Significance
Natural killer (NK) cells are lymphocytes that kill virus-infected and tumor cells as well as activate adaptive immunity through cytokine production. Although several transcription factors have been identified as having roles in NK cell development, little is known about the transcriptional control of these transcription factors and the epigenetic control of NK cell development. In this study we identified the importance of MYSM1, a histone H2A deubiquitinase, for NK cell maturation. We found that MYSM1-mediated epigenetic alterations control the expression of an important NK cell transcription factor, inhibitor of DNA-binding protein (ID2). This study unfolds the regulatory events of key transcription factors of NK cell development such as nuclear factor IL-3 and ID2 through an epigenetic mechanism.
Keywords: NFIL3, histone deubiquitination
Abstract
Histone modifications play critical roles in regulating immunity; however, little is known about the epigenetic control of natural killer (NK) cell development. Here, we found that NK cell development is severely impaired in mice deficient in the histone H2A deubiquitinase MYSM1. We demonstrated that MYSM1 is required for NK cell maturation but not for NK lineage specification and commitment. We also found that MYSM1 intrinsically controls this NK cell maturation. Mechanistic studies revealed that the expression of transcription factor, inhibitor of DNA-binding protein (ID2), a critical factor for NK cell development, is impaired in Mysm1−/− NK cells. MYSM1 interacts with nuclear factor IL-3 (NFIL3, also known as E4BP4), a critical factor for mouse NK cell development, and the recruitment of nuclear factor Il-3 to the ID2 locus is dependent on MYSM1. Further, we observed that MYSM1 is involved in maintaining an active chromatin at the ID2 locus to promote NK cell development. Hence this study demonstrates the critical epigenetic regulation of NK cell development by the histone H2A deubiquitinase MYSM1 through the transcriptional control of transcription factors important for NK cell development.
Natural killer (NK) cells are lymphocytes that play critical roles in adaptive and innate immune responses. They can recognize virus-infected and cancerous cells through their multiple surface-expressed activatory and inhibitory receptors and lyse them through a cytotoxic effect (1). Natural killing occurs through the release of granzyme- and perforin-containing cytoplasmic granules through a metabolically active process. Not only is the NK response in the innate immune system rapid; it also produces a distinct set of cytokines such as IFN- γ, TNF-α, IL-10, 1L-5, and 1L-13 or chemokines such as MIP-1α and -β and RANTES, which can further elicit an adaptive immune response (2). Together, these functional activities of NK cells help eliminate the susceptible targets in multiple ways and help amplify the inflammatory response.
NK cells develop from the common lymphoid progenitors (CLPs), as do B cells and T cells. The primary site of NK cell development is bone marrow, although some evidence showing the presence of immature NK cells in the liver and thymus suggests that NK cells also may develop at these sites (2). NK cell development in the bone marrow is defined primarily by the stepwise expression of CD122 (IL-2 and IL-15 receptor-β chain), NK1.1 (activating NK receptor), and DX5 (integrin α2) (3, 4). CD122+NK1.1−DX5−Lin−cells originally were described as NK progenitors (NKPs), but recently it has been shown that this population also exhibits a T-cell potential in a notch-dependent manner both in vivo and in vitro (5). For convenience, CD122+NK1.1−DX5−Lin−cells still are referred to as NKPs in this study. Based on a refined analysis of markers expressed on these progenitors [CD27, IL-7 receptor (IL-7R) and CD244], NKPs enriched for NK cell potential known as refined NKPs (rNKPs) and an intermediate stage between NKPs and CLPs known as pre-NKPs have been identified recently (6). Acquisition of NK1.1 occurs at the immature NK (iNK) cell stage, at a time when multiple NK receptors including NKp46, a preferential marker expressed in NK cells and conserved in mammals, begin to express (7, 8). This onset of NKp46 expression marks the irreversible engagement of cells into the NK cell lineage, because NK1.1+NKp46 − cells still can give rise to both NK and T cells, but NK1.1+NKp46+ cells cannot (5, 8). Cells then transition into mature NK cells (mNK) with the sequential acquisition of DX5, CD11B, and KLRG1 expression and down-regulation of c-KIT, CD27, and CD51 expression (3, 4). Many transcription factors play key roles at different stages of NK cell development. Transcription factors such as ID2 andID3 control the development of mature NK cells from their precursors (9), whereas GATA-3, T-bet, Eomes, and IRF2 are involved in generating functional NK cells that can exit bone marrow and enter peripheral tissues to perform their function (10). However, unlike the mechanisms in T and B lymphocytes, the molecular mechanisms that regulate the transcription of these key transcription factors during NK cell development remain poorly defined.
Protein mono- or polyubiquitination plays a critical role in a variety of cellular processes, including protein degradation, the cell cycle, protein trafficking, signal transduction, and transcriptional regulation (11). Polyubiquitination of a protein usually is associated with protein degradation; however, although it was discovered in 1975, monoubiquitination of histones remains a poorly studied area (11). Among the four core histones, H2A at K119 (5–15% of the total H2A) and H2B at K120 were found to be monoubiquitinated (11). Recently, it was reported that two RING-type ubiquitin E3 ligases, RNF8 and RNF168, modify H2A at a previously unknown site on H2A (K13 or K15) and have roles in the DNA-damage response (12). There is evidence that monoubiquitination of histone proteins can influence the activation of transcription positively and negatively. Moreover, studies of the H2B ubiquitination state revealed that deubiquitinated H2B is required for the progression of transcription elongation (13). An H2A ubiquitinase, 2A-HUB, functions as an elongation inhibitor in an N-CoR/HDAC1/3 corepressor complex to inhibit the expression of chemokine genes (14). Another H2A ubiquitinase, Ring1B/Ring2, is a core component of the polycomb repressive complex 1 and is well known for its role in the regulation of hematopoiesis and other cellular processes such as ES cell maintenance, differentiation, regulation of gene expression during embryogenesis, and others (15). Ring1B also enhances the survival of Th2 effector cells by inhibiting apoptosis (16). Zhou et al. (14) identified MYSM1 (the abbreviation stands for “Myb-like, SWIRM, and MPN domains-containing protein 1”), a histone H2A deubiquitinase, and reported that its activity in H2A deubiquitination is required for the activation of several target genes in prostate cancer cells. In our recent study, we found that MYSM1 plays a critical role in early B-cell commitment and development by de-repressing the transcription of EBF1 and other transcription factors by orchestrating histone modifications and transcription factor recruitment to the Ebf1 locus (17, 18). In this study, we demonstrate that MYSM1 has an important and intrinsic role in the maturation of NK cells but not in NK lineage specification and early development. We further delineate the underlying mechanism by which MYSM1 controls target gene transcription and NK cell maturation.
Results
Reduction in the Frequency and Number of NK Cells in Mysm1−/− Mice.
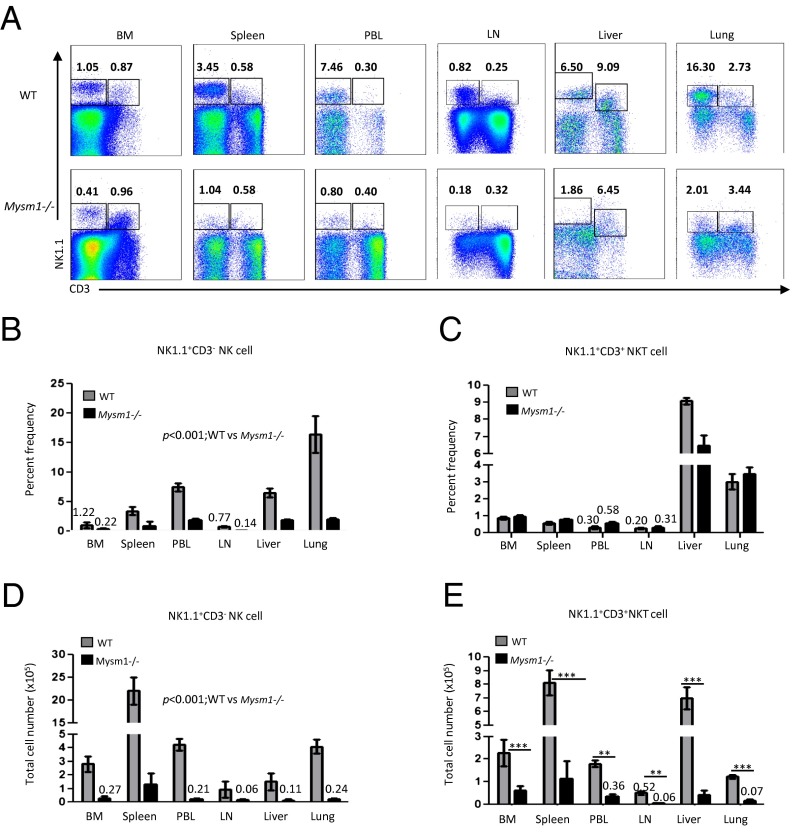
When systemically examining hematopoiesis of Mysm1-deficient mice (Mysm1-KO first-floxed mice), in addition to the severe reduction in peripheral B cells in Mysm1−/− mice reported in our recent study (17, 18), we found a drastic reduction in both the percentage and number of NK1.1+CD3− NK cells in various lymphoid tissues of Mysm1−/− mice as compared with their WT littermates (Fig. 1 A, B, and D). Jiang et al. (18) reported the reduction in MYSM1 protein and mRNA levels in various tissues and cell lineages such as B cells, T cells, and hematopoietic stem cells (HSCs). Using real-time PCR, we confirmed that MYSM1 transcript levels were reduced ∼8- to10-fold in the NK cells of Mysm1−/− mice as compared with WT controls (SI Appendix, Fig. S1A). There was a threefold reduction in the percentages of NK cells in the bone marrow, the primary site of NK cell development, and the percentages of peripheral NK cells were reduced by three- to ninefold in the spleen, blood, lymph nodes (pooled inguinal, axillary and cervical nodes), liver, and lung (Fig. 1 A and B).
Fig. 1.
Reduction in the frequency and number of NK cells in Mysm1−/− mice. (A) Representative flow cytometric profiles of NK1.1+CD3− NK cells or NK1.1+CD3+ NKT cells in the bone marrow (BM) pooled from femurs and tibias, spleen, peripheral blood (PBL), lymph node (LN), liver, and lung of WT and Mysm1−/− mice. Numbers indicate the percentages of cells in each quadrant. (B and C) Quantification of the average percentages of NK and NKT cells in the indicated organs of WT and Mysm1−/− mice. (D and E). Average of total number of NK and NKT cells in the indicated organs. Data shown are mean ± SEM of six or more mice per group and are representative of at least three independent experiments. **P < 0.01; ***P < 0.001.
In contrast, there were no significant differences in the frequency of NK1.1+CD3+ NK T cells (NKT cells) in Mysm1−/− mice and their WT littermates (Fig. 1 A, C, and E). We examined T cells and found that, unlike NK cells, in Mysm1−/− mice the frequency of CD4+ T cells was not altered significantly and that the frequency of the CD8+ T cells was slightly decreased in spleen but in the peripheral blood was comparable to that in WT littermates (SI Appendix, Fig. S1 B and C). We found some alterations in the thymic T-cell development, with reduced frequency of double-negative 2 cells in Mysm1−/− mice (SI Appendix, Fig. S1D); however, the frequency of double-positive (DP) CD4+CD8+ thymocytes was reduced only slightly in these mice, and the frequency of the single-positive CD4 and CD8 thymocytes was roughly similar in WT and Mysm1−/− mice (SI Appendix, Fig. S1 A and B). We noticed that the frequency of Lin−CD122+DX5+IL-7R+ thymic NK cells also was not compromised significantly in Mysm1−/− mice (SI Appendix, Fig. S2 A and B).
The absolute numbers of NK1.1+CD3+ NKT cells, CD4+ T cells, and CD8+ T cells (Fig. 1E and SI Appendix, Fig. S1E) were significantly lower in Mysm1−/− mice than in their WT littermates because of a severe reduction in total cell numbers in the Mysm1−/− mice (17, 18). Nevertheless, in contrast to NKT and T lymphocytes (Fig. 1E and SI Appendix, Fig. S1E), the drastic reduction in the number of NK1.1+CD3− NK cells in the various lymphoid tissues (Fig. 1D) results from both the reduction in the frequency of these cells and the reduction in the total number of cells in the Mysm1−/− mice compared with their WT littermates. Together, these data suggest that, in addition to B-cell lineage, MYSM1 is necessary for the generation of NK cells but not NKT and T lymphocytes.
NK Cell Maturation Is Defective but NK Lineage Commitment Is Normal in Mysm1−/− Mice.
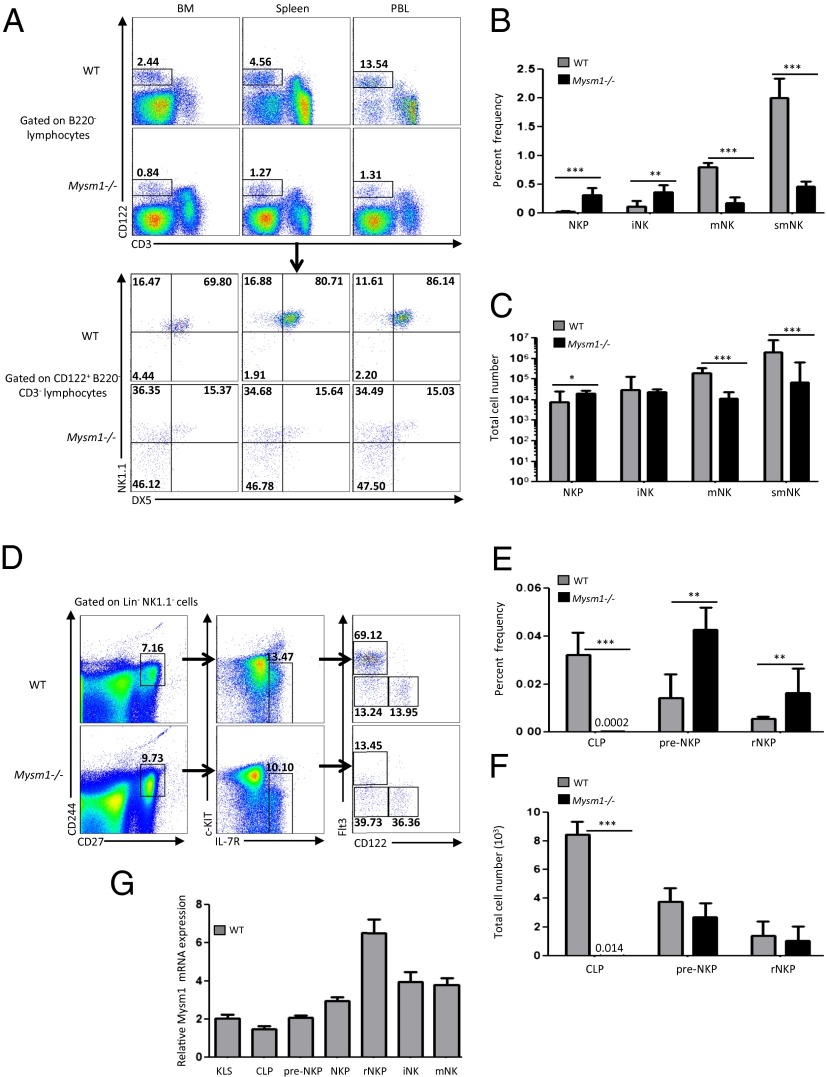
To dissect in detail the effect of MYSM1 deficiency on NK cell development, we assessed the surface expression of CD122, NK1.1, and DX5 on hematopoietic cells to gate out precisely the different stages of NK cell development. The three major developmental stages of NK cells are defined as NKP (Lin−CD122+NK1.1−DX5−), iNK (Lin−CD122+NK1.1+DX5−), and mNK (Lin−CD122+NK1.1+DX5+) (3, 19). The frequency of mNK cells was reduced drastically (approximately five- to sixfold), whereas the frequency of NKP and iNK cells was increased in both the bone marrow and peripheral tissues of Mysm1−/− mice (Fig. 2 A and B). The absolute number of cells was increased at the NKP stage and was not much affected at the iNK stage but was reduced severely at the mNK stage of development in the bone marrow of Mysm1−/− mice as compared with WT controls (Fig. 2C). The number of Lin−CD122+NK1.1+DX5+ mature NK cells was reduced by 17-fold in the bone marrow and by 29-fold in the spleen because of the severely reduced frequency of mNK cells in Mysm1−/− mice as compared with their WT littermates (Fig. 2 B and C).
Fig. 2.
NK cell maturation is defective, but commitment to the NK lineage is normal in Mysm1−/− mice. (A) Representative flow cytometric analyses of WT and Mysm1−/− mice using NK1.1 and DX5 to identify NKP (NK1.1− DX5−), iNK (NK1.1+DX5−), and mNK (NK1.1+DX5+) cells (Lower) subgated from CD122+CD3−B220− lymphocytes (Upper) in the bone marrow pooled from femurs and tibias, spleen, and peripheral blood of WT and Mysm1−/− mice. Numbers indicate percentages of cells in each quadrant. (B) Average percentages of NKP, iNK, and mNK cells in the bone marrow and spleen (smNK) of WT and Mysm1−/− mice. (C) Average of total cell numbers of NKP, iNK and mNK cells in the bone marrow and spleen (smNK) of WT and Mysm1−/− mice. Data shown are mean ± SEM of six or more mice per group and are representative of at least three independent experiments. (D) Representative flow cytometric analyses of WT and Mysm1−/− bone marrow using Flt3 and CD122 to identify CLPs (FLT3+CD122-), pre-NKPs (FLT3−CD122−), and rNKPs (FLT3−CD122+) in Lin−c-KIT−CD27+CD244+IL7R+ cells. Numbers indicate the percentages of cells in each quadrant. (E) Average percentages of CLPs, pre-NKPs, and rNKPs in the bone marrow of WT and Mysm1−/− mice. (F) Average of total numbers of CLPs, pre-NKPs, and rNKPs in the bone marrow of WT and Mysm1−/− mice. Data shown are mean ± SEM of five mice per group and are representative of four independent experiments. (G) Expression of MYSM1 mRNA at each NK developmental stage and in the progenitors of NK cells: Lin−c-KIT−CD27+CD244+IL-7R+Flt3−CD122− pre-NKPs; Lin−CD122+NK1.1−DX5− NKPs, Lin−c-KIT−CD27+CD244+IL-7R+Flt3−CD122+ rNKPs; Lin−CD122+NK1.1+DX5− iNK cells; Lin−CD122+NK1.1+DX5+ mNK cells; Lin−c-KIT−CD27+CD244+IL-7R+Flt3+CD122− CLPs; and Lin−cKit+Sca1+ KLS cells. All cells were sorted from bone marrow pooled from WT femurs and tibias. MYSM1 expression was normalized to GAPDH. Data are representative of five independent experiments; shown are mean ± SEM of triplicate determinations from one of the experiments. In B, C, E, and F, *P < 0.05; **P < 0.01; ***P < 0.001.
Previously, impairment in the development of B lymphocytes at the early stages of their differentiation has been demonstrated in the Mysm1−/− mice (17, 18). To test if MYSM1 regulates the commitment of lymphoid progenitors to an NK fate, as seen in B cells, we evaluated the NK precursor compartment of WT and Mysm1−/− mice. Lin−IL-7R+cKitintSca1+ cells, commonly defined as CLPs, can be fractionated into Flt3+ and Flt3− CLPs. Flt3+ CLPs were severely affected in the Mysm1−/− mice (SI Appendix, Fig. S3) because of the loss of Flt3+ Lin−c-KIT+Sca1+ (KLS) subsets, as also observed and reported by Nijnik et al. (17). However, to identify precisely the CLPs that can differentiate into NKPs, Fathman et al. (6) recently developed a strategy to distinguish CLPs, pre-NKPs, and rNKPs based on a refined analysis of markers that are expressed on these subsets (6). CD27 and CD244 are cell-surface markers that are expressed on CLPs and maintained on mature NK cells (6). Additionally, on commitment to the NK lineage and with loss of the potential for differentiating into B, T, and dendritic cell (DC) lineages, Flt3+ CLPs lose Flt3 expression and sequentially acquire CD122 expression (6). Within the Lin−CD27+CD244+IL-7R+ subset, differential expression of Flt3 and CD122 and the NK lineage bias exhibited by these cells in functional assays distinguished three progenitor subsets: Flt3+CD122− (CLPs), Flt3−CD122−(pre-NKPs), and Flt3−CD122+ (rNKPs). In our analysis using these markers, we found a severe reduction in the frequency and total number of CLPs (defined as Lin−c-KIT−CD27+CD244+IL-7R+Flt3+CD122− cells); in contrast the frequency of pre-NKPs (defined as Lin−c-KIT−CD27+CD244+IL-7R+Flt3−CD122− cells) and of rNKPs (defined as Lin−c-KIT−CD27+CD244+IL-7R+Flt3−CD122+ cells) was increased significantly (threefold) in the bone marrow of Mysm1−/− mice as compared with WT controls (Fig. 2 D–F). The absolute numbers of pre-NKPs and rNKPs were similar in Mysm1−/− and WT mice (Fig. 2E). Collectively, our data show that the reduction of Flt3+ CLPs in the Mysm1−/− mice did not compromise the generation of Flt3− CLPs comprising pre-NKPs and rNKPs (Fig. 2 D–F and SI Appendix, Fig. S3).
Real-time PCR analyses of sorted WT KLS cells, CLPs, pre-NKPs, rNKPs, NKPs, iNK cells, and mNK cells showed that higher levels of MYSM1 mRNA expressed by rNKP and by iNK and mNK cells than by their progenitors (Fig. 2G). The sharp increase in the mRNA levels at the rNKP stage suggests that the later stages of NK cell development are dependent on this onset of MYSM1 expression and is consistent with the blockage observed in NK cell maturation rather in NK lineage commitment in Mysm1−/− mice (Fig. 2G).
Together, these data demonstrate a blockage in the maturation of NK cells during NK cell development in Mysm1−/− mice, whereas the development of NKP and iNK cells from their lymphoid precursors is not compromised. Thus, unlike its critical role in early B-cell commitment, MYSM1 is required for the maturation of NK cells but not for the cells’commitment to the NK lineage or for their transition to iNK cells.
Mysm1−/− NK Cells Are Phenotypically Immature.
The reduction observed in the number of mNK cell subsets but not of iNK cells in the bone marrow and peripheral organs (Fig. 2 A–C) raised the question whether the NK cells that egress from the Mysm1−/− bone marrow to the peripheral organs are mainly from the iNK cell subsets. To test this possibility, we assessed the expression of a panel of well-characterized cell-surface antigens expressed on mature and immature WT and Mysm1−/− NK cells.
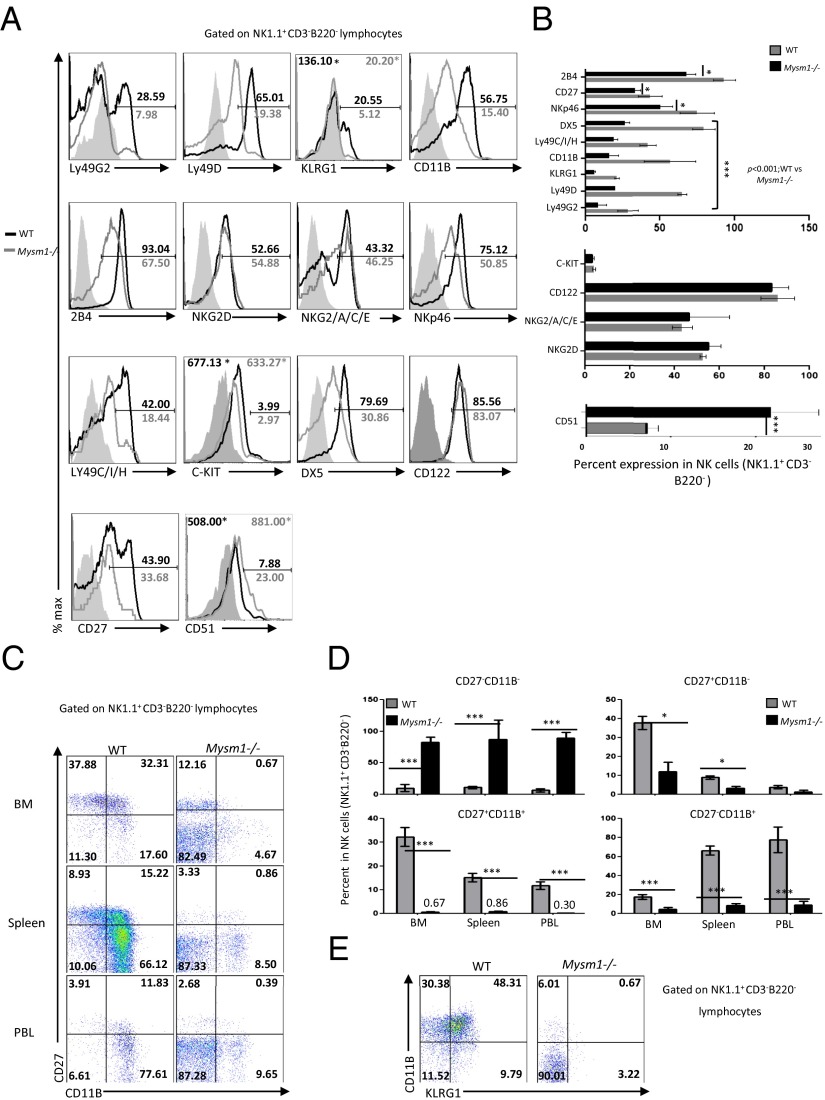
The most mature NK cells express the markers DX5, CD11B, and KLRG1 but not the immature markers c-KIT, CD27, and CD51 (3, 20–22). Flow cytometric analysis of a panel of NK developmental markers in the Mysm1−/− and WT splenic NK1.1+CD3−B220− NK cells (Fig. 3 A and B) revealed that Mysm1−/− NK cells express lower levels of mature markers such as DX5, CD11B, and KLRGI, but the levels of the immature markers were increased (CD51) or unaltered (c-KIT) compared with WT cells (Fig. 3 A and B). However, CD27 expression was decreased in the Mysm1−/− NK cells compared with WT cells (Fig. 3 A and B).
Fig. 3.
Mysm1−/− NK cells are phenotypically immature. (A) Representative flow cytometric histograms of NK developmental markers gated on splenic NK1.1+B220−CD3− NK cells of WT and Mysm1−/− mice. Numbers near the gate indicate percentages of cells within the positive gate. Numbers against the asterisk indicate relative median fluorescence intensities (MFIs) of specific markers above the autofluorescence of isotype controls in the WT and Mysm1−/− mice. (B) Quantification of the average percentages of the developmental markers in the splenic NK cells (NK1.1+B220−CD3−) that are decreased (Top), unaltered (Middle), or increased (Bottom) in the Mysm1−/− mice vs. WT controls. Data shown are mean ± SEM of three mice per group and are representative of at least three independent experiments. *P < 0.05; ***P < 0.001. (C) Representative flow cytometric profiles of the four-stage NK cell development: DN CD27−CD11B−, CD27+CD11B−, DP CD27+CD11B+, and CD27−CD11B+ expression on NK1.1+B220−CD3− NK cells in bone marrow pooled from femurs and tibias, spleen, and peripheral blood of WT and Mysm1−/− mice. Numbers indicate percentages of cells in each quadrant. Data shown are mean ± SEM of five mice per group and are representative of at least three independent experiments. (D) Quantification of the average percentages of DN, CD27+CD11B−, DP, and CD27−CD11B+ cells in the NK (NK1.1+B220−CD3−) populations in the indicated organs. *P < 0.05; ***P < 0.001. (E) Representative flow cytometric plots of KLRG1 vs. CD11B expression on NK1.1+B220−CD3− lymphocytes in the spleens of WT and Mysm1−/− mice. Numbers indicate percentages of cells in each quadrant. Data shown are mean ± SEM of five mice per group and are representative of at least three independent experiments.
Hayakawa et al. (20) proposed that NK cell maturation can be subdivided into four stages based on the expression levels of CD27 and CD11B; in order of development these stages are double negative (DN) (CD27−CD11B−); CD27+CD11B−; DP (CD27+CD11B+); and CD27−CD11B+. In the bone marrow and peripheral organs of Mysm1−/−mice, there were significant decreases in the frequency of the CD27−CD11B+ (approximately four- to eightfold) and CD27+CD11B+ NK cell subsets (∼18- to 48-fold) and an increase of ∼8- to 13-fold in the frequency of CD27−CD11B− NK cell subsets compared with WT tissues (Fig. 3 C and D). CD27+CD11B− NK cell subsets also were decreased in the Mysm1−/− mice (except in the blood) but less severely than the DP and CD27−CD11B+ subsets (Fig. 3 C and D). Our analyses revealed that Mysm1−/− NK cells were composed predominantly of the DN subsets, whereas the WT NK cells were mainly DP and CD27−CD11B+ NK cells (Fig. 3 C and D). Narni-Mancinelli et al. (8) suggested that the engagement of cells into the NK cell lineage occurs after the acquisition of NK1.1 and before the expression of NKp46. Our analysis of NK1.1 vs. NKp46 on CD122+B220−CD3− bone marrow lymphocytes showed that NK1.1+NKp46+ cells were reduced by only ∼1.5-fold in the Mysm1−/− mice (SI Appendix, Fig. S4). However, the development of mature NK cell subsets gated from NK1.1+NKp46+ lymphocytes, such as the NKp46+DX5+, CD27+CD11B+, or CD27−CD11B+ NK cell subsets, was severely affected in the Mysm1−/− mice. We also compared the expression of CD11B versus KLRGI (the most mature NK cell marker) and found that in Mysm1−/− mice the NK cell compartment was predominately CD11B−KLRG1− NK cells and that the pool of mature (CD11B+KLRG1−, CD11B+KLRG1+, and CD11B−KLRG1+) NK cells was severely compromised compared with that of the WT controls (Fig. 3E). Together, these data support our finding by showing that MYSM1 is required for the maturation of NK cells specifically during the onset of NKP46 and during the transition to mature NK cells but not for commitment to the NK lineage or for the transition to immature Lin−CD122+NK1.1+NKp46− NK cells.
Activatory receptors on NK cells are necessary for target recognition and induction of NK cell-mediated cytolysis (23). The expression of the activatory receptors 2B4, Ly49D, NKp46, LY49H, and CD11b was lower in Mysm1−/− mice than in WT controls (Fig. 3 A and B). However, the expression of NKG2D, an activatory receptor, remained unaltered in the Mysm1−/− NK cells compared with WT cells (Fig. 3 A and B). For complete NK cell maturation, NK cell education through the recognition of the self-MHC class I molecule by the inhibitory receptors (LY49C/I, LY49G2, NKG2A/C/E) is necessary, and this process occurs during the transition from the iNK to the mNK cell stage. We analyzed the expression of these inhibitory receptors and found a significant decrease in the levels of Ly49G2 and LY49C/I in the Mysm1−/− NK cells compared with WT cells; however, the NKG2A/C/E levels remained unaltered (Fig. 3 A and B).
Taken together, the severe decreases in the levels of many mature developmental markers, including the activatory and inhibitory receptors, demonstrate the predominantly immature nature of MYSM1-deficient NK cells.
MYSM1 Is Intrinsically Required for NK Cell Maturation in Vivo.
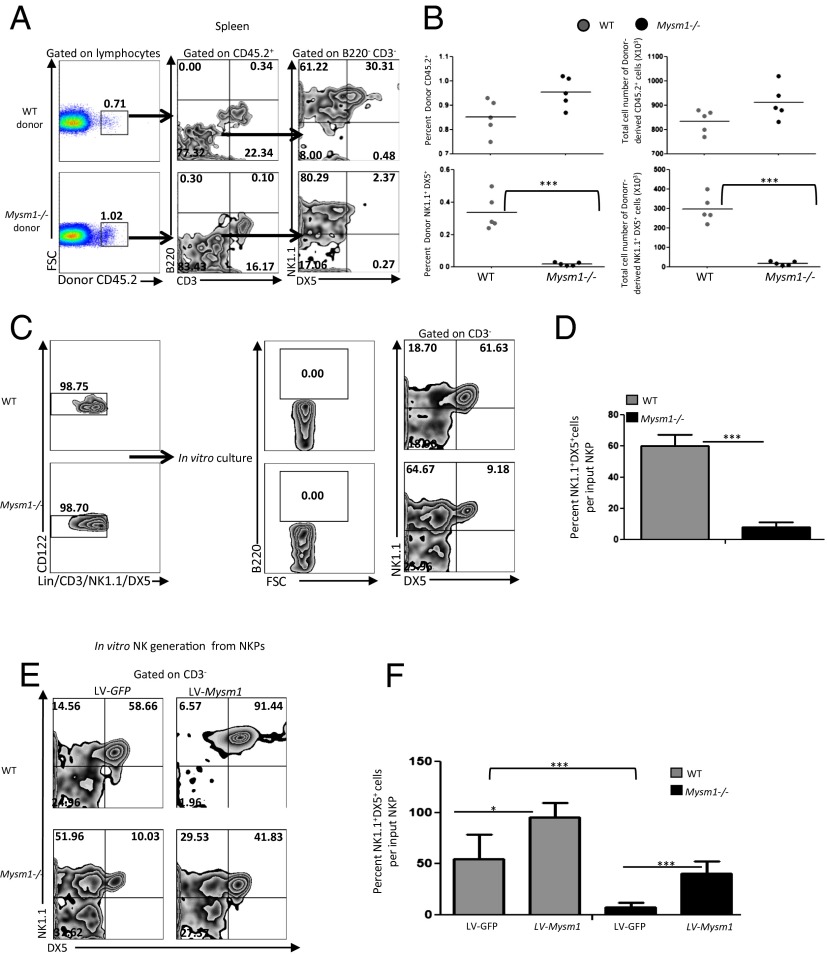
Our data demonstrate a defect in the Mysm1−/− mNK development but not in the commitment to the NK lineage or in the transition to immature NK cells (Fig. 2 A–F). To investigate whether the defective NK cell maturation in vivo comes from the microenvironment or from a phenomenon intrinsic to NK progenitors, we performed transplantation assays using NKPs as recently described (5). To account for bias that might arise from the reduced numbers of Mysm1−/− HSC progenitors as compared with WT cells (17), we did not transplant bone marrow cells. Instead, we transplanted equal numbers of NKPs to establish a more direct and accurate comparison between the maturation of WT and Mysm1−/− NK cells from their committed NK progenitors. Lin−CD122+NK1.1−DX5− NKPs sorted to high purity (SI Appendix, Fig. S11) from CD45.2 bone marrow of WT and Mysm1−/− mice were injected i.v. into sublethally irradiated CD45.1 WT B6.SJL-PtprcPep3/BoyJ recipients. Flow cytometric analysis of spleen 21 d after transplantation showed similar frequency and number of total CD45.2 chimerism in the recipient mice reconstituted with WT or Mysm1−/− donor cells (Fig. 4 A and B). Also, donor-derived reconstitution of T cells was similar in the recipient mice reconstituted with WT or Mysm1−/− donor cells, and, as expected, transplanted NKPs did not contribute to detectable B-cell reconstitution (Fig. 4A). CD45.2 immature NK (B220−CD3−NK1.1+DX5−) chimerism was evident in the recipient mice that received WT donor cells and was increased in the recipient mice that received Mysm1−/− donor cells (Fig. 4A). However the reconstitution of mature (B220−CD3−NK1.1+DX5+) NK cells occurred only with the WT donor cells and was severely defective in the recipient mice that received Mysm1−/− donor cells (Fig. 4 A and B), indicating that MYSM1 has an intrinsic role in NK cell maturation. We also used a mixed-chimera model in which we transplanted equal numbers of WT CD45.1 and WT or Mysm1−/− CD45.2 NKPs into sublethally irradiated CD45.1 WT recipients (SI Appendix, Fig. S5A). The presence of host CD45.1 cells in the sublethally irradiated recipient mice prevented a side-by-side comparison of the ability of CD45.1 and CD45.2 cells to reconstitute NK cells. Nevertheless, the findings that WT CD45.2 NKPs contributed to a normal NK cell reconstitution and that Mysm1−/− CD45.2 NKPs showed defective NK cell maturation in the recipient mice were consistent with the results of the NKP chimera study, reaffirming the conclusion that Mysm1 is an intrinsic factor for the maturation of NK cells in vivo.
Fig. 4.
MYSM1 is intrinsically required for NK cell maturation. (A and B). Sublethally irradiated 10- to 12-wk-old WT B6.SJL-PtprcPep3/BoyJ recipient mice were transplanted with 1,500 Lin−CD122+NK1.1−DX5− NKPs sorted from the bone marrow of 9- to 12-wk-old CD45.2 WT and Mysm1−/− mice. Donor-derived lineage reconstitution was evaluated in the spleen 21d after transplantation by FACS. (A) Representative FACS plots of donor-derived CD45.2 NK (NK1.1+DX5+CD3−B220−) reconstitution in the spleen. The specific gates stated above the plots and are indicated by arrows. (B) Percentages and total numbers of donor-derived CD45.2 cells and donor-derived NK cells (NK1.1+DX5+CD3−B220−) from the spleens of mice transplanted with WT or Mysm1−/− NKPs. NKPs were injected into five individual mice per group in each experiment. Data shown are representative of one of two experiments and are mean ± SEM of five or more mice per group in each experiment. (C and D) In vitro NK differentiation assays. Ten thousand FACS-sorted Lin−CD122+NK1.1−DX5− NKPs from 12-wk-old WT and Mysm1−/− mice were cultured on OP9 stroma with the cytokines KL, IL-7 (for the first week only), FL, IL-2, and IL-15 for 14 d; then cells were harvested and evaluated for NK cell outgrowth by FACS. 7AAD was used to exclude dead cells. (C) Representative flow cytometric profiles of NK cells (NK1.1+DX5+CD3−) generated from WT and KO NKPs; numbers indicate the percentages of cells in each quadrant. (D) Mean proportions of NK1.1+DX5+CD3− NK cells generated in vitro per input WT and Mysm1−/− NKPs. Data shown are mean ± SEM of three independent experiments. (E and F) In vitro MYSM1 rescue assays. FACs-sorted WT and Mysm1−/− CD122+Lin−NK1.1−DX5− NKPs from the bone marrow were transduced with a recombinant lentiviral vector LV-Mysm1 or a control lentiviral vector LV-GFP, and 10,000 transduced cells were transferred to an OP9 cell coculture and subjected to NK cell generation for 14 d, as explained above. Then cells were harvested and assessed for NK cell outgrowth by flow cytometric analysis. 7AAD was used to exclude dead cells. (E) Representative flow cytometric profiles of NK cells (NK1.1+DX5+CD3−) generated from WT NKPs, Mysm1−/− NKPs, WT NKPs in which MYSM1 was overexpressed, or Mysm1−/− NKPs rescued with MYSM1; numbers indicate the percentages of cells in each quadrant. (F) Mean proportions of NK1.1+DX5+CD3− NK cells generated in vitro per input respective NKPs as indicated. Data shown are mean ± SEM of three independent experiments. *P < 0.05; ***P < 0.001 in B, D, and F.
MYSM1 Is Intrinsically Required for NK Cell Maturation in Vitro.
To confirm further the intrinsic role of MYSM1 in NK cell maturation, we performed in vitro NK cell-generation assays, once again from NKPs (SI Appendix, Fig. S11) and not from hematopoietic progenitors to exclude the possibility that defective KLS (Lin−c-KIT+Sca1+) differentiation at earlier stages of hematopoiesis (17) might impact mNK cell generation in the Mysm1-deficient mice. We used the protocol developed by Nozad Charoudeh et al. (5) in which as few as 1–10 NKPs cocultured on OP9 stroma cell lines for 14 d in the presence of KIT ligand (KL), Fms-like tyrosinekinase-3 ligand (FL), IL-7 (for the first week only), IL-2, and IL-15 gave rise to a large number of NK cell clones. As is consistent with the in vivo results, NKPs did not produce any detectable B220+ B cells (Fig. 4C). However, there was a severe defect in the development of Mysm1−/− Lin−CD122+NK1.1−DX5− NKPs into NK1.1+DX5+ mNK cells, whereas WT NKPs differentiated normally into mNK cells in the presence of IL-15 (Fig. 4 C and D).
Because our data showed no defect in the Mysm1−/− cells’ commitment to the NK lineage in vivo (Fig. 2 A–F), we expected to see a trend similar to that seen with NK cell generation from Mysm1−/− NKPs even when Mysm1−/− KLS cells were used as inputs, and the results confirmed this expectation (SI Appendix, Fig. S5 B and C). The fact that both KLS- and NKP-derived NK cells suffered a maturation defect reconfirms our previous finding that the defective KLS differentiation at the early stages of hematopoiesis (17) did not contribute to the impaired development of mature NK cells in Mysm1−/− mice (Fig. 2 A–F). Together, these results recapitulated the in vivo phenotype of Mysm1−/− mice.
To test further the direct relationship between MYSM1 and NK cell development, we extended the in vitro system and carried out a rescue assay of Mysm1−/− NKPs using a recombinant lentiviral vector (LV) expressing MYSM1 (18). NKPs sorted to high purity (SI Appendix, Fig. S11) were transduced with LV-Mysm1, and the transduced cells were subjected to NK cell differentiation in vitro. Fig. 4 E and F shows that forced expression of MYSM1 in vitro rescued the defective ability of Mysm1−/− NKPs to generate NK1.1+DX5+ mNK cells. In contrast, transduction with control LV-GFP failed to rescue the developmental defect of Mysm1−/− NKPs (Fig. 4 E and F). Furthermore, ectopic expression of MYSM1 in WT NKPs increased the proportion of mature NK cells produced (Fig. 4 E and F), showing that MYSM1 overexpression can boost NK cell development in vitro. Collectively, these data demonstrate the dependence of NK cell maturation on MYSM1.
IL-15 is a critical factor for NK cell development and homeostasis, and both IL-15−/− and IL-15R−/− mice were defective in their production of mature NK cells (24). We were curious to know how MYSM1 responds to IL-15 and hence checked the outgrowth of NK cells from WT NKPs in which MYSM1 was overexpressed in the absence of IL-15. Interestingly, the overexpression of MYSM1 in WT NKPs significantly increased the generation of NK cells even in the absence of exogenous IL-15 (SI Appendix, Fig. S5 D and E). Furthermore, upon IL-15 stimulation, the level of MYSM1 mRNA was increased significantly both in the WT NK cells derived in vitro from NKPs in which Mysm1 was overexpressed (SI Appendix, Fig. S5F) and in the ex vivo-cultured freshly sorted Lin−CD3−CD122+NK1.1+ NK cells (SI Appendix, Fig. S5G). These data suggest that MYSM1 functions downstream of IL-15 during NK cell development.
Thus, both our in vivo and in vitro data indicate that NK maturation requires the expression of cell-intrinsic MYSM1.
MYSM1 Is Required for ID2 Transcription.
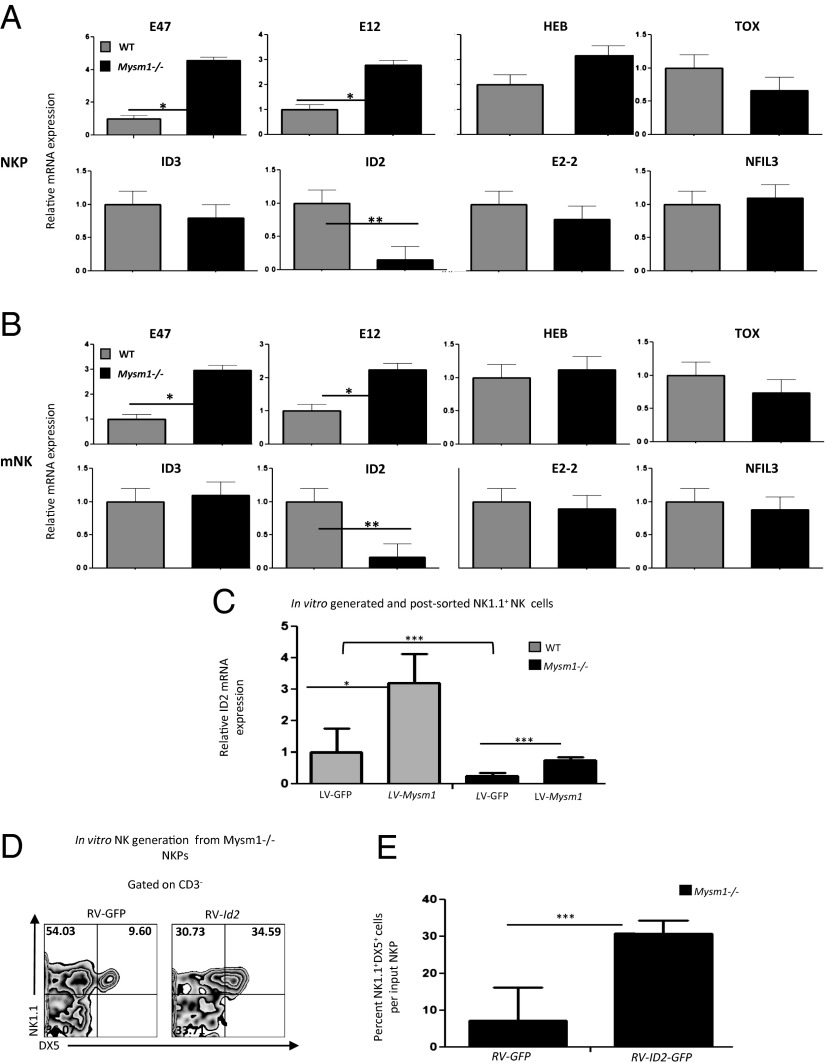
Given that MYSM1, a histone H2A deubiquitinase, functions as a transcriptional regulator (14, 18), we set out to assess by quantitative RT-PCR (qRT-PCR) assays whether the expression of any genes involved in NK cell development (10) was altered in Mysm1−/− mature NK cells and their progenitors. We detected a marked (approximately sixfold) reduction in the levels of ID2 mRNA and an increase in the levels of E2A mRNA in the sorted Mysm1−/− mNK cells(CD122+Lin−NK1.1+DX5+) and NKPs (CD122+Lin−NK1.1−DX5−) compared with the levels in WT cells. The levels of TBET mRNA and GATA3 mRNA also were decreased, but less severely than ID2 mRNA, in Mysm1−/− mNK cells as compared with WT cells (Fig. 5 A and B and SI Appendix, Fig. S6).
Fig. 5.
MYSM1 is required for ID2 transcription. (A and B) Real-time PCR analyses of a panel of NK development transcription factors in NKPs (CD122+Lin−NK1.1−DX5−) (A) and mNKs (CD122+Lin−NK1.1+DX5+) (B) sorted from bone marrow pooled from the femurs and tibias of 10–15 Mysm1−/− and WT mice. Data are mean ± SEM of triplicate determinations from one of two or more independent experiments. *P < 0.05; **P < 0.01. (C) ID2 mRNA levels of in vitro-derived and postsorted NK1.1+ NK cells from WT or Mysm1−/− NKPs transduced with or without LV-Mysm1 and control LV-GFP vectors. mRNA expressions were normalized to GAPDH, and values are presented as relative expression with that of the mRNA levels of NK cells derived from LV-GFP–transduced WT NKPs (values set to 1). Data are mean ± SEM of triplicate determinations from one of two independent experiments. ***P < 0.001. (D and E) Forced expression of ID2 rescued defective NK cell maturation. FACS-sorted WT and Mysm1−/− CD122+Lin−NK1.1−DX5− NKPs from the bone marrow were transduced with a recombinant retroviral vector RV-Id2 or a control retroviral vector RV-GFP, and 10,000 transduced cells were transferred to OP9 cell coculture and subjected to NK cell generation for 14 d as explained above. Then cells were harvested and assessed for NK cell outgrowth by flow cytometric analysis. 7AAD was used to exclude dead cells. (D) Representative flow cytometric profiles of NK cells (NK1.1+DX5+CD3−) generated from Mysm1−/− NKPs or Mysm1−/− NKPs transduced with ID2; numbers indicate the percentages of cells in each quadrant. (E) Mean proportions of NK1.1+DX5+CD3− NK cells generated in vitro per input NKPs as indicated. Data shown are mean ± SEM of three independent experiments. ***P < 0.001.
ID2 and ID3 are both expressed in NKPs, but ID2 is the predominant transcription factor in mature NK cells (9, 25). In the absence of ID2, NKP and iNK cell development is not affected, likely because of the functional compensation of ID3 protein, which is highly expressed during these stages of NK cell development. As they do in the absence of MYSM1, NK cells fail to mature in the absence of ID2 (9). The earlier onset of expression for MYSM1 mRNA (which peaks in rNKPs) compared with ID2 expression (which peaks in iNK cells) is consistent with the hypothesis that ID2 transcription is dependent on MYSM1 during NK cell development (Fig. 2G and SI Appendix, Fig. S7). These data combined with the significant reduction in the mRNA levels of ID2 in the Mysm1−/− NK cells (Fig. 5 A and B) indicate that MYSM1 is required for ID2 transcription.
We then sought to test if MYSM1 controls ID2 transcription during NK cell development. To do so, we transduced Mysm1−/− NKPs with LV-Mysm1 or control LV-GFP and transferred them to an OP9 coculture system in the presence of NK cell-conditioned medium. After 14 d, the level of ID2 mRNA in vitro-generated and postsorted NK1.1+ cells was examined by qRT-PCR. Fig. 5C shows that forced expression of MYSM1, but not of the GFP control, significantly increased the ID2 mRNA levels in the Mysm1−/− NK cells generated in vitro, indicating that ID2 is directly or indirectly transcriptionally regulated by MYSM1.
Next, we further tested if the defective expression of ID2 in Mysm1−/− NK cells is indeed one of the possible causes of the observed defective NK maturation. To do so, we transduced WT and Mysm1−/− NKPs with a retrovirus (RV) expressing ID2 protein and examined NK cell generation in vitro for 2 wk using assays similar to that described above. Forced ID2 expression rescued the defect in NK cell maturation in the Mysm1−/− NKPs (Fig. 5 D and E).
Collectively, these data demonstrate that MYSM1 is required for ID2 transcription and that defective ID2 transcription in Mysm1−/− NK cells is likely to be one cause of their defective NK cell maturation.
MYSM1 Associates with the Id2 Locus.
To gain further insight into the molecular mechanisms controlling ID2 transcription, we used ChIP assays to examine whether MYSM1 protein is associated with the Id2 locus.
To obtain a sufficient number of cells, we used Lin−CD3−CD122+NK1.1+ NK cells (comprising mature and immature NK cells) sorted from the bone marrow and spleen cells of WT or Mysm1−/− mice for all our ChIP assays (SI Appendix, Fig. S11). Because ID2 expression is required at the later stages of NK cell development and homeostasis (9), the transcriptional regulation of ID2 should be evident in mature splenic and bone marrow NK cells. In agreement with this notion, Ramirez et al. (26) recently demonstrated ETS-1–mediated ID2 regulation using sorted splenic mNK cells. A panel of PCR primer pairs was constructed spanning the promoter region encompassing different transcription factor binding sites (1 kb upstream transcription start site), the initiator sequence and the first coding exon of Id2 (1 kb downstream transcription start site) (Fig. 6A). As a negative control, we chose an intergenic region that is not known to contain any regulatory sequences, ∼3.8-kb downstream of the ID2 locus (27). Although they were sorted to 98–99% purity (SI Appendix, Fig. S11), we validated our chromatin preparations by checking for the enrichment of known interactions in NK cell-specific genes. To this end, we used two controls. RUNX3 binds the proximal Nkp46 promoter in a NK cell-specific manner (28). Although, NKp46 expression is reduced by ∼1.5-fold in Mysm1−/− NK cells, a substantial level of residual NKp46 was present (Fig. 3D). We speculated that RUNX3 binding of the proximal Nkp46 promoter was intact because RUNX3 expression was unaffected by MYSM1 deletion (SI Appendix, Fig. S6), and we confirmed this notion using a ChIP assay (SI Appendix, Fig. S8). RUNX3 also can bind to the Cd122 promoter (28, 29), whose expression is unaltered in Mysm1−/− mice (Fig. 3A); we used this assay as a second control to validate our chromatin preparation (SI Appendix, Fig. S8). Anti-IgG was used as a negative control for the antibody of interest used in the immunoprecipitations, unless otherwise indicated.
Fig. 6.
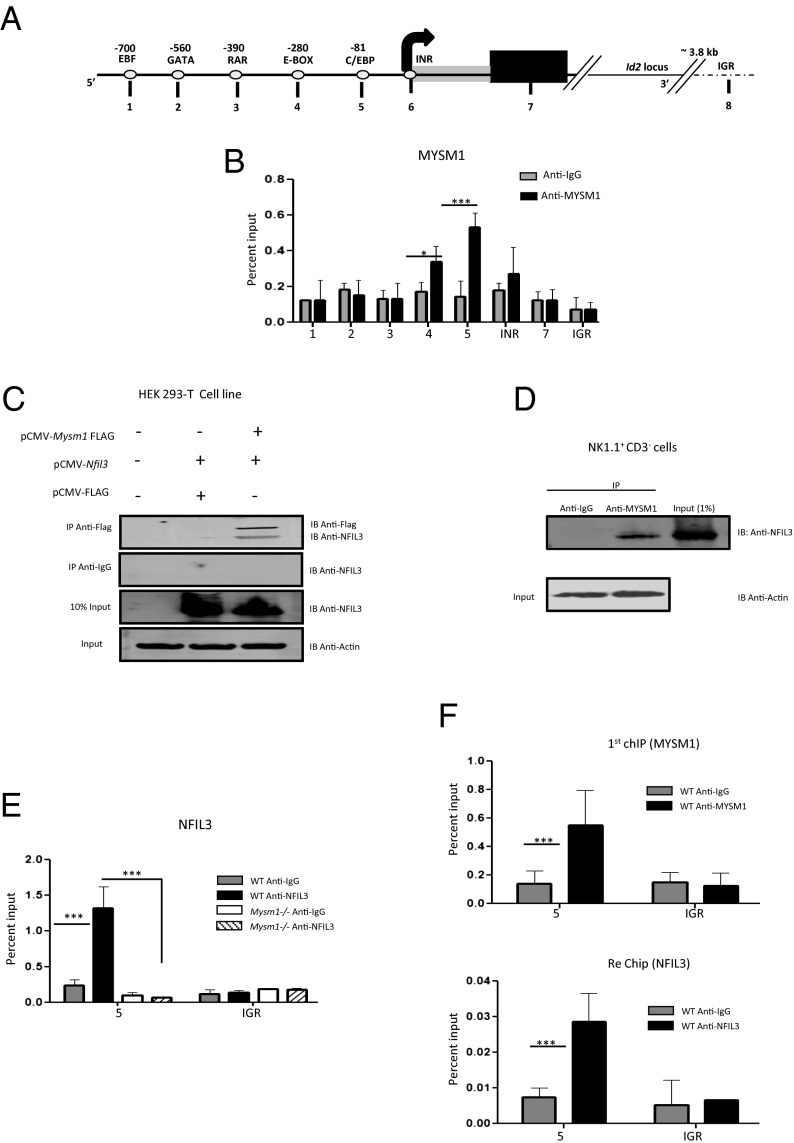
MYSM1 associates with the Id2 locus. (A) Schematic diagram of the Id2 locus: promoter encompassing different transcription factor binding sites (small circles; numbers above small circles denote the position of transcription factor binding sites from the transcription start site in base pairs), the transcription imitation site (INR), the first exonic region (black), and an intergenic region (IGR) downstream Id2 gene. The positions of the primers used for ChIP assays are indicated by numbers. (B) ChIP assays of WT NK cells (Lin−CD3−CD122+NK1.1+) comprising mature and immature NK cells from the pooled bone marrow and spleen sorted using anti-MYSM1 antibody. The precipitated DNA was analyzed by qRT-PCR using the primers indicated along the Id2 locus in A. The relative amount of immunoprecipitated DNA is presented as a percentage of input DNA. The binding of MYSM1 was determined by comparing the ChIP fractions with control IgG. The IGR region served as a negative control. Data are mean ± SEM of triplicate determinations from one of three independent experiments. (C–F) MYSM1 interacts with NFIL3, and this interaction is critical for their recruitment to the ID2 locus. (C) Coimmunoprecipitation assays. Cell lysates from HEK293T cells cotransfected with pCMV-Nfil3 and pCMV-FLAG-Mysm1 or pCMV-FLAG expression vectors were precipitated with an anti-Flag antibody or anti-IgG (negative control), and precipitated proteins were analyzed by Western blot by probing with anti-NFIL3 antibody. Ten percent of the input was loaded. (D) Cell lysates of 2 × 107 WT NK1.1+CD3− cells were incubated with anti-MYSM1 antibody and anti-IgG antibody (negative control), and immunoprecipitated proteins were analyzed by Western blot by probing with anti-NFIL3 antibody. One percent of the input was loaded. Actin was used as a control to confirm equal starting amounts of protein for immunoprecipitation. Data are representative of two (D) or three (C) independent experiments. (E) ChIP assays of WT and Mysm1−/− NK cells (Lin−CD3−CD122+NK1.1+) sorted using anti-NFIL3 and anti-IgG antibodies. The precipitated DNA was analyzed by qRT-PCR using the primers indicated along the Id2 locus in A. The relative amount of immunoprecipitated DNA is presented as a percentage of input DNA. The binding of NFIL3 was determined by comparing the ChIP fractions with control IgG and also was compared with the WT immunoprecipitated chromatin fractions. The IGR region served as a negative control. Data are mean ± SEM of triplicate determinations from one of three independent experiments. (F) Sequential ChIP assays of sorted WT NK cells (Lin−CD3−CD122+NK1.1+) using anti-MYSM1, anti-NFIL3, and the corresponding negative control antibodies (IgG). The precipitated chromatin from the first ChIP with anti-MYSM1 was reprecipitated with antibody against second protein NFIL3 and IgG control. The precipitated DNA was analyzed by qRT- PCR using the primers indicated along the Id2 locus in A. The relative amount of immunoprecipitated DNA is presented as a percentage of input DNA. The binding of MYSM1 and NFIL3 was determined by comparing the ChIP fractions with the corresponding first ChIP or second ChIP control IgGs. The IGR region served as a negative control. Data are mean ± SEM of triplicate determinations from one of two independent experiments. *P < 0.05; ***P < 0.001 in B, E, and F.
Chromatin from the sorted cells was immunoprecipitated with either anti-MYSM1 or anti-IgG, and the precipitated DNA was tested for the enrichment of Id2 gene. In comparison with the IgG control, there was a significant enrichment of Id2 gene using primer pair 5 (Fig. 6A) (comprising the C/EBP binding site) and less significantly using primer pair 4 (Fig. 6A) (comprising the E-BOX binding site). No enrichment was observed in the intergenic region (Fig. 6B), suggesting that MYSM1 is associated directly with the ID2 locus in NK cells. This result further supports our previous finding that MYSM1 regulates ID2 transcription and does so through a direct association. However, at this point we did not know whether MYSM1 is selectively targeted to the Id2 regulatory elements in NK cells through its direct DNA-binding activity mediated via its N-terminal SANT domain (30) or with the help of other DNA-binding proteins and regulators of Id2 gene.
MYSM1 Interacts with NFIL3, and This Interaction Is Critical for Their Recruitment to the Id2 Locus.
To gain further insight on how MYSM1 activates ID2 transcription, we examined whether MYSM1 is required for the recruitment of regulators of Id2 gene transcription. We used immunoprecipitation assays with co-overexpressed proteins in 293-T cells to investigate whether MYSM1 specifically facilitates the recruitment of any transcription factors related to NK cell development, such as NFIL3, TOX, KLF4, ETS-1, or EOMES (31). Interestingly, NFIL3 protein, which is essential for NK cell maturation through its activation of ID2 transcription (32), readily coimmunoprecipitated with MYSM1 when overexpressed in 293-T cells that were transfected with pCMV-Mysm1-FLAG but not in the cells that were transfected with the pCMV-FLAG (Fig. 6C). In our experimental settings, none of the other proteins coimmunoprecipitated with MYSM1. Also, endogenous MYSM1 was strongly associated with endogenous NFIL3 in the NK1.1+CD3− NK cells pooled from WT spleens and bone marrow (Fig. 6D).
Next we used ChIP assays of WT and Mysm1−/− Lin−CD3−CD122+NK1.1+ NK cells to test whether interaction between MYSM1 and NFIL3 is required to recruit these proteins to the Id2 locus. We found that NFIL3 indeed is associated with the promoter region of the Id2 locus in WT NK cells at the C/EBP binding site (primer 5) (Fig. 6E). The Id2 promoter encompasses two C/EBP binding sites: CbE2 (−81 to −73) and CbE3 (−73 to −65) (33), and our finding was consistent with earlier reports that suggested the consensus C/EBP binding site (ATTGC/GCAAT) as one of the high-affinity binding sites for bZIP factors such as NFIL3 (E4BP4) (34, 35). However, the association of transcription factor NFIL3 with the Id2 locus was lost in the absence of Mysm1 and in the intergenic region (Fig. 6E). This result indicates that MYSM1 has an important role in recruiting NFIL3 to the Id2 locus in NK cells.
To confirm further the corecruitment and colocalization of MYSM1 and NFIL3 at the Id2 locus, we performed sequential two-step ChIP assays with WT Lin−CD3−CD122+NK1.1+ NK cells. To do so, chromatin first was immunoprecipitated with anti-MYSM1 antibody, followed by a second immunoprecipitation with anti-NFIL3. MYSM1 and NFIL3 were found to be colocalized at the promoter region of the Id2 locus in the WT NK cells (Fig. 6F). Colocalization again was enriched at the C/EBP binding site (primer 5); this result explains, at least in part, how MYSM1 may be targeted to the Id2 locus through its interaction with the DNA-binding NFIL3 protein. The association of these factors at the Id2 locus was not detectable in the intergenic region (Fig. 6F).
MYSM1 also has its own DNA-binding activity (30), and it is possible that this activity or its concerted action with the DNA-binding NFIL3 protein may help bring it closer to the DNA-regulatory elements of Id2 locus. However, the fact that NFIL3’s association is lost in absence of MYSM1 shows that MYSM1 and its interaction with NFIL3 are critical for the stable association of NFIL3 at the Id2 locus during NK cell development. Collectively these results suggest the existence of some sort of concerted molecular mechanism in which NFIL3 may help in targeting MYSM1 to Id2 promoter and MYSM1 in turn enhances NFIL3 binding by inducing localized alterations in the nucleosome structures of the Id2 locus.
Id2 Locus of the Mysm1−/− NK Cells Is Poised in Its Repressed State.
Cell development is controlled by the activation of a specific genetic program that is particular to a specific stage of cell development and by the repression of the genetic program corresponding to the previous cellular state. This process is regulated at the transcriptional level, to a large extent by transcription factors that can dictate the epigenetic signature of a specific locus (36). MYSM1 previously was reported to regulate target gene transcription by deubiquitinating histone K119, by coordinating with other histone modifications, and by subsequently recruiting transcription factors to the target locus (14, 18). We performed ChIP assays to examine whether the chromatin context of the Id2 locus is altered in Mysm1−/− NK cells, consistent with its reduced transcript levels. We determined the chromatin state of the locus in the MYSM1/NFIL3-C/EBP binding site, in the transcription start site, in the first-coding exon of the Id2 locus, and in the negative control intergenic region 3′ of the Id2 locus (SI Appendix, Supplementary Results and Fig. S9A).
Discussion
Although the use of KO mice has enabled the identification of transcription factors for NK cell development, unveiling the regulation of events that control NK cell development still remains a challenge. Specifically, there are no studies on epigenetic regulation of transcription factors required for NK cell development.
In this study we have identified the essential and intrinsic role of a histone H2A deubiquitinase, MYSM1, in NK cell maturation. Our study outlines this role of MYSM1 in three parts. (i) MYSM1 intrinsically controls the maturation of NK cells downstream of IL-15 signaling but is dispensable for the commitment to the NK lineage. (ii) Our mechanistic studies show that MYSM1 interacts with NFIL3, and this interaction is critical for the recruitment of these proteins to the Id2 locus for the activation of its transcription during NK cell development; the expression of the transcription factor ID2 is impaired in MYSM1-deficient NK cells. (iii) Our mechanistic study also demonstrate that, in the absence of MYSM1, the Id2 locus is poised in its repressed state for future activation and that MYSM1-mediated epigenetic alterations may move its chromatin from a poised to an activated state promoting NK cell development.
Our mechanistic study revealed that the activity of MYSM1 in promoting the transcription of ID2 is essential for the development of mature NK cells. Several lines of evidence support this conclusion (Figs. 5 and 6): (i) the expression of ID2 was reduced significantly in Mysm1−/− mNK cells and NKPs; (ii) the forced expression of MYSM1 rescued the defective expression of ID2 in Mysm1−/− NK cells; (iii) the association of MYSM1 with the ID2 locus was detected by ChIP assays; (iv) altered histone modifications and paused RNA Pol II occupancy demonstrate the existence of a poised Id2 locus in the Mysm1−/− NK cells; and (v), the forced expression of ID2 rescued the defective development of mature NKs from Mysm1−/− NK precursors. In addition to the evidence presented in this study, our conclusion is supported further by the similarity of the NK cell phenotypes of Mysm1−/− and Id2 −/− mice. Both these KO mice showed impaired maturation of NK cells, but the early stages of development remained unaffected (9). The development of thymic NK cells was not compromised in either Id2 −/− or Mysm1−/− mice (SI Appendix, Fig. S2). We also observed fewer and smaller lymph nodes and Peyer’s patches in Mysm1−/− mice (SI Appendix, Fig. S10), which resembled the defective development of lymph nodes and Peyer’s patches in Id2 −/− mice (9). Thus, the blocking of NK cell maturation observed in Mysm1−/− mice is likely to be caused by the defective expression of ID2 in Mysm1−/− mNK cells and their precursors.
Although our study did not identify much reduction in the transcript levels of many known transcription factors involved in NK development [except for ID2, T-BET, and GATA3 (Fig. 5 A and B and SI Appendix, Fig. S6)], we cannot exclude the possibility that MYSM1 may directly or indirectly regulate additional genes apart from Id2 during NK cell development. For example, we observed a loss of Flt3 expression in CLPs (Fig. 2 D and F and SI Appendix, Fig. S3 A–C), and our data are consistent with the findings of Nijnik et al. (17), who demonstrated a loss of Flt3+ KLS cells in Mysm1−/− mice. Previously, Flt3 signaling has been implicated in the development and/or maintenance of Flt3+ MPPs (multipotent progenitors), CLPs, and B-, T-, NK-, and DC- lineage cells (37, 38). However, two independent groups recently demonstrated the down-regulation of Flt3 and up-regulation of CD122 as sequential steps in NK cell commitment from Flt3+ CLPs (6, 39). Our data, in fact, show that the reduction of Flt3+ CLPs in Mysm1−/− mice did not compromise the generation of pre-NKPs and rNKPs (Fig. 2 D and F and SI Appendix, Fig. S3 A–C). Another group showed similar evidence that the reduction of Flt3+ CLPs in hoxa9−/− mice did not compromise their ability to generate NKPs or even mature NKs cells (40). However, the precise role of MYSM1 in regulating NK cell development through Flt3-dependent or independent regulatory circuits may need more rigorous investigation.
Our further investigation into how MYSM1 is targeted selectively to the Id2 locus suggests that MYSM1 interacts with the DNA-binding NFIL3 protein and that the two are co-recruited to the Id2 locus (Fig. 6 C–F). We also found that this interaction is critical for the recruitment of NFIL3 to the Id2 locus (Fig. 6F), possibly because of the MYSM1-induced chromatin alterations that could enhance NFIL3 binding in the Id2 locus. However, it is interesting that Nfil3−/− mice showed defective NK cell development at both the iNK and mNK stages of development (32), whereas the NK cells in Mysm1−/− mice were defective only during the transition from iNK to mNK cells (Fig. 2 A–C). It is unclear why MYSM1 is not involved in NK cell development during the transition from NKP to iNK cells. It may be that organized epigenetic signals for the coordination of NFIL3 and MYSM1 are composed only at the later stages of NK cell development. What other proteins help in targeting MYSM1 to the Id2 locus and whether NFIL3 requires any epigenetic factors other than MYSM1 to be recruited to the Id2 locus during the early stages of NK cell development are questions that need more investigation. Clearly, an interesting focus of future studies will be to understand fully how MYSM1 interacts with different partners for selective activation of its target genes in different cell lineages or at different developmental stages of cells from the same lineage.
Specific chromatin markers keep master regulators of differentiation silent but poised for further induction (41). Our data indicate that the Id2 gene is poised in its repressed state for future activation and that MYSM1-mediated epigenetic alterations may shift its chromatin from a poised to an activated state. These data converge well with a previous report that showed RNA polymerase pausing to be one of the down-stream consequences of histone H2A monoubiquitination (42). Although ID2 is not essential for early NK cell development, and Mysm1−/− mice show defects only in NK cell maturation and not in commitment to the NK lineage, the expression of ID2 is one of the first indications of NK cell lineage specification (9). It will be interesting to identify the precise developmental point at which MYSM1 composes the epigenetic signals for the induction of the Id2 gene for NK lineage specification and development.
Nevertheless, this study demonstrates that MYSM1 has an important and intrinsic role in NK cell development through an epigenetic control of ID2 transcription, which is critical for NK cell development.
Methods
Detailed descriptions of techniques used in our study are provided in SI Appendix, Supplementary Materials and Methods.
Animals.
Mysm1-deficient mice (Mysm1-KO first-floxed mice) were generated through a KO-first strategy (43). All animal breeding and experiments were approved and performed in accordance with the University of Southern California Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank Xiao-Xia Jiang, Quan Nguyen, Tao Wang, Peter Yates, Suzi Sanchez, Lindsey Jones, Hae Jung Won, and other members of the S.-Y.C. laboratory for valuable technical assistance and helpful suggestions. We also thank Omid Akbari, Hye-Ra Lee, Jae U Jung, Judd Rice, Michael Stallcup, Peter Jones, and other colleagues at the University of Southern California. We thank Christopher A. Klug for providing valuable reagents and help. This work was supported by National Institutes of Health Grants R01CA090427, AI084811, CA116677, and AI068472 (to S.-Y.C.) and CA100841 and AI08185 (to X.F.H.), and by a Leukemia & Lymphoma Society Specialized Center of Research Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308888110/-/DCSupplemental.
References
- 1.Smyth MJ, Hayakawa Y, Takeda K, Yagita H. New aspects of natural-killer-cell surveillance and therapy of cancer. Nat Rev Cancer. 2002;2(11):850–861. doi: 10.1038/nrc928. [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama WM, Kim S. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol Rev. 2006;214:143–154. doi: 10.1111/j.1600-065X.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- 3.Kim S, et al. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. 2002;3(6):523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- 4.Rosmaraki EE, et al. Identification of committed NK cell progenitors in adult murine bone marrow. Eur J Immunol. 2001;31(6):1900–1909. doi: 10.1002/1521-4141(200106)31:6<1900::aid-immu1900>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 5.Nozad Charoudeh H, et al. Identification of an NK/T cell-restricted progenitor in adult bone marrow contributing to bone marrow- and thymic-dependent NK cells. Blood. 2010;116(2):183–192. doi: 10.1182/blood-2009-10-247130. [DOI] [PubMed] [Google Scholar]
- 6.Fathman JW, et al. Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood. 2011;118(20):5439–5447. doi: 10.1182/blood-2011-04-348912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vosshenrich CA, et al. Roles for common cytokine receptor gamma-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J Immunol. 2005;174(3):1213–1221. doi: 10.4049/jimmunol.174.3.1213. [DOI] [PubMed] [Google Scholar]
- 8.Narni-Mancinelli E, et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc Natl Acad Sci USA. 2011;108(45):18324–18329. doi: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204(5):1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boos MD, Ramirez K, Kee BL. Extrinsic and intrinsic regulation of early natural killer cell development. Immunol Res. 2008;40(3):193–207. doi: 10.1007/s12026-007-8006-9. [DOI] [PubMed] [Google Scholar]
- 11.Komander D, Clague MJ, Urbé S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 12.Mattiroli F, et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150(6):1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Chandrasekharan MB, Huang F, Sun Z-W. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci USA. 2009;106(39):16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou W, et al. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29(1):69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luis NM, Morey L, Di Croce L, Benitah SA. Polycomb in stem cells: PRC1 branches out. Cell Stem Cell. 2012;11(1):16–21. doi: 10.1016/j.stem.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Majewski IJ, et al. Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood. 2010;116(5):731–739. doi: 10.1182/blood-2009-12-260760. [DOI] [PubMed] [Google Scholar]
- 17.Nijnik A, et al. Sanger Institute Microarray Facility; Sanger Mouse Genetics Project The critical role of histone H2A-deubiquitinase Mysm1 in hematopoiesis and lymphocyte differentiation. Blood. 2012;119(6):1370–1379. doi: 10.1182/blood-2011-05-352666. [DOI] [PubMed] [Google Scholar]
- 18.Jiang X-X, et al. Control of B cell development by the histone H2A deubiquitinase MYSM1. Immunity. 2011;35(6):883–896. doi: 10.1016/j.immuni.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Santo JP. Natural killer cell developmental pathways: A question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 20.Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol. 2006;176(3):1517–1524. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- 21.Huntington ND, et al. NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J Immunol. 2007;178(8):4764–4770. doi: 10.4049/jimmunol.178.8.4764. [DOI] [PubMed] [Google Scholar]
- 22.Robbins SH, Tessmer MS, Mikayama T, Brossay L. Expansion and contraction of the NK cell compartment in response to murine cytomegalovirus infection. J Immunol. 2004;173(1):259–266. doi: 10.4049/jimmunol.173.1.259. [DOI] [PubMed] [Google Scholar]
- 23.Moretta A, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19(1):197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy MK, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191(5):771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci USA. 2001;98(9):5164–5169. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez K, et al. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 2012;36(6):921–932. doi: 10.1016/j.immuni.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang L-J, et al. (2012) Coordinate regulation of Bcl11b activity in thymocytes by the MAPK pathways and protein sumoylation. J Biol Chem 287(32):26971–26988. [DOI] [PMC free article] [PubMed]
- 28.Lai CB, Mager DL. Role of runt-related transcription factor 3 (RUNX3) in transcription regulation of natural cytotoxicity receptor 1 (NCR1/NKp46), an activating natural killer (NK) cell receptor. J Biol Chem. 2012;287(10):7324–7334. doi: 10.1074/jbc.M111.306936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohno S-i, et al. Runx proteins are involved in regulation of CD122, Ly49 family and IFN-γ expression during NK cell differentiation. Int Immunol. 2008;20(1):71–79. doi: 10.1093/intimm/dxm120. [DOI] [PubMed] [Google Scholar]
- 30.Yoneyama M, et al. Structural and functional differences of SWIRM domain subtypes. J Mol Biol. 2007;369(1):222–238. doi: 10.1016/j.jmb.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 31. Luevano ME, Madrigal A, Saudemont A (2012) Transcription factors involved in the regulation of natural killer cell development and function: An update. Front Immunol 3:319. [DOI] [PMC free article] [PubMed]
- 32.Gascoyne DM, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10(10):1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 33.Karaya K, et al. Regulation of Id2 expression by CCAAT/enhancer binding protein β. Nucleic Acids Res. 2005;33(6):1924–1934. doi: 10.1093/nar/gki339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haas NB, Cantwell CA, Johnson PF, Burch JB. DNA-binding specificity of the PAR basic leucine zipper protein VBP partially overlaps those of the C/EBP and CREB/ATF families and is influenced by domains that flank the core basic region. Mol Cell Biol. 1995;15(4):1923–1932. doi: 10.1128/mcb.15.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, Liu J, Jo M, Curry TE., Jr A role for nuclear factor interleukin-3 (NFIL3), a critical transcriptional repressor, in down-regulation of periovulatory gene expression. Mol Endocrinol. 2011;25(3):445–459. doi: 10.1210/me.2010-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu Rev Biochem. 2006;75(1):243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 37.McKenna HJ, et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95(11):3489–3497. [PubMed] [Google Scholar]
- 38.Cheng M, et al. Distinct and overlapping patterns of cytokine regulation of thymic and bone marrow-derived NK cell development. J Immunol. 2009;182(3):1460–1468. doi: 10.4049/jimmunol.182.3.1460. [DOI] [PubMed] [Google Scholar]
- 39.Carotta S, Pang SHM, Nutt SL, Belz GT. Identification of the earliest NK-cell precursor in the mouse BM. Blood. 2011;117(20):5449–5452. doi: 10.1182/blood-2010-11-318956. [DOI] [PubMed] [Google Scholar]
- 40.Gwin K, Dolence JJ, Shapiro MB, Medina KL. Differential requirement for Hoxa9 in the development and differentiation of B, NK, and DC-lineage cells from Flt3+ multipotential progenitors. BMC Immunol. 2013;14(1):5. doi: 10.1186/1471-2172-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oguro H, et al. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell. 2010;6(3):279–286. doi: 10.1016/j.stem.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Stock JK, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9(12):1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 43.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474(7351):337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.