Significance
Bacteria use regulatory modules called two-component systems to respond to changes in their surrounding environment. Bacteria have evolved ways to insulate each two-component system, thereby preventing unwanted cross-talk. Here we describe an example where partners of distinct two-component systems show remarkable cross-specificity for each other. Loss of the quorum-sensing Escherichia coli (Qse)BC sensor QseC leads to robust cross-interaction of its cognate partner QseB with the polymyxin resistance (Pmr)AB two-component system. This cross-interaction in the absence of the cognate sensor is detrimental, severely attenuating pathogen virulence. Our findings suggest that (i) robust cross-talk between noncognate partners is possible and (ii) this interaction can be manipulated for developing antivirulence strategies against uropathogenic E. coli and potentially other QseBC−PmrAB-bearing pathogens.
Keywords: UPEC, signal transduction, phosphorylation, regulatory networks
Abstract
Bacterial two-component systems (TCSs) mediate specific responses to distinct conditions and/or stresses. TCS interactions are highly specific between cognate partners to avoid unintended cross-talk. Although cross-talk between a sensor kinase and a noncognate response regulator has been previously demonstrated, the majority of reported interactions have not been robust. Here, we report that in the case of the quorum-sensing Escherichia coli (Qse)BC TCS, absence of the cognate sensor QseC leads to robust, constitutive activation of the QseB response regulator by the noncognate polymyxin resistance (Pmr) sensor kinase PmrB. Remarkably, the noncognate PmrB exhibits a kinetic preference for QseB that is similar to QseC. However, although PmrB readily phosphorylates QseB in vitro, it is significantly less efficient at dephosphorylating QseB, compared with QseC, thereby explaining the increased levels of active QseB in the qseC mutant. In addition to PmrB activating QseB on the protein level, we found that the PmrA response regulator contributes to qseB transcription in the absence of QseC and PmrA specifically binds the qseBC promoter, indicative of a direct regulation of qseBC gene transcription by PmrAB under physiological conditions. Addition of ferric iron in the growth medium of wild-type uropathogenic E. coli induced the expression of qseBC in a PmrB-dependent manner. Taken together, our findings suggest that (i) robust cross-talk between noncognate partners is possible and (ii) this interaction can be manipulated for the development of antivirulence strategies aimed at targeting uropathogenic Escherichia coli and potentially other QseBC−PmrAB-bearing pathogens.
Two component-systems (TCSs) allow bacteria to translate extracellular signals into gene expression patterns that are beneficial for survival in a given environment. They are typically composed of a membrane-embedded sensor histidine kinase and a cytoplasmic cognate response regulator (RR) (1). The sensor kinase intercepts the environmental stimulus, becomes autophosphorylated at a conserved histidine residue and then transfers the phosphoryl group to a conserved aspartate residue on the cognate RR (1). Phosphorylation induces RR conformational changes that influence its regulatory activity (1, 2). In most cases, the sensor protein is bifunctional, acting as a kinase and a phosphatase, thus catalyzing both the phosphorylation and dephosphorylation of the RR (1, 3, 4). This bifunctional ability enables the sensor to tightly control RR activity and relayed output.
Bacteria harbor several TCSs, each mediating specific responses to distinct conditions and/or stresses (5). Despite their similarities, TCS interactions are highly specific, to convey desired signals and avoid unintended cross-talk (5). Quorum-sensing Escherichia coli (Qse)BC is found in several important human pathogens, and deletion of qseC leads to misregulation of gene expression and/or virulence attenuation (6–9). We have previously reported that the ΔqseC-related defects in pathogenic E. coli are a result of constitutive activation of the QseB RR, resulting in increased qseB expression, aberrant regulation of conserved metabolic pathways, and down-regulation of virulence-associated genes (8, 9). In this study, we determined the source of QseB phosphorylation, using uropathogenic E. coli (UPEC) as a model organism. Transposon mutagenesis, coupled with biochemical analyses, revealed that the polymyxin resistance (Pmr) sensor PmrB is the QseB phosphodonor in the absence of QseC. In addition, we found that the PmrB cognate RR, PmrA, directly controls qseBC expression. Addition of the PmrAB activating signal, ferric iron, induced qseBC gene expression in the wild-type UPEC strain in a PmrB-dependent manner that appears to involve QseB and PmrA.
Results
Transposon Mutagenesis Identifies Suppressors of the qseC Deletion.
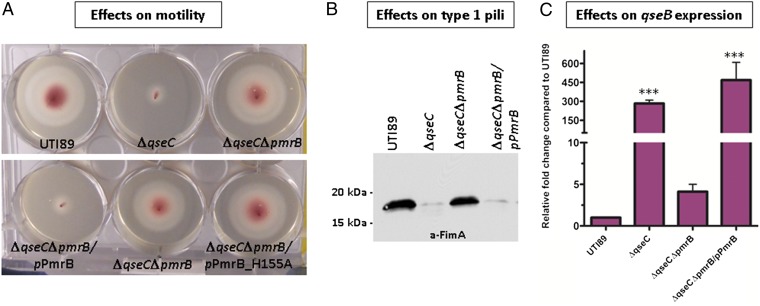
To identify the primary QseB activating source, we performed a suppressor screen using a library of UTI89ΔqseC transposon mutants and a phenotypic assay that assesses curli production based on colony morphology. Deletion of qseC down-regulates expression of curli adhesive fibers (9); thus, when grown on Yeast Extract-Casamino Acids (YESCA) agar, supplemented with Congo Red dye (CR) (10), UTI89ΔqseC forms white and smooth colonies indicative of diminished curli production, in contrast to WT UTI89, which exhibits a red, dry, and rough morphotype (9). Screening the 53,000 mutants comprising the UTI89ΔqseC transposon library (10× genome coverage) on YESCA-CR agar identified 37 suppressor mutants with WT or near-WT morphology. Subsequent transposition mapping of the mutants with WT morphology identified seven unique genes that, when disrupted, led to restoration of curli production (Fig. S1A). Among those, qseB served as a robust internal control, as we have previously shown that UTI89ΔqseBC produces WT levels of curli (9). The other identified targets included the PmrB (also known as BasS) histidine kinase, the SdiA regulatory protein, the cold-shock DEAD box protein A, the hypothetical YfiR protein, as well as FadD and CyoA, involved in respiration (Fig. S1A). UTI89ΔqseC is nonmotile and exhibits reduced type 1 pili expression (8, 9). Given that disruption of the true QseB phosphodonor should suppress all ΔqseC-related defects, we also tested for restoration of motility and type 1 pili expression in each of the seven identified transposon mutants. Western blot analysis probing for the major type 1 pili subunit FimA and motility assays revealed that only disruption of either qseB or pmrB fully restored UTI89ΔqseC FimA expression and motility to WT levels (Fig. S1 B and C).
Disruption of pmrB in the Absence of QseC Suppresses the ΔqseC Phenotypes.
We generated a clean pmrB deletion in UTI89ΔqseC to verify that the effects we saw with Tn::pmrB were specific to pmrB disruption and not an artifact of transposition. The resulting mutant, UTI89ΔqseCΔpmrB, behaved like the corresponding transposon mutant, being restored for motility, curli, and type 1 pili expression (Fig. 1 A and B). Quantitative (q)PCR analysis showed that qseB transcript levels in UTI89ΔqseCΔpmrB were similar to WT UTI89, indicating that disruption of pmrB abrogates the increased qseB transcription observed in UTI89ΔqseC (Fig. 1C). Complementation of UTI89ΔqseCΔpmrB with pmrB under its native promoter (plasmid pPmrB) restored qseB transcription to the levels observed in UTI89ΔqseC and resulted in ΔqseC phenotypes (Fig. 1).
Fig. 1.
Deletion of pmrB in UTI89ΔqseC restores wild-type phenotypes. (A) Motility and (B) FimA Western blot assays demonstrating that UTI89ΔqseCΔpmrB exhibits WT motility and expresses WT levels of type 1 pili. Motility experiments include the effects of inactivating PmrB kinase activity (UTI89ΔqseCΔpmrB/pPmrB_H155A). Motility diameters were measured after a 7 h incubation at 37 °C. Experiments were repeated at least five times. A representative experiment is shown. (C) Relative-fold change of qseB transcript in WT UTI89, UTI89ΔqseC, UTI89ΔqseCΔpmrB, and UTI89ΔqseCΔpmrB/pPmrB measured by qPCR. Quantitative real-time PCR was executed in triplicate, using multiplexed TaqMan minor groove binder chemistry. Error bars represent SEM. Statistical analyses were performed by two-tailed unpaired t test, where ***P ≤ 0.0005.
UTI89ΔqseC is severely attenuated in vivo (9, 11). We tested the ability of UTI89ΔqseCΔpmrB to establish acute urinary tract infection (UTI) in our mouse model of acute infection (12) by assessing bladder colony-forming units (CFU) and formation of intracellular bacterial communities (IBCs) (12) at 6 h and 16 h post infection. UTI89ΔqseCΔpmrB established acute infection as efficiently as WT UTI89 (Fig. S2).
PmrB Exhibits Kinetic Preference to QseB in a Unidirectional Fashion.
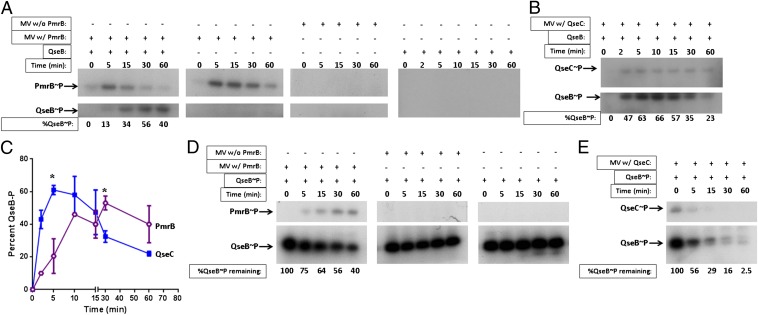
If PmrB phosphorylates QseB in the absence of QseC, then ablating the PmrB kinase activity by site-directed mutagenesis should yield the same outcome as the pmrB deletion. We thus altered the PmrB phosphoaccepting histidine (H155) to alanine. The resulting construct, pPmrB_H155A, was introduced to UTI89ΔqseCΔpmrB, and the generated strain UTI89ΔqseCΔpmrB/pPmrB_H155A was tested for motility. In contrast to the nonmotile UTI89ΔqseCΔpmrB/pPmrB, UTI89ΔqseCΔpmrB/pPmrB_H155A exhibited WT motility (Fig. 1A). Subsequent in vitro phosphotransfer assays confirmed that PmrB phosphorylates QseB (Fig. 2A). We observed that PmrB-mediated phosphorylation of purified QseB could be observed as early as 5 min (Fig. 2A, Left), indicating a kinetic preference for a noncognate partner that is very rare in nature. The cognate QseC was, as expected, superior in its capacity to phosphorylate greater amounts of QseB within the same time frame, as indicated by kinetic analysis (Fig. 2 B and C).
Fig. 2.
PmrB phosphorylates QseB in the absence of QseC. Phosphotransfer assays with (A) PmrB- or (B) QseC-enriched MV and purified QseB. Percent QseB∼P was calculated by determining the peak intensity in each lane, compared with t = 0 using ImageJ. Loss of greytone intensity in each lane was compared with t = 0, converted to a percentage and subtracted from 100 to obtain the gain in intensity that represents increase in QseB∼P. All gels were exposed for the same amount of time, and all experiments were performed with the same amounts of QseC or PmrB MV. Center Left in A demonstrates PmrB autokinase activity in the absence of QseB. Center Right in A depicts a mock phosphotransfer assay using MV without PmrB to verify that phosphotransfer to QseB occurs specifically by PmrB. Right in A shows phosphotransfer in the presence of QseB alone to rule out the possibility of QseB auto-phosphorylation. A representative of five independent experiments is shown. (C) Graph depicting the rate of QseB phosphorylation by QseC (blue) or PmrB (purple). Percent intensities over time were calculated as described in A. Results are the average of two combined, independent experiments. Statistical analysis was performed by unpaired t test, with Welch’s correction; *P < 0.03. (D and E) Phosphatase assays with PmrB-enriched MV and in vitro phosphorylated QseB∼P. Percent QseB∼P was calculated based on peak intensity analysis of each band normalized to the sample at t = 0, using ImageJ software. A representative of three independent experiments is shown.
We then tested the hypothesis that PmrB cannot dephosphorylate QseB as efficiently as QseC. We performed a time course, assessing dephosphorylation of in vitro phosphorylated QseB (QseB∼P) incubated with PmrB-enriched membrane vesicles (MV), MV lacking PmrB, or QseC-enriched MV. Using ImageJ software, we determined that, after 5 min of incubation, 25% of QseB was dephosphorylated by PmrB. After 60 min of incubation, 40% of QseB remained phosphorylated (Fig. 2D, Left). In striking contrast, 44% of QseB was dephosphorylated after 5 min of incubation in the presence of QseC, and by 60 min of incubation, only 2.5% of QseB∼P remained phosphorylated (Fig. 2E). Incubation of QseB∼P with MV lacking PmrB or QseC or incubation of QseB in the absence of MV altogether did not impact the QseB phosphorylation state (Fig. 2D, Center and Right), indicating that QseB∼P does not get dephosphorylated by other kinases/phosphatases in the MV and it does not undergo significant spontaneous dephosphorylation. Thus, although PmrB can phosphorylate QseB at rates relatively comparable to QseC, it is significantly less efficient in its ability to dephosphorylate QseB∼P, resulting in the disproportionate activation of QseB in the qseC mutant.
The PmrA Response Regulator Regulates qseBC.
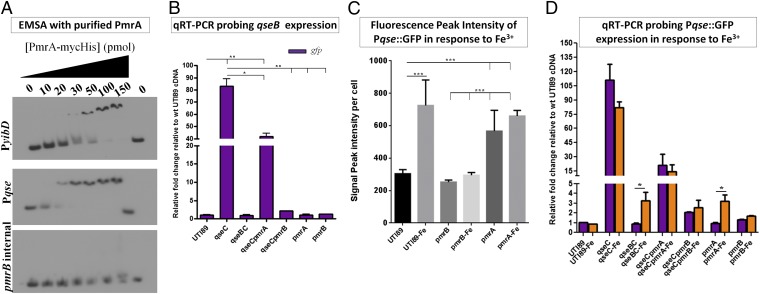
The fact that PmrB exhibits strong kinetic preference toward QseB prompted us to assess whether PmrAB and QseBC are physiologically linked. Scanning the qseBC promoter region revealed the presence of a PmrA binding consensus [5′-(C/T)TTAA(G/T)-N5-(C/T)TTAA(G/T)-3′] (13–15) (Fig. S3), overlapping the previously reported QseB-dependent promoter (16). We therefore tested if PmrA directly binds to the qseBC promoter using electromobility shift assays. As a positive control, we tested PmrA binding to the yibD promoter, a previously identified direct PmrA target (17). As a negative control, we used an internal region of the pmrB gene sequence. Using a previously established protocol (17) with increasing concentrations of PmrA, we detected binding to qseBC promoter at concentrations lower than those observed (and reported (17)) for yibD (Fig. 3A). Competition assays with unlabeled qseBC promoter DNA, titrated PmrA binding from the 32P-labeled qseBC promoter fragments, validating specificity of PmrA for this sequence (Fig. S4). To assess the contribution of PmrA to qseBC gene expression, we used a previously constructed qseBC promoter-GFP reporter plasmid, Pqse::GFP (9) in combination with qPCR. We had previously shown that, although Pqse::GFP was robustly active in UTI89ΔqseC, there was no significant promoter activity recorded in WT UTI89 or UTI89ΔqseBC during growth in either LB or N-minimal media (9). Here, we constructed a nonpolar pmrA deletion in UTI89 and UTI89ΔqseC, introduced Pqse::GFP in the resulting mutants, and sampled gfp steady-state transcript and GFP fluorescence as a proxy to qseBC expression. This allowed us to assess qseBC promoter activity in a panel of mutants, including those lacking qseB (in which a qseB-specific probe would not be informative). Consistent with the above observations, qseBC expression in UTI89ΔqseCΔpmrB was similar to WT UTI89 and UTI89ΔqseBC and was significantly lower than UTI89ΔqseC (Fig. 3B and Fig. S5). However, deletion of pmrA in the absence of QseC (UTI89ΔqseCΔpmrA) resulted in a twofold reduction of gfp steady-state transcript (Fig. 3B and Fig. S5), indicating that PmrA contributes to qseBC transcription. Given that PmrA regulates pmrAB expression, we tested whether the 50% reduction in qseB expression observed in UTI89ΔqseC upon deletion of pmrA stemmed from reduced transcription of pmrB. We sampled pmrB expression in the absence of PmrA using qPCR, and found that pmrB transcript levels in fact increased in the pmrA mutants under the tested conditions (Fig. S6). Interestingly, deletion of qseB in the pmrA deletion background suppressed pmrB up-regulation (Fig. S6), strongly suggesting the presence of cross-regulation between the two TCSs. Taken together, these data indicate that in the absence of QseC, qseBC up-regulation comes from a possible synergistic effect of aberrantly activated QseB∼P by PmrB and the activity of PmrA (the phosphorylation state for this function remains unknown) on the qseBC promoter. In addition, it appears that there is bidirectional cross-regulation between QseBC and PmrAB.
Fig. 3.
QseBC and PmrAB are physiologically interconnected. (A) Electromobility shift assays (EMSA) with purified PmrA and the qseBC promoter. The yibD promoter is a known PmrA direct target and was used as a positive control (17); an internal region of the pmrB gene was amplified and used as a negative control for binding. (B) Relative-fold change of gfp driven by the qseBC promoter in WT UTI89, UTI89ΔqseC, UTI89ΔqseBC, UTI89ΔqseCΔpmrA, UTI89ΔqseCΔpmrB, UTI89ΔpmrA, and UTI89ΔpmrB measured by qRT-PCR. (C) Graph depicting GFP intensity in bacterial cells harboring PqseBC::GFP in the presence of ferric iron, the PmrAB activating signal. Peak intensity measurements were taken for 50 bacterial cells per strain from three independent microscopy experiments. Error graphs in both graphs indicate the geometric mean with 95% confidence interval. Statistical analysis was performed using two-tailed unpaired t test, where ***P < 0.0001. (D) Relative-fold change of gfp driven by the qseBC promoter in response to Fe3+ measured by qPCR.
The PmrAB Activating Signal Induces qseBC Expression.
The presence of a PmrA binding site in the qseBC promoter suggests that QseBC is part of the PmrAB regulon. We thus tested the effects of ferric iron (Fe3+), the PmrAB activating signal (18), on the expression of qseBC in WT UPEC. We have shown that Pqse::GFP is silent under normal growth conditions in WT UTI89 (ref. 9, Fig. S5). Addition of 100μM FeCl3 in the culture media, uniformly induced gfp expression in WT UTI89/Pqse::GFP in a manner that depended on PmrB (Fig. 3C). Interestingly, deletion of pmrA resulted in low-level constitutive activation of the qseBC promoter (Fig. 3C). The qPCR analysis of RNA samples taken from the same panel of the qse/pmr mutants validated the fluorescence observations, but captured no significant changes in the steady-state gfp transcript in FeCl3-treated WT UTI89, despite the observed fluorescence recorded (Fig. 3D). The transcription surge in response to signal reception drops as quickly as 20 min after TCS activation under physiological conditions (19, 20). The samples tested here with microscopy and qPCR were collected after 18 h static incubation at 37 °C in the presence or absence of FeCl3, in accordance with the growth conditions used throughout our studies. It is thus possible that the signal-mediated transcription surge was missed, while the GFP protein levels remained stable in the WT strain. Similarly, the observed elevation of gfp steady-state transcript in UTI89ΔqseC and UTI89ΔqseCΔpmrA was not altered by the presence of Fe3+, indicating that in the absence of QseC, PmrAB-mediated induction of qseBC occurs irrespective of signal, or that any surge in transcription was not captured in the particular samples. Interestingly, a reproducible 3.5-fold increase in steady-state transcript was observed for UTI89ΔpmrA and UTI89ΔqseBC in response to Fe3+ (Fig. 3D). If PmrB activates both QseB and PmrA in response to Fe3+, this observation may be the result of functional redundancy between the two RRs that allows signal-dependent promoter activation.
Over 500 genes are differentially expressed in the absence of QseC, due to aberrant qseB induction (8). Deletion of pmrB in the qseC mutant suppresses qseB induction and restores WT phenotypes (Fig. 1 and Fig. S1). Expression of qseBC is only reduced twofold in UTI89ΔqseCΔpmrA (Fig. 3B). Examining the virulence phenotypes of UTI89ΔqseCΔpmrA revealed that UTI89ΔqseCΔpmrA expressed WT levels of type 1 pili (Fig. S7), and regained motility (Fig. S7), but was defective in YESCA biofilm formation, compared with WT UTI89 and UTI89ΔqseCΔpmrB (Fig. S7). These data suggest that deletion of pmrA is not sufficient to suppress all of the ΔqseC phenotypes. Based on these observations, QseBC is subject to PmrB control under physiological conditions in a manner that appears to involve PmrA and QseB.
Discussion
Numerous reports demonstrate the importance of the kinase activity of TCS sensor proteins, and highlight that loss of the sensor leads to an inactive response regulator that can no longer modulate gene expression. However, few examples stress the significance of sensor phosphatase activity, despite the fact that RR dephosphorylation is required to reset the signal transduction baseline state upon signal depletion and to prevent aberrant RR activity in the absence of signal (2).
We have previously reported that in UPEC, the absence of the QseC sensor leads to aberrant and robust phosphorylation of the cognate RR QseB by a noncognate sensor or phosphodonor molecule (9). This aberrant cross-talk imparts pleiotropic gene expression changes that impact metabolic processes and attenuate virulence (8, 9). Our work highlighted the importance of a bifunctional sensor and demonstrated the detrimental effects that can occur in the absence of the sensor. Russo and Silhavy were among the first to point out that sensor phosphatase activity is instrumental in maintaining a balanced ratio of phosphorylated and unphosphorylated RR, by showing that EnvZ phosphatase mutants exhibited aberrant osmolarity phenotypes, indicative of an overactive OmpR RR (21, 22). In these studies, EnvZ retained its kinase activity. In agreement with these studies, we have previously shown that the QseC phosphatase domain is critical in maintaining optimal phosphorylated QseB levels and gene expression (9). Siryaporn and Goulian recently demonstrated that although phosphotransfer from a noncognate sensor to a RR can occur, noncognate RR dephosphorylation is disfavored and highly restricted to the cognate pair (23).
Amemura et al., Wanner, and Wanner et al. were among the first to demonstrate that cross-regulation among noncognate partners exists, by showing that CreC (formerly known as PhoM) robustly phosphorylates the noncognate PhoB (24–26). This study documents another strong interaction between a RR and a noncognate sensor: PmrB readily phosphorylates QseB; however, PmrB phosphatase activity toward QseB is slow, thus explaining the observed QseB activation when QseC is not there. Interestingly, unlike other systems in which cross-regulation has been reported (24–26), we show that although weak, there is cross-phosphatase activity between PmrB and QseB. Another intriguing aspect of our findings is that cross-talk is not promiscuous, as no other sensor was identified in our screen and deletion of pmrB in the qseC mutant entirely suppressed QseB activation. It is thus possible that the interacting interfaces of PmrB and QseC share common features that enable similar protein−protein interactions with the QseB RR. This could be a result of the two TCSs coevolving because they are physiologically connected, or it could be due to the fact that QseBC and PmrAB recently arose by duplication and have not sufficiently diverged yet. Sequence analysis indicates that PmrB is the closest homolog of QseC in UTI89, with 37% sequence identity and 70% coverage. An investigation by Skerker et al. previously demonstrated the presence of a subset of coevolving residues that confer sensor−RR specificity (27). Evaluating sequence similarities and investigating protein−protein interactions between PmrB−QseB and QseC−QseB will provide further insights into why their cross-interactions are so robust.
In addition to identifying PmrB and investigating its phosphotransfer kinetics in relation to QseC, we have discovered that in the absence of QseC, the PmrB cognate RR, PmrA contributes to aberrant expression of the qseBC operon, possibly by augmenting qseB gene expression. Whereas the qseCpmrB double mutant suppresses aberrant qseB expression back to WT levels, deletion of pmrA in UTI89ΔqseC only results in a 50% reduction of qseB steady-state transcript (Fig. 3B). These data, combined with the ability of PmrA to bind the qseBC promoter point toward a potential synergistic effect of the two RR on qseBC promoter activation in the absence of QseC. Absence of PmrA may also alter the kinase/phosphatase kinetics of PmrB to QseB, thereby contributing to the observed reduction in qseB transcript. Microarray analysis has captured differentially expressed genes in UTI89ΔqseC that are normally under PmrA control (13) (Table S1). These genes may be affected both by PmrA and QseB, by QseB alone, or there may be a surge in PmrA activity in the absence of QseC that leads to differential expression of these targets. Our current data demonstrate that UTI89ΔqseCΔpmrA is restored for motility, and type 1 pili expression, but remains partially defective for biofilm formation in YESCA media (Fig. S7). Given the massive transcriptional shift observed in UTI89ΔqseC, additional global analyses probing the transcriptome of UTI89ΔqseCΔpmrA are warranted to expand on these observations. These possibilities open unique avenues of study, investigating not only contacts between PmrA and QseB, but also potential—yet unexplored—interactions of QseC with PmrA.
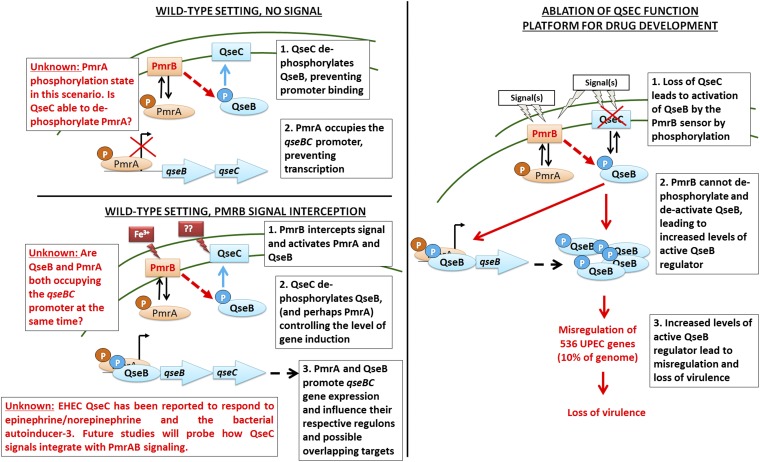
This work has also uncovered physiological links between PmrAB and QseBC; elevated ferric iron induces qseBC in a manner that depends on PmrB (Fig. 3 C and D). However, loss of PmrA leads to constitutive, low-level induction of qseBC that is abolished in the absence of QseB. Based on these findings, we have developed the following working model (Fig. 4); in the wild-type setting, QseC is present, dephosphorylating QseB (and perhaps PmrA) and preventing QseB binding to the promoter. This allows PmrA to bind the promoter but is not sufficient to promote transcription. Signal reception by PmrB leads to phosphorylation of PmrA and QseB and changes the affinity of the RR for the qseBC promoter and/or allows synergistic binding (and transcription) that is controlled by the phosphatase activity of QseC. Loss of QseC function tips this delicate balance toward more phosphorylated QseB, leading to elevated qseBC transcription, more QseB∼P, and global gene deregulation, as we previously reported. Thus, it may be possible to derail pathogen gene expression by identifying QseC inhibitors that will favor PmrB−QseB cross-talk. Notably, EHEC QseC has been reported to respond to epinephrine/norepinephrine and the bacterial autoinducer-3. Future studies probing how QseC signals integrate with PmrAB signaling will address this gap.
Fig. 4.
Model of PmrAB-QseBC interactions. (Left) Representation of our working model with regards to PmrAB-QseBC physiological cross-interactions. Although we have begun unraveling the contribution of ferric iron in mediating QseBC/PmrAB responses, the role of the QseC previously reported signals remains elusive. (Right) Demonstration of the consequences of losing QseC (or QseC function).
Materials and Methods
Transposon Mutagenesis.
Electrocompetent UTI89ΔqseC cells and transposon mutagenesis were performed as previously described (28). Kanamycin resistant transposants were patch-plated on YESCA (1 g yeast extract, 10 g casamino acids, 20 g agar/L) agar supplemented with CR, and incubated at 30 °C for 48 h. Each YESCA-CR plate included streaks of WT UTI89 and UTI89ΔqseC as positive and negative controls. Mutations were mapped using the multiple-round PCR procedure of Ducey and Dyer (29, 30) and primers Inv-1 or Inv-2 (29, 30). Amplicons were purified (Qiagen) and sequenced using the KAN-2 FP-1 and KAN-2 RP-1 primers (Epicentre).
Strains and Constructs.
UTI89ΔqseC, UTI89ΔqseB and UTI89ΔqseBC were created previously (9). UTI89ΔqseCΔpmrB, UTI89ΔqseCΔpmrA, and UTI89ΔpmrA were created using λ Red Recombinase (31) and primers listed on Table S2. The pPmrB was created in pTrc99A (Invitrogen), by cloning pmrB (UTI89_C4706) downstream of its native promoter. Site-directed mutagenesis was performed on pPmrB using Pfu Ultra DNA polymerase (Stratagene). UTI89ΔqseC/pQseC-mycHis was previously constructed (9). The pmrB gene was amplified from pPmrB or pPmrB_H155A, cloned into pBADmyc-HisA (Invitrogen) and electroporated into UTI89ΔqseCΔpmrB. The transcriptional reporter plasmid pPqse::GFP was previously constructed (9).
Immunoblots, HA, and Phase Assays.
For type 1 pili, bacteria were incubated in LB media at 37 °C for 4 h under shaking conditions, subcultured (1:1,000) in fresh LB media, and incubated statically at 37 °C for 18 h. Immunoblots (using antibody raised against type 1 pili), HA, and phase assays were performed on normalized cells (OD600 = 1) as previously described (9), with the exception of the Western blot in Fig. S7; in this blot, Alexa Fluor 680 donkey antirabbit IgG (Life Technologies) was diluted 1:10,000 in Odyssey blocking buffer (Li-Cor Biosciences) with 0.1% Tween-20, and applied for 30 min. The blot was then imaged on an Odyssey infrared imaging system (Li-Cor Biosciences) in the 700-nm channel.
Motility Assays.
Motility assays were performed as previously described (32). Motility was evaluated by measuring the motility diameters. Experiments were performed 5 times.
RNA Extraction and qPCR.
RNA was extracted using the RNeasy kit (Qiagen), DNase-treated, and reverse transcribed using Turbo DNase I (Ambion) and SuperScript II Reverse Transcriptase (Invitrogen). For qseB qPCR, WT UTI89, UTI89∆qseC and UTI89∆qseC∆pmrB cDNA was generated from RNA samples of cells grown statically at 37 °C for 18 h in LB. For gfp qPCR, samples for RNA extraction and reverse transcription were obtained from cells grown statically at 37 °C for 18 h in N-minimal media with and without 100 μM FeCl3. Quantitative real-time PCR was executed in triplicate on an ABI StepOne Plus Real-Time PCR machine, using multiplexed TaqMan minor groove binder chemistry. Two dilutions of cDNA (25 ng and 6.25 ng) were analyzed per isolate for each experimental condition. Abundance of gfp (probe: 5′6FAM-ACGTGCTGAAGTCAAG3′) and qseB transcripts (probe: 5′6FAM-TGCGCGAATGGCGA-3′) in UTI89 isogenic mutant strains was calculated using the ΔΔCT method, with each transcript signal normalized to the abundance of the rrsH (probe: 5′VIC-CGTTAATCGGAATTACTG3′) internal control and comparison with the normalized transcript levels of WT UTI89. Abundance of pmrB transcripts (probe: 5′6FAM-ACGCCCATTGCCA) was calculated as above but normalized to the abundance of the gyrB internal control (probe: 5′VIC-ACGAACTGCTGGCGGA). DNase-treated RNA samples not subjected to reverse transcription were analyzed by qPCR in parallel as a negative control.
Phosphotransfer Assays.
QseB purification, generation of QseC- and PmrB-enriched membranes, and execution of phosphotransfer assays were performed as described in ref. 9. Band intensities corresponding to QseB∼P over time were quantified using ImageJ software and normalized to QseB∼P at time = 0. All experiments were repeated at least three times.
Electromobility Shift Assays.
PmrA was cloned into pBAD/myc-His A (Invitrogen), and expression was induced with 0.1% arabinose. PmrA was affinity-purified using a HIS-Select nickel affinity column (Sigma), followed by anion exchange chromatography through a Hi Trap Q HP column (GE Healthcare). PCR-generated amplicons for yibD promoter (109 bp), qseBC promoter (105 bp), and pmrB (104 bp) were P32 end-labeled. Varying amounts of PmrA at 10 20, 30, 50, 100, and 150 pmol were incubated with 0.5 μL of labeled DNA in 20-μL final volume for 20 min at room temperature. The binding buffer used was 20 mM Tri-HCl pH = 7.4, 5 mM MgCl2, 5 mM KCl, 10% glycerol 25 micrograms/ml BSA.
Supplementary Material
Acknowledgments
We thank Dr. Eduardo Groisman and Dr. Wonsik Yeo for provision of reagents for phosphatase assays, Dr. Christina Stallings and Leslie Weiss for providing reagents and laboratory space for the phosphotransfer assays, and Dr. Timothy L. Cover for providing the AB Step One Plus platform for qPCR. This work was supported by National Institutes of Health Grants P50 DK64540 and R01 AI48689 (to S.J.H.) and APS Development Fund 1-040520-9211 (to M.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315320110/-/DCSupplemental.
References
- 1.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goulian M. Two-component signaling circuit structure and properties. Curr Opin Microbiol. 2010;13(2):184–189. doi: 10.1016/j.mib.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 5.Procaccini A, Lunt B, Szurmant H, Hwa T, Weigt M. Dissecting the specificity of protein-protein interaction in bacterial two-component signaling: Orphans and crosstalks. PLoS ONE. 2011;6(5):e19729. doi: 10.1371/journal.pone.0019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bearson BL, Bearson SM. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb Pathog. 2008;44(4):271–278. doi: 10.1016/j.micpath.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Bearson BL, Bearson SM, Lee IS, Brunelle BW. The Salmonella enterica serovar Typhimurium QseB response regulator negatively regulates bacterial motility and swine colonization in the absence of the QseC sensor kinase. Microb Pathog. 2010;48(6):214–219. doi: 10.1016/j.micpath.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Hadjifrangiskou M, et al. A central metabolic circuit controlled by QseC in pathogenic Escherichia coli. Mol Microbiol. 2011;80(6):1516–1529. doi: 10.1111/j.1365-2958.2011.07660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kostakioti M, Hadjifrangiskou M, Pinkner JS, Hultgren SJ. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol Microbiol. 2009;73(6):1020–1031. doi: 10.1111/j.1365-2958.2009.06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kostakioti M, et al. Distinguishing the contribution of type 1 pili from that of other QseB-misregulated factors when QseC is absent during urinary tract infection. Infect Immun. 2012;80(8):2826–2834. doi: 10.1128/IAI.00283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Justice SS, et al. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci USA. 2004;101(5):1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamayo R, Prouty AM, Gunn JS. Identification and functional analysis of Salmonella enterica serovar Typhimurium PmrA-regulated genes. FEMS Immunol Med Microbiol. 2005;43(2):249–258. doi: 10.1016/j.femsim.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Wösten MM, Groisman EA. Molecular characterization of the PmrA regulon. J Biol Chem. 1999;274(38):27185–27190. doi: 10.1074/jbc.274.38.27185. [DOI] [PubMed] [Google Scholar]
- 15.Kato A, Latifi T, Groisman EA. Closing the loop: The PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc Natl Acad Sci USA. 2003;100(8):4706–4711. doi: 10.1073/pnas.0836837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke MB, Sperandio V. Transcriptional autoregulation by quorum sensing Escherichia coli regulators B and C (QseBC) in enterohaemorrhagic E. coli (EHEC) Mol Microbiol. 2005;58(2):441–455. doi: 10.1111/j.1365-2958.2005.04819.x. [DOI] [PubMed] [Google Scholar]
- 17.Tamayo R, Ryan SS, McCoy AJ, Gunn JS. Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar Typhimurium. Infect Immun. 2002;70(12):6770–6778. doi: 10.1128/IAI.70.12.6770-6778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wösten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. A signal transduction system that responds to extracellular iron. Cell. 2000;103(1):113–125. doi: 10.1016/s0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 19.Shin D, Lee EJ, Huang H, Groisman EA. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science. 2006;314(5805):1607–1609. doi: 10.1126/science.1134930. [DOI] [PubMed] [Google Scholar]
- 20.Yeo WS, et al. Intrinsic negative feedback governs activation surge in two-component regulatory systems. Mol Cell. 2012;45(3):409–421. doi: 10.1016/j.molcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russo FD, Silhavy TJ. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol. 1991;222(3):567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 22.Russo FD, Silhavy TJ. The essential tension: Opposed reactions in bacterial two-component regulatory systems. Trends Microbiol. 1993;1(8):306–310. doi: 10.1016/0966-842x(93)90007-e. [DOI] [PubMed] [Google Scholar]
- 23.Siryaporn A, Goulian M. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol Microbiol. 2008;70(2):494–506. doi: 10.1111/j.1365-2958.2008.06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amemura M, Makino K, Shinagawa H, Nakata A. Cross talk to the phosphate regulon of Escherichia coli by PhoM protein: PhoM is a histidine protein kinase and catalyzes phosphorylation of PhoB and PhoM-open reading frame 2. J Bacteriol. 1990;172(11):6300–6307. doi: 10.1128/jb.172.11.6300-6307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wanner BL. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol. 1992;174(7):2053–2058. doi: 10.1128/jb.174.7.2053-2058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wanner BL, Wilmes MR, Young DC. Control of bacterial alkaline phosphatase synthesis and variation in an Escherichia coli K-12 phoR mutant by adenyl cyclase, the cyclic AMP receptor protein, and the phoM operon. J Bacteriol. 1988;170(3):1092–1102. doi: 10.1128/jb.170.3.1092-1102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skerker JM, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133(6):1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadjifrangiskou M, et al. Transposon mutagenesis identifies uropathogenic Escherichia coli biofilm factors. J Bacteriol. 2012;194(22):6195–6205. doi: 10.1128/JB.01012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ducey TF, Carson MB, Orvis J, Stintzi AP, Dyer DW. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J Bacteriol. 2005;187(14):4865–4874. doi: 10.1128/JB.187.14.4865-4874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anriany Y, Sahu SN, Wessels KR, McCann LM, Joseph SW. Alteration of the rugose phenotype in waaG and ddhC mutants of Salmonella enterica serovar Typhimurium DT104 is associated with inverse production of curli and cellulose. Appl Environ Microbiol. 2006;72(7):5002–5012. doi: 10.1128/AEM.02868-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol. 2003;4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright KJ, Seed PC, Hultgren SJ. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun. 2005;73(11):7657–7668. doi: 10.1128/IAI.73.11.7657-7668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.