Significance
In a developing embryo, individual cells need to “know” where they are to do the right thing. How much do they know, and where is this knowledge written down? Here, we show that these questions can be made mathematically precise. In the fruit fly embryo, information about position is thought to be encoded by the concentration of particular protein molecules, and we measure this information, in bits. Just four different kinds of molecules are almost enough to specify the identity of every cell along the long axis of the embryo, and we argue that the way in which this information is distributed reflects an optimization principle, maximizing the information available from a limited number of molecules.
Keywords: gene regulatory networks, embryonic development, optimization
Abstract
Cells in a developing embryo have no direct way of “measuring” their physical position. Through a variety of processes, however, the expression levels of multiple genes come to be correlated with position, and these expression levels thus form a code for “positional information.” We show how to measure this information, in bits, using the gap genes in the Drosophila embryo as an example. Individual genes carry nearly two bits of information, twice as much as would be expected if the expression patterns consisted only of on/off domains separated by sharp boundaries. Taken together, four gap genes carry enough information to define a cell’s location with an error bar of  along the anterior/posterior axis of the embryo. This precision is nearly enough for each cell to have a unique identity, which is the maximum information the system can use, and is nearly constant along the length of the embryo. We argue that this constancy is a signature of optimality in the transmission of information from primary morphogen inputs to the output of the gap gene network.
along the anterior/posterior axis of the embryo. This precision is nearly enough for each cell to have a unique identity, which is the maximum information the system can use, and is nearly constant along the length of the embryo. We argue that this constancy is a signature of optimality in the transmission of information from primary morphogen inputs to the output of the gap gene network.
Building a complex, differentiated body requires that individual cells in the embryo make decisions, and ultimately adopt fates, that are appropriate to their position. There are wildly diverging models for how cells acquire this “positional information” (1), but there is general consensus that they encode positional information in the expression levels of various key genes. A classic example is provided by anterior/posterior patterning in the fruit fly, Drosophila melanogaster, where a small set of gap genes and then a larger set of pair rule and segment polarity genes are involved in the specification of the body plan (2). These genes have expression levels that vary systematically along the body axis, forming a blueprint for the segmented body of the developed larva that we can “read” within hours after the start of development (3).
Although there is consensus that particular genes carry positional information, less is known quantitatively about how much information is being represented by the expression levels in individual cells. Do the broad, smooth expression profiles of the gap genes, for example, provide enough information to specify the exact pattern of development, cell by cell, along the anterior/posterior axis? How much information does the whole embryo use in making this pattern? Answering these questions is important, in part, because we know that crucial molecules involved in the regulation of gene expression are present at low concentrations and even low absolute copy numbers, so that expression is noisy (4–10), and this noise must limit the transmission of information (11–14). Is it possible, as suggested theoretically (15–18), that the information transmitted through these regulatory networks is close to the physical limits set by the irreducible randomness of counting individual molecular events? To answer this and other questions, we need to measure positional information quantitatively, in bits. We do this here using the gap genes in Drosophila as an example.
There are many ways in which positional information could be represented during the process of development. Cells could make decisions based on the integration of signals over time or by comparing their internal states with those of their neighbors. Eventually, the internal state of each individual cell must carry enough information to specify that cell’s fate, but it is not clear at what point in development this happens. Thus, when we look at the gap genes during the 14th nuclear cycle after fertilization, there is no guarantee that their expression levels will carry all the information that cells eventually will acquire, either from maternal inputs or via communication with their neighbors. Because our experimental methods give us access to snapshots of gene expression levels, however, we will start by asking how much positional information is carried by local measurements in individual cells at a moment in time. These expression levels themselves reflect an integration of many inputs over space and time (9, 19), but these molecular mechanisms do not influence the definition or measurement of the information that the expression levels carry.
Quantifying Information
In the early stages of development, different cells have essentially the same morphology, at least in the bulk of the embryo, away from the poles. Thus, if we do not look at the expression levels of the relevant genes, we have no information about the position of the cell; it could be anywhere along the anterior/posterior axis of the embryo. Mathematically, this is equivalent to saying that, a priori, the position x of the cell is drawn from a distribution of possibilities,  . Once we observe the expression level g, we still do not know the precise position x of the cell, but our uncertainty is greatly reduced. In Fig. 1, we illustrate this idea using the gap gene hunchback (hb). Expression levels of hb vary systematically along the anterior/posterior axis of the Drosophila embryo, but these expression levels also are variable across cells in the same position, both within a single embryo and across multiple embryos. Thus, if we make a “slice” through the expression profile at some particular level g, we cannot point uniquely to the position x of the nucleus in which the Hb protein has that exact concentration. Instead, there is a range of positions that are consistent with the value of g, and we can summarize this range of possibilities by the conditional probability distribution,
. Once we observe the expression level g, we still do not know the precise position x of the cell, but our uncertainty is greatly reduced. In Fig. 1, we illustrate this idea using the gap gene hunchback (hb). Expression levels of hb vary systematically along the anterior/posterior axis of the Drosophila embryo, but these expression levels also are variable across cells in the same position, both within a single embryo and across multiple embryos. Thus, if we make a “slice” through the expression profile at some particular level g, we cannot point uniquely to the position x of the nucleus in which the Hb protein has that exact concentration. Instead, there is a range of positions that are consistent with the value of g, and we can summarize this range of possibilities by the conditional probability distribution,  , that a cell with expression level g will be found at position x. For all values of g that occur in the embryo, we see that this conditional distribution is narrower or more concentrated than the nearly uniform distribution
, that a cell with expression level g will be found at position x. For all values of g that occur in the embryo, we see that this conditional distribution is narrower or more concentrated than the nearly uniform distribution 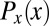 .
.
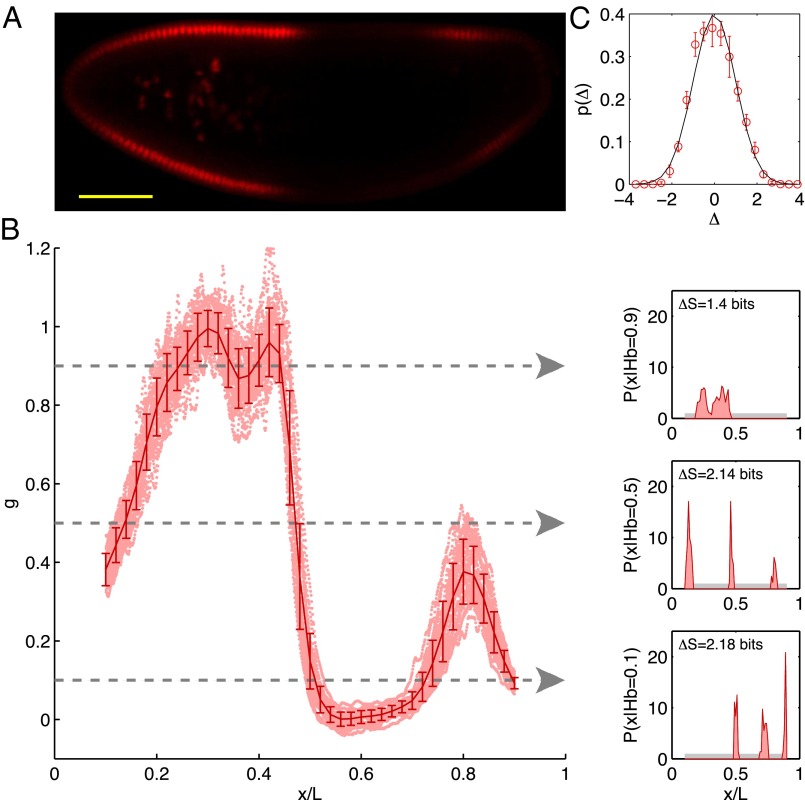
Fig. 1.
Positional information carried by the expression of Hb. (A) Optical section through the midsagittal plane of a Drosophila embryo with immunofluorescence staining against Hb protein. (Scale bar = 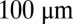 .) (B) Normalized dorsal profiles of fluorescence intensity, which we identify as Hb expression level g, from 24 embryos (light red dots) selected in a 38- to 48-min time interval after the beginning of nuclear cycle 14. Position x along the anterior/posterior axis is normalized by the length L of the embryo;
.) (B) Normalized dorsal profiles of fluorescence intensity, which we identify as Hb expression level g, from 24 embryos (light red dots) selected in a 38- to 48-min time interval after the beginning of nuclear cycle 14. Position x along the anterior/posterior axis is normalized by the length L of the embryo; 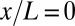 corresponds to the anterior end of the embryo, and
corresponds to the anterior end of the embryo, and 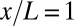 corresponds to the posterior end. Means
corresponds to the posterior end. Means 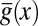 and SDs
and SDs 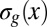 are plotted in darker red. Considering all points with g = 0.1, 0.5, or 0.9 (Left), yields the conditional distributions with probability densities
are plotted in darker red. Considering all points with g = 0.1, 0.5, or 0.9 (Left), yields the conditional distributions with probability densities 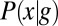 (Right). Note that these distributions are much more sharply concentrated than the uniform distribution
(Right). Note that these distributions are much more sharply concentrated than the uniform distribution 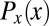 shown in light gray; correspondingly, the entropies
shown in light gray; correspondingly, the entropies 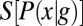 are very much smaller than the entropy
are very much smaller than the entropy 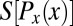 . For each g, we note the reduction of uncertainty in x by reading out g,
. For each g, we note the reduction of uncertainty in x by reading out g, 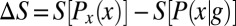 . (C) Variations in expression level around the mean at each position, estimated by the distribution of normalized relative expression, given by
. (C) Variations in expression level around the mean at each position, estimated by the distribution of normalized relative expression, given by 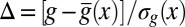 (red circles with SEMs). The solid line is a zero mean/unit variance Gaussian.
(red circles with SEMs). The solid line is a zero mean/unit variance Gaussian.
The probability distributions 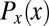 and
and 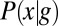 provide the ingredients we need to make a mathematically precise version of the qualitative statement that “the expression level g of a gene provides information about the position x of the cell.” Crucially, the foundational result of information theory is that there is only one way of doing this that is consistent with simple and plausible requirements, for example, that independent signals give additive information (20–22).
provide the ingredients we need to make a mathematically precise version of the qualitative statement that “the expression level g of a gene provides information about the position x of the cell.” Crucially, the foundational result of information theory is that there is only one way of doing this that is consistent with simple and plausible requirements, for example, that independent signals give additive information (20–22).
For any probability distribution, we can define an entropy S, which is the same quantity that appears in statistical mechanics and thermodynamics; for the two distributions here,
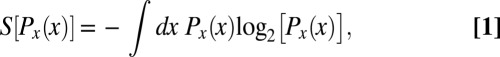 |
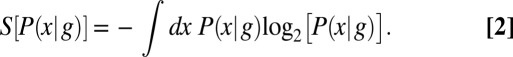 |
For example, if we measure x from 0 to L along the length of the embryo, then a uniform distribution of cells corresponds to  , and this has the maximum possible entropy
, and this has the maximum possible entropy 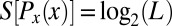 . The intuition that the conditional distribution
. The intuition that the conditional distribution 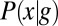 is narrower or more concentrated than
is narrower or more concentrated than 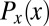 is quantified by the fact that the entropy
is quantified by the fact that the entropy  is smaller than
is smaller than 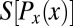 , and this reduction in entropy is exactly the information that observing g provides about x, measured here in bits. As an example, if observing the expression level g tells us, with complete certainty, that the cell is located in a small region of size
, and this reduction in entropy is exactly the information that observing g provides about x, measured here in bits. As an example, if observing the expression level g tells us, with complete certainty, that the cell is located in a small region of size  , then the gain in information is
, then the gain in information is 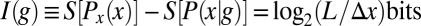 . Notice that entropies of continuous variables, such as position, depend on our choice of units, while the information, being the difference of entropies, is independent of this choice (22).
. Notice that entropies of continuous variables, such as position, depend on our choice of units, while the information, being the difference of entropies, is independent of this choice (22).
If we look at one cell and observe expression level g, then we gain information
However, when we choose a cell at random, we will see an expression level g drawn from the distribution 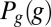 . The average information that this expression level provides about position is then
. The average information that this expression level provides about position is then
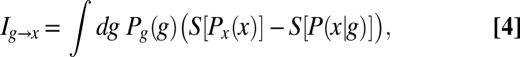 |
 |
where 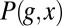 is the joint probability of observing a cell at x with expression level g, and we have rearranged the terms to emphasize the symmetry: Information that the expression level provides about the position of the cell is, on average, the same as the information that the position of the cell provides about the expression level,
is the joint probability of observing a cell at x with expression level g, and we have rearranged the terms to emphasize the symmetry: Information that the expression level provides about the position of the cell is, on average, the same as the information that the position of the cell provides about the expression level, 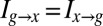 . This average information is called the mutual information between g and x. Again, we emphasize that this measure of information is not one among many equally good possibilities; rather, it is unique (20).
. This average information is called the mutual information between g and x. Again, we emphasize that this measure of information is not one among many equally good possibilities; rather, it is unique (20).
Because information is mutual, we can also write  in terms of the distribution of expression levels g that we find in cells at a particular position,
in terms of the distribution of expression levels g that we find in cells at a particular position, 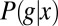 ,
,
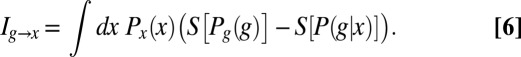 |
This emphasizes that the amount of information that can be conveyed is limited both by the overall dynamic range of expression levels, which determines 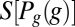 , and by the variability or noise in expression levels at a fixed position, which is measured by
, and by the variability or noise in expression levels at a fixed position, which is measured by 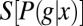 . It will be useful that the distribution of expression levels at each point,
. It will be useful that the distribution of expression levels at each point, 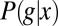 , is approximately Gaussian, as shown in Fig. 1C.
, is approximately Gaussian, as shown in Fig. 1C.
In what follows, we will use Eq. 6 to make a “direct” measurement of information, whereas Eq. 4 invites us to try and “decode” the information carried by the expression levels to recover estimates of the position x of each cell. Each approach has a natural generalization to the case where information is conveyed not by the expression level of one gene but by the combined expression levels of multiple genes 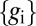 , and we will explore this as well. It is important to emphasize that the number of bits of information carried by the gene expression levels has meaning independent of the mechanisms by which this coding is established. Thus, at one extreme, it could be that each cell sets its expression levels independently in response to some primary morphogen [e.g., Bicoid in the Drosophila embryo (23–25)] whereas at the other extreme, the spatial patterns of expression could arise entirely from communication between neighboring cells, in a Turing-like mechanism (26, 27). In these different extremes, the precise value of the positional information places different quantitative constraints on the underlying mechanisms; however, in all cases, the number of available bits tells us about the reliability and complexity of the pattern that can be constructed from the local expression levels alone.
, and we will explore this as well. It is important to emphasize that the number of bits of information carried by the gene expression levels has meaning independent of the mechanisms by which this coding is established. Thus, at one extreme, it could be that each cell sets its expression levels independently in response to some primary morphogen [e.g., Bicoid in the Drosophila embryo (23–25)] whereas at the other extreme, the spatial patterns of expression could arise entirely from communication between neighboring cells, in a Turing-like mechanism (26, 27). In these different extremes, the precise value of the positional information places different quantitative constraints on the underlying mechanisms; however, in all cases, the number of available bits tells us about the reliability and complexity of the pattern that can be constructed from the local expression levels alone.
Information Carried by Single Gap Genes
Estimating the mutual information that one gene expression level provides about position requires, from Eq. 6, that we obtain a good estimate of the conditional distribution 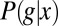 . Using immunofluorescent staining, we can measure g vs. x along the anterior/posterior axis of single Drosophila embryos, and by making such measurements on multiple embryos, as shown in Fig. 1, we obtain many samples of the expression level at corresponding positions; from these samples, we can then build up an estimate of the distribution
. Using immunofluorescent staining, we can measure g vs. x along the anterior/posterior axis of single Drosophila embryos, and by making such measurements on multiple embryos, as shown in Fig. 1, we obtain many samples of the expression level at corresponding positions; from these samples, we can then build up an estimate of the distribution  . Armed with this estimate, we can use Eq. 6 to compute the positional information. To be sure that the answer is meaningful, we have to address a number of technical issues (28).
. Armed with this estimate, we can use Eq. 6 to compute the positional information. To be sure that the answer is meaningful, we have to address a number of technical issues (28).
First, as explained at the outset, we would like to measure the information carried by a snapshot of the expression levels, so we need to make measurements on embryos at a well-defined time, and we use the length of the cellularization membrane as a precisely calibrated proxy for time (29–32). We choose this time to be the window from 38 to 48 min after the start of nuclear cycle 14, because we have seen that gap gene expression levels are at a plateau in this window. We also confine our attention to the central 80% of the anterior/posterior axis, because quantitative imaging at the poles is more difficult and because there are additional genes associated specifically with terminal patterning, and we make measurements along the dorsal edge of the midsagittal plane.
Second, Fig. 1 shows that the SD of expression levels typically is less than 10% of the maximum expression level. To draw convincing quantitative conclusions, then, we must be sure that our measurements have accuracy much better than this, lest we confuse experimental error for real noise and variability in the system. As discussed by Dubuis et al. (28), the intensity of immunostaining is linear in protein concentration over the relevant dynamic range (also ref. 9), and errors can be minimized by careful attention to the orientation and age of the embryos. By comparing large numbers of embryos stained in a single batch, we find that there is little or no sign of errors due to variations in the efficiency of staining, which means we can avoid previously troubling issues surrounding the normalization of profiles across embryos (details are provided in Materials and Methods). When the dust settles, our experimental or measurement errors are below 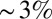 of the maximal expression level, and hence well below the observed noise levels (28). Note that measurement errors will always reduce the information, and so our estimate defines lower bounds on the information carried by the real biological signals.
of the maximal expression level, and hence well below the observed noise levels (28). Note that measurement errors will always reduce the information, and so our estimate defines lower bounds on the information carried by the real biological signals.
Finally, as has been addressed in other contexts (Materials and Methods), care is required to be sure that the finite number of samples we collect is sufficient to get a reliable estimate of 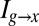 ; however, once we have control over the potential systematic errors, the statistical errors in our measurements are very small. Analysis of the data in Fig. 1 shows that the expression level of Hb provides
; however, once we have control over the potential systematic errors, the statistical errors in our measurements are very small. Analysis of the data in Fig. 1 shows that the expression level of Hb provides 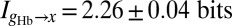 of information about the position of a cell along the middle 80% of the anterior/posterior axis. We can repeat this analysis for the gap genes krüppel (Kr), giant (Gt), and knirps (Kni), in addition to Hb, and we find
of information about the position of a cell along the middle 80% of the anterior/posterior axis. We can repeat this analysis for the gap genes krüppel (Kr), giant (Gt), and knirps (Kni), in addition to Hb, and we find 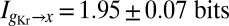 ,
, 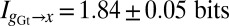 , and
, and 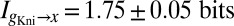 .
.
In all cases, the expression of a single gene carries much more than one bit of information; indeed, it carries more nearly two bits. The conventional view of the gap genes is that they are characterized by domains of expression, with boundaries, and the sharpness of the boundary often is taken as a measure of precision. However, if the patterns of expression were perfect on/off domains with infinitely sharp boundaries, then the expression level could provide at most one bit of information about position. Our result that gap genes provide nearly two bits of information about position demonstrates that intermediate expression levels are sufficiently reproducible from embryo to embryo that they carry significant amounts of positional information, and that the view of domains and boundaries misses almost half of this information.
How Much Information Does the Embryo Use?
At best, every nucleus could be labeled with a unique identity, so that with N nuclei, the embryo could make use of  bits (21). Along the anterior/posterior axis, we can count nuclei in a single midsagittal slice through the embryo, and in the middle
bits (21). Along the anterior/posterior axis, we can count nuclei in a single midsagittal slice through the embryo, and in the middle 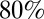 of the embryo, where the images are clearest, we have
of the embryo, where the images are clearest, we have 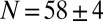 along the dorsal side and
along the dorsal side and 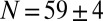 along the ventral side, where the error bars represent SDs across a population of 57 embryos in nuclear cycle 14; this corresponds to
along the ventral side, where the error bars represent SDs across a population of 57 embryos in nuclear cycle 14; this corresponds to 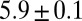 bits of information. However, do individual cells, in fact, “know” their identity? More precisely, are the elements of the anterior/posterior pattern specified with single-cell resolution?
bits of information. However, do individual cells, in fact, “know” their identity? More precisely, are the elements of the anterior/posterior pattern specified with single-cell resolution?
Several experiments suggest that elements of the body plan in the larval fly that emerges from the embryo can be traced to identifiable rows of cells along the anterior/posterior axis (33), which is consistent with the idea that at least some single rows of cells have a reproducible identity. Quantitatively, we can ask about the reproducibility of various pattern elements in early development, elements that appear not long after the expression patterns of the gap genes are established. A classic case is the cephalic furrow, which can be observed in live embryos and is known to have a position along the anterior/posterior axis that is reproducible with an accuracy of 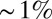 of the embryo length (34).
of the embryo length (34).
Is the cephalic furrow special, or can the embryo more generally position pattern elements with 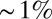 accuracy? The striped patterns of pair rule gene expression allow us to ask about the position of multiple pattern elements, seven peaks and six troughs of expression along the anterior/posterior axis. As shown in Fig. 2, all these elements have positions that are reproducible to within
accuracy? The striped patterns of pair rule gene expression allow us to ask about the position of multiple pattern elements, seven peaks and six troughs of expression along the anterior/posterior axis. As shown in Fig. 2, all these elements have positions that are reproducible to within 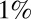 of the embryo length. This strongly suggests that all cells know their position along the anterior/posterior axis with a precision
of the embryo length. This strongly suggests that all cells know their position along the anterior/posterior axis with a precision 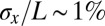 .
.
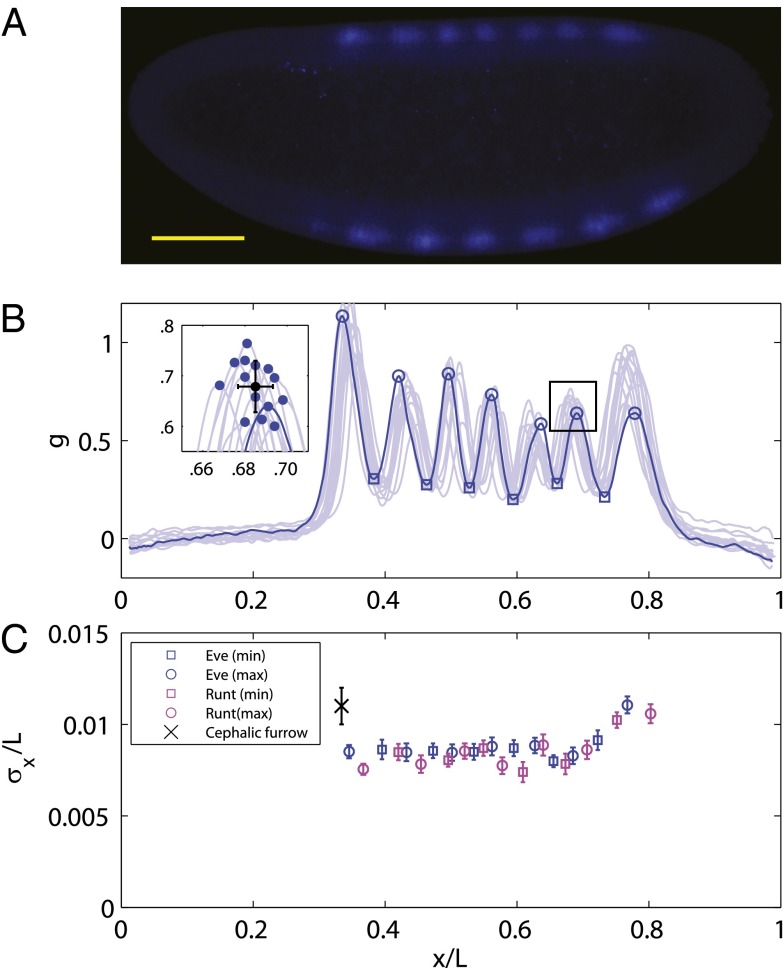
Fig. 2.
Reproducibility of multiple pattern elements along the anterior/posterior axis. (A) Optical section through the midsagittal plane of a Drosophila embryo with immunofluorescence staining against even-skipped (Eve) protein. (Scale bar =  .) (B) Normalized dorsal profiles of fluorescence intensity from 12 embryos selected in a 45- to 55-min time window after the beginning of nuclear cycle 14 (light blue lines); the dorsal profile of the top panel embryo is shown in darker blue. (Inset) Zooming in on a single peak, we can measure the SD of both the expression level and position of this element in the pattern. (C) Summary of results from such measurements on Eve (blue) and Runt (magenta), plotting the SD of the position
.) (B) Normalized dorsal profiles of fluorescence intensity from 12 embryos selected in a 45- to 55-min time window after the beginning of nuclear cycle 14 (light blue lines); the dorsal profile of the top panel embryo is shown in darker blue. (Inset) Zooming in on a single peak, we can measure the SD of both the expression level and position of this element in the pattern. (C) Summary of results from such measurements on Eve (blue) and Runt (magenta), plotting the SD of the position 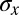 as a function of the mean position
as a function of the mean position 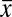 , together with a similar measurement on the reproducibility of the cephalic furrow (33). Note that all the elements are positioned with 1% accuracy or better.
, together with a similar measurement on the reproducibility of the cephalic furrow (33). Note that all the elements are positioned with 1% accuracy or better.
The distance between neighboring nuclei is 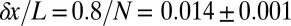 of the embryo’s length. If cells know their position with
of the embryo’s length. If cells know their position with  accuracy, this error is smaller than the internuclear spacing, suggesting that every cell indeed has a specified position. However, this is not quite right, because errors are probabilistic and probability distributions have tails. Specifically, if the best we (or the cells) can do is to specify positions with an error that has an SD of
accuracy, this error is smaller than the internuclear spacing, suggesting that every cell indeed has a specified position. However, this is not quite right, because errors are probabilistic and probability distributions have tails. Specifically, if the best we (or the cells) can do is to specify positions with an error that has an SD of 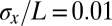 , and the errors come from a Gaussian distribution, then there is a probability
, and the errors come from a Gaussian distribution, then there is a probability 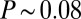 that we will be off by
that we will be off by 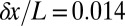 or more in one direction. This confusion means that the reproducibility of pattern elements in Fig. 2 provides evidence for individual nuclei having access to
or more in one direction. This confusion means that the reproducibility of pattern elements in Fig. 2 provides evidence for individual nuclei having access to 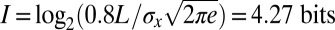 of information (22), although more may be available, as discussed below.
of information (22), although more may be available, as discussed below.
Decoding the Information Carried by Multiple Genes
Do the four gap genes, taken together, carry enough information to specify position with 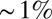 accuracy? To answer this question, we need to know not just the distribution of expression levels for single genes at each point x along the anterior/posterior axis but the joint distribution of all the expression levels. The major difficulty in such an experiment is to avoid spectral cross-talk among the different fluorescence signals, but for the experiments shown in Fig. 3, we have shown that cross-talk is
accuracy? To answer this question, we need to know not just the distribution of expression levels for single genes at each point x along the anterior/posterior axis but the joint distribution of all the expression levels. The major difficulty in such an experiment is to avoid spectral cross-talk among the different fluorescence signals, but for the experiments shown in Fig. 3, we have shown that cross-talk is 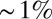 or less (28, 30), and, as noted in Materials and Methods, modest amounts of cross-talk actually do not change our estimate of
or less (28, 30), and, as noted in Materials and Methods, modest amounts of cross-talk actually do not change our estimate of 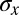 or the information. Given that we can sample the joint distribution of expression levels, how do we estimate the information that these expression levels carry?
or the information. Given that we can sample the joint distribution of expression levels, how do we estimate the information that these expression levels carry?
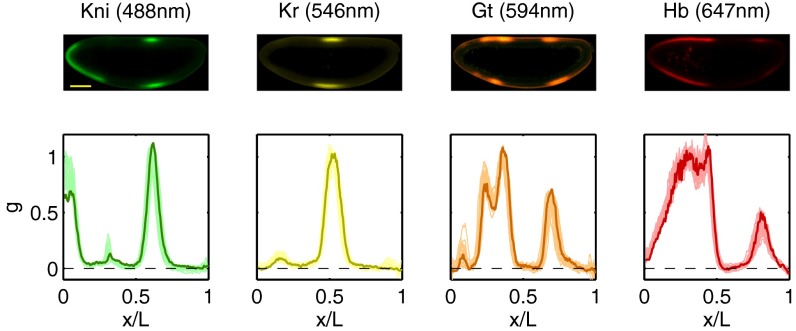
Fig. 3.
Simultaneous measurements on four gap genes reproduced, in part, from the work of Dubuis et al. (28). (Upper) Optical sections through the midsagittal plane of a single Drosophila embryo with immunofluorescence staining against Kni (green), Kr (yellow), Gt (orange), and Hb (red), with fluorescence excitation wavelengths in parentheses. (Scale bar = 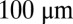 .) (Lower) Normalized expression levels along the dorsal edge for 24 embryos in a 38- to 48-min time interval after the start of nuclear cycle 14 (light colors); the sample embryo is highlighted (dark colors).
.) (Lower) Normalized expression levels along the dorsal edge for 24 embryos in a 38- to 48-min time interval after the start of nuclear cycle 14 (light colors); the sample embryo is highlighted (dark colors).
We observe the expression levels 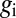 , with
, with  . At each point x, there are average values of these expression levels
. At each point x, there are average values of these expression levels  , and across an ensemble of embryos, there are fluctuations
, and across an ensemble of embryos, there are fluctuations 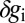 . Let us assume that these fluctuations have a Gaussian distribution. If we look just at one gene, this means that the statistics of the fluctuations are described completely by the mean and the variance
. Let us assume that these fluctuations have a Gaussian distribution. If we look just at one gene, this means that the statistics of the fluctuations are described completely by the mean and the variance 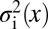 , so that if we look at the same position x in many embryos, we will see a distribution of expression levels
, so that if we look at the same position x in many embryos, we will see a distribution of expression levels
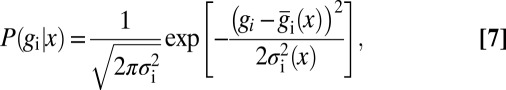 |
and this is in reasonable agreement with the data, as shown for the case of Hb in Fig. 1C (results for other genes are similar). If we look at many genes simultaneously, we have not just the variances of each gene but the correlations or covariances among the genes, which define a matrix 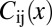 . The joint distribution of expression levels at one point is then
. The joint distribution of expression levels at one point is then
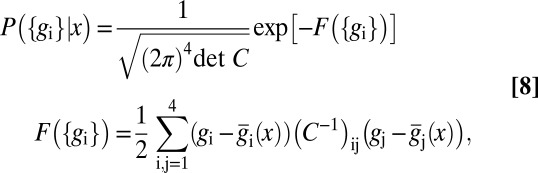 |
where  denotes the inverse of the matrix C and
denotes the inverse of the matrix C and 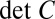 denotes its determinant. We can estimate all the elements of the covariance matrix, at every position x, in the usual way, averaging over samples taken from multiple embryos.
denotes its determinant. We can estimate all the elements of the covariance matrix, at every position x, in the usual way, averaging over samples taken from multiple embryos.
As an aside, we note that most of the significant off-diagonal elements of the covariance matrix are negative. For example, if the expression level of Hb happens to be a bit above average at one point in a single embryo, then the expression of Kr will be a bit below average at that same point. Presumably, this reflects the mutually repressive interactions among the gap genes (35–37).
The distribution 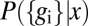 characterizes the measurements that we can make as an outside observer of the embryo. However, a single nucleus does not have access to the position x; rather, the whole idea of positional information is that this position is encoded in the expression levels. To assess the quality of this code, we can try to read it, asking for the distribution of positions that are consistent with a particular set of expression levels that we might observe. By Bayes’ rule, this can be written as
characterizes the measurements that we can make as an outside observer of the embryo. However, a single nucleus does not have access to the position x; rather, the whole idea of positional information is that this position is encoded in the expression levels. To assess the quality of this code, we can try to read it, asking for the distribution of positions that are consistent with a particular set of expression levels that we might observe. By Bayes’ rule, this can be written as
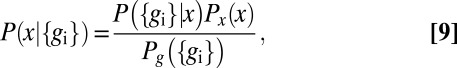 |
where 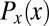 is, as before, the (nearly uniform) distribution of cell positions and
is, as before, the (nearly uniform) distribution of cell positions and 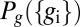 is the (joint) distribution of expression levels averaged over all cells in the embryo.
is the (joint) distribution of expression levels averaged over all cells in the embryo.
If the noise levels are small, then 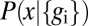 will be sharply peaked at some
will be sharply peaked at some 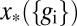 , which is the best estimate of the position, given the expression levels. Expanding around this estimate, the distribution is approximately Gaussian,
, which is the best estimate of the position, given the expression levels. Expanding around this estimate, the distribution is approximately Gaussian,
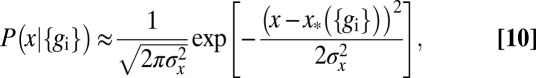 |
where the error in our position estimate is defined by
 |
All the terms in Eq. 11 are experimentally accessible.
Eq. 11 tells us the precision with which expression levels encode position: Observing the expression levels 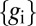 allows us (or the cell) to specify position, at best, with an “error bar”
allows us (or the cell) to specify position, at best, with an “error bar”  ; this error could be different at different points in the embryo, so we really should write
; this error could be different at different points in the embryo, so we really should write 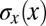 . Checking our intuition, we see that this error bar is smaller when the variability in expression is smaller (smaller C), when the mean slopes of the expression levels are larger (larger
. Checking our intuition, we see that this error bar is smaller when the variability in expression is smaller (smaller C), when the mean slopes of the expression levels are larger (larger 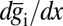 ), or when we can sum over more genes. We can define a similar quantity based on measurements of a single gene,
), or when we can sum over more genes. We can define a similar quantity based on measurements of a single gene,
 |
and this construction is shown schematically in Fig. 4 A and B in the case of Hb. Note that when 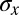 is small, we can justify our approximation that
is small, we can justify our approximation that 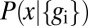 is sharply peaked, but when
is sharply peaked, but when 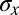 becomes large, it is more rigorous simply to say that we do not have much information about x rather than trying to give a more quantitative interpretation.
becomes large, it is more rigorous simply to say that we do not have much information about x rather than trying to give a more quantitative interpretation.
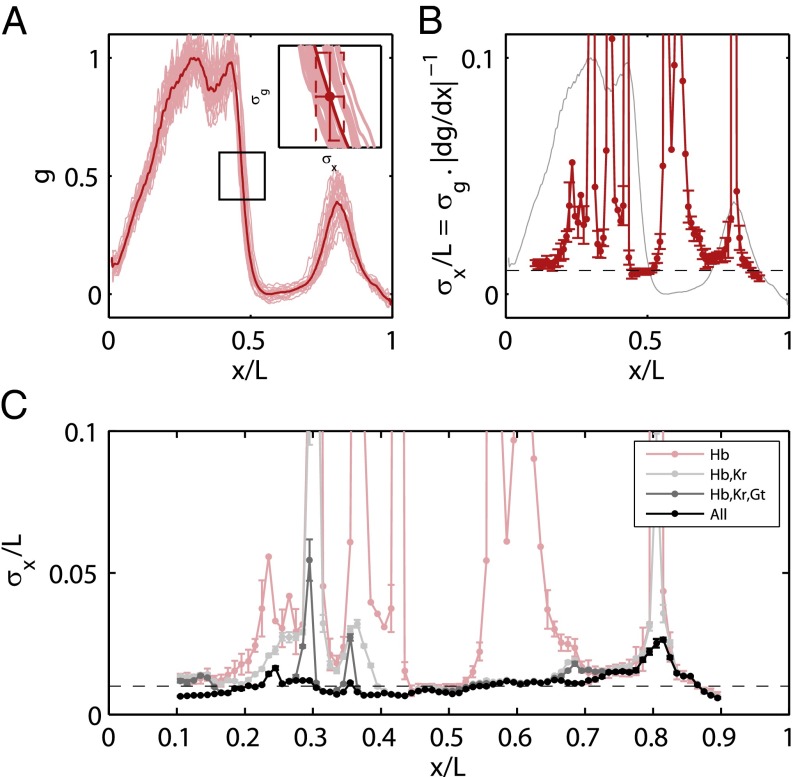
Fig. 4.
Positional error as a function of position. (A) Geometrical interpretation of the positional error for a single gene (Hb) at a given position. From Eq. 12, 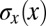 is proportional to the reproducibility of the profiles and is inversely proportional to the derivative of the mean profile. (B) Positional error based on the expression of Hb alone (red; mean ± SEM from bootstrapping) compared with the mean profile (gray). (C) Positional error based on combinations of gap genes, from Eq. 11. Note that once we combine information from all the gap genes, the net positional error is nearly constant and equal to 1% along the entire anterior/posterior axis.
is proportional to the reproducibility of the profiles and is inversely proportional to the derivative of the mean profile. (B) Positional error based on the expression of Hb alone (red; mean ± SEM from bootstrapping) compared with the mean profile (gray). (C) Positional error based on combinations of gap genes, from Eq. 11. Note that once we combine information from all the gap genes, the net positional error is nearly constant and equal to 1% along the entire anterior/posterior axis.
Analyzing the spatial profiles and variability of gene expression as suggested by Eq. 11, we obtain the estimates of 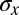 shown in Fig. 4C. Remarkably, the reliability of position estimates based on the four gap genes is
shown in Fig. 4C. Remarkably, the reliability of position estimates based on the four gap genes is 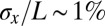 (compare with dashed line), almost precisely equal to the observed reproducibility with which pattern elements are positioned along the anterior/posterior axis. This is strong evidence that the gap genes, taken together, carry the information needed to specify the full pattern. Further, this positional accuracy is almost constant along the length of the embryo, which again is consistent with what we see in Fig. 2. This constancy emerges in a nontrivial way from the expression profiles, the noise levels, and the correlation structure of the noise. If we try to make estimates based on one gene, we can reach
(compare with dashed line), almost precisely equal to the observed reproducibility with which pattern elements are positioned along the anterior/posterior axis. This is strong evidence that the gap genes, taken together, carry the information needed to specify the full pattern. Further, this positional accuracy is almost constant along the length of the embryo, which again is consistent with what we see in Fig. 2. This constancy emerges in a nontrivial way from the expression profiles, the noise levels, and the correlation structure of the noise. If we try to make estimates based on one gene, we can reach 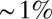 accuracy only in a very limited region of the embryo, but the detailed structure of the spatial profiles ensures that these signals can be combined to give nearly constant accuracy.
accuracy only in a very limited region of the embryo, but the detailed structure of the spatial profiles ensures that these signals can be combined to give nearly constant accuracy.
If the errors in estimating position really are Gaussian, as in Eq. 10, then we can substitute into Eq. 4 to show that 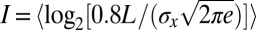 , where L is the length of the embryo, and
, where L is the length of the embryo, and  denotes an average over position. Computing this average, we have
denotes an average over position. Computing this average, we have 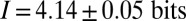 . Alternatively, we can use the distribution of expression levels at each position, Eq. 8, to compute the information directly as in Eq. 6, and we find
. Alternatively, we can use the distribution of expression levels at each position, Eq. 8, to compute the information directly as in Eq. 6, and we find 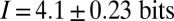 . The agreement between these estimates supports our approximations and gives us confidence that the measurement of
. The agreement between these estimates supports our approximations and gives us confidence that the measurement of 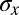 in Fig. 4 really does characterize the encoding of positional information by the gap genes.
in Fig. 4 really does characterize the encoding of positional information by the gap genes.
Thus, the gap genes carry enough information for each nucleus to know its position with an error bar 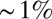 of the embryo’s length, and this is equal to the variability in localization of features that emerge in later stages of development. On the other hand, as noted above, this is not quite enough to specify the position of every nucleus uniquely. Is it possible that more information is “hiding” in the expression profiles? In particular, if the noise in neighboring cells is correlated, the errors in specifying relative positions (e.g., that one cell is more posterior than another) could be much smaller than the errors in specifying absolute positions. As a first step, we can ask how much information the expression levels of the gap genes provide about position measured from a “center of mass” that we compute from the whole spatial profile, rather than position in the fixed coordinate system that starts with
of the embryo’s length, and this is equal to the variability in localization of features that emerge in later stages of development. On the other hand, as noted above, this is not quite enough to specify the position of every nucleus uniquely. Is it possible that more information is “hiding” in the expression profiles? In particular, if the noise in neighboring cells is correlated, the errors in specifying relative positions (e.g., that one cell is more posterior than another) could be much smaller than the errors in specifying absolute positions. As a first step, we can ask how much information the expression levels of the gap genes provide about position measured from a “center of mass” that we compute from the whole spatial profile, rather than position in the fixed coordinate system that starts with 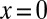 at the anterior end of the embryo. This relative positional information is
at the anterior end of the embryo. This relative positional information is 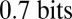 larger than the absolute positional information; although the data are very preliminary, we see hints of a similar gain of information about relative position for the peaks of Eve expression in Fig. 2. These results indicate that, through spatial comparisons, there may be enough information available to specify each cell’s identity.
larger than the absolute positional information; although the data are very preliminary, we see hints of a similar gain of information about relative position for the peaks of Eve expression in Fig. 2. These results indicate that, through spatial comparisons, there may be enough information available to specify each cell’s identity.
More Than One Bit per Gene?
The positional information carried by single gap genes is more nearly two bits than one, as described above, suggesting that spatial variations in gene expression define much more than on/off expression domains. However, when we combine information from different genes, redundancy among the spatial profiles of the different genes limits the information gain, with the result that the total information from four genes still is more than four bits, but not that much more. Perhaps almost all this information could be captured by a network that recognizes only on and off states of each gene, without resolving intermediate expression levels. How can we tell if the continuous gradations of expression are truly significant?
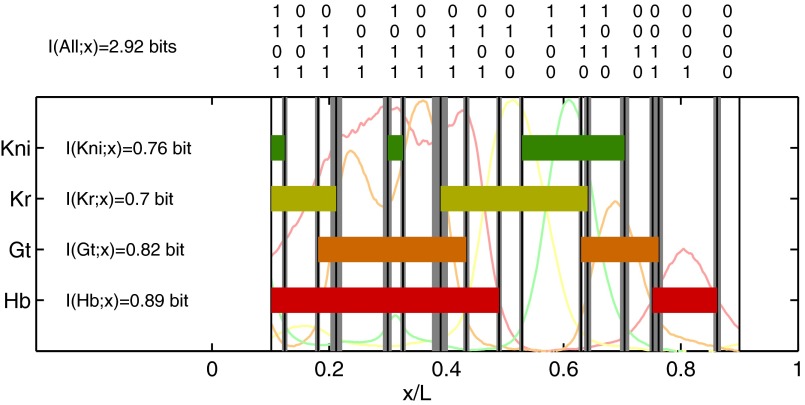
Suppose that the mechanisms that respond to the gap genes are limited to distinguishing only on and off states. The definition of “on” (“off”) is that the expression level is above (below) some threshold, which could be different for each gene, and to be fair we should imagine that these thresholds can be adjusted to capture as much positional information as possible. Instead of the state of each cell being defined by a set of continuous expression levels 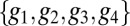 , the state would be given by a four-bit binary word, as in Fig. 5. At best, these words could convey four bits of positional information, but the actual information will be less because, given the spatial profiles, there is no set of on/off thresholds that will use all the 16 possible words equally often; there is an extra loss of information because of noise and variability across embryos. The result is that the maximum information that can be conveyed in such a binary scheme is
, the state would be given by a four-bit binary word, as in Fig. 5. At best, these words could convey four bits of positional information, but the actual information will be less because, given the spatial profiles, there is no set of on/off thresholds that will use all the 16 possible words equally often; there is an extra loss of information because of noise and variability across embryos. The result is that the maximum information that can be conveyed in such a binary scheme is 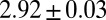 bits. Further, this information is distributed very inhomogeneously along the length of the anterior/posterior axis so that some binary words point to regions of the embryo that are defined within
bits. Further, this information is distributed very inhomogeneously along the length of the anterior/posterior axis so that some binary words point to regions of the embryo that are defined within 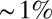 of the total length, whereas others (e.g., 0011, 1100, 0001) define domains as large as
of the total length, whereas others (e.g., 0011, 1100, 0001) define domains as large as 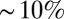 of L. Thus, mechanisms that ignore intermediate expression levels would lose a substantial fraction of the available positional information, as has been suggested from very different arguments (38).
of L. Thus, mechanisms that ignore intermediate expression levels would lose a substantial fraction of the available positional information, as has been suggested from very different arguments (38).
Fig. 5.
Binary view of the gap gene system. For each gap gene i, we quantize the expression level so that the gene is on (1) if 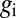 is greater than a threshold
is greater than a threshold 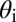 and off (0) otherwise. Here, we show the resulting domains of gene expression (dark color bars), as well as the fluctuations of their borders (in gray) for a set of thresholds that maximizes the total information carried by the binary variables (
and off (0) otherwise. Here, we show the resulting domains of gene expression (dark color bars), as well as the fluctuations of their borders (in gray) for a set of thresholds that maximizes the total information carried by the binary variables ( ,
, 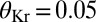 ,
, 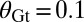 ,
, 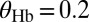 ). For reference, the mean profiles are plotted in dim colors in the background. (Left) Information carried by the quantized profiles of the individual genes is shown. The joint pattern of gap gene activity at each position is represented by a four-digit binary code (shown above, with the bits representing Kni, Kr, Gt, and Hb from top to bottom), and the total information encoded jointly by the on/off variables is computed as explained in the main text.
). For reference, the mean profiles are plotted in dim colors in the background. (Left) Information carried by the quantized profiles of the individual genes is shown. The joint pattern of gap gene activity at each position is represented by a four-digit binary code (shown above, with the bits representing Kni, Kr, Gt, and Hb from top to bottom), and the total information encoded jointly by the on/off variables is computed as explained in the main text.
Signature of Optimization?
The discussion thus far concerns the amount of information that actually is transmitted by the levels of gap gene expression. However, we know that the capacity to transmit information is strictly limited by the available numbers of molecules, and that significant increases in information capacity would require vastly more than proportional increases in these numbers (11). Given these limitations, however, cells can still make more or less efficient use of the available capacity. To maximize efficiency, the input/output relations and noise characteristics of the regulatory network must be matched to the distribution of input transcription factor concentrations (15). This matching principle has a long history in the analysis of neural coding (39–41), and it has been suggested that the regulation of Hb by Bicoid might provide an example of this principle (15). Here, we consider the generalization of this argument to the gap gene network as a whole.
If we imagine that there is a single primary morphogen, then the expression levels of the different gap genes, taken together, can be thought of as encoding the concentration c of this morphogen. By analogy with Eq. 11, these expression levels can be decoded with some accuracy 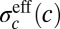 , which itself depends on the mean local concentration. The key result of ref. 15 is that when noise levels are small, all the symbols in the code should be used in proportion to their reliability, or in inverse proportion to their variability. Thus, if we point to a cell at random, we should see that the concentration of the primary morphogen is drawn from a distribution
, which itself depends on the mean local concentration. The key result of ref. 15 is that when noise levels are small, all the symbols in the code should be used in proportion to their reliability, or in inverse proportion to their variability. Thus, if we point to a cell at random, we should see that the concentration of the primary morphogen is drawn from a distribution
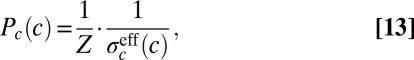 |
where the constant Z is chosen to normalize the distribution. However, the input is a morphogen, so its variation is connected with the physical position x of cells along the embryo: We should have 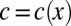 . Then, if the cells are distributed uniformly along the length of the embryo, the probability that we find a cell at x is just
. Then, if the cells are distributed uniformly along the length of the embryo, the probability that we find a cell at x is just 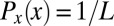 , and hence
, and hence
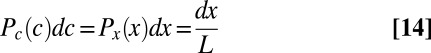 |
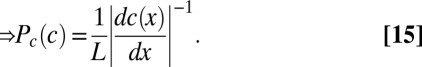 |
We have two expressions for the distribution of input transcription factor concentrations: Eq. 15, which expresses the role of the input as morphogen, encoding position x, and Eq. 13, which expresses the solution to the problem of optimizing information transmission through the network that responds to the input. Putting these expressions together, we have
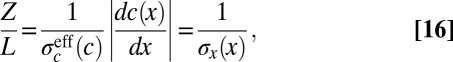 |
where, in the last step, we recognize the equivalent positional noise 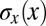 by analogy with Eqs. 11 and 12. Thus, optimizing information transmission predicts that the positional uncertainty
by analogy with Eqs. 11 and 12. Thus, optimizing information transmission predicts that the positional uncertainty 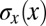 will be constant along the length of the embryo, as observed in Fig. 4C. Details are provided in Materials and Methods.
will be constant along the length of the embryo, as observed in Fig. 4C. Details are provided in Materials and Methods.
To measure the closeness of the embryo’s approach to optimality, we can compare the observed positional information with the maximum 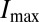 , which could be obtained if the embryo could adjust the distribution of nuclear positions
, which could be obtained if the embryo could adjust the distribution of nuclear positions 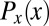 to match the positional error
to match the positional error 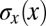 perfectly. In other words, if we take the measured positional errors as given, what is the capacity
perfectly. In other words, if we take the measured positional errors as given, what is the capacity 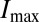 of the gap gene system to carry positional information, and what fraction of this capacity is achieved by the embryo? The result, from ref. 15, is that
of the gap gene system to carry positional information, and what fraction of this capacity is achieved by the embryo? The result, from ref. 15, is that
 |
The observed information transmission is 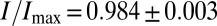 , within a few percent of the optimum.
, within a few percent of the optimum.
Discussion
The final result of embryonic development appears precise and reproducible. Less is known about the degree of this precision, and about the time at which precision first becomes apparent. Our central result is that in the early Drosophila embryo, the patterns of gap gene expression provide enough information to specify the positions of individual cells with a precision of 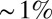 along the anterior/posterior axis. This is the same precision with which subsequent pattern elements are specified, from the pair rule expression stripes through the cephalic furrow, so that all the required information is available from a local, instantaneous readout of the gap genes.
along the anterior/posterior axis. This is the same precision with which subsequent pattern elements are specified, from the pair rule expression stripes through the cephalic furrow, so that all the required information is available from a local, instantaneous readout of the gap genes.
The precise value of the information that we observe is also interesting. It corresponds to being able to locate any nucleus with an error bar that is smaller than the distance to its neighbor, but the total number of bits is not quite large enough to specify the position of every cell uniquely. The difference is that when we make an estimate with error bars, the estimate comes from a distribution with tails, and the (small) overlap of the tails of these distributions means that one cannot quite identify every cell. It is possible that cells, in fact, do not quite have unique identities or that these identities emerge only later in development. Alternatively, although the gap genes encode position with an error bar, the difference between positions coded by expression levels in neighboring cells could have a much smaller error bar, and we have preliminary evidence for this idea. Although further experiments are required to settle this issue, we find it remarkable that the gap gene expression levels carry so much information, such that an enormously precise pattern is available very early in development.
The fact that precision is available early does not mean that there is no enhancement of precision by subsequent processes. In particular, because the joint distribution of expression levels does not fill the full space of possibilities, it would be possible for the embryo to recognize a large error, and perhaps to correct it, with no additional inputs. The question of whether the embryo achieves such an error-correcting code (21) for positional information is completely open.
The information that gene expression levels can carry about position is limited by noise. In particular, both because the concentrations of transcription factors are low and because the absolute copy numbers of the output proteins are small, there are physical sources of noise that cannot be reduced without the embryo investing more resources in making these molecules. Given these limits, it still is possible to transmit more information through the gap gene network by “matching” the distribution of input signals to the noise characteristics of the network. Although this matching condition is generally complicated, in the limit that the noise is small, it can be expressed very simply: The density of cells along the anterior/posterior axis should be inversely proportional to the precision with which we can infer position by decoding the signals carried in the gap gene expression levels. Because cells are almost uniformly distributed at this stage of development, this predicts that an optimal network would have a uniform precision, and this is what we find. This uniformity emerges despite the complex spatial dependence of all the ingredients, and thus seems likely to be a signature of selection for optimal information transmission.
Materials and Methods
Experiments.
To allow simultaneous imaging of proteins encoded by all four gap genes, polyclonal antibodies were generated (Panigen, Inc., Blanchardville, WI) in mice, rats, and guinea pigs against His-Trx-tagged full length Hb, Kni, and Gt fusion proteins (42); procedures were under the approval of Princeton University's Institutional Animal Care and Use Committee, Protocol No. 1798A to E.F.W. To image Kr protein, we use a rabbit anti-Kr antibody generated by Chris Rushlow (New York University). Fixation and staining were done as described by Dubuis et al. (28); details of the imaging, profile extraction, and staging (embryo age determination during nuclear cycle 14) are described by Dubuis et al. (28). We draw attention to the discussion of experimental errors in the study of Dubuis et al. (28), because this issue is especially important for our analysis.
Analysis.
Measurements on the expression profiles of a single gene in multiple embryos provide many samples of the joint distribution 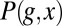 . To compute the mutual information between g and x, we discretize the two continuous axes into a number of bins; along the g axis, we use these bins adaptively so that the histogram of g in these bins is nearly flat. We then take the (normalized) counts in each bin as an estimate of the probability, compute the information, and examine the dependence on the number of bins and the number of samples. Following refs. 43 and 44, we search for the expected systematic dependencies and extrapolate to the limit where the number of bins and samples both become large. We can obtain an upper bound on the information by assuming that the conditional distribution
. To compute the mutual information between g and x, we discretize the two continuous axes into a number of bins; along the g axis, we use these bins adaptively so that the histogram of g in these bins is nearly flat. We then take the (normalized) counts in each bin as an estimate of the probability, compute the information, and examine the dependence on the number of bins and the number of samples. Following refs. 43 and 44, we search for the expected systematic dependencies and extrapolate to the limit where the number of bins and samples both become large. We can obtain an upper bound on the information by assuming that the conditional distribution 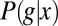 is Gaussian, and we can obtain an approximation to the information by taking this Gaussian approximation through to the construction of
is Gaussian, and we can obtain an approximation to the information by taking this Gaussian approximation through to the construction of 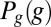 ; all these estimates agree within error bars. With simultaneous measurements of expression levels for multiple genes, we can estimate the information that they carry jointly. The difficulty is that the space of expression levels is now much larger but our number of samples is not. Having calibrated the Gaussian approximation against more direct calculations for single genes (above), we can use this approximation in the case of multiple genes, using Eq. 8 directly in the multidimensional generalization of Eq. 5; we use a Monte Carlo method to evaluate these integrals numerically and estimate errors by a bootstrap method. Means and covariance matrices are calculated from our multiple samples of joint expression levels in the usual way. Importantly, if the signals that we observe are invertible linear combinations of the true signals, as might happen, for example, because of a small amount of cross-talk among the different imaging channels, then the invariance of the information to coordinate transformations tells us that this will not change our estimate. The other path to the analysis of multiple genes is through the computation of
; all these estimates agree within error bars. With simultaneous measurements of expression levels for multiple genes, we can estimate the information that they carry jointly. The difficulty is that the space of expression levels is now much larger but our number of samples is not. Having calibrated the Gaussian approximation against more direct calculations for single genes (above), we can use this approximation in the case of multiple genes, using Eq. 8 directly in the multidimensional generalization of Eq. 5; we use a Monte Carlo method to evaluate these integrals numerically and estimate errors by a bootstrap method. Means and covariance matrices are calculated from our multiple samples of joint expression levels in the usual way. Importantly, if the signals that we observe are invertible linear combinations of the true signals, as might happen, for example, because of a small amount of cross-talk among the different imaging channels, then the invariance of the information to coordinate transformations tells us that this will not change our estimate. The other path to the analysis of multiple genes is through the computation of 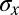 , as described in the discussion leading to Eq. 11. Here, too, we have to be careful about the dependence of our estimates on the number of samples that we include in our analysis, and quoted results are extrapolated as by Strong et al. (43) and Slonim et al. (44). In the discussion leading to Fig. 5, we set thresholds to quantize the expression levels and then estimate the mutual information between the four-bit words and the position x; the results we show are for the settings of the four thresholds that maximize the information.
, as described in the discussion leading to Eq. 11. Here, too, we have to be careful about the dependence of our estimates on the number of samples that we include in our analysis, and quoted results are extrapolated as by Strong et al. (43) and Slonim et al. (44). In the discussion leading to Fig. 5, we set thresholds to quantize the expression levels and then estimate the mutual information between the four-bit words and the position x; the results we show are for the settings of the four thresholds that maximize the information.
Derivation of Optimality Condition.
To derive Eq. 13, consider the case where information flows from a single input transcription factor (e.g., Bicoid) to a set of K output genes (the gap genes). The concentration of the input is c, and the output genes have expression levels 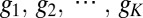 (16–18). Different cells in the embryo experience different values of c, depending on their position, and if we choose a cell at random, it sees a concentration drawn from the distribution
(16–18). Different cells in the embryo experience different values of c, depending on their position, and if we choose a cell at random, it sees a concentration drawn from the distribution 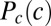 . The network responds to this input, generating expression levels that are drawn from the distribution
. The network responds to this input, generating expression levels that are drawn from the distribution 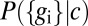 ; it will also be useful to define the (joint) distribution of output expression levels,
; it will also be useful to define the (joint) distribution of output expression levels,
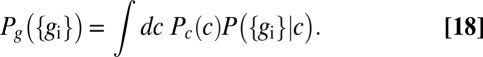 |
The information that flows from input to output can be written, as in Eq. 4, as
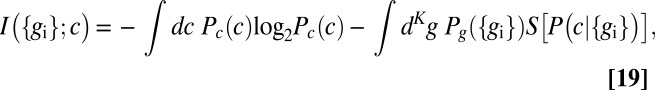 |
where, from Bayes’ rule, we have
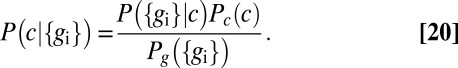 |
The transmitted information  depends both on the characteristics of the gene network, expressed as
depends both on the characteristics of the gene network, expressed as 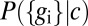 , and on the distribution of input signals,
, and on the distribution of input signals,  . In particular, noise associated with the finite number of available molecules is encoded by the details of
. In particular, noise associated with the finite number of available molecules is encoded by the details of 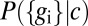 . Given these constraints, it still is possible to maximize information transmission by the proper choice of the input distribution (20, 21). In general, this optimization is a hard problem, but we can make progress if we assume that the noise is small, and we will argue that this is a good approximation.
. Given these constraints, it still is possible to maximize information transmission by the proper choice of the input distribution (20, 21). In general, this optimization is a hard problem, but we can make progress if we assume that the noise is small, and we will argue that this is a good approximation.
In Eq. 19, we need to take an average over the full distribution of output expression levels,  . This distribution is broadened by two effects. First, the inputs c are varying, and the outputs vary in response. Second, even when the input c is fixed, the outputs
. This distribution is broadened by two effects. First, the inputs c are varying, and the outputs vary in response. Second, even when the input c is fixed, the outputs 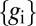 vary because of noise. We assume that noise is small in the sense that the first effect is much larger than the second, so that we can average over outputs by assuming that the output is always equal to its average value,
vary because of noise. We assume that noise is small in the sense that the first effect is much larger than the second, so that we can average over outputs by assuming that the output is always equal to its average value, 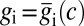 , and then average over the input c. In this approximation,
, and then average over the input c. In this approximation,
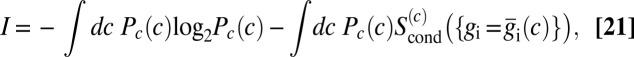 |
where 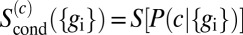 . To find the distribution of inputs that maximizes the information, we introduce, as usual, a Lagrange multiplier to fix the normalization of
. To find the distribution of inputs that maximizes the information, we introduce, as usual, a Lagrange multiplier to fix the normalization of 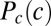 and solve
and solve
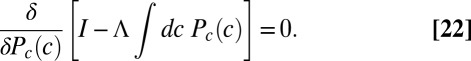 |
The result is
 |
where is Z chosen to normalize the distribution. If the noise is also approximately Gaussian—given knowledge of the gene expression levels 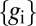 , we know the input concentration to within some error bar
, we know the input concentration to within some error bar 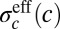 , which itself depends on the actual value of the input—then
, which itself depends on the actual value of the input—then 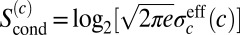 and
and
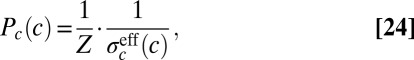 |
corresponding to Eq. 13. The system can optimize information transmission by using the symbols c in proportion to their reliability (15).
The noise in the system can be summarized by 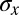 itself, which is smaller than the distances over which the output of any single gap gene varies significantly. Thus, in retrospect, the effective noise really is small, as assumed above, which justifies the approximation leading to Eq. 23. This derivation can be generalized to cases where there are multiple independent morphogen inputs, each varying along x.
itself, which is smaller than the distances over which the output of any single gap gene varies significantly. Thus, in retrospect, the effective noise really is small, as assumed above, which justifies the approximation leading to Eq. 23. This derivation can be generalized to cases where there are multiple independent morphogen inputs, each varying along x.
Acknowledgments
We thank F. Liu, M. Petkova, and R. Samanta for help with the experiments; V. Hakim for helpful discussions; and the referees for their thoughtful comments on the manuscript. This work was supported, in part, by National Science Foundation Grants PHY-0957573 and CCF-0939370; National Institutes of Health Grants P50GM071508, R01GM077599, and R01GM097275; the Howard Hughes Medical Institute; the W. M. Keck Foundation, and Searle Scholar Award 10-SSP-274 (to T.G.).
Footnotes
The authors declare no conflict of interest.
See QnAs on page 16288.
References
- 1.Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969;25(1):1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 2.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence PA. The Making of a Fly: The Genetics of Animal Design. Oxford: Blackwell; 1992. [Google Scholar]
- 4.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 5.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31(1):69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 6.Blake WJ, KAErn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422(6932):633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 7.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307(5717):1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 8.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123(6):1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 9.Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130(1):153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tkačik G, Gregor T, Bialek W. The role of input noise in transcriptional regulation. PLoS ONE. 2008;3:e2774. doi: 10.1371/journal.pone.0002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tkačik G, Callan CG, Jr, Bialek W. Information capacity of genetic regulatory elements. Phys Rev E. 2007;78:011910. doi: 10.1103/PhysRevE.78.011910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziv E, Nemenman I, Wiggins CH. Optimal signal processing in small stochastic biochemical networks. PLoS ONE. 2007;2(10):e1077. doi: 10.1371/journal.pone.0001077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Ronde WH, Tostevin F, ten Wolde PR. Effect of feedback on the fidelity of information transmission of time-varying signals. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82(3 Pt 1):031914. doi: 10.1103/PhysRevE.82.031914. [DOI] [PubMed] [Google Scholar]
- 14.Tkačik G, Walczak AM. Information transmission in genetic regulatory networks: A review. J Phys Condens Matter. 2011;23(15):153102. doi: 10.1088/0953-8984/23/15/153102. [DOI] [PubMed] [Google Scholar]
- 15.Tkačik G, Callan CG, Jr, Bialek W. Information flow and optimization in transcriptional regulation. Proc Natl Acad Sci USA. 2008;105(34):12265–12270. doi: 10.1073/pnas.0806077105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tkačik G, Walczak AM, Bialek W (2009) Optimizing information flow in small genetic networks. I. Phys Rev E 80:031920. [DOI] [PMC free article] [PubMed]
- 17.Walczak AM, Tkačik G, Bialek W. Optimizing information flow in small genetic networks. II: Feed-forward interaction. Phys Rev E. 2009;81:041905. doi: 10.1103/PhysRevE.81.041905. [DOI] [PubMed] [Google Scholar]
- 18.Tkačik G, Walczak AM, Bialek W. Optimizing information flow in small genetic networks. III. A self-interacting gene. Phys Rev E. 2011;85:041903. doi: 10.1103/PhysRevE.85.041903. [DOI] [PubMed] [Google Scholar]
- 19.Erdmann T, Howard M, ten Wolde PR. Role of spatial averaging in the precision of gene expression patterns. Phys Rev Lett. 2009;103(25):258101. doi: 10.1103/PhysRevLett.103.258101. [DOI] [PubMed] [Google Scholar]
- 20.Shannon CE. A mathematical theory of communication. Bell Sys Tech J. 1948 27:379–423, 623–656. [PubMed] [Google Scholar]
- 21.Cover TM, Thomas JA. Elements of Information Theory. New York: Wiley; 1991. [Google Scholar]
- 22.Bialek W. Biophysics: Searching for Principles. Princeton: Princeton Univ Press; 2012. [Google Scholar]
- 23.Driever W, Nüsslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54(1):83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 24.Driever W, Nüsslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54(1):95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- 25.Ephrussi A, St Johnston D. Seeing is believing: The bicoid morphogen gradient matures. Cell. 2004;116(2):143–152. doi: 10.1016/s0092-8674(04)00037-6. [DOI] [PubMed] [Google Scholar]
- 26.Turing AM. The chemical basis of morphogenesis. Philos Trans R Soc Lond B Biol Sci. 1952;237:33–72. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meinhardt H. Models of Biological Pattern Formation. New York: Academic; 1982. [Google Scholar]
- 28.Dubuis JO, Samanta R, Gregor T. Accurate measurements of dynamics and reproducibility in small genetic networks. Mol Syst Biol. 2013;9:639. doi: 10.1038/msb.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myasnikova E, Samsonova A, Kozlov K, Samsonova M, Reinitz J. Registration of the expression patterns of Drosophila segmentation genes by two independent methods. Bioinformatics. 2001;17(1):3–12. doi: 10.1093/bioinformatics/17.1.3. [DOI] [PubMed] [Google Scholar]
- 30. Dubuis JO (2012) Quantifying positional information during early embryonic development. PhD dissertation (Princeton University, Princeton)
- 31.Lecuit T, Samanta R, Wieschaus EF. slam encodes a developmental regulator of polarized membrane growth during cleavage of the Drosophila embryo. Dev Cell. 2002;2(4):425–436. doi: 10.1016/s1534-5807(02)00141-7. [DOI] [PubMed] [Google Scholar]
- 32.Merrill PT, Sweeton D, Wieschaus E. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. Development. 1988;104(3):495–509. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- 33.Gergen JP, Coulter D, Wieschaus EF. Segmental pattern and blastoderm cell identities. In: Gall JG, editor. Gametogenesis and the Early Embryo. New York: Liss; 1986. pp. 195–220. [Google Scholar]
- 34.Liu F, Morrison AH, Gregor T. Dynamic interpretation of maternal inputs by the Drosophila segmentation gene network. Proc Natl Acad Sci USA. 2013;110(17):6724–6729. doi: 10.1073/pnas.1220912110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraut R, Levine M. Spatial regulation of the gap gene giant during Drosophila development. Development. 1991;111(2):601–609. doi: 10.1242/dev.111.2.601. [DOI] [PubMed] [Google Scholar]
- 36.Kosman D, Small S. Concentration-dependent patterning by an ectopic expression domain of the Drosophila gap gene knirps. Development. 1997;124(7):1343–1354. doi: 10.1242/dev.124.7.1343. [DOI] [PubMed] [Google Scholar]
- 37.Clyde DE, et al. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. 2003;426(6968):849–853. doi: 10.1038/nature02189. [DOI] [PubMed] [Google Scholar]
- 38.Yu D, Small S. Precise registration of gene expression boundaries by a repressive morphogen in Drosophila. Curr Biol. 2008;18(12):868–876. doi: 10.1016/j.cub.2008.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barlow HB. Sensory mechanisms, the reduction of redundancy, and intelligence. In: Blake DV, Uttley AM, editors. Proceedings of the Symposium on the Mechanization of Thought Processes. vol 2. London: HM Stationery Office; 1959. pp. 537–574. [Google Scholar]
- 40.Laughlin SB. A simple coding procedure enhances a neuron’s information capacity. Z Naturforsch C. 1981;36(9-10):910–912. [PubMed] [Google Scholar]
- 41.Brenner N, Bialek W, de Ruyter van Steveninck R. Adaptive rescaling maximizes information transmission. Neuron. 2000;26(3):695–702. doi: 10.1016/s0896-6273(00)81205-2. [DOI] [PubMed] [Google Scholar]
- 42.Kosman D, Small S, Reinitz J. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev Genes Evol. 1998;208(5):290–294. doi: 10.1007/s004270050184. [DOI] [PubMed] [Google Scholar]
- 43.Strong SP, Koberle R, de Ruyter van Steveninck RR, Bialek W. Entropy and information in neural spike trains. Phys Rev Lett. 1998;80:197–200. [Google Scholar]
- 44. Slonim N, Atwal GS, Tkačik G, Bialek W (2005) Estimating mutual information and multi-information in large networks. arXiv:cs.IT/0502017.