | rMOR-1 | 4 | LENLEAETAPLP |
| rMOR-1A | 3b | VCAF | |
| rMOR-1B1 | 5a | KIDLF | |
| rMOR-1B2 | 5ab | EPQSVET | |
| rMOR-1C1 | 5abc | PALAVSVAQIFTGYPSPTHGEKPCKSYRDRPRPCGRTWSLKSRAESNVENHFHCGAALIYNNVNFI | |
| rMOR-1C2 | 5abcd | PALAVSVAQIFTGYPSPTHGEKPCKSYRDRPRPCGRTWSLKSRAESNVENHFHCGAALIYNNELKIGPVSWLQMPAHVLVRPW | |
| rMOR-1D | 5abcde | T | |
| rMOR-1H1/H2/i1/i2/i3 | 4 | LENLEAETAPLP | |
| rMOR-1P | 15 | GAEL |
Splicing in the rat shows a similar pattern to that in mice (Fig. 17) (Pasternak et al., 2004; Pan, 2005; Xu et al., 2011). The first variant reported was rMOR-1B, in which exon 4 was replaced by exon 5, leading to a different set of C-terminal amino acids (Zimprich et al., 1995). The mouse also contains an exon 5 homolog, leading to mMOR-1B. However, alternative splicing in exon 5 generates five different variants in the mouse (mMOR-1B1 through mMOR-1B5), two in rats (rMOR-1B1 and rMOR-1B2), and five in humans (hMOR-1B1 through hMOR-1B5) with different C-terminal amino acid sequences depending upon which splice site within exon 5 is used (Pan, 2005; Pan et al., 2005b).
mMOR-1A, rMOR-1A, and mMOR-1O are intron-retention variants with silent donor splice sites in exons 3a and 7a, respectively. Splicing at exon 18 provides yet another example of alternative 3′−splice sites that generate two different C-terminal variants, mMOR-1V and mMOR-1W (Doyle et al., 2007a).
The exon composition of rat rMOR-1C1 is identical to that in the mouse, with a high homology at the amino acid level for the 30 amino acids encoded by rat exon 7 compared with the mouse exon 7 (83%) and human exon O (67%) (Pasternak et al., 2004). However, the amino acid composition of the rat exon 8 differs from the mouse exon 8, with exons 8 and 9 in the rat in rMOR-1C1 yielding 35 amino acids compared with only 22 in the mouse homolog. The rat also differs from the mouse with the isolation of a second variant, rMOR-1C2, containing the same exon composition as rMOR-1C1 except for an alternative splice site upstream of exon 9a that results in insertion of exon 9b (Pasternak et al., 2004). Therefore, both variants share identical protein sequences through exon 8, but differ in the sequences encoded by exon 9a (four amino acids) in rMOR-1C1 or exon 9b (21 amino acids) in rMOR-1C2. rMOR-1D has a splicing pattern resembling mMOR-1D, but translation of the rat exon 8 predicts a single threonine residue (Pasternak et al., 2004). The splice pattern of rMOR-1P also is similar to that in the mouse.
2. Human.
The human splicing patterns are similar to those in rodents, with alternative 3′-splicing, intron-retention and exon skipping (Fig. 18). hMOR-1A was the first splice variant isolated (Bare et al., 1994), leading to the identification of its homolog in both mice and rats. This, in turn, led to the subsequent cloning of a range of human variants, most from human neuroblastoma cell lines (Cadet et al., 2003; Pan et al., 2003, 2005a; Choi et al., 2006; Chou et al., 2006; Shabalina et al., 2009; Xu et al., 2009, 2011). As in rodents, most of the variants are generated through the exon 1 promoter and produce full-length G protein-coupled receptor variants in which exon 4 is replaced with a variety of alternative exons. As in rodents, the human exon 5 has five alternative 3′-splice sites (5a, 5b, 5c, 5d, and 5e), generating five C-terminal variants (hMOR-1B1–hMOR-1B5). Although highly homologous, the predicted amino acid sequence of the human exon 5a contains an extra 13 amino acids beyond the five amino acids that are identical to those predicted from the mouse and rat exon 5a. Unlike hMOR-1B1, the nucleotide sequences and predicted amino acid sequences from human exons 5b, 5c, 5d, and 5e and hMOR-1B2, hMOR-1B3, hMOR-1B4, and hMOR-1B5 also differ from the mouse homologs.
The human exon O, a homolog of the mouse exon 7a and the rat exon 7, is located 128 kb downstream of exon 4, a distance very close to the 131 and 120 kb between exon 4 and the exon 7 homologs in mouse and rat, respectively. The predicted sequence of the 30 amino acids in the human exon O share ∼65% identity with the corresponding sequences predicted from exon 7a in mMOR-1O or mMOR-1C and exon 7 from rMOR-1C1 (Pan et al., 1999, 2003; Pasternak et al., 2004). One difference between the human splicing and that in rodents is that the downstream exons 8 and 9 present in rodents have not yet been identified in the human OPRM1 gene.
Exon X in humans is located ∼2 kb downstream of exon 3, a distance similar to exon 15 in the mouse (Pan et al., 2005b). Despite some homology between exon X and exon 15 at the nucleotide level, their predicted amino acids differ significantly. The insertion of exon Y between exons 3 and 5c distinguishes hMOR-1Y from hMOR-1B3 (Pan et al., 2005b).
There also is an exon 11-associated full-length rat variant, rMOR1H2. Translation from the exon 11a AUG predicts a novel full length, 7TM N-terminal variant containing an additional 50 amino acids at the N terminus compared with the original rMOR- 1 (Xu et al., 2011), a structure similar to that of hMOR-1i. In vitro translation studies reveal that the exon 11a AUG is the preferential initiation site rather than the exon 1a AUG. When expressed in CHO cells, the additional 50 amino acids in rMOR-1H2 do not affect opioid binding affinity, but they do alter agonist-induced G protein activation, with an increased potency in agonist stimulated [35S]GTPγS binding and no change in maximal levels (Xu et al., 2011).
Similar to the rat, there is an exon 11-associated full-length human variant generated by 5′ splicing, hMOR-1i (Xu et al., 2009). Similar to the rat rMOR-1H2, the human variant hMOR-1i has an extended N terminus due to the loss of 105 bases (exon 1b) spliced out from exon 1. Translation from exon 11 predicts a small peptide because of early termination. However, the ATG from exon 1c, which is 279 bases upstream of the initiation ATG in hMOR-1, predicts a novel protein sequence of 93 amino acids fused immediately upstream to those encoded in hMOR-1, leading to a greatly elongated N-terminal length, with the remainder of the sequence identical to hMOR-1. The additional N-terminal sequence does not contain any predicted glycosolation sites or transmembrane regions, consistent with a classic GPCR structure. The generation of the protein has been confirmed with in vitro translation where proteins translated from the upstream AUG in exon 1c predominate over those from the typical AUG in exon 1a. In these studies, two bands comparable to products using the first and second AUGs of exon 1c are seen. When expressed in CHO cells, the extra 93 amino acids in hMOR-1i do not affect opioid binding affinities but significantly change agonist-induced G protein activation. Whereas the potencies of several opiates (morphine and buprenorphine) for stimulation of [35S]GTPγS in hMOR-1i is similar to that in MOR-1, the potency of β-endorphin, dynorphin A, and M6G is reduced by 3- to 4-fold (Xu et al., 2009).
B. Truncated MOR-1 Variants
1. Exon 11-Associated 6TM Variants.
In addition to the 3′ splicing that leads to full-length variants with 7TM domains, two classes of truncated variants have been identified in rodents and humans. These include variants containing exon 1 that encode a single transmembrane (1TM) protein and exon 11-associated variants that predict a 6TM protein. As noted above, exon 11 is located approximately 30 kb upstream of exon 1 and is under the control of its own promoter, which is distinct from the promoter responsible for generating the exon 1-associated full-length variants.
In the mouse, exon 11 and its variants were initially identified using 5′−RACE (rapid amplification of cDNA ends) with primers located in exon 2 (Fig. 16) (Pan et al., 2001; Pan, 2005). These studies yielded an additional three exons, exons 12, 13, and 14, as well as nine exon 11-associated splice variants in which exon 11 is the first exon. Exon 12 is located ∼28 kb upstream of exon 1, just 2 kb downstream from exon 11, whereas exons 13 and 14 are located 35 and 36 kb downstream of exon 1, respectively.
The mouse has three exon 11-associated variants that contain exons 1, 2, 3, and 4 (mMOR-1H, mMOR-1i, and mMOR-1J). Translation from the AUG in exon 1 in these variants generates the same protein as MOR-1 itself. Translation from exon 11, on the other hand, yields a short protein (<10 kDa) due to early termination of translation. Although both sets of proteins can be documented using in vitro translation, the smaller peptides have not been identified within the brain, suggesting that the major product of these variants is the same protein as MOR-1 but under the control of a different promoter (Pan et al., 2001).
The remainder of the mouse exon 11-associated variants generates 6TM proteins that also undergo 3′ splicing. Three proteins, mMOR-1G, mMOR-1M, and mMOR-1N, have exon 11 spliced directly to exon 2, with differing C termini (Table 14). Whereas mMOR-1G has a terminal exon 4, mMOR-1M has exons 7/8/9, the same 3′ composition as mMOR-1C, and mMOR-1N has exons 8/9, the same 3′ composition as mMOR-1D (Pan et al., 2001, 2005b). Western blots using an antiserum against exon 11 suggest that these variants are expressed in mouse brain (Abbadie et al., 2004). Although mMOR-1K and mMOR-1L also lack exon 1, inclusions of small exons between exons 11 and 2 (exon 13 in mMOR-1K and exon 14 in mMOR-1L) lead to early termination within the small exons when using the exon 11 AUG and short peptides (<10 kDa). However, they may still be translated starting with a methionine residue at the beginning of exon 2, which leads to a 6TM protein identical to that of mMOR-1G, with the exception of the lack of the exon 11 coding sequence. This is analogous to the mu3 receptor proposed by Stefano and co-workers (Cadet et al., 2003).
TABLE 14.
Amino acid sequences resulting from 5′ splicing of OPRM1
The exon 11 sequences are immediately upstream of exon 2, as noted. The N-terminal fusion sequences exon 11-associated splice variants are found in the rat and human variants, as noted. In the human variant hMOR-1i, translation is initiated at the AUG at the beginning of exon 1c, which encodes 49 aa,and the proceeds through exon 1a, with an additional 44 aa generated by the exon 1a sequence immediately upstream of the AUG used to initiate translation in MOR-1. In the rat variant rMOR-1H2, the additional sequence comes from exon 11a, which is joined to exon 1. Translation in the full exon 11a/b leads to a truncated protein corresponding to only the first 7 aa shown for exon 11a alone. The * designates the termination of translation due to a stop codon, and the indicated peptide is the complete product.
| Species | Exon | Variant | Amino Acid Sequence |
|---|---|---|---|
| Exon 11 sequences | |||
| Mouse | Exon 11 | mMOR-1G/M/N | MMEAFSKSAFQKLRQRDGNQEGKSYLR |
| Human | Exon 11a/b | hMOR-1G1 | MMRAKSISTKAGKPSRFIWKKILL* |
| Exon 11a | hMOR-1G2 | MMRAKSISTKAGKPSR | |
| Rat | Exon 11a | rMOR-1G2 | MGSGPML |
| Exon 11a/b | rMOR-1G1 | MGSGPML* | |
| N-terminal fusion sequences | |||
| Human | Exon 1c/a | hMOR-1i | MCLHRRVPSEETYSLDRFAQNPPLFPPPSLPASESRMAHAPLLQRCGAARTGFCKKQQELWQRRKEAAEALGTRKVSVLLATSHSGARPAVST |
| Rat | Exon 11a | rMOR-1H2 | MGSGPMLAGPCKNLTEPRAAVRGRGWGAWNPKSLSALSYSLPSPQQAFST |
Similar variants have been identified in rat (Fig. 17) (Xu et al., 2011). After mapping a mouse exon 11 homolog in the rat to a locus approximately 21 kb upstream of exon 1, a distance similar to that in the mouse and humans, we isolated several rat exon 11-associated variants (Xu et al., 2011). Although rMOR-1G1 and rMOR-1G2 have similar splicing patterns, they use alternative 5′ splice sites in exon 11. By use of the exon 11a AUG as the translational start codon, rMOR-1G2 encodes a 6TM protein. The predicted proteins from rMOR-1G1 are more complicated. rMOR-1G1 contains exons 11a/11b/2/3/4. If translation is initiated from the AUG in exon 11a, a small peptide of only seven amino acids is predicted based upon the stop codon in exon 11b. However, rMOR-1G1 might still be able to generate a 6TM protein by initiating translation from the first AUG in exon 2, a situation analogous to mMOR-1K, mMOR-1L, hMOR-1G1, hMOR-1K, and the mu3 receptor.
Five rat variants contain exons 11, 1, 2, 3, and 4, with alternative splicing among exons 11a, 11b, 1a, 1b, and 1c generating different transcripts (Xu et al., 2011). Despite their different 5′nucleotide sequences, rMOR-1H1, rMOR-1i1, rMOR-1i2, and rMOR-1i3 all predict the same protein sequence as the original rMOR-1 when using AUG in exon 1a as the translational start codon. Translation in these variants from the exon 11a AUG predicts short proteins because of early termination of translation. Thus, four exon 11-containing transcripts encode the identical rMOR-1 protein as the original rMOR-1, a situation similar to the mouse with mMOR-1H, mMOR-1i, and mMOR-1J.
5′ Splicing of the human OPRM1 gene generates similar truncated variants (Fig. 18). Searching the human genome database reveals a mouse exon 11 homolog 28 kb upstream of exon 1 with the subsequent isolation of four full-length variants, hMOR-1G1, hMOR-1G2, hMOR-1i, and hMOR-1H by RT-PCR cloning (Xu et al., 2009). Splicing from two alternative 5′ splice sites in the human exon 11 (exons 11a and 11b) directly to exon 2 generates two 6TM variants hMOR-1G1 and hMOR-1G2, each lacking exon 1, which encodes TM1. Translation from exon 11a in hMOR-1G1 leads to termination of translation in exon 11b, but use of the AUG from exon 2 predicts a six TM protein, as in the mouse and rat variants.
Mu3 is a 6TM protein proposed by Stefano and colleagues (Cadet et al., 2003). It reportedly contains exons 2 and 3, followed by a new exon of 149 bp and a portion of exon 4 (202 bp). Translation from the AUG within exon 2 predicts a 6TM protein similar to hMOR-1G1, with the exception of the replacement of the C-terminal 12 amino acids encoded by exon 4 by 26 amino acids encoded by the new exon. Although there is a consensus splice site upstream of the new exon, the exact splice sites between the new exon and exon 4 are not clear because of ambiguous sequences. When expressed in COS-1 cells, the mu3 clone shows only moderate binding affinity toward morphine (Ki 29 nM), naltrexone (Ki 31 nM), and naloxone (Ki 39 nM) using a [3H]dihydromorphine binding assay and displays functional properties similar to their earlier report (Cadet et al., 2003).
hMOR-1K contains an exon homologous to the mouse exon 13 (Shabalina et al., 2009). Located 3 kb upstream of exon 2, the human exon 13 has no predicted coding sequence. However, the use of the AUG within exon 2 predicts a 6TM protein identical to that of hMOR-1G1. It is noteworthy that a SNP located upstream of exon 13 contains a conserved internal ribosome entry site and influences expression of hMOR-1K at both the mRNA and protein levels. Together, there are at least four human 6TM variants indicated as follows: hMOR-1G1, hMOR-1G2, hMOR-1K, and mu3.
2. Single Transmembrane Domain Variants.
In addition to the 6TM truncated proteins, a series of single transmembrane domain variants have been identified in mice, rats, and humans (Figs. 16–18) (Du et al., 1996, 1997; Kvam et al., 2004; Pan, 2005; Pan and Pasternak, 2011; Xu et al., 2013). The transmembrane domain of all of them corresponds to TM1, which is encoded by exon 1. It is hard to imagine how these proteins could function independently as a receptor. However, they potentially are of interest. First, they contain TM1, the transmembrane domain missing in the 6TM variants, thus raising the interesting question of whether they could complement the 6TM variants to form a functional “full-length” receptor containing 7TM regions. This is particularly intriguing given that co-immunoprecipitation studies show that 1TM1 and 6TM variants can physically associate, although there is not yet evidence demonstrating that the complex comprises a functional receptor. Second are some findings by He et al. (2011). As discussed earlier, their construct containing the first TM of MOR-1 coupled to TAT (MORTM1-TAT) physically associates with DOR-1, thereby dissociating the MOR-1/DOR-1 heterodimers. In vivo, MORTM1-TAT increases morphine’s analgesic responsiveness and prevents morphine-induced analgesic tolerance. Thus, the 1TM variants may have interesting modulatory roles through influencing dimerization. More recent studies demonstrate the ability of the 1TM variants to enhance the expression of the full-length variant MOR-1 by a chaperone function, minimizing receptor turnover, and thereby play an important role in analgesia (section VIII.C.2) (Xu et al., 2013).
Five single TM mouse variants have been isolated, four of which are generated by exon skipping and are associated with the exon 1 promoter (Table 15) (Du et al., 1996, 1997; Kvam et al., 2004; Pan, 2005; Pan and Pasternak, 2011; Xu et al., 2013). Translation from the exon 1 AUG in mMOR-1Q, mMOR-1R, and mMOR-1Z, which lack exon 2, leads to termination of translation within exon 3 because of a frame shift. Thus, all three predict a single protein with one transmembrane domain consisting of the same N terminus and TM1 domain of MOR-1 from exon 1, followed by a carboxyl tail of 127 amino acids encoded by exon 3. The additional exons downstream of exon 3 contained within these variants are within the 3′-UTR. The rat contains a similar 1TM variant, rMOR-1Z.
TABLE 15.
Single TM variants of OPRM1: Amino acid sequences downstream of exon 1
The amino acid sequences downstream of exon 1 for the single TM variants are presented.
| Species | Variant | Amino Acid Sequence Downstream of Exon 1 |
|---|---|---|
| Mouse | mMOR-1Q/R/Z | VHRLHPHVLSSHMVLGEPAQNLCLHLRLHHAGPHHHCVLWTDDLTTQECPHAVGLQRKGQEPAQDHPDGAGGRGCIYCLLDPHPHLCHHQSTDHDSRNHFPDCFLALLHCLGLHKQLPEPSSLCVPG |
| mMOR-1S | S | |
| mMOR-1T | RSHMELPLYLPRLPSLHWSK | |
| Human | hMOR-1S | S |
| hMOR-1Z | FHRLYTNILSSNLVLGKPAEDLCFHLRLHYASAHHYRVLWTDDLAPQECPHALWLQRKGQESSKDHQDGAGGGGCVHRLLDSHSHLRHH | |
| SV1 | RYSWFVIGGPEGRRKQRRLGEDKRARGCGEKG | |
| SV2 | SSWF | |
| Rat | rMOR-1S | S |
| rMOR-1Z | VHRLHPHVLPPNLVLGEPAQNLCLYLRFHHADPHHHCVLRPDDLTTQERSHAIGLQRKGQESAQDHPDGAGGRGCIYRLLDPHPHLRHHQSADHDSRNHISDRFLALLHCFGLHEQLPESSSLRLPG |
The fourth exon 1-associated single TM variant, mMOR-1S, is generated by splicing from exon 1 directly to exon 4, skipping exons 2 and 3. Translation from the exon 1 AUG in mMOR-1S terminates immediately upon reaching exon 4 because of a reading-frame shift, producing a shorter C terminus than the variants lacking only exon 2. The rat contains a similar 1TM variant, rMOR-1S.
An exon 11-associated variant, mMOR-1T, has an exon composition similar to mMOR-1H, with the exception of the insertion of exon 16 between exons 1a and 2 (Pan, 2005). Translation from the exon 1 AUG terminates in exon 16, generating a single TM protein comprising 95 amino acids from exon 1 containing the N terminus and TM domain followed by an extension of the C terminus of 20 amino acids from exon 16. Initiating translation from the exon 11 AUG yields a short protein of 84 amino acids identical to mMOR-1H.
Four similar single TM variants have been isolated from humans (Choi et al., 2006; Du et al., 1997). Similar to its rodent homolog, hMOR-1S lacks exons 2 and 3, with exon 1 spliced directly to exon 4. It was first identified from the SH-SY-5Y neuroblastoma cell line, and these studies observed a mRNA abundance similar to hMOR-1 itself (Du et al., 1997). Similar to the mouse, hMOR-1Z skips exon 3, leading to a single TM variant containing TM1 with translation terminated within exon 3. The SV1 and SV2 variants were identified in human NMB cells, in which exon B or exons A/B are inserted between exons 1 and 2, respectively (Choi et al., 2006), also generating a single TM variant with termination of translation in exon B or A. Functionally, SV1 and SV2 reportedly modulate hMOR-1 binding when coexpressed, lowering [3H]diprenorphine binding.
C. Characterization of MOR-1 Splice Variants
The existence of the variants alone does not ensure that they are pharmacologically relevant, but several lines of evidence support their functional significance.
1. Regional Distribution of MOR-1 Variants.
Northern blots of MOR-1 yield different banding patterns depending upon the exon sequences used to probe the mRNA (Thompson et al., 1993; Mansour et al., 1994a; Raynor et al., 1995; Pan et al., 2001), supporting the concept of alternative splicing of the OPRM1 genes. In situ hybridization approaches examined the distribution of MOR-1 mRNA in the brain in a more anatomically relevant way (Mansour et al., 1994a; Minami et al., 1995; Zhu et al., 1998; Peckys and Landwehrmeyer, 1999). Looking back at these studies, interpreting them leaves a number of questions because most exons are shared among more than one variant, meaning that the hybridization probes may be labeling more than one species of mRNA, depending upon the actual probe sequence. Semiquantitative RT-PCR reveals marked regional differences for many of the variants, in contrast to the relatively universal distribution of mMOR-1 mRNA (Fig. 20) (Pan et al., 1999, 2000, 2001). For example, in mouse brain, the thalamus has high expression levels of mMOR-1C with little expression of mMOR-1D and mMOR-1E mRNAs (Pan et al., 1999). The striatum and hypothalamus highly express mMOR-1E mRNA, whereas mMOR-1D mRNA levels are higher in the cortex, brain stem, and periaqueductal gray (Pan et al., 1999). At the time of these studies, only the full-length variants were known. Now it is clear that some of the probes will amplify more than one variant because of 5′ splicing. In human brain, hMOR-1G2 is highly expressed in the prefrontal cortex, piriform cortex, nucleus accumbens, and pons but not in the temporal cortex and spinal cord (Xu et al., 2009). Although semiquantitative RT-PCR has a number of disadvantages and the quantification is only approximate, these findings imply that the OPRM1 gene undergoes region-specific splicing.
Fig. 20.
Regional mRNA splice variants. The regional distribution of the indicated splice variants was determined using RT-PCR, with β2-microglobulin as an internal control. Adapted from Pan et al. (1999, 2001).
Assessing the regional distribution of the variants at the protein level is not simple because of the combination of both 3′ and 5′ splicing. Autoradiography was used extensively in early studies, and the distributions were helpful in understanding opioid action. However, autoradiography is limited by its inability to distinguish among the full-length MOR-1 variants because they have similar binding affinities for the radiolabeled drugs. Greater accuracy can be achieved using imunohistochemistry (Arvidsson et al., 1995b; Ding et al., 1996; Mansour et al., 1995b; Moriwaki et al., 1996). However, this also has its limitations. The initial studies used antisera generated against exon 4. Although this antiserum labels MOR-1, it also labels truncated splice variants that also express the exon 4 epitope and will not label the other 3′-spliced variants. Finally, there has been much discussion on the pitfalls of immunohistochemistry and the potential of labeling unrelated proteins. Thus, these studies also must be interpreted cautiously.
Even with the above caveats, it is interesting to examine the immunohistochemical distribution of a number of variants using antisera against C-terminal epitopes or against exon 11 (Abbadie et al., 2000b,c, 2004; Zhang et al., 2006). For example, an antisera against the epitope of the C terminus in the exon 7/8/9 variants differ from the labeling using antisera against an exon 4 epitope in the medial eminence, thalamic nuclei, and nucleus ambiguus (Abbadie et al., 2000c). Even when variants are expressed in the same regions, such as the dorsal horn of the spinal cord, confocal microscopy reveals that the two antisera label different cell populations (Abbadie et al., 2000c). Another antiserum raised against a mouse exon 8/9 epitope uniquely labels the dentate gurus, the mossy fibers of the hippocampal formation, and the nucleus of the solitary tract (Abbadie et al., 2000b), whereas an antiserum raised against an exon 3/5a epitope predominantly labels the olfactory bulb (Schulz et al., 1998). The exon 4 epitope is predicted in eight mouse variants, four of which generate the identical protein as mMOR-1 (mMOR-1, mMOR-1H, mMOR-1i, and mMOR-1J), whereas the others can potentially generate truncated 6TM variants (mMOR-1G, mMOR-1i, and mMOR-1J). Similarly, the epitope in the exon 7/8/9 variants might label either mMOR-1C and/or mMOR-1M, whereas the epitope in the exon 8/9 variant could label either mMOR-1D and/or mMOR-1N. Distinguishing among these variants will be challenging.
It is interesting to note the apparent mismatch between mRNA and protein levels. For example, expression of mMOR-1C and mMOR-1M mRNA is lower than mMOR-1 mRNA (Pan et al., 1999, 2001). Yet, immunostaining of the epitope in the mouse exons 7/8/9 variants is readily seen and appears similar to that of mMOR-1 itself (Abbadie et al., 2000c). Although this may simply reflect the sensitivity of the two antisera, there may be differences in the relationship between mRNA and protein levels, possibly because of protein and/or mRNA stability issues.
Exon 4 and the exon 7/8/9 containing variants were also compared ultrastructurally, as well as their association with the peptide CGRP (Abbadie et al., 2001). The exon 7/8/9-LI in the superficial laminae of the spinal cord is localized almost exclusively presynaptically and colocalizes with calcitonin gene-related peptide (CGPR), whereas the labeling of the exon 4 epitope displays an equal pre- and postsynaptic distribution and does not show colocalization with CGPR.
2. Biochemical Characterization of MOR-1 Splice Variants.
With all the full-length variants containing the identical TM domains that define the binding pocket, one question is whether their C-terminal differences affect their affinities and/or selectivities for mu opioids. More intriguing, these C-terminal differences may affect biased signaling. The importance of β-arrestins in GPCR signaling has been extensively reviewed (Raehal et al., 2011; Reiter et al., 2012). It is noteworthy that the mu-opioid receptor was the first to reveal biased agonism (Bohn et al., 1999; Groer et al., 2007; Violin and Lefkowitz, 2007). Recently, Violin and colleagues reported the synthesis of a G protein-biased ligand, TRV130 ([(3-methoxythiophen-2-yl)methyl]({2-[(9R)-9-(pyridin-2-yl)-6-oxaspiro[4.5]decan-9-yl]ethyl})amine), with potent analgesic actions and fewer potential side effects, presumably attributed to its biased agonism (Dewire et al., 2013). Thus, biased agonism is likely to play a more important role in opioid pharmacology in the future. We do not yet know whether biased signaling will change for a drug when examined in different C-terminal splice variants, but considering the wide array of structural differences generated by 3′ splicing, it seems likely that they will. If so, this will add yet another layer of complexity to our understanding of opioid action.
When expressed in CHO cells, the full-length variants display similar affinities for the mu opiates and are mu selective, the one exception being mMOR-1B4 (Pan et al., 1999, 2000, 2003, 2005a,b; Pasternak et al., 2004; Xu et al., 2009). Small differences are seen among the variants for some of the endogenous opioid peptides. For example, both β-endorphin and dynorphin A show over 4- to 5-fold higher affinity for mMOR-1D than mMOR-1. On the other hand, hMOR-1 demonstrates a 2-fold higher affinity than hMOR-1X (Pan et al., 2003). The reasons for these differences are not clear because the binding pockets are identical, but differences in the C termini may produce subtle conformational changes, either alone or through their ability to recruit other proteins to the receptor complex.
Mu receptors are inhibitory, as seen by their ability to inhibit stimulated adenylyl cyclase activity (Blume et al., 1979; Law et al., 1981; Childers, 1991; Knapp et al., 1995; Standifer et al., 1996; Standifer and Pasternak, 1997). As with other GPCRs, mu agonists induce G protein activation, as demonstrated by stimulation of [35S]GTPγS binding (Childers, 1991). Although all the agonists stimulate [35S]GTPγS binding in the full-length variants, there are differences among the variants when testing individual drugs for both potency (i.e., EC50 values) and efficacy (i.e., maximal stimulation) (Abbadie et al., 2000c; Bolan et al., 2004; Pasternak et al., 2004; Pan et al., 2005a,b). Furthermore, the correlation between binding affinity (Ki) and potency (EC50) in the [35S]GTPγS binding assays is weak. For example, β-endorphin stimulates [35S]GTPγS binding over 20-fold more potently in rMOR-1 than in rMOR-1C1 despite its similar binding affinity (Pasternak et al., 2004), implying differences in their intrinsic activities because maximal [35S]GTPγS binding is seen at different receptor occupancies. There also is no obvious correlation between potency and efficacy. Finally, functional differences also are observed for the full length 5′-splice variants hMOR-1i and rMOR-1H2 with their extended N termini in agonist-induced [35S]GTPγS binding assays (Pan et al., 2003; Xu et al., 2009).
Mu-opioid receptors primarily couple to inhibitory G proteins, such as Gαi/Gαo, inhibiting adenylyl cyclase and decreasing intracellular cAMP levels, although they can have stimulatory effects under certain circumstances (Crain et al., 1988; Cruciani et al., 1993; Crain and Shen, 2000b). As with the stimulation of [35S]GTPγS binding, both potency (IC50 values) and efficacy (maximal inhibition) for a range of opioids differ among the full-length 3′-splice variants when examined for inhibition of forskolin-stimulated cAMP accumulation (Pan, 2005). However, the [35S]GTPγS binding results do not always correspond with those from the adenylyl cyclase assays. For example, β-endorphin is the more potent in [35S]GTPγS binding assays but more efficacious in adenylyl cyclase assays (Pan, 2005; Pan et al., 2005b). Although these differences may be attributed to a wide range of factors, including different assay conditions, they raise intriguing questions regarding the role of biased agonism and 3′ splicing.
MOR-1 trafficking has been extensively studied (Keith et al., 1996, 1998; Sternini et al., 1996; Whistler et al., 1999; Fan et al., 2005; Von Zastrow, 2011). As noted above, the inability of morphine to induce rapid internalization of MOR-1 is not universal among the MOR-1 splice variants (Abbadie and Pasternak, 2001; Tanowitz and Von Zastrow, 2003; Tanowitz et al., 2008). The efficient postendocytic sorting of MOR-1 is dependent upon a sequence termed the MOR1-derived recycling sequence (MRS), located at the tip of the C terminus and encoded by exon 4 (Tanowitz and Von Zastrow, 2003). Several C-terminal variants, including mMOR-1B, mMOR-1D, and mMOR-1E, lack the MRS because of alternative 3′ splicing and show inefficient receptor recycling after DAMGO-induced endocytosis in HEK293 cells, further supporting the functional roles of C-terminal splicing (Tanowitz et al., 2008). mMOR-1D and mMOR-1E display robust morphine-induced receptor phosphorylation and internalization compared with the lack of responsiveness of mMOR-1 (Koch et al., 2001). The inability of morphine to internalize mMOR-1C in this study contrasts with the internalization of mMOR-1C in the brain when the drug was given intracerebroventricularly (Abbadie and Pasternak, 2001), raising the interesting possibility that trafficking may be dependent upon the cell type in which the receptor is being expressed. It is important to remember that morphine will interact with all the full-length splice variants, so its inability to internalize one may not be predictive of its overall in vivo actions, which probably represent the summation of its actions at many variants simultaneously.
Similar to many receptors, phosphorylation is thought to be important in mu-opioid receptor regulation, having been implicated in a range of actions, including desensitization, tolerance, dependence, and internalization and biased agonism (Koch et al., 1998; Law et al., 2000; Deng et al., 2001; Von Zastrow et al., 2003; Wang, 2003; Bohn et al., 2004; Waldhoer et al., 2004; Violin and Lefkowitz, 2007; International HapMap3 Consortium, 2010; Law, 2011; Reiter et al., 2012). Most phosphorylation sites are contained within the intracellular loops and C terminus in regions encoded by exons 2 and 3. However, consensus sequences have suggested potential sites for β-adrenergic receptor kinase, protein kinase C, casein kinase, tyrosine kinase, and cAMP- and cGMP-dependent protein kinases within the alternatively spliced exons downstream from exon 3 (Pan et al., 1999, 2003, 2005b; Zimprich et al., 1995; Pasternak et al., 2004; Doyle et al., 2007a,b). Threonine 394, located at the C-terminal tip of MOR-1, is an excellent example. Encoded by exon 4, it is a major residue for DAMGO-induced receptor phosphorylation of rMOR-1, as shown by the loss of 90% of DAMGO-induced phosphorylation with a T394A mutation (Deng et al., 2000) and its correlation with the elimination of DAMGO-induced adenylyl cyclase superactivation (Wang et al., 2007). Serine 375, also located at the C terminus, can be phosphorylated by mu agonists, such as DAMGO, fentanyl, etonitazene, and morphine. However, in a knock-in mouse expressing the S375A mutant mu receptor, analgesic tolerance to high-efficacy agonists, including fentanyl and etonitazene, is attenuated, whereas morphine tolerance is unaffected, suggesting that phosphorylation may differentially mediate the actions of various mu agonists (Grecksch et al., 2011) Thus, 3′ alternative splicing clearly affects phosphorylation patterns and internalization of the various variants.
3. Role of Alternative Splicing of MOR-1 in Opioid Analgesia.
A number of approaches have correlated the various cloned opioid receptors with analgesia. The first approach involved antisense, followed by the generation of a series of knockout mice. Each approach has its advantages and disadvantages. Antisense approaches can be used in a mature animal, avoiding potential compensatory developmental changes. However, it is only a partial knockdown, and when dealing with administration in the brain its targets are diffusion limited. Antisense mapping provides a valuable approach in assessing the potential role of splice variants functionally. It was initially demonstrated with the delta-opioid receptor DOR-1 shortly after it was cloned (Standifer et al., 1994) and has been used to explore the effects of splice variants of neuronal nitric oxide on opioid analgesia (Kolesnikov et al., 1997, 2009; Pasternak and Pan, 2000; Pasternak and Kolesnikov, 2005). It provides a means of selectively targeting different regions of the mRNA, enabling the selective downregulation of only those splice variants containing the target sequence. It is even possible to target the splice site junction between two exons and selectively target a variant without a unique exon (Kolesnikov et al., 1997; Xu et al., 2013).
Knockouts have a complete loss of the targeted sequences, but their absence during development may lead to compensatory systems, making behavioral analysis, in particular, difficult to interpret. This can be addressed with conditional knockouts, but most models used to study opioid function are traditional knockouts. Knockouts often will eliminate all gene products, depending upon the constructs, making it very difficult to assess the individual contributions of splice variants. However, this is not always the case. Some knockouts continue to express splice variants not containing the targeted regions, as first reported with a knockout of neuronal NOS (Huang et al., 1993; Eliasson et al., 1997). Neuronal NOS undergoes extensive splicing. The knockout mouse targeted exon 2, but a variant that lacks exon 2, neuronal NOSβ, is still expressed in the animal. Thus, targeting one exon within a gene does not necessarily eliminate all gene expression. As discussed below, a similar situation is seen in several MOR-1 knockout mice.
a. Full-length variants
i. Antisense mapping
The first demonstration of the functional significance of MOR-1 in morphine analgesia was presented at a symposium in November of 1993 honoring William Martin (Uhl et al., 1994) in which an antisense against exon 1 of MOR-1 administered into the periaqueductal gray of the rat blocked morphine analgesia (Rossi et al., 1994a). Antisense approaches also provided a means to document the functional significance of the other opioid receptor families (Adams et al., 1994; Bilsky et al., 1994; Chien et al., 1994; Lai et al., 1994a,b; Pan et al., 1994,1995; Rossi et al., 1994a; Standifer et al., 1994; Tseng et al., 1994; Cha et al., 1995; Chen et al., 1995). Antisense approaches have now been used against a wide range of opioid functions, but the focus here will be on analgesia.
Antisense mapping of MOR-1 in both mice (Rossi et al., 1995a, 1996) and rats (Rossi et al., 1994a, 1995b) reveals very interesting findings (Table 16). Supraspinally, targeting exons 1 and 4 lowers morphine analgesia without impacting the effectiveness of morphine-6β-glucuronide (M6G). Conversely, probes targeting exons 2 and 3 decrease M6G analgesia but not that of morphine. Thus, these early studies imply that both morphine and M6G analgesia are mediated through the mu-opioid receptor gene OPRM1 but through different variants. A more recent study examined the effect of downregulating mMOR-1S using an antisense targeting the exon 1/exon 4 splice junction. Decreasing the levels of the 1TM variant lowered the levels of MOR-1 expression, consistent with the chaperone function of the 1TM variant, and decreased morphine’s analgesic response (Xu et al., 2013). Thus, it is possible that the decreased analgesia seen with the antisense probes targeting either exon 1 or exon 4 might be acting by targeting the full-length 7TM variant MOR-1 or the 1TM variant.
TABLE 16.
Antisense mapping MOR-1 exons
Antisense mapping involved designing antisense oligodeoxynucleotides (DNA) of approximately 20–25 bases that targeted the specific indicated exons were administered either supraspinally or spinally and their effect on analgesia determined. Compiled from the literature (Rossi et al., 1994a, 1995a,b; Kolesnikov et al., 1996b; Pan et al., 1999; Pasternak and Pan, 2000; Pasternak, 2001; Abbadie et al., 2002).
| Targeted Exon |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 6 | 7 | 8 | 9 | 10 | |
| Supraspinal | |||||||||
| Morphine | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes |
| M6G | No | Yes | Yes | No | No | No | No | No | No |
| Spinal | |||||||||
| Morphine | No | No | No | Yes | ND | ND | ND | ND | ND |
| Peripheral (topical) | |||||||||
| Morphine | Yes | No | No | Yes | ND | ND | ND | ND | ND |
ND, not determined.
Early work showed that supraspinal, but not spinal, morphine analgesia is blocked by naloxonazine, implying a different mechanism of action for the two sites (Paul et al., 1989a). Antisense studies also reveal a difference between supraspinal and spinal morphine analgesia, with antisense probes targeting exon 4 blocking both sites, whereas the exon 1 antisense blocks only supraspinal morphine analgesia. Antisense approaches also demonstrate the importance of variants containing exons 6, 7, 8, 9, and 10 in systemic morphine analgesia (Pan et al., 1999, 2000). Because these exons encode a range of different splice variants, these results imply that morphine analgesia cannot be explained by a single receptor. Presumably, each variant involved contributes to the overall effect. With all of them contributing to a common response, it is easy to see how removal of any one decreases activity.
ii. Knockout models
Knockouts of the OPRM1 gene using various gene targeting strategies have explored mu receptor function, including an exon 2 disruption (Matthes et al., 1996), deletion of the coding region of exon 1 (Schuller et al., 1999), deletion of both the coding and promoter regions of exon 1 (Sora et al., 1997), deletion of both exons 2 and 3 (Loh et al., 1998), and deletion of exon 11 (Pan et al., 2009) as recently reviewed (Ansonoff et al., 2011). Each model is unique, and comparing results among them provides interesting insights into morphine analgesia.
Disrupting exons 1, 2, and/or 3 leads to a virtually complete loss of morphine function, indicating that morphine responses depend upon the exon 1-associated full-length variants. The exon 1 knockout mouse generated by Pintar and colleagues (Schuller et al., 1999) is particularly interesting. Although exon 1 is disrupted in this model, the other exons are still expressed, as evidenced by the persistence of an exon 2–3 band on PCR and immunohistochemically using an antisera against the exon 7/8/9 epitope. As with the other knockouts that target exons 1, 2, and/or 3, morphine is inactive in the Pintar mice. However, heroin, 6-acetylmorphine (the active metabolite of heroin), and morphine-6β-glucuronide retain full analgesic activity, although with a slightly lower potency (Fig. 21).
Fig. 21.
Opiate analgesia in an exon 1 MOR-1 knockout mouse. With use of the radiant heat tailflick assay, dose-response curves were carried out in wild-type and exon 1 MOR-1 knockout mouse with the indicated opiate. Adapted from Schuller et al. (1999).
Assessing heroin and morphine-6β-glucuronide actions with the other knockout models gives additional insights. Mice with disruption of exons 2 and/or 3 lose sensitivity toward heroin analgesia, implying that variants containing these exons are a component of their target. However, heroin and morphine-6β-glucuronide reportedly are inactive in an exon 1 knockout model generated by Uhl and colleagues, in which both the exon and at least a portion of the promoter were deleted (Kitanaka et al., 1998). The reasons underlying these differences between the two exon 1 knockout animals remain unclear. The Pintar mice still express the exon 11-associated variants, but it is not known whether the exon 1 knockout animals generated by Uhl, which have lost a larger region of the gene, still express them. In addition to a possible loss of promoter activity, the elimination of a longer stretch of the gene might influence the overall splicing of the gene, leading to a more wide-spread disruption of splicing and loss of variants.
The exon 11 knockout mice reveal a very different pharmacology. Unlike the others, eliminating the exon 11-associated splice variants has little effect on morphine analgesia (Table 17). However, the exon 11-associated variants are important in the actions of some mu opioids, as seen by the decreased analgesic potency of heroin and morphine-6β-glucuronide and several other mu drugs (Table 17). These observations suggest a role for both the exon 1-associated and the exon 11-associated variants in mu analgesia, with some mu drugs acting through one or the other or both sets of variants.
TABLE 17.
Opioid analgesia in an exon 11 MOR-1 knockout mouse
Analgesia was assessed in either wild-type C57 (WT C57) or in mice with a disruption of exon 11 (exon 11 KO). Results (means ± S.E.M. of independent determinations) are from the literature (Pan et al., 2009; Majumdar et al., 2011).
| Drug | Analgesic ED50 |
Shift | |
|---|---|---|---|
| WT C57 | Exon 11 KO | ||
| mg/kg | |||
| Morphine | 1.58 ± 0.17 | 2.6 ± 0.5 | 1.6 |
| Methadone | 1.53 ± 0.06 | 1.8 ± 0.14 | 1.2 |
| IBNtxA | 0.53 ± 0.07 | > 20 | >35 |
| NalBzoH | 22 ± 5.3 | >100 | >5 |
| Nalbuphine | 47 ± 15 | >200 | >4 |
| Levorphanol | 0.1 ± 0.01 | 0.72 ± 0.13 | 7 |
| Butorphanol | 5.9 ± 3.9 | >100 | >15 |
b. Truncated variants: 6TM
Questions regarding the functional significance of the truncated exon 11-associated truncated variants have been raised. As noted above, loss of these variants in the exon 11 knockout mouse impairs the analgesic actions of several mu opiates, but not morphine (Table 17). The functional significance of these splice variants has been further strengthened by the actions of a series of unique analgesics based upon 3′-iodobenzoyl-6β-naltrexamide (IBNtxA) (Fig. 22) (Majumdar et al., 2011, 2012). Despite being a naltrexone derivative, this ligand is 10-fold more potent an analgesic than morphine. Additional testing reveals a very interesting pharmacology (Fig. 23). Despite its profound analgesic actions, it has no respiratory depression, displays no evidence of physical dependence with chronic dosing, and shows no cross-tolerance with morphine. In addition, it displays no reward behavior in a conditioned place preference model and has minimal effects on gastrointestinal transit. Thus, its pharmacological profile has many advantages over traditional opiates. However, its mechanism(s) of action have proven even more intriguing.
Fig. 22.
Structure of IBNtxA.
Fig. 23.
In vivo pharmacology of IBNtxA. (A) IBNtxA analgesia was assessed in wild-type (WT) or triple knockout (KO) mice with a disruption of exon 1 in MOR-1, DOR-1, and KOR-1. ED50 values: WT 0.22 mg/kg (0.13, 0.32); Triple KO 0.39 mg/kg (0.15, 0.58). (B) Analgesia was assessed with a fixed dose of IBNtxA (0.5 mg/kg) in wild-type, triple knockout (see above) and exon 11 knockout mice. (C) Respiratory rates were determined following saline or either IBNtxA (2.5 mg/kg) or morphine (20 mg/kg) at doses 5-fold greater than their analgesic ED50. (D) Conditioned place preference was carried out with saline, IBNtxA, or morphine in a 2-compartment. In contrast to morphine, IBNtxA did not produce place preference. Adapted from Majumdar et al. (2011).
To assess the possibility that the drug might be acting through either delta or kappa1 receptors, it was examined in a triple knockout mouse generated from Pintar’s exon 1 MOR-1, DOR-1, and KOR-1 knockout mice (Fig. 23). Traditional opioids are inactive in this animal, but IBNtxA retains full activity. However, IBNtxA is inactive in the exon 11 MOR-1 knockout mouse, implicating an exon 11-associated splice variant in its actions. Thus, the knockout studies indicate that the target of this drug is generated by the mu-opioid receptor gene OPRM1, based upon the loss of effect with the exon 11 knockout, whereas the other knockouts indicate that the target does not involve any exon 1-containing MOR-1 variants or delta or kappa1 receptors. Together, these observations imply that the target contains one of the truncated 6TM exon 11-associated variants.
A radiolabeled version of the drug, [125I]IBNtxA, identifies an opioid binding site in the triple knockout mice that is lost in both the exon 11 (Majumdar et al., 2011) and in the exon 2 (Matthes et al., 1996) knockout mice (Fig. 24). The selectivity of the binding site is very unusual (Table 18). Although many mu drugs, such as morphine and DAMGO display very poor affinity for the site, as do selective kappa1 (U50,488H) and delta [[d-Pen2,d-Pen5]enkephalin or DPDPE] compounds, the binding is readily competed by a large number of established opioid analgesics, including many that have been used clinically. The selectivity profile is reminiscent of the kappa3 binding site proposed over 20 years ago (Clark et al., 1989), although some differences exist.
Fig. 24.
[125I]BNtxA binding. (A) Saturation studies were carried out with [125I]BNtxA mouse brain membranes from either wild-type mice with blockers or triple KO mice. (B) [125I]BNtxA binding was determined in E1, E11, or E2 MOR-1 KO mice in the presence of mu (CTAP), kappa1, U50,488H (kappa1), or delta (DPDPE) blockers, whereas the triple KO mice were assayed without blockers. Adapted from Majumdar et al. (2011).
TABLE 18.
Binding selectivity of [125I]BNtxA binding in a mu/delta/kappa1 knockout mouse
[125I]BNtxA binding was performed in triple knockout mice generated by the Pintar laboratory with disruptions of exon 1 of MOR-1, as well as DOR-1 and KOR-1. These animals have no demonstrable [3H]DAMGO, [3H]U69,593, or [3H]DPDPE binding. Results are from Majumdar et al. (2011).
| Inactive Drugs | Ki | Active Drugs | Ki |
|---|---|---|---|
| nM | nM | ||
| Mu Selective | Antagonists | ||
| Morphine | >1000 | β-Funaltrexamine | 36 ± 5.3 |
| CTAP | >1000 | Naloxone | 51.9 ± 1.4 |
| DAMGO | >1000 | Naltrexone | 20.5 ± 1.8 |
| Oxymorphone | >1000 | Diprenorphine | 2.2 ± 0.71 |
| Oxycodone | >1000 | Levallorphan | 0.34 ± 0.02 |
| Morphine-6-glucuronide | >1000 | Benzomorphans | |
| β-Endorphin | >1000 | Ketocyclazocine | 0.04 ± 0.01 |
| Meperidine | >1000 | (−)-SKF10,047 | 13.5 ± 1.6 |
| Kappa1 Selective | (−)Ethylketocyclazocine | 0.21 ± 0.11 | |
| U50,488H | >1000 | Cyclazocine | 1.8 ± 0.67 |
| DynorphinA | >1000 | kappa3 | |
| α-Neoendorphin | >1000 | Naloxone benzoylhydrazone | 0.59 ± 0.15 |
| Delta Selective | Nalorphine | 3.7 ± 1.45 | |
| DADLE | >1000 | Levorphanol* | 8.8 ± 2.5 |
| DPDPE | >1000 | Buprenorphine* | 1.8 ± 0.93 |
| SNC80 | Nalbuphine* | 3.47 ± 1.18 | |
| Butorphanol* | 2.9 ± 1.55 |
Identifying the [125I]IBNtxA binding site at the molecular level is not yet complete, but there are a several insights. First, the binding levels in brain are not influenced by loss of the exon 1-associated variants. Second, binding is lost in the exon 11 knockout and also in the exon 2 knockout (Fig. 24) (Majumdar et al., 2011). These observations are consistent with those looking at analgesia and indicate that the binding site includes a 6TM exon 11-associated variant containing exon 2 and not exon 1. Humans, mice, and rats all have these variants (section VIII.B.1). When expressed alone, the 6TM variants display poor affinity for any opioids, including [125I]BNtxA. However, when coexpressed with another G protein-coupled receptor, high-affinity [125I]BNtxA binding is seen, presumably because of the formation of heterodimers containing the 6TM variant (Table 19). The natural partners of the heterodimer are not yet known. However, there is the interesting possibility that these 6TM/E11 heterodimers represent a class of receptors rather than a single one.
TABLE 19.
Binding of [125I]BNtxA in transfected HEK cells
[125I]BNtxA binding was determined in HEK cells transfected with either the 6TM variant mMOR-1G, ORL1, or both. No binding was detectable in the cells transfected with either of the proteins alone. However, high-affinity binding was determined in cells cotransfected. From the Majumdar et al. (2011).
| Transfection | [125I]BNtxA Binding KD |
|---|---|
| nM | |
| mMOR-1G alone | No binding detected |
| ORL1 alone | No binding detected |
| mMOR-1G + ORL1 | 0.24 ± 0.03 |
c. Truncated variants: 1TM
The last group of splice variants includes the single transmembrane proteins (Figs. 16 and 17). Although they were first reported shortly after the initial cloning of MOR-1 (Du et al., 1996, 1997), their functional significance was unclear. This was particularly intriguing because the abundance of the single TM mRNA is quite high in neuroblastoma cells. More recent studies confirm the relevance of these variants (Xu et al., 2013).
Selective downregulation of the single TM variant mMOR-1S using an antisense approach diminishes morphine analgesia in vivo. Although the single TM variants are incapable of binding opioids alone, they have a chaperone-like action, physically associating and stabilizing the full-length variant MOR-1 in the endoplasmic reticulum. The single TM protein slows the turnover of the full-length MOR-1, thereby elevating its expression levels and increasing morphine’s potency (Xu et al., 2013).
IX. The Future
The opioid field is among the oldest in pharmacology. Yet, it continually provides surprises, often just when we think we understand the system. For close to a century, it was assumed that morphine and other opiates acted through a single mechanism and receptor, only to find from the morphine/nalorphine combination studies in the 1950s that more than one class exists. The biochemical identification of the receptors was anticipated, but the discovery of the endogenous peptides was surprising, particularly the prediction of a host of additional putative opioids derived from processing of the larger precursors. Indeed, we still do not have a firm understanding of the roles for many of these additional putative opioid peptides, although one, BAM-22, labels a distinct family of sensory neuron-specific G protein-coupled receptors (Baird et al., 1982; Lembo et al., 2002; Grazzini et al., 2004; Hong et al., 2004). Whether the others also will have receptors identified remains to be seen.
Although clinical observations long ago clearly implied that more than one receptor is responsible for mu actions, a conclusion supported by traditional pharmacological studies, the number, and the complexity of splice variants of the mu-opioid receptor gene OPRM1 was unanticipated. The newest insights come with the truncated MOR-1 variants. Although known for over decade, their functional significance is only now becoming apparent. The concept of GPCR dimerization is well established in pharmacology and heterodimers between the traditional opioid receptors leading to targets with unique pharmacological profiles have been well documented. In addition to the mu/delta dimers already discussed, the KOR-1/DOR-1 dimer corresponds to the pharmacologically defined unique kappa2 receptor (Jordan and Devi, 1999; Zukin et al., 1988), whereas MOR-1 can dimerize with ORL1 (KOR-3) to form a functionally distinct receptor (Pan et al., 2002). The truncated 6TM variants appear to generate novel receptor targets with greatly advantageous pharmacological profiles, which was not anticipated. The possibility of a family of heterodimers generated by truncated MOR-1 splice variants opens the door to a whole family of new receptor targets. Even the single TM variants are relevant, opening questions of the significance of truncated variants in GPCRs in general. Single TM GPCR variants are not restricted to the opioid receptor family. Approximately 7% human GPCR genes, other than the olfactory receptor ones, have one or more potential single TM domain splice variants predicted by searching the genomic databases. Although their expression has yet to be verified experimentally, they might serve similar functional roles in other GPCR families. Overall, one is struck with the complexity of the mu-opioid system, confirming the observation that “the only simple things in science are scientists.”
Acknowledgments
The authors thank William Schmidt, Charles Inturrisi, Susruta Majumdar, Joel Schrock, and Gina Marone for their suggestions and comments.
Abbreviations
- β-FNA
β-funaltrexamine
- 6TM
truncated six transmembrane receptor 1
- 7TM
full-length traditional seven transmembrane receptor
- bp
base pair
- BAM-22
bovine adrenal medulla 22 protein (Tyr-Gly-Gly-Phe-Met-Arg-Arg-Val-Gly-Arg-Pro-Glu-Trp-Trp-Met-Asp-Tyr-Gln-Lys-Arg-Tyr-Gly)
- CHO
Chinese hamster ovary
- CTAP
H-d-Phe-Cys-Tyr-d-Trp-Orn-Thr-Pen-Thr-NH2
- CTOP
H-d-Phe-Cys-Tyr-d-Trp-Orn-Thr-Pen-Thr-NH2
- DAMGO
[d-Ala2,MePhe4,Gly(ol)5]enkephalin
- DPDPE
[d-Pen2,d-Pen5]enkephalin
- DOR-1
delta-opioid receptor
- E
embryonic day
- E1
exon 1 associated
- E11
exon 11 associated
- GPCR
G protein-coupled receptor
- HEK
human embryonic kidney
- IBNtxA
3′-iodobenzoyl-6β-naltrexamide
- IL
interleukin
- kb
kilobase pair
- KOR-1
kappa1-opioid receptor
- M6G
morphine-6β-glucuronide
- MK-801
(5S,10R)-(+)-5-methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine maleate or dizocilpine
- MOR-1
the cloned mu1 receptor
- NF-κB
nuclear factor-κB
- NMD
nonsense-mediated mRNA degradation
- norBNI
nor-binaltorphimine
- NOS
nitric-oxide synthase
- NRSE
neurorestrictive suppressor element
- Pgp
P-glycoprotein
- TM
truncated single transmembrane receptor
- NMDA
N-methyl-d-aspartate
- NSAID
nonsteroidal anti-inflammatory drugs
- RT-PCR
reverse-transcription polymerase chain reaction
- STAT
signal transducers and activators of transcription type
- TAPS
Tyr-d-Arg-Phe-Sar
- TAT
Tat peptide derived from the transactivator of transcription of human immunodeficiency virus
- TRV130
[(3-methoxythiophen-2-yl)methyl]({2-[(9R)-9-(pyridin-2-yl)-6-oxaspiro[4.5]decan-9-yl]ethyl})amine
- U50,488H
2-(3,4-dichlorophenyl)-N-methyl-N-[(1R,2R)-2-pyrrolidin-1-ylcyclohexyl]acetamide
- U69,593
methyl-2-phenyl-N-[(5R,7S,8S)-7-(pyrrolidin-1-yl)-1-oxaspiro[4.5]dec-8-yl]acetamide
- UTR
untranslated region
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Pasternak and Pan.
Footnotes
This work was supported in part by the National Institutes of Health National Institute on Drug Abuse [Grants DA00220, DA02615, DA06241, DA07242] (to G.W.P.) and [Grants DA013997 and DA029244] (to X.Y.P.).
References
- Abbadie C, Gultekin SH, Pasternak GW. (2000a) Immunohistochemical localization of the carboxy terminus of the novel mu opioid receptor splice variant MOR-1C within the human spinal cord. Neuroreport 11:1953–1957 [DOI] [PubMed] [Google Scholar]
- Abbadie C, Pan Y-X, Drake CT, Pasternak GW. (2000b) Comparative immunohistochemical distributions of carboxy terminus epitopes from the mu-opioid receptor splice variants MOR-1D, MOR-1 and MOR-1C in the mouse and rat CNS. Neuroscience 100:141–153 [DOI] [PubMed] [Google Scholar]
- Abbadie C, Pan Y-X, Pasternak GW. (2000c) Differential distribution in rat brain of mu opioid receptor carboxy terminal splice variants MOR-1C-like and MOR-1-like immunoreactivity: evidence for region-specific processing. J Comp Neurol 419:244–256 [DOI] [PubMed] [Google Scholar]
- Abbadie C, Pan Y-X, Pasternak GW. (2004) Immunohistochemical study of the expression of exon11-containing mu opioid receptor variants in mouse brain. Neuroscience 127:419–430 [DOI] [PubMed] [Google Scholar]
- Abbadie C, Pasternak GW. (2001) Differential in vivo internalization of MOR-1 and MOR-1C by morphine. Neuroreport 12:3069–3072 [DOI] [PubMed] [Google Scholar]
- Abbadie C, Pasternak GW, Aicher SA. (2001) Presynaptic localization of the carboxy-terminus epitopes of the mu opioid receptor splice variants MOR-1C and MOR-1D in the superficial laminae of the rat spinal cord. Neuroscience 106:833–842 [DOI] [PubMed] [Google Scholar]
- Abbadie C, Rossi GC, Orciuolo A, Zadina JE, Pasternak GW. (2002) Anatomical and functional correlation of the endomorphins with mu opioid receptor splice variants. Eur J Neurosci 16:1075–1082 [DOI] [PubMed] [Google Scholar]
- Abdelhamid EE, Sultana M, Portoghese PS, Takemori AE. (1991) Selective blockage of delta opioid receptors prevents the development of morphine tolerance and dependence in mice. J Pharmacol Exp Ther 258:299–303 [PubMed] [Google Scholar]
- Abood LG, Salem N, MacNeil M, Butler M. (1978) Phospholipid changes in synaptic membranes by lipolytic enzymes and subsequent restoration of opiate binding with phosphatidylserine. Biochim Biophys Acta 530:35–46 [DOI] [PubMed] [Google Scholar]
- Adams JU, Chen X, DeRiel JK, Adler MW, Liu-Chen L-Y. (1994) Intracerebroventricular treatment with an antisense oligodeoxynucleotide to kappa-opioid receptors inhibited kappa-agonist-induced analgesia in rats. Brain Res 667:129–132 [DOI] [PubMed] [Google Scholar]
- Andoh T, Yageta Y, Konno M, Yamaguchi-Miyamoto T, Takahata H, Nojima H, Nemoto H, Kuraishi Y. (2008) Evidence for separate involvement of different mu-opioid receptor subtypes in itch and analgesia induced by supraspinal action of opioids. J Pharmacol Sci 106:667–670 [DOI] [PubMed] [Google Scholar]
- Andria ML, Simon EJ. (2001) Identification of a neurorestrictive suppressor element (NRSE) in the human mu-opioid receptor gene. Brain Res Mol Brain Res 91:73–80 [DOI] [PubMed] [Google Scholar]
- Ansonoff M, Wen T, Pintar JE. (2011) Genetic studies of opioid receptor function in mice, in The Opiate Receptors (Pasternak GW, ed, ed) pp 341–388, Springer, New York [Google Scholar]
- Aquilante CL, Letrent SP, Pollack GM, Brouwer KLR. (2000) Increased brain P-glycoprotein in morphine tolerant rats. Life Sci 66:PL47–PL51 [DOI] [PubMed] [Google Scholar]
- Archer S, Albertson NF, Harris LS, Pierson AK, Bird JG, Keats AS, Telford J, Papadopoulos CN. (1962) Narcotic antagonists as analgesics. Science 137:541–543 [DOI] [PubMed] [Google Scholar]
- Archer S, Glick SD, Bidlack JM. (1996) Cyclazocine revisited. Neurochem Res 21:1369–1373 [DOI] [PubMed] [Google Scholar]
- Arnér S, Rawal N, Gustafsson LL. (1988) Clinical experience of long-term treatment with epidural and intrathecal opioids—a nationwide survey. Acta Anaesthesiol Scand 32:253–259 [DOI] [PubMed] [Google Scholar]
- Arvidsson U, Dado RJ, Riedl M, Lee J-H, Law PY, Loh HH, Elde R, Wessendorf MW. (1995a) delta-Opioid receptor immunoreactivity: distribution in brainstem and spinal cord, and relationship to biogenic amines and enkephalin. J Neurosci 15:1215–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson U, Riedl M, Chakrabarti S, Lee J-H, Nakano AH, Dado RJ, Loh HH, Law P-Y, Wessendorf MW, Elde R. (1995b) Distribution and targeting of a mu-opioid receptor (MOR1) in brain and spinal cord. J Neurosci 15:3328–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood TK, Findlay JBC. (1994) Fingerprinting G-protein-coupled receptors. Protein Eng 7:195–203 [DOI] [PubMed] [Google Scholar]
- Atweh SF, Kuhar MJ. (1977a) Autoradiographic localization of opiate receptors in rat brain. I. Spinal cord and lower medulla. Brain Res 124:53–67 [DOI] [PubMed] [Google Scholar]
- Atweh SF, Kuhar MJ. (1977b) Autoradiographic localization of opiate receptors in rat brain. II. The brain stem. Brain Res 129:1–12 [DOI] [PubMed] [Google Scholar]
- Atweh SF, Kuhar MJ. (1977c) Autoradiographic localization of opiate receptors in rat brain. III. The telencephalon. Brain Res 134:393–405 [DOI] [PubMed] [Google Scholar]
- Aydar E, Palmer CP, Klyachko VA, Jackson MB. (2002) The sigma receptor as a ligand-regulated auxiliary potassium channel subunit. Neuron 34:399–410 [DOI] [PubMed] [Google Scholar]
- Babey AM, Kolesnikov Y, Cheng J, Inturrisi CE, Trifilletti RR, Pasternak GW. (1994) Nitric oxide and opioid tolerance. Neuropharmacology 33:1463–1470 [DOI] [PubMed] [Google Scholar]
- Baird A, Ling N, Böhlen P, Benoit R, Klepper R, Guillemin R. (1982) Molecular forms of the putative enkephalin precursor BAM-12P in bovine adrenal, pituitary, and hypothalamus. Proc Natl Acad Sci USA 79:2023–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardo MT, Bhatnagar RK, Gebhart GF. (1981) Opiate receptor ontogeny and morphine-induced effects: influence of chronic footshock stress in preweanling rats. Brain Res 227:487–495 [DOI] [PubMed] [Google Scholar]
- Bare LA, Mansson E, Yang D. (1994) Expression of two variants of the human μ opioid receptor mRNA in SK-N-SH cells and human brain. FEBS Lett 354:213–216 [DOI] [PubMed] [Google Scholar]
- Baron A, Shuster L, Eleftheriou BE, Bailey DW. (1975) Opiate receptors in mice: genetic differences. Life Sci 17:633–640 [DOI] [PubMed] [Google Scholar]
- Bart G, Heilig M, LaForge KS, Pollak L, Leal SM, Ott J, Kreek MJ. (2004) Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol Psychiatry 9:547–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart G, Kreek MJ, Ott J, LaForge KS, Proudnikov D, Pollak L, Heilig M. (2005) Increased attributable risk related to a functional mu-opioid receptor gene polymorphism in association with alcohol dependence in central Sweden. Neuropsychopharmacology 30:417–422 [DOI] [PubMed] [Google Scholar]
- Basbaum AI, Bautista DM, Scherrer G, Julius D. (2009) Cellular and molecular mechanisms of pain. Cell 139:267–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbaum AI, Julius D. (2006) Toward better pain control. Sci Am 294:60–67 [DOI] [PubMed] [Google Scholar]
- Bayerer B, Stamer U, Hoeft A, Stuber F. (2007) Genomic variations and transcriptional regulation of the human μ-opioid Receptor gene. Eur J Pain 11:421–427 [DOI] [PubMed] [Google Scholar]
- Beckett AH. (1956) Analgesics and their antagonists: some steric and chemical considerations. I. The dissociation constants of some tertiary amines and synthetic analgesics, the conformations of methadone-type compounds. J Pharm Pharmacol 8:848–859 [DOI] [PubMed] [Google Scholar]
- Beckett AH, Casy AF. (1954) Synthetic analgesics: stereochemical considerations. J Pharm Pharmacol 6:986–1001 [DOI] [PubMed] [Google Scholar]
- Beckett AH, Casy AF. (1965) Analgesics and their antagonists: biochemical aspects and structure-activity relationships. Prog Med Chem 4:171–218 [DOI] [PubMed] [Google Scholar]
- Beckett AH, Casy AF, Harper NJ. (1956a) Analgesics and their antagonists: some steric and chemical considerations. III. The influence of the basic group on the biological response. J Pharm Pharmacol 8:874–883, discussion, 883–884 [PubMed] [Google Scholar]
- Beckett AH, Casy AF, Harper NJ, Phillips PM. (1956b) Analgesics and their antagonists: some steric and chemical considerations. II. The influence of the basic group on physico-chemical properties and the activity of methadone and thiambutene-type compounds. J Pharm Pharmacol 8:860–873 [PubMed] [Google Scholar]
- Beecher HK. (1946) Pain in men wounded in battle. Ann Surg 123:96–105 [PMC free article] [PubMed] [Google Scholar]
- Beecher HK. (1960) Increased stress and effectiveness of placebos and “active” drugs. Science 132:91–92 [DOI] [PubMed] [Google Scholar]
- Befort K, Costigan M, Woolf CJ. (2001a) Differential gene expression—how to find new analgesic targets. Curr Opin Investig Drugs 2:396–398 [PubMed] [Google Scholar]
- Befort K, Filliol D, Décaillot FM, Gavériaux-Ruff C, Hoehe MR, Kieffer BL. (2001b) A single nucleotide polymorphic mutation in the human mu-opioid receptor severely impairs receptor signaling. J Biol Chem 276:3130–3137 [DOI] [PubMed] [Google Scholar]
- Belknap JK, Mogil JS, Helms ML, Richards SP, O’Toole LA, Bergeson SE, Buck KJ. (1995) Localization to chromosome 10 of a locus influencing morphine analgesia in crosses derived from C57BL/6 and DBA/2 strains. Life Sci 57:PL117–PL124 [DOI] [PubMed] [Google Scholar]
- Ben-Eliyahu S, Marek P, Vaccarino AL, Mogil JS, Sternberg WF, Liebeskind JC. (1992) The NMDA receptor antagonist MK-801 prevents long-lasting non-associative morphine tolerance in the rat. Brain Res 575:304–308 [DOI] [PubMed] [Google Scholar]
- Bentley KW, Hardy DG. (1967) Novel analgesics and molecular rearrangements in the morphine-thebaine group. 3. Alcohols of the 6,14-endo-ethenotetrahydrooripavine series and derived analogs of N-allylnormorphine and -norcodeine. J Am Chem Soc 89:3281–3292 [DOI] [PubMed] [Google Scholar]
- Berezniuk I, Fricker LD. (2011) Endogenous opioids, in The Opiate Receptors (Pasternak GW, ed, ed) pp 93–120, Springer, New York [Google Scholar]
- Berkowitz BA, Way EL. (1971) Analgesic activity and central nervous system distribution of the optical isomers of pentazocine in the rat. J Pharmacol Exp Ther 177:500–508 [PubMed] [Google Scholar]
- Berrettini WH, Ferraro TN, Alexander RC, Buchberg AM, Vogel WH. (1994) Quantitative trait loci mapping of three loci controlling morphine preference using inbred mouse strains. Nat Genet 7:54–58 [DOI] [PubMed] [Google Scholar]
- Berrettini WH, Lerman CE. (2005) Pharmacotherapy and pharmacogenetics of nicotine dependence. Am J Psychiatry 162:1441–1451 [DOI] [PubMed] [Google Scholar]
- Besse D, Lombard MC, Zajac JM, Roques BP, Besson JM. (1990) Pre- and postsynaptic distribution of mu, delta and kappa opioid receptors in the superficial layers of the cervical dorsal horn of the rat spinal cord. Brain Res 521:15–22 [DOI] [PubMed] [Google Scholar]
- Bilsky EJ, Bernstein RN, Pasternak GW, Hruby VJ, Patel D, Porreca F, Lai J. (1994) Selective inhibition of [D-Ala2, Glu4]deltorphin antinociception by supraspinal, but not spinal, administration of an antisense oligodeoxynucleotide to an opioid delta receptor. Life Sci 55:PL37–PL43 [DOI] [PubMed] [Google Scholar]
- Birdsall NJM, Hulme EC. (1976) C fragment of lipotropin has a high affinity for brain opiate receptors. Nature 260:793–795 [DOI] [PubMed] [Google Scholar]
- Bloomfield SS, Barden TP, Mitchell J. (1983) Metkephamid and meperidine analgesia after episiotomy. Clin Pharmacol Ther 34:240–247 [DOI] [PubMed] [Google Scholar]
- Blumberg H, Dayton HB, George M, Rapaport DN. (1961) N-allylnoroxymorphone: a potent narcotic antagonist. Fed Proc 20:311 [Google Scholar]
- Blume AJ. (1978a) Interaction of ligands with the opiate receptors of brain membranes: regulation by ions and nucleotides. Proc Natl Acad Sci USA 75:1713–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume AJ. (1978b) Opiate binding to membrane preparations of neuroblastoma x glioma hybrid cells NG108-15: effects of ions and nucleotides. Life Sci 22:1843–1852 [DOI] [PubMed] [Google Scholar]
- Blume AJ, Lichtshtein D, Boone G. (1979) Coupling of opiate receptors to adenylate cyclase: requirement for Na+ and GTP. Proc Natl Acad Sci USA 76:5626–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmuhl M. (1948) Uber eine neue klasse von analgetisch wirkenden. Verbindungen Ann Chem 561:52 [Google Scholar]
- Bodnar RJ. (2000) Supraspinal circuitry mediating opioid antinociception: antagonist and synergy studies in multiple sites. J Biomed Sci 7:181–194 [DOI] [PubMed] [Google Scholar]
- Bodnar RJ, Pasternak GW, Mann PE, Paul D, Warren R, Donner DB. (1989) Mediation of anorexia by human recombinant tumor necrosis factor through a peripheral action in the rat. Cancer Res 49:6280–6284 [PubMed] [Google Scholar]
- Bodnar RJ, Paul D, Pasternak GW. (1990) Proglumide selectively potentiates supraspinal mu 1 opioid analgesia in mice. Neuropharmacology 29:507–510 [DOI] [PubMed] [Google Scholar]
- Bodnar RJ, Paul D, Pasternak GW. (1991) Synergistic analgesic interactions between the periaqueductal gray and the locus coeruleus. Brain Res 558:224–230 [DOI] [PubMed] [Google Scholar]
- Bodnar RJ, Williams CL, Lee SJ, Pasternak GW. (1988) Role of mu 1-opiate receptors in supraspinal opiate analgesia: a microinjection study. Brain Res 447:25–34 [DOI] [PubMed] [Google Scholar]
- Boettcher C, Fellermeier M, Boettcher C, Dräger B, Zenk MH. (2005) How human neuroblastoma cells make morphine. Proc Natl Acad Sci USA 102:8495–8500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn LM, Dykstra LA, Lefkowitz RJ, Caron MG, Barak LS. (2004) Relative opioid efficacy is determined by the complements of the G protein-coupled receptor desensitization machinery. Mol Pharmacol 66:106–112 [DOI] [PubMed] [Google Scholar]
- Bohn LM, Gainetdinov RR, Lin FT, Lefkowitz RJ, Caron MG. (2000) Mu-opioid receptor desensitization by beta-arrestin-2 determines morphine tolerance but not dependence. Nature 408:720–723 [DOI] [PubMed] [Google Scholar]
- Bohn LM, Gainetdinov RR, Sotnikova TD, Medvedev IO, Lefkowitz RJ, Dykstra LA, Caron MG. (2003) Enhanced rewarding properties of morphine, but not cocaine, in beta(arrestin)-2 knock-out mice. J Neurosci 23:10265–10273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn LM, Lefkowitz RJ, Caron MG. (2002) Differential mechanisms of morphine antinociceptive tolerance revealed in (beta)arrestin-2 knock-out mice. J Neurosci 22:10494–10500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn LM, Lefkowitz RJ, Gainetdinov RR, Peppel K, Caron MG, Lin FT. (1999) Enhanced morphine analgesia in mice lacking beta-arrestin 2. Science 286:2495–2498 [DOI] [PubMed] [Google Scholar]
- Bolan EA, Pan YX, Pasternak GW. (2004) Functional analysis of MOR-1 splice variants of the mouse mu opioid receptor gene Oprm. Synapse 51:11–18 [DOI] [PubMed] [Google Scholar]
- Bolan EA, Tallarida RJ, Pasternak GW. (2002) Synergy between mu opioid ligands: evidence for functional interactions among mu opioid receptor subtypes. J Pharmacol Exp Ther 303:557–562 [DOI] [PubMed] [Google Scholar]
- Bolognesi ML, Ojala WH, Gleason WB, Griffin JF, Farouz-Grant F, Larson DL, Takemori AE, Portoghese PS. (1996) Opioid antagonist activity of naltrexone-derived bivalent ligands: importance of a properly oriented molecular scaffold to guide “address” recognition at kappa opioid receptors. J Med Chem 39:1816–1822 [DOI] [PubMed] [Google Scholar]
- Bond C, LaForge KS, Tian MT, Melia D, Zhang SW, Borg L, Gong JH, Schluger J, Strong JA, Leal SM, et al. (1998) Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci USA 95:9608–9613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner G, Meng F, Akil H. (2000) Selectivity of mu-opioid receptor determined by interfacial residues near third extracellular loop. Eur J Pharmacol 403:37–44 [DOI] [PubMed] [Google Scholar]
- Börner C, Wöltje M, Höllt V, Kraus J. (2004) STAT6 transcription factor binding sites with mismatches within the canonical 5′-TTC...GAA-3′ motif involved in regulation of delta- and mu-opioid receptors. J Neurochem 91:1493–1500 [DOI] [PubMed] [Google Scholar]
- Bot G, Blake AD, Li SX, Reisine T. (1998) Fentanyl and its analogs desensitize the cloned mu opioid receptor. J Pharmacol Exp Ther 285:1207–1218 [PubMed] [Google Scholar]
- Brodsky M, Elliott K, Hynansky A, Inturrisi CE. (1994) Strain differences in the analgesic potency of morphine and in mu opioid receptor MRNA levels in adult male mice. Regul Pept 54:33–34 [Google Scholar]
- Brodsky M, Elliott K, Hynansky A, Inturrisi CE. (1995a) CNS levels of mu opioid receptor (MOR-1) mRNA during chronic treatment with morphine or naltrexone. Brain Res Bull 38:135–141 [DOI] [PubMed] [Google Scholar]
- Brodsky M, Elliott K, Hynansky A, Jenab S, Inturrisi CE. (1995b) Quantitation of mu-opioid receptor (MOR-1) mRNA in selected regions of the rat CNS. Neuroreport 6:725–729 [DOI] [PubMed] [Google Scholar]
- Bunzow JR, Saez C, Mortrud M, Bouvier C, Williams JT, Low M, Grandy DK. (1994) Molecular cloning and tissue distribution of a putative member of the rat opioid receptor gene family that is not a mu, δ or kappa opioid receptor type. FEBS Lett 347:284–288 [DOI] [PubMed] [Google Scholar]
- Bunzow JR, Zhang G, Bouvier C, Saez C, Ronnekleiv OK, Kelly MJ, Grandy DK. (1995) Characterization and distribution of a cloned rat mu-opioid receptor. J Neurochem 64:14–24 [DOI] [PubMed] [Google Scholar]
- Burkhardt C, Frederickson RCA, Pasternak GW. (1982) Metkephamid (Tyr-D-ala-Gly-Phe-N(Me)Met-NH2), a potent opioid peptide: receptor binding and analgesic properties. Peptides 3:869–871 [DOI] [PubMed] [Google Scholar]
- Búzás B, Izenwasser S, Portoghese PS, Cox BM. (1994) Evidence for delta opioid receptor subtypes regulating adenylyl cyclase activity in rat brain. Life Sci 54:PL101–PL106 [DOI] [PubMed] [Google Scholar]
- Cadet P, Mantione KJ, Stefano GB. (2003) Molecular identification and functional expression of mu 3, a novel alternatively spliced variant of the human mu opiate receptor gene. J Immunol 170:5118–5123 [DOI] [PubMed] [Google Scholar]
- Calimlim JF, Wardell WM, Sriwatanakul K, Lasagna L, Cox C. (1982) Analgesic efficacy of parenteral metkephamid acetate in treatment of postoperative pain. Lancet 1:1374–1375 [DOI] [PubMed] [Google Scholar]
- Callaghan R, Riordan JR. (1993) Synthetic and natural opiates interact with P-glycoprotein in multidrug-resistant cells. J Biol Chem 268:16059–16064 [PubMed] [Google Scholar]
- Campbell ND. (2006) “A new deal for the drug addict”: the Addiction Research Center, Lexington, Kentucky. J Hist Behav Sci 42:135–157 [DOI] [PubMed] [Google Scholar]
- Campbell ND. (2010) The history of a public science: How the Addiction Research Center became the NIDA Intramural Research Program. Drug Alcohol Depend 107:108–112 [DOI] [PubMed] [Google Scholar]
- Campbell ND, Olsen JP, Walden L. (2008) The Narcotic Farm: The Rise and Fall of America's First Prison for Drug Addicts, Abrams, New York [Google Scholar]
- Cha XY, Xu H, Ni Q, Partilla JS, Rice KC, Matecka D, Calderon SN, Porreca F, Lai J, Rothman RB. (1995) Opioid peptide receptor studies. 4. Antisense oligodeoxynucleotide to the delta opioid receptor delineates opioid receptor subtypes. Regul Pept 59:247–253 [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Liu NJ, Gintzler AR. (2010) Formation of mu-/kappa-opioid receptor heterodimer is sex-dependent and mediates female-specific opioid analgesia. Proc Natl Acad Sci USA 107:20115–20119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Sultana M, Portoghese PS, Takemori AE. (1993) Differential antagonism by naltrindole-5′-isothiocyanate on [3H]DSLET and [3H]DPDPE binding to striatal slices of mice. Life Sci 53:1761–1765 [DOI] [PubMed] [Google Scholar]
- Chang A, Emmel DW, Rossi GC, Pasternak GW. (1998) Methadone analgesia in morphine-insensitive CXBK mice. Eur J Pharmacol 351:189–191 [DOI] [PubMed] [Google Scholar]
- Chang K-J, Cuatrecasas P. (1979) Multiple opiate receptors. Enkephalins and morphine bind to receptors of different specificity. J Biol Chem 254:2610–2618 [PubMed] [Google Scholar]
- Chavkin C, McLaughlin JP, Celver JP. (2001) Regulation of opioid receptor function by chronic agonist exposure: constitutive activity and desensitization. Mol Pharmacol 60:20–25 [DOI] [PubMed] [Google Scholar]
- Chen XH, Adams JU, Geller EB, DeRiel JK, Adler MW, Liu-Chen L-Y. (1995) An antisense oligodeoxynucleotide to mu-opioid receptors inhibits mu-opioid receptor agonist-induced analgesia in rats. Eur J Pharmacol 275:105–108 [DOI] [PubMed] [Google Scholar]
- Chen Y, Fan Y, Liu J, Mestek A, Tian M, Kozak CA, Yu L. (1994) Molecular cloning, tissue distribution and chromosomal localization of a novel member of the opioid receptor gene family. FEBS Lett 347:279–283 [DOI] [PubMed] [Google Scholar]
- Chen Y, Mestek A, Liu J, Hurley JA, Yu L. (1993a) Molecular cloning and functional expression of a mu-opioid receptor from rat brain. Mol Pharmacol 44:8–12 [PubMed] [Google Scholar]
- Chen Y, Mestek A, Liu J, Yu L. (1993b) Molecular cloning of a rat kappa opioid receptor reveals sequence similarities to the μ and δ opioid receptors. Biochem J 295:625–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov HI, Woods LA. (1965) Central nervous system distribution and metabolism of C14-morphine during morphine-induced feline mania. J Pharmacol Exp Ther 149:146–155 [PubMed] [Google Scholar]
- Cherny N, Ripamonti C, Pereira J, Davis C, Fallon M, McQuay H, Mercadante S, Pasternak G, Ventafridda V, Expert Working Group of the European Association of Palliative Care Network (2001) Strategies to manage the adverse effects of oral morphine: an evidence-based report. J Clin Oncol 19:2542–2554 [DOI] [PubMed] [Google Scholar]
- Chien C-C, Brown G, Pan Y-X, Pasternak GW. (1994) Blockade of U50,488H analgesia by antisense oligodeoxynucleotides to a kappa-opioid receptor. Eur J Pharmacol 253:R7–R8 [DOI] [PubMed] [Google Scholar]
- Chien C-C, Pasternak GW. (1993) Functional antagonism of morphine analgesia by (+)-pentazocine: evidence for an anti-opioid sigma 1 system. Eur J Pharmacol 250:R7–R8 [DOI] [PubMed] [Google Scholar]
- Chien C-C, Pasternak GW. (1994) Selective antagonism of opioid analgesia by a sigma system. J Pharmacol Exp Ther 271:1583–1590 [PubMed] [Google Scholar]
- Chien C-C, Pasternak GW. (1995a) (-)-Pentazocine analgesia in mice: interactions with a σ receptor system. Eur J Pharmacol 294:303–308 [DOI] [PubMed] [Google Scholar]
- Chien C-C, Pasternak GW. (1995b) Sigma antagonists potentiate opioid analgesia in rats. Neurosci Lett 190:137–139 [DOI] [PubMed] [Google Scholar]
- Childers SR. (1991) Opioid receptor-coupled second messenger systems. Life Sci 48:1991–2003 [DOI] [PubMed] [Google Scholar]
- Childers SR, Pasternak GW. (1982) Naloxazone, a novel opiate antagonist: irreversible blockade of rat brain opiate receptors in vitro. Cell Mol Neurobiol 2:93–103 [Google Scholar]
- Childers SR, Snyder SH. (1978) Guanine nucleotides differentiate agonist and antagonist interactions with opiate receptors. Life Sci 23:759–761 [DOI] [PubMed] [Google Scholar]
- Choi HS, Hwang CK, Kim CS, Song KY, Law PY, Loh HH, Wei LN. (2008a) Transcriptional regulation of mouse mu opioid receptor gene in neuronal cells by poly(ADP-ribose) polymerase-1. J Cell Mol Med 12 (6A):2319–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Hwang CK, Kim CS, Song KY, Law PY, Wei LN, Loh HH. (2005) Transcriptional regulation of mouse mu opioid receptor gene: Sp3 isoforms (M1, M2) function as repressors in neuronal cells to regulate the mu opioid receptor gene. Mol Pharmacol 67:1674–1683 [DOI] [PubMed] [Google Scholar]
- Choi HS, Kim CS, Hwang CK, Song KY, Wang W, Qiu Y, Law PY, Wei LN, Loh HH. (2006) The opioid ligand binding of human mu-opioid receptor is modulated by novel splice variants of the receptor. Biochem Biophys Res Commun 343:1132–1140 [DOI] [PubMed] [Google Scholar]
- Choi HS, Song KY, Hwang CK, Kim CS, Law PY, Wei LN, Loh HH. (2008b) A proteomics approach for identification of single strand DNA-binding proteins involved in transcriptional regulation of mouse mu opioid receptor gene. Mol Cell Proteomics 7:1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou R, Fanciullo GJ, Fine PG, Adler JA, Ballantyne JC, Davies P, Donovan MI, Fishbain DA, Foley KM, Fudin J, et al. American Pain Society-American Academy of Pain Medicine Opioids Guidelines Panel (2009) Clinical guidelines for the use of chronic opioid therapy in chronic noncancer pain. J Pain 10:113–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou WY, Wang CH, Liu PH, Liu CC, Tseng CC, Jawan B. (2006) Human opioid receptor A118G polymorphism affects intravenous patient-controlled analgesia morphine consumption after total abdominal hysterectomy. Anesthesiology 105:334–337 [DOI] [PubMed] [Google Scholar]
- Cicero TJ. (1980) Effects of exogenous and endogenous opiates on the hypothalamic—pituitary—gonadal axis in the male. Fed Proc 39:2551–2554 [PubMed] [Google Scholar]
- Clark AJ. (1933) The Mode of Action of Drugs on Cells, Williams and Wilkins, Baltimore [Google Scholar]
- Clark JA, Liu L, Price M, Hersh B, Edelson M, Pasternak GW. (1989) Kappa opiate receptor multiplicity: evidence for two U50,488-sensitive kappa 1 subtypes and a novel kappa 3 subtype. J Pharmacol Exp Ther 251:461–468 [PubMed] [Google Scholar]
- Clendeninn NJ, Petraitis M, Simon EJ. (1976) Ontological development of opiate receptors in rodent brain. Brain Res 118:157–160 [DOI] [PubMed] [Google Scholar]
- Clouet DH, Williams N. (1973) Localization in brain particulate fractions of narcotic analgesic drugs administered intracisternally to rats. Biochem Pharmacol 22:1283–1293 [DOI] [PubMed] [Google Scholar]
- Cobos EJ, Entrena JM, Nieto FR, Cendán CM, Del Pozo E. (2008) Pharmacology and therapeutic potential of sigma(1) receptor ligands. Curr Neuropharmacol 6:344–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier HO. (1980) Cellular site of opiate dependence. Nature 283:625–629 [DOI] [PubMed] [Google Scholar]
- Collier HO, Roy AC. (1974) Morphine-like drugs inhibit the stimulation of E prostaglandins of cyclic AMP formation by rat brain homogenate. Nature 248:24–27 [DOI] [PubMed] [Google Scholar]
- Cowan A, Lewis JW, Macfarlane IR. (1977) Agonist and antagonist properties of buprenorphine, a new antinociceptive agent. Br J Pharmacol 60:537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan A, Zhu XZ, Porreca F. (1985) Studies in vivo with ICI 174864 and [D-Pen2, D-Pen5]enkephalin. Neuropeptides 5:311–314 [DOI] [PubMed] [Google Scholar]
- Cox BM. (2011) Pharmacology of opioid drugs, in The Opiate Receptors (Pasternak GW, ed), pp 23–58, Springer, New York [Google Scholar]
- Cox BM, Ginsburg M, Osman OH. (1968) Acute tolerance to narcotic analgesic drugs in rats. Br Pharmacol Chemother 33:245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BM, Opheim KE, Teschemacher H, Goldstein A. (1975) A peptide-like substance from pituitary that acts like morphine. 2. Purification and properties. Life Sci 16:1777–1782 [DOI] [PubMed] [Google Scholar]
- Cox BM, Osman OH. (1969) The role of protein synthesis inhibition in the prevention of morphine tolerance. Br J Pharmacol 35:373–374 [PMC free article] [PubMed] [Google Scholar]
- Cox BM, Weinstock M. (1966) The effect of analgesic drugs on the release of acetylcholine from electrically stimulated guinea-pig ileum. Br Pharmacol Chemother 27:81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle JT, Pert CB. (1976) Ontogenetic development of [3H]naloxone binding in rat brain. Neuropharmacology 15:555–560 [DOI] [PubMed] [Google Scholar]
- Crain SM, Shen K. (2000a) Enhanced analgesic potency and reduced tolerance of morphine in 129/SvEv mice: evidence for a deficiency in GM1 ganglioside-regulated excitatory opioid receptor functions. Brain Res 856:227–235 [DOI] [PubMed] [Google Scholar]
- Crain SM, Shen KF. (2000b) Antagonists of excitatory opioid receptor functions enhance morphine’s analgesic potency and attenuate opioid tolerance/dependence liability. Pain 84:121–131 [DOI] [PubMed] [Google Scholar]
- Crain SM, Shen K-F. (1990) Opioids can evoke direct receptor-mediated excitatory effects on sensory neurons. Trends Pharmacol Sci 11:77–81 [DOI] [PubMed] [Google Scholar]
- Crain SM, Shen K-F, Chalazonitis A. (1988) Opioids excite rather than inhibit sensory neurons after chronic opioid exposure of spinal cord-ganglion cultures. Brain Res 455:99–109 [DOI] [PubMed] [Google Scholar]
- Creese I, Pasternak GW, Pert CB, Snyder SH. (1975) Discrimination by temperature of opiate agonist and antagonist receptor binding. Life Sci 16:1837–1842 [DOI] [PubMed] [Google Scholar]
- Creese I, Snyder SH. (1975) Receptor binding and pharmacological activity of opiates in the guinea-pig intestine. J Pharmacol Exp Ther 194:205–219 [PubMed] [Google Scholar]
- Cruciani RA, Dvorkin B, Morris SA, Crain SM, Makman MH. (1993) Direct coupling of opioid receptors to both stimulatory and inhibitory guanine nucleotide-binding proteins in F-11 neuroblastoma-sensory neuron hybrid cells. Proc Natl Acad Sci USA 90:3019–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvejic S, Devi LA. (1997) Dimerization of the delta opioid receptor: implication for a role in receptor internalization. J Biol Chem 272:26959–26964 [DOI] [PubMed] [Google Scholar]
- D'Amour FE, Smith DL. (1941) A method for determining loss of pain sensation. J Pharmacol Exp Ther 72:74–79 [Google Scholar]
- Darlison MG, Greten FR, Harvey RJ, Kreienkamp HJ, Stühmer T, Zwiers H, Lederis K, Richter D. (1997) Opioid receptors from a lower vertebrate (Catostomus commersoni): sequence, pharmacology, coupling to a G-protein-gated inward-rectifying potassium channel (GIRK1), and evolution. Proc Natl Acad Sci USA 94:8214–8219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C(1875) Insectivorius Plants, New York. [Google Scholar]
- Davis AM, Inturrisi CE. (1999) d-Methadone blocks morphine tolerance and N-methyl-D-aspartate-induced hyperalgesia. J Pharmacol Exp Ther 289:1048–1053 [PubMed] [Google Scholar]
- Davis MP. (2012) Twelve reasons for considering buprenorphine as a frontline analgesic in the management of pain. J Support Oncol 10:209–219 [DOI] [PubMed] [Google Scholar]
- Delfs JM, Kong H, Mestek A, Chen Y, Yu L, Reisine T, Chesselet M-F. (1994a) Expression of mu opioid receptor mRNA in rat brain: an in situ hybridization study at the single cell level. J Comp Neurol 345:46–68 [DOI] [PubMed] [Google Scholar]
- Delfs JM, Yu L, Reisine T, Chesselet M-F. (1994b) The distribution and regulation of mu opioid receptor MRNA in rat basal ganglia. Regul Pept 54:79–80 [Google Scholar]
- Deng HB, Yu Y, Pak Y, O’Dowd BF, George SR, Surratt CK, Uhl GR, Wang JB. (2000) Role for the C-terminus in agonist-induced mu opioid receptor phosphorylation and desensitization. Biochemistry 39:5492–5499 [DOI] [PubMed] [Google Scholar]
- Deng HB, Yu YK, Wang HY, Guang W, Wang JB. (2001) Agonist-induced μ opioid receptor phosphorylation and functional desensitization in rat thalamus. Brain Res 898:204–214 [DOI] [PubMed] [Google Scholar]
- de Stevens G. (1965) Analgetics, Academic Press, New York [Google Scholar]
- DeWire SM, Yamashita DS, Rominger DH, Liu G, Cowan CL, Graczyk TM, Chen XT, Pitis PM, Gotchev D, Yuan C, et al. (2013) A G protein-biased ligand at the μ-opioid receptor is potently analgesic with reduced gastrointestinal and respiratory dysfunction compared with morphine. J Pharmacol Exp Ther 344:708–717 [DOI] [PubMed] [Google Scholar]
- Diego L, Atayee R, Helmons P, Hsiao G, von Gunten CF. (2011) Novel opioid antagonists for opioid-induced bowel dysfunction. Expert Opin Investig Drugs 20:1047–1056 [DOI] [PubMed] [Google Scholar]
- Diego L, Atayee R, Helmons P, von Gunten CF. (2009) Methylnaltrexone: a novel approach for the management of opioid-induced constipation in patients with advanced illness. Expert Rev Gastroenterol Hepatol 3:473–485 [DOI] [PubMed] [Google Scholar]
- Dietrich G, Gaibelet G, Capeyrou R, Butour JL, Pontet F, Emorine LJ. (1998) Implication of the first and third extracellular loops of the mu-opioid receptor in the formation of the ligand binding site: a study using chimeric mu-opioid/angiotensin receptors. J Neurochem 70:2106–2111 [DOI] [PubMed] [Google Scholar]
- Ding YQ, Kaneko T, Nomura S, Mizuno N. (1996) Immunohistochemical localization of mu-opioid receptors in the central nervous system of the rat. J Comp Neurol 367:375–402 [DOI] [PubMed] [Google Scholar]
- Ding Z, Raffa RB. (2009) Identification of an additional supraspinal component to the analgesic mechanism of action of buprenorphine. Br J Pharmacol 157:831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dole VP, Nyswander ME, Kreek MJ. (1966) Narcotic blockade. Arch Intern Med 118:304–309 [PubMed] [Google Scholar]
- Donnerer J, Cardinale G, Coffey J, Lisek CA, Jardine I, Spector S. (1987) Chemical characterization and regulation of endogenous morphine and codeine in the rat. J Pharmacol Exp Ther 242:583–587 [PubMed] [Google Scholar]
- Donnerer J, Oka K, Brossi A, Rice KC, Spector S. (1986) Presence and formation of codeine and morphine in the rat. Proc Natl Acad Sci USA 83:4566–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley CT, Kaplan RA, Chung NN, Schiller PW, Bidlack JM, Houghten RA. (1995) Six highly active mu-selective opioid peptides identified from two synthetic combinatorial libraries. Pept Res 8:124–137 [PubMed] [Google Scholar]
- Doyle GA, Rebecca Sheng X, Lin SS, Press DM, Grice DE, Buono RJ, Ferraro TN, Berrettini WH. (2007a) Identification of three mouse mu-opioid receptor (MOR) gene (Oprm1) splice variants containing a newly identified alternatively spliced exon. Gene 388:135–147 [DOI] [PubMed] [Google Scholar]
- Doyle GA, Sheng XR, Lin SS, Press DM, Grice DE, Buono RJ, Ferraro TN, Berrettini WH. (2007b) Identification of five mouse mu-opioid receptor (MOR) gene (Oprm1) splice variants containing a newly identified alternatively spliced exon. Gene 395:98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreborg S, Sundström G, Larsson TA, Larhammar D. (2008) Evolution of vertebrate opioid receptors. Proc Natl Acad Sci USA 105:15487–15492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y-L, Elliot K, Pan Y-X, Pasternak GW, Inturrisi CE. (1997) A splice variant of the mu opioid receptor is present in human SHSY-5Y cells. Soc Neurosci 23:1206 [Google Scholar]
- Du Y-L, Pan Y-X, Pasternak GW, Inturrisi CE. (1996) Identification of a Novel Splice Variant of the Mouse Mu Opioid Receptor. Soc Neurosci 22:1766 [Google Scholar]
- Duka T, Wüster M, Schubert P, Stoiber R, Herz A. (1981) Selective localization of different types of opiate receptors in hippocampus as revealed by in vitro autoradiography. Brain Res 205:181–186 [DOI] [PubMed] [Google Scholar]
- Dum JE, Herz A. (1981) In vivo receptor binding of the opiate partial agonist, buprenorphine, correlated with its agonistic and antagonistic actions. Br J Pharmacol 74:627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant PAC, Yaksh TL. (1986) Epidural injections of bupivacaine, morphine, fentanyl, lofentanil, and DADL in chronically implanted rats: a pharmacologic and pathologic study. Anesthesiology 64:43–53 [DOI] [PubMed] [Google Scholar]
- Ehrlich P. (1913) Chemotherapeutics: Scientific Principles, Methods and Results. Lancet 2:445–451 [Google Scholar]
- Eliasson MJL, Blackshaw S, Schell MJ, Snyder SH. (1997) Neuronal nitric oxide synthase alternatively spliced forms: prominent functional localizations in the brain. Proc Natl Acad Sci USA 94:3396–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott JA, Horton E, Fibuch EE. (2011) The endocrine effects of long-term oral opioid therapy: a case report and review of the literature. J Opioid Manag 7:145–154 [DOI] [PubMed] [Google Scholar]
- Elliott K, Minami N, Kolesnikov YA, Pasternak GW, Inturrisi CE. (1994) The NMDA receptor antagonists, LY274614 and MK-801, and the nitric oxide synthase inhibitor, NG-nitro-L-arginine, attenuate analgesic tolerance to the mu-opioid morphine but not to kappa opioids. Pain 56:69–75 [DOI] [PubMed] [Google Scholar]
- Elliott KJ, Brodsky M, Hyanansky A, Foley KM, Inturrisi CE. (1995) Dextromethorphan shows efficacy in experimental pain (nociception) and opioid tolerance. Neurology 45(12, Suppl 8)S66–S68 [DOI] [PubMed] [Google Scholar]
- El Maarouf A, Kolesnikov Y, Pasternak G, Rutishauser U. (2011) Removal of polysialylated neural cell adhesion molecule increases morphine analgesia and interferes with tolerance in mice. Brain Res 1404:55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer GI, Pieper JO, Negus SS, Woods JH. (1998) Genetic variance in nociception and its relationship to the potency of morphine-induced analgesia in thermal and chemical tests. Pain 75:129–140 [DOI] [PubMed] [Google Scholar]
- Entrena JM, Cobos EJ, Nieto FR, Cendán CM, Baeyens JM, Del Pozo E. (2009a) Antagonism by haloperidol and its metabolites of mechanical hypersensitivity induced by intraplantar capsaicin in mice: role of sigma-1 receptors. Psychopharmacology (Berl) 205:21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entrena JM, Cobos EJ, Nieto FR, Cendán CM, Gris G, Del Pozo E, Zamanillo D, Baeyens JM. (2009b) Sigma-1 receptors are essential for capsaicin-induced mechanical hypersensitivity: studies with selective sigma-1 ligands and sigma-1 knockout mice. Pain 143:252–261 [DOI] [PubMed] [Google Scholar]
- Eppler CM, Hulmes JD, Wang J-B, Johnson B, Corbett M, Luthin DR, Uhl GR, Linden J. (1993) Purification and partial amino acid sequence of a μ opioid receptor from rat brain. J Biol Chem 268:26447–26451 [PubMed] [Google Scholar]
- Erez M, Takemori AE, Portoghese PS. (1982) Narcotic antagonistic potency of bivalent ligands which contain beta-naltrexamine. Evidence for bridging between proximal recognition sites. J Med Chem 25:847–849 [DOI] [PubMed] [Google Scholar]
- Escriva H, Manzon L, Youson J, Laudet V. (2002) Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol Biol Evol 19:1440–1450 [DOI] [PubMed] [Google Scholar]
- Evans CJ, Keith DE, Jr, Morrison H, Magendzo K, Edwards RH. (1992) Cloning of a delta opioid receptor by functional expression. Science 258:1952–1955 [DOI] [PubMed] [Google Scholar]
- Fairbanks CA, Wilcox GL. (1999) Spinal antinociceptive synergism between morphine and clonidine persists in mice made acutely or chronically tolerant to morphine. J Pharmacol Exp Ther 288:1107–1116 [PubMed] [Google Scholar]
- Fan T, Varghese G, Nguyen T, Tse R, O’Dowd BF, George SR. (2005) A role for the distal carboxyl tails in generating the novel pharmacology and G protein activation profile of mu and delta opioid receptor hetero-oligomers. J Biol Chem 280:38478–38488 [DOI] [PubMed] [Google Scholar]
- Faris PL, Komisaruk BR, Watkins LR, Mayer DJ. (1983) Evidence for the neuropeptide cholecystokinin as an antagonist of opiate analgesia. Science 219:310–312 [DOI] [PubMed] [Google Scholar]
- Fichna J, Janecka A, Costentin J, Do Rego JC. (2007) The endomorphin system and its evolving neurophysiological role. Pharmacol Rev 59:88–123 [DOI] [PubMed] [Google Scholar]
- Fields HL, Emson PC, Leigh BK, Gilbert RFT, Iversen LL. (1980) Multiple opiate receptor sites on primary afferent fibres. Nature 284:351–353 [DOI] [PubMed] [Google Scholar]
- Fields HL, Martin JB. (2001) Pain: pathophysiology and management, in Harrison's Principles of Internal Medicine (Braunwald E, Hauser SL, Fauci AS, Longo DL, Kasper DL, Jameson JL, eds, ed) pp 55–60, McGraw-Hill, New York [Google Scholar]
- Foley KM. (1993) Changing concepts of tolerance to opioids. What the cancer patient has taught us, in Current and Emerging Issues in Cancer Pain: Research and Practice (Chapman CR, Foley KM, eds, ed) pp 331–350, Raven Press, New York [Google Scholar]
- Foley KM. (1996) Controlling the pain of cancer. Sci Am 275:164–165 [DOI] [PubMed] [Google Scholar]
- Foote RW, Maurer R. (1982) Autoradiographic localization of opiate kappa-receptors in the guinea-pig brain. Eur J Pharmacol 85:99–103 [DOI] [PubMed] [Google Scholar]
- Fraser HF, Harris LS. (1967) Narcotic and narcotic antagonist analgesics. Annu Rev Pharmacol 7:277–300 [DOI] [PubMed] [Google Scholar]
- Frederickson RC, Smithwick EL, Shuman R, Bemis KG. (1981) Metkephamid, a systemically active analog of methionine enkephalin with potent opioid alpha-receptor activity. Science 211:603–605 [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. (2003) The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 63:1256–1272 [DOI] [PubMed] [Google Scholar]
- Freye E, Schmidhammer H, Latasch L. (2000) 14-methoxymetopon, a potent opioid, induces no respiratory depression, less sedation, and less bradycardia than sufentanil in the dog. Anesth Analg 90:1359–1364 [DOI] [PubMed] [Google Scholar]
- Fukuda K, Kato S, Mori K, Nishi M, Takeshima H, Iwabe N, Miyata T, Houtani T, Sugimoto T. (1994) cDNA cloning and regional distribution of a novel member of the opioid receptor family. FEBS Lett 343:42–46 [DOI] [PubMed] [Google Scholar]
- Fukuda K, Terasako K, Kato S, Mori K. (1995) Identification of the amino acid residues involved in selective agonist binding in the first extracellular loop of the delta- and mu-opioid receptors. FEBS Lett 373:177–181 [DOI] [PubMed] [Google Scholar]
- Fürst Z, Búzás B, Friedmann T, Schmidhammer H, Borsodi A. (1993) Highly potent novel opioid receptor agonist in the 14-alkoxymetopon series. Eur J Pharmacol 236:209–215 [DOI] [PubMed] [Google Scholar]
- Gaddum JH. (1937) The quantitative effects of antagonistic drugs. J Physiol 89:7p–9p [Google Scholar]
- Galetta S, Ling GSF, Wolfin L, Pasternak GW. (1982) Receptor binding and analgesic properties of oxymorphazone. Life Sci 31:1389–1392 [DOI] [PubMed] [Google Scholar]
- Gamse R, Holzer P, Lembeck F. (1979) Indirect evidence for presynaptic location of opiate receptors on chemosensitive primary sensory neurones. Naunyn Schmiedebergs Arch Pharmacol 308:281–285 [DOI] [PubMed] [Google Scholar]
- Garcin F, Coyle JT. (1977) Effects of perinatal 6-hydroxydopamine treatment on opiate receptor distribution in adult brain. Commun Psychopharmacol 1:283–290 [PubMed] [Google Scholar]
- Garfield E. (1983). Current Comments. Current Contents 16:5–14 [Google Scholar]
- Gates M, Tschudi G. (1956) The synthesis of morphine. J Am Chem Soc 78:1380–1393 [Google Scholar]
- Gear RW, Miaskowski C, Gordon NC, Paul SM, Heller PH, Levine JD. (1999) The kappa opioid nalbuphine produces gender- and dose-dependent analgesia and antianalgesia in patients with postoperative pain. Pain 83:339–345 [DOI] [PubMed] [Google Scholar]
- George SR, Fan T, Xie Z, Tse R, Tam V, Varghese G, O’Dowd BF. (2000) Oligomerization of mu- and delta-opioid receptors. Generation of novel functional properties. J Biol Chem 275:26128–26135 [DOI] [PubMed] [Google Scholar]
- Georges F, Normand E, Bloch B, Le Moine C. (1998) Opioid receptor gene expression in the rat brain during ontogeny, with special reference to the mesostriatal system: an in situ hybridization study. Brain Res Dev Brain Res 109:187–199 [DOI] [PubMed] [Google Scholar]
- Gintzler AR, Gershon MD, Spector S. (1978) A nonpeptide morphine-like compound: immunocytochemical localization in the mouse brain. Science 199:447–448 [DOI] [PubMed] [Google Scholar]
- Gintzler AR, Rothman TP, Gershon MD. (1980) Ontogeny of opiate mechanisms in relation to the sequential development of neurons known to be components of the guinea pig’s enteric nervous system. Brain Res 189:31–48 [DOI] [PubMed] [Google Scholar]
- Gintzler AR, Schnell SA, Gupta DS, Liu NJ, Wessendorf MW. (2008) Relationship of spinal dynorphin neurons to delta-opioid receptors and estrogen receptor alpha: anatomical basis for ovarian sex steroid opioid antinociception. J Pharmacol Exp Ther 326:725–731 [DOI] [PubMed] [Google Scholar]
- Giros B, Pohl M, Rochelle JM, Seldin MF. (1995) Chromosomal localization of opioid peptide and receptor genes in the mouse. Life Sci 56:PL369–PL375 [DOI] [PubMed] [Google Scholar]
- Goldstein A. (1976) Opioid peptides endorphins in pituitary and brain. Science 193:1081–1086 [DOI] [PubMed] [Google Scholar]
- Goldstein A, Barrett RW, James IF, Lowney LI, Weitz CJ, Knipmeyer LL, Rapoport H. (1985) Morphine and other opiates from beef brain and adrenal. Proc Natl Acad Sci USA 82:5203–5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A, Lowney LI, Pal BK. (1971) Stereospecific and nonspecific interactions of the morphine congener levorphanol in subcellular fractions of mouse brain. Proc Natl Acad Sci USA 68:1742–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A, Tachibana S, Lowney LI, Hunkapiller M, Hood L. (1979) Dynorphin-(1-13), an extraordinarily potent opioid peptide. Proc Natl Acad Sci USA 76:6666–6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes I, Filipovska J, Jordan BA, Devi LA. (2002) Oligomerization of opioid receptors. Methods 27:358–365 [DOI] [PubMed] [Google Scholar]
- Gomes I, Gupta A, Filipovska J, Szeto HH, Pintar JE, Devi LA. (2004) A role for heterodimerization of mu and delta opiate receptors in enhancing morphine analgesia. Proc Natl Acad Sci USA 101:5135–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes I, Jordan BA, Gupta A, Trapaidze N, Nagy V, Devi LA. (2000) Heterodimerization of mu and delta opioid receptors: A role in opiate synergy. J Neurosci 20:RC110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RR, Pasternak GW. (1985) Visualization of mu1 opiate receptors in rat brain by using a computerized autoradiographic subtraction technique. Proc Natl Acad Sci USA 82:6667–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RR, Snyder SH. (1982a) Autoradiographic localization of kappa opiate receptors to deep layers of the cerebral cortex may explain unique sedative and analgesic effects. Life Sci 31:1291–1294 [DOI] [PubMed] [Google Scholar]
- Goodman RR, Snyder SH. (1982b) Kappa opiate receptors localized by autoradiography to deep layers of cerebral cortex: relation to sedative effects. Proc Natl Acad Sci USA 79:5703–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RR, Snyder SH, Kuhar MJ, Young WS., 3rd (1980) Differentiation of delta and mu opiate receptor localizations by light microscopic autoradiography. Proc Natl Acad Sci USA 77:6239–6243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman AL, Elliott KJ, Inturrisi CE. (1997) The d- and l-isomers of methadone bind to the non-competitive site on the N-methyl-D-aspartate (NMDA) receptor in rat forebrain and spinal cord. Neurosci Lett 223:5–8 [DOI] [PubMed] [Google Scholar]
- Grabow TS, Hurley RW, Banfor PN, Hammond DL. (1999) Supraspinal and spinal delta(2) opioid receptor-mediated antinociceptive synergy is mediated by spinal alpha(2) adrenoceptors. Pain 83:47–55 [DOI] [PubMed] [Google Scholar]
- Granados-Soto V, Kalcheva I, Hua XY, Newton A, Yaksh TL. (2000) Spinal PKC activity and expression: role in tolerance produced by continuous spinal morphine infusion. Pain 85:395–404 [DOI] [PubMed] [Google Scholar]
- Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka BK. (2012) Structure of the δ-opioid receptor bound to naltrindole. Nature 485:400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazzini E, Puma C, Roy MO, Yu XH, O’Donnell D, Schmidt R, Dautrey S, Ducharme J, Perkins M, Panetta R, et al. (2004) Sensory neuron-specific receptor activation elicits central and peripheral nociceptive effects in rats. Proc Natl Acad Sci USA 101:7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grecksch G, Just S, Pierstorff C, Imhof AK, Gluck L, Doll C, Lupp A, Becker A, Koch T, Stumm R, Hollt V, Schulz S. (2011) Analgesic tolerance to high-efficacy agonists but not to morphine is diminished in phosphorylation-deficient S375A mu-opioid receptor knock-in mice. J Neurosci 31:13890–13896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gris P, Gauthier J, Cheng P, Gibson DG, Gris D, Laur O, Pierson J, Wentworth S, Nackley AG, Maixner W, et al. (2010) A novel alternatively spliced isoform of the mu-opioid receptor: functional antagonism. Mol Pain 6:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobe N, Lamshöft M, Orth RG, Dräger B, Kutchan TM, Zenk MH, Spiteller M. (2010) Urinary excretion of morphine and biosynthetic precursors in mice. Proc Natl Acad Sci USA 107:8147–8152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groer CE, Schmid CL, Jaeger AM, Bohn LM. (2011) Agonist-directed interactions with specific beta-arrestins determine mu-opioid receptor trafficking, ubiquitination, and dephosphorylation. J Biol Chem 286:31731–31741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groer CE, Tidgewell K, Moyer RA, Harding WW, Rothman RB, Prisinzano TE, Bohn LM. (2007) An opioid agonist that does not induce mu-opioid receptor—arrestin interactions or receptor internalization. Mol Pharmacol 71:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulland JM, Robinson R. (1925) Constitution of codeine and thebaine. Mem Proc Manchester Lit Phil Soc 69:79–86 [Google Scholar]
- Gulya K, Pelton JT, Hruby VJ, Yamamura HI. (1986) Cyclic somatostatin octapeptide analogues with high affinity and selectivity toward mu opioid receptors. Life Sci 38:2221–2229 [DOI] [PubMed] [Google Scholar]
- Gupta A, Mulder J, Gomes I, Rozenfeld R, Bushlin I, Ong E, Lim M, Maillet E, Junek M, Cahill CM, et al. (2010) Increased abundance of opioid receptor heteromers after chronic morphine administration. Sci Signal 3:ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutstein HB, Mansour A, Watson SJ, Akil H, Fields HL. (1998) Mu and kappa opioid receptors in periaqueductal gray and rostral ventromedial medulla. Neuroreport 9:1777–1781 [DOI] [PubMed] [Google Scholar]
- Gutstein HB, Trujillo KA. (1993) MK-801 inhibits the development of morphine tolerance at spinal sites. Brain Res 626:332–334 [DOI] [PubMed] [Google Scholar]
- Hahn EF, Carroll-Buatti M, Pasternak GW. (1982) Irreversible opiate agonists and antagonists: the 14-hydroxydihydromorphinone azines. J Neurosci 2:572–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn EF, Itzhak Y, Nishimura S, Johnson N, Pasternak GW. (1985) Irreversible opiate agonists and antagonists. III. Phenylhydrazone derivatives of naloxone and oxymorphone. J Pharmacol Exp Ther 235:846–850 [PubMed] [Google Scholar]
- Hahn EF, Pasternak GW. (1982) Naloxonazine, a potent, long-lasting inhibitor of opiate binding sites. Life Sci 31:1385–1388 [DOI] [PubMed] [Google Scholar]
- Han W, Kasai S, Hata H, Takahashi T, Takamatsu Y, Yamamoto H, Uhl GR, Sora I, Ikeda K. (2006) Intracisternal A-particle element in the 3′ noncoding region of the mu-opioid receptor gene in CXBK mice: a new genetic mechanism underlying differences in opioid sensitivity. Pharmacogenet Genomics 16:451–460 [DOI] [PubMed] [Google Scholar]
- Han X, Lamshöft M, Grobe N, Ren X, Fist AJ, Kutchan TM, Spiteller M, Zenk MH. (2010) The biosynthesis of papaverine proceeds via (S)-reticuline. Phytochemistry 71:1305–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanner M, Moebius FF, Flandorfer A, Knaus HG, Striessnig J, Kempner E, Glossmann H. (1996) Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proc Natl Acad Sci USA 93:8072–8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins KN, Knapp RJ, Lui GK, Gulya K, Kazmierski W, Wan YP, Pelton JT, Hruby VJ, Yamamura HI. (1989) [3H]-[H-D-Phe-Cys-Tyr-D-Trp-Orn-Thr-Pen-Thr-NH2] ([3H]CTOP), a potent and highly selective peptide for mu opioid receptors in rat brain. J Pharmacol Exp Ther 248:73–80 [PubMed] [Google Scholar]
- Hazum E, Chang KJ, Cuatrecasas P, Pasternak GW. (1981a) Naloxazone irreversibly inhibits the high affinity binding of [125I]D-ala2-D-leu5-enkephalin. Life Sci 28:2973–2979 [DOI] [PubMed] [Google Scholar]
- Hazum E, Chang K-J, Cuatrecasas P. (1979) Specific nonopiate receptors for beta-endorphin. Science 205:1033–1035 [DOI] [PubMed] [Google Scholar]
- Hazum E, Sabatka JJ, Chang K-J, Brent DA, Findlay JWA, Cuatrecasas P. (1981b) Morphine in cow and human milk: could dietary morphine constitute a ligand for specific morphine (μ) receptors? Science 213:1010–1012 [DOI] [PubMed] [Google Scholar]
- He L, Lee NM. (1998) Delta opioid receptor enhancement of mu opioid receptor-induced antinociception in spinal cord. J Pharmacol Exp Ther 285:1181–1186 [PubMed] [Google Scholar]
- He S-Q, Zhang ZN, Guan J-S, Liu H-R, Zhao B, Wang HB, Li Q, Yang H, Luo J, Li ZY, et al. (2011) Facilitation of μ-opioid receptor activity by preventing δ-opioid receptor-mediated codegradation. Neuron 69:120–131 [DOI] [PubMed] [Google Scholar]
- Herrero-Turrion MJ, Rodríguez RE. (2008) Bioinformatic analysis of the origin, sequence and diversification of mu opioid receptors in vertebrates. Mol Phylogenet Evol 49:877–892 [DOI] [PubMed] [Google Scholar]
- Herz A, Albus K, Metys J, Schubert P, Teschemacher H. (1970) On the central sites for the antinociceptive action of morphine and fentanyl. Neuropharmacology 9:539–551 [DOI] [PubMed] [Google Scholar]
- Heyman JS, Williams CL, Burks TF, Mosberg HI, Porreca F. (1988) Dissociation of opioid antinociception and central gastrointestinal propulsion in the mouse: studies with naloxonazine. J Pharmacol Exp Ther 245:238–243 [PubMed] [Google Scholar]
- Hiller JM, Pearson J, Simon EJ. (1973) Distribution of stereospecific binding of the potent narcotic analgesic etorphine in the human brain: predominance in the limbic system. Res Commun Chem Pathol Pharmacol 6:1052–1062 [PubMed] [Google Scholar]
- Hoffmann O, Plesan A, Wiesenfeld-Hallin Z. (1998) Genetic differences in morphine sensitivity, tolerance and withdrawal in rats. Brain Res 806:232–237 [DOI] [PubMed] [Google Scholar]
- Holaday JW, Pasternak GW, Faden AI. (1983) Naloxazone pretreatment modifies cardiorespiratory, temperature, and behavioral effects of morphine. Neurosci Lett 37:199–204 [DOI] [PubMed] [Google Scholar]
- Hoehe MR, Kopke K, Wendel B, Rohde K, Flachmeier C, Kidd KK, Berrettini WH, Church GM. (2000) Sequence Variability and Candidate Gene Analysis in Complex Disease: Association of Mu Opioid Receptor Gene Variation With Substance Dependence. Hum Mol Genet 9:2895–2908 [DOI] [PubMed] [Google Scholar]
- Homberg JR, Mul JD, de Wit E, Cuppen E. (2009) Complete knockout of the nociceptin/orphanin FQ receptor in the rat does not induce compensatory changes in mu, delta and kappa opioid receptors. Neuroscience 163:308–315 [DOI] [PubMed] [Google Scholar]
- Hong Y, Dai P, Jiang J, Zeng X. (2004) Dual effects of intrathecal BAM22 on nociceptive responses in acute and persistent pain—potential function of a novel receptor. Br J Pharmacol 141:423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkanen A, Vilamo L, Wegelius K, Sarviharju M, Hyytiä P, Korpi ER. (1996) Alcohol drinking is reduced by a mu 1- but not by a delta-opioid receptor antagonist in alcohol-preferring rats. Eur J Pharmacol 304:7–13 [DOI] [PubMed] [Google Scholar]
- Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang SR. (2008) Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu Rev Pharmacol Toxicol 48:393–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan P, Tallarida RJ, Haaseth RC, Matsunaga TO, Hruby VJ, Porreca F. (1992) Antinociceptive interactions of opioid delta receptor agonists with morphine in mice: supra- and sub-additivity. Life Sci 50:1535–1541 [DOI] [PubMed] [Google Scholar]
- Houde RW, Wallenstein SL. (1956) Clinical studies of morphine-nalorphine combinations. Fed Proc 15:440–441 [Google Scholar]
- Huang P, Chen C, Xu W, Yoon SI, Unterwald EM, Pintar JE, Wang Y, Chong PL, Liu-Chen LY. (2008) Brain region-specific N-glycosylation and lipid rafts association of the rat mu opioid receptor. Biochem Biophys Res Commun 365:82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PL, Dawson TM, Bredt DS, Snyder SH, Fishman MC. (1993) Targeted disruption of the neuronal nitric oxide synthase gene. Cell 75:1273–1286 [DOI] [PubMed] [Google Scholar]
- Hug CC, Jr, Oka T. (1971) Uptake of dihydromorphine-3H by synaptosomes. Life Sci I 10:201–213 [DOI] [PubMed] [Google Scholar]
- Hughes J. (1975) Isolation of an endogenous compound from the brain with pharmacological properties similar to morphine. Brain Res 88:295–308 [DOI] [PubMed] [Google Scholar]
- Hughes J, Smith TW, Kosterlitz HW, Fothergill LA, Morgan BA, Morris HR. (1975) Identification of two related pentapeptides from the brain with potent opiate agonist activity. Nature 258:577–580 [DOI] [PubMed] [Google Scholar]
- Hunger A, Kebrle J, Rossi A, Hoffmann K. (1957) Synthese basisch substituierter, analgetisch wirksamer Benzimidazol-Derivate. Synthesis of analgesically active benzimidazole derivatives with basic substitutions Experientia 13:400–401 [DOI] [PubMed] [Google Scholar]
- Hwang CK, Song KY, Kim CS, Choi HS, Guo XH, Law PY, Wei LN, Loh HH. (2007) Evidence of endogenous mu opioid receptor regulation by epigenetic control of the promoters. Mol Cell Biol 27:4720–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CK, Song KY, Kim CS, Choi HS, Guo XH, Law PY, Wei LN, Loh HH. (2009) Epigenetic programming of mu-opioid receptor gene in mouse brain is regulated by MeCP2 and Brg1 chromatin remodelling factor. J Cell Mol Med 13 (9B):3591–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S, Han W, Kasai S, Hata H, Sora I, Ikeda K. (2005) Characterization of the 3′ untranslated region of the human mu-opioid receptor (MOR-1) mRNA. Gene 364:139–145 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Ide S, Han W, Hayashida M, Uhl GR, Sora I. (2005) How individual sensitivity to opiates can be predicted by gene analyses. Trends Pharmacol Sci 26:311–317 [DOI] [PubMed] [Google Scholar]
- Ingoglia NA, Dole VP. (1970) Localization of d- and l-methadone after intraventricular injection into rat brains. J Pharmacol Exp Ther 175:84–87 [PubMed] [Google Scholar]
- International HapMap3 Consortium (2010) A map of human genome variation from population-scale sequencing. Nature 467:1061–1073. [DOI] [PMC free article] [PubMed]
- Inturrisi CE. (2002) Clinical pharmacology of opioids for pain. Clin J Pain 18(4, Suppl)S3–S13 [DOI] [PubMed] [Google Scholar]
- Inturrisi CE, Max MB, Foley KM, Schultz M, Shin SU, Houde RW. (1984) The pharmacokinetics of heroin in patients with chronic pain. N Engl J Med 310:1213–1217 [DOI] [PubMed] [Google Scholar]
- Inturrisi CE, Schultz M, Shin S, Umans JG, Angel L, Simon EJ. (1983) Evidence from opiate binding studies that heroin acts through its metabolites. Life Sci 33 (Suppl 1):773–776 [DOI] [PubMed] [Google Scholar]
- Inturrisi CE, Yoburn BC, Portenoy RK, Foley KM. (1996) Species dependent formation of morphine-6-glucuronide (M-6-G) from morphine (MOR). Committee Problems Drug Dependence 174:157 [Google Scholar]
- Irwin S, Houde RW, Bennett DR, Hendershot LC, Seevers MH. (1951) The effects of morphine methadone and meperidine on some reflex responses of spinal animals to nociceptive stimulation. J Pharmacol Exp Ther 101:132–143 [PubMed] [Google Scholar]
- Isbell H. (1977) The search for a nonaddicting analgesic: has it been worth it? The twenty-third Oscar B. Hunter Memorial Award in Therapeutics lecture. Clin Pharmacol Ther 22:377–384 [DOI] [PubMed] [Google Scholar]
- Jacobson AE, May EL, Sargent LJ. (1970) Analgetics, in Medicinal Chemistry (Part II) (Burger A, ed, ed Wiley Interscience, New York [Google Scholar]
- Jacox A, Carr DB, Payne R. (1994) Management of Cancer Pain: Clinical Practice Guidelines, US Department of Health and Human Services, Rockville, MD [Google Scholar]
- Janik J, Callahan P, Rabii J. (1992) The role of the mu(1) opioid receptor subtype in the regulation of prolactin and growth hormone secretion by Beta-endorphin in female rats: studies with naloxonazine. J Neuroendocrinol 4:701–708 [DOI] [PubMed] [Google Scholar]
- Janssen PA, Hellerback J, Schnider O, Besendorf L, Pellmont B. (1960) Diphenylpropylamines, morphinans, in Synthetic Analgesics Part I, Pergamon Press, New York [Google Scholar]
- Janssen PA, Hellerback J, Schnider O, Besendorf L, Pellmont B. (1966) Diphenylpropylamines, morphinans, in Synthetic Analgesics Part II, Pergamon, New York [Google Scholar]
- Janssen PA, Jageneau AH. (1957) A new series of potent analgesics: dextro 2:2-diphenyl-3-methyl-4-morpholino-butyrylpyrrolidine and related amides. I. Chemical structure and pharmacological activity. J Pharm Pharmacol 9:381–400 [PubMed] [Google Scholar]
- Janssen PA, Jageneau AH, Demoen PJ, Van De Westeringh C, De Canniere JH, Raeymaekers AH, Wouters MS, Sanczuk S, Hermans BK. (1959) Compounds related to pethidine—II. Mannich bases derived from various esters of 4-carboxy-4-phenylpiperidine and acetophenones. J Med Pharm Chem 1:309–317 [DOI] [PubMed] [Google Scholar]
- Ji RR, Zhang Q, Law PY, Low HH, Elde R, Hökfelt T. (1995) Expression of mu-, delta-, and kappa-opioid receptor-like immunoreactivities in rat dorsal root ganglia after carrageenan-induced inflammation. J Neurosci 15:8156–8166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Takemori AE, Sultana M, Portoghese PS, Bowen WD, Mosberg HI, Porreca F. (1991) Differential antagonism of opiate delta antinociception by [D-Ala2,Cys6]enkaphalin and naltrindole-5′-iosothiocyanate: evidence for subtypes. J Pharmacol Exp Ther 257:1069–1075 [PubMed] [Google Scholar]
- Jordan BA, Devi LA. (1999) G-protein-coupled receptor heterodimerization modulates receptor function. Nature 399:697–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius D, Basbaum AI. (2001) Molecular mechanisms of nociception. Nature 413:203–210 [DOI] [PubMed] [Google Scholar]
- Kabli N, Martin N, Fan T, Nguyen T, Hasbi A, Balboni G, O’Dowd BF, George SR. (2010) Agonists at the δ-opioid receptor modify the binding of µ-receptor agonists to the µ-δ receptor hetero-oligomer. Br J Pharmacol 161:1122–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiko RF, Foley KM, Grabinski PY, Heidrich G, Rogers AG, Inturrisi CE, Reidenberg MM. (1983) Central nervous system excitatory effects of meperidine in cancer patients. Ann Neurol 13:180–185 [DOI] [PubMed] [Google Scholar]
- Kamei J, Saitoh A, Suzuki T, Misawa M, Nagase H, Kasuya Y. (1995) Buprenorphine exerts its antinociceptive activity via mu 1-opioid receptors. Life Sci 56:PL285–PL290 [DOI] [PubMed] [Google Scholar]
- Kamei J, Sodeyama M, Tsuda M, Suzuki T, Nagase H. (1997) Antinociceptive effect of buprenorphine in M1-opioid receptor deficient CXBK mice. Life Sci 60:PL333–PL337 [DOI] [PubMed] [Google Scholar]
- Kaufman DL, Keith DE, Jr, Anton B, Tian J, Magendzo K, Newman D, Tran TH, Lee DS, Wen C, Xia Y-R, et al. (1995) Characterization of the murine μ opioid receptor gene. J Biol Chem 270:15877–15883 [DOI] [PubMed] [Google Scholar]
- Keith D, Jr, Maung T, Anton B, Evans C. (1994) Isolation of CDNA clones homologous to opioid receptors. Regul Pept 54:143–144 [Google Scholar]
- Keith DE, Anton B, Murray SR, Zaki PA, Chu PC, Lissin DV, Monteillet-Agius G, Stewart PL, Evans CJ, von Zastrow M. (1998) mu-Opioid receptor internalization: opiate drugs have differential effects on a conserved endocytic mechanism in vitro and in the mammalian brain. Mol Pharmacol 53:377–384 [PubMed] [Google Scholar]
- Keith DE, Murray SR, Zaki PA, Chu PC, Lissin DV, Kang L, Evans CJ, von Zastrow M. (1996) Morphine activates opioid receptors without causing their rapid internalization. J Biol Chem 271:19021–19024 [DOI] [PubMed] [Google Scholar]
- Kekuda R, Prasad PD, Fei YJ, Leibach FH, Ganapathy V. (1996) Cloning and functional expression of the human type 1 sigma receptor (hSigmaR1). Biochem Biophys Res Commun 229:553–558 [DOI] [PubMed] [Google Scholar]
- Kelly LE, Rieder M, van den Anker J, Malkin B, Ross C, Neely MN, Carleton B, Hayden MR, Madadi P, Koren G. (2012) More codeine fatalities after tonsillectomy in North American children. Pediatrics 129:e1343–e1347 [DOI] [PubMed] [Google Scholar]
- Kent JL, Pert CB, Herkenham M. (1981) Ontogeny of opiate receptors in rat forebrain: visualization by in vitro autoradiography. Brain Res 254:487–504 [DOI] [PubMed] [Google Scholar]
- Kepler KL, Standifer KM, Paul D, Kest B, Pasternak GW, Bodnar RJ. (1991) Gender effects and central opioid analgesia. Pain 45:87–94 [DOI] [PubMed] [Google Scholar]
- Kest B, Beczkowska I, Franklin SO, Lee CE, Mogil JS, Inturrisi CE. (1998a) Differences in delta opioid receptor antinociception, binding, and mRNA levels between BALB/c and CXBK mice. Brain Res 805:131–137 [DOI] [PubMed] [Google Scholar]
- Kest B, Lee CE, McLemore GL, Inturrisi CE. (1996) An antisense oligodeoxynucleotide to the delta opioid receptor (DOR-1) inhibits morphine tolerance and acute dependence in mice. Brain Res Bull 39:185–188 [DOI] [PubMed] [Google Scholar]
- Kest B, McLemore GL, Sadowski B, Mogil JS, Belknap JK, Inturrisi CE. (1998b) Acute morphine dependence in mice selectively-bred for high and low analgesia. Neurosci Lett 256:120–122 [DOI] [PubMed] [Google Scholar]
- Kest B, Wilson SG, Mogil JS. (1999) Sex differences in supraspinal morphine analgesia are dependent on genotype. J Pharmacol Exp Ther 289:1370–1375 [PubMed] [Google Scholar]
- Kiefel JM, Rossi GC, Bodnar RJ. (1993) Medullary μ and δ opioid receptors modulate mesencephalic morphine analgesia in rats. Brain Res 624:151–161 [DOI] [PubMed] [Google Scholar]
- Kieffer BL, Befort K, Gaveriaux-Ruff C, Hirth CG. (1992) The delta-opioid receptor: isolation of a cDNA by expression cloning and pharmacological characterization. Proc Natl Acad Sci USA 89:12048–12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian AK, Schuster CR, House JT, Sholl S, Connors M, Wainer BH. (1981) A non-peptide morphine-like compound from brain. Life Sci 28:811–817 [DOI] [PubMed] [Google Scholar]
- Kim CS, Hwang CK, Choi HS, Song KY, Law PY, Wei LN, Loh HH. (2004a) Neuron-restrictive silencer factor (NRSF) functions as a repressor in neuronal cells to regulate the mu opioid receptor gene. J Biol Chem 279:46464–46473 [DOI] [PubMed] [Google Scholar]
- Kim FJ, Kovalyshyn I, Burgman M, Neilan C, Chien CC, Pasternak GW. (2010) Sigma 1 receptor modulation of G-protein-coupled receptor signaling: potentiation of opioid transduction independent from receptor binding. Mol Pharmacol 77:695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Kim JW, Song JY, Park S, Lee HJ, Chung JH. (2004b) Association of polymorphisms in nicotinic acetylcholine receptor alpha 4 subunit gene (CHRNA4), mu-opioid receptor gene (OPRM1), and ethanol-metabolizing enzyme genes with alcoholism in Korean patients. Alcohol 34:115–120 [DOI] [PubMed] [Google Scholar]
- Kim SG, Kim CM, Kang DH, Kim YJ, Byun WT, Kim SY, Park JM, Kim MJ, Oslin DW. (2004c) Association of functional opioid receptor genotypes with alcohol dependence in Koreans. Alcohol Clin Exp Res 28:986–990 [DOI] [PubMed] [Google Scholar]
- King M, Su W, Chang A, Zuckerman A, Pasternak GW. (2001a) Transport of opioids from the brain to the periphery by P-glycoprotein: peripheral actions of central drugs. Nat Neurosci 4:268–274 [DOI] [PubMed] [Google Scholar]
- King MA, Bradshaw S, Chang AH, Pintar JE, Pasternak GW. (2001b) Potentiation of opioid analgesia in dopamine2 receptor knock-out mice: evidence for a tonically active anti-opioid system. J Neurosci 21:7788–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MA, Pan Y-X, Mei J, Chang A, Xu J, Pasternak GW. (1997a) Enhanced kappa opioid analgesia by antisense targeting the Σ1 receptor. Eur J Pharmacol 331:R5–R6 [DOI] [PubMed] [Google Scholar]
- King MA, Rossi GC, Chang AH, Williams L, Pasternak GW. (1997b) Spinal analgesic activity of orphanin FQ/nociceptin and its fragments. Neurosci Lett 223:113–116 [DOI] [PubMed] [Google Scholar]
- King MA, Su W, Nielan CL, Chang AH, Schütz J, Schmidhammer H, Pasternak GW. (2003) 14-Methoxymetopon, a very potent mu-opioid receptor-selective analgesic with an unusual pharmacological profile. Eur J Pharmacol 459:203–209 [DOI] [PubMed] [Google Scholar]
- Kitanaka N, Sora I, Kinsey S, Zeng ZZ, Uhl GR. (1998) No heroin or morphine 6β-glucuronide analgesia in mu-opioid receptor knockout mice. Eur J Pharmacol 355:R1–R3 [DOI] [PubMed] [Google Scholar]
- Knapp RJ, Malatynska E, Collins N, Fang L, Wang JY, Hruby VJ, Roeske WR, Yamamura HI. (1995) Molecular biology and pharmacology of cloned opioid receptors. FASEB J 9:516–525 [DOI] [PubMed] [Google Scholar]
- Ko JL, Chen HC, Loh HH. (2002) Differential promoter usage of mouse mu-opioid receptor gene during development. Brain Res Mol Brain Res 104:184–193 [DOI] [PubMed] [Google Scholar]
- Ko JL, Liu HC, Minnerath SR, Loh HH. (1998) Transcriptional regulation of mouse mu-opioid receptor gene. J Biol Chem 273:27678–27685 [DOI] [PubMed] [Google Scholar]
- Ko JL, Minnerath SR, Loh HH. (1997) Dual promoters of mouse mu-opioid receptor gene. Biochem Biophys Res Commun 234:351–357 [DOI] [PubMed] [Google Scholar]
- Koch T, Kroslak T, Averbeck M, Mayer P, Schröder H, Raulf E, Höllt V. (2000) Allelic variation S268P of the human mu-opioid receptor affects both desensitization and G protein coupling. Mol Pharmacol 58:328–334 [DOI] [PubMed] [Google Scholar]
- Koch T, Schulz S, Pfeiffer M, Klutzny M, Schröder H, Kahl E, Höllt V. (2001) C-terminal splice variants of the mouse mu-opioid receptor differ in morphine-induced internalization and receptor resensitization. J Biol Chem 276:31408–31414 [DOI] [PubMed] [Google Scholar]
- Koch T, Schulz S, Schröder H, Wolf R, Raulf E, Höllt V. (1998) Carboxyl-terminal splicing of the rat mu opioid receptor modulates agonist-mediated internalization and receptor resensitization. J Biol Chem 273:13652–13657 [DOI] [PubMed] [Google Scholar]
- Kolakowski LF., Jr (1994) GCRDb: a G-protein-coupled receptor database. Receptors Channels 2:1–7 [PubMed] [Google Scholar]
- Kolesnikov Y, Cristea M, Oksman G, Torosjan A, Wilson R. (2004) Evaluation of the tail formalin test in mice as a new model to assess local analgesic effects. Brain Res 1029:217–223 [DOI] [PubMed] [Google Scholar]
- Kolesnikov Y, Jain S, Wilson R, Pasternak GW. (1996a) Peripheral kappa 1-opioid receptor-mediated analgesia in mice. Eur J Pharmacol 310:141–143 [DOI] [PubMed] [Google Scholar]
- Kolesnikov Y, Jain S, Wilson R, Pasternak GW. (1998) Lack of morphine and enkephalin tolerance in 129/SvEv mice: evidence for a NMDA receptor defect. J Pharmacol Exp Ther 284:455–459 [PubMed] [Google Scholar]
- Kolesnikov Y, Pasternak GW. (1999) Topical opioids in mice: analgesia and reversal of tolerance by a topical N-methyl-D-aspartate antagonist. J Pharmacol Exp Ther 290:247–252 [PubMed] [Google Scholar]
- Kolesnikov YA, Chereshnev I, Criesta M, Pan YX, Pasternak GW. (2009) Opposing actions of neuronal nitric oxide synthase isoforms in formalin-induced pain in mice. Brain Res 1289:14–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov YA, Chereshnev I, Pasternak GW. (2000) Analgesic synergy between topical lidocaine and topical opioids. J Pharmacol Exp Ther 295:546–551 [PubMed] [Google Scholar]
- Kolesnikov YA, Cristea M, and Pasternak GW (2003a) Analgesic synergy between topical morphine and butamben in mice. Anesth Analg 97:1103–1107 table. [DOI] [PubMed]
- Kolesnikov YA, Ferkany J, Pasternak GW. (1993a) Blockade of mu and kappa 1 opioid analgesic tolerance by NPC17742, a novel NMDA antagonist. Life Sci 53:1489–1494 [DOI] [PubMed] [Google Scholar]
- Kolesnikov YA, Jain S, Wilson R, Pasternak GW. (1996b) Peripheral morphine analgesia: synergy with central sites and a target of morphine tolerance. J Pharmacol Exp Ther 279:502–506 [PubMed] [Google Scholar]
- Kolesnikov YA, Maccechini M-L, Pasternak GW. (1994) 1-Aminocyclopropane carboxylic acid (ACPC) prevents mu and delta opioid tolerance. Life Sci 55:1393–1398 [DOI] [PubMed] [Google Scholar]
- Kolesnikov YA, Pan YX, Babey AM, Jain S, Wilson R, Pasternak GW. (1997) Functionally differentiating two neuronal nitric oxide synthase isoforms through antisense mapping: evidence for opposing NO actions on morphine analgesia and tolerance. Proc Natl Acad Sci USA 94:8220–8225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov YA, Pick CG, Ciszewska G, Pasternak GW. (1993b) Blockade of tolerance to morphine but not to kappa opioids by a nitric oxide synthase inhibitor. Proc Natl Acad Sci USA 90:5162–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov YA, Pick CG, Pasternak GW. (1992) NG-nitro-L-arginine prevents morphine tolerance. Eur J Pharmacol 221:399–400 [DOI] [PubMed] [Google Scholar]
- Kolesnikov YA, Wilson RS, Pasternak GW. (2003b) The synergistic analgesic interactions between hydrocodone and ibuprofen. Anesth Analg 97:1721–1723 [DOI] [PubMed] [Google Scholar]
- Kosterlitz HW, Robinson JA. (1957a) Inhibition of the peristaltic reflex of the isolated guinea-pig ileum. J Physiol 136:249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosterlitz HW, Robinson JA. (1957b) Inhibition of the peristaltic reflex of the isolated guinea-pig ileum. J Physiol 136:249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovelowski CJ, Bian D, Hruby VJ, Lai J, Ossipov MH, Porreca F. (1999) Selective opioid δ agonists elicit antinociceptive supraspinal/spinal synergy in the rat. Brain Res 843:12–17 [DOI] [PubMed] [Google Scholar]
- Kozak CA, Filie J, Adamson MC, Chen Y, Yu L. (1994) Murine chromosomal location of the μ and kappa opioid receptor genes. Genomics 21:659–661 [DOI] [PubMed] [Google Scholar]
- Kraus J. (2009) Regulation of mu-opioid receptors by cytokines. Front Biosci (Schol Ed) 1:164–170 Schol Ed [DOI] [PubMed] [Google Scholar]
- Kraus J, Borner C, Giannini E, Hickfang K, Braun H, Mayer P, Hoehe MR, Ambrosch A, Konig W, Hollt V. (2001) Regulation of mu-opioid receptor gene transcription by interleukin-4 and influence of an allelic variation within a STAT6 transcription factor binding site. J Biol Chem 276:43901–43908 [DOI] [PubMed] [Google Scholar]
- Kraus J, Börner C, Giannini E, Höllt V. (2003) The role of nuclear factor kappaB in tumor necrosis factor-regulated transcription of the human mu-opioid receptor gene. Mol Pharmacol 64:876–884 [DOI] [PubMed] [Google Scholar]
- Kraus J, Horn G, Zimprich A, Simon T, Mayer P, Höllt V. (1995) Molecular cloning and functional analysis of the rat μ opioid receptor gene promoter. Biochem Biophys Res Commun 215:591–597 [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Bart G, Lilly C, LaForge KS, Nielsen DA. (2005a) Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol Rev 57:1–26 [DOI] [PubMed] [Google Scholar]
- Kreek MJ, LaForge KS. (2007) Stress responsivity, addiction, and a functional variant of the human mu-opioid receptor gene. Mol Interv 7:74–78 [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. (2005b) Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nat Neurosci 8:1450–1457 [DOI] [PubMed] [Google Scholar]
- Kroslak T, Laforge KS, Gianotti RJ, Ho A, Nielsen DA, Kreek MJ. (2007) The single nucleotide polymorphism A118G alters functional properties of the human mu opioid receptor. J Neurochem 103:77–87 [DOI] [PubMed] [Google Scholar]
- Krueger H, Eddy NB, Sumwalt M. (1941) The Pharmacology of the Opium Alkaloids, Federal Security Agency, U.S. Public Health Service, Washington, D.C. [Google Scholar]
- Kuehn BM. (2013) FDA: No codeine after tonsillectomy for children. JAMA 309:1100. [DOI] [PubMed] [Google Scholar]
- Kuhar MJ, Pert CB, Snyder SH. (1973) Regional distribution of opiate receptor binding in monkey and human brain. Nature 245:447–450 [DOI] [PubMed] [Google Scholar]
- Kupferberg HJ, Way EL. (1963) Pharmacologic basis for the increased sensitivity of the newborn rat to morphine. J Pharmacol Exp Ther 141:105–112 [PubMed] [Google Scholar]
- Kvam TM, Baar C, Rakvåg TT, Kaasa S, Krokan HE, Skorpen F. (2004) Genetic analysis of the murine mu opioid receptor: increased complexity of Oprm gene splicing. J Mol Med (Berl) 82:250–255 [DOI] [PubMed] [Google Scholar]
- Lacroix-Fralish ML, Mogil JS. (2009) Progress in genetic studies of pain and analgesia. Annu Rev Pharmacol Toxicol 49:97–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaForge KS, Yuferov V, Kreek MJ. (2000) Opioid receptor and peptide gene polymorphisms: potential implications for addictions. Eur J Pharmacol 410:249–268 [DOI] [PubMed] [Google Scholar]
- Lai J, Bilsky EJ, Bernstein RN, Rothman RB, Pasternak GW, Porreca F. (1994a) antisense oligodeoxynucleotide to the cloned delta opioid receptor selectively inhibits supraspinal, but not spinal, antinociceptive effects of [D-Ala2, Glu4]deltorphin. Regul Pept 54:159–160 [Google Scholar]
- Lai J, Bilsky EJ, Rothman RB, Porreca F. (1994b) Treatment with antisense oligodeoxynucleotide to the opioid δ receptor selectively inhibits δ 2-agonist antinociception. Neuroreport 5:1049–1052 [DOI] [PubMed] [Google Scholar]
- Lamotte C, Pert CB, Snyder SH. (1976) Opiate receptor binding in primate spinal cord: distribution and changes after dorsal root section. Brain Res 112:407–412 [DOI] [PubMed] [Google Scholar]
- Lange DG, Roerig SC, Fujimoto JM, Wang RI. (1980) Absence of cross-tolerance to heroin in morphine-tolerant mice. Science 208:72–74 [DOI] [PubMed] [Google Scholar]
- Langley JN. (1909) On the contraction of muscle, chiefly in relation to the presence of; receptive’ substances: Part IV. The effect of curari and of some other substances on the nicotine response of the sartorius and gastrocnemius muscles of the frog. J Physiol 39:235–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larhammar D, Dreborg S, Larsson TA, Sundström G. (2009) Early duplications of opioid receptor and Peptide genes in vertebrate evolution. Ann N Y Acad Sci 1163:451–453 [DOI] [PubMed] [Google Scholar]
- Lariviere WR, Wilson SG, Laughlin TM, Kokayeff A, West EE, Adhikari SM, Wan Y, Mogil JS. (2002) Heritability of nociception. III. Genetic relationships among commonly used assays of nociception and hypersensitivity. Pain 97:75–86 [DOI] [PubMed] [Google Scholar]
- Lasagna L. (1964) The clinical evaluation of morphine and its substitutes as analgesics. Pharmacol Rev 16:47–83 [PubMed] [Google Scholar]
- Lasagna L. (1984) Historical overview, in Analgesics: Neurochemical, Behavioral and Clinical Perspectives (Kuhar MJ, Pasternak GW, eds, ed) pp 1–8, Raven Press Inc, New York, NY [Google Scholar]
- Lasagna L, Beecher HK. (1954) The analgesic effectiveness of nalorphine and nalorphine-morphine combinations in man. J Pharmacol Exp Ther 112:356–363 [PubMed] [Google Scholar]
- Law P-Y. (2011) Opioid receptor signal transduction mechanisms, in The Opiate Receptors (Pasternak GW, ed, ed) pp 195–238, Springer, New York [Google Scholar]
- Law PY, Erickson-Herbrandson LJ, Zha QQ, Solberg J, Chu J, Sarre A, Loh HH. (2005) Heterodimerization of mu- and delta-opioid receptors occurs at the cell surface only and requires receptor-G protein interactions. J Biol Chem 280:11152–11164 [DOI] [PubMed] [Google Scholar]
- Law PY, Fischer G, Loh HH, Herz A. (1979) Inhibition of specific opiate binding to synaptic membrane by cerebroside sulfatase. Biochem Pharmacol 28:2557–2562 [DOI] [PubMed] [Google Scholar]
- Law PY, Harris RA, Loh HH, Way EL. (1978) Evidence for the involvement of cerebroside sulfate in opiate receptor binding: Studies with Azure A and jimpy mutant mice. J Pharmacol Exp Ther 207:458–468 [PubMed] [Google Scholar]
- Law P-Y, Loh HH. (1999) Regulation of opioid receptor activities. J Pharmacol Exp Ther 289:607–624 [PubMed] [Google Scholar]
- Law PY, Wong YH, Loh HH. (2000) Molecular mechanisms and regulation of opioid receptor signaling. Annu Rev Pharmacol Toxicol 40:389–430 [DOI] [PubMed] [Google Scholar]
- Law PY, Wu J, Koehler JE, Loh HH. (1981) Demonstration and characterization of opiate inhibition of the striatal adenylate cyclase. J Neurochem 36:1834–1846 [DOI] [PubMed] [Google Scholar]
- Le Bars D, Gozariu M, Cadden SW. (2001) Animal models of nociception. Pharmacol Rev 53:597–652 [PubMed] [Google Scholar]
- Leander JD. (1987) Buprenorphine has potent kappa opioid receptor antagonist activity. Neuropharmacology 26:1445–1447 [DOI] [PubMed] [Google Scholar]
- Lee C, Atanelov L, Modrek B, Xing Y. (2003) ASAP: the Alternative Splicing Annotation Project. Nucleic Acids Res 31:101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Akera T, Stolman S, Brody TM. (1975) Saturable binding of dihydromorphine and naloxone to rat brain tissue in vitro. J Pharmacol Exp Ther 194:583–592 [PubMed] [Google Scholar]
- Lee PW, Wu S, Lee YM. (2004) Differential expression of mu-opioid receptor gene in CXBK and B6 mice by Sp1. Mol Pharmacol 66:1580–1584 [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Caron MG, Michel T, Stadel JM. (1982) Mechanisms of hormone receptor-effector coupling: the beta-adrenergic receptor and adenylate cyclase. Fed Proc 41:2664–2670 [PubMed] [Google Scholar]
- Lefkowitz RJ, Caron MG, Stiles GL. (1984) Mechanisms of membrane-receptor regulation. Biochemical, physiological, and clinical insights derived from studies of the adrenergic receptors. N Engl J Med 310:1570–1579 [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Mullikin D, Caron MG. (1976) Regulation of beta-adrenergic receptors by guanyl-5′-yl imidodiphosphate and other purine nucleotides. J Biol Chem 251:4686–4692 [PubMed] [Google Scholar]
- Lembo PM, Grazzini E, Groblewski T, O’Donnell D, Roy MO, Zhang J, Hoffert C, Cao J, Schmidt R, Pelletier M, et al. (2002) Proenkephalin A gene products activate a new family of sensory neuron—specific GPCRs. Nat Neurosci 5:201–209 [DOI] [PubMed] [Google Scholar]
- Lerman C, Wileyto EP, Patterson F, Rukstalis M, Audrain-McGovern J, Restine S, Shields PG, Kaufmann V, Redden D, Benowitz N, et al. (2004) The functional mu opioid receptor (OPRM1) Asn40Asp variant predicts short-term response to nicotine replacement therapy in a clinical trial. Pharmacogenomics J 4:184–192 [DOI] [PubMed] [Google Scholar]
- Lewenstein MJ and Fishman J (1966) Morphine derivative, U.S. Patent 3254088. [Google Scholar]
- Lewis JW. (1985) Buprenorphine. Drug Alcohol Depend 14:363–372 [DOI] [PubMed] [Google Scholar]
- Li CH, Chung D. (1976) Primary structure of human beta-lipotropin. Nature 260:622–624 [DOI] [PubMed] [Google Scholar]
- Li CH, Chung D, Doneen BA. (1976) Isolation, characterization and opiate activity of beta-endorphin from human pituitary glands. Biochem Biophys Res Commun 72:1542–1547 [DOI] [PubMed] [Google Scholar]
- Li J, Huang P, Chen C, de Riel JK, Weinstein H, Liu-Chen LY. (2001) Constitutive activation of the mu opioid receptor by mutation of D3.49(164), but not D3.32(147): D3.49(164) is critical for stabilization of the inactive form of the receptor and for its expression. Biochemistry 40:12039–12050 [DOI] [PubMed] [Google Scholar]
- Li JG, Chen CG, Yin JL, Rice K, Zhang Y, Matecka D, de Riel JK, DesJarlais RL, Liu-Chen LY. (1999) ASP147 in the third transmembrane helix of the rat μ opioid receptor forms ion-pairing with morphine and naltrexone. Life Sci 65:175–185 [DOI] [PubMed] [Google Scholar]
- Li S, Zhu J, Chen C, Chen Y-W, Deriel JK, Ashby B, Liu-Chen L-Y. (1993) Molecular cloning and expression of a rat kappa opioid receptor. Biochem J 295:629–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Carr LG. (1997) Transcription of the mouse mu-opioid receptor gene is regulated by two promoters. Brain Res 769:372–374 [DOI] [PubMed] [Google Scholar]
- Liang Y, Mestek A, Yu L, Carr LG. (1995) Cloning and characterization of the promoter region of the mouse μ opioid receptor gene. Brain Res 679:82–88 [DOI] [PubMed] [Google Scholar]
- Liang YB, Carr LG. (1996) Identification of an octamer-1 transcription factor binding site in the promoter of the mouse mu-opioid receptor gene. J Neurochem 67:1352–1359 [DOI] [PubMed] [Google Scholar]
- Ling GSF, MacLeod JM, Lee S, Lockhart SH, Pasternak GW. (1984) Separation of morphine analgesia from physical dependence. Science 226:462–464 [DOI] [PubMed] [Google Scholar]
- Ling GSF, Simantov R, Clark JA, Pasternak GW. (1986) Naloxonazine actions in vivo. Eur J Pharmacol 129:33–38 [DOI] [PubMed] [Google Scholar]
- Ling GSF, Spiegel K, Lockhart SH, Pasternak GW. (1985) Separation of opioid analgesia from respiratory depression: evidence for different receptor mechanisms. J Pharmacol Exp Ther 232:149–155 [PubMed] [Google Scholar]
- Ling GSF, Spiegel K, Nishimura SL, Pasternak GW. (1983) Dissociation of morphine’s analgesic and respiratory depressant actions. Eur J Pharmacol 86:487–488 [DOI] [PubMed] [Google Scholar]
- Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, IJzerman AP, et al. (2012) Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XY, Liu ZC, Sun YG, Ross M, Kim S, Tsai FF, Li QF, Jeffry J, Kim JY, Loh HH, et al. (2011) Unidirectional cross-activation of GRPR by MOR1D uncouples itch and analgesia induced by opioids. Cell 147:447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chen LY, Phillips CA. (1987) Covalent labeling of mu opioid binding site by [3H]beta-funaltrexamine. Mol Pharmacol 32:321–329 [PubMed] [Google Scholar]
- Liu-Chen L-Y, Chen C, Phillips CA. (1993) Beta-[3H]funaltrexamine-labeled mu-opioid receptors: species variations in molecular mass and glycosylation by complex-type, N-linked oligosaccharides. Mol Pharmacol 44:749–756 [PubMed] [Google Scholar]
- Loh HH, Liu HC, Cavalli A, Yang W, Chen YF, Wei LN. (1998) mu Opioid receptor knockout in mice: effects on ligand-induced analgesia and morphine lethality. Brain Res Mol Brain Res 54:321–326 [DOI] [PubMed] [Google Scholar]
- Lord JAH, Waterfield AA, Hughes J, Kosterlitz HW. (1977) Endogenous opioid peptides: multiple agonists and receptors. Nature 267:495–499 [DOI] [PubMed] [Google Scholar]
- Lötsch J, Tegeder I, Angst MS, Geisslinger G. (2000) Antinociceptive effects of morphine-6-glucuronide in homozygous MDR1a P-glycoprotein knockout and in wildtype mice in the hotplate test. Life Sci 66:2393–2403 [DOI] [PubMed] [Google Scholar]
- Luke MC, Hahn EF, Price M, Pasternak GW. (1988) Irreversible opiate agonists and antagonists: V. Hydrazone and acylhydrazone derivatives of naltrexone. Life Sci 43:1249–1256 [DOI] [PubMed] [Google Scholar]
- Lundin LG, Larhammar D, Hallböök F. (2003) Numerous groups of chromosomal regional paralogies strongly indicate two genome doublings at the root of the vertebrates. J Struct Funct Genomics 3:53–63 [PubMed] [Google Scholar]
- Lupardus PJ, Wilke RA, Aydar E, Palmer CP, Chen Y, Ruoho AE, Jackson MB. (2000) Membrane-delimited coupling between sigma receptors and K+ channels in rat neurohypophysial terminals requires neither G-protein nor ATP. J Physiol 526:527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutfy K, Cowan A. (2004) Buprenorphine: a unique drug with complex pharmacology. Curr Neuropharmacol 2:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz RA, Cruciani RA, Costa T, Munson PJ, Rodbard D. (1984) A very high affinity opioid binding site in rat brain: demonstration by computer modeling. Biochem Biophys Res Commun 122:265–269 [DOI] [PubMed] [Google Scholar]
- Macht DI. (1915) The history of opium and some its preparations and alkaloids. JAMA LXIV:477–481 [Google Scholar]
- Mague SD, Isiegas C, Huang P, Liu-Chen LY, Lerman C, Blendy JA. (2009) Mouse model of OPRM1 (A118G) polymorphism has sex-specific effects on drug-mediated behavior. Proc Natl Acad Sci USA 106:10847–10852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, Grinnell S, Le Rouzic V, Burgman M, Polikar L, Ansonoff M, Pintar J, Pan YX, Pasternak GW. (2011) Truncated G protein-coupled mu opioid receptor MOR-1 splice variants are targets for highly potent opioid analgesics lacking side effects. Proc Natl Acad Sci USA 108:19778–19783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, Subrath J, Le Rouzic V, Polikar L, Burgman M, Nagakura K, Ocampo J, Haselton N, Pasternak AR, Grinnell S, et al. (2012) Synthesis and evaluation of aryl-naloxamide opiate analgesics targeting truncated exon 11-associated μ opioid receptor (MOR-1) splice variants. J Med Chem 55:6352–6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. (2012) Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature 485:321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann PE, Arjune D, Romero MT, Pasternak GW, Hahn EF, Bodnar RJ. (1988a) Differential sensitivity of opioid-induced feeding to naloxone and naloxonazine. Psychopharmacology (Berl) 94:336–341 [DOI] [PubMed] [Google Scholar]
- Mann PE, Pasternak GW, Hahn EF, Curreri G, Lubin E, Bodnar RJ. (1988b) Comparison of chronic naloxone and naloxonazine effects upon food intake and maintenance of body weight in rats. Neuropharmacology 27:349–355 [DOI] [PubMed] [Google Scholar]
- Mansour A, Fox CA, Akil H, Watson SJ. (1995a) Opioid-receptor mRNA expression in the rat CNS: anatomical and functional implications. Trends Neurosci 18:22–29 [DOI] [PubMed] [Google Scholar]
- Mansour A, Fox CA, Burke S, Akil H, Watson SJ. (1995b) Immunohistochemical localization of the cloned μ opioid receptor in the rat CNS. J Chem Neuroanat 8:283–305 [DOI] [PubMed] [Google Scholar]
- Mansour A, Fox CA, Burke S, Meng F, Thompson RC, Akil H, Watson SJ. (1994a) Mu, delta, and kappa opioid receptor mRNA expression in the rat CNS: an in situ hybridization study. J Comp Neurol 350:412–438 [DOI] [PubMed] [Google Scholar]
- Mansour A, Fox CA, Burke S, Watson SJ. (1994b) Immunohistochemical Localization of the Mu Opioid Receptors. Regul Pept 54:179–180 [Google Scholar]
- Mansour A, Khachaturian H, Lewis ME, Akil H, Watson SJ. (1987) Autoradiographic differentiation of mu, delta, and kappa opioid receptors in the rat forebrain and midbrain. J Neurosci 7:2445–2464 [PMC free article] [PubMed] [Google Scholar]
- Martin WR. (1963) Analgesic and antipyretic drugs, in Physiological Pharmacology (Root WS, Hofman FG, eds, ed) pp 275–312, Academic Press, New York [Google Scholar]
- Martin WR. (1967) Opioid antagonists. Pharmacol Rev 19:463–521 [PubMed] [Google Scholar]
- Martin WR, Eades CG, Thompson JA, Huppler RE, Gilbert PE. (1976) The effects of morphine- and nalorphine- like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther 197:517–532 [PubMed] [Google Scholar]
- Matthes HWD, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, Befort K, Dierich A, Le Meur M, Dollé P, et al. (1996) Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature 383:819–823 [DOI] [PubMed] [Google Scholar]
- Mattia A, Vanderah T, Mosberg HI, Porreca F. (1991) Lack of antinociceptive cross-tolerance between [D-Pen2, D-Pen5]enkephalin and [D-Ala2]deltorphin II in mice: evidence for delta receptor subtypes. J Pharmacol Exp Ther 258:583–587 [PubMed] [Google Scholar]
- Maurer R, Cortés R, Probst A, Palacios JM. (1983) Multiple opiate receptor in human brain: an autoradiographic investigation. Life Sci 33 (Suppl 1):231–234 [DOI] [PubMed] [Google Scholar]
- Maurer R, Gaehwiler BH, Buescher HH, Hill RC, Roemer D. (1982) Opiate antagonistic properties of an octapeptide somatostatin analog. Proc Natl Acad Sci USA 79:4815–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer DJ, Mao J, Price DD. (1995) The development of morphine tolerance and dependence is associated with translocation of protein kinase C. Pain 61:365–374 [DOI] [PubMed] [Google Scholar]
- Mayer P, Höllt V. (2006) Pharmacogenetics of opioid receptors and addiction. Pharmacogenet Genomics 16:1–7 [DOI] [PubMed] [Google Scholar]
- Mayer P, Schulzeck S, Kraus J, Zimprich A, Höllt V. (1996) Promoter region and alternatively spliced exons of the rat mu-opioid receptor gene. J Neurochem 66:2272–2278 [DOI] [PubMed] [Google Scholar]
- McCawley EL, Hart ER, Marsh DF. (1941) The preparation of N-allylnormorphine. J Am Chem Soc 63:314 [Google Scholar]
- Mei J, Pasternak GW. (2001) Molecular cloning and pharmacological characterization of the rat sigma1 receptor. Biochem Pharmacol 62:349–355 [DOI] [PubMed] [Google Scholar]
- Mei J, Pasternak GW. (2007) Modulation of brainstem opiate analgesia in the rat by sigma 1 receptors: a microinjection study. J Pharmacol Exp Ther 322:1278–1285 [DOI] [PubMed] [Google Scholar]
- Melief EJ, Miyatake M, Carroll FI, Béguin C, Carlezon WA, Jr, Cohen BM, Grimwood S, Mitch CH, Rorick-Kehn L, Chavkin C. (2011) Duration of action of a broad range of selective κ-opioid receptor antagonists is positively correlated with c-Jun N-terminal kinase-1 activation. Mol Pharmacol 80:920–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Xie G-X, Thompson RC, Mansour A, Goldstein A, Watson SJ, Akil H. (1993) Cloning and pharmacological characterization of a rat kappa opioid receptor. Proc Natl Acad Sci USA 90:9954–9958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min BH, Augustin LB, Felsheim RF, Fuchs JA, Loh HH. (1994) Genomic structure analysis of promoter sequence of a mouse μ opioid receptor gene. Proc Natl Acad Sci USA 91:9081–9085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami M, Maekawa K, Yabuuchi K, Satoh M. (1995) Double in situ hybridization study on coexistence of mu-, delta- and kappa-opioid receptor mRNAs with preprotachykinin A mRNA in the rat dorsal root ganglia. Brain Res Mol Brain Res 30:203–210 [DOI] [PubMed] [Google Scholar]
- Minami M, Nakagawa T, Seki T, Onogi T, Aoki Y, Katao Y, Katsumata S, Satoh M. (1996) A single residue, Lys108, of the delta-opioid receptor prevents the mu-opioid-selective ligand [D-Ala2,N-MePhe4,Gly-ol5]enkephalin from binding to the delta-opioid receptor. Mol Pharmacol 50:1413–1422 [PubMed] [Google Scholar]
- Minami M, Onogi T, Toya T, Katao Y, Hosoi Y, Maekawa K, Katsumata S, Yabuuchi K, Satoh M. (1994) Molecular cloning and in situ hybridization histochemistry for rat mu-opioid receptor. Neurosci Res 18:315–322 [DOI] [PubMed] [Google Scholar]
- Minami M, Satoh M. (1995) Molecular biology of the opioid receptors: structures, functions and distributions. Neurosci Res 23:121–145 [DOI] [PubMed] [Google Scholar]
- Mitchell JM, Lowe D, Fields HL. (1998) The contribution of the rostral ventromedial medulla to the antinociceptive effects of systemic morphine in restrained and unrestrained rats. Neuroscience 87:123–133 [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Bowen WD, Portoghese PS, Takemori AE. (1994) Lack of involvement of delta-1 opioid receptors in the development of physical dependence on morphine in mice. J Pharmacol Exp Ther 270:37–39 [PubMed] [Google Scholar]
- Mogil JS. (1999) The genetic mediation of individual differences in sensitivity to pain and its inhibition. Proc Natl Acad Sci USA 96:7744–7751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogil JS, Bailey AL. (2010) Sex and gender differences in pain and analgesia. Prog Brain Res 186:141–157 [DOI] [PubMed] [Google Scholar]
- Mogil JS, Chesler EJ, Wilson SG, Juraska JM, Sternberg WF. (2000a) Sex differences in thermal nociception and morphine antinociception in rodents depend on genotype. Neurosci Biobehav Rev 24:375–389 [DOI] [PubMed] [Google Scholar]
- Mogil JS, Kest B, Sadowski B, Belknap JK. (1996) Differential genetic mediation of sensitivity to morphine in genetic models of opiate antinociception: influence of nociceptive assay. J Pharmacol Exp Ther 276:532–544 [PubMed] [Google Scholar]
- Mogil JS, Marek P, Flodman P, Spence MA, Sternberg WF, Kest B, Sadowski B, Liebeskind JC. (1995) One or two genetic loci mediate high opiate analgesia in selectively bred mice. Pain 60:125–135 [DOI] [PubMed] [Google Scholar]
- Mogil JS, Marek P, O’Toole LA, Helms ML, Sadowski B, Liebeskind JC, Belknap JK. (1994) Mu-opiate receptor binding is up-regulated in mice selectively bred for high stress-induced analgesia. Brain Res 653:16–22 [DOI] [PubMed] [Google Scholar]
- Mogil JS, Yu L, Basbaum AI. (2000b) Pain genes?: natural variation and transgenic mutants. Annu Rev Neurosci 23:777–811 [DOI] [PubMed] [Google Scholar]
- Mollereau C, Parmentier M, Mailleux P, Butour JL, Moisand C, Chalon P, Caput D, Vassart G, Meunier JC. (1994) ORL1, a novel member of the opioid receptor family. Cloning, functional expression and localization. FEBS Lett 341:33–38 [DOI] [PubMed] [Google Scholar]
- Morgan MM, Heinricher MM, Fields HL. (1992) Circuitry linking opioid-sensitive nociceptive modulatory systems in periaqueductal gray and spinal cord with rostral ventromedial medulla. Neuroscience 47:863–871 [DOI] [PubMed] [Google Scholar]
- Moriwaki A, Wang JB, Svingos A, van Bockstaele E, Cheng P, Pickel V, Uhl GR. (1996) μ Opiate receptor immunoreactivity in rat central nervous system. Neurochem Res 21:1315–1331 [DOI] [PubMed] [Google Scholar]
- Moskowitz AS, Goodman RR. (1985a) Autoradiographic analysis of mu1, mu2, and delta opioid binding in the central nervous system of C57BL/6BY and CXBK (opioid receptor-deficient) mice. Brain Res 360:108–116 [DOI] [PubMed] [Google Scholar]
- Moskowitz AS, Goodman RR. (1985b) Autoradiographic distribution of mu1 and mu2 opioid binding in the mouse central nervous system. Brain Res 360:117–129 [DOI] [PubMed] [Google Scholar]
- Moulin DE, Ling GSF, Pasternak GW. (1988) Unidirectional analgesic cross-tolerance between morphine and levorphanol in the rat. Pain 33:233–239 [DOI] [PubMed] [Google Scholar]
- Mulé SJ, Casella G, Clouet DH. (1975) The specificity of binding of the narcotic agonist etorphine in synaptic membranes of rat brain in vivo. Psychopharmacology (Berl) 44:125–129 [DOI] [PubMed] [Google Scholar]
- Munson PJ. (1983) Experimental artifacts and the analysis of ligand binding data: results of a computer simulation. J Recept Res 3:249–259 [DOI] [PubMed] [Google Scholar]
- Munson PJ, Cruciani RA, Lutz RA, Rodbard D. (1984) New methods for characterization of complex receptor systems involving 3 or more binding sites: application to brain opiate receptors. J Recept Res 4:339–355 [DOI] [PubMed] [Google Scholar]
- Navon S, Lajtha A. (1970) Uptake of morphine in particulate fractions from rat brain. Brain Res 24:534–536 [DOI] [PubMed] [Google Scholar]
- Neilan CL, King MA, Rossi G, Ansonoff M, Pintar JE, Schiller PW, Pasternak GW. (2003) Differential sensitivities of mouse strains to morphine and [Dmt1]DALDA analgesia. Brain Res 974:254–257 [DOI] [PubMed] [Google Scholar]
- Newman LC, Sands SS, Wallace DR, Stevens CW. (2002) Characterization of mu, kappa, and delta opioid binding in amphibian whole brain tissue homogenates. J Pharmacol Exp Ther 301:364–370 [DOI] [PubMed] [Google Scholar]
- Newman LC, Wallace DR, Stevens CW. (2000) Selective opioid receptor agonist and antagonist displacement of [3H]naloxone binding in amphibian brain. Eur J Pharmacol 397:255–262 [DOI] [PubMed] [Google Scholar]
- Nishi M, Takeshima H, Mori M, Nakagawara K, Takeuchi T. (1994) Structure and chromosomal mapping of genes for the mouse kappa-opioid receptor and an opioid receptor homologue (MOR-C). Biochem Biophys Res Commun 205:1353–1357 [DOI] [PubMed] [Google Scholar]
- Nitsche JF, Schuller AG, King MA, Zengh M, Pasternak GW, Pintar JE. (2002) Genetic dissociation of opiate tolerance and physical dependence in delta-opioid receptor-1 and preproenkephalin knock-out mice. J Neurosci 22:10906–10913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg G. (1984) Pharmacokinetic aspects of spinal morphine analgesia. Acta Anaesthesiol Scand Suppl 79:1–38 [DOI] [PubMed] [Google Scholar]
- Ohkawa S, DiGiacomo B, Larson DL, Takemori AE, Portoghese PS. (1997) 7-Spiroindanyl derivatives of naltrexone and oxymorphone as selective ligands for δ opioid receptors. J Med Chem 40:1720–1725 [DOI] [PubMed] [Google Scholar]
- Ohno S. (1999) Gene duplication and the uniqueness of vertebrate genomes circa 1970-1999. Semin Cell Dev Biol 10:517–522 [DOI] [PubMed] [Google Scholar]
- Ossipov MH, Lopez Y, Bian D, Nichols ML, Porreca F. (1997) Synergistic antinociceptive interactions of morphine and clonidine in rats with nerve-ligation injury. Anesthesiology 86:196–204 [DOI] [PubMed] [Google Scholar]
- Paakkari P, Paakkari I, Feuerstein G, Sirén A-L. (1992) Evidence for differential opioid mu 1- and mu 2-receptor-mediated regulation of heart rate in the conscious rat. Neuropharmacology 31:777–782 [DOI] [PubMed] [Google Scholar]
- Paakkari P, Paakkari I, Vonhof S, Feuerstein G, Sirén A-L. (1993) Dermorphin analog Tyr-D-Arg2-Phe-sarcosine-induced opioid analgesia and respiratory stimulation: the role of mu 1-receptors? J Pharmacol Exp Ther 266:544–550 [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. (2000) Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289:739–745 [DOI] [PubMed] [Google Scholar]
- Pan L, Xu J, Yu R, Xu MM, Pan YX, Pasternak GW. (2005a) Identification and characterization of six new alternatively spliced variants of the human mu opioid receptor gene, Oprm. Neuroscience 133:209–220 [DOI] [PubMed] [Google Scholar]
- Pan YX. (2002) Identification and characterization of a novel promoter of the mouse mu opioid receptor gene (Oprm) that generates eight splice variants. Gene 295:97–108 [DOI] [PubMed] [Google Scholar]
- Pan YX. (2005) Diversity and complexity of the mu opioid receptor gene: alternative pre-mRNA splicing and promoters. DNA Cell Biol 24:736–750 [DOI] [PubMed] [Google Scholar]
- Pan Y-X, Bolan E, Pasternak GW. (2002) Dimerization of morphine and orphanin FQ/nociceptin receptors: generation of a novel opioid receptor subtype. Biochem Biophys Res Commun 297:659–663 [DOI] [PubMed] [Google Scholar]
- Pan Y-X, Cheng J, Xu J, Pasternak GW. (1994) Cloning, expression and classification of a kappa3-related opioid receptor using antisense oligodeoxynucleotides. Regul Pept 54:217–218 [Google Scholar]
- Pan Y-X, Cheng J, Xu J, Rossi GC, Jacobson E, Ryan-Moro J, Brooks AI, Dean GE, Standifer KM, Pasternak GW. (1995) Cloning and functional characterization through antisense mapping of a kappa 3-related opioid receptor. Mol Pharmacol 47:1180–1188 [PubMed] [Google Scholar]
- Pan Y-X, Pasternak GW. (2011) Molecular Biology of Mu Opioid Receptors, in The Opiate Receptors (Pasternak GW, ed, ed) pp 121–160, Springer, New York [Google Scholar]
- Pan YX, Mei JF, Xu J, Wan BL, Zuckerman A, Pasternak GW. (1998a) Cloning and characterization of a mouse sigma1 receptor. J Neurochem 70:2279–2285 [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Bolan E, Chang A, Mahurter L, Rossi G, Pasternak GW. (2000) Isolation and expression of a novel alternatively spliced mu opioid receptor isoform, MOR-1F. FEBS Lett 466:337–340 [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Bolan E, Moskowitz HS, Xu M, Pasternak GW. (2005b) Identification of four novel exon 5 splice variants of the mouse mu-opioid receptor gene: functional consequences of C-terminal splicing. Mol Pharmacol 68:866–875 [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Bolan EA, Abbadie C, Chang A, Zuckerman A, Rossi GC, Pasternak GW. (1999) Identification and characterization of three new alternatively spliced mu-opioid receptor isoforms. Mol Pharmacol 56:396–403 [DOI] [PubMed] [Google Scholar]
- Pan Y-X, Xu J, Mahurter L, Bolan EA, Xu MM, Pasternak GW. (2001) Generation of the mu opioid receptor (MOR-1) protein by three new splice variants of the Oprm gene. Proc Natl Acad Sci USA 98:14084–14089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YX, Xu J, Mahurter L, Xu M, Gilbert AK, Pasternak GW. (2003) Identification and characterization of two new human mu opioid receptor splice variants, hMOR-1O and hMOR-1X. Biochem Biophys Res Commun 301:1057–1061 [DOI] [PubMed] [Google Scholar]
- Pan Y-X, Xu J, Rossi GC, Leventhal L, Wan B-L, Zuckerman A, Pasternak GW. (1998b) Cloning and expression of a novel splice variant of the mouse mu-opioid receptor (MOR-1) gene. Soc Neurosci 24:524 [Google Scholar]
- Pan YX, Xu J, Xu M, Rossi GC, Matulonis JE, Pasternak GW. (2009) Involvement of exon 11-associated variants of the mu opioid receptor MOR-1 in heroin, but not morphine, actions. Proc Natl Acad Sci USA 106:4917–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasanisi F, Sloan L, Rubin PC. (1985) Cardiovascular properties of metkephamid, a delta opioid receptor agonist, in man. Clin Sci (Lond) 68:209–213 [DOI] [PubMed] [Google Scholar]
- Pasternak DA, Pan L, Xu J, Yu R, Xu MM, Pasternak GW, Pan YX. (2004) Identification of three new alternatively spliced variants of the rat mu opioid receptor gene: dissociation of affinity and efficacy. J Neurochem 91:881–890 [DOI] [PubMed] [Google Scholar]
- Pasternak GW. (1981) Opiate, enkephalin, and endorphin analgesia: relations to a single subpopulation of opiate receptors. Neurology 31:1311–1315 [DOI] [PubMed] [Google Scholar]
- Pasternak GW. (1994) Anti-opioid activity of sigma1 systems. Regul Pept 54:219–220 [Google Scholar]
- Pasternak GW. (2001) The pharmacology of mu analgesics: from patients to genes. Neuroscientist 7:220–231 [DOI] [PubMed] [Google Scholar]
- Pasternak GW. (2004) Multiple opiate receptors: déjà vu all over again. Neuropharmacology 47 (Suppl 1):312–323 [DOI] [PubMed] [Google Scholar]
- Pasternak GW. (2007) When it comes to opiates, just say NO. J Clin Invest 117:3185–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak GW. (2010) Molecular insights into mu opioid pharmacology: From the clinic to the bench. Clin J Pain 26 (Suppl 10):S3–S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak GW, Bodnar RJ, Clark JA, Inturrisi CE. (1987) Morphine-6-glucuronide, a potent mu agonist. Life Sci 41:2845–2849 [DOI] [PubMed] [Google Scholar]
- Pasternak GW, Childers SR, Snyder SH. (1980a) Naloxazone, a long-acting opiate antagonist: effects on analgesia in intact animals and on opiate receptor binding in vitro. J Pharmacol Exp Ther 214:455–462 [PubMed] [Google Scholar]
- Pasternak GW, Childers SR, Snyder SH. (1980b) Opiate analgesia: evidence for mediation by a subpopulation of opiate receptors. Science 208:514–516 [DOI] [PubMed] [Google Scholar]
- Pasternak GW, Gintzler AR, Houghten RA, Ling GS, Goodman RR, Spiegel K, Nishimura S, Johnson N, Recht LD. (1983) Biochemical and pharmacological evidence for opioid receptor multiplicity in the central nervous system. Life Sci 33 (Suppl 1):167–173 [DOI] [PubMed] [Google Scholar]
- Pasternak GW, Goodman R, Snyder SH. (1975a) An endogenous morphine-like factor in mammalian brain. Life Sci 16:1765–1769 [DOI] [PubMed] [Google Scholar]
- Pasternak GW, Hahn EF. (1980) Long-acting opiate agonists and antagonists: 14-hydroxydihydromorphinone hydrazones. J Med Chem 23:674–676 [DOI] [PubMed] [Google Scholar]
- Pasternak GW, Kolesnikov Y. (2005) The NMDA/nitric oxide synthase cascade in opioid analgesia and tolerance, in Contemporary Clinical Neuroscience: Glutamate and Addiction (Herman BH, Frankenheim J, Litten R, Sheridan PH, Weight FF, Zukin SR, eds, ed) pp 409–416, Humana Press, Inc., Totowa [Google Scholar]
- Pasternak GW, Pan YX. (2000) Antisense mapping: assessing functional significance of genes and splice variants. Methods Enzymol 314:51–60 [DOI] [PubMed] [Google Scholar]
- Pasternak GW, Pan YX. (2011) Mix and match: heterodimers and opioid tolerance. Neuron 69:6–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak GW, Simantov R, Snyder SH. (1976) An endogenous morphine-like factor, in Tissue Responses to Addictive Drugs (Ford DH, Clouet DH, eds) pp 103–122, Spectrum Publications, New York [Google Scholar]
- Pasternak GW, Snowman AM, Snyder SH. (1975b) Selective enhancement of [3H]opiate agonist binding by divalent cations. Mol Pharmacol 11:735–744 [PubMed] [Google Scholar]
- Pasternak GW, Snyder SH. (1974a) Opiate receptor binding: effects of enzymatic treatments. Mol Pharmacol 10:183–193 [PubMed] [Google Scholar]
- Pasternak GW and Snyder SH (1974b) The effect of enzymatic treatments on 3H-naloxone binding. Proc Comm Drug Dependence 370-375.
- Pasternak GW and Snyder SH (1975a) An endogenous morphine-like factor in mammalian brain. Proc Comm Drug Dependence 460-470. [DOI] [PubMed]
- Pasternak GW, Snyder SH. (1975b) Identification of novel high affinity opiate receptor binding in rat brain. Nature 253:563–565 [DOI] [PubMed] [Google Scholar]
- Pasternak GW, Snyder SH. (1975c) Opiate receptor binding: enzymatic treatments and discrimination between agonists and antagonists. Mol Pharmacol 11:735–74454869 [Google Scholar]
- Pasternak GW, Wilson HA, Snyder SH. (1975c) Differential effects of protein-modifying reagents on receptor binding of opiate agonists and antagonists. Mol Pharmacol 11:340–351 [PubMed] [Google Scholar]
- Paton WDM. (1957) The action of morphine and related substances on contraction and on acetylcholine output of coaxially stimulated guinea-pig ileum. Br Pharmacol Chemother 12:119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattinson KT. (2008) Opioids and the control of respiration. Br J Anaesth 100:747–758 [DOI] [PubMed] [Google Scholar]
- Paul D, Bodnar RJ, Gistrak MA, Pasternak GW. (1989a) Different μ receptor subtypes mediate spinal and supraspinal analgesia in mice. Eur J Pharmacol 168:307–314 [DOI] [PubMed] [Google Scholar]
- Paul D, Pasternak GW. (1988) Differential blockade by naloxonazine of two μ opiate actions: analgesia and inhibition of gastrointestinal transit. Eur J Pharmacol 149:403–404 [DOI] [PubMed] [Google Scholar]
- Paul D, Standifer KM, Inturrisi CE, Pasternak GW. (1989b) Pharmacological characterization of morphine-6β-glucuronide, a very potent morphine metabolite. J Pharmacol Exp Ther 251:477–483 [PubMed] [Google Scholar]
- Pavlovic ZW, Bodnar RJ. (1998) Opioid supraspinal analgesic synergy between the amygdala and periaqueductal gray in rats. Brain Res 779:158–169 [DOI] [PubMed] [Google Scholar]
- Payne R. (1987) Role of epidural and intrathecal narcotics and peptides in the management of cancer pain. Med Clin North Am 71:313–327 [DOI] [PubMed] [Google Scholar]
- Payne R, Pasternak GW. (1992) Pain, in Principles of Drug Therapy in Neurology (Johnston MV, Macdonald RL, Young AB, eds, ed) pp 268–301, F.A. Davis, Philadelphia [Google Scholar]
- Peckys D, Landwehrmeyer GB. (1999) Expression of mu, kappa, and delta opioid receptor messenger RNA in the human CNS: a 33P in situ hybridization study. Neuroscience 88:1093–1135 [DOI] [PubMed] [Google Scholar]
- Pelton JT, Gulya K, Hruby VJ, Duckles S, Yamamura HI. (1985a) Somatostatin analogs with affinity for opiate receptors in rat brain binding assay. Peptides 6 (Suppl 1):159–163 [DOI] [PubMed] [Google Scholar]
- Pelton JT, Gulya K, Hruby VJ, Duckles SP, Yamamura HI. (1985b) Conformationally restricted analogs of somatostatin with high mu-opiate receptor specificity. Proc Natl Acad Sci USA 82:236–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergolizzi J, Aloisi AM, Dahan A, Filitz J, Langford R, Likar R, Mercadante S, Morlion B, Raffa RB, Sabatowski R, et al. (2010) Current knowledge of buprenorphine and its unique pharmacological profile. Pain Pract 10:428–450 [DOI] [PubMed] [Google Scholar]
- Pert A, Yaksh TL. (1974) Sites of morphine induced analgesia in the primate brain: relation to pain pathways. Brain Res 80:135–140 [DOI] [PubMed] [Google Scholar]
- Pert CB, Aposhian D, Snyder SH. (1974a) Phylogenetic distribution of opiate receptor binding. Brain Res 75:356–361 [DOI] [PubMed] [Google Scholar]
- Pert CB, Kuhar MJ, Snyder SH. (1975) Autoradiograhic localization of the opiate receptor in rat brain. Life Sci 16:1849–1853 [DOI] [PubMed] [Google Scholar]
- Pert CB, Kuhar MJ, Snyder SH. (1976a) Opiate receptor: autoradiographic localization in rat brain. Proc Natl Acad Sci USA 73:3729–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pert CB, Pasternak GW, Snyder SH. (1973) Opiate agonists and antagonists discriminated by receptor binding in brain. Science 182:1359–1361 [DOI] [PubMed] [Google Scholar]
- Pert CB, Pert A, Chang JK, Fong BTW. (1976b) (D-Ala2)-Met-enkephalinamide: a potent, long-lasting synthetic pentapeptide analgesic. Science 194:330–332 [DOI] [PubMed] [Google Scholar]
- Pert CB, Snowman AM, Snyder SH. (1974b) Localization of opiate receptor binding in synaptic membranes of rat brain. Brain Res 70:184–188 [DOI] [PubMed] [Google Scholar]
- Pert CB, Snyder SH. (1973) Opiate receptor: demonstration in nervous tissue. Science 179:1011–1014 [DOI] [PubMed] [Google Scholar]
- Pick CG, Cheng J, Paul D, Pasternak GW. (1991) Genetic influences in opioid analgesic sensitivity in mice. Brain Res 566:295–298 [DOI] [PubMed] [Google Scholar]
- Pick CG, Nejat RJ, Pasternak GW. (1993) Independent expression of two pharmacologically distinct supraspinal mu analgesic systems in genetically different mouse strains. J Pharmacol Exp Ther 265:166–171 [PubMed] [Google Scholar]
- Pick CG, Peter Y, Schreiber S, Weizman R. (1997) Pharmacological characterization of buprenorphine, a mixed agonist-antagonist with kappa 3 analgesia. Brain Res 744:41–46 [DOI] [PubMed] [Google Scholar]
- Poeaknapo C, Schmidt J, Brandsch M, Dräger B, Zenk MH. (2004) Endogenous formation of morphine in human cells. Proc Natl Acad Sci USA 101:14091–14096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl J. (1915) Ueber Das N-Allylnorcodeine, Einin Antagonisten Des Morphins; Z Exp Pathol Therap 17:370–382 [Google Scholar]
- Porreca F, Heyman JS, Mosberg HI, Omnaas JR, Vaught JL. (1987) Role of mu and delta receptors in the supraspinal and spinal analgesic effects of [D-Pen2, D-Pen5]enkephalin in the mouse. J Pharmacol Exp Ther 241:393–400 [PubMed] [Google Scholar]
- Portoghese PS. (1965) A new concept on the mode of interaction of narcotic analgesics with receptors. J Med Chem 8:609–616 [DOI] [PubMed] [Google Scholar]
- Portoghese PS. (1966) Stereochemical factors and receptor interactions associated with narcotic analgesics. J Pharm Sci 55:865–887 [DOI] [PubMed] [Google Scholar]
- Portoghese PS. (1970) Relationships between stereostructure and pharmacological activities. Annu Rev Pharmacol 10:51–76 [DOI] [PubMed] [Google Scholar]
- Portoghese PS, Larson DL, Sayre LM, Fries DS, Takemori AE. (1980) A novel opioid receptor site directed alkylating agent with irreversible narcotic antagonistic and reversible agonistic activities. J Med Chem 23:233–234 [DOI] [PubMed] [Google Scholar]
- Portoghese PS, Larson DL, Sayre LM, Yim CB, Ronsisvalle G, Tam SW, Takemori AE. (1986) Opioid agonist and antagonist bivalent ligands. The relationship between spacer length and selectivity at multiple opioid receptors. J Med Chem 29:1855–1861 [DOI] [PubMed] [Google Scholar]
- Portoghese PS, Lipkowski AW, Takemori AE. (1987) Binaltorphimine and nor-binaltorphimine, potent and selective kappa-opioid receptor antagonists. Life Sci 40:1287–1292 [DOI] [PubMed] [Google Scholar]
- Portoghese PS, Moe ST, Takemori AE. (1993) A selective delta 1 opioid receptor agonist derived from oxymorphone. Evidence for separate recognition sites for delta 1 opioid receptor agonists and antagonists. J Med Chem 36:2572–2574 [DOI] [PubMed] [Google Scholar]
- Portoghese PS, Sultana M, Takemori AE. (1988) Naltrindole, a highly selective and potent non-peptide delta opioid receptor antagonist. Eur J Pharmacol 146:185–186 [DOI] [PubMed] [Google Scholar]
- Portoghese PS, Sultana M, Takemori AE. (1990) Naltrindole 5′-isothiocyanate: a nonequilibrium, highly selective delta opioid receptor antagonist. J Med Chem 33:1547–1548 [DOI] [PubMed] [Google Scholar]
- Raehal KM, Bohn LM. (2011) The role of beta-arrestin2 in the severity of antinociceptive tolerance and physical dependence induced by different opioid pain therapeutics. Neuropharmacology 60:58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raehal KM, Schmid CL, Groer CE, Bohn LM. (2011) Functional selectivity at the μ-opioid receptor: implications for understanding opioid analgesia and tolerance. Pharmacol Rev 63:1001–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa RB, Burmeister JJ, Yuvasheva E, Pergolizzi JV., Jr (2012) QTc interval prolongation by d-propoxyphene: what about other analgesics? Expert Opin Pharmacother 13:1397–1409 [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. (2011) Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477:549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. (2004) A polymorphism of the mu-opioid receptor gene (OPRM1) and sensitivity to the effects of alcohol in humans. Alcohol Clin Exp Res 28:1789–1795 [DOI] [PubMed] [Google Scholar]
- Raynor K, Kong H, Mestek A, Bye LS, Tian M, Liu J, Yu L, Reisine T. (1995) Characterization of the cloned human mu opioid receptor. J Pharmacol Exp Ther 272:423–428 [PubMed] [Google Scholar]
- Reisine T, Pasternak GW. (1996) Opioid analgesics and antagonists, in Goodman & Gilman's: The Pharmacological Basis of Therapeutics (Hardman JG, Limbird LE, eds, ed) pp 521–556, McGraw-Hill, New York [Google Scholar]
- Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. (2012) Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu Rev Pharmacol Toxicol 52:179–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith MEA, Sershen H, Vadasz C, Lajtha A. (1981) Strain differences in opiate receptors in mouse brain. Eur J Pharmacol 74:377–380 [DOI] [PubMed] [Google Scholar]
- Robson PJ. (1983) Clinical review of parenteral meptazinol. Postgrad Med J 59 (Suppl 1):85–92 [PubMed] [Google Scholar]
- Roerig SC, O’Brien SM, Fujimoto JM, Wilcox GL. (1984) Tolerance to morphine analgesia: decreased multiplicative interaction between spinal and supraspinal sites. Brain Res 308:360–363 [DOI] [PubMed] [Google Scholar]
- Rossi GC, Brown GP, Leventhal L, Yang K, Pasternak GW. (1996) Novel receptor mechanisms for heroin and morphine-6 β-glucuronide analgesia. Neurosci Lett 216:1–4 [DOI] [PubMed] [Google Scholar]
- Rossi GC, Pan Y-X, Brown GP, Pasternak GW. (1995a) Antisense mapping the MOR-1 opioid receptor: evidence for alternative splicing and a novel morphine-6 β-glucuronide receptor. FEBS Lett 369:192–196 [DOI] [PubMed] [Google Scholar]
- Rossi GC, Pan Y-X, Cheng J, Pasternak GW. (1994a) Blockade of morphine analgesia by an antisense oligodeoxynucleotide against the mu receptor. Life Sci 54:PL375–PL379 [DOI] [PubMed] [Google Scholar]
- Rossi GC, Pasternak GW, Bodnar RJ. (1993) Synergistic brainstem interactions for morphine analgesia. Brain Res 624:171–180 [DOI] [PubMed] [Google Scholar]
- Rossi GC, Pasternak GW, Bodnar RJ. (1994b) Mu and delta opioid synergy between the periaqueductal gray and the rostro-ventral medulla. Brain Res 665:85–93 [DOI] [PubMed] [Google Scholar]
- Rossi GC, Standifer KM, Pasternak GW. (1995b) Differential blockade of morphine and morphine-6 β-glucuronide analgesia by antisense oligodeoxynucleotides directed against MOR-1 and G-protein α subunits in rats. Neurosci Lett 198:99–102 [DOI] [PubMed] [Google Scholar]
- Rossi GC, Su W, Leventhal L, Su H, Pasternak GW. (1997) Antisense mapping DOR-1 in mice: further support for delta receptor subtypes. Brain Res 753:176–179 [DOI] [PubMed] [Google Scholar]
- Rothman R, Westfall TC. (1983) Further evidence for an opioid receptor complex. J Neurobiol 14:341–351 [DOI] [PubMed] [Google Scholar]
- Rothman RB, Bykov V, de Costa BR, Jacobson AE, Rice KC, Brady LS. (1990) Interaction of endogenous opioid peptides and other drugs with four kappa opioid binding sites in guinea pig brain. Peptides 11:311–331 [DOI] [PubMed] [Google Scholar]
- Rothman RB, Bykov V, Xue BG, Xu H, De Costa BR, Jacobson AE, Rice KC, Kleinman JE, Brady LS. (1992) Interaction of opioid peptides and other drugs with multiple kappa receptors in rat and human brain. Evidence for species differences. Peptides 13:977–987 [DOI] [PubMed] [Google Scholar]
- Rothman RB, Westfall TC. (1982a) Allosteric coupling between morphine and enkephalin receptors in vitro. Mol Pharmacol 21:548–557 [PubMed] [Google Scholar]
- Rothman RB, Westfall TC. (1982b) Morphine allosterically modulates the binding of [3H]leucine enkephalin to a particulate fraction of rat brain. Mol Pharmacol 21:538–547 [PubMed] [Google Scholar]
- Rowbotham MC, Twilling L, Davies PS, Reisner L, Taylor K, Mohr D. (2003) Oral opioid therapy for chronic peripheral and central neuropathic pain. N Engl J Med 348:1223–1232 [DOI] [PubMed] [Google Scholar]
- Rutherford JM, Wang J, Xu H, Dersch CM, Partilla JS, Rice KC, Rothman RB. (2008) Evidence for a mu-delta opioid receptor complex in CHO cells co-expressing mu and delta opioid peptide receptors. Peptides 29:1424–1431 [DOI] [PubMed] [Google Scholar]
- Sánchez-Fernández C, Nieto FR, González-Cano R, Artacho-Cordón A, Romero L, Montilla-García A, Zamanillo D, Baeyens JM, Entrena JM, Cobos EJ. (2013) Potentiation of morphine-induced mechanical antinociception by σ₁ receptor inhibition: role of peripheral σ₁ receptors. Neuropharmacology 70:348–358 [DOI] [PubMed] [Google Scholar]
- Schaumann W. (1955) The paralysing action of morphine on the guinea-pig ileum. Br Pharmacol Chemother 10:456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumann W. (1957) Inhibiiton by morphine ofthe release of acetylcholine from the intestine of the guinea-pig. Br J Pharmacol 12:115–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller PW (1999) Peptide and peptidomimetic ligands of opiod receptors. [DOI] [PubMed]
- Schiller PW. (2005) Opioid peptide-derived analgesics. AAPS J 7:E560–E565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller PW, Nguyen TM, Chung NN, Lemieux C. (1989) Dermorphin analogues carrying an increased positive net charge in their “message” domain display extremely high mu opioid receptor selectivity. J Med Chem 32:698–703 [DOI] [PubMed] [Google Scholar]
- Schiller PW, Nguyen TMD, Weltrowska G, Wilkes BC, Marsden BJ, Lemieux C, Chung NN. (1992) Differential stereochemical requirements of μ vs. δ opioid receptors for ligand binding and signal transduction: development of a class of potent and highly δ-selective peptide antagonists. Proc Natl Acad Sci USA 89:11871–11875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkel AH, Smit JJ, van Tellingen O, Beijnen JH, Wagenaar E, van Deemter L, Mol CA, van der Valk MA, Robanus-Maandag EC, te Riele HP, et al. (1994) Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell 77:491–502 [DOI] [PubMed] [Google Scholar]
- Schmidhammer H, Schratz A, Mitterdorfer J. (1990) synthesis and biological evaluation of the 14-alkoxymorphinans. 8. 14-methoxymetopon, an extremely potent opioid agonist. Helv Chim Acta 73:1784–1787 [Google Scholar]
- Schubert P, Höllt V, Herz A. (1975) Autoradiographic evaluation of the intracerebral distribution of 3-H-etorphine in the mouse brain. Life Sci 16:1855–1856 [DOI] [PubMed] [Google Scholar]
- Schuller AG, King MA, Zhang J, Bolan E, Pan YX, Morgan DJ, Chang A, Czick ME, Unterwald EM, Pasternak GW, et al. (1999) Retention of heroin and morphine-6 beta-glucuronide analgesia in a new line of mice lacking exon 1 of MOR-1. Nat Neurosci 2:151–156 [DOI] [PubMed] [Google Scholar]
- Schulz S, Schreff M, Koch T, Zimprich A, Gramsch C, Elde R, Höllt V. (1998) Immunolocalization of two mu-opioid receptor isoforms (MOR1 and MOR1B) in the rat central nervous system. Neuroscience 82:613–622 [DOI] [PubMed] [Google Scholar]
- Seeman P, Chau-Wong M, Moyyen S. (1972) The membrane binding of morphine, diphenylhydantoin, and tetrahydrocannabinol. Can J Physiol Pharmacol 50:1193–1200 [DOI] [PubMed] [Google Scholar]
- Seki T, Minami M, Nakagawa T, Ienaga Y, Morisada A, Satoh M. (1998) DAMGO recognizes four residues in the third extracellular loop to discriminate between mu- and kappa-opioid receptors. Eur J Pharmacol 350:301–310 [DOI] [PubMed] [Google Scholar]
- Serturner FWA. (1805). Trommsdorf's J der Pharmazie 13:234 [Google Scholar]
- Seth P, Leibach FH, Ganapathy V. (1997) Cloning and structural analysis of the cDNA and the gene encoding the murine type 1 sigma receptor. Biochem Biophys Res Commun 241:535–540 [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Zaykin DV, Gris P, Ogurtsov AY, Gauthier J, Shibata K, Tchivileva IE, Belfer I, Mishra B, Kiselycznyk C, et al. (2009) Expansion of the human mu-opioid receptor gene architecture: novel functional variants. Hum Mol Genet 18:1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K-F, Crain SM. (1992) Chronic selective activation of excitatory opioid receptor functions in sensory neurons results in opioid ‘dependence’ without tolerance. Brain Res 597:74–83 [DOI] [PubMed] [Google Scholar]
- Shen K-F, Crain SM. (1994) Antagonists at excitatory opioid receptors on sensory neurons in culture increase potency and specificity of opiate analgesics and attenuate development of tolerance/dependence. Brain Res 636:286–297 [DOI] [PubMed] [Google Scholar]
- Shimomura K, Kamata O, Ueki S, Ida S, Oguri K. (1971) Analgesic effect of morphine glucuronides. Tohoku J Exp Med 105:45–52 [DOI] [PubMed] [Google Scholar]
- Shorr J, Foley K, Spector S. (1978) Presence of a non-peptide morphine-like compound in human cerebrospinal fluid. Life Sci 23:2057–2062 [DOI] [PubMed] [Google Scholar]
- Simon EJ, Hiller JM, Edelman I. (1973) Stereospecific binding of the potent narcotic analgesic (3H) Etorphine to rat-brain homogenate. Proc Natl Acad Sci USA 70:1947–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder SH, Matthysse S. (1975) Opiate Receptor Mechanisms, MIT Press, Boston: [PubMed] [Google Scholar]
- Snyder SH, Pasternak GW, Pert CB. (1975) Opiate receptor mechanisms, in Handbook of Psychopharmacology (Iverson L, Iverson S, Snyder SH, eds, ed) pp 329–360, Plenum Press, New York [Google Scholar]
- Snyder SH, Pert CB, Pasternak GW. (1974) The opiate receptor. Ann Intern Med 81:534–540 [DOI] [PubMed] [Google Scholar]
- Sofuoglu M, Portoghese PS, Takemori AE. (1993) 7-Benzylidenenaltrexone (BNTX): a selective delta 1 opioid receptor antagonist in the mouse spinal cord. Life Sci 52:769–775 [DOI] [PubMed] [Google Scholar]
- Sora I, Takahashi N, Funada M, Ujike H, Revay RS, Donovan DM, Miner LL, Uhl GR. (1997) Opiate receptor knockout mice define μ receptor roles in endogenous nociceptive responses and morphine-induced analgesia. Proc Natl Acad Sci USA 94:1544–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel K, Kalb R, Pasternak GW. (1983) Analgesic activity of tricyclic antidepressants. Ann Neurol 13:462–465 [DOI] [PubMed] [Google Scholar]
- Spiegel K, Kourides IA, Pasternak GW. (1982a) Prolactin and growth hormone release by morphine in the rat: different receptor mechanisms. Science 217:745–747 [DOI] [PubMed] [Google Scholar]
- Spiegel K, Kourides IA, Pasternak GW. (1982b) Different receptors mediate morphine-induced prolactin and growth hormone release. Life Sci 31:2177–2180 [DOI] [PubMed] [Google Scholar]
- Spiegel K, Pasternak GW. (1984) Meptazinol: a novel Mu-1 selective opioid analgesic. J Pharmacol Exp Ther 228:414–419 [PubMed] [Google Scholar]
- Standifer KM, Chien C-C, Wahlestedt C, Brown GP, Pasternak GW. (1994) Selective loss of δ opioid analgesia and binding by antisense oligodeoxynucleotides to a δ opioid receptor. Neuron 12:805–810 [DOI] [PubMed] [Google Scholar]
- Standifer KM, Pasternak GW. (1997) G proteins and opioid receptor-mediated signalling. Cell Signal 9:237–248 [DOI] [PubMed] [Google Scholar]
- Standifer KM, Rossi GC, Pasternak GW. (1996) Differential blockade of opioid analgesia by antisense oligodeoxynucleotides directed against various G protein α subunits. Mol Pharmacol 50:293–298 [PubMed] [Google Scholar]
- Stefano GB, Kream RM, Zukin RS. (1980) Demonstration of stereospecific opiate binding in the nervous tissue of the marine mollusc Mytilus edulis. Brain Res 181:440–445 [DOI] [PubMed] [Google Scholar]
- Stefano GB, Scharrer B, Assanah P. (1982) Demonstration, characterization and localization of opioid binding sites in the midgut of the insect Leucophaea maderae (Blattaria). Brain Res 253:205–212 [DOI] [PubMed] [Google Scholar]
- Stein C, Schäfer M, Machelska H. (2003) Attacking pain at its source: new perspectives on opioids. Nat Med 9:1003–1008 [DOI] [PubMed] [Google Scholar]
- Stein C, Yassouridis A. (1997) Peripheral morphine analgesia. Pain 71:119–121 [PubMed] [Google Scholar]
- Sternini C, Spann M, Anton B, Keith DE, Jr, Bunnett NW, von Zastrow M, Evans C, Brecha NC. (1996) Agonist-selective endocytosis of μ opioid receptor by neurons in vivo. Proc Natl Acad Sci USA 93:9241–9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CW. (2009) The evolution of vertebrate opioid receptors. Front Biosci (Landmark Ed) 14:1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub (1911) Eine empfindliche biologische reaktion auf morphin. Deutsche Med Wehnschr 37:1462.
- Su W, Pasternak GW. (2013) The role of multidrug resistance-associated protein in the blood-brain barrier and opioid analgesia. Synapse 67:609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surratt CK, Johnson PS, Moriwaki A, Seidleck BK, Blaschak CJ, Wang JB, Uhl GR. (1994) -μ opiate receptor. Charged transmembrane domain amino acids are critical for agonist recognition and intrinsic activity. J Biol Chem 269:20548–20553 [PubMed] [Google Scholar]
- Sutters KA, Miaskowski C, Taiwo YO, Levine JD. (1990) Analgesic synergy and improved motor function produced by combinations of mu-delta- and mu-kappa-opioids. Brain Res 530:290–294 [DOI] [PubMed] [Google Scholar]
- Tan EC, Tan CH, Karupathivan U, Yap EP. (2003) Mu opioid receptor gene polymorphisms and heroin dependence in Asian populations. Neuroreport 14:569–572 [DOI] [PubMed] [Google Scholar]
- Tanowitz M, Hislop JN, von Zastrow M. (2008) Alternative splicing determines the post-endocytic sorting fate of G-protein-coupled receptors. J Biol Chem 283:35614–35621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanowitz M, von Zastrow M. (2003) A novel endocytic recycling signal that distinguishes the membrane trafficking of naturally occurring opioid receptors. J Biol Chem 278:45978–45986 [DOI] [PubMed] [Google Scholar]
- Terenius L. (1973) Stereospecific interaction between narcotic analgesics and a synaptic plasm a membrane fraction of rat cerebral cortex. Acta Pharmacol Toxicol (Copenh) 32:317–320 [DOI] [PubMed] [Google Scholar]
- Terenius L, Wahlström A. (1975) Search for an endogenous ligand for the opiate receptor. Acta Physiol Scand 94:74–81 [DOI] [PubMed] [Google Scholar]
- Terry CE, Pellens M. (1928) The Opium Problem, Bureau of Social Hygiene, Inc, New York [Google Scholar]
- Terskiy A, Wannemacher KM, Yadav PN, Tsai M, Tian B, Howells RD. (2007) Search of the human proteome for endomorphin-1 and endomorphin-2 precursor proteins. Life Sci 81:1593–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschemacher H, Opheim KE, Cox BM, Goldstein A. (1975) A peptide-like substance from pituitary that acts like morphine. I. Isolation. Life Sci 16:1771–1775 [DOI] [PubMed] [Google Scholar]
- Thompson RC, Mansour A, Akil H, Watson SJ. (1993) Cloning and pharmacological characterization of a rat μ opioid receptor. Neuron 11:903–913 [DOI] [PubMed] [Google Scholar]
- Tiseo PJ, Inturrisi CE. (1993) Attenuation and reversal of morphine tolerance by the competitive N-methyl-D-aspartate receptor antagonist, LY274614. J Pharmacol Exp Ther 264:1090–1096 [PubMed] [Google Scholar]
- Tiseo PJ, Thaler HT, Lapin J, Inturrisi CE, Portenoy RK, Foley KM. (1995) Morphine-6-glucuronide concentrations and opioid-related side effects: a survey in cancer patients. Pain 61:47–54 [DOI] [PubMed] [Google Scholar]
- Tive LA, Ginsberg K, Pick CG, Pasternak GW. (1992) Kappa 3 receptors and levorphanol-induced analgesia. Neuropharmacology 31:851–856 [DOI] [PubMed] [Google Scholar]
- Trendelenburg P. (1917) Physiologische und pharmkologische versuche uber die dunndarmperistaltik. Naunyn-Schmiedeberg's Arch Path Pharmacol 81:55–128 [DOI] [PubMed] [Google Scholar]
- Trendelenburg U. (1957) The action of morphine on the superior cervical ganglion and on the nictitating membrane of the cat. Br Pharmacol Chemother 12:79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KA, Akil H. (1991) Inhibition of morphine tolerance and dependence by the NMDA receptor antagonist MK-801. Science 251:85–87 [DOI] [PubMed] [Google Scholar]
- Trujillo KA, Akil H. (1994) Inhibition of opiate tolerance by non-competitive N-methyl-D-aspartate receptor antagonists. Brain Res 633:178–188 [DOI] [PubMed] [Google Scholar]
- Tseng LF, Collins KA, Kampine JP. (1994) Antisense oligodeoxynucleotide to a delta-opioid receptor selectively blocks the spinal antinociception induced by delta-, but not mu- or kappa-opioid receptor agonists in the mouse. Eur J Pharmacol 258:R1–R3 [DOI] [PubMed] [Google Scholar]
- Uhl GR, Childers S, Pasternak GW. (1994) An opiate-receptor gene family reunion. Trends Neurosci 17:89–93 [DOI] [PubMed] [Google Scholar]
- Uhl GR, Sora I, Wang ZJ. (1999) The μ opiate receptor as a candidate gene for pain: polymorphisms, variations in expression, nociception, and opiate responses. Proc Natl Acad Sci USA 96:7752–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulens C, Van Boven M, Daenens P, Tytgat J. (2000) Interaction of p-fluorofentanyl on cloned human opioid receptors and exploration of the role of Trp-318 and His-319 in mu-opioid receptor selectivity. J Pharmacol Exp Ther 294:1024–1033 [PubMed] [Google Scholar]
- Unna K. (1943) Antagonistic Effect of N-Allylnormorphine Upon Morphine. J Pharmacol Exp Ther 79:27–31 [Google Scholar]
- Van Praag D, Simon EJ. (1966) Studies on the intracellular distribution and tissue binding of dihydromorphine-7,8-H3 in the rat. Proc Soc Exp Biol Med 122:6–11 [DOI] [PubMed] [Google Scholar]
- Violin JD, Lefkowitz RJ. (2007) Beta-arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol Sci 28:416–422 [DOI] [PubMed] [Google Scholar]
- Voelker R. (2012) Children’s deaths linked with postsurgical codeine. JAMA 308:963. [DOI] [PubMed] [Google Scholar]
- Von Braun J. (1916). Unter Suchungen Uber Morphium-Alkalvide. III Mitteilung. Ber Deut Chem Ges 49:977–989 [Google Scholar]
- Von Zastrow M. (2011) Opioid receptor trafficking, in The Opiate Receptors (Pasternak GW, ed, ed) pp 389–406, Springer, New York [Google Scholar]
- von Zastrow M, Svingos A, Haberstock-Debic H, Evans C. (2003) Regulated endocytosis of opioid receptors: cellular mechanisms and proposed roles in physiological adaptation to opiate drugs. Curr Opin Neurobiol 13:348–353 [DOI] [PubMed] [Google Scholar]
- Vonhof S, Barone FC, Price WJ, Paakkari P, Millison JH, Feuerstein GZ, Sirén AK. (2001) Receptor binding and biological activity of the dermorphin analog Tyr-D-Arg(2)-Phe-Sar (TAPS). Eur J Pharmacol 416:83–93 [DOI] [PubMed] [Google Scholar]
- Waksman G, Hamel E, Fournié-Zaluski MC, Roques BP. (1986) Autoradiographic comparison of the distribution of the neutral endopeptidase “enkephalinase” and of mu and delta opioid receptors in rat brain. Proc Natl Acad Sci USA 83:1523–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald A. (2012) Motility disorders of the colon and rectum. Curr Opin Gastroenterol 28:52–56 [DOI] [PubMed] [Google Scholar]
- Waldhoer M, Bartlett SE, Whistler JL. (2004) Opioid receptors. Annu Rev Biochem 73:953–990 [DOI] [PubMed] [Google Scholar]
- Wallenstein SL. (1984) The evaluation of analgesics in man, in Analgesics: Neurochemical, Behavioral and Clinical Perspectives (Kuhar MJ, Pasternak GW, eds, ed) pp 235–255, Raven Press Inc, New York [Google Scholar]
- Wamsley JK, Zarbin MA, Young WS, 3rd, Kuhar MJ. (1982) Distribution of opiate receptors in the monkey brain: an autoradiographic study. Neuroscience 7:595–613 [DOI] [PubMed] [Google Scholar]
- Wang D, Quillan JM, Winans K, Lucas JL, Sadée W. (2001) Single nucleotide polymorphisms in the human mu opioid receptor gene alter basal G protein coupling and calmodulin binding. J Biol Chem 276:34624–34630 [DOI] [PubMed] [Google Scholar]
- Wang HY, Guang W, Barbier E, Shapiro P, Wang JB. (2007) Mu opioid receptor mutant, T394A, abolishes opioid-mediated adenylyl cyclase superactivation. Neuroreport 18:1969–1973 [DOI] [PubMed] [Google Scholar]
- Wang JB. (2003) Study of opioid receptor phosphorylation using cell-labeling method with 32P-orthrophosphate. Methods Mol Med 84:47–51 [DOI] [PubMed] [Google Scholar]
- Wang JB, Imai Y, Eppler CM, Gregor P, Spivak CE, Uhl GR. (1993) μ opiate receptor: cDNA cloning and expression. Proc Natl Acad Sci USA 90:10230–10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JB, Imai Y, Johnson PS, Walther D, Stein C, Schaefer M, Wu JM, Wang WF, Moriwaki A, Uhl GR. (1994a) Human μ receptor: gene structure, expression, and m/kappa chimeras that define nontransmembrane domains influencing peptide binding affinities. Regul Pept 54:317–320 [Google Scholar]
- Wang JB, Johnson PS, Imai Y, Persico AM, Ozenberger BA, Eppler CM, Uhl GR. (1994b) cDNA cloning of an orphan opiate receptor gene family member and its splice variant. FEBS Lett 348:75–79 [DOI] [PubMed] [Google Scholar]
- Wang JB, Johnson PS, Persico AM, Hawkins AL, Griffin CA, Uhl GR. (1994c) Human μ opiate receptor. cDNA and genomic clones, pharmacologic characterization and chromosomal assignment. FEBS Lett 338:217–222 [DOI] [PubMed] [Google Scholar]
- Wang WW, Shahrestanifar M, Jin J, Howells RD. (1995) Studies on μ and δ opioid receptor selectivity utilizing chimeric and site-mutagenized receptors. Proc Natl Acad Sci USA 92:12436–12440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Barker K, Shi S, Diaz M, Mo B, Gutstein HB. (2012) Blockade of PDGFR-β activation eliminates morphine analgesic tolerance. Nat Med 18:385–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins LR, Kinscheck IB, Mayer DJ. (1984) Potentiation of opiate analgesia and apparent reversal of morphine tolerance by proglumide. Science 224:395–396 [DOI] [PubMed] [Google Scholar]
- Watson B, Meng F, Akil H. (1996) A chimeric analysis of the opioid receptor domains critical for the binding selectivity of mu opioid ligands. Neurobiol Dis 3:87–96 [DOI] [PubMed] [Google Scholar]
- Way EL, Loh HH, Shen F. (1968) Morphine tolerance, physical dependence, and synthesis of brain 5-hydroxytryptamine. Science 162:1290–1292 [DOI] [PubMed] [Google Scholar]
- Wei LN, Loh HH. (2011) Transcriptional and epigenetic regulation of opioid receptor genes: present and future. Annu Rev Pharmacol Toxicol 51:75–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijlard J and Erickson AE(1942) N-Allylnormorphine. J Am Chem Soc 64:869-870. [Google Scholar]
- Weitz CJ, Faull KF, Goldstein A. (1987) Synthesis of the skeleton of the morphine molecule by mammalian liver. Nature 330:674–677 [DOI] [PubMed] [Google Scholar]
- Weitz CJ, Lowney LI, Faull KF, Feistner G, Goldstein A. (1986) Morphine and codeine from mammalian brain. Proc Natl Acad Sci USA 83:9784–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel B, Hoehe MR. (1998) The human μ opioid receptor gene: 5′ regulatory and intronic sequences. J Mol Med (Berl) 76:525–532 [DOI] [PubMed] [Google Scholar]
- Whistler JL, Chuang HH, Chu P, Jan LY, von Zastrow M. (1999) Functional dissociation of μ opioid receptor signaling and endocytosis: implications for the biology of opiate tolerance and addiction. Neuron 23:737–746 [DOI] [PubMed] [Google Scholar]
- Wilke RA, Mehta RP, Lupardus PJ, Chen Y, Ruoho AE, Jackson MB. (1999) Sigma receptor photolabeling and sigma receptor-mediated modulation of potassium channels in tumor cells. J Biol Chem 274:18387–18392 [DOI] [PubMed] [Google Scholar]
- Wilson HA, Pasternak GW, Snyder SH. (1975) Differentiation of opiate agonist and antagonist receptor binding by protein modifying reagents. Nature 253:448–450 [DOI] [PubMed] [Google Scholar]
- ilson SG, Bryant CD, Lariviere WR, Olsen MS, Giles BE, Chesler EJ, Mogil JS. (2003a) The heritability of antinociception II: pharmacogenetic mediation of three over-the-counter analgesics in mice. J Pharmacol Exp Ther 305:755–764 [DOI] [PubMed] [Google Scholar]
- Wilson SG, Smith SB, Chesler EJ, Melton KA, Haas JJ, Mitton B, Strasburg K, Hubert L, Rodriguez-Zas SL, Mogil JS. (2003b) The heritability of antinociception: common pharmacogenetic mediation of five neurochemically distinct analgesics. J Pharmacol Exp Ther 304:547–559 [DOI] [PubMed] [Google Scholar]
- Wolozin BL, Pasternak GW. (1981) Classification of multiple morphine and enkephalin binding sites in the central nervous system. Proc Natl Acad Sci USA 78:6181–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DT, Horng JS. (1973) Stereospecific interaction of opiate narcotics in binding of 3H-dihydromorphine to membranes of rat brain. Life Sci 13:1543–1556 [DOI] [PubMed] [Google Scholar]
- Wood PL, Pasternak GW. (1983) Specific mu 2 opioid isoreceptor regulation of nigrostriatal neurons: in vivo evidence with naloxonazine. Neurosci Lett 37:291–293 [DOI] [PubMed] [Google Scholar]
- Woolfe G and MacDonald AD (1943) The evaluation of the analgesic action of pethidine hydrochloride (Demerol). J Pharmacol Exp Ther 80:300-307. [Google Scholar]
- Woolfolk DR, Holtzman SG. (1995) Rat strain differences in the potentiation of morphine-induced analgesia by stress. Pharmacol Biochem Behav 51:699–703 [DOI] [PubMed] [Google Scholar]
- Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, et al. (2012) Structure of the human κ-opioid receptor in complex with JDTic. Nature 485:327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Hwang CK, Yao S, Law PY, Loh HH, Wei LN. (2005) A major species of mouse mu-opioid receptor mRNA and its promoter-dependent functional polyadenylation signal. Mol Pharmacol 68:279–285 [DOI] [PubMed] [Google Scholar]
- Xu H, Lu YF, Partilla JS, Zheng QX, Wang JB, Brine GA, Carroll FI, Rice KC, Chen KX, Chi ZQ, et al. (1999a) Opioid peptide receptor studies, 11: involvement of Tyr148, Trp318 and His319 of the rat mu-opioid receptor in binding of mu-selective ligands. Synapse 32:23–28 [DOI] [PubMed] [Google Scholar]
- Xu J, Xu M, Brown T, Rossi GC, Hurd YL, Inturrisi CE, Pasternak GW, Pan YX. (2013) Stabilization of the μ-opioid receptor by truncated single transmembrane splice variants through a chaperone-like action. J Biol Chem 288:21211–21227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xu M, Hurd YL, Pasternak GW, Pan YX. (2009) Isolation and characterization of new exon 11-associated N-terminal splice variants of the human mu opioid receptor gene. J Neurochem 108:962–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xu M, Pan YX. (2006) Characterizing exons 11 and 1 promoters of the mu opioid receptor (Oprm) gene in transgenic mice. BMC Mol Biol 7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xu M, Rossi GC, Pasternak GW, Pan YX. (2011) Identification and characterization of seven new exon 11-associated splice variants of the rat μ opioid receptor gene, OPRM1. Mol Pain 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Chen CG, Huang P, Li J, de Riel JK, Javitch JA, Liu-Chen LY. (2000) The conserved cysteine 7.38 residue is differentially accessible in the binding-site crevices of the mu, delta, and kappa opioid receptors. Biochemistry 39:13904–13915 [DOI] [PubMed] [Google Scholar]
- Xu W, Ozdener F, Li JG, Chen CG, de Riel JK, Weinstein H, Liu-Chen LY. (1999b) Functional role of the spatial proximity of Asp114(2.50) in TMH 2 and Asn332(7.49) in TMH 7 of the μ opioid receptor. FEBS Lett 447:318–324 [DOI] [PubMed] [Google Scholar]
- Xu Y, Carr LG. (2001) Transcriptional regulation of the human mu opioid receptor (hMOR) gene: evidence of positive and negative cis-acting elements in the proximal promoter and presence of a distal promoter. DNA Cell Biol 20:391–402 [DOI] [PubMed] [Google Scholar]
- Xue J-C, Chen C, Zhu J, Kunapuli S, DeRiel JK, Yu L, Liu-Chen L-Y. (1994) Differential binding domains of peptide and non-peptide ligands in the cloned rat kappa opioid receptor. J Biol Chem 269:30195–30199 [PubMed] [Google Scholar]
- Yamamoto H, Yamamoto T, Sagi N, Klenerová V, Goji K, Kawai N, Baba A, Takamori E, Moroji T. (1995) Sigma ligands indirectly modulate the NMDA receptor-ion channel complex on intact neuronal cells via sigma 1 site. J Neurosci 15:731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung JC, Rudy TA. (1980) Multiplicative interaction between narcotic agonisms expressed at spinal and supraspinal sites of antinociceptive action as revealed by concurrent intrathecal and intracerebroventricular injections of morphine. J Pharmacol Exp Ther 215:633–642 [PubMed] [Google Scholar]
- Young WS, 3rd, Kuhar MJ. (1979) A new method for receptor autoradiography: [3H]opioid receptors in rat brain. Brain Res 179:255–270 [DOI] [PubMed] [Google Scholar]
- Young WS, 3rd, Wamsley JK, Zarbin MA, Kuhar MJ. (1980) Opioid receptors undergo axonal flow. Science 210:76–78 [DOI] [PubMed] [Google Scholar]
- Zadina JE, Hackler L, Ge LJ, Kastin AJ. (1997) A potent and selective endogenous agonist for the mu-opiate receptor. Nature 386:499–502 [DOI] [PubMed] [Google Scholar]
- Zelcer S, Kolesnikov YA, Kovalyshyn I, Pasternak DA, Pasternak GW. (2005) Selective potentiation of opioid analgesia by nonsteroidal anti-inflammatory drugs. Brain Res 1040:151–156 [DOI] [PubMed] [Google Scholar]
- Zhang A-Z, Pasternak GW. (1981) Ontogeny of opioid pharmacology and receptors: high and low affinity site differences. Eur J Pharmacol 73:29–40 [DOI] [PubMed] [Google Scholar]
- Zhang PS, Johnson PS, Zöllner C, Wang WF, Wang ZJ, Montes AE, Seidleck BK, Blaschak CJ, Surratt CK. (1999) Mutation of human μ opioid receptor extracellular “disulfide cysteine” residues alters ligand binding but does not prevent receptor targeting to the cell plasma membrane. Brain Res Mol Brain Res 72:195–204 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Pan YX, Kolesnikov Y, Pasternak GW. (2006) Immunohistochemical labeling of the mu opioid receptor carboxy terminal splice variant mMOR-1B4 in the mouse central nervous system. Brain Res 1099:33–43 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang D, Johnson AD, Papp AC, Sadée W. (2005) Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem 280:32618–32624 [DOI] [PubMed] [Google Scholar]
- Zhu YX, Hsu MS, Pintar JE. (1998) Developmental expression of the mu, kappa, and δ opioid receptor mRNAs in mouse. J Neurosci 18:2538–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YX, King MA, Schuller AGP, Nitsche JF, Reidl M, Elde RP, Unterwald E, Pasternak GW, Pintar JE. (1999) Retention of supraspinal δ-like analgesia and loss of morphine tolerance in δ opioid receptor knockout mice. Neuron 24:243–252 [DOI] [PubMed] [Google Scholar]
- Zimprich A, Simon T, Höllt V. (1995) Cloning and expression of an isoform of the rat μ opioid receptor (rMOR1B) which differs in agonist induced desensitization from rMOR1. FEBS Lett 359:142–146 [DOI] [PubMed] [Google Scholar]
- Zong J, Pollack GM. (2000) Morphine antinociception is enhanced in mdr1a gene-deficient mice. Pharm Res 17:749–753 [DOI] [PubMed] [Google Scholar]
- Zuckerman A, Bolan E, de Paulis T, Schmidt D, Spector S, Pasternak GW. (1999) Pharmacological characterization of morphine-6-sulfate and codeine-6-sulfate. Brain Res 842:1–5 [DOI] [PubMed] [Google Scholar]
- Zukin RS, Eghbali M, Olive D, Unterwald EM, Tempel A. (1988) Characterization and visualization of rat and guinea pig brain kappa opioid receptors: evidence for kappa 1 and kappa 2 opioid receptors. Proc Natl Acad Sci USA 85:4061–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]