Significance
The Middle East respiratory syndrome coronavirus (MERS-CoV) is the latest emerged coronavirus causing severe respiratory disease with a high case fatality rate in humans. To better understand the disease caused by MERS-CoV, we developed a rhesus macaque model. Infection of rhesus macaques with MERS-CoV resulted in the rapid development of a transient pneumonia, with MERS-CoV replication largely restricted to the lower respiratory tract. This affinity of MERS-CoV for the lungs partly explains the severity of the disease observed in humans. The MERS-CoV rhesus macaque model will be instrumental in developing and testing vaccine and treatment options for an emerging viral pathogen with pandemic potential.
Keywords: emerging infectious disease, DPP4
Abstract
In 2012, a novel betacoronavirus, designated Middle East respiratory syndrome coronavirus or MERS-CoV and associated with severe respiratory disease in humans, emerged in the Arabian Peninsula. To date, 108 human cases have been reported, including cases of human-to-human transmission. The availability of an animal disease model is essential for understanding pathogenesis and developing effective countermeasures. Upon a combination of intratracheal, ocular, oral, and intranasal inoculation with 7 × 106 50% tissue culture infectious dose of the MERS-CoV isolate HCoV-EMC/2012, rhesus macaques developed a transient lower respiratory tract infection. Clinical signs, virus shedding, virus replication in respiratory tissues, gene expression, and cytokine and chemokine profiles peaked early in infection and decreased over time. MERS-CoV caused a multifocal, mild to marked interstitial pneumonia, with virus replication occurring mainly in alveolar pneumocytes. This tropism of MERS-CoV for the lower respiratory tract may explain the severity of the disease observed in humans and the, up to now, limited human-to-human transmission.
In June of 2012, a novel betacoronavirus, associated with severe respiratory disease in humans emerged in the Middle East (1, 2), which is closely related to betacoronaviruses circulating in bats (3, 4). The first isolate of Middle East respiratory coronavirus (MERS-CoV) (5), HCoV-EMC/2012, was obtained from a patient with a fatal pneumonia and acute renal failure. To date, 107 additional human cases have been identified, of which 49 were fatal (6). Aside from cases in Saudi Arabia, Qatar, Jordan, and the United Arab Emirates, imported cases have been identified in the United Kingdom, Germany, France, Tunisia, and Italy (6). Although no information is available on the source or route of primary transmission of MERS-CoV, human-to-human transmission has been recorded (7–9). Clinical data on human cases of MERS-CoV infection are currently sparse, but it appears that this virus mainly causes severe lower respiratory tract disease, occasionally accompanied by renal disease. The severity of disease distinguishes MERS-CoV from other coronaviruses circulating in the human population, HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1, which are generally associated with upper respiratory tract infections. Instead, MERS-CoV appears to be more similar to the severe respiratory disease caused by severe acute respiratory syndrome (SARS)-CoV.
In vitro studies have shown that MERS-CoV replicates efficiently in nonciliated cells in the primary human airway epithelium (10), and in ex vivo human lung cultures MERS-CoV replicated in bronchial, bronchiolar, and alveolar epithelial cells (11), in line with the observed respiratory disease in humans. The recently defined receptor for MERS-CoV, dipeptidylpeptidase 4 (DPP4), is generally expressed in endothelial and epithelial cells and has been shown to be present on cultured human nonciliated bronchiolar epithelium cells (12), providing further information on the respiratory tropism of MERS-CoV.
Animal models that recapitulate human disease are essential for understanding pathologic processes involved in disease progression. Moreover, these models are instrumental for the development of prophylactic and therapeutic countermeasures. We have previously shown that rhesus macaques inoculated with a high dose of MERS-CoV isolate HCoV-EMC/2012 developed a respiratory disease reminiscent of that observed in humans (13). To increase our understanding of the pathogenesis of MERS-CoV in the absence of clinical and pathological data from human patients, we present herein a more detailed analysis of the extent of virus replication, the histopathological changes in the respiratory tract and changes in systemic (peripheral blood mononuclear cell, PBMC) and local (lung tissue) gene expression of MERS-CoV–infected rhesus macaques.
Results
Clinical Signs and Hematological Changes in Animals Inoculated with MERS-CoV.
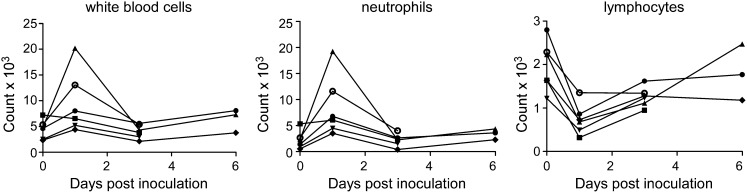
Six rhesus macaques were inoculated with 7 × 106 TCID50 (50% tissue culture infectious dose) of MERS-CoV isolate HCoV-EMC/2012 (13). Clinical signs upon inoculation with HCoV-EMC/2012 were transient and varied between animals, as specified in Table S1. Analysis of blood samples collected 0, 1, 3, and 6 d postinfection (dpi) indicated increased white blood cell counts in five of six animals on 1 dpi (Fig. 1). Further analysis indicated that this was the result of an increase in the number of neutrophils. Moreover, there was a reduction in the number of lymphocytes. By 3 dpi, blood cell counts had returned to numbers comparable to those before inoculation (Fig. 1).
Fig. 1.
Hematological changes in rhesus macaques inoculated with MERS-CoV. Total number of white blood cells (Left), neutrophils (Center), and lymphocytes (Right) were determined in blood samples obtained from animals 0, 1, 3, and 6 dpi. Of note, CoV-2, CoV-4, and CoV-6 were euthanized 3 dpi, and thus no samples are available for 6 dpi. Each line represents one animal: CoV-1 (●), CoV-2 (■), CoV-3 (▲), CoV-4 (▼), CoV-5 (♦), CoV-6 (○).
Viral RNA in Swabs and Bronchoalveolar Lavages.
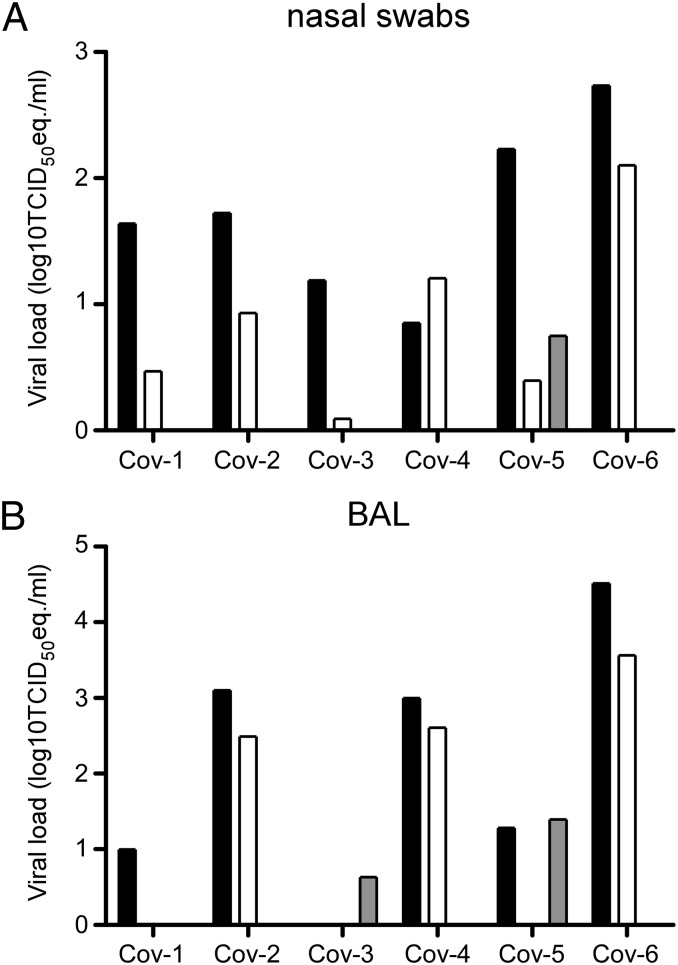
Nasal, oropharyngeal, urogenital, and rectal swabs were obtained from the animals 1, 3, and 6 dpi. On 1 and 3 dpi, nasal swabs from all animals were positive for the presence of viral RNA by quantitative RT-PCR (qRT-PCR); only one animal had a positive nasal swab on day 6 postinfection (Fig. 2A). Oropharyngeal swabs obtained from CoV-3 on days 3 and 6 postinfection and obtained from CoV-6 on day 6 postinfection were positive. All urogenital and rectal swabs were negative with the exception of a urogenital swab collected from CoV-2 on day 1 postinfection. Bronchoalveolar lavages (BAL) were also performed 1, 3, and 6 dpi and analyzed for the presence of viral RNA. BAL obtained 1 dpi were positive by qRT-PCR in five of six animals; 3 dpi three of six BALs were positive, and 6 dpi two of three BALs contained viral RNA (Fig. 2B).
Fig. 2.
Virus shedding in rhesus macaques inoculated with MERS-CoV. Nasal swabs (A) and BAL fluid (B) were collected 1, 3, and 6 dpi. Of note, CoV-2, CoV-4, and CoV-6 were euthanized 3 dpi and thus no samples are available for 6 dpi. RNA was extracted and viral load was determined as TCID50 equivalents by qRT-PCR. TCID50 equivalents were extrapolated from standard curves generated by adding dilutions of RNA extracted from a HCoV-EMC/2012 stock with known virus titer in parallel to each run. Animal numbers are indicated on the x axis. Black bars indicate viral load at 1 dpi, white bars indicate viral load at 3 dpi, and gray bars indicate viral load at 6 dpi.
Widespread Dissemination of Virus Through the Respiratory Tract.
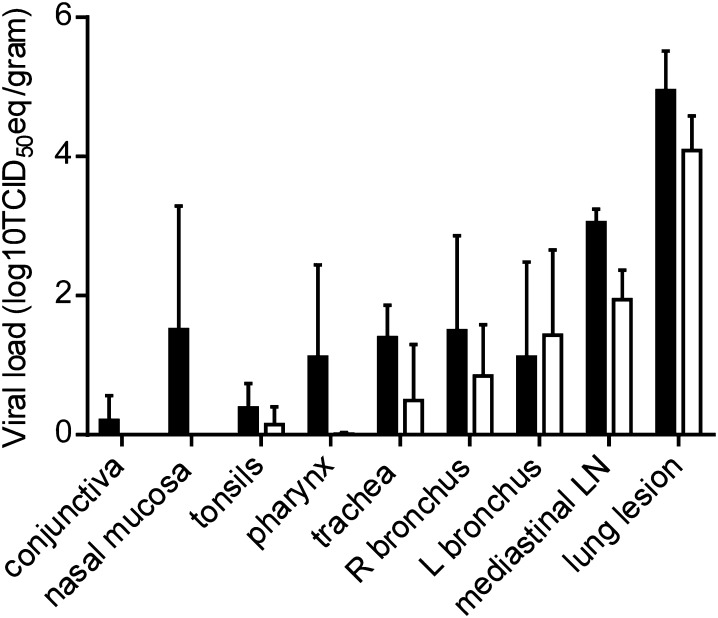
We have previously reported the presence of viral RNA and infectious virus throughout the lungs of inoculated macaques, with viral load decreasing between 3 and 6 dpi (13). In addition to the different lung lobes, we have previously analyzed several other tissues, including tissues of the upper respiratory tract, lung lesions, and kidney (for complete list, see Materials and Methods). Although there was some variation between the different macaques with regard to the presence of viral RNA, we could consistently detect the virus by qRT-PCR in the nasal mucosa, trachea, and mediastinal lymph nodes on 3 dpi (Fig. 3). Furthermore, we could detect viral RNA in conjunctiva, tonsils, oronasopharynx, and in the left and right bronchus. Viral loads were lower in these tissues by 6 dpi and viral RNA could no longer be detected in the nasal mucosa or conjunctiva at that time (Fig. 3). Viral RNA could not be detected in kidney or bladder tissue samples.
Fig. 3.
Viral load in respiratory tissues of rhesus macaques inoculated with MERS-CoV. Rhesus macaques were euthanized on day 3 (black bars) and day 6 (white bars) postinfection and tissue samples were collected. RNA was extracted and viral load was determined as TCID50 equivalents by qRT-PCR. TCID50 equivalents were extrapolated from standard curves generated by adding dilutions of RNA extracted from a HCoV-EMC/2012 stock with known virus titer in parallel to each run. Geometric mean viral loads were calculated; error bars represent SD. R, right; L, left; LN, lymph node.
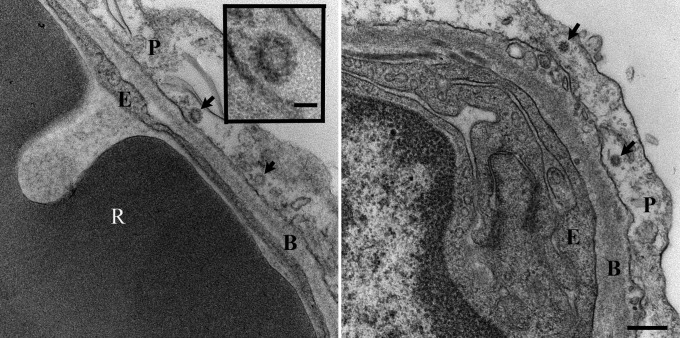
To further prove that virus replication occurred in the lungs of MERS-CoV–inoculated rhesus macaques, transmission electron microscopy was performed on lung lesion samples collected 6 dpi. At this time point, coronavirus particles could be detected in alveolar pneumocytes (Fig. 4). Moreover, qRT-PCR analysis of RNA extracted from the right lower lung lobe and lung lesions obtained 3 and 6 dpi, indicated the transcription of subgenomic mRNA (ORF5) as well as ORF1 RNA in samples collected from all animals and at both time points (Fig. S1), another indicator of active virus replication in the lower respiratory tract.
Fig. 4.
Virus replication in the lungs of MERS-CoV inoculated rhesus macaques. Transmission electron microscopy was performed on lung lesion samples collected from MERS-CoV–inoculated macaques at 6 dpi. Virus particles (arrows) are visible in type I pneumocytes (Left and Right). (Scale bar, Right, 250 nm; Inset, 50 nm). B, basement membrane; E, endothelial cell; P, pneumocytes; R, red blood cell.
Pathology of MERS-CoV in the Respiratory Tract.
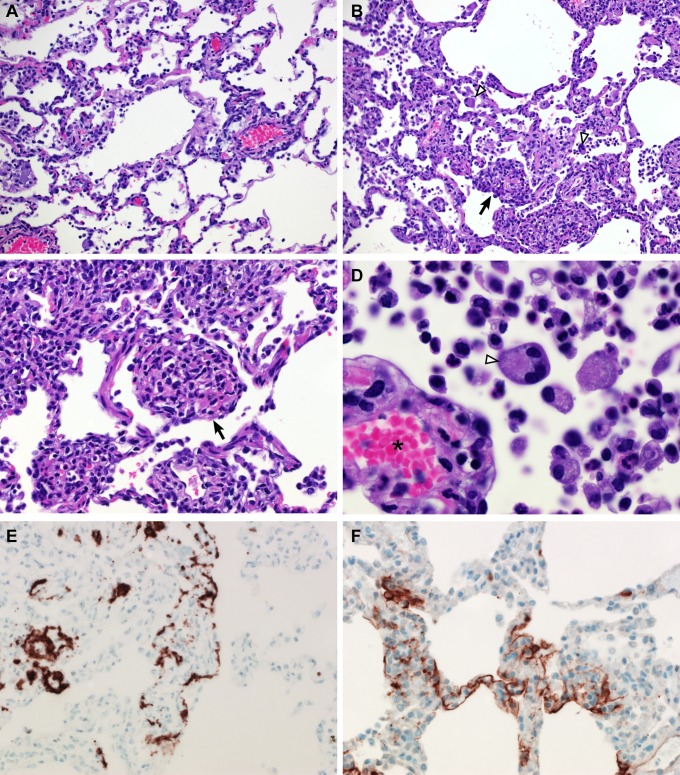
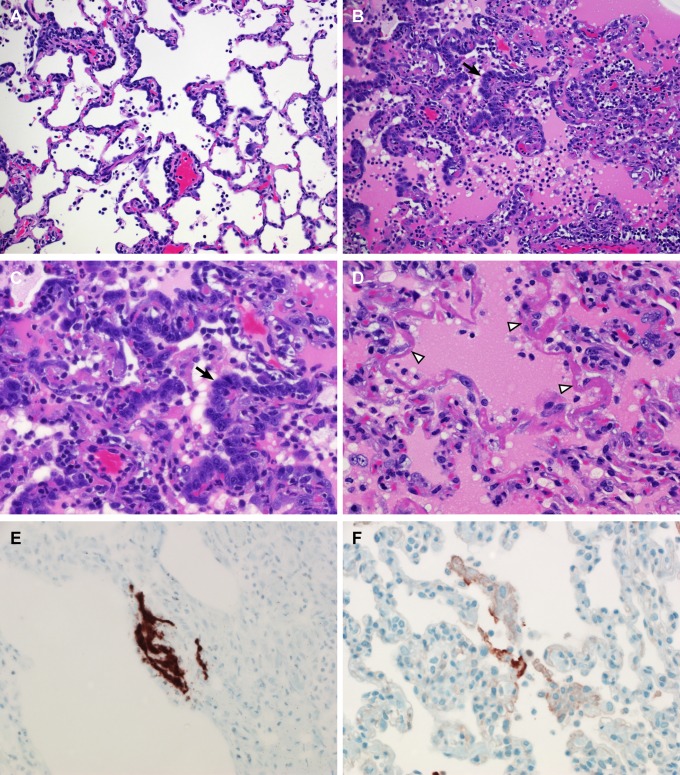
Upon necropsy of animals 3 and 6 dpi, the areas of the individual lung lobes displaying lesions were scored. Lung lobes revealed varying degrees of gross lesions, ranging from 0 to 75% (Fig. S2). Histological lesions in MERS-CoV–infected macaques were limited to the lungs. On day 3 postinfection, lesions were characterized as multifocal, mild (Fig. 5A) to marked (Fig. 5B) interstitial pneumonia. The pneumonia was characterized by thickening of alveolar septae by edema fluid and fibrin, and small to moderate numbers of macrophages and fewer neutrophils. Alveoli contained moderate numbers of pulmonary macrophages and neutrophils; many of these alveolar macrophages formed multinucleate syncytia (Fig. 5D). One animal euthanized 3 dpi also displayed lesions similar to those described as bronchiolitis obliterans organizing pneumonia (Fig. 5C). This organizing pneumonia consisted of aggregations of fibrin (Fig. S3), macrophages, and sloughed pulmonary epithelium that fill and occlude small airways, an early lesion of bronchiolitis obliterans organizing pneumonia that has yet to develop the characteristic dense collagenous stroma, and marked infiltration of fibroblasts that would occur if the lesion was given time to mature. Perivascular infiltrates of inflammatory cells were observed multifocally within and adjacent to affected areas of the lung. Animals necropsied at 6 dpi demonstrated moderate (Fig. 6A) to marked (Fig. 6B) changes in the lungs with type II pneumocyte hyperplasia (Fig. 6C), abundant alveolar edema, and fibrin with formation of hyaline membranes (Fig. 6D).
Fig. 5.
Histopathological changes at 3 dpi in rhesus macaques inoculated with MERS-CoV. Rhesus macaques were euthanized on day 3 postinfection and lung tissue was collected and stained with H&E. (A) Mild acute interstitial pneumonia demonstrated by mild thickening of alveolar septae by small numbers of neutrophils, macrophages, fibrin, and edema. Small numbers of inflammatory cells and fibrin also extend into alveolar spaces. (B) Marked interstitial pneumonia with bronchiolitis obliterans (arrow). Airways are filled with organizing fibrin, necrotic debris, collagen and fibroblasts. There are numerous multinucleate syncytial cells scattered throughout the alveoli (arrowhead). (C) Bronchiolitis obliterans organizing pneumonia. (D) Alveolar capillary (asterisk) bounded by degenerate, vacuolated type I pulmonary epithelium. The alveolar space contains inflammatory cells and a multinucleate syncytial cell (arrowhead). (E) ISH demonstrates viral RNA predominantly within alveolar pneumocytes. (F) Anticoronaviral IHC reveals viral antigen within alveolar pneumocytes. (Magnification: A and B, 200×; C, E, and F, 400×; D, 1,000×.)
Fig. 6.
Histopathological changes at 6 dpi in rhesus macaques inoculated with MERS-CoV. Rhesus macaques were euthanized at 6 dpi and lung tissue was collected and stained with H&E. (A) Mild acute interstitial pneumonia demonstrated by mild thickening of alveolar septae by small numbers of neutrophils, macrophages, fibrin, and edema. Small numbers of inflammatory cells and fibrin also extend into alveolar spaces. (B) Marked interstitial pneumonia with extensive type II pneumocyte hyperplasia (arrow). Alveolar spaces are flooded with edema fluid, fibrin, and inflammatory cells. (C) Type II pneumocyte hyperplasia (arrow) with alveolar flooding edema and fibrin. (D) Hyaline membranes line alveolar space (arrowhead). (E) ISH demonstrates viral RNA predominantly within alveolar pneumocytes. (F) Anticoronaviral IHC reveals viral antigen within alveolar pneumocytes. (Magnification: A and B, 200×; C–F, 400×.)
MERS-CoV Replicates in Alveolar Pneumocytes.
At 3 dpi, in situ hybridization (ISH) demonstrated viral RNA multifocally in the lungs of all monkeys. The signal was found in spindle to polygonal cells along alveolar septae (Fig. 5E). This location and the cellular morphology suggested that virus replication occurs mainly in the pulmonary epithelium. Viral RNA was not detected in endothelial cells. Immunohistochemistry (IHC) yielded similar results for the presence of viral antigen (Fig. 5F). Additionally, there were rare round mononuclear cells and stellate cells within the cortex of the mediastinal lymph nodes that were positive for both viral RNA by ISH and viral antigen by IHC. These cells were morphologically consistent with cells of the macrophage or dendritic cell lineage. Viral RNA and viral antigen could still be detected at 6 dpi (Fig. 6 E and F).
To confirm that virus replication occurred mainly in alveolar pneumocytes, fluorescent IHC was performed using anticytokeratin and anticoronavirus antibodies. Viral antigen was predominantly found in alveolar pneumocytes on 3 as well as 6 dpi (Fig. S4 A and B). This distribution of viral antigen in type I and type II pneumocytes corresponded with the distribution of the receptor for MERS-CoV, DPP4 (Fig. S4 C and D) as indicated by IHC performed on the lungs of infected animals.
Analysis of Gene Expression in Lungs and PBMC.
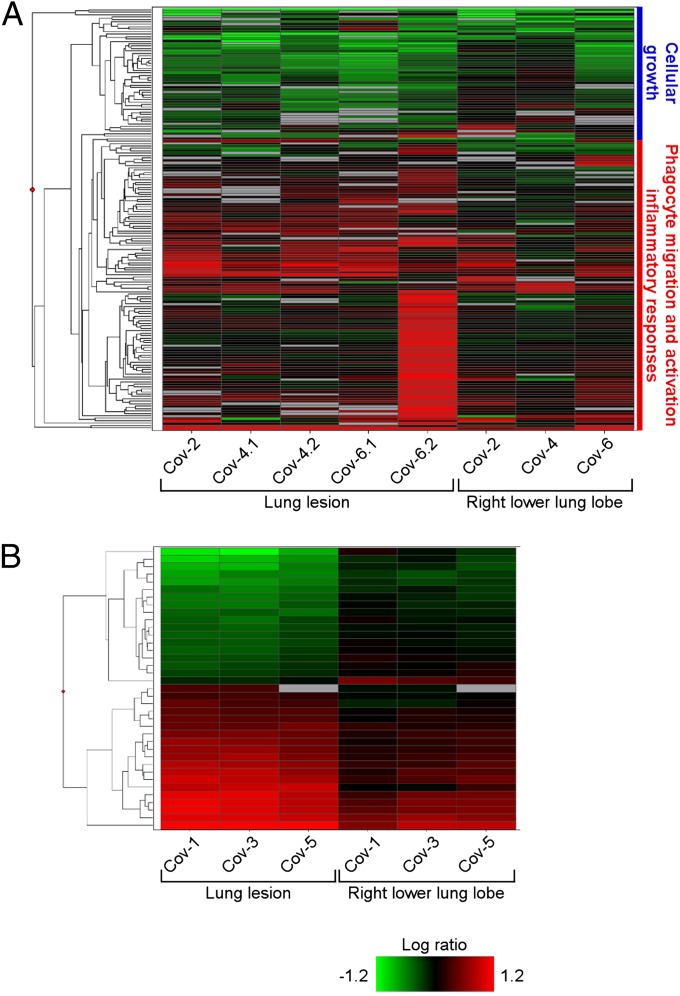
To assess genes that may play a significant role in disease pathology, we performed a microarray analysis on lung tissue and PBMC. A Welch’s t test (P < 0.01, fold-change ≥ 2) was used to compare profiles of significant differentially expressed genes (DEG) from the right lower lung lobe to observable gross lesions, relative to uninfected controls. The right lower lobe was selected, as qPCR demonstrated that viral genomes were present in those samples, allowing us to compare sites of histopathologic injury to the background transcriptional profiles of sites exposed to virus. On day 3, we observed a total of 173 DEG (Fig. 7A and Table S2), although the expression was highly variable between animals and sample sites. This variability is likely multifactorial, driven by differences between individual animals, such as levels of virus in the lung and at lesion sites, distribution of virus receptors, lung sizes, age, and distribution and quantity of susceptible cells. Using ingenuity pathway analysis (IPA), we determined that the majority of DEG were associated with antiviral immunity, inflammation, and chemotaxis. Notably, genes such as IL-6, chemokine C-X-C ligand 1 (CXCL1), and matrix metalloproteinase 9 (MMP9) associated with proinflammatory processes and recruitment of inflammatory cells were significantly up-regulated in the lesions compared with the right lower lung lobes. By day 6, we identified only 37 DEG distinguishing right lower lung lobes from lesions (Fig. 7B and Table S3), and these were not grouped in any heavily enriched functional categories. Additionally, by 6 dpi the lung lesions showed a higher magnitude of gene expression compared with the right lower lung lobe samples. This finding may reflect the resolving clinical status of the animals at day 6 postinfection, and indicate that substantial differences in gene expression occur at sites of virus-induced injury rather than throughout the lung.
Fig. 7.
Microarray analysis of gene expression in lungs of rhesus macaques inoculated with MERS-CoV. The heatmap shows log10 ratios to uninfected controls of differentially expressed genes in the right lung lower lobe and lung lesion samples as determined by Welch’s t test (P < 0.01, fold-change ≥ 2), and grouped by hierarchical clustering. A representation is shown of 173 differentially expressed genes from individual animals and sample sites at 3 dpi (A) and of 37 differentially expressed genes from individual animals and sample sites at 6 dpi (B). Functional analysis was performed on subgroups of the day 3 differentially expressed genes, and enriched functional categories identified by IPA are shown in the blue and red vertical bars to the right of the heatmap in A. Differentially expressed genes are specified in Table S2 (3 dpi) and Table S3 (6 dpi).
We sought to determine if MERS-CoV–induced gene expression dynamics can be observed in PBMC and lung tissue and lesion samples from infected macaques, and change over time. Using singular value decomposition-coupled multidimensional scaling, we identified changes in gene expression occurring in parallel in both organs (Fig. S5). In PBMC a significant response to MERS-CoV infection is observed only on day 1 postinfection. In lungs a significant global response was found only 3 dpi (Fig. S5). By 6 dpi, the gene-expression dynamics in both organs return close to an uninfected or baseline state. Furthermore, functional analysis with IPA reveals that MERS-CoV infection in macaques leads to very rapid innate immune activation (14) through pattern-recognition receptors. Leukocyte activation is evident in blood and lungs, as well as evidence of a robust but self-limiting inflammatory response. Importantly, innate immune activation occurs very rapidly after infection, and is subsequently also rapidly controlled, indicating quick resolution.
Cytokine and Chemokine Profiles in MERS-CoV–Infected Animals.
Plasma levels of 23 cytokines and chemokines were determined in serum samples obtained at 0, 1, 3, and 6 dpi. Transient differences were only observed for IL-1RA, IL-2, IL-6, IL-8, IL-12, IL-13, IL-15, IFN-γ, and monocyte chemotactic protein-1 (MCP-1). AT 1 dpi serum levels of IL-1RA, IL-2, IL-13, IL-15, INF-γ, and MCP-1 were significantly higher than at 0 dpi and all levels returned to baseline by 6 dpi (Fig. S6). Although the serum level of IL-6 was higher and serum levels of IL-8 and IL-12 were lower at 1 dpi, these differences were not statistically significant.
Discussion
The emergence of MERS-CoV in 2012 was the second introduction in 10 y of a novel coronavirus, causing severe respiratory disease, into the human population. Both the severity of the respiratory disease and the genetic similarity to betacoronaviruses isolated from bats are reminiscent of the emergence of SARS-CoV in 2003, which resulted in 8,422 human cases with 916 deaths in over 30 countries (15). To understand the disease caused by MERS-CoV in humans, we developed a nonhuman primate model, which recapitulates the respiratory disease observed in mild or moderate human cases but not the occasionally occurring kidney disease.
In this rhesus macaque model, virus shedding as indicated by qRT-PCR occurred predominantly via the nose and, to a limited extent, the throat. In nasal swabs and BAL, viral loads were highest day 1 postinfection and decreased over time. However, at 6 dpi, two of three animals were still shedding virus from the respiratory tract. Although MERS-CoV was detected in the upper respiratory tract and the lymphoid tissue draining the lungs, replication of MERS-CoV was most prominent in the lower respiratory tract. MERS-CoV replicated predominantly in type I and II pneumocytes in the alveoli. These two cell types form the main component of the architecture of the alveolar space around the terminal bronchioles. This predominant replication of MERS-CoV in alveoli may explain the limited amount of virus shedding observed in our rhesus macaque model. In addition, the fact that human-to-human transmission so far seems limited to a few family clusters in Saudi Arabia (6), the United Kingdom (7), and Tunisia and nosocomial transmission in Jordan, Saudi Arabia (6), and France (9) might be explained by the propensity of the novel coronavirus to replicate deep down in the lower respiratory tract. Recently, DPP4 was identified as the receptor for MERS-CoV (12). DPP4 is an evolutionarily conserved type II transmembrane glycoprotein expressed on endothelial and epithelial cells, in lungs and kidney. Currently, the exact distribution of DPP4 throughout the human respiratory tract is unknown, although DPP4 expression has been observed in vitro in human bronchiolar epithelial cells (12). In mouse lungs, DPP4 is present on type I and type II pneumocytes in the alveoli, with expression on type I pneumocytes more abundant than on type II pneumocytes (16). The distribution of DPP4 is in agreement with our observation of MERS-CoV virus replication in the alveoli and suggests a role for DPP4 in tissue tropism of the novel coronavirus. A more thorough analysis of DPP4 distribution in the human airway is needed to examine the apparent alveolar restriction of virus replication and whether efficient human-to-human transmission might be restricted by the receptor distribution, similar to what has been described for highly pathogenic avian influenza A/H5N1 viruses (17, 18). No virus replication was observed in kidney samples from MERS-CoV–infected rhesus macaques. This finding could indicate that the renal failure reported in some of the human cases might be a result of hypoxia rather than virus replication in the kidney, or of other underlying, preexisting medical conditions.
Currently, few clinical and no pathological data are available on the human cases of MERS-CoV. In addition, the data that are available might be biased toward severe cases in need of hospitalization, as there is no information on the prevalence of MERS-CoV in the general population. Recent reports have revealed that the majority of the severe human MERS-CoV cases have underlying medical conditions, potentially negatively affecting outcome of disease in these patients (2, 8, 9, 19). As such, a direct comparison of the pathogenesis of the novel coronavirus observed in the rhesus macaque model to that in humans should be approached cautiously. However, the transient nature of disease observed in rhesus macaques suggests that this model does not exactly recapitulate the disease observed in the fatal cases of MERS-CoV, likely because of the health status of these animals compared with severe human MERS cases. Rather, the MERS-CoV–associated disease observed in rhesus macaques, and the variation observed between individual animals, may be more similar to the recent, less-severe cases of MERS-CoV identified in the United Kingdom and Saudi Arabia. Interestingly, the disease displayed in the rhesus macaques is reminiscent of SARS-CoV in nonhuman primates, because a wide disease spectrum was reported in the different SARS-CoV studies in nonhuman primates (20–27).
In the case of SARS-CoV, the ability to cause severe disease in a macaque model was associated with an exacerbated innate host response in aged animals (10–19 y old) (28). The majority of human cases of MERS-CoV have been middle aged and aged male individuals (39–73 y old) (6). In the rhesus macaques inoculated with MERS-CoV, no difference in severity of disease could be attributed to age (6–12 y old, young adult) or sex, but low animal numbers used here do not allow a firm conclusion and we cannot exclude that severe disease could be age dependent in the MERS-CoV model described herein.
Proinflammatory cytokines are likely to play an important role in the observed acute lung injury observed in the fatal human cases. The up-regulation of the expression levels of proinflammatory cytokines and chemokines, such as IL-6, CXCL1, and MMP9 in the rhesus macaques and the associated cytokine and chemokine levels in serum samples may have resulted in chemotaxis and activation of neutrophils. Indeed, increased numbers of neutrophils were observed in the blood of all animals on 1 dpi, in accordance with what has been reported for the first human case (2).
The transient, self-limiting nature of MERS-CoV infection in rhesus macaques was displayed at various levels within the model. Transient patterns were observed for virus shedding, viral loads in tissues, neutrophilia, type II pneumocyte hyperplasia, regulation of host gene expression, and production of cytokines. The controlled IFN-mediated innate immune response was very rapid and resolved within the first 3 d. Correlation of gene-expression dynamics between PBMC and lung-tissue samples also suggests that circulating profiles may have utility as a means of assessing host responses to infection without requiring an invasive tissue biopsy.
Taken together, our study provides data on the extent and duration of respiratory disease caused by the novel coronavirus in rhesus macaques. MERS-CoV primarily affects the epithelium of the lower respiratory tract in a transient manner. This tropism of MERS-CoV for the lower respiratory tract explains both the severity of the disease observed in humans and the limited number of cases of human-to-human transmission to date.
Materials and Methods
Ethics Statement.
All animal experiments were approved by the Institutional Animal Care and Use Committee of the Rocky Mountain Laboratories, and performed following the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC) by certified staff in an AAALAC-approved facility.
The work with infectious MERS-CoV was approved under biosafety level-3 conditions by the Institutional Biosafety Committee. Sample inactivation was performed according to standard operating procedures approved by the Institutional Biosafety Committee for removal of specimens from high containment.
Virus and Cells.
MERS-CoV isolate HCoV-EMC/2012 was kindly provided by the Viroscience Laboratory, Erasmus Medical Center, Rotterdam, The Netherlands, and propagated in VeroE6 cells in DMEM (Sigma) supplemented with 2% (vol/vol) FCS (Logan), 1 mM l-glutamine (Lonza), 50 U/mL penicillin, and 50 μg/mL streptomycin (Gibco).
Animal Study and Sample Collection.
Six rhesus macaques (four males, two females; age 6–10 y, weight 5–10 kg) were inoculated with 7 × 106 TCID50 of MERS-CoV isolate HCoV-EMC/2012, as described previously (13). For detailed information, see SI Materials and Methods.
Histopathology, ISH, and IHC.
Histopathology, ISH, and IHC were performed on macaque tissues. For detailed information, see SI Materials and Methods.
Transmission Electron Microscopy.
Transmission Electron Microscopy was performed essentially as described previously (29). For detailed information, see SI Materials and Methods.
RNA Extraction.
RNA was extracted from swabs, BAL, and whole blood samples using the QiaAmp Viral RNA kit (Qiagen) according to the manufacturer’s instructions. Tissues (30 mg) were homogenized in RLT buffer and RNA was extracted using the RNeasy kit (Qiagen).
Quantitative PCR.
For detection of viral RNA, we used the one-step real-time RT-PCR upE assay (30); for the detection of mRNA, we used an ORF1- and ORF5-specific SYBR green assay. See SI Materials and Methods for detailed information.
Microarray Data and Functional Analysis.
RNA extracted from lung tissue samples and PBMC was run on Agilent Rhesus macaque 4 × 44 K microarrays (Agilent Technologies) using the manufacturer’s one-color analysis protocol. Raw array data were uploaded to Genedata Analyst 7.6 (Genedata) and further analyzed. Raw microarray data have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database under GEO accession no. GSE44542, and are also available to the public at http://viromics.washington.edu. For detailed information, see SI Materials and Methods.
Serum Cytokine and Chemokine Analysis.
Concentrations of cytokines and chemokines in serum were measured using the Non-Human Primate Cytokine MILLIPLEX map 23-plex kit (Millipore) according to the manufacturer’s instructions. See SI Materials and Methods for more information.
Supplementary Material
Acknowledgments
We thank Drs. Bart Haagmans and Ron Fouchier at the Erasmus Medical Center for providing Middle East respiratory coronavirus isolate HCoV-EMC/2012; Ralph Baric at the University of North Carolina at Chapel Hill for subgenomic RNA primer sequences; Laura Baseler, Tina Thomas, Dan Long, and Rebecca Rosenke for histopathology; the staff of the Rocky Mountain Veterinary Branch for assistance with the animal experiments; and Anita Mora for figure preparation. This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health; A.L.R., A.G.B., A.O., and M.G.K. are funded through Grant U54 AI081680, National Institute of Allergy and Infectious Diseases Regional Centers of Excellence, National Institutes of Health/National Institute of Allergy and Infectious Diseases Contract HHSN272200800060C, and National Institutes of Health Grant P51OD010425 (Washington National Primate Center).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE44542) and are also available to the public at http://viromics.washington.edu.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310744110/-/DCSupplemental.
References
- 1.van Boheemen S, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6):pii e00473-12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Annan A, et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19(3):456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony SJ, et al. Coronaviruses in bats from Mexico. J Gen Virol. 2013;94(Pt 5):1028–1038. doi: 10.1099/vir.0.049759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Groot RJ, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J Virol. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. WHO (2013) Coronavirus infections. Available at www.who.int/csr/don/archive/disease/coronavirus_infections/en/index.html. Accessed August 30, 2013.
- 7.Health Protection Agency UK Novel Cornoavirus Investigation Team Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Euro Surveill. 2013;18(11):20427. doi: 10.2807/ese.18.11.20427-en. [DOI] [PubMed] [Google Scholar]
- 8.Assiri A, et al. KSA MERS-CoV Investigation Team Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369(5):407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guery B, et al. MERS-CoV study group Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: A report of nosocomial transmission. Lancet. 2013;381(9885):2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kindler E, et al. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. MBio. 2013;4(1):e00611-12. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan RW, et al. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol. 2013;87(12):6604–6614. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raj VS, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munster VJ, de Wit E, Feldmann H. Pneumonia from human coronavirus in a macaque model. N Engl J Med. 2013;368(16):1560–1562. doi: 10.1056/NEJMc1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacquelin B, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119(12):3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peiris JS, Yuen KY, Osterhaus AD, Stöhr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349(25):2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 16.Mentzel S, Dijkman HB, Van Son JP, Koene RA, Assmann KJ. Organ distribution of aminopeptidase A and dipeptidyl peptidase IV in normal mice. J Histochem Cytochem. 1996;44(5):445–461. doi: 10.1177/44.5.8627002. [DOI] [PubMed] [Google Scholar]
- 17.Shinya K, et al. Avian flu: Influenza virus receptors in the human airway. Nature. 2006;440(7083):435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 18.van Riel D, et al. H5N1 virus attachment to lower respiratory tract. Science. 2006;312(5772):399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 19.Drosten C, et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13(9):745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouchier RA, et al. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature. 2003;423(6937):240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenough TC, et al. Pneumonitis and multi-organ system disease in common marmosets (Callithrix jacchus) infected with the severe acute respiratory syndrome-associated coronavirus. Am J Pathol. 2005;167(2):455–463. doi: 10.1016/S0002-9440(10)62989-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haagmans BL, Osterhaus AD. Nonhuman primate models for SARS. PLoS Med. 2006;3(5):e194. doi: 10.1371/journal.pmed.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuiken T, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawler JV, et al. Cynomolgus macaque as an animal model for severe acute respiratory syndrome. PLoS Med. 2006;3(5):e149. doi: 10.1371/journal.pmed.0030149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAuliffe J, et al. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology. 2004;330(1):8–15. doi: 10.1016/j.virol.2004.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin C, et al. An animal model of SARS produced by infection of Macaca mulatta with SARS coronavirus. J Pathol. 2005;206(3):251–259. doi: 10.1002/path.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowe T, et al. Macaque model for severe acute respiratory syndrome. J Virol. 2004;78(20):11401–11404. doi: 10.1128/JVI.78.20.11401-11404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smits SL, et al. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 2010;6(2):e1000756. doi: 10.1371/journal.ppat.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munster VJ, et al. Rapid Nipah virus entry into the central nervous system of hamsters via the olfactory route. Sci Rep. 2012;2:736. doi: 10.1038/srep00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corman VM, et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17(39):pii 20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.