Significance
CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1) is a well-conserved multifunctional protein in both plants and animals. Depending on the context, COP1 can have distinct roles, such as an oncogene or a tumor suppressor in mammalian cells. In light regulation of plant development, COP1 is a central repressor under far-red and visible light whereas it is a positive regulator under UV-B. However, how COP1 positively regulates UV-B signaling remains largely unknown. In this study, we demonstrate that the UV-B–induced reorganization of COP1 complexes achieves a functional switch of COP1 from repressing to promoting photomorphogenesis.
Keywords: light signaling, protein complex, posttranscriptional regulation
Abstract
The evolutionarily conserved CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1) is a RING and WD40 protein that functions as a substrate receptor of CULLIN4–DAMAGED DNA BINDING PROTEIN 1 (CUL4–DDB1)–based E3 ubiquitin ligases in both plants and animals. In Arabidopsis, COP1 is a central repressor of photomorphogenesis in the form of COP1–SUPPRESSOR OF PHYA (SPA) complex(es). CUL4–DDB1–COP1–SPA suppresses the photomorphogenic program by targeting the transcription factor ELONGATED HYPOCOTYL 5 for degradation. Intriguingly, under photomorphogenic UV-B light, COP1 reverses its repressive role and promotes photomorphogenesis. However, the mechanism by which COP1 is functionally switched is still obscure. Here, we demonstrate that UV-B triggers the physical and functional disassociation of the COP1–SPA core complex(es) from CUL4–DDB1 and the formation of a unique complex(es) containing the UV-B receptor UV RESISTANCE LOCUS 8 (UVR8). The establishment of this UV-B–dependent COP1 complex(es) is associated with its positive modulation of ELONGATED HYPOCOTYL 5 stability and activity, which sheds light on the mechanism of COP1’s promotive action in UV-B–induced photomorphogenesis.
In response to light and darkness, plant seedlings establish light-grown and dark-grown phenotypes via a series of developmental changes, termed photomorphogenesis and skotomorphogenesis, respectively. CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) is a known RING E3 ubiquitin ligase that has been evolutionally conserved from plants to humans (1, 2). It was originally identified by genetic screens for seedlings of Arabidopsis thaliana that exhibit constitutive photomorphogenesis in darkness (1, 3), as a key member of the pleiotropic CONSTITUTIVE PHOTOMORPHOGENIC/DE-ETIOLATED/FUSCA (COP/DET/FUS) gene family. These COP/DET/FUS proteins biochemically contribute to three entities: the COP1-SUPRESSOR OF PHYA (SPA) complex(es), the COP9 signalosome (CSN), and the COP10–DET1–Damaged DNA Binding Protein 1 (DDB1) (CDD) complex. COP1–SPA, independent of CDD but in concert with CULLIN4–DDB1 (CUL4–DDB1), targets photomorphogenesis promoting transcription factors including ELONGATED HYPOCOTYL 5 (HY5) for the ubiquitin–proteasome system-mediated degradation, so as to repress the traditional photomorphogenesis triggered by far-red and visible light (4, 5).
Intriguingly, in contrast to their antagonistic roles in the traditional photomorphogenesis, COP1 and HY5 both take positive parts in low-fluence and long-wavelength UV-B–induced photomorphogenesis. This response is initiated by the UV-B receptor UV RESISTANCE LOCUS 8 (UVR8) which absorbs UV-B through its internal chromophore tryptophan residues (6, 7). UVR8 then monomerizes to interact with the UV-B–inducible protein COP1 for downstream signaling (8–10). The physical manifestations of this process include hypocotyl shortening, anthocyanin accumulation, and tolerance against damaging UV-B. The loss of either COP1 or HY5 has previously been shown to result in decreased activation of UV-B–responsive genes, impaired photomorphogenesis, and defective UV-B acclimation (11, 12). Both COP1 and HY5 localize in the nucleus under UV-B (11). The UV-B–induced HY5 expression is largely dependent on UVR8 and COP1 (8, 11, 13), and HY5 undertakes a positive feedback on COP1 by targeting the COP1 promoter for the UV-B–mediated activation (9). Little is known, however, regarding how COP1 takes promotive action, particularly the changes of its biochemical nature toward HY5 in this UV-B–specific signaling.
Here we demonstrate that in response to photomorphogenic UV-B, the COP1–SPA core complex(es) functionally disassociates with CUL4–DDB1 and recruits UVR8 to establish UVR8– COP1–SPA complex(es). This UV-B–induced machinery is associated with the positive role of COP1 toward HY5 in facilitating HY5 stability and activity. The functional switch from degrading to stabilizing HY5 enables COP1 to act as a positive regulator in UV-B–induced photomorphogenesis.
Results and Discussion
COP1 and CUL4 Display Functional and Physical Disassociation in UV-B–Induced Photomorphogenesis.
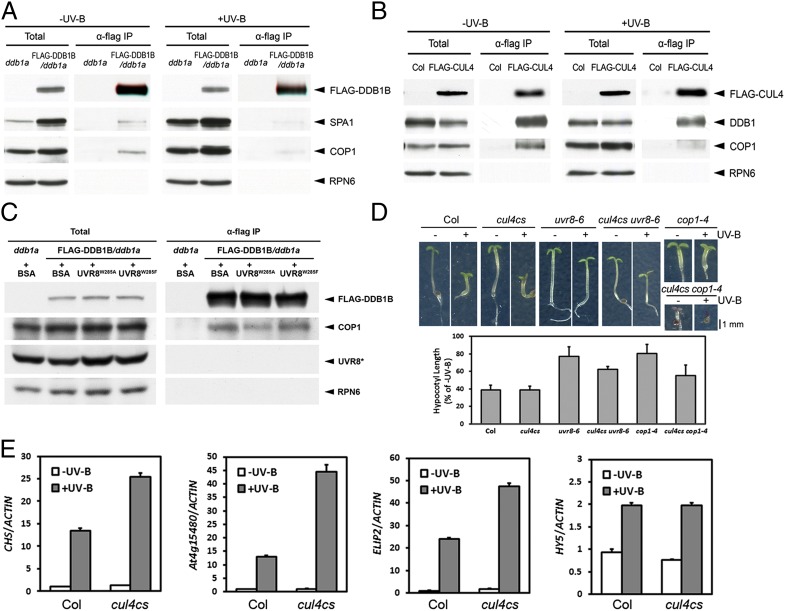
We have shown previously that as substrate receptors, COP1–SPA complexes are constituents of the CUL4–based E3 ligases to mediate the repression of photomorphogenesis in darkness (5). Therefore, we examined whether COP1 and CUL4–DDB1 still exist in the same complex under photomorphogenic UV-B. Although more COP1 and SPA1 proteins were found to accumulate in UV-B–treated seedlings, less COP1 and SPA1 proteins were coimmunoprecipitated with FLAG-DDB1B (Fig. 1A). Similarly, a smaller portion of COP1 was detected to be coimmunoprecipitated with FLAG-CUL4 in UV-B–treated seedlings (Fig. 1B). A decrease in both the transgenic and endogenous DDB1 abundance was also observed (Fig. 1 A and B). These results indicate that photomorphogenic UV-B might provoke the disassembly of the CUL4–DDB1–COP1–SPA E3 apparatus, which might be partly ascribed to the decline in the DDB1 protein level. It is also possible that UVR8 might compete with DDB1 for binding COP1, because the UV-B–stimulated monomerized UVR8 interacts with COP1 via COP1’s WD40 domain that also mediates DDB1–COP1 interaction (5). We thus examined this possibility by adding recombinant UVR8 mutant proteins into coimmunoprecipitation reactions. We found that FLAG-DDB1B coimmunoprecipitated a reduced level of COP1 protein in the presence of the at least partly monomeric UVR8W285A that could interact with COP1 (6, 7, 10). However, compared with UVR8W285A, the recombinant dimeric UVR8W285F that is unable to physically interact with COP1 (6, 7, 10), as well as BSA, displayed a relatively high level of the association between FLAG-DDB1B and COP1 (Fig. 1C). This result accordingly suggests that in response to UV-B, monomerized UVR8 might sequester COP1 from DDB1, which contributes to the disassembly of the CUL4–DDB1–COP1–SPA in addition to the declined DDB1 protein level.
Fig. 1.
COP1 and CUL4 display functional and physical disassociation in UV-B–induced photomorphogenesis. (A) Coimmunoprecipitation (co-IP) assays using 4-d-old ddb1a and FLAG-DDB1B/ddb1a seedlings grown under –UV-B and +UV-B (RPN6, the loading and unprecipitated control). (B) Co-IP assays using 4-d-old Columbia (Col) and FLAG-CUL4 seedlings grown under –UV-B and +UV-B. (C) Co-IP assays by adding BSA, UVR8W285A, and UVR8W285F in 4-d-old ddb1a and FLAG-DDB1B/ddb1a seedlings grown under –UV-B. UVR8*, the endogenous and exogenous UVR8 proteins. (D) Photomorphogenic UV-B–induced hypocotyl growth of Arabidopsis seedlings. (Upper) Phenotypes of 4-d-old seedlings of indicated genotypes grown under –UV-B and +UV-B. (Lower) Quantification of relative hypocotyl length. (E) The expression of photomorphogenic UV-B–responsive genes in 4-d-old Col and cul4cs seedlings grown under –UV-B and +UV-B.
Next we analyzed whether CUL4 might genetically work together with UVR8 or COP1 to promote UV-B–induced photomorphogenesis. In contrast to the uvr8 and cop1 mutants, the cosuppression mutant of CUL4, cul4cs (14), displayed no obvious defect in the UV-B–induced inhibition of hypocotyl elongation (Fig. 1D), suggesting that CUL4 is not essentially required for the response. Upon further examination, we found that compared with the WT counterpart, cul4cs resulted in the increased expression of UV-B–inducible genes with the exception of HY5 (Fig. 1E). Moreover, cul4cs was able to partially suppress the hyporesponsiveness to UV-B of uvr8-6 and cop1-4 in terms of hypocotyl growth (Fig. 1D). Thus, the phenotypic and molecular analysis reveals that contrary to UVR8 and COP1, CUL4 is a negative regulator in UV-B–induced photomorphogenesis.
UVR8–COP1–SPA Complexes Are UV-B– and COP1–Dependent.
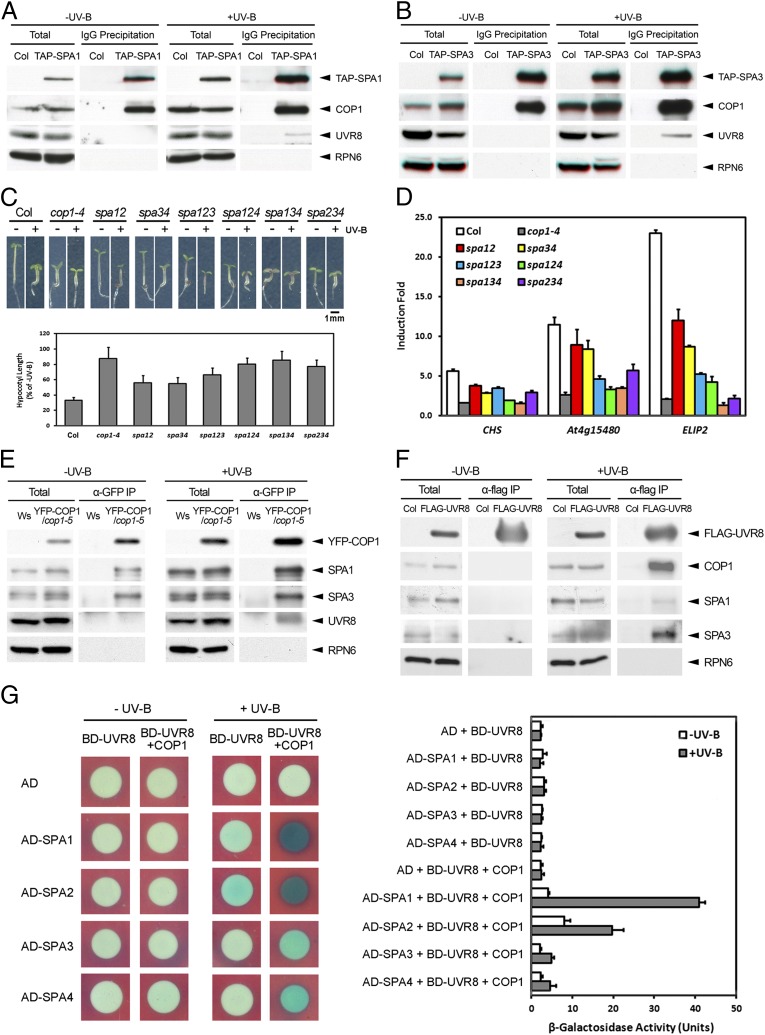
In contrast to the disassociation of CUL4–DDB1–COP1–SPA complexes, we found that TAP-SPA proteins constitutively interacted with COP1 in both −UV-B– and +UV-B–grown seedlings (Fig. 2 A and B), suggesting UV-B has little impact on the COP1–SPA core complexes. The tight association between SPA proteins and COP1 prompted us to investigate the role of SPA proteins in UV-B–induced photomorphogenesis. Because the four SPA family members function redundantly (15), we examined the spa double and triple mutants under UV-B. Similar to cop1-4, the spa mutants were less sensitive than WT to the UV-B–mediated inhibition of hypocotyl elongation (Fig. 2C). Furthermore, the UV-B–induced expression of the tested genes was also compromised in these spa mutants (Fig. 2D). These results suggest that, like COP1, the SPA proteins also act as positive regulators in this UV-B–specific signaling. Given that the COP1–SPA complexes physically and functionally disassociate from CUL4–DDB1 upon UV-B treatment, and their respective mutants share similarities in the phenotypes and the expression profiles of UV-B–responsive genes, we tested whether COP1, SPAs, and UVR8 constitute a unique UV-B–induced complex(es). Through a series of coimmunoprecipitation (co-IP) assays, we observed that COP1 and SPAs associated with UVR8 in a UV-B–dependent manner in vivo (Fig. 2 A, B, E, and F). The interaction between COP1 and UVR8 has been shown to occur via the WD40 domain of COP1 (8, 10). Although each SPA harbors a similar WD40 domain (5), the interaction indicated by β-galactosidase activity was hardly detected in each pair of SPA and UVR8 in our yeast two-hybrid assays (Fig. 2G), suggesting no direct contact between SPAs and UVR8. Based on this observation, along with the fact that the COP1 could independently interact with either SPAs (15, 16) or UVR8 (8, 10), we proposed that COP1 might bridge SPAs and UVR8. To substantiate this hypothesis, we developed a yeast three-hybrid system which consisted of SPAs fused with the activation domain (AD-SPAs), UVR8 fused with the DNA binding domain (BD-UVR8), and an untagged COP1. In the assays, the interactions between SPAs and UVR8 were only detected when COP1 was present and the UV-B irradiation was provided (Fig. 2G). These results indicate that COP1 does bridge SPAs and UVR8 in the formation of the UV-B–induced UVR8–COP1–SPA complex(es).
Fig. 2.
UVR8–COP1–SPA complexes are UV-B– and COP1–dependent. (A and B) Co-IP assays using 4-d-old Col, TAP-SPA1 (A) and TAP-SPA3 (B) seedlings grown under –UV-B and +UV-B. (C) Photomorphogenic UV-B–induced hypocotyl growth of spa mutant seedlings. (Upper) Phenotypes of 4-d-old seedlings of indicated genotypes grown under –UV-B and +UV-B. (Lower) Quantification of relative hypocotyl length. (D) The expression of photomorphogenic UV-B–responsive genes in 4-d-old Col, cop1-4, and spa mutant seedlings. (E and F) Co-IP assays using 4-d-old Wassileskija (Ws), YFP-COP1/cop1-5 (E), Col, and FLAG-UVR8 (F) seedlings grown under –UV-B and +UV-B. (G) The interaction among SPA, COP1, and UVR8 in yeast. (Left) Yeast three-hybrid assays expressing indicated proteins under –UV-B and +UV-B. (Right) Quantification of β-galactosidase activity of yeast grown under –UV-B and +UV-B for 16 h.
Stability of HY5 Requires Photomorphogenic UV-B and UVR8–COP1–SPA Complex(es).
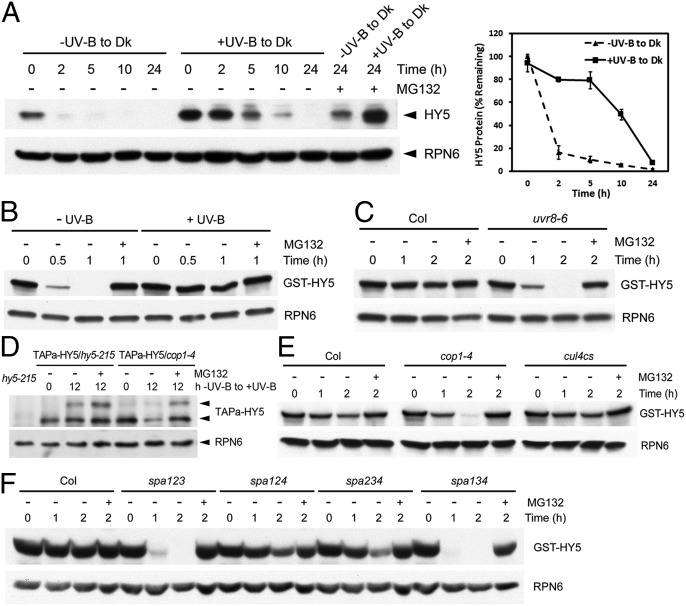
Opposite to their roles in far-red and visible light regions, COP1 and SPAs do not act as repressors in UV-B–induced photomorphogenesis. We thus examined their roles on HY5, one of their degradation targets in darkness as well as a key player in UV-B–specific responses. We noticed that in WT seedlings, the mRNA level of HY5 was elevated within 1 h of exposure to +UV-B and then fell back (Fig. S1A), and HY5 protein abundance increased continuously over the course of 12 h of UV-B treatment (Fig. S1B). To exclude the effect from transcription regulation by the native promoter of HY5, we examined a version of HY5 that was fused with an alternative tandem affinity purification tag (TAPa-HY5) and driven by the constitutive 35S promoter. Again, we detected both the long-term and rapid accumulation of TAPa-HY5 protein doublet under UV-B irradiation (Fig. S1 C and D). This suggests that HY5 is posttranscriptionally up-regulated by photomorphogenic UV-B. We next examined whether UV-B affected the stability of HY5. Using the seedlings first grown under −UV-B or +UV-B and then transferred to darkness to induce HY5 degradation, we detected more HY5 protein in +UV-B–grown seedlings than in −UV-B–grown counterparts at each time point of 10-h darkness treatment. The analysis of the HY5 degradation kinetics also showed that the decrease in the HY5 protein level of +UV-B-to-darkness–treated seedlings was much slower than that of −UV-B-to-darkness–treated counterparts, with the degradation half-life of 1.20 ± 0.08 h and 9.70 ± 1.65 h, respectively (Fig. 3A). The difference in the rates cannot be attributed to transcription as the HY5 mRNA levels were similar in both groups (Fig. S2). These results suggest that UV-B enhances the stability of HY5. Further, the degradation of the recombinant GST-fused HY5 (GST-HY5) was reconstituted in a cell-free assay in the −UV-B–grown WT seedlings, whereas it was impaired in the +UV-B–grown counterparts (Fig. 3B). As expected, the null mutation of UVR8 also led to an obvious faster turnover of GST-HY5 (Fig. 3C), which supports the notion that HY5 is stabilized by UV-B signaling.
Fig. 3.
The stability of HY5 requires photomorphogenic UV-B and UVR8–COP1–SPA complex(es). (A) The effect of photomorphogenic UV-B on HY5 stability. (Left) Immunoblot analysis using Col seedlings grown under –UV-B or +UV-B for 4 d and then transferred to darkness for the indicated time treated with or without 50 µM MG132. (Right) Percentage of remaining HY5 protein normalized to the loading control RPN6. (B) Cell-free degradation of recombinant GST-HY5 in 4-d-old Col seedlings grown under –UV-B and +UV-B. (C) Cell-free degradation of recombinant GST-HY5 in 4-d-old +UV-B–grown Col and uvr8-6 seedlings. (D) The effect of COP1 on HY5 stability under photomorphogenic UV-B. Immunoblot analysis using TAPa-HY5/hy5-215 and TAPa-HY5/cop1-4 seedlings grown under –UV-B for 4 d and then transferred to +UV-B for the indicated time treated with or without 50 µM MG132. (E and F) Cell-free degradation of recombinant GST-HY5 in 4-d-old +UV-B–grown Col, cop1-4, cul4cs (E), and spa (F) seedlings.
In contrast to the fact that COP1 mediates HY5 degradation in darkness (4), a range of evidence indicates that COP1 positively modulates HY5 function under UV-B. First, we found that the UV-B–induced accumulation of endogenous HY5 protein was abolished in the cop1-4 mutant (Fig. S3A) but enhanced in cul4cs (Fig. S3B). Second, the protein level of TAPa-HY5 in cop1-4 failed to rise after 12 h of UV-B irradiation, but the accumulation of TAPa-HY5 protein can be restored by the treatment of proteasome inhibitor MG132 (Fig. 3D). Third, GST-HY5 exhibited a higher rate of degradation in UV-B–treated cop1-4 than WT and cul4cs (Fig. 3E). Collectively, these results not only suggest that in the absence of COP1, HY5 is susceptible to the ubiquitin-proteasome–mediated degradation, but also indicate that there might be an alternative E3 ubiquitin ligase responsible for HY5 degradation induced by photomorphogenic UV-B. Like cop1-4 and uvr8-6, the spa triple mutants all exhibited a defective UV-B–induced increase in HY5 abundance (Fig. S3 C and D) and reduced stability of GST-HY5 (Fig. 3 C and F). These data suggest that the UV-B–induced UVR8–COP1–SPA complex(es) plays an active role in the stabilization of HY5, and each component of this complex(es) is required for the promotion of HY5 stability.
COP1 and UVR8 in the Photomorphogenic UV-B Signaling Are Important for HY5 Activity.
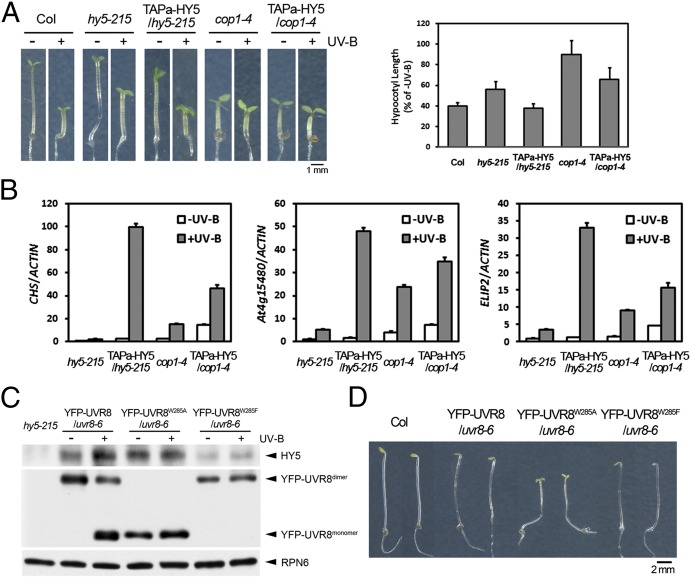
Based on the above biochemical evidence, we examined genetically whether the UVR8–COP1–SPA complex(es) positively modulated HY5 function under photomorphogenic UV-B. First, we examined HY5 activity in the absence of COP1 in vivo. The overexpression of TAPa-HY5 in cop1-4 failed both to accomplish hypocotyl shortening (Fig. 4A) and to fully induce the expression of several putative HY5 target genes (11) under UV-B (Fig. 4B). Second, to further validate our hypothesis that HY5 stabilization is mediated by UV-B signaling, e.g., UV-B–induced monomerized UVR8 and its interaction with the core COP1–SPA complex(es), we generated transgenic lines expressing yellow fluorescent protein (YFP)-fused UVR8WT, UVR8W285A, and UVR8W285F in the uvr8-6 mutant background. The transgenic seedlings expressing UVR8W285A exhibited constitutively monomerized UVR8 and relatively high HY5 protein levels under both –UV-B and +UV-B. In contrast, UVR8W285F led to constitutively dimerized UVR8 and relatively low HY5 protein levels in both –UV-B– and +UV-B–grown seedlings (Fig. 4C). The difference in the HY5 protein levels did not show a positive correlation with that in the HY5 mRNA levels among these three lines (Fig. S4). In addition, we observed that YFP-UVR8W285A/uvr8-6, rather than YFP-UVR8W285F/uvr8-6, displayed constitutive photomorphogenesis with open cotyledons in darkness (Fig. 4D). These results consistently support a conclusion that the activated photomorphogenic UV-B signaling sustains the formation of UVR8–COP1–SPA complex(es) and in turn constitutively facilitates HY5 stability and activity in darkness.
Fig. 4.
COP1 and UVR8 in the photomorphogenic UV-B signaling are important for HY5 activity. (A) Photomorphogenic UV-B–induced hypocotyl growth of HY5 overexpressors in cop1-4 mutant. (Left) Phenotype of 4-d-old seedlings of indicated genotypes grown under –UV-B and +UV-B. (Right) Quantification of relative hypocotyl length. (B) The expression of photomorphogenic UV-B–responsive genes in the seedlings shown in A. (C) The effects of UVR8 point mutations on UVR8 and HY5 proteins. Immunoblot analysis using 4-d-old YFP-UVR8/uvr8-6, YFP-UVR8W285A/uvr8-6, and YFP-UVR8W285F/uvr8-6 seedlings grown under –UV-B and +UV-B. (D) Phenotypes of 4-d-old dark-grown Col, YFP-UVR8/uvr8-6, YFP-UVR8W285A/uvr8-6, and YFP-UVR8W285F/uvr8-6 seedlings.
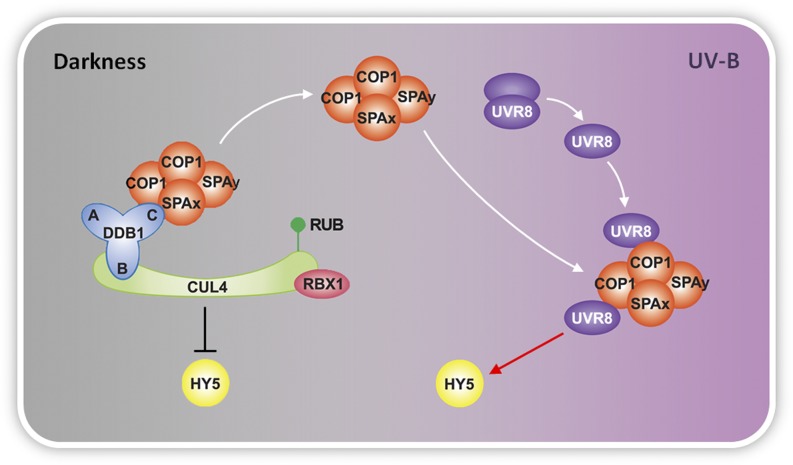
Taken together, our results demonstrate that in response to photomorphogenic UV-B, the CUL4–based COP1–SPA E3 apparatus tends to diminish, while the UVR8–COP1–SPA complex(es) takes over. Such a complex conversion is associated with a promotion of HY5 stability and activity (Fig. 5). In photomorphogenesis triggered by far-red and visible light, the association of phytochromes and cryptochromes with COP1 and the exclusion of COP1 from the nucleus mediate rapid and slow alterations of the repressive role of COP1, respectively (1, 2). Nevertheless, in UV-B–induced photomorphogenesis that is independent of phytochromes and cryptochromes (9, 11), the colocalization of COP1 and UVR8 in the nucleus (8) enables the rapid formation of the UVR8–containing COP1 complex(es) to fulfill the promotive activity of COP1. Therefore, UVR8 differs from phytochromes and cryptochromes in the functional coordination with COP1. Furthermore, our study also suggests that the COP1–SPA core complex(es) can interact with distinct light-specific components as the light environment varies. The reorganization from the CUL4–DDB1–COP1–SPA E3 apparatus to UVR8–COP1–SPA complex(es) upon UV-B irradiation achieves a functional switch of COP1 from repressing to promoting photomorphogenesis. Therefore, this mechanism also serves as an example that limited protein-encoding capacity is elaborately used to adapt diversified functional requirements in an organism.
Fig. 5.
A model for the function of COP1 complexes toward HY5 in darkness and under photomorphogenic UV-B. White arrows indicate the organization of protein complexes. Black bars indicate negative regulation. The red arrow indicates positive regulation.
Materials and Methods
Plant Materials and Growth Conditions.
The WT A. thaliana used in this study are of the Columbia (Col) and Wassileskija (Ws) ecotypes. Some of the mutants and transgenic lines used in this study were described previously: cop1-4 (17); cul4cs and cul4cs cop1-4 (14); ddb1a and FLAG-DDB1B/ddb1a (18); FLAG-CUL4 (14); TAP-SPA1 (16); TAP-SPA3 (15); spa12, spa123, spa124, and spa134 (19); spa34 (20); spa234 (21); uvr8-6 (8); hy5-215 (22); and TAPa-HY5/hy5-215 (23).
For the YFP-COP1/cop1-5 transgenic line, the full-length COP1 open reading frame (ORF) was cloned into the pENTR/d-TOPO vector (Invitrogen) and introduced into the plant binary vector pEarleyGate 104 (24) under the 35S promoter using Gateway LR Clonase enzyme mix (Invitrogen). For the FLAG-UVR8 transgenic line, a KpnI/SacI fragment containing full-length UVR8 ORF was cloned into the plant binary vector pF3PZPY122 (25). Then a BamHI/SacI fragment with the inserted DNA was subcloned into the plant binary vector pJIM 19 (KAN) under the 35S promoter. For the YFP-UVR8/uvr8-6 transgenic line, the full-length UVR8 ORF was cloned into the pENTR/d-TOPO vector (Invitrogen) and introduced into the plant binary vector pEarleyGate 104 using Gateway LR Clonase enzyme mix (Invitrogen). Then two restriction endonuclease sites, AflII and AatII, were inserted flanking the 35S promoter in the resulted construct, and the 35S promoter was subsequently replaced by an AflII/AatII fragment containing the native UVR8 promoter (26). The vectors for the YFP-UVR8W285A/uvr8-6 and YFP-UVR8W285F/uvr8-6 transgenic lines were generated using the QuikChange site-directed mutagenesis kit (Stratagene). The primers are listed in Table S1. These transgenic lines were prepared using the floral-dipping method (27).
The Arabidopsis materials were grown as described previously (9). The seeds were surface-sterilized and sown on solid Murashige and Skoog medium supplemented with 1% sucrose for biochemical assays or with 0.3% sucrose for phenotypic analysis and cold-treated at 4 °C for 4 d. Then for photomorphogenic UV-B treatment, seedlings were grown at 22 °C under continuous white light (3 μmol·m−2·s−1, measured by LI-250 Light Meter, LI-COR Biosciences) supplemented with Philips TL20W/01RS narrowband UV-B tubes (1.5 μmol·m−2·s−1, measured by TN-340 UV-B Light Meter, TAINA) under a 350-nm cutoff (half-maximal transmission at 350 nm) filter ZUL0350 (−UV-B; Asahi Spectra) or a 300-nm cutoff (half-maximal transmission at 300 nm) filter ZUL0300 (+UV-B; Asahi Spectra).
Hypocotyl Measurement.
Relative hypocotyl length was measured as previously described (9). For each line grown under −UV-B or +UV-B for 4 d, hypocotyl length was analyzed in three biological replicates. In each replicate, at least 30 Arabidopsis seedlings were measured. The relative hypocotyl length was presented as the percentage of the hypocotyl length under +UV-B with respect to that under −UV-B (percent of −UV-B).
Quantitative Real-Time PCR.
Total RNA was extracted from 4-d-old Arabidopsis seedlings grown under −UV-B or +UV-B using the RNeasy plant mini kit (Qiagen). Reverse transcription was performed using the SuperScript II first-strand cDNA synthesis system (Invitrogen) according to the manufacturer’s instructions. Real-time qPCR analysis was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) with a Bio-Rad CFX96 real-time PCR detection system. Each experiment was repeated with three independent samples, and RT-PCR reactions were performed in three technical replicates for each sample. The primers are listed in Table S1.
Coimmunoprecipitation Assays and Immunoblot Analysis.
1 mg of total proteins was extracted from 4-d-old Arabidopsis seedlings in protein extraction buffer containing 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% (vol/vol) glycerol, 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× complete protease inhibitor mixture (Roche). The extracts were incubated with 25 µL anti-FLAG–conjugated Agarose (Sigma-Aldrich), 30 µL IgG Sepharose 6 Fast Flow (General Electric Healthcare), or 8 µL anti-GFP antibodies (Invitrogen) coupled with 25 µL Dynabeads Protein G (Invitrogen) for 3 h at 4 °C under the same condition (−UV-B or +UV-B) as where the seedlings were grown. Then the agarose/Sepharose/dynabeads was washed three times by protein extraction buffer. Next the precipitates were eluted into 100 mM Glycine (pH 2.5) and 100 mM NaCl, immediately neutralized by 2 M Tris⋅HCl (pH 9.0) and 100 mM NaCl, and finally concentrated using Strataresin (Stratagene) before immunoblot analysis. For the coimmunoprecipitation assays added with exogenous proteins, 200 ng of BSA, UVR8W285A, or UVR8W285F was added into 1 mg of plant proteins before the incubation with 25 µL anti-FLAG–conjugated Agarose. Recombinant UVR8W285A and UVR8W285F proteins (6) are gifts from Yigong Shi (Tsinghua University, China).
For the test of UVR8 dimer/monomer status, total proteins were extracted from 4-d-old Arabidopsis seedlings grown under –UV-B or +UV-B in protein extraction buffer containing 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Tween 20, 1 mM PMSF, and 1× complete protease inhibitor mixture (Roche). The cell extracts were then kept on ice under exactly the same condition (–UV-B or +UV-B) as where the seedlings were grown for 30 min. Added with 4× loading buffer containing 250 mM Tris⋅HCl (pH 6.8), 2% SDS, 20% β-mercaptoethanol, 40% glycerol, and 0.5% bromophenol blue, the samples were subjected to immunoblot analysis without boiling.
Primary antibodies used in this study were anti-FLAG (Sigma-Aldrich), anti-SPA1, and anti-SPA3 (15); anti-COP1 and anti-RPN6 (14); anti-Myc (Sigma-Aldrich), anti-GFP (Invitrogen),and anti-HY5 (4); and anti-GST (Sigma-Aldrich) antibodies. Anti-UVR8 antibodies were raised as previously described (8). The peptide CGDISVPQTDVKRVRI was synthesized to generate the polyclonal antibodies in a rabbit. Three biological replicates were used for the quantification of the HY5 protein levels in –UV-B-to-darkness and +UV-B-to-darkness transition assays by ImageJ (http://rsb.info.nih.gov/ij/).
Yeast Three-Hybrid Assay.
The LexA-based yeast two-hybrid system (Clontech) was modified by introducing a third plasmid expressing COP1. To express COP1 without any domain fusion in yeast, a KpnI/EcoRI fragment containing full-length COP1 ORF was cloned into pGAD-T7 (Clontech). The respective combinations were cotransformed into the yeast strain EGY48 (Clontech) containing the reporter plasmid p8op::LacZ. Transformants were selected on SD/−His/−Trp/−Leu/−Ura plates and then transferred to SD/Gal/Raf/−His/−Trp/−Leu/−Ura plates supplemented with X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and BU salt under –UV-B (3 μmol·m−2·s−1 of white light) and +UV-B (3 μmol·m−2·s−1 of white light and 1.5 μmol·m−2·s−1 of UV-B) for blue color development. Quantitative β-galactosidase activity was assayed using four biological replicates with the yeast β-galactosidase assay kit (Thermo Scientific).
Cell-Free Degradation Assay.
Cell-free degradation assay was performed as previously described (4). Total proteins were extracted from 4-d-old Arabidopsis seedlings grown under −UV-B or +UV-B in degradation buffer containing 25 mM Tris⋅HCl (pH 7.5), 10 mM NaCl, 10 mM MgCl2, 1 mM PMSF, 5 mM DTT, and 10 mM ATP. In each assay, 100 ng of recombinant GST-HY5 protein was incubated in 500 µg of total proteins at 22 °C under the same condition (−UV-B or +UV-B) as where the seedlings were grown, and the aliquots were harvested at different time points. The proteasome inhibitor MG132 (Calbiochem) was selectively added as indicated.
Supplementary Material
Acknowledgments
We thank Haodong Chen and Abigail Coplin for their suggestions on the manuscript and Yang Yang for her technical assistance. This work is supported by National Institute of Health Grant GM47850 (to X.W.D.), and in part by the Ministry of Science and Technology of China (2012CB910900); the State Key Laboratory of Protein and Plant Gene Research at Peking University; a grant from the Next-Generation BioGreen 21 Program (PJ00901003), Rural Development Administration, Republic of Korea; China Postdoctoral Science Foundation Grants 2013M530009 (to X.H.), 2012M510266 (to X.O.), 2013T60032 (to X.O.); and Major Project of Education Department in Sichuan 13ZA0253 (to X.O.). X.H. and X.O. were Monsanto Fellows during their visit to Yale University, and they are supported by the Postdoctoral Fellowship at Center for Life Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316622110/-/DCSupplemental.
References
- 1.Yi C, Deng XW. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 2005;15(11):618–625. doi: 10.1016/j.tcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17(10):584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Schwechheimer C, Deng XW. The COP/DET/FUS proteins-regulators of eukaryotic growth and development. Semin Cell Dev Biol. 2000;11(6):495–503. doi: 10.1006/scdb.2000.0203. [DOI] [PubMed] [Google Scholar]
- 4.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405(6785):462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, et al. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22(1):108–123. doi: 10.1105/tpc.109.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu D, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484(7393):214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 7.Christie JM, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335(6075):1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Favory JJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28(5):591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X, et al. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell. 2012;24(11):4590–4606. doi: 10.1105/tpc.112.103994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizzini L, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332(6025):103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 11.Oravecz A, et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006;18(8):1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown BA, et al. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA. 2005;102(50):18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulm R, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA. 2004;101(5):1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, et al. Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell. 2006;18(8):1991–2004. doi: 10.1105/tpc.106.043224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu D, et al. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell. 2008;20(9):2307–2323. doi: 10.1105/tpc.107.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saijo Y, et al. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003;17(21):2642–2647. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNellis TW, von Arnim AG, Deng XW. Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: Evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell. 1994;6(10):1391–1400. doi: 10.1105/tpc.6.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, et al. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell. 2008;20(1):152–167. doi: 10.1105/tpc.107.055418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laubinger S, Fittinghoff K, Hoecker U. The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in arabidopsis. Plant Cell. 2004;16(9):2293–2306. doi: 10.1105/tpc.104.024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laubinger S, Hoecker U. The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J. 2003;35(3):373–385. doi: 10.1046/j.1365-313x.2003.01813.x. [DOI] [PubMed] [Google Scholar]
- 21.Fittinghoff K, et al. Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J. 2006;47(4):577–590. doi: 10.1111/j.1365-313X.2006.02812.x. [DOI] [PubMed] [Google Scholar]
- 22.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11(22):2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubio V, et al. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 2005;41(5):767–778. doi: 10.1111/j.1365-313X.2004.02328.x. [DOI] [PubMed] [Google Scholar]
- 24.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45(4):616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 25.Feng S, et al. The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell. 2003;15(5):1083–1094. doi: 10.1105/tpc.010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiserli E, Jenkins GI. UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell. 2007;19(8):2662–2673. doi: 10.1105/tpc.107.053330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weigel D, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122(4):1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.