Since livestock were first domesticated, humans have been selecting particular phenotypes as mating stock for each new generation. As a result, humans have been selecting for combinations of alleles that result in animals that better meet our needs: increased production, disease resistance, docility, and so forth. As purpose-bred animals became standard in animal agriculture, breeds began to diverge such that a mating produced between differently purposed breeds (dairy vs. beef, layers vs. broilers) resulted in offspring that were less desirable than individuals from either breed. As causative mutations are discovered that affect agriculturally important traits, their utility is limited if they are exclusive to a singly purposed breed. As an example, a disease resistance gene from a beef-type breed is limited to beef animals because the loss of milk production in a dairy-type breed prevents the crossbred animal from being economically viable. In PNAS, Tan et al. remedy this problem from a technical point of view (1). However, do technical views really matter? The impact of Tan et al. depends on perceptions.
As regulatory agencies across the globe have unsuccessfully searched for real-world safety issues with genetically engineered (GE) food animals, and peripheral groups have predicted various forms of calamity from GE animals, genetic engineers have trudged forward. Shortly after the production of the first transgenic mammal that displayed a phenotype of potential agricultural utility (2), techniques were adapted for production of transgenic livestock (3). Over the years, several examples of transgenic animals have emerged that have agricultural potential: more efficient fish (4), sows with greater milk production for their piglets (5), mastitis-resistant cows (6), goats that produce milk that can prevent diarrhea (7), and chickens that do not propagate influenza (8). However, because there is no direct evidence that any regulatory agency on the planet is capable of approving a GE animal for food production, it has been difficult to move forward.
The basic premise for regulation appears to be that any genotype produced by breeding is safe, and that any genotype produced intentionally via recombinant DNA technologies cannot be allowed to go to market. Because most GE food animals have been based thus far on the introduction of either extra copies of genes or additions of genes that could not be naturally inherited, genetic engineers have conceded that the engineered genotypes are different from historic genotypes. However, unique genotypes do not routinely trigger regulatory evaluation. The fact is that every animal produced by natural mating (excluding identical twins) also has a unique genotype, and therefore is also different from every historic genotype that has ever been consumed as food.
In PNAS, Tan et al. (1) not only demonstrate that livestock genomes can be efficiently edited with precision; they also demonstrate that genotypes, which could have arisen from natural mating, can be produced efficiently and intentionally. The value of this technique in agriculture is that it facilitates movement of alleles between phenotypically distinct, purpose-bred animals without the loss of the breed characteristics. In one example, an allele that produces a hornless phenotype in a beef breed (9) was created in cultured cells from a dairy breed. The engineered genotype is indistinguishable from a dairy genotype that could have been produced through decades of backcrossing. However, with this strategy the goal is accomplished in a single generation and does not require the temporary but long-lasting loss of all of the characteristics that make a dairy cow a productive milk producer. The hornless phenotype is valued to prevent injury or damage to animals, personnel, facilities, and equipment. Because of the danger presented by horns, most dairy animals are dehorned shortly after birth.
In addition to the multitude of allelic introgressions that are likely to be made for agricultural reasons, Tan et al. (1) also demonstrate that subtle modifications can be made to livestock for biomedical applications. The tools that allow one allele to be converted to another can also be used to convert a functional allele to one that has been disrupted or has characteristics of a human gene. This ability is not new. Genetic engineers have been introducing dominant human disease genes into livestock since 1997 (10) and making gene knockouts in livestock for biomedical applications since 2002 (11). In fact, the general methods of Tan et al. and similar methods have been previously demonstrated in livestock (12–15). However, Tan et al. (1) demonstrate that gene knockouts can be routine and inexpensive. Furthermore, the authors show a general robustness of the technology. It remains to be seen as to whether granting agency reviewers will accept livestock as viable alternatives when traditional model organisms fail to recapitulate human symptoms. However, it is clear that for some human disorders, livestock can more accurately reproduce observations in humans in response to causative stimuli than can traditional model organisms (16). For interventions that address symptoms and for preclinical trails, models that reproduce human responses with high fidelity are paramount.
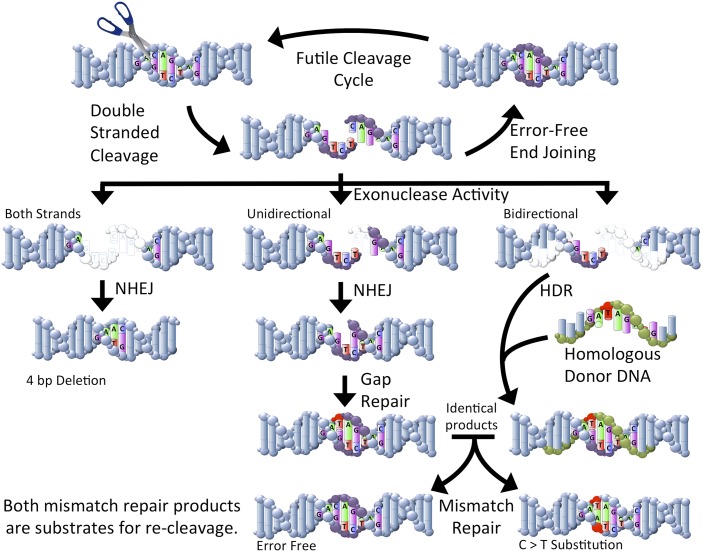
At the core of Tan et al.’s work (1) is a GE strategy first demonstrated in 2001 in Xenopus (17). That is, enzymes can be designed to cleave the genome at precise locations. In response to a genome cleavage event, DNA repair mechanisms attempt to remedy the breakage. However, DNA repair does not operate with absolute fidelity; there can be errors as a result of DNA repair (Fig. 1). If these efforts are
Tan et al. illustrate that production of livestock models for biomedical applications can be efficient and that genome editing can be robust.
being applied to cultured cells, the resulting cells can be used for somatic cell nuclear transfer to reconstruct embryos, and animals can be produced that have the recovered genotype. The introgressed allele can then be propagated through breeding.
Fig. 1.
Simplified representation of the repair of double-strand breaks. Sequence-specific endonucleases can be designed to cleave genome targets (represented by scissors). In general, cleaved sites can be rejoined to correct the double-strand break. This cycle can continue until the nuclease is no longer present or until an error occurs during repair that prevents recleavage. Exonuclease activity can truncate the free ends (bases that have been removed are shown in white). When both strands are truncated, deletions can be produced by nonhomologous end-joining (NHEJ). The change in local spacing produces a sequence that cannot be recleaved (Left pathway). If exonuclease activity removes bases from one strand, a gap can be created after NHEJ. The missing bases can be replaced by polymerase activity (Center pathway). Although this process generally proceeds with high fidelity, errors can occur (C > T error shown in red). If homologous DNA is present (shown in green), homology-directed repair (HDR) can result in the introduction of specific modifications (C > T modification shown in red). The result shown here (Right pathway) produces a product that is identical to the error shown for gap repair. After further processing through mismatch repair, the C > T substitution can be corrected or can retain the substitution. In either case, the resulting products may be recleaved. Larger modifications can be made than shown here. In addition, larger repair templates can be used to facilitate homologous recombination.
However, Tan et al. (1) also demonstrate that this process can be biased toward production of the desired genetic modification. In the example above, the DNA is repaired through a process called nonhomologous end-joining. However, there are other DNA repair mechanisms that can use the second allele of a gene or a closely related gene. When homologous sequence is available, the cell can use the sequence information found on the uninterrupted gene to fill in any gaps that may have resulted from initial DNA cleavage. This homology-directed repair provides a mechanism to introduce DNA sequence into gaps. Tan et al. provided an exogenous sequence that was homologous to the cleavage site over most of the length of the fragment, but also contained a specific difference representing a predetermined modification. At a variable but logistically practical rate, the cells used the homologous sequence to repair the cleavage and thus introduced a designed modification. Because Tan et al. targeted multiple genome locations, this work clearly demonstrates that designed nucleases can be robustly applied to livestock species to edit the genome with precision.
It is has been evident that there are practical reasons to genetically engineer livestock for agriculture and biomedicine. It is now obvious that genome editing can be used to efficiently produce precise changes to livestock, including intentional construction of genotypes that could have been produced from mating. Will animals produced from gene editing be regulated differently than those produced by sex? Are there scientifically demonstrable reasons to consider gene-edited animals from a different perspective than natural offspring? At a minimum, Tan et al. (1) illustrate that production of livestock models for biomedical applications can be efficient and that genome editing can be robust. Perhaps, for genotypes that can be produced by breeding and alleles that could have arisen via natural mutation, Tan et al. also illustrate a strategy to begin to address real-world problems in animal agriculture. Will regulatory agencies agree?
Footnotes
The author declares no conflict of interest.
See companion article on page 16526.
References
- 1.Tan W, et al. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci USA. 2013;110:16526–16531. doi: 10.1073/pnas.1310478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmiter RD, et al. Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes. Nature. 1982;300(5893):611–615. doi: 10.1038/300611a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammer RE, et al. Production of transgenic rabbits, sheep and pigs by microinjection. Nature. 1985;315(6021):680–683. doi: 10.1038/315680a0. [DOI] [PubMed] [Google Scholar]
- 4.Du SJ, et al. Growth enhancement in transgenic Atlantic salmon by the use of an “all fish” chimeric growth hormone gene construct. Biotechnology (N Y) 1992;10(2):176–181. doi: 10.1038/nbt0292-176. [DOI] [PubMed] [Google Scholar]
- 5.Bleck GT, White BR, Miller DJ, Wheeler MB. Production of bovine alpha-lactalbumin in the milk of transgenic pigs. J Anim Sci. 1998;76(12):3072–3078. doi: 10.2527/1998.76123072x. [DOI] [PubMed] [Google Scholar]
- 6.Wall RJ, et al. Genetically enhanced cows resist intramammary Staphylococcus aureus infection. Nat Biotechnol. 2005;23(4):445–451. doi: 10.1038/nbt1078. [DOI] [PubMed] [Google Scholar]
- 7.Maga EA, et al. Production and processing of milk from transgenic goats expressing human lysozyme in the mammary gland. J Dairy Sci. 2006;89(2):518–524. doi: 10.3168/jds.S0022-0302(06)72114-2. [DOI] [PubMed] [Google Scholar]
- 8.Lyall J, et al. Suppression of avian influenza transmission in genetically modified chickens. Science. 2011;331(6014):223–226. doi: 10.1126/science.1198020. [DOI] [PubMed] [Google Scholar]
- 9.Allais-Bonnet A, et al. Novel insights into the bovine polled phenotype and horn ontogenesis in Bovidae. PLoS ONE. 2013;8(5):e63512. doi: 10.1371/journal.pone.0063512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petters RM, et al. Genetically engineered large animal model for studying cone photoreceptor survival and degeneration in retinitis pigmentosa. Nat Biotechnol. 1997;15(10):965–970. doi: 10.1038/nbt1097-965. [DOI] [PubMed] [Google Scholar]
- 11.Dai Y, et al. Targeted disruption of the alpha1,3-galactosyltransferase gene in cloned pigs. Nat Biotechnol. 2002;20(3):251–255. doi: 10.1038/nbt0302-251. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe M, et al. Knockout of exogenous EGFP gene in porcine somatic cells using zinc-finger nucleases. Biochem Biophys Res Commun. 2010;402(1):14–18. doi: 10.1016/j.bbrc.2010.09.092. [DOI] [PubMed] [Google Scholar]
- 13.Hauschild J, et al. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci USA. 2011;108(29):12013–12017. doi: 10.1073/pnas.1106422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlson DF, et al. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109(43):17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon DN, et al. Production of biallelic CMP-Neu5Ac hydroxylase knock-out pigs. Sci Rep. 2013;3:1981. doi: 10.1038/srep01981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers CS, et al. Production of CFTR-null and CFTR-DeltaF508 heterozygous pigs by adeno-associated virus-mediated gene targeting and somatic cell nuclear transfer. J Clin Invest. 2008;118(4):1571–1577. doi: 10.1172/JCI34773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bibikova M, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21(1):289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]