Significance
The crustacean, Hyalella azteca, is commonly used in environmental monitoring to test the toxicity of water or sediment. We show that among three laboratory cultures and seven wild populations of H. azteca, there is a more than 550-fold variation in sensitivity to widely used pyrethroid insecticides. Some individuals have attained resistance by mutations in the voltage-gated sodium channel, the target site for pyrethroid toxicity. Similar mutations have been found in agricultural pests targeted by pyrethroids, but this study indicates that runoff of terrestially applied urban and agricultural pesticides has been sufficient to induce resistance in a nontarget aquatic species on multiple, independent occasions. Our results have far-reaching implications for biomonitoring programs in general and especially those relying on H. azteca.
Abstract
Use of pesticides can have substantial nonlethal impacts on nontarget species, including driving evolutionary change, often with unknown consequences for species, ecosystems, and society. Hyalella azteca, a species complex of North American freshwater amphipods, is widely used for toxicity testing of water and sediment and has frequently shown toxicity due to pyrethroid pesticides. We demonstrate that 10 populations, 3 from laboratory cultures and 7 from California water bodies, differed by at least 550-fold in sensitivity to pyrethroids. The populations sorted into four phylogenetic groups consistent with species-level divergence. By sequencing the primary pyrethroid target site, the voltage-gated sodium channel, we show that point mutations and their spread in natural populations were responsible for differences in pyrethroid sensitivity. At least one population had both mutant and WT alleles, suggesting ongoing evolution of resistance. Although nonresistant H. azteca were susceptible to the typical neurotoxic effects of pyrethroids, gene expression analysis suggests the mode of action in resistant H. azteca was not neurotoxicity but was oxidative stress sustained only at considerably higher pyrethroid concentrations. The finding that a nontarget aquatic species has acquired resistance to pesticides used only on terrestrial pests is troubling evidence of the impact of chronic pesticide transport from land-based applications into aquatic systems. Our findings have far-reaching implications for continued uncritical use of H. azteca as a principal species for monitoring and environmental policy decisions.
Although evolution of pesticide resistance in pest species has been a major area of investigation, evolutionary impacts of pesticide pollution on nontarget species are poorly understood but may hold important consequences for affected species and their ecosystems (1, 2). Acquisition of pesticide resistance may be especially troublesome when it occurs in nontarget species used in biomonitoring programs that inform public policy. One such species is the amphipod, Hyalella azteca, widely used for sediment toxicity testing and sometimes for water testing (3, 4). There are many instances of sediment or water toxicity to H. azteca due to pyrethroids, which are widely used insecticides in urban and agricultural settings (4–6). However, we find wild populations of H. azteca in some pyrethroid-contaminated water bodies from which water or sediment samples cause acute mortality to laboratory-cultured populations. One possibility is that wild populations have evolved resistance, as has been reported in target pests (7, 8).
The picture is further complicated by the fact that H. azteca is actually a species complex. Genetic analyses of what are nominally H. azteca have suggested the presence of 33 species in the Great Basin of California and Nevada (9), 7 species in Wisconsin and parts of Canada (10), 4 species in Michigan (11), and 2 species in Oregon (12). Genetic differences among individuals within a single population of H. azteca led to differences in survival when exposed to contaminants (13). The possibility of differential contaminant sensitivity among species within the complex has not been fully appreciated by the ecotoxicological community, yet it could have implications for the many monitoring programs that rely on this organism.
In the present study, we demonstrate wide variation in pyrethroid sensitivity among populations of H. azteca. We contrast sensitivity of three laboratory-cultured and seven wild-collected populations and interpret differences in light of phylogenetic relationships, emergence of resistance, and contrasting gene expression patterns among the populations.
Results
Sources of H. azteca Investigated and Relationship to Pyrethroid Exposure.
Natural populations of H. azteca were obtained from seven sites in California (Table 1). Using sediment pyrethroid concentrations as an integrated measure of exposure, the sites represent a broad range of prior exposure. Two sites with minimal surrounding development (Blodgett and Laguna) contained bifenthrin barely above the 1-ng/g reporting limit. Chualar Creek contained no measurable pyrethroids, although agricultural pyrethroid use is widespread in the watershed (14). The organophosphate insecticide chlorpyrifos was present at 248 ng/g at the site. Among four urban creeks, Morrison Creek sediments had no measurable pyrethroids, perhaps because the site was in a reach of the creek along an airfield, although there was extensive residential development and therefore likely pyrethroid use, both upstream and downstream. The Pleasant Grove Creek site had 6.8 ng/g bifenthrin, approximately one-third the median lethal concentration (LC50) for laboratory-cultured H. azteca (15), and sediment toxicity attributed to pyrethroids has been reported 2 km downstream (16). Mosher Slough and Grayson Creek contained pyrethroids at least twice the LC50 for laboratory-cultured H. azteca.
Table 1.
Collecting sites for the wild H. azteca populations, including the organic content and total pyrethroid concentration in the sediments, measured at the time the animals were collected
| Site and coordinates | Land use surrounding sampling site | Sample date | Sediment organic carbon (%) | Pyrethroid concentration (ng/g)* |
| Blodgett Reservoir, Sacramento, CA 38.5176°N, 121.2109°W |
Undeveloped grasslands | Nov. 19, 2010 | 2.7 | 1.3 |
| Laguna Lake, Petaluma, CA 38.2064°N, 122.7636°W |
Grasslands with scattered homes and agriculture | July 3, 2010 | 0.2 | 1.8 |
| Pleasant Grove Creek, Roseville, CA 38.8052°N, 121.3069°W |
Open space with residential use 1 km upstream | June 1, 2010; March 13, 2013 | 2.9† | 6.8† |
| Morrison Creek, Sacramento, CA 38.5271°N, 121.3274°W |
Airfield with residential use 5 km upstream | Oct. 19, 2010 | 1.6 | Not detected (<1) |
| Mosher Slough, Stockton, CA 38.0325°N, 121.3654°W |
Commercial and residential land use | June 11, 2010 | 3.4 | 57.6 |
| Chualar Creek, Chualar, CA 36.5583°N, 121.5296°W |
Agriculture | Aug. 18. 2010 | 1.1 | Not detected (<1) |
| Grayson Creek, Concord, CA 37.9665°N, 122.0666°W |
Commercial and residential land use | Aug. 12, 2010 | 1.2 | 50.7 |
Total sediment pyrethroids were comprised entirely of bifenthrin except Mosher Slough (35.5 ng/g bifenthrin, 5.9 ng/g cyfluthrin, 4.2 ng/g cyhalothrin, 5.4 ng/g cypermethrin, 6.6 ng/g permethrin) and Grayson Creek (26.6 ng/g bifenthrin, 7.7 ng/g cyfluthrin, 3.6 ng/g cypermethrin, 12.8 ng/g permethrin).
Chemistry data from the June 2010 collecting event.
For comparison with wild populations, laboratory-cultured H. azteca were obtained from the University of California Berkeley (UCB), Southern Illinois University (SIU), and Chesapeake Cultures, a commercial supplier. Two of these cultures (UCB and SIU) can be traced to an initial collection of wild H. azteca from Oregon (SI Appendix, Fig. S1). The original source of animals for the third culture could not be established.
Populations Examined Consist of Four Distinct Species Groups.
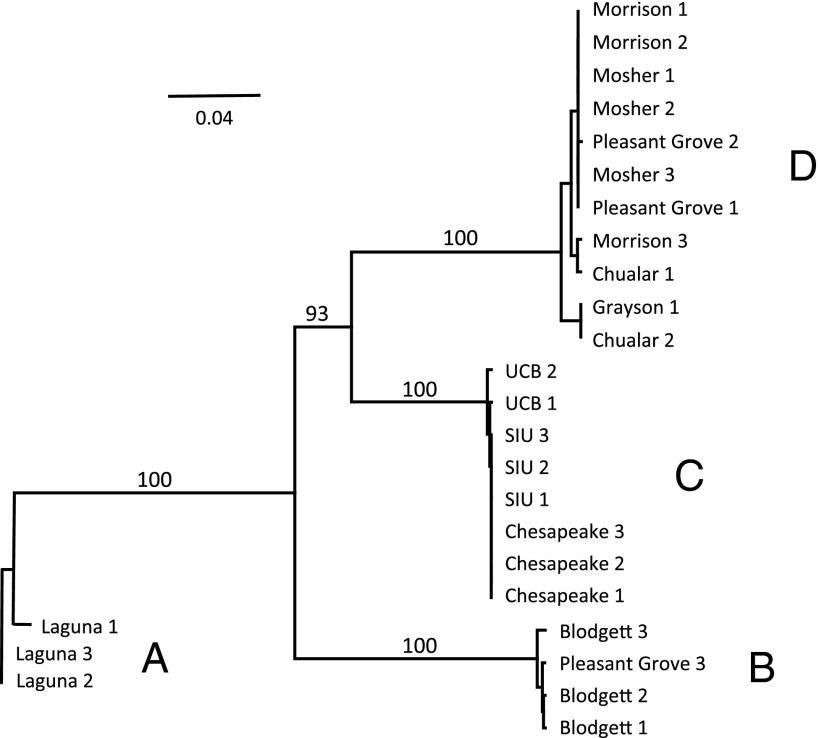
Because H. azteca is a species complex, we investigated genetic diversity of wild and laboratory populations by evaluating nucleotide sequences in the mitochondrial gene cytochrome c oxidase subunit I (COI) and the 28S nuclear ribosomal large subunit rRNA gene (28S rDNA). Maximum likelihood analysis of a 695-bp fragment of COI revealed four distinct clades separated by substantial genetic divergence (Fig. 1; SI Appendix, Table S1). The same four clades were recovered with high bootstrap support in analysis of the 28S rDNA gene (SI Appendix, Fig. S2), based on a 674 nucleotide region for all individuals except specimens in clade B (472–519 nucleotides). These results are consistent with the presence of four distinct species. All laboratory-cultured organisms fell within clade C. Field-collected animals fell within clades A, B, and D, with organisms from any one site usually appearing in only a single clade. The exception was Pleasant Grove Creek, containing both clades B and D.
Fig. 1.
Maximum likelihood phylogram based on COI mtDNA sequence data. Numbers on branches indicate branch support based on 1,000 bootstrap replicates. Group designations A–D indicate putative species. Analysis was conducted with PhyML implemented in Geneious using a GTR model of nucleotide substitution. (Scale bar, number of substitutions per site.)
Sensitivity to Pyrethroids Varies Across Populations.
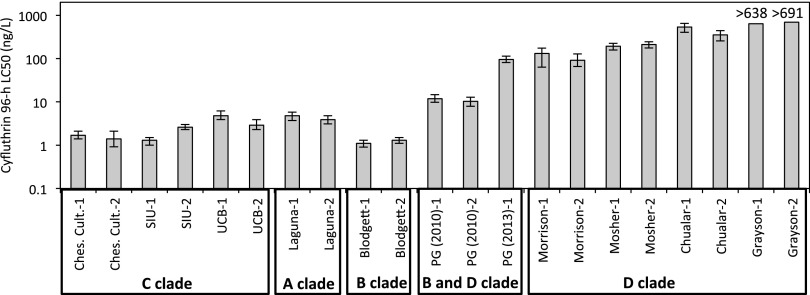
Pyrethroid sensitivity was quantified by determining each population’s 96-h LC50 in water-only exposures to cyfluthrin (Fig. 2; SI Appendix, Table S2). When enough individuals were available, LC50 values for the pyrethroid bifenthrin were also determined. Results parallel those for cyfluthrin and are provided in SI Appendix, Table S2. Cyfluthrin sensitivity was similar among the three laboratory cultures (clade C), with cyfluthrin LC50s from 1.3 to 4.8 ng/L. Wild populations from the two sites with little prior pyrethroid exposure had sensitivity comparable to the cultures: Blodgett Reservoir animals (clade B) had LC50s of 1.1–1.3 ng/L, and Laguna Lake amphipods (clade A) had LC50s of 3.9–4.8 ng/L.
Fig. 2.
Cyfluthrin sensitivity of H. azteca from the various clades, with error bars showing the 95% CI of the LC50. The LC50 values were typically determined in two independent trials with both results reported. All data were adjusted to reflect actual, rather than nominal, initial concentrations. LC50 was not determinable for Grayson Creek because the highest concentrations tested (values shown) produced only 33% mortality in both trials. PG, Pleasant Grove; Ches. Cult., Chesapeake Cultures.
Wild populations from sites with the highest pyrethroid exposures as indicated by sediment concentrations (Mosher and Grayson) or with pyrethroid use nearby in the watershed (Morrison and Chualar) showed extraordinary pyrethroid resistance. H. azteca from the Morrison, Mosher, and Chualar sites had cyfluthrin LC50s of 92–535 ng/L. Grayson Creek animals were even more resistant, with only 33% mortality at the highest concentration tested (691 ng/L). All of these populations were composed of animals belonging to clade D. When resistant animals were cultured in the laboratory for up to 3 mo, there was no loss of resistance in the F1 generation (SI Appendix).
Chualar Creek was of particular interest because the concentration of the organophosphate insecticide chlorpyrifos in sediments was 248 ng/g, which is five times the 10-d LC50 for laboratory-cultured H. azteca. When tested with the UCB culture, these sediments caused 97% mortality, and chlorpyrifos was likely responsible because addition of an organophosphate-hydrolyzing enzyme reduced mortality to 22% (5). Therefore, the population in Chualar Creek was resistant not only to a pyrethroid (Fig. 2) but to an organophosphate as well.
Determination of LC50 values for B and D clade populations that co-occur in Pleasant Grove Creek was complicated because we cannot visually distinguish these species and relied on gene sequencing of the group used for toxicity testing to provide a post hoc interpretation of LC50 results. Species assignment was based on a 195-bp region of COI containing 33 diagnostic nucleotide loci. We determined pyrethroid sensitivity in H. azteca from Pleasant Grove Creek in both 2010 and 2013. In 2010, 21 individuals from the group used for toxicity testing were sequenced, with 57% and 43% belonging to clades B and D, respectively. In 2010, there was only slight pyrethroid resistance (cyfluthrin LC50s of 10.3–11.8 ng/L). Only one individual of 60 survived at the highest concentration tested of 56 ng/L, a concentration at which there was at least 50% survival and often >90% in the more pyrethroid-resistant populations. Given the roughly equal proportions of clades B and D, our interpretation is that one or both species likely had slightly elevated resistance, but only a very small percentage of either species exhibited substantially elevated resistance. In 2013, amphipods were collected at the same Pleasant Grove Creek site as the 2010 sample, but earlier in the year (March vs. June). Juveniles were absent, so while the smallest individuals available were used, they were larger than those used in 2010 testing. Fourteen individuals from the group used for toxicity testing were 93% clade B and 7% clade D. The bias toward clade B was likely a result of the overall smaller size of clade B individuals and absence of juveniles from either clade. Toxicity testing with 2013 animals showed much higher resistance (cyfluthrin LC50 of 96 ng/L). There was almost complete survival (93%) at the 56-ng/L concentration that caused near complete mortality in the 2010 tests (SI Appendix, Fig. S3). Genotyping of all 16 individuals surviving the highest cyfluthrin treatments (106–847 ng/L) indicated that all were clade B. We interpret these results as indicating the clade B population in Pleasant Grove Creek had substantially elevated pyrethroid resistance in 2013 and that there is a suggestion of an increase in resistance of the clade B population between 2010 and 2013, although the latter interpretation must be viewed with caution. Moreover, these results indicate that pyrethroid resistance is not consistent across all populations within a clade. Although clade D populations exhibited resistance at other locations, clade D in Pleasant Grove Creek showed little or no resistance, at least in 2010, the only occasion when it was present in appreciable numbers in toxicity trials. In contrast, the clade B population in Pleasant Grove Creek exhibited high resistance, whereas that same clade in Blodgett Reservoir lacked resistance.
Tolerance Across Species Groups Is Correlated with Mutations in the Voltage-Gated Sodium Channel.
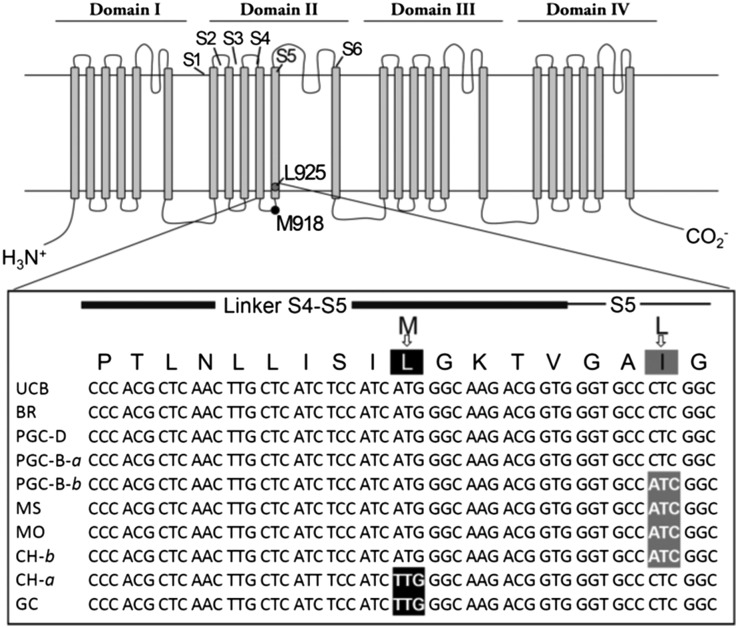
The primary target site for pyrethroids is the voltage-gated sodium channel (VGSC) of the peripheral and central nervous system. Pyrethroids impede closing of the channel, causing hyperexcitability and repetitive firing. Eventually nerve cells become exhausted, resulting in incapacitation of the organism and the knockdown phenotype (7). The VGSC gene (vgsc) consists of four domains, each with six transmembrane domains (Fig. 3). To investigate possible mutations responsible for pyrethroid resistance, we sequenced four segments of the vgsc where mutations conferring pyrethroid resistance are known to occur in multiple species (8), including domain IS6, a segment of the linker region between domain I and II, domain II, and domain IIIS6 (SI Appendix, Fig. S4). Primers were designed (SI Appendix, Table S3) to amplify corresponding segments of the vgsc and applied to two or three individuals (three for domain II) from the clade C culture at UCB and clade B or D field populations. Sequencing of these products revealed that amino acid sequences were generally highly conserved; however, amino acid substitutions within domain II were apparent (SI Appendix, Fig. S4). Substitutions included M918L, found in Grayson Creek and some Chualar Creek individuals (both clade D). Substitution L925I was found in other Chualar Creek individuals, as well as Morrison Creek and Mosher Slough (all clade D), and some Pleasant Grove Creek individuals (clade B) (Fig. 3; complete sequences in SI Appendix, Fig. S5). These mutations were only present in populations with decreased sensitivity to cyfluthrin, suggesting a role in pyrethroid resistance.
Fig. 3.
VGSC with mutations found in domain II associated with pyrethroid resistance. General structure of the VGSC is shown above with positions of the point mutations found in the present study noted. Close-up of partial amino acid and nucleotide sequences of domain II of the vgsc reveals amino acid substations in different H. azteca populations. The positions of mutations are highlighted in black (M918L) and dark gray (L925I). Amino acid positions are numbered according to the housefly VGSC I channel protein (8). UCB, University of California Berkeley culture; BR, Blodgett Reservoir; PGC, Pleasant Grove Creek (with clade noted and both alleles for clade B); MS, Mosher Slough; MO, Morrison Creek; CH, Chualar Creek (with both alleles shown); GC, Grayson Creek.
Additional amino acids substitutions were found in domain II and in domain IIIS6. However, these mutations were either only found in a few individuals from one population (I936F and K1557R) or found in a few individuals from both high and low resistance populations (K1578N). Therefore, their presence was not well correlated with the pyrethroid resistance of the populations.
Within a Single Population, Pyrethroid Resistance Is Tightly Coupled to the Presence of the L925I Mutation.
We investigated the frequency of mutant alleles within Pleasant Grove Creek animals, which contained both clade B and D H. azteca and animals with and without the L925I mutation. In the 2013 toxicity test, 93% of control animals were from clade B, with the allele harboring the L925I mutation present in 5 of 13 clade B individuals genotyped. One of these individuals was heterozygous (Table 2). All H. azteca surviving at the highest cyfluthrin concentrations (106–847 ng/L; all above the LC50) belonged to clade B, and all contained the L925I substitution. The fact that the L925I mutation was in only 38% of individuals initially used for testing, but in 100% of those surviving the high cyfluthrin concentrations, provides further support that this mutation confers pyrethroid resistance. Sequencing of additional H. azteca not used in toxicity testing was unable to detect the L925I mutation in clade D individuals from Pleasant Grove Creek (10 individuals from 2013 examined). In addition, we were unable to detect presence of the L925I mutation in the 2010 collection (Table 2) when the population showed minimal pyrethroid resistance, again consistent with the mutation’s role in resistance.
Table 2.
Species composition and allele frequencies at Pleasant Grove Creek in 2010 and 2013
| Year/condition | Clade B |
Clade D |
||
| L925 WT | L925I | L925 WT | L925I | |
| 2010/unexposed | 12 (100%) | 0 (0%) | 9 (100%) | 0 (0%) |
| 2013/unexposed | 8 (62%) | 5 (38%)* | 1 (100%) | 0 (0%) |
| 2013/exposed | 0 (0%) | 16 (100%) | 0 (0%) | 0 (0%) |
Individual amphipods were collected from Pleasant Grove Creek in 2010 and 2013 for toxicity bioassays. At the conclusion of the toxicity bioassays, surviving amphipods were collected from the unexposed control (2010 and 2013) or from the highest cyfluthrin exposures, 106–847 ng/L (exposed; 2013 only). The number of individuals with the L925 or L925I genotype from each clade is shown, with the corresponding allele frequency within that clade provided in parentheses.
One of the clade B individuals from 2013 shown here contained both the WT and L925I alleles.
Gene Expression Results Suggest an Alternate Mechanism of Toxicity in Resistant Individuals.
To better understand how vgsc mutations affect the mechanism of pyrethroid toxicity, gene expression analysis using an oligonucleotide microarray for H. azteca was done with the UCB culture and the most resistant natural population: Grayson Creek. Individuals from both populations were challenged with exposure to cyfluthrin at 0.4 ng/L [the no observed effect concentration (NOEC) of the UCB population], and the Grayson Creek population was challenged at 170 ng/L (NOEC for that population). Direct comparison of microarray signal intensities between the two populations was not possible because the level of species divergence would likely result in differential hybridization to microarray probes (17). Therefore, microarray results were compared between cyfluthrin treatments and untreated controls from the same populations, allowing us to identify genes induced or repressed in each population by exposure.
In UCB animals at 0.4 ng/L, 126 sequences were differentially expressed relative to the unexposed UCB treatment. When Grayson Creek animals were exposed to 0.4 ng/L, no genes were differentially expressed, and exposed animals displayed a similar expression pattern as unexposed controls (SI Appendix, Fig. S6). However, when exposure of Grayson Creek animals was increased to 170 ng/L, 1,321 sequences were differentially expressed (SI Appendix, Table S4).
Gene ontology terms were mapped using Blast2Go. Of the total 1,447 differentially expressed genes, 429 were successfully mapped to GO terms and 355 were annotated. Many affected genes in the UCB 0.4-ng/L samples were consistent with the known mechanism of pyrethroid toxicity, showing involvement in neurological system processes, synapse organization, and transmission of nerve impulses (SI Appendix, Table S5 and Fig. S6). In contrast, there was a very different gene expression response in the Grayson Creek 170 ng/L–exposed H. azteca. Instead of genes involved in nerve transmission, genes affected were involved in stress response: oxidation/reduction [cytochrome P450s (CYPs), glutathione S-transferases (GSTs), other oxidases], heat shock proteins, and cellular metabolic enzymes.
Discussion
Pyrethroid Resistance Varies Across Species Groups and Natural Populations of H. azteca.
Phylogenetic analysis provides evidence that our H. azteca belong to four distinct species. Previous studies of the H. azteca species complex have documented similarly deep levels of mtDNA divergence among populations and reproductive isolation between similarly divergent groups (10–12). Four populations that exhibited high pyrethroid resistance (Morrison, Mosher, Chualar, and Grayson) fell within clade D, and one resistant population (Pleasant Grove) fell within clade B.
Several lines of evidence indicate reduced pyrethroid sensitivity observed in H. azteca is due to adaptive resistance acquired through mutations in domain II of the vgsc. The presence of two amino acid substitutions, M918L or L925I, are highly correlated with resistance across populations and within a single population containing both sensitive and resistant individuals. The M918L mutation was first observed in pyrethroid-resistant cotton aphids, Aphis gossypii (18), whereas the L925I mutation was first identified in resistant whiteflies, Bemisia tabaci (19), and has been characterized extensively in several insects (8). Sites M918 and L925 are two of several residues critical for pyrethroid binding to the VGSC (20), and in vitro assays have shown mutations at these sites retain VGSC gate function in the presence of pyrethroids (21, 22). An amino acid substitution at M918 to a non–sulfur-containing residue reduces susceptibility to pyrethroids (7), and disassociation of pyrethroids from a mutated M918 is 200 times faster than WT (21). The L925I mutation is considered conservative because of the similarity in structure of leucine and isoleucine; however, this small change is sufficient to reduce pyrethroid binding (20, 22).
Further evidence for target site resistance comes from the results of gene expression analyses. At the lower concentration of cyfluthrin (0.4 ng/L), animals from a nonresistant population expressed genes associated with neurological processes. However, in the resistant Grayson Creek population, there was no gene expression response because cyfluthrin could not bind to the VGSC. Toxicity experienced by resistant animals at the higher concentration (170 ng/L) was distinct from the primary mechanism of pyrethroid action and instead invoked processes associated with oxidative stress, including oxidation-reduction and GST activity. Interestingly, this mode is similar to pyrethroid toxicity in mammals. Because the mammalian sodium channel ortholog (e.g., rat brain IIA) is insensitive to pyrethroids (23), toxicity in mammals can occur through a secondary mode of action related to glutathione depletion and oxidative damage (24, 25).
Multiple Origins of Resistance and Rapid Selection.
Because resistant H. azteca span clades B and D and WT individuals were found within these clades, it appears these mutations are recently derived. Additionally, resistance arose independently at least three times, as evidenced by alleles with either of two different mutations (M918L or L925I) and the L925I mutation present in two different clades. Convergent evolution and constraints on possible amino acid substitutions resulted in the repeated selection of the two different mutations across populations. Mutations at M918 and L925 have been selected multiple times within insects (8), even within a single species. The L925I mutation arose independently in two sympatric, but reproductively isolated, biotypes of white flies (19).
In Pleasant Grove Creek, where clades B and D were present, we observed an order of magnitude increase in pyrethroid resistance between 2010 and 2013. Resistance alleles were not detectable in 2010, although they could have been present in clade B at low frequency (<8%). Resampling in 2013 revealed that the allele frequency had climbed to 38% within clade B, and it played a role in decreased sensitivity of the population, as all individuals surviving cyfluthrin treatment contained the L925I mutation. These results suggest that clade B H. azteca at Pleasant Grove Creek are experiencing selective pressure from pyrethroid exposure and are undergoing rapid selection for resistance.
The appearance of resistance in a nontarget aquatic species, presumably as a result of incidental exposure to agents used for terrestrial pests, provides troubling evidence of emergent effects of pesticide use on natural populations (1). Resistant organisms were found at every urban and agricultural site sampled, but not at the two sites with minimal surrounding development. These results imply that pyrethroid resistance may be common in natural populations of H. azteca in areas of pyrethroid contamination, and such areas exist throughout the United States (6). Although insecticide resistance has been often documented in target species or other species living in the target’s environment, there are only a few reports of resistance in nontarget aquatic species exposed through runoff or spray drift (26, 27).
Although emergence of resistance demonstrates the adaptive capacity of biota to withstand anthropogenic stressors, this adaptation may come at a cost to genetic and biological diversity (2, 28). Pyrethroid resistance can reduce overall fitness (29) directly through reduced efficiency of the VGSC and associated metabolic costs (30) or indirectly through genetic hitchhiking (31). In addition, under strong selective pressures of pesticide exposure, genetic diversity may become depleted by “genetic erosion,” as selection of a few adapted organisms creates genetic bottlenecks (32). Reduced genetic diversity results in populations without the genetic variation necessary to tolerate other stressors associated with other pollutants, climate change, or natural ecosystem changes (33).
Implications for Environmental Assessment.
The impetus for this study was our observation of wild H. azteca in waters determined to be acutely toxic when tested in the laboratory. Our findings of resistance mutations in a species complex that toxicologists often treat as a single species explain our observations and have significant ramifications to the current use of H. azteca in toxicity testing. All of the cultures we tested have, at one or more times in their history, been supplemented with local wild-caught animals (SI Appendix, Fig. S1). Given the extensive diversification that has occurred in the H. azteca complex, addition of local animals to a culture almost certainly introduces a different species and could introduce resistance alleles. Either result undermines comparability of organisms among toxicity testing laboratories on which modern environmental risk assessment depends.
A critical unanswered question is whether the many laboratories performing testing with H. azteca are all using the same species. One investigation of cultures at six laboratories, based on allozyme analysis of 16 loci, found sufficient divergence to suggest the presence of three species (34). Our phylogenetic work with three cultures suggests they are all of the same species and at least two of the three cultures can be traced to a common source. Although these two cultures have been separated from this common source for 17 y, they have not experienced any divergence in pyrethroid sensitivity. However, our results strongly argue for an urgent need to determine the origin and genetic characteristics of the many cultures now in use for toxicity testing, because data generated from them are widely used to support regulatory decision-making.
Finding pesticide resistance in an aquatic species raises the question of how common this phenomenon may be in other species, potentially distorting conclusions drawn from bioassessment surveys. Bioassessments interpret habitat quality based on composition of the resident fauna, and such conclusions often serve as the basis for regulatory action. For example, the presence of H. azteca in Pleasant Grove Creek, and the presumption that this wild population was highly sensitive to pyrethroids based on data from laboratory-cultured animals, has been used to imply that pyrethroids are not a significant stressor in that system (35). Without actual testing of contaminant sensitivity and genetic sequencing methods, the presence of mutations conferring resistance in wild populations is likely to go unrecognized. Molecular tools, including Taqman genotyping assays, could be developed for H. azteca to characterize the prevalence and geographic distribution of pyrethroid resistance in these important biomonitoring organisms. If resistance alleles correlate with presence of pyrethroids in the environment, as our current results suggest, molecular genotyping assays could provide biomarkers for pyrethroid contamination. We suspect that contaminant resistance, as a general phenomenon, may be far more common than is generally recognized, and merits considerable further study.
Materials and Methods
Further details on all methods used can be found in SI Appendix.
Species Identification.
Reference specimens from all laboratory and field populations were sent to Adam Baldinger, Harvard Museum of Comparative Zoology, for confirmation that they fell within H. azteca s.l., based on morphology.
Toxicity Testing.
Animals of comparable size were used in comparing pyrethroid sensitivity between populations from the collection sites. Stacked screens were used to obtain animals 1.8–2.5 mm in length (excluding antennae), which generally corresponds to an age of 7–14 d (3). Test organisms were exposed for 96 h to laboratory water spiked with a range of bifenthrin or cyfluthrin concentrations. At test completion, survival at each concentration was quantified and used to determine the LC50 for each population. All populations were tested within 2 d of collection, but animals from Grayson and Chualar Creeks were also held in the laboratory for extended periods in pyrethroid-free conditions. Juveniles produced while in laboratory culture were collected and tested for cyfluthrin sensitivity after 25 d in culture (both creeks) and after 96 d (Chualar only).
Chemical Analyses.
After preparation of pyrethroid-spiked waters, samples were liquid:liquid extracted using dichloromethane following Environmental Protection Agency (EPA) method 3510C. Extracts were cleaned with a dual layer graphitized black carbon and primary/secondary amine column and analyzed on an Agilent 6890 gas chromatograph with microelectron capture detector (Agilent Technologies). Sediment samples from field sites were extracted using a matrix-dispersive accelerated solvent method using a Dionex 200, with clean-up and analysis as for the water samples.
Phylogenetic Sequencing.
The mitochondrial gene COI and the nuclear 28S rDNA gene were PCR-amplified. PCR products were gel purified and sequenced using an ABI 3730 automated sequencer (Applied Biosystems). Sequences were aligned using Geneious (36), and evaluation of relationships among aligned sequences used maximum likelihood analysis implemented in PhyML (37). For COI, genetic distances among groups were quantified as the Kimura 2-parameter distance (38), and calculated in MEGA 5 (39).
VGSC Sequencing.
Seven sequences were identified as partial sequences of the vgsc through a tblastx homology search to the Musca domestica X96668. Analyses of potential mutations were conducted using three individuals each from all collected populations, with more extensive analysis (61 individuals) at Pleasant Grove Creek. PCR was performed to amplify four targeted areas of the vgsc gene using the primers provided in SI Appendix, Table S3, and the products were cloned into plasmids using TA cloning (Invitrogen) for sequencing.
Microarray Construction.
H. azteca from a commercial vendor were exposed to sublethal concentrations of cadmium chloride (0.5 µg/L), atrazine (20 µg/L), and PCB Aroclor 1254 (0.5 µg/L). Detailed protocols for RNA extraction, cDNA synthesis, and 454 transcriptomics sequencing, assembly, and sequencing analysis can be found in Meyer et al. (40). A 135k oligonucleotide microarray was designed using the H. azteca 454 sequence data on a Nimblegen platform (12 × 135k; Roche Nimblegen). The microarray platform has been deposited to the Gene Expression Omnibus (GEO) with accession number GPL17458. Sequencing results including total number of reads and annotated sequences can be found in SI Appendix, Table S6.
Transcriptomics.
Animals were preserved at the conclusion of toxicity tests in RNAlater (Ambion) and frozen until further analysis. Four replicate RNA samples consisting of 6–10 individuals per treatment were reverse-transcribed using Sigma TransPlex Whole Transcriptome Amplification Kit, which produces amplified cDNA. Amplified cDNA was labeled using Nimblegen’s one-color DNA labeling kit (cy3 only) and hybridized to the 135k oligonucleotide microarray (GEO accession no. GSE48943). Comparisons were performed between genes differentially expressed by pyrethroid exposure in the UCB or Grayson Creek populations and their respective unexposed controls.
Supplementary Material
Acknowledgments
We thank Adam Baldinger of the Harvard Museum of Comparative Zoology for taxonomic identification of our specimens. G.A.W. was supported by the US National Science Foundation, and D.P.W. was supported by the Delta Science Program. Sodium channel sequencing and microarray studies were supported through the University of Massachusetts, Boston faculty startup funds to H.C.P. Transcriptome sequencing of H. azteca libraries and microarray construction were funded through an Illinois-Indiana Sea Grant to M.S.S. and J.K.C.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GAJQ00000000.1, GAJP00000000.1, JX678289, KF596727–KF596752, and KF596753–KF596771), and the data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GPL17458 and GSE48943).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302023110/-/DCSupplemental.
References
- 1.Bickham JW. The four cornerstones of evolutionary toxicology. Ecotoxicology. 2011;20(3):497–502. doi: 10.1007/s10646-011-0636-y. [DOI] [PubMed] [Google Scholar]
- 2.Medina MH, Correa JA, Barata C. Micro-evolution due to pollution: Possible consequences for ecosystem responses to toxic stress. Chemosphere. 2007;67(11):2105–2114. doi: 10.1016/j.chemosphere.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 3.USEPA . Methods for Measuring the Toxicity and Bioaccumulation of Sediment-Associated Contaminants with Freshwater Invertebrates. EPA Publication 600/R-99/064. Washington, DC: US Environmental Protection Agency; 2000. [Google Scholar]
- 4.Weston DP, Lydy MJ. Urban and agricultural sources of pyrethroid insecticides to the Sacramento-San Joaquin Delta of California. Environ Sci Technol. 2010;44(5):1833–1840. doi: 10.1021/es9035573. [DOI] [PubMed] [Google Scholar]
- 5.Weston DP, Ding Y, Zhang M, Lydy MJ. Identifying the cause of sediment toxicity in agricultural sediments: The role of pyrethroids and nine seldom-measured hydrophobic pesticides. Chemosphere. 2013;90(3):958–964. doi: 10.1016/j.chemosphere.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 6.Hladik ML, Kuivila KM. Pyrethroid insecticides in bed sediments from urban and agricultural streams across the United States. J Environ Monit. 2012;14(7):1838–1845. doi: 10.1039/c2em10946h. [DOI] [PubMed] [Google Scholar]
- 7.Davies TG, Field LM, Usherwood PN, Williamson MS. DDT, pyrethrins, pyrethroids and insect sodium channels. IUBMB Life. 2007;59(3):151–162. doi: 10.1080/15216540701352042. [DOI] [PubMed] [Google Scholar]
- 8.Rinkevich FD, Du Y, Dong K. Diversity and convergence of sodium channel mutations involved in resistance to pyrethroids. Pestic Biochem Physiol. 2013;106(3):93–100. doi: 10.1016/j.pestbp.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witt JDS, Threloff DL, Hebert PDN. DNA barcoding reveals extraordinary cryptic diversity in an amphipod genus: Implications for desert spring conservation. Mol Ecol. 2006;15(10):3073–3082. doi: 10.1111/j.1365-294X.2006.02999.x. [DOI] [PubMed] [Google Scholar]
- 10.Witt JDS, Hebert PDN. Cryptic species diversity and evolution in the amphipod genus Hyalella within central glaciated North America: A molecular phylogenetic approach. Can J Fish Aquat Sci. 2000;57(4):687–698. [Google Scholar]
- 11.Wellborn GA, Cothran RD, Bartholf S. Life history and allozyme diversification in regional ecomorphs of the Hyalella azteca (Crustacea: Amphipoda) species complex. Biol J Linn Soc Lond. 2005;8(2):161–175. [Google Scholar]
- 12.Wellborn GA, Broughton RE. Diversification on an ecologically constrained adaptive landscape. Mol Ecol. 2008;17(12):2927–2936. doi: 10.1111/j.1365-294X.2008.03805.x. [DOI] [PubMed] [Google Scholar]
- 13.Duan Y, Guttman SI, Oris JT, Huang X, Burton GA. Genotype and toxicity relationships among Hyalella azteca: II. Acute exposure to fluoranthene-contaminated sediment. Environ Toxicol Chem. 2000;19(5):1422–1426. [Google Scholar]
- 14. Ng CM, Weston DP, You J, Lydy MJ (2008) Patterns of pyrethroid contamination and toxicity in agricultural and urban stream segments. Synthetic Pyrethroids: Occurrence and Behavior in Aquatic Environments, ed Gan J, Spurlock F, Hendley P, Weston D (Oxford Univ Press, New York), pp 355–369.
- 15.Amweg EL, Weston DP, Ureda N. (2005) Use and toxicity of pyrethroid pesticides in the Central Valley, CA, USA. Environ Toxicol Chem 24(4):966–972, and erratum in 24(5):1300–1301. [DOI] [PubMed]
- 16.Weston DP, Holmes RW, You J, Lydy MJ. Aquatic toxicity due to residential use of pyrethroid insecticides. Environ Sci Technol. 2005;39(24):9778–9784. doi: 10.1021/es0506354. [DOI] [PubMed] [Google Scholar]
- 17.Buckley BA. Comparative environmental genomics in non-model species: Using heterologous hybridization to DNA-based microarrays. J Exp Biol. 2007;210(Pt 9):1602–1606. doi: 10.1242/jeb.002402. [DOI] [PubMed] [Google Scholar]
- 18.Carletto J, Martin T, Vanlerberghe-Masutti F, Brévault T. Insecticide resistance traits differ among and within host races in Aphis gossypii. Pest Manag Sci. 2010;66(3):301–307. doi: 10.1002/ps.1874. [DOI] [PubMed] [Google Scholar]
- 19.Alon M, et al. Multiple origins of pyrethroid resistance in sympatric biotypes of Bemisia tabaci (Hemiptera: Aleyrodidae) Insect Biochem Mol Biol. 2006;36(1):71–79. doi: 10.1016/j.ibmb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 20.O’Reilly AO, et al. Modelling insecticide-binding sites in the voltage-gated sodium channel. Biochem J. 2006;396(2):255–263. doi: 10.1042/BJ20051925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vais H, et al. Mutations of the para sodium channel of Drosophila melanogaster identify putative binding sites for pyrethroids. Mol Pharmacol. 2003;64(4):914–922. doi: 10.1124/mol.64.4.914. [DOI] [PubMed] [Google Scholar]
- 22.Usherwood PN, Vais H, Khambay BP, Davies TG, Williamson MS. Sensitivity of the Drosophila para sodium channel to DDT is not lowered by the super-kdr mutation M918T on the IIS4-S5 linker that profoundly reduces sensitivity to permethrin and deltamethrin. FEBS Lett. 2005;579(28):6317–6325. doi: 10.1016/j.febslet.2005.09.096. [DOI] [PubMed] [Google Scholar]
- 23.Warmke JW, et al. Functional expression of Drosophila para sodium channels. Modulation by the membrane protein TipE and toxin pharmacology. J Gen Physiol. 1997;110(2):119–133. doi: 10.1085/jgp.110.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kale M, Rathore N, John S, Bhatnagar D. Lipid peroxidative damage on pyrethroid exposure and alterations in antioxidant status in rat erythrocytes: A possible involvement of reactive oxygen species. Toxicol Lett. 1999;105(3):197–205. doi: 10.1016/s0378-4274(98)00399-3. [DOI] [PubMed] [Google Scholar]
- 25.Giray B, Gürbay A, Hincal F. Cypermethrin-induced oxidative stress in rat brain and liver is prevented by vitamin E or allopurinol. Toxicol Lett. 2001;118(3):139–146. doi: 10.1016/s0378-4274(00)00277-0. [DOI] [PubMed] [Google Scholar]
- 26.Brausch JM, Smith PN. Pesticide resistance from historical agricultural chemical exposure in Thamnocephalus platyurus (Crustacea: Anostraca) Environ Pollut. 2009;157(2):481–487. doi: 10.1016/j.envpol.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Vinson SB, Boyd CE, Ferguson DE. Resistance to DDT in the mosquito fish, Gambusia affinis. Science. 1963;139(3551):217–218. doi: 10.1126/science.139.3551.217. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann AA, Willi Y. Detecting genetic responses to environmental change. Nat Rev Genet. 2008;9(6):421–432. doi: 10.1038/nrg2339. [DOI] [PubMed] [Google Scholar]
- 29.Berticat C, et al. Costs and benefits of multiple resistance to insecticides for Culex quinquefasciatus mosquitoes. BMC Evol Biol. 2008;8:104. doi: 10.1186/1471-2148-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Park Y, Adams ME. Functional and evolutionary consequences of pyrethroid resistance mutations in S6 transmembrane segments of a voltage-gated sodium channel. Biochem Biophys Res Commun. 2000;278(3):516–521. doi: 10.1006/bbrc.2000.3832. [DOI] [PubMed] [Google Scholar]
- 31.Cutter AD, Payseur BA. Genomic signatures of selection at linked sites: Unifying the disparity among species. Nat Rev Genet. 2013;14(4):262–274. doi: 10.1038/nrg3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Straalen NM, Timmermans M. Genetic variation in toxicant-stressed populations: An evaluation of the “genetic erosion” hypothesis. Hum Ecol Risk Assess. 2002;8(5):983–1002. [Google Scholar]
- 33.Reed DH, Frankham R. Correlation between fitness and genetic diversity. Conserv Biol. 2003;17(1):230–237. [Google Scholar]
- 34.Duan Y, Guttman SI, Oris JT. Genetic differentiation among laboratory populations of Hyalella azteca: Implications for toxicology. Environ Toxicol Chem. 1997;16(4):691–695. [Google Scholar]
- 35.Hall LW, Killen WD, Anderson RD, Alden RW. The influence of physical habitat, pyrethroids, and metals on benthic community condition in an urban and residential stream in California. Hum Ecol Risk Assess. 2009;15(3):526–553. [Google Scholar]
- 36. Drummond AJ, et al. (2012) Geneious v5.6. Available at http://www.geneious.com. Accessed June 22, 2012.
- 37.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 38.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 39.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer E, et al. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics. 2009;10:219. doi: 10.1186/1471-2164-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.