Significance
Large parts of eukaryotic genomes are composed of transposons. Mammalian genomes use DNA methylation to silence these genomic parasites. A class of small RNAs called Piwi-interacting RNAs (piRNAs) is used to specifically guide the DNA methylation machinery to the transposon DNA elements. How germ cells make piRNAs is not entirely understood. We identify a mouse protein and demonstrate its importance for transposon silencing. We find that the protein collaborates with other factors already implicated in piRNA production. Moreover, the protein is required for piRNA production and assembly of the nuclear silencing complex. Physiological importance of the protein is highlighted by the fact that male mice lacking the protein are infertile. This study will greatly benefit the field of germ-cell biology.
Keywords: helicase, DNA methylation, spermatogenesis
Abstract
Piwi-interacting RNAs (piRNAs) are gonad-specific small RNAs that provide defense against transposable genetic elements called transposons. Our knowledge of piRNA biogenesis is sketchy, partly due to an incomplete inventory of the factors involved. Here, we identify Tudor domain-containing 12 (TDRD12; also known as ECAT8) as a unique piRNA biogenesis factor in mice. TDRD12 is detected in complexes containing Piwi protein MILI (PIWIL2), its associated primary piRNAs, and TDRD1, all of which are already implicated in secondary piRNA biogenesis. Male mice carrying either a nonsense point mutation (reproductive mutant 23 or repro23 mice) or a targeted deletion in the Tdrd12 locus are infertile and derepress retrotransposons. We find that TDRD12 is dispensable for primary piRNA biogenesis but essential for production of secondary piRNAs that enter Piwi protein MIWI2 (PIWIL4). Cell-culture studies with the insect ortholog of TDRD12 suggest a role for the multidomain protein in mediating complex formation with other participants during secondary piRNA biogenesis.
Repetitive mobile genomic elements called transposons are a potential source of mutations causing genome instability. They are particularly active in the germ line as mobilization can be inherited, allowing them to spread in the population. To counter this threat, animal germ lines have evolved a dedicated class of 24- to 30-nucleotide (nt)-long small RNAs called Piwi-interacting RNAs (piRNAs) (1–3). In mice, the piRNA pathway is mainly active in the male germ line where all of the three Piwi proteins (MILI, MIWI, and MIWI2) are expressed. Nuclear MIWI2 is implicated in establishing transcriptional silencing in embryonic germ cells by deposition of DNA methylation marks on target transposon loci (4, 5). Cytoplasmic MILI and MIWI have a role in maintaining repression by direct cleavage of transposon transcripts using their endonucleolytic (Slicer) cleavage activity (6–8).
Biogenesis of piRNAs is only beginning to be understood, but two biochemically distinct pathways can be discerned. Primary biogenesis describes conversion of long, single-stranded transcripts arising from genomic loci called piRNA clusters (up to 100 kilobases long) into 24- to 30-nt piRNAs that associate with MILI and MIWI (9). On the other hand, biogenesis of MIWI2-bound piRNAs is indirect, whereby MILI-mediated slicer cleavage of a target is proposed to initiate production of a new secondary piRNA (4, 5, 7). This arrangement allows germ cells to monitor activity of transposons and adaptively respond to it by guiding MIWI2 to their genomic loci. Events that follow the initial MILI cleavage of a target are unknown, partly because of an incomplete knowledge of the components involved. Here, we identify Tudor domain-containing 12 (TDRD12) in mouse Piwi complexes and demonstrate its in vivo role in secondary piRNA biogenesis and transposon silencing.
Results
TDRD12 Is a Component of the MILI Ribonucleoprotein Complex.
Piwi proteins are posttranslationally modified at their N termini by symmetrical dimethyl arginine (sDMA) marks which serve as ligands for tudor domains found in tudor domain-containing (TDRD) proteins (10). Previous proteomic studies of mouse Piwi complexes have identified almost every member of the TDRD family (6, 11, 12). Uniquely, TDRD12 (also called ES-cell associated transcript 8, ECAT8) (13) was never detected. TDRD12 is composed of a central helicase and two flanking tudor domains, along with a C-terminal CS domain (as present in CHORD-SGT1 proteins) (Fig. 1A). It is highly conserved and represented by a single gene in insects to human. The Drosophila genome on the other hand has three members: Yb, Brother of Yb (BoYb), and Sister of Yb (SoYb), all of which are implicated in transposon control. Additionally, Yb is shown to be essential for primary piRNA biogenesis in the ovarian somatic follicle cells (14, 15). We wished to examine the involvement of TDRD12 in the mouse piRNA pathway.
Fig. 1.
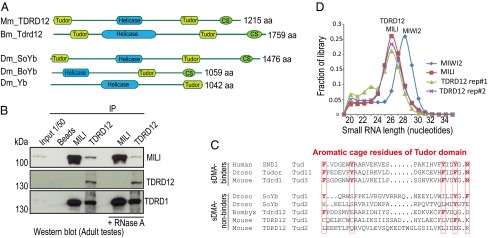
TDRD12 associates with mouse secondary piRNA biogenesis factors. (A) Domain organization of TDRD12 proteins in mouse (Mm), Drosophila (Dm), and Bombyx (Bm). (B) Immunoprecipitated (IP) complexes were probed by Western analysis for the indicated proteins. +RNaseA, treatment with RNaseA. (C) Sequence alignment of tudor domain from proteins demonstrated to bind symmetrical dimethyl arginine (sDMA), and those from TDRD12 proteins. Aromatic cage residues essential for binding sDMA are highlighted in red. (D) The read-length distribution in small RNA libraries from indicated proteins.
Like most members of the TDRD family, Tdrd12 expression is restricted to the mouse gonads, and its expression domain extends from embryonic to the adult stages in mouse testes (Fig. S1A). We examined its presence in purified MILI complexes isolated from adult testes (Fig. 1B). Consistent with previous Piwi proteomics data (11, 16), TDRD12 was not detected in the MILI immunoprecipitates whereas TDRD1, an established interacting partner of MILI, was present (Fig. 1B and Fig. S1 B and C). Nevertheless, direct immunoprecipitation of TDRD12 identified MILI and TDRD1 as components, suggesting that only a subpopulation of MILI is in complex with TDRD12, possibly explaining why it went undetected previously.
Interaction between MILI and TDRD1 is mediated via recognition of sDMA marks on MILI by the aromatic cage within tudor domains of TDRD1 (17). However, a sequence alignment of tudor domains from known sDMA binders and those from TDRD12 orthologs reveals that amino acid residues critical for constructing such an aromatic cage are absent in the TDRD12 proteins (Fig. 1C). This predicted inability to recognize modified arginine was confirmed by isothermal calorimetry (ITC) measurements (Fig. S1 D and E). Thus, direct interaction between MILI and TDRD12, if any, should be independent of the methylation status of MILI. Interestingly, RNase treatment reduced the recovery of MILI in TDRD12 complexes, indicating the importance of RNA for complex formation (Fig. 1B and Fig. S1C). The sensitivity of TDRD12-MILI complex towards RNaseA prompted us to ask whether MILI-bound small RNAs are present in the complex. We prepared two independent deep-sequencing libraries of small RNAs present in TDRD12 complexes from embryonic day 18.5 (E18.5) testes, where MILI and MIWI2 are coexpressed. The TDRD12 reads were found to have the same length profile (∼26 nt) as MILI-bound piRNAs, but not that of the coexpressed MIWI2 (∼28 nt) (Fig. 1D). Based on further analyses of nucleotide preferences (Fig. S2A), and mappings to transposon consensus and embryonic clusters, the small RNAs in TDRD12 complexes can be identified as those that normally guide MILI in fetal germ cells. Presence of TDRD1 was not affected by the RNase treatment (Fig. 1B), indicating its recruitment via protein–protein interactions with an unknown partner within the complex, as TDRD1 and TDRD12 did not interact directly (Fig. S2B). Taken together, TDRD12 exists in a biochemical complex containing TDRD1 and the MILI piRNP, linking it to the mouse piRNA pathway.
Roles of Tdrd12 Domains.
To gain further insight into the function of TDRD12, we wished to examine the importance of its multiple domains in vivo. As mammalian cell-culture models suitable for piRNA studies are unavailable, we used the Bombyx mori BmN4 insect cell-culture system (18). BmN4 cells express two Piwi proteins, Siwi and Ago3, which display all known features of the piRNA pathway found in the Drosophila germ line. Important for our study, Siwi binds primary piRNAs whereas Ago3 accumulates secondary piRNAs, very much analogous to mouse Piwi proteins MILI and MIWI2, respectively.
We mined a polyA+ trascriptome library to identify Bombyx mori Tdrd12 (BmTdrd12) as the sole Tdrd12 representative in BmN4 cells (Fig. 2A and Experimental Procedures). Using specific antibodies to the endogenous BmTdrd12 (Fig. S3A), we demonstrate its presence in a complex with endogenous Siwi, but not Ago3 (Fig. 2B). The complex formation between Siwi and BmTdrd12 is further confirmed with tagged Piwi proteins (Fig. 2C). Deep sequencing of small RNAs in HA-BmTdrd12 and endogenous BmTdrd12 complexes supports the biochemical association data by identifying small RNAs that share sequence features (Fig. 2D) and transposon-mapping characteristics that are similar to Siwi-bound piRNAs (Fig. S3B). Thus, similar to mouse TDRD12, the Bombyx ortholog is also in complex with the primary piRNA-bound Piwi protein (MILI in mouse testes or Siwi in BmN4), validating BmN4 cells as a useful system for molecular dissections. Using deletion versions (Fig. 2A), we map the interaction domain for Siwi to the second tudor domain of HA-BmTdrd12 (Fig. 2E).
Fig. 2.
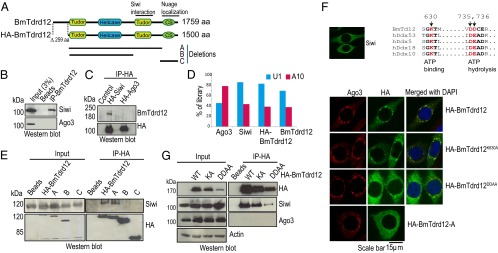
Roles of Tdrd12 domains in Piwi interaction and nuage localization. (A) Domain organization of Bombyx Tdrd12 (BmTdrd12). An HA-tagged version lacking the N-terminal region (259 aa) was used for cell-culture studies (Experimental Procedures). Other deletion versions used in this study are indicated. (B) Immunoprecipitated (IP) endogenous BmTdrd12 associates with endogenous Piwi protein Siwi. (C) HA-tagged Siwi, but not HA-Ago3, associates with endogenous BmTdrd12. (D) The nucleotide bias for a uridine at first position (1U) or adenosine at 10th position (A10) of reads in small RNA libraries is shown. (E) Analysis of HA-BmTdrd12 deletion versions for association with endogenous Siwi. (F) Subcellular localization of HA-BmTdrd12 and its mutants. Alignment of helicase domains from indicated proteins showing critical residues required for ATP binding or hydrolysis. Residues mutated in HA-BmTdrd12 are highlighted in red. (G) Helicase domain mutations in HA-BmTdrd12 do not affect association with endogenous Siwi.
Piwi proteins display distinct localization patterns in BmN4 cells. Siwi is distributed throughout the cytoplasm whereas Ago3 is sequestered in perinuclear cytoplasmic granules called nuage (19). Unexpectedly, although HA-BmTdrd12 interacts with Siwi, it is found colocalized with Ago3 in the nuage (Fig. 2F). Deletion of the CS domain (construct A) abolished this localization (Fig. 2F). All other deletion constructs (constructs B and C) were also found to be diffused in the cytoplasm. Next, to examine the importance of the helicase domain, we introduced mutations (20) to perturb ATP binding (K→A) or hydrolysis (DD→AA). Mutation of the ATPase motif resulted in a substantial displacement of the protein from the nuage whereas mutation of the ATP-binding motif did not (Fig. 2F). Importantly, all helicase point mutants (Fig. 2G) and the deletion version lacking the CS domain (Fig. 2E) showed interaction with Siwi, indicating that their association can occur in the cytoplasm before nuage localization. In sum, these cell-culture experiments reveal contributions of BmTdrd12 domains to interaction with the Piwi protein and nuage localization.
Tdrd12 Is Essential for Spermatogenesis.
To examine the in vivo role of TDRD12, we first examined the repro23 point mutant which was generated by N-ethyl-N-nitrosourea (ENU)-induced mutagenesis. The repro 23 mutation was previously mapped to a region containing several genes, including Tdrd12 (21). In this study, we identify it as a nonsense mutation in exon 8 of the Tdrd12 gene, truncating the coding sequence (Fig. 3A). The homozygous repro23 mutant mice display male infertility (21). The male specific infertility of repro23 mutant mice suggests a potential role for TDRD12 in mouse spermatogenesis. To directly verify its role, we engineered a targeted disruption of the mouse Tdrd12 locus (Fig. S4 A–C). Animals of all genotypes are viable and females are fertile, but homozygous Tdrd12 mutant males displayed infertility and had atrophied testes (Fig. 3B). Histological examination of Tdrd12 mutant testes reveals seminiferous tubules that are narrow with large vacuolated spaces, devoid of late-stage germ cells. In contrast, the testes from heterozygous littermates contain germ cells of all developmental stages: meiotic spermatocytes, postmeiotic round spermatids, and elongating spermatids (Fig. 3C). The germ-cell defect in the mutant is apparent early in development, and spermatocytes fail to proceed beyond the prepachytene stage (Fig. S4D). The XY body, a γ-H2AX-marked silent chromatin domain containing unpaired X and Y chromosomes in late zygotene to pachytene spermatocytes, is absent in the mutant (Fig. S4E). Finally, postmeiotic round spermatids and their gene products are also not detected (Fig. S4F). Thus, germ-cell development falters during meiosis in the zygotene–pachytene transition, and the cells go into apoptosis, as indicated by the high number of TUNEL-positive cells in P20 and adult mutant testes (Fig. S4G). These results illustrate an essential role of Tdrd12 for normal spermatogenesis, a hallmark of all mouse piRNA pathway mutants.
Fig. 3.
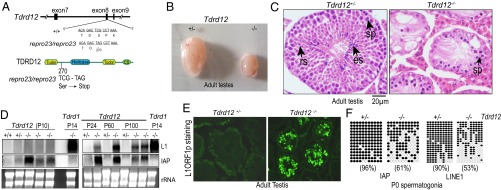
Tdrd12 mutant male mice are infertile and display derepression of retrotransposons. (A) The nonsense C-to-A nucleotide substitution in exon 8 of Tdrd12 in repro23 mice leads to truncation of the coding sequence. (B) Atrophied testes of homozygous (−/−) Tdrd12 mutants compared with their heterozygous (+/−) littermates. (C) Hematoxylin/eosin staining of adult testes sections from Tdrd12 animals. es, elongating spermatids; rs, round spermatids; sp, spermatocytes. (Scale bar: 20 µm.) (D) Northern analysis for LINE1 (L1) and IAP retrotransposons in Tdrd12 mutants. Only L1 elements are activated in Tdrd1 mutants. Loading control is provided by ethidium bromide staining of ribosomal RNA (rRNA). (E) Immunofluorescence detection of L1ORF1p in adult Tdrd12 mutant testis. (F) Promoter CpG DNA methylation (indicated as filled circles) on transposon promoters quantified (in percentages) by bisulfite sequencing.
Retrotransposons Are Derepressed in the Tdrd12 Mutants.
The germ-cell defect in piRNA pathway mutants is believed to be a consequence of deregulated transposon control in the germ line. Consistently, derepression of LTR intracisternal A particle (IAP) elements is already observed in 10-d-old (P10) mutant animals (Fig. 3D), at a time point when the germ-cell defects are not histologically discernible (Fig. S4D). Non-LTR LINE1 (L1) activation was delayed, with robust levels of L1 mRNA detected only in P24 testes, but the levels of both transposons remain high in aged (P60 and P100) animals (Fig. 3D). L1 activation was further confirmed in adult mutants by immunofluorescence detection of L1ORF1p (22), a protein product from active L1 elements (Fig. 3E). Furthermore, we also confirm derepression of retrotransposons in the repro23 point mutant (Fig. S4H).
Mammalian genomes use DNA methylation as a mechanism to silence transposons at the level of transcription (23). In male mice, DNA methylation takes place during a developmental window immediately before birth, when DNA methylation marks are laid down de novo. We examined the DNA methylation status by bisulfite sequencing using genomic DNA from purified spermatogonia. In Tdrd12 mutants, promoter methylation on CpG dinucleotides was reduced for both L1 (53%) and IAP (61%) elements compared with their heterozygous littermates (∼90%), suggesting a defect in their de novo methylation (Fig. 3F). Although repeat elements are the major beneficiaries of de novo DNA methylation, imprinted genomic loci also receive silencing marks during this period. Examination of methylation on three imprinted loci in Tdrd12 mutants revealed no changes (Fig. S5A), underscoring the specificity of the piRNA pathway in targeting repeat elements in the fetal male germ line. Thus, TDRD12 is essential for germ cells in maintaining control over endogenous retrotransposons.
TDRD12 Is Required for Biogenesis of MIWI2 piRNAs.
Deposition of DNA methylation marks on repeat elements is linked to the nuclear arm of the piRNA pathway, a key player of which is the nuclear Piwi protein MIWI2. Functionality of MIWI2 is dependent on production and incorporation of its small RNA guides via secondary processing (4). To examine the integrity of piRNA biogenesis, we purified total small RNAs (14-40 nt) from P0 (newborn) testes and subjected them to deep sequencing. As expected for mouse testes small RNA libraries, the read-length distribution profiles reveal two major peaks expected for miRNAs (19-22 nt) and piRNAs (24-30 nt) in both the Tdrd12 mutant and heterozygous littermates, with overall level of piRNAs slightly reduced in the mutant (Fig. 4A). Examined over IAP consensus sequence, much of this reduction is accounted for by antisense-oriented reads (Fig. 4B). The reduction in antisense-oriented reads is further strengthened by the observation that ratio of antisense to sense piRNAs over both LINE1 and IAP was reduced in the Tdrd12 mutant (Fig. 4C). Because MILI- and MIWI2-bound RNAs contribute to total small RNA populations in P0 testes, we directly determined the impact on these individual populations by immunoprecipitations. In the control testes, both MILI and MIWI2 are present in complex with distinctly sized small RNAs (Fig. 4D). In contrast, only MILI was found to associate with piRNAs in the Tdrd12 mutant (three independent experiments), suggesting a defect in biogenesis of piRNAs that normally associate with MIWI2 (Fig. 4D and Fig. S5B). Because loading of MIWI2 with piRNAs is a prerequisite for its nuclear import, unloaded MIWI2 remains stranded in the cytoplasm of E17.5 germ cells (gonocytes) in the Tdrd12 mutant (Fig. 4E).
Fig. 4.
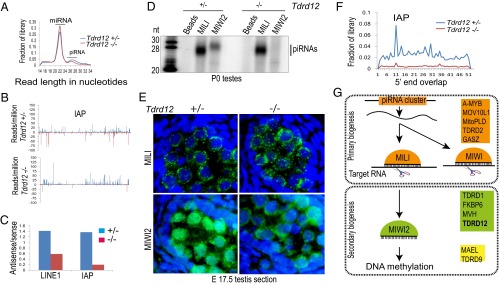
TDRD12 is required for biogenesis of MIWI2-bound secondary piRNAs. (A) Read-length distribution profile of testes total small RNAs from indicated animals. (B) Normalized (to microRNAs) density of reads mapping to IAP transposon consensus from P0 total small RNA libraries. Note the reduction of antisense reads in the Tdrd12 mutant. (C) Ratio of antisense/sense piRNAs mapping LINE1 or IAP consensus. (D) Association of piRNAs with Piwi proteins examined by immunoprecipitation and 5′-end labeling from newborn pups (P0). RNA markers (nucleotides, nt) are shown. (E) Immunofluorescence detection of proteins in embryonic testes from indicated Tdrd12 genotypes. Images (green) are shown along with DAPI staining for DNA. (Scale: Width of each panel is 70 μm.) Note the loss of nuclear MIWI2 staining in Tdrd12 mutants. (F) Plot showing 5′-end overlap of opposing reads on IAP consensus in total small RNA libraries. Note the reduction in signal at position 10 (Ping-Pong signature) in the Tdrd12 mutant. (G) Current knowledge on factors implicated in the mouse piRNA pathway, and placement of TDRD12 as a secondary piRNA biogenesis factor.
Primary piRNAs associating with MILI are proposed to guide its slicer activity for biogenesis of MIWI2 piRNAs (4, 7). An outcome of this pathway is the detected 10-nt overlap between 5′ ends of MILI- and MIWI2-bound piRNAs (4). A similar signature is also evident in total small RNA populations from the Tdrd12 hetrozygous animals but, consistent with the loss of MIWI2-bound piRNAs, is absent in the Tdrd12 mutants (Fig. 4F). As shown in Fig. 4D, biogenesis of piRNAs associating with MILI appeared normal in the lack of Tdrd12 (Fig. 4D). To examine piRNAs at the molecular level, two independent libraries of MILI-bound piRNAs were prepared from Tdrd12 mutant or control P0 animals and subjected to deep sequencing. Analysis indicates unchanged genome annotation profiles (Fig. S5C) and signature 1U-bias of primary piRNAs in the mutant (Fig. S5D). The reads distribution on transposon consensus sequences and piRNA clusters is also not affected (Fig. S5 E and F). Furthermore, immunoprecipitations confirm that primary biogenesis continues to supply piRNAs for MILI in postnatal (P10 and P14) Tdrd12 mutants (Fig. S5G). These analyses indicate that primary piRNA-loaded MILI remains competent to initiate secondary biogenesis but that, in the absence of TDRD12, downstream events fail.
Factors implicated in piRNA biogenesis occupy distinct cytoplasmic granules in germ cells called nuages. In mouse fetal germ cells, they appear as electron-dense structures between mitochondria, the so-called intermitochondrial cement (24, 25). Our repeated efforts to localize TDRD12 failed, possibly due to the unsuitability of our antibodies for immunofluorescence or due to the low abundance of the protein. Nevertheless, we show that subcellular localization of MILI and TDRD1 are not affected in the Tdrd12 mutant (Fig. 4E and Fig. S4I). Furthermore, electron micrographs also reveal the normal presence of the intermitochondrial cement (in three biological replicates), suggesting the absence of any defect in granule assemblies in the mutant germ cells (Fig. S5H). In the light of the above data, we propose a direct role for TDRD12 in the machinery that generates the secondary piRNAs associating with MIWI2.
Discussion
Biogenesis of piRNAs is only beginning to be understood, and progress has to be made on two fronts: identification of the complete set of factors involved and molecular mechanisms used by them. Here, we identified TDRD12 as a unique mouse piRNA biogenesis factor. The ability to produce piRNAs rests with a few hundred genomic regions called piRNA clusters, some of which are up to 100 kilobases in length. The piRNA clusters are transcribed by RNA polymerase II to give rise to long, single-stranded transcripts that are 5′-capped and polyadenylated, with some undergoing splicing, too (9). At least for a subset of these clusters (those producing intergenic piRNAs), the transcription factor A-MYB is shown to directly recognize upstream DNA elements to initiate transcription (9) (Fig. 4G). The transcripts are then exported to the cytoplasm where they undergo primary processing that is likely to include fragmentation by unknown nuclease(s) followed by loading of their 5′ ends into the Piwi proteins (MILI or MIWI). The protruding 3′ ends of such piRNA intermediates are then trimmed by an exonuclease (tentatively termed as Trimmer) (26) to the mature size as determined by the footprint of the Piwi protein. Finally, the 3′ end is modified by the 2′-O-methyltransferase HEN1 to complete the process, resulting in primary piRNAs with a predominant preference for a 5′ uridine (1U-bias). Currently, the putative RNA helicase MOV10L1 (27, 28) and the single-stranded endonuclease MitoPLD (29–31) are implicated in the early steps, but the exact substrates and products of these factors are not known. TDRD2 (32) is shown to be essential for recruitment of the Trimmer, as piRNAs with extended 3′ ends (and properly methylated) accumulate in MILI in Tdrd2 mutant mice. GASZ (33) is a structural component of perinuclear cytoplasmic granules variably called nuage or intermitochondrial cement where most piRNA pathway factors accumulate. Loss of Gasz results in reduced piRNA levels and destabilization of the (unloaded) MILI. Primary piRNA-loaded MILI and MIWI slice target transcripts to ensure transposon silencing.
In fetal mouse germ cells, primary piRNA-guided MILI endonuclease action on a target is proposed to create the 5′ end of new secondary piRNA entering MIWI2 (4, 7). Piwi slicer activity is mechanistically similar to that mediated by Ago proteins in the RNA-induced silencing complex (RISC) (6, 34). However, unlike in the RISC where both fragments are degraded, one of the MILI-generated cleavage fragments is believed to mature as a new secondary piRNA (4, 7). How the cleavage fragment is safely transferred to MIWI2 is not known. Additional factors that are essential for secondary biogenesis include the RNA helicase MVH (35), Hsp90 cochaperone Fkbp6 (19), and TDRD1 (11, 16). It is possible that RNA helicases like MVH and TDRD12 might facilitate RNP remodeling required for inter-Piwi exchange of piRNA intermediates.
Our biochemical studies indicate that TDRD12 functions together with other secondary biogenesis factors like the MILI piRNP and TDRD1 (Fig. 1). Nevertheless, we cannot rule out the presence of unloaded MIWI2 within this complex. Within this complex, the tudor domains of TDRD12 are likely to engage MILI in a methylation-independent manner (Fig. 1C). Although a direct comparison of the mouse and Bombyx systems is not possible, the second tudor domain of mouse TDRD12 could be mediating the interaction with MILI. Furthermore, sensitivity of MILI-TDRD12 association to RNase treatment (Fig. 1B) raises the possibility that RNAs present in the complex could be targets of MILI-bound primary piRNAs, and thus substrates for secondary biogenesis. Finally, our studies with the insect ortholog of mouse TDRD12 revealed a role for the CS domain in nuage localization. The CS domain is a reported protein–protein interaction domain, which some cochaperones use to recruit the molecular chaperones HSP90 and HSP70 (36, 37). We explored this experimentally, but our experiments do not support such a role for the CS domain from TDRD12 (Fig. S2 C and D). It is possible that interaction with unknown nuage components might allow its retention in the granules. Future live-cell imaging and structural studies will be important in uncovering the exact molecular function of this multidomain protein in piRNA biogenesis.
Experimental Procedures
The cDNA for mouse TDRD12 (Uniprot accession no. Q9CWU0) was amplified from adult mouse testes and inserted into a mammalian expression vector. The cDNA for Bombyx mori Tdrd12 (BmTdrd12) was amplified from BmN4 cell total RNA and cloned into a vector suitable for expression in BmN4 cells. Tdrd12 mice were maintained in C57BL/6;129S4/SvJae mixed genetic background. Detailed information on materials and methods is provided in SI Experimental Procedures. Illumina deep sequencing data are deposited with Gene Expression Omnibus (GEO) (GSE50544), and the Tdrd12 mutant mouse is available from the RIKEN Bio Resource Center (accession no. RBRC02326). All small RNA libraries and primers used in this study are listed in Table S1 and S2, respectively.
Supplementary Material
Acknowledgments
We acknowledge the kind gift of antibody from Sandra Martin (L1ORF1p). We thank Anna Adamiok, Charlotta Funaya, Andres Palencia, and Nikos Mathioudakis for assistance with experiments, and Jafar Sharif and Kohzoh Mitsuya for bisulfite data. S.C. is grateful to Norio Nakatsuji and is supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan. T.K. is funded by the Japan Society for the Promotion of Science. R.R.P. is supported by European Molecular Biology Laboratory (EMBL) Interdisciplinary Postdoctoral and European Molecular Biology Organization Long-Term Fellowships. This work was funded by a European Research Council Starting Grant (pisilence) from the European Union (to R.S.P.). The R.S.P. laboratory is supported by the EMBL. We also thank EMBL Genomics, and Protein Expression Core Facilities for assistance.
Footnotes
Conflict of interest statement: S.Y. is a member without salary of the scientific advisory boards of iPierian, iPS Academia Japan, Megakaryon Corporation, and Retina Institute Japan.
Data deposition: The Illumina deep sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE50544).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316316110/-/DCSupplemental.
References
- 1.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136(4):656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12(4):246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 4.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31(6):785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22(7):908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reuter M, et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480(7376):264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 7.De Fazio S, et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480(7376):259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- 8.Di Giacomo M, et al. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell. 2013;50(4):601–608. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 9.Li XZ, et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell. 2013;50(1):67–81. doi: 10.1016/j.molcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010;24(7):636–646. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vagin VV, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23(15):1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C, et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci USA. 2009;106(48):20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 14.Handler D, et al. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011;30(19):3977–3993. doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito K, et al. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24(22):2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reuter M, et al. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16(6):639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 17.Mathioudakis N, et al. The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors. RNA. 2012;18(11):2056–2072. doi: 10.1261/rna.034181.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaoka S, et al. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA. 2009;15(7):1258–1264. doi: 10.1261/rna.1452209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiol J, et al. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell. 2012;47(6):970–979. doi: 10.1016/j.molcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 20.Linder P, Jankowsky E. From unwinding to clamping: The DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12(8):505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 21.Asano Y, et al. Characterization and linkage mapping of an ENU-induced mutant mouse with defective spermatogenesis. Exp Anim. 2009;58(5):525–532. doi: 10.1538/expanim.58.525. [DOI] [PubMed] [Google Scholar]
- 22.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: Implications for transposition. Mol Cell Biol. 1994;14(4):2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 24.Aravin AA, et al. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5(12):e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: Germinal granules in mammals. Mol Cell Endocrinol. 2009;306(1-2):17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011;43(6):1015–1022. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 27.Zheng K, et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci USA. 2010;107(26):11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frost RJ, et al. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci USA. 2010;107(26):11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe T, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011;20(3):364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491(7423):279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimasu H, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491(7423):284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 32.Saxe JP, Chen M, Zhao H, Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32(13):1869–1885. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5(9):e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006):343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 35.Kuramochi-Miyagawa S, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24(9):887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchner J, Weikl T, Bügl H, Pirkl F, Bose S. Purification of Hsp90 partner proteins Hop/p60, p23, and FKBP52. Methods Enzymol. 1998;290:418–429. doi: 10.1016/s0076-6879(98)90035-0. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi A, Casais C, Ichimura K, Shirasu K. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA. 2003;100(20):11777–11782. doi: 10.1073/pnas.2033934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


