Significance
The infrequent detection of circulating tumor cells (CTCs) has hindered their clinical implication and their potential use in the sense of a “liquid biopsy” for cancer diagnosis and therapy. Hypothesizing that the limited blood volume commonly used for CTC analysis (1–10 mL) accounts for variable detection rates, we used leukapheresis to screen large blood volumes for CTCs. This enabled a more reliable detection of CTCs at high frequency even in nonmetastatic cancer patients. Thus, diagnostic leukapheresis may facilitate the routine clinical use of CTCs as biomarkers for personalized medicine. Combined with technologies for single-cell molecular genetics or cell biology, it may significantly improve prediction of therapy response and monitoring, especially in early systemic cancer.
Keywords: minimal residual disease, single-cell analysis, metastasis, comparative genomic hybridization, gastrointestinal cancer
Abstract
Circulating tumor cells (CTCs) are promising biomarkers for diagnosis and therapy in systemic cancer. However, their infrequent and unreliable detection, especially in nonmetastatic cancer, currently impedes the clinical use of CTCs. Because leukapheresis (LA) targets peripheral blood mononuclear cells, which have a similar density to CTCs, and usually involves processing the whole circulating blood, we tested whether LA could substantially increase CTC detection in operable cancer patients. Therefore, we screened LA products generated from up to 25 L of blood per patient in two independent studies, and found that CTCs can be detected in more than 90% of nonmetastatic breast cancer patients. Interestingly, complete white blood cell sampling enabled determining an upper level for total CTC numbers of about 100,000 cells (median, 7,500 CTCs) per patient and identified a correlation of CTC numbers with anatomic disease spread. We further show that diagnostic leukapheresis can be easily combined with the US Food and Drug Administration-approved CellSearch system for standardized enumeration of CTCs. Direct comparison with 7.5 mL of blood revealed a significantly higher CTC frequency in matched LA samples. Finally, genomic single-cell profiling disclosed highly aberrant CTCs as therapy-escaping variants in breast cancer. In conclusion, LA is a clinically safe method that enabled a reliable detection of CTCs at high frequency even in nonmetastatic cancer patients, and might facilitate the routine clinical use of CTCs as in the sense of a liquid biopsy. Combined with technologies for single-cell molecular genetics or cell biology, it may significantly improve prediction of therapy response and monitoring of early systemic cancer.
Circulating tumor cells (CTCs) hold promise as relevant biomarkers for early detection of systemic cancer spread, surrogate markers for therapy monitoring, and for direct access to the molecular characteristics of early and advanced metastatic disease (1, 2). However, in the past 20 y, their infrequent and unreliable detection has prevented clinical routine use of CTCs. The search for CTCs in 1–10 mL of peripheral blood by various methods has generated controversial results. For example, CTCs were detected in nonmetastatic patients [Union for International Cancer Control (UICC) stage M0 (nonmetastatic)] between 1 and 100,000 cells per mL of blood in 5–100% of patients (3–8). Although reliable methods for single-cell genomics have been available for more than 10 y (9), most of the studies did not provide direct genetic proof of malignancy for the detected cells, raising doubts about some of the reported results. Successful experiments using immunomagnetic enrichment of cancer cells expressing the membranous epithelial cell adhesion molecule (EpCAM) protein (10) led to the development of the US Food and Drug Administration (FDA)-approved CellSearch system, which has become the most widely used standard for CTC detection (11). With this method, baseline CTC counts (≥3 or ≥5 CTCs per 7.5-mL blood sample, depending on the tumor type) are observed in 26–49% of patients with metastatic cancer (12). The detection rate is even lower in nonmetastatic cancer (5–24%), usually with a median count of only one CTC per 7.5 mL (5, 9, 11, 13). Although 20–30 mL of blood was used in some CellSearch studies in nonmetastatic cancer (13–15) and more recently developed CTC detection devices claimed higher detection rates (16), all available systems failed to detect CTCs at an acceptable rate and yield in a large fraction of patients, especially in M0 cancer patients. Therefore, the clinical use of CTCs is currently limited.
We reasoned that this problem could be overcome by substantially increasing the analyzed peripheral blood (PB) volume. To this end, leukapheresis (LA) is a standard clinical method that is frequently used to isolate mononuclear cells (MNCs) from blood for various applications including stem cell harvest. LA enables the extracorporeal continuous density-based cell separation of MNCs with a density of 1.055–1.08 g/mL from several liters of total processed blood (TPB) volume. Because one to three blood volumes are commonly processed in adults and because the density of epithelial cells falls in the optimal range of LA, we reasoned that CTCs could be collected together with MNCs and enriched in LA products. The high processed blood volume would then reduce sampling error of venipuncture. To test our hypothesis, we analyzed two independent LA sample sets for CTC prevalence: a historical sample collective from M0 breast cancer patients, and LA samples from a prospective validation study in gastrointestinal cancer and breast cancer of different disease stages.
Results
Cytokeratin-Positive Cells Are Frequent in Products Generated from High-Volume LAs.
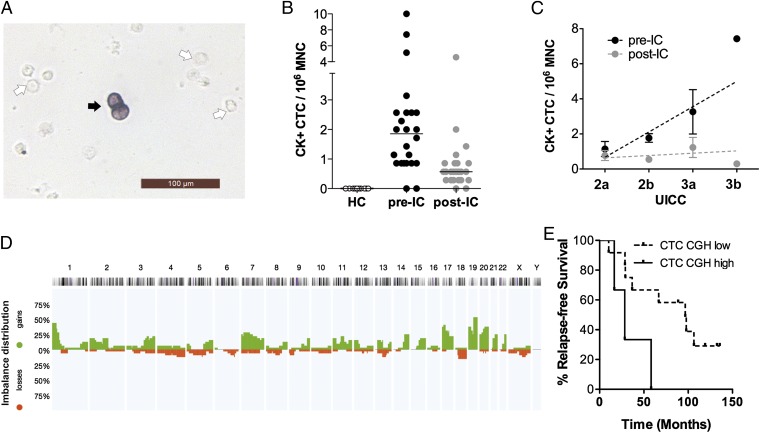
In a first set of experiments, we screened historical LA products for CTCs that were harvested after resection of the primary tumor in a controlled clinical trial (AM-01) (17) testing a regimen of high-dose chemotherapy for nonmetastatic breast cancer (patient cohort 1; Table S1). We applied immunohistochemistry to slides prepared from 48 LA samples of 24 breast cancer patients using the anti-cytokeratin antibody A45-B/B3 directed against the cytokeratins (CKs) 8, 18, and 19 for CTC detection. For each patient, one LA sample had been collected before induction of chemotherapy (pre-IC) and one after induction of chemotherapy (post-IC). In total, we detected CK-positive cells in 91.7% (44/48) of LA samples, which we designated CK+CTCs (Fig. 1A). In contrast, we could not detect CK+ cells in LA samples from 10 healthy female donors (Fig. 1B). The median CK+CTC count in LA samples isolated from cancer patients was 0.86 per 106 MNCs (range, 0.3–10 CK+CTCs), a concentration about 20-fold higher than typically reported for the analysis of peripheral blood (13). Interestingly, the pre-IC samples displayed significantly higher CK+CTC counts than matched post-IC samples (1.86 vs. 0.57; Wilcoxon test, P < 0.001). Furthermore, we noted a trend that advanced tumor stages correlated positively with CK+CTC counts in the pre-IC samples (Spearman r = 0.39, P = 0.06) but not in the post-IC samples (Spearman r = –0.004, P = 0.98) (Fig. 1C).
Fig. 1.
CK+CTCs are highly prevalent in LA samples and display genomic alterations characteristic for cancer cells. (A) Immunocytochemistry of an LA product. The black arrow points to a CK-positive cell, and the white arrows point to examples for CK-negative cells. (B) Number of CK-positive cells in healthy G-CSF–treated controls (HC; n = 10) in breast cancer cases (M0) before induction of chemotherapy (pre-IC, n = 24) and after induction of therapy (post-IC, n = 24), respectively. The horizontal black bars indicate the median. (C) Correlation of the number of CK+ cells with the UICC stage. The slope of the line for the pre-IC samples (n = 24) was 1.444 [95% confidence interval (CI), 0.4317–2.456] and the correlation coefficient (r2) was 0.285. The slope deviated significantly from zero (P = 0.0073). For post-IC samples (n = 24), the slope of the line was 0.133 (95% CI, 0.3416–0.6074; r2 = 0.015). The slope did not deviate significantly from zero (P = 0.57). The dots illustrate the mean value; error bars indicate the SEM. The regression line is shown as a dotted line. (D) Horizontal view of the chromosomal gains (above the horizontal line in green) and losses (below the horizontal line in red) detected by CGH in 32 of the 65 single CK-positive cells isolated from cohort 1. The mean aberration number of CK+CTCs with detectable CGH alterations was 5.5 (range, 1–25). (E) Survival of CTC-positive cases with higher (>10) vs. lower (≤10) mean number of chromosomal aberrations.
Highly Aberrant CK+CTCs in Transplanted LA Products Are Associated with Early Metastasis.
Next, we wanted to confirm the malignant nature of CK+CTCs and performed single-cell comparative genomic hybridization (CGH) to assess chromosomal gains and losses characteristic for cancer cells. We successfully isolated and analyzed 65 single CK+CTCs from 19 of the 24 patients with LA samples. The observed chromosomal alterations (Fig. 1D) were generally consistent with breast cancer [compared with those in the Progenetix database (18); n > 2,000], and comprised gains at chromosomes 3q, 8q, and 17q and losses at chromosomes 4 and 8p. Notably, 51% of the CK+CTCs (n = 33) displayed a balanced CGH profile without chromosomal gains or losses. Despite this, we detected aberrant cells in 17 of the 19 (89%) patients in which we could isolate CK+CTCs, validating the malignant nature of the CK+CTCs in most analyzed patients. Because it was previously shown that highly aberrant disseminated tumor cells (DTCs) isolated from bone marrow indicated metastatic disease in breast cancer (19), we tested whether the extent of chromosomal alteration of CK+CTCs confers risk for disease progression. Our analysis revealed that >10 alterations per CTC in the transplanted (post-IC) apheresis product were associated with significantly shorter metastasis-free survival (log-rank test, P = 0.023; Fig. 1E, Fig. S1, and Table S2).
Validating the High CTC Frequency in LA Products of M0 Patients in a Prospective Study.
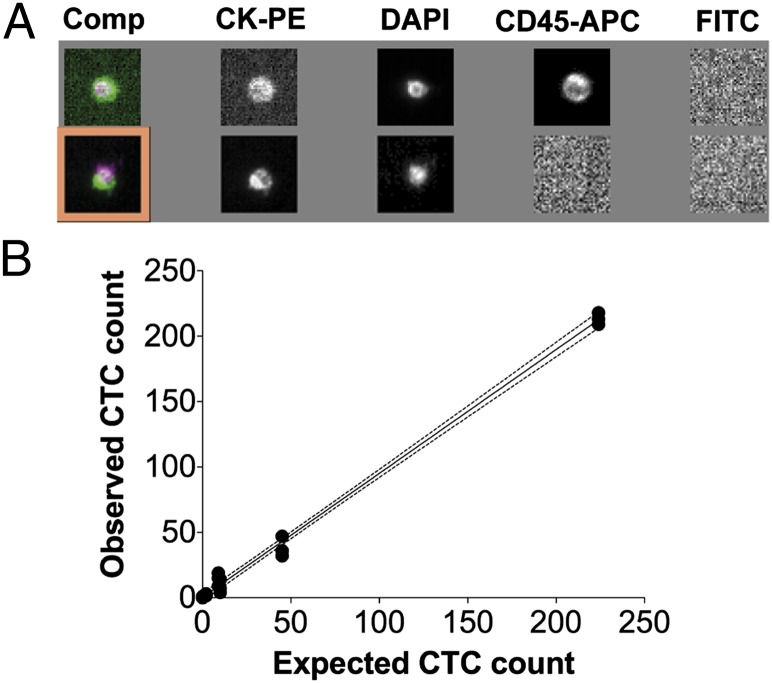
We then performed a prospective study to directly compare the CTC prevalence in fresh LA products with matched PB samples using the standardized CellSearch CTC isolation system. The CellSearch assay enriches EpCAM-positive cells and identifies CTCs by positive cytokeratin staining in the absence of the pan-leukocyte marker CD45 (CK+/CD45– cells). Application of this strict criterion for CTC identification was important, because we noted a high background of CK+/CD45+ double-positive cells in healthy donors (Fig. 2A). We deem it likely that such cells result from false-positive CK+ staining of leukocytes, especially in LA samples with high white blood cell counts. Excluding CK+/CD45+ double-positive cells, only one single CK+/CD45– cell was observed in LA samples from 14 healthy donors (Fig. 3A). We next tested tumor cell recovery of the CellSearch system for LA products using healthy donor samples spiked with varying numbers of SK-BR-3 breast cancer cells. This experiment revealed a linear detection rate across the entire tested analytical range (Fig. 2B and Table S3).
Fig. 2.
Adapting the CellSearch system to DLA products. (A) Representative examples for a dual positive cell (Upper) as frequently observed in DLA products and a CK+/CD45– cell determined to be a CTC (Lower). Comp, composite image of PE, DAPI and APC; PE, phycoerythrin; APC, allophycocyanin. (B) Recovery of known numbers of spiked SK-BR-3 cells (n = 0, 2, 9, 10, 45, and 224) within the background of DLA products (1–2 × 108 white blood cells in a volume of 7.5 mL) from healthy donors. Three independent experiments were done, and each filled circle represents an individual data point (n = 18). The dotted lines above and below the regression line (straight black line) display the 95% confidence interval (slope, 0.947; 95% CI, 0.915–0.979; r2 = 0.996).
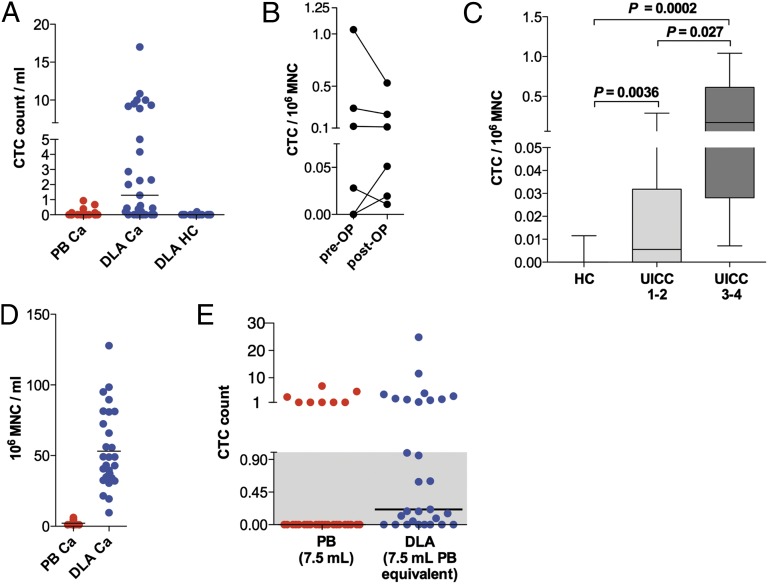
Fig. 3.
Prospective study to validate the high prevalence of CTCs in diagnostic leukapheresis products using standardized CTC detection. (A) Number of CTCs detected via CellSearch per mL in DLA and PB samples (Ca, cancer, n = 29; HC, healthy control, n = 14). (B) DLA-CTC counts per 106 MNCs before (pre-OP) and after surgery (post-OP) (n = 6). (C) CTC numbers detected in HC (n = 14), UICC1–2 samples (pre-OP; n = 15), and UICC3–4 samples (pre-OP/pretherapy; n = 5). Patients who received neoadjuvant therapy were excluded. P values were calculated with Mann–Whitney tests. Whiskers represent total range. (D) MNCs per mL in peripheral blood samples (n = 29) versus DLA samples from cancer patients (n = 29). (E) CTC counts in 7.5 mL of PB equivalent calculated from DLA data (blue) vs. 7.5 mL of PB sample (red). Gray highlights the values below the detection limit set at 1.0 for 7.5 mL of PB equivalents that were scored as CTC-negative. Horizontal lines indicate the median.
Having established the suitability of the CellSearch system for LA products, we prospectively performed 29 diagnostic LA (DLA) in 23 patients with different cancer types, including breast, colorectal, esophageal, and pancreatic cancers (patient cohort 2; Table 1). Just before starting the DLA, we drew 7.5 mL of PB and subjected both sample types to the CellSearch assay. Strikingly, we detected CTCs in 21 of the 29 DLA samples (72%), whereas only 8 of the 29 (28%) PB samples (Fisher’s exact test, P = 0.0014) were positive for CTCs. The CTC count per mL was also markedly higher in DLA samples compared with their matched PB samples (Fig. 3A; Mann–Whitney test, P < 0.0001). Importantly, we could confirm the high CTC detection rate of 90% for breast cancer-derived DLA samples, similar to the first patient cohort (CTC positivity, DLA 9/10 vs. PB 3/10; Fisher’s exact test, P = 0.0198). Investigating postoperative DLA in a subset of patients (n = 6) confirmed that CTCs persist after tumor resection (Fig. 3B). Despite the relatively small number of performed postoperative DLAs in cohort 2, the CTC prevalence in this subset was significantly different when tested against the healthy control patients (6/0 vs. 1/14; Fisher’s exact test, P < 0.001). Next, we asked whether the CTCs detected in DLA products by CellSearch reflect the tumor stage. As before with pre-IC breast cancer samples, the CTC frequency was correlated to the UICC stage (Spearman r = 0.45, Spearman correlation, P = 0.03) (Fig. 3C).
Table 1.
UICC stages and CTC numbers detected in PB and DLA samples of cohort 2
| Patient no./DLA no. | Diagnosis | UICC stage | PB samples |
DLA samples |
|||
| CTCs per 7.5 mL | CTCs per 106 MNCs* | CTCs in analyzed sample | CTCs per DLA product | CTCs per 106 MNCs* | |||
| 1/1 | BC | 1a | 0 | 0.00 | 1 | 24 | 0.01 |
| 2/2 | BC | 2a | 1 | 0.07 | 0 | 0 | 0.00 |
| 2/3 | BC | 1 | 0.05 | 5 | 150 | 0.05 | |
| 3/4 | BC | 2a | 0 | 0.00 | 0 | 0 | 0.00 |
| 4/5 | BC | 2a | 0 | 0.00 | 5 | 109 | 0.03 |
| 4/6 | BC | 0 | 0.00 | 2 | 35 | 0.01 | |
| 5/7 | BC | 2b | 1 | 0.03 | 3 | 108 | 0.03 |
| 6/8 | BC | 2b | 0 | 0.00 | 13 | 1,148 | 0.29 |
| 6/9 | BC | 0 | 0.00 | 11 | 678 | 0.23 | |
| 7/10 | BC | 3a | 0 | 0.00 | 18 | 400 | 1.04 |
| 7/11 | BC | 0 | 0.00 | 51 | 884 | 0.53 | |
| 8/12 | PAC | 1 | 0 | 0.00 | 0 | 0 | 0.00 |
| 9/13 | PAC | 2b | 0 | 0.00 | 1 | 9 | 0.01 |
| 10/14 | PDAC | 2b | 0 | 0.00 | 2 | 4 | 0.01 |
| 11/15 | PDAC | 2b | 0 | 0.00 | 0 | 0 | 0.00 |
| 12/16 | PDAC | 2b | 0 | 0.00 | 0 | 0 | 0.00 |
| 13/17 | PDAC | 2b | 1 | 0.05 | 14 | 467 | 0.09 |
| 14/18 | PDAC | 2b | 7 | 0.40 | 6 | 300 | 0.12 |
| 14/19 | PDAC | 0 | 0.00 | 12 | 563 | 0.11 | |
| 15/20 | PDAC | 4 | 5 | 0.23 | 32 | 240 | 0.18 |
| 16/21 | PDAC | 4 | 1 | 0.05 | 1 | 8 | 0.01 |
| 17/22 | PDAC | 4 | 0 | 0.00 | 4 | 110 | 0.05 |
| 18/23 | PDAC | 4 | 3 | 0.24 | 20 | 500 | 0.17 |
| 19/24 | CRC | 2a | 0 | 0.00 | 2 | 77 | 0.01 |
| 20/25 | ESCC | 1a | 0 | 0.00 | 0 | 0 | 0.00 |
| 20/26 | ESCC | 0 | 0.00 | 2 | 17 | 0.02 | |
| 21/27 | ESCC | NA | 0 | 0.00 | 0 | 0 | 0.00 |
| 22/28 | EAC | NA | 0 | 0.00 | 8 | 134 | 0.08 |
| 23/29 | EAC | NA | 0 | 0.00 | 0 | 0 | 0.00 |
For cases in which pre- and postoperative DLAs were perfomed UICC information is provided only once. BC, breast cancer; CRC, colorectal cancer; EAC, adenocarcinoma of the esophagus; ESCC, squamous cell carcinoma of the esophagus; NA, no UICC stage is available because of neoadjuvant therapy (no. 21, yT0N0M0; no. 22, yT0N1M0; no. 23, yT0N0M0); PAC, ampullary cancer; PDAC, pancreatic ductal adenocarcinoma.
Values provided here are rounded to two decimal places.
Finally, we checked whether the high CTC detection rate in DLA products could be attributed mainly to the increased blood volume that was screened. Because MNCs are the main cell population isolated by DLA (Fig. 3D), we used this population as a reference and calculated the CTC number per 106 MNCs for each matched PB–DLA sample pair. Thereby, we determined a median peripheral blood volume of 60.2 mL as equivalent to the DLA volume that we screened (median, 2.3 mL) in the CellSearch assay. Then, we extrapolated from our DLA results the number of CTCs in 7.5 mL of PB and found a high congruency to the observed counts in the empirical PB sample (8/29 vs. 10/29; Fisher’s exact test, P = 0.79; Fig. 3E). This indicates that CTCs are concentrated together with the MNC population (Fig. 3D) during DLA.
Assuming a homogeneous distribution of CTCs in DLA products, we calculated the total number of CTCs per patient. The median blood volume of cohort 2 patients comprised 4.5 L, and on average we collected MNCs from 62% of the blood volume. The total number of CTCs in the circulation ranged from 29 to 13,102 in CTC-positive patients of cohort 2 (median, 613; calculations are provided in SI Materials and Methods). In cohort 1, total CTC numbers can be estimated to range from 2,455 to 81,854 (median, 7,364).
Discussion
CTCs hold great promise for diagnosis and therapy of systemic cancer as a “liquid biopsy,” but low detection rates are thus far hindering their clinical routine use. Standard CTC blood tests usually analyze a restricted volume of 1–10 mL of PB. Our study reveals that such low blood volumes are insufficient. Instead, large blood volumes need to be screened for a reliable detection of CTCs, especially in nonmetastatic cancer patients. CTCs are concentrated along with the targeted MNC population during DLA, which significantly enhances the CTC detection frequency. The implementation of DLA as a marker-independent preanalytic CTC enrichment step into clinical workflows was unproblematic, and all patients tolerated the DLA procedure quite well.
The fraction of the DLA product that we analyzed in the CellSearch assay was equivalent to a PB volume of around 60 mL (theoretically eight CellSearch assays per patient). Notably, our data perfectly fit hypothetical detection rates that were predicted for a volume of 50–75 mL of PB when calculated from CellSearch assays (20). Another factor that might contribute to a more reliable CTC detection by DLA is that the final LA products were generated from at least more than half of the whole circulating blood volume (cohort 2), thereby reducing the sampling error of a simple venipuncture. Eifler et al. suggested by their proof-of-concept study using apheresis products spiked with cancer cell lines that an additional elutriation step might even further enhance CTC recovery after high-volume screening for CTCs by apheresis (21).
However, because of the high blood volumes screened and because DLA is not known to destroy cells, we think our approach provides a reliable upper estimate for cytokeratin-positive and cytokeratin/EpCAM double-positive CTCs, at least for nonmetastatic patients. The total number of CTCs in the circulation ranged from 29 to 13,102 in CTC-positive patients of cohort 2 (median, 613) and from 2,455 to 81,854 (median, 7,364) in cohort 1. This discrepancy may reflect the differences of cohort 1 in comparison with cohort 2: a significantly higher median TPB processed (15.1 vs. 2.7 L; Mann–Whitney test, P < 0.0001), the administration of growth factors for stem cell mobilization, and the use of an EpCAM-independent CTC detection assay. However, these numbers are in striking contrast to previous reports (22) that found a range from to 2.2 × 104 to 3.7 × 106 CTCs in patients with different types of metastatic cancer and in localized prostate cancer [a median of 2.2 × 105 CTCs given 50 CTCs per mL (16)], although that approach used EpCAM enrichment and cytokeratin detection as we did. Importantly, only around 1 mL of PB was investigated in these studies. Our data put a caveat on these claims of excessive CTC numbers in M0 patients (4, 22), particularly because LA is not known to damage or destroy cells. Therefore, DLA provides an upper estimate for minimal residual disease (MRD) in the circulation, comprising usually less than 10,000 CTCs per patient, and we deem it unlikely that CTC numbers are several-fold higher. Whether CTCs increase on average 100-fold in M1-stage patients, as suggested by other approaches (22, 23), needs to be explored. Among our patients, M1-stage patients harbored about 10-fold more CTCs than M0-stage patients.
The reliable and high CTC detection rates in LA products allowed us to gain insight into MRD in M0-stage breast cancer patients. Notably, we could observe in this MRD situation decreasing CTC counts over time. Whether the dropping CTC counts were a direct consequence of the adjuvant chemotherapy, for example, as described for CTCs in peripheral blood by Bozionellou et al. (24), or reflect the natural course of MRD (25) remains unclear; however, this question should be addressed in future DLA studies comparing patients treated with and without adjuvant systemic therapies. Further, our data indicate that CTC numbers correspond to anatomical disease spread even weeks after resection of the primary tumor in locally advanced breast cancer. The missing correlation in the case of post-IC samples may then reflect individual therapy responses of the residual disease, blurring the footprint of the original disease stage. From our comprehensive genomic analysis of single CTCs, we learned that CTCs with more than 10 chromosomal aberrations are associated with early relapse. Therefore, highly aberrant CTCs may either inform about a more advanced evolutionary stage of minimal residual cancer that readily progresses to metastasis or that those highly aberrant CTCs, once retransplanted, might drive disease relapse after high-dose chemotherapy. Obviously, these conclusions are drawn from a limited dataset and warrant further validation by future studies in larger cohorts. However, consistent with the latter thought are observations that after high-dose chemotherapy, transplantation of purified LA-CD34+ stem cells resulted in significantly longer survival of metastatic breast cancer patients (26). On the other hand, about 50% of CK+CTCs showed a balanced metaphase CGH profile. Although the origin of these cells currently remains unclear, this finding is reminiscent of MRD in bone marrow, where likewise 50% of cytokeratin-positive DTCs were found to harbor normal karyotypes (19) but were subsequently shown to display breast cancer-associated loss of heterozygosity and amplifications when investigated at higher resolution (27). It is therefore tempting to speculate that genomically nonprogressed DTCs can be mobilized from the bone marrow during hematopoietic stem cell harvest.
In conclusion, we show that commonly analyzed blood volumes of 1–10 mL are too low to detect rare CTCs but that the clinically safe DLA reliably enables detection of CTCs in M0-stage cancer patients. The high CTC numbers observed in leukapheresis products might enable a more comprehensive biological and functional characterization of CTCs to determine cells with metastatic capacity among the heterogeneous CTC population. The clinical value of DLA, which is more invasive than a simple venipuncture, is clearly related to the further molecular and functional characterization of CTCs in the context of personalized molecular therapies. We envision that screening high volumes of blood will enable a true liquid biopsy for solid cancers and opens the possibility of using CTCs as biomarkers to guide and monitor systemic therapies even in the adjuvant therapy setting.
Materials and Methods
Historical LA Samples from M0 Breast Cancer Patients: Cohort 1.
We retrospectively studied retained cryopreserved samples of LA products from 24 patients participating in the West Germany Study Group AM-01 trial (17). This phase III trial compared an intensive double-cycle high-dose chemotherapy regimen with an accelerated conventionally dosed regimen as an adjuvant therapy in nonmetastatic high-risk breast cancer. The experimental arm underwent two LAs for stem cell support after the primary tumor was completely resected, one before the first administration of chemotherapy as a backup procedure and the second apheresis after the administration of the first two cycles of epirubicin and cyclophosphamide as induction therapy. The median TPB of the LAs was 15.1 L (range, 7.7 L–25.2 L). LA was performed via a central catheter, and stem cell mobilization was augmented by growth-factor support (G-CSF; filgrastim 10 µg/kg body weight) (17). For our study, we identified 24 cases for which remaining LA-retained cell samples, stored in liquid nitrogen, from both time points were available (Table S1). LA procedures were performed between November 1995 and April 1998. The median postsurgical time period was 18 d (range, 13–28 d) for the first LA sample and 76 d (range, 61–105 d) for the second sample. In addition, 10 retained cryopreserved LA samples from unrelated sex-matched healthy G-CSF–mobilized donors were prepared and analyzed according to the protocols used in the cancer patients. The study was approved by the ethics committee of the Heinrich Heine University Düsseldorf. All patients/healthy donors gave written informed consent to use surplus material of LA products not needed for clinical use for medical research. The cryopreserved samples were rapidly thawed in a 37 °C water bath, diluted 1:1 RPMI 1640/15% (vol/vol) FCS containing dornase alfa (Pulmozyme; Hoffmann-La Roche) at 250 U/mL, and incubated for 5 min to prevent clotting. After the incubation, the samples were slowly transferred to 30 mL of RPMI 1640 medium to wash the cells. After a washing step with PBS, 2 mM EDTA, 0.5% inactivated BSA, the cells were centrifuged at 400 × g for 5 min (without a break). MNCs were resuspended at a cell dilution of 1 × 106 cells per mL of PBS and were placed on positively charged glass slides (Menzel) at a density of 250,000 cells per 227 mm2. After sedimentation for 45 min, the slides were dried overnight and stored at –20 °C until further analysis. The detection, isolation, and characterization of CK+ cells were done as previously described (19, 28). A more detailed description is provided in SI Materials and Methods.
Prospective Study for CTC Enrichment by Diagnostic LA: Cohort 2.
To validate our data from cohort 1 in an independent patient cohort, we performed a prospective study. We determined the CTC detection rate in fresh DLA products and compared it with the CTC detection rate in peripheral blood immediately drawn before DLA. For this we used the FDA-approved CellSearch assay for CTC detection in PB and DLA samples. Patients with clinically diagnosed breast cancer, pancreatic cancer, and other gastrointestinal cancer types (Table 1) were included in the study. In cancer patients scheduled for surgery, we conducted DLA at least 1 d before. DLA after surgery was carried out around the 14th postoperative day. To increase patient comfort and clinical applicability, the DLA differed from the LA procedure for hematopoietic stem cell harvesting in cohort 1 as follows: (i) no growth-factor support was used in cohort 2; (ii) instead of a central catheter, only peripheral vein access was used to process peripheral blood; and (iii) the median processed blood volume of 2.6 L (range, 0.8–6.2) was significantly lower compared with cohort 1 (Mann–Whitney U test, P < 0.001). We also included 14 healthy donors whose LA samples were prepared identically by collecting MNCs using cell separator COBE Spectra software version 7.0, MNC program (Terumo BCT). Blood was anticoagulated with citrate dextrose solution A (ACD-A solution; Fresenius HemoCare) at a ratio starting from 12:1 up to 20:1. DLA products were adjusted to at least a 12:1 ACD-A ratio (with ACD-A). The prospective study was approved by the ethics committee of the Heinrich Heine University Hospital Düsseldorf. All participating patients and healthy donors gave written informed consent.
CellSearch Analysis of Fresh Diagnostic LA Samples.
All CTCs of cohort 2 were enumerated with the CellSearch system (Veridex) in PB and DLA samples by trained personnel. DLA and PB samples were collected in CellSave Preservative Tubes, maintained at room temperature until use, and processed within 96 h using the CellSearch Circulating Tumor Cell Kit and CellTracks Auto Prep System. CTCs were identified using Cell Tracks Analyzer II strictly according to CellSearch criteria. PB samples (7.5 mL) were drawn immediately prior to leukapheresis and were further processed as recommended by the manufacturer. From each cancer patient, 1–6 mL (median, 2.3 mL) of the fresh DLA product and 7.5 mL of PB were subjected in parallel to CellSearch analysis. Up to 2 × 108 white blood cells (WBCs) of each patient’s DLA product were brought up to a final volume of 4–8 mL with CellSearch Circulating Tumor Cell Kit Dilution Buffer before analysis and processed as a control within the CellTracks Auto Prep System.
To determine CTC recovery rates from DLA products, up to 2 × 108 WBCs from healthy donors were spiked with ∼225, 45, 10, 9, 2, and 0 fixed cells of breast cancer cell line SK-BR-3 (Table S3). At least three independent experiments were done for each SK-BR-3 concentration. In parallel, an aliquot of SK-BR-3 cells in dilution buffer was analyzed per run and used as a reference for calculating expected values.
Statistical Analysis.
To test the equality of two binomial proportions, Fisher’s exact test was performed. The significance of differences between groups with a nonparametric data distribution was analyzed with the Mann–Whitney U test for two independent groups. For comparison of paired nonparametric data, we used the Wilcoxon test. CK+CTC counts were correlated with UICC stages of the patients by linear regression analysis.
We used the R software package (www.r-project.org) to compute the estimated Kaplan–Meier–based mean relapse-free survival based on the continuous covariate “CGH aberration per CTC” (29) (SI Materials and Methods). Differences in relapse-free survival regarding >10 chromosomal aberrations were analyzed with the log-rank test.
To validate the results of cohort 1, we conducted a prospective validation study. Assuming a prevalence of CK-positive cells in healthy volunteers, apheresis products of 5% (observed in 0% of healthy controls of cohort 1) and in cancer patients of 95%, seven individuals per group should detect this difference at a significance level of 0.01 (two-sided) and with a power of 95% (30). Thus, we studied 29 diagnostic leukapheresis products from 23 clinically diagnosed cancer patients and compared this with 14 healthy controls in our validation study (cohort 2).
Differences between groups were considered significant if the P value was <0.05 in a two-tailed test. If not otherwise stated, the statistical data analysis was carried out using Prism 5 (GraphPad Software), SPSS 19 (IBM), and Excel (Microsoft).
Supplementary Material
Acknowledgments
We thank J. Wettke for his philanthropic support of our study and S. Seidschner for excellent technical assistance. This research was supported by a grant from the Krebsgesellschaft North Rhine-Westphalia (NRW) (to J.C.F., D.N., W.J., W.T.K., and N.H.S.), the Krebsstiftung NRW (to D.N. and N.H.S.), and in part by a grant from the Deutsche Forschungsgemeinschaft (STO 464/2-2 to N.H.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313594110/-/DCSupplemental.
References
- 1.Kaiser J. Cancer’s circulation problem. Science. 2010;327(5969):1072–1074. doi: 10.1126/science.327.5969.1072. [DOI] [PubMed] [Google Scholar]
- 2.Kling J. Beyond counting tumor cells. Nat Biotechnol. 2012;30(7):578–580. doi: 10.1038/nbt.2295. [DOI] [PubMed] [Google Scholar]
- 3.Riethdorf S, Pantel K. Advancing personalized cancer therapy by detection and characterization of circulating carcinoma cells. Ann N Y Acad Sci. 2010;1210:66–77. doi: 10.1111/j.1749-6632.2010.05779.x. [DOI] [PubMed] [Google Scholar]
- 4.Pachmann K, et al. Monitoring the response of circulating epithelial tumor cells to adjuvant chemotherapy in breast cancer allows detection of patients at risk of early relapse. J Clin Oncol. 2008;26(8):1208–1215. doi: 10.1200/JCO.2007.13.6523. [DOI] [PubMed] [Google Scholar]
- 5.Thalgott M, et al. Detection of circulating tumor cells in different stages of prostate cancer. J Cancer Res Clin Oncol. 2013;139(5):755–763. doi: 10.1007/s00432-013-1377-5. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, et al. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin Cancer Res. 2012;18(20):5701–5710. doi: 10.1158/1078-0432.CCR-12-1587. [DOI] [PubMed] [Google Scholar]
- 7.Rahbari NN, et al. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology. 2010;138(5):1714–1726. doi: 10.1053/j.gastro.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Rahbari NN, et al. Compartmental differences of circulating tumor cells in colorectal cancer. Ann Surg Oncol. 2012;19(7):2195–2202. doi: 10.1245/s10434-011-2178-1. [DOI] [PubMed] [Google Scholar]
- 9.Klein CA, et al. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc Natl Acad Sci USA. 1999;96(8):4494–4499. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Racila E, et al. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci USA. 1998;95(8):4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cristofanilli M, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 12.Danila DC, Pantel K, Fleisher M, Scher HI. Circulating tumors cells as biomarkers: Progress toward biomarker qualification. Cancer J. 2011;17(6):438–450. doi: 10.1097/PPO.0b013e31823e69ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucci A, et al. Circulating tumour cells in non-metastatic breast cancer: A prospective study. Lancet Oncol. 2012;13(7):688–695. doi: 10.1016/S1470-2045(12)70209-7. [DOI] [PubMed] [Google Scholar]
- 14.Lalmahomed ZS, et al. Circulating tumor cells and sample size: The more, the better. J Clin Oncol. 2010;28(17):e288–e289. doi: 10.1200/JCO.2010.28.2764. author reply e290. [DOI] [PubMed] [Google Scholar]
- 15.Franken B, et al. Circulating tumor cells, disease recurrence and survival in newly diagnosed breast cancer. Breast Cancer Res. 2012;14(5):R133. doi: 10.1186/bcr3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: Approaches to isolation and characterization. J Cell Biol. 2011;192(3):373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nitz UA, et al. West German Study Group Comparison of rapidly cycled tandem high-dose chemotherapy plus peripheral-blood stem-cell support versus dose-dense conventional chemotherapy for adjuvant treatment of high-risk breast cancer: Results of a multicentre phase III trial. Lancet. 2005;366(9501):1935–1944. doi: 10.1016/S0140-6736(05)67784-7. [DOI] [PubMed] [Google Scholar]
- 18.Baudis M, Cleary ML. Progenetix.net: An online repository for molecular cytogenetic aberration data. Bioinformatics. 2001;17(12):1228–1229. doi: 10.1093/bioinformatics/17.12.1228. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt-Kittler O, et al. From latent disseminated cells to overt metastasis: Genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci USA. 2003;100(13):7737–7742. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coumans FAW, Ligthart ST, Uhr JW, Terstappen LWMM. Challenges in the enumeration and phenotyping of CTC. Clin Cancer Res. 2012;18(20):5711–5718. doi: 10.1158/1078-0432.CCR-12-1585. [DOI] [PubMed] [Google Scholar]
- 21.Eifler RL, et al. Enrichment of circulating tumor cells from a large blood volume using leukapheresis and elutriation: Proof of concept. Cytometry B Clin Cytom. 2011;80(2):100–111. doi: 10.1002/cyto.b.20560. [DOI] [PubMed] [Google Scholar]
- 22.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y-F, et al. Circulating tumor cells: Advances in detection methods, biological issues, and clinical relevance. J Cancer Res Clin Oncol. 2011;137(8):1151–1173. doi: 10.1007/s00432-011-0988-y. [DOI] [PubMed] [Google Scholar]
- 24.Bozionellou V, et al. Trastuzumab administration can effectively target chemotherapy-resistant cytokeratin-19 messenger RNA-positive tumor cells in the peripheral blood and bone marrow of patients with breast cancer. Clin Cancer Res. 2004;10(24):8185–8194. doi: 10.1158/1078-0432.CCR-03-0094. [DOI] [PubMed] [Google Scholar]
- 25.Janni W, et al. The persistence of isolated tumor cells in bone marrow from patients with breast carcinoma predicts an increased risk for recurrence. Cancer. 2005;103(5):884–891. doi: 10.1002/cncr.20834. [DOI] [PubMed] [Google Scholar]
- 26.Müller AMS, et al. Long-term outcome of patients with metastatic breast cancer treated with high-dose chemotherapy and transplantation of purified autologous hematopoietic stem cells. Biol Blood Marrow Transplant. 2012;18(1):125–133. doi: 10.1016/j.bbmt.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schardt JA, et al. Genomic analysis of single cytokeratin-positive cells from bone marrow reveals early mutational events in breast cancer. Cancer Cell. 2005;8(3):227–239. doi: 10.1016/j.ccr.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Stoecklein NH, et al. Direct genetic analysis of single disseminated cancer cells for prediction of outcome and therapy selection in esophageal cancer. Cancer Cell. 2008;13(5):441–453. doi: 10.1016/j.ccr.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Gentleman R, Crowley J. Graphical methods for censored data. J Am Stat Assoc. 1991;86(415):678–683. [Google Scholar]
- 30.Faul F, Erdfelder E, Buchner A, Lang A-G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav Res Methods. 2009;41(4):1149–1160. doi: 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.