Significance
Selective breeding has long been practiced to enrich for desirable DNA variation that influences livestock traits. We demonstrate that genetic variants can be directly introgressed into livestock genomes using a modified transcription activator-like effector nuclease system. The transient exposure of livestock cells to sequence-targeted editors stimulates homology-directed repair to levels that eliminate the need for transgene-dependent selection. Use of oligonucleotide template enables efficient single nucleotide changes to the genome and permits the transmission of both natural and novel DNA sequence variants into naïve livestock breeds and species. Gene editing offers a powerful method for accelerating the genetic improvement of livestock for food and also for developing swine as a resource for regenerative medicine and models of human disease.
Abstract
We have expanded the livestock gene editing toolbox to include transcription activator-like (TAL) effector nuclease (TALEN)- and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-stimulated homology-directed repair (HDR) using plasmid, rAAV, and oligonucleotide templates. Toward the genetic dehorning of dairy cattle, we introgressed a bovine POLLED allele into horned bull fibroblasts. Single nucleotide alterations or small indels were introduced into 14 additional genes in pig, goat, and cattle fibroblasts using TALEN mRNA and oligonucleotide transfection with efficiencies of 10–50% in populations. Several of the chosen edits mimic naturally occurring performance-enhancing or disease- resistance alleles, including alteration of single base pairs. Up to 70% of the fibroblast colonies propagated without selection harbored the intended edits, of which more than one-half were homozygous. Edited fibroblasts were used to generate pigs with knockout alleles in the DAZL and APC genes to model infertility and colon cancer. Our methods enable unprecedented meiosis-free intraspecific and interspecific introgression of select alleles in livestock for agricultural and biomedical applications.
Meeting global challenges presented by climate change and burgeoning populations requires development and application of biotechnologies that enhance productivity while reducing environmental footprints and improving animal welfare (1, 2). Since the creation of the first transgenic pig (3), more than 180 trials have been recorded to genetically engineer livestock in various ways (2). Until recently, most were accomplished by random insertions of expression cassettes, which suffered from low efficiency and unpredictable expression, or homologous recombination (efficiency <1 in 104) with linked selection markers.
Three recent technologies—zinc finger nucleases (4), transcription activator-like (TAL) effector nucleases (TALENs) (5), and clustered regularly interspaced short palindromic repeats/CRISPR- associated endonuclease cas9 (CRISPR/Cas9) (6–8)—have been used to disrupt gene function by introducing small insertions and/or deletions (indels) into genomes of species mediated by nonhomologous end-joining (NHEJ). In particular, TALEN-induced gene disruption has been demonstrated in various species ranging from model organisms (9–12) to crops (13, 14), farm animals (15), and humans (16–18). However, indels introduced by NHEJ are variable in size and sequence, which make screening for functionally disrupted clones arduous (15) and do not allow for precise alterations. TALEN- or CRISPR/cas9-mediated homology-directed repair (HDR) supports the introduction of defined nucleotide changes; however, so far this has only been demonstrated in lower eukaryotic models, including yeast (19), zebrafish (20), and, very recently, mice (21, 22).
Here we report precise, high-frequency editing of 15 genes in pig, goat, and cattle genomes. In many cases, the gene edits are indistinguishable from alleles that exist within a species or clade and represent the first demonstration of marker-free, nonmeiotic allele introgression. This work demonstrates that precise, high-efficiency gene editing can be achieved in commercially important loci in livestock for agricultural or biomedical purposes.
Results
Introgression of the Celtic POLLED Allele into the Genome of Horned Dairy Cattle Breeds.
We previously reported an efficient system for deriving TALEN-mediated NHEJ knockouts in livestock fibroblasts and embryos (15), and theorized that similar methods would be suitable for derivation of cells with HDR-mediated genetic changes. Accordingly, we used TALENs to stimulate HDR with plasmid and recombinant adeno-associated virus (rAAV) donor templates designed to introduce a naturally occurring 11-bp deletion or the Belgian Blue mutation (23–25) at position 821 (821del11) of growth differentiation factor 8 (GDF8) (SI Appendix, Fig. S1). Individual colonies with the intended mutation were recovered in 1% of colonies treated with the plasmid template and in 13% of colonies treated with the rAAV template (SI Appendix, Fig. S1D). These results inspired us to attempt introgression of more complex alleles into livestock.
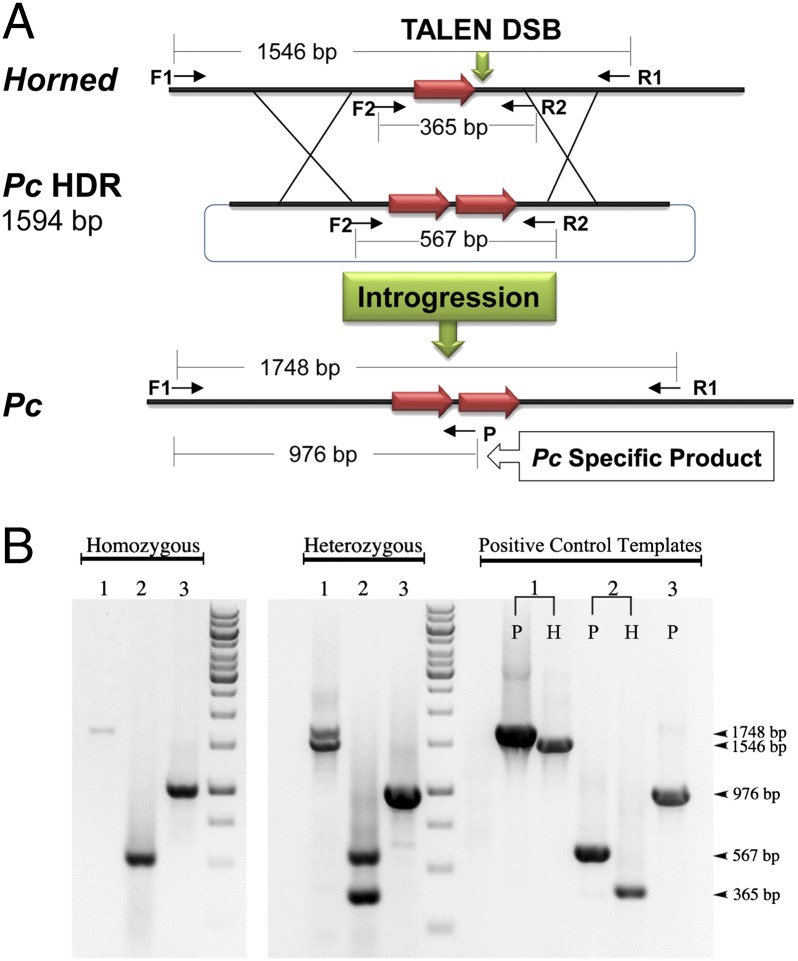
To protect the welfare of dairy farm operators and cattle, horns are routinely manually removed from the majority of dairy cattle in the United States and Europe. Dehorning is painful, elicits a temporary elevation in animal stress, and adds expense to animal production (26). Moreover, despite the intent of protecting animals from subsequent injury, some view the practice as inhumane. Some beef breeds are naturally horn-free (e.g., Angus), a dominant trait referred to as POLLED (27). Two allelic variants conferring polledness have been identified on chromosome 1 (28). Meiotic introgression of the POLLED trait into horned dairy breeds can be accomplished by traditional crossbreeding, but the genetic merit of the resulting animals would rank lower owing to the admixture of unselected (inferior) alleles for net merit (i.e., milk production) into the population. We undertook the nonmeiotic introgression of the Celtic POLLED allele (duplication of 212 bp that replaces 10 bp), referred to as Pc, into fibroblasts derived from horned dairy bulls. We constructed a plasmid HDR template containing a 1,594-bp fragment including the Pc allele from the Angus breed (Fig. 1A). TALENs were designed such that they could cleave the HORNED allele but leave the Pc allele unaffected. In addition, after finding that one pair of TALENs delivered as mRNA had similar activity as plasmid DNA (SI Appendix, Fig. S2), we chose to deliver TALENs as mRNA to eliminate the possible genomic integration of TALEN expression plasmids. Five of 226 colonies (2%) passed each PCR test shown to confirm introgression of Pc (Fig. 1B). Three of the five clones were homozygous for Pc introgression and were confirmed by sequencing (SI Appendix, Table S1).
Fig. 1.
TALEN-mediated introgression of Polled. (A) Strategy for introgressing the Pc allele into Holstein (HORNED) cells. The Pc allele is a tandem repeat of 212 bp (red arrow) with a 10-bp deletion (not shown). TALENs were developed to specifically target the HORNED allele (green vertical arrow), which could be repaired by homologous recombination using the Pc HDR plasmid. Primer sets used in B are depicted. (B) Representative images of colonies with homozygous or heterozygous introgression of Pc. Three primer sets, indicated by number, were used for positive classification of candidate colonies: set 1, F1+R1; set 2, F2+R2; and set 3, F1+P (Pc-specific). Amplicons generated using positive control templates (P, plasmid template containing a sequence-verified Pc 1,748-bp insert between primers F1 and R1; H, Holstein bull genomic DNA) are shown. The identity of the PCR products was confirmed by sequencing of F1+R1 amplicons.
Reliable Oligonucleotide-Templated Allele Introgression in Pig, Goat, and Cattle Genomes.
Although plasmid templates are effective for introgression of Pc and GDF8 alleles, many desirable alleles correspond to single nucleotide polymorphisms (SNPs) or small indels that ideally do not rely on the use of double-stranded DNA templates that could randomly integrate into the genome. In addition, although introgression of the GDF8 821del11 allele with rAAV was efficient, viral methods are unlikely to be popular with consumers of animal protein. Recent studies have demonstrated that single-stranded oligonucleotides (oligos) 40–100 nt in length could serve as effective templates for HDR when stimulated by double-strand breaks (DSBs) in DNA (20, 21, 29–31). Accordingly, we sought to determine whether this approach could stimulate HDR in primary fibroblasts from pigs and cattle, cellular substrates suitable for the creation of gene-edited animals by cloning. We designed oligo templates to overlap their cognate TALEN-binding sites and to introduce a 4-bp indel into the spacer region for restriction fragment length polymorphism (RFLP) analysis. Primary fibroblasts were transfected with plasmid- or mRNA-encoded TALENs plus oligo templates and incubated for 3 d at either 30 °C or 37 °C.
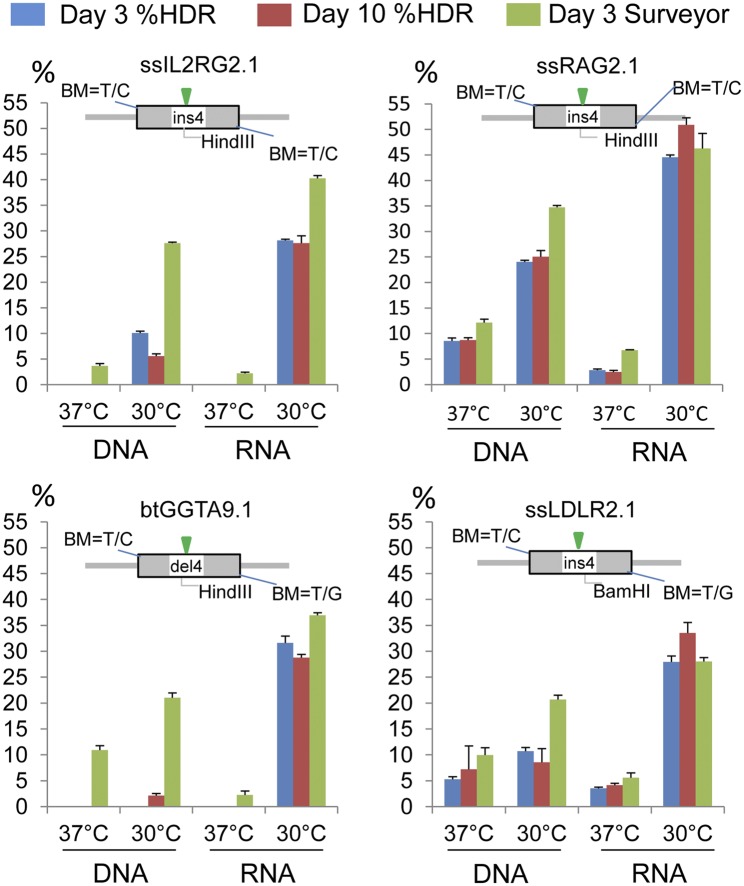
We found that TALENs delivered as mRNA consistently outperformed plasmid in cells incubated at 30 °C, as suggested by the Surveyor assay (Fig. 2). Despite appreciable levels of TALEN activity measured by Surveyor, HDR was consistently more than twofold higher when TALENs were delivered as mRNA rather than as plasmids. We speculated that this may have been a kinetic issue: TALENs from mRNA were translated more rapidly, allowing use of the template before oligo degradation. However, a time-course experiment showed little difference in the onset of HDR between TALENs encoded by plasmid versus those by mRNA (SI Appendix, Fig. S3). Among the replicates using TALEN mRNA at 30 °C, the levels of cumulative mutation and total HDR were similar, suggesting the majority of mutant alleles correspond to the intended introgression.
Fig. 2.
An mRNA source of TALENs stimulates efficient and consistent HDR using an oligo donor. Each chart displays results of targeting a specific locus in fibroblasts (e.g., ssIL2RG; “ss” for Sus scrofa and “bt” for Bos taurus) using oligo donor templates and TALENs delivered as plasmid DNA or mRNA. (Insets) Diagrams of the oligo templates, in which the shaded boxes represent the TALEN-binding site and the spacers are shown in white. Each oligo contains either a 4-bp insertion (ins4) or deletion (del4) that introduces a novel restriction site for RFLP analysis. Presumptive BMs replace the conserved −1 thymidine (relative to the TALEN-binding site) with the indicated nucleotide. Fibroblasts were transfected with either TALEN-encoding plasmids (3 µg) or mRNA (1 µg) along with 3 µM of their cognate oligo-homologous template (SI Appendix, Table S4). Cells were then incubated at 37 °C or 30 °C for 3 d before expansion at 37 °C until day 10. TALEN activity was measured by the Surveyor assay at day 3 (Day3 Surveyor), and HDR was measured at days 3 and 10 by RFLP analysis (Day3 %HDR and Day10 %HDR). Each bar displays the average and SEM from three replicates.
In our previous study, NHEJ-induced indels declined by 50–90% after extended culture without the aid of selection (15). In contrast, HDR levels at four loci were roughly equivalent when measured at day 3 and day 10 without selective enrichment, indicating that these HDR indel alleles were stable in culture (Fig. 2). The consistently high rate (25–50%) and stability of gene edits at all four loci suggest that edited cells should be recoverable by dilution cloning without selective enrichment. Indeed, analysis of ∼650 colonies for intended indel alleles in eight separate loci revealed a recovery rate of 10–64% (average, 45%), with up to 32% of the colonies homozygous for the edit (Table 1).
Table 1.
Frequencies of recovery of colonies with HDR alleles
| Reagent | ID | Species | Mutation type | Nucleotidechange | Amino acid change | Day3%HDR | HDR+ (%) | Biallelic HDR+ (%) |
| TALEN | ssLDLR2.1* | Pig ♀ | Ins/FS | 141 (ins4) | 47∆PTC | 38 | 55/184 (30) | 4/184 (2) |
| TALEN | ssDAZL3.1† | Pig ♂ | Ins/FS | 173 (ins4) | 57∆PTC | 25 | 34/92 (37) | 8/92 (9) |
| TALEN | ssAPC14.2† | Pig ♂ | Ins/FS | 2703 (ins4) | 902∆PTC | 48 | 22/40 (55) | 4/40 (10) |
| TALEN | ssTp53 | Pig ♂ | Ins/FS | 463 (ins4) | 154∆PTC | 22 | 42/71 (59) | 12/71 (17) |
| TALEN | ssRAG2.1 | Pig ♂ | Ins/FS | 228 (ins4) | 76∆PTC | 47 | 32/77 (42) | 13/77 (17) |
| TALEN | btRosa1.2‡ | Cow ♂ | Ins/mloxP | Ins34 | NA | 45 | 14/22 (64) | 7/22 (32) |
| TALEN | ssSRY3.2 | Pig ♂ | Ins/mloxP | Ins34 | NA | 30 | ND | ND |
| TALEN | ssKissR3.2 | Pig ♂ | Ins/FS | 322 (ins6) 323 (del2) | 107∆PTC | 53 | 57/96 (59) | 17/96 (18) |
| TALEN | btGDF83.1 | Cow ♂ | del/FS | 821 (del11) | FS | ∼10 | 7/72 (10) | 2/72 (3) |
| TALEN | ssEIF4GI14.1 | Pig ♂ | SNPs | G2014A T2017C C2019T | N672D L673F | 52 | 68/102 (67) | 40/102 (39) |
| TALEN | btGDF83.6N | Cow ♂ | SNPs | G938A T945C | C313Y | 18 | 8/94 (9) | 3/94 (3) |
| TALEN | btGDF83.6N§ | Cow ♂ | SNP | G938A | C313Y | NA | 7/105 (7) | 2/105 (2) |
| TALEN | ssP65.8 | Pig ♂ | SNP | T1591C | S531P | 18 | 6/40 (15) | 3/40 (8) |
| TALEN | ssP65.8R | Pig ♂ | SNP | T1591C | S531P | 7 | 9/63 (14) | 5/63 (8) |
| TALEN | ssGDF83.6§ | Pig ♂ | SNP | G938A | C313Y | NA | 3/90 (3) | 1/90 (1) |
| TALEN | caFecB6.1 | Goat ♂ | SNP | A747G | Q249R | 17 | 17/72 (24) | 3/72 (4) |
| TALEN | caCLPG1.1 | Goat ♂ | SNP | A→G | Extragenic | 4 | ND | ND |
| CRISPR | ssP65 G1s | Pig ♂ | SNP | T1591C | S531P | 6 | 6/96 (6) | 2/96 (2) |
| CRISPR | ssP65 G2a | Pig ♂ | SNP | T1591C | S531P | 5 | 2/45 (4) | 0/45 |
| CRISPR | ssAPC14.2 G1a | Pig ♂ | Ins/FS | 2703 (ins4) | 902∆PTC | 32 | ND | ND |
ss, Sus scrofa; bt, Bos taurus; ca, Capra aegagrus; FS, frame shift; NA, not applicable; ND, not determined; PTC, premature termination codon; R, replicate.
Homozygous null LDLR−/− could not be propagated in culture.
HDR+ colonies from these replicates were used to generate founder animals by cloning (Fig. 5).
A modified loxP site (mloxP) was inserted into btROSA26 and ssSRY; positive colonies were identified by PCR.
Only the target SNP was introduced. A triprimer PCR combined with Sanger sequencing was used to identify positive colonies (SI Appendix, Table S2).
Differential Stability of Gene Edits.
We were extremely successful in isolating individual colonies with custom indel alleles; however, introgression of SNPs presented a greater challenge. Both day 3 levels of HDR (7–18%) and the isolation of cellular clones with the intended SNP alleles (3–15%) in cattle and swine GDF8 or pig p65 were significantly lower compared with indel HDR (day 3 level, 10–53%; clone isolation, 10–64%) (Table 1). We hypothesized that indels likely were more stable than SNP because introduction of indels into the TALEN spacer region would be expected to reduce recleavage of the locus, consistent with known constraints on TALEN spacer length (17). Accordingly, the introgression of large insertions (loxP) was extremely efficient and stable, and comparison of HDR frequencies with oligos within the same locus suggested that even a 4-bp insertion allele was more efficient than SNP alleles (SI Appendix, Fig. S4). Sequence analysis also revealed that nearly one-half of the isolated SNP-positive colonies for GDF8 or pig p65 harbored concomitant indels expected to change TALEN spacing (SI Appendix, Table S2). Regardless, we were able to recover colonies with homozygous conversion of G938A in GDF8 (both pigs and cattle) and T1591C in pig p65 at up to nearly a 5% level without any additional changes to the locus (Table 1 and SI Appendix, Table S2). We also introgressed SNP alleles for the sheep FecB and Callipyge loci into the goat genome. This ability to precisely alter a single nucleotide is significant and unprecedented.
For comparison, we designed CRISPR gRNAs that overlapped the T1591C site of p65, and we evaluated introgression using the two platforms. Despite efficient production of DSBs at the intended site, CRISPR/Cas9-mediated HDR was <6% at day 3 and below the limit of detection at day 10 (SI Appendix, Fig. S5). In addition, recovery of modified clones was lower with CRISPR-mediated HDR than with TALENs, even though the TALENs cut 35 bp away from the SNP site (Table 1). Analysis of CRISPR/Cas9-induced targeting at a second locus, ssAPC14.2, was much more efficient, but still did not reach the level of HDR induced by TALENs at this site (∼30% vs. 60%; SI Appendix, Fig. S6).
Strategies for Stabilizing Introgressed SNP Alleles.
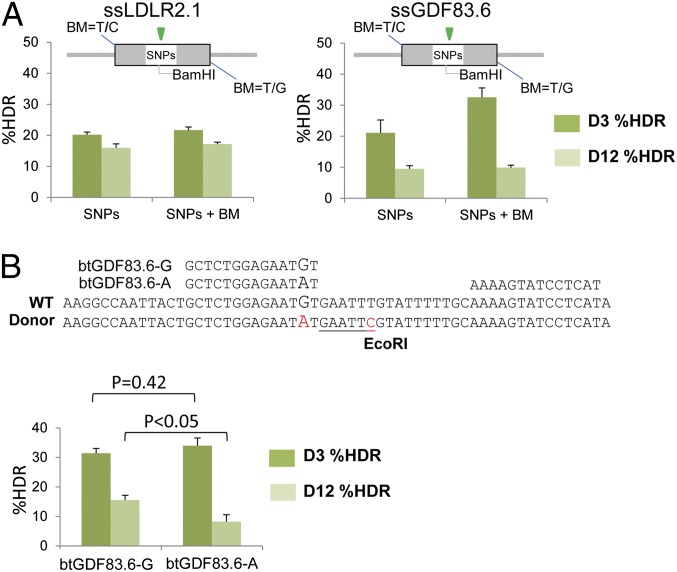
Given the conservation of the 5′-thymidine nucleotide immediately preceding TAL-binding sites (32, 33), we reasoned that altering these bases in the oligo [known as blocking mutations (BMs)] would inhibit recleavage of edited alleles. Surprisingly, we found that BMs had no significant impact on the maintenance of SNP alleles at the pig LDLR or GDF8 loci (Fig. 3A). This suggests that either the conversion tract for oligo-templated HDR is quite short and does not incorporate the BM or that altering the 5′-thymidine does not completely abolish TALEN activity.
Fig. 3.
SNP introgression using oligo donors. (A) The influence of BMs on maintenance of HDR alleles was evaluated. Each oligo was designed to introduce the same SNPs/restriction sites with or without BMs. HDR was quantified by RFLP assay in transfected populations initially cultured at 30 °C for 3 d and then maintained at 37 °C up to day 12. Values are averaged (n = 3). (B) Introgression of myostatin C313Y into Wagyu fibroblasts. The C313Y missense mutation is caused by a G-to-A transition (indicated by oversized text) at nucleotide 938 of bovine myostatin (23–25). The HDR template also includes a T-to-C transition (red) to introduce a novel EcoRI site for RFLP screening. Two left TALENs were designed against the locus: btGDF83.6-G, targeting the WT alelle (Wt), and btGDF83.6-A targeting the mutant allele (C313Y). The two share a common right TALEN. Transfection, culture, and measurement were conducted as described above. The average and SEM values for btGDF83.6-G (n = 30) and btGDF83.6-A (n = 5) represent 12 and 3 biological replicates, respectively. A two-sided Student t test was used to compare averages between groups; P values are indicated.
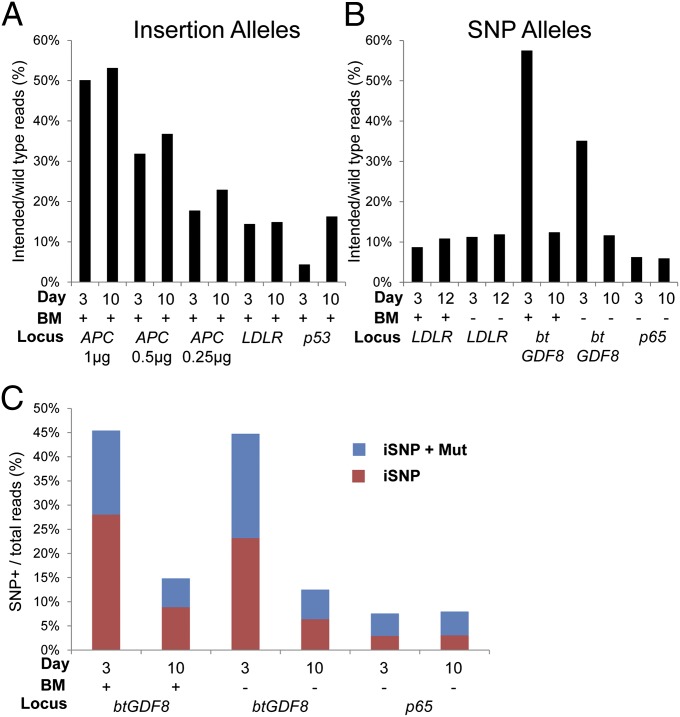
To examine this effect in greater detail, we conducted Illumina next-generation sequencing of 200- to 250-bp amplicons flanking the target sites from populations of cells transfected with oligos and TALEN mRNA. The results from five loci in pigs and cattle show that insertion alleles generally were more prevalent and stable in the population (Fig. 4). Whereas BMs had little influence on the preservation of intended alleles in culture, there was a slight bias toward incorporation of BMs in SNP-edited alleles compared with insertional edits (SI Appendix, Fig. S7). Consistent with our colony analysis, reads sorted on the basis of incorporating the intended SNP (iSNP), G938A, or T1591C conversion in btGDF8 and p65 revealed that nearly one-half of the reads with the iSNP had an additional mutation (iSNP+Mut) (Fig. 4B), the majority of which were indels (SI Appendix, Figs. S7 and S8). The majority of iSNP btGDF8 reads with indels in the spacer also contained one or both BMs (SI Appendix, Fig. S8), demonstrating that modification of the conserved 5′-thymidine was not able to suppress recleavage and subsequent indel generation. Thus, this base must be less critical to TALEN binding than suggested by conservation, and provides a molecular basis for the inability of BMs to preserve alleles as described above.
Fig. 4.
Sequence analysis of TALEN-stimulated HDR alleles. PCR amplicons flanking the target site (200–250 bp total) derived from TALEN mRNA and oligo-transfected cell populations were sequenced by Illumina sequencing. The total read count ranged from 10,000 to 400,000 per sample. (A and B) The counts of perfect, intended HDR reads versus the WT reads are plotted for insertion (A) and SNP alleles (B). The target locus, time point, and whether or not BMs were included in the oligo are indicated. (C) Reads from btGDF8 and p65 populations were sorted for incorporation of the target SNP and then classified into intended (iSNP only) vs. those with the target SNP plus an additional mutation (iSNP+Mut) and plotted against the total number of reads.
Another strategy to reduce recutting of the SNP edits is to design TALENs such that their binding sites overlap the target SNPs. We evaluated the influence of such repeat variable diresidue (RVD)/nucleotide mismatches for introgression of G938A SNP into cattle GDF8. Two pairs of TALENs were generated, one pair that bound the WT “G” allele (btGDF83.6-G) and another that bound the intended “A” allele (btGDF83.6-A) (Fig. 3B). HDR with each TALEN pair was similar at day 3, whereas levels measured at day 12 were significantly higher using the TALENs that bound the WT G allele, indicating that recleavage was more prevalent with btGDF83.6-A, which targets the repaired allele perfectly. Different RVD/nucleotide mismatches likely will have a greater influence on maintenance of HDR alleles, because the Asn-Asn (NN) RVD used for the WT G TALENs is able to bind both G and A nucleotides. For modification of porcine EIF4GI, we found that three RVD/nucleotide mismatches were sufficient for protection of the HDR edit; nearly 70% of isolated colonies contained an edited allele, more than half of which were homozygotes (Table 1 and SI Appendix, Fig. S9). Thus, the intentional alteration of the target locus to resist recleavage is an effective strategy for preserving edits.
Ultimately, gene editing is a dynamic process. TALEN cleavage and recleavage are in flux with repair by NHEJ, HDR with an oligo template, and HDR with the sister chromatid as a template. We hypothesized that the observed loss of SNP alleles might be reduced by extending the hypothermic treatment, slowing cell proliferation long enough to outlast the burst of TALEN activity from TALEN mRNA transfection. Indeed, this extension almost tripled the level of SNP HDR-edited alleles recovered after extended culture (SI Appendix, Fig. S10).
Production of Biomedical Model Pigs with Gene-Edited Alleles.
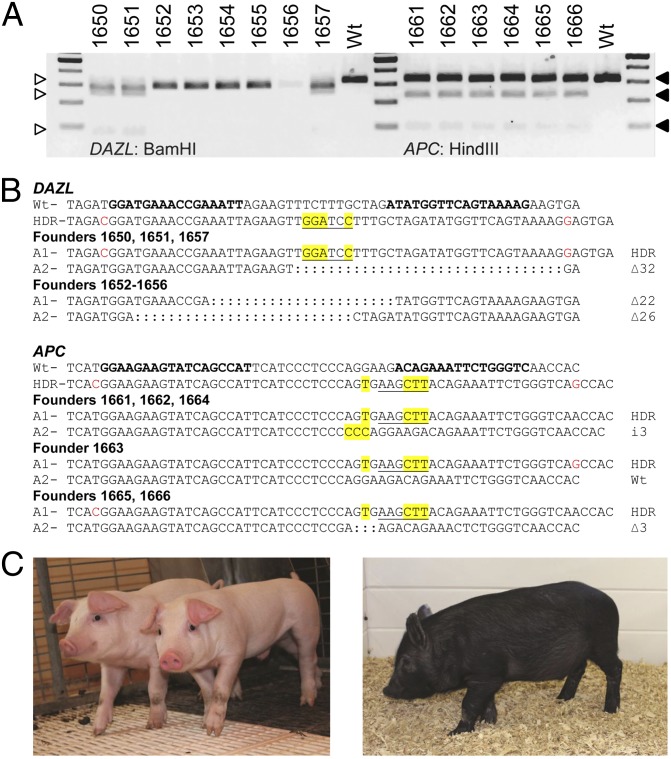
We chose two gene-edited loci in the porcine genome, deleted in azoospermia-like (DAZL) and adenomatous polyposis coli (APC), to carry through to live animals. Colonies with HDR- or NHEJ-edited alleles of DAZL or APC were pooled for cloning by chromatin transfer (CT). Each pool yielded two pregnancies from three transfers, of which one pregnancy each was carried to term. A total of eight piglets were born from DAZL-modified cells, each of which reflected genotypes of the chosen colonies consistent with either the HDR allele (founders 1650, 1651, and 1657) or deletions resulting from NHEJ (Fig. 5A). Three of the DAZL piglets (founders 1655–1657) were stillborn. Of the six piglets from APC-modified cells, one was stillborn, three died within 1 wk, and another died after 3 wk, leaving only founder 1661 alive. The lack of correlation between genotype and survival suggests that the early deaths were related to cloning rather than to gene edits (34). All six APC piglets were heterozygous for the intended HDR-edited allele, and all but one piglet had either an in-frame insertion or deletion of 3 bp on the second allele (Fig. 5 A and B). The remaining animals are being raised for phenotypic analyses of spermatogenesis arrest (DAZL−/− founders) or development of colon cancer (APC+/− founders).
Fig. 5.
Cloned pigs with HDR alleles of DAZL and APC. (A) RFLP analysis of cloned piglets derived from DAZL- and APC-modified landrace and Ossabaw fibroblasts, respectively. Expected RFLP products for DAZL founders are 312, 242, and 70 bp (open triangles), and those for APC are 310, 221, and 89 bp (filled triangles). The difference in size of the 312-bp band between WT and DAZL founders reflects the expected deletion alleles. (B) Sequence analysis confirming the presence of the HDR allele in three of eight DAZL founders, and in six of six APC founders. BMs in the donor templates (HDR) are displayed in red, and inserted bases are highlighted in yellow. The bold text in the top WT sequence indicates the TALEN-binding sites. (C) Photographs of DAZL (Left) and APC (Right) founder animals.
Discussion
The data presented in Table 1 demonstrate that combining mRNAs encoding TALENs and oligo templates for directing HDR achieves several key benchmarks for a precise genome-editing strategy: (i) Only target nucleotides were changed, and mRNA transfection avoids unintended integration of plasmid DNA; (ii) selection markers are unnecessary, an important factor in the acceptance of edited livestock for human consumption; (iii) gene edits were efficient, ranging from approximately 10% for SNPs to >50% for some larger alterations; and (iv) the method was highly reliable; at each locus tested, targeted alteration of 16 of 16 sites (15 genes) was achieved. The high efficiency and precision reported here are astonishing compared with what was feasible only a few years ago (2).
There are two concerns with gene editing: stabilizing the changes at the targeted site and avoiding modification of unintended sites. Regarding the first concern, we found evidence that HDR edits directing single bp changes (i.e., SNPs) could be lost (Figs. 3 and 4B). Based on the prediction that a thymidine preceding the targeted DNA sequence influences TAL binding (32, 33), we attempted to block recleavage of introgressed alleles by introducing BMs; however, BMs did not prevent TALEN activity and recleavage of edited alleles (Fig. 3 and SI Appendix, Figs. S7 and S8). In contrast, introduction of multiple SNPs or additional sequences (Fig. 2 and SI Appendix, Fig. S9) resulted in more stable HDR edits. Extension of hypothermic culture also resulted in the stabilization of introgressed SNP alleles. Because hypothermia slows cell proliferation primarily by prolonging the G1 phase of the cell cycle (35), this treatment may differentially favor oligo-HDR versus sister chromatid-templated repair in a cell cycle-dependent manner. Regardless of the mechanism, this approach offers a straightforward strategy for recovering cells with precise introgression of SNP alleles. As to the second concern, the frequency of modification at unintended sites is exceedingly low and in the context of livestock breeding will be lost by independent segregation in subsequent generations (2).
Gene Editing for Single-Step Introgression of Valuable Alleles into Livestock.
Various objectives were achieved by precise gene editing (Table 1). Knockout of genes of biomedical relevance was accomplished by interrupting coding sequences with 4-bp indels. This strategy was very reliable and generally resulted in HDR edits in approximately 40% of the clones (range, 26–60%), up to one-third of which were homozygotes. At similar frequencies, we integrated a modified loxP(mloxP) site into ROSA26, a presumptive safe harbor locus, and SRY loci in cattle and pigs that can serve as a landing pad for insertion of novel sequences in livestock via recombinase-mediated cassette exchange (36, 37). Previously, only NHEJ edits had been demonstrated for the Y chromosome of livestock (15); however, TALENs are clearly suitable for direct stimulation of knockout/knockin, a capability that we are currently exploiting in livestock and that was recently demonstrated in mice (38).
Of more immediate value, we achieved efficient nonmeiotic introgression of native alleles between species or breeds. Interspecies introgression is impossible by breeding, and intraspecies crossbreeding for allele introgression is either insufficient or deleterious to other performance features owing to the genetic admixture. Our nonmeiotic introgression includes the double-muscling mutations of GDF8 [SNP G938A from Piedmontese cattle (23, 25) and 821del11 (23–25) from Belgian Blue cattle] into the genome of Wagyu cattle and Landrace pigs. For improvement of animal welfare, we transferred the Pc allele for polledness from a beef-producing breed into cells from horned dairy cattle. We also transferred a candidate SNP allele for African swine fever virus resilience T1591C of p65 (39) from warthog to the genome of conventional swine cells and introgressed sheep SNPs responsible for elevated fecundity (FecB; BMPR-IB) (40) and parent-of-origin dependent muscle hypertrophy (Callipyge) (41) into the goat genome. Up to now, nonmeiotic allele introgression has not been possible without selective enrichment, and our efficiencies are at least 105-fold greater than those obtained previously with selection (42, 43). Such high levels of unselected single-allele introgression suggest the feasibility of altering multiple alleles in a single generation of farm animals, decreasing the impact of long generation intervals on genetic improvement.
Implications for Animal Breeding and Human Medicine.
High-throughput genome sequencing coupled with detailed phenotyping provides unprecedented opportunities for identifying specific alleles that affect livestock performance, and for locus-centric or whole-genome–based selection for improvement of animal genetics (44). Our results suggest that gene editing can be incorporated into selection programs to accelerate genetic improvement when selective breeding is either inefficient or impossible. Nonmeiotic introgression provides a genetic method for precisely crossbreeding for large effect alleles without compromising the genetic merit of indigenous or purpose-bred populations by whole-genome admixtures (2). By allowing such single-generation introgression of select alleles, it will accelerate genetic improvement based on local ecologies and needs that may derive from climate change and emerging diseases.
We used customized endonucleases to generate live animals with precise edits at two independent loci. Pigs edited to disrupt the DAZL gene can serve as a model for studying the restoration of human fertility by germ cell transplantation or for producing genetically modified offspring by transfer of genetically modified germline stem cells, as has been demonstrated in pigs (45), goats (46), and rodents (47, 48). Gene-edited alleles of APC also could provide a size-relevant model of colon cancer for preclinical evaluation of therapeutics, surgical intervention, or detection modalities. These results, coupled with our previous findings (15), demonstrate relative groundbreaking ease in introducing genetic modifications that mimic natural polymorphisms or human disease alleles into livestock.
Our work also has implications for personalized medicine, demonstrating the ability to develop precisely engineered large animals to serve as tailored biomedical models for testing drug, device, and cellular therapeutics, and potentially as resources for xenogeneic and autologous therapeutic cells and organs. Finally, the precision and high efficiency that we have achieved in altering either single nucleotides or small sequences in livestock fibroblasts is likely to be applicable to the correction of disease-causing mutations in patient-derived fibroblasts, a resource for conversion to induced pluripotent stem cells for use in cellular therapy (49), without the use of exogenous selection markers.
Materials and Methods
Detailed information is provided in SI Appendix and our previous paper (15). In brief, TALENs were assembled using the Golden Gate assembly protocol and library (50). TALEN-encoding mRNA was synthesized in vitro using the mMESSAGE mMACHINE T3 Kit (Ambion). The CRISPR/Cas9 endonucleases were generated based on the Church laboratory system and methods (8). The transfections were conducted using the Neon transfection system (Invitrogen) unless stated otherwise. The frequency of total mutations in a TALEN- or CRISPR/Cas9-transfected population was assessed by the Surveyor Mutation Detection Kit (Transgenomic), and rates of mutation (Day3 Surveyor) was calculated by the Guschin method (51); %HDR was evaluated by RFLP or PCR. The colonies listed in Table 1 were obtained by dilution cloning from corresponding populations without drug selection. Population samples were sequenced on a Illumina MiSeq sequencer after PCR amplification. The RVD sequences of TALENs, oligo sequences for HDR, and PCR primers are listed in SI Appendix, Tables S1, S3, S4, S5, and S6. Pigs were cloned by CT under contract with Minitube of America under its Animal Welfare Assurance no. A4520/01.
Supplementary Material
Acknowledgments
We thank our colleagues at the University of Minnesota’s Center for Genome Engineering for helpful discussions and comments. We also thank Drs. Jared Decker and Jerry Taylor for Angus DNA, Dr. Brian Crooker for dairy cattle cells, Dr. Irina Polejaeva for goat cells, and Dr. Michael Sturek for Ossabaw swine cells. This work was supported by Grant 1R43RR033149-01A1 from the National Institutes of Health and Biotechnology Risk Assessment Program and by Competitive Grant 2012-33522-19766 from the US Department of Agriculture, National Institute of Food and Agriculture.
Footnotes
Conflict of interest statement: D.F.C., C.A.L., D.A.W., P.B.H., and S.C.F. either have equity in and/or work for Recombinetics, Inc., a biotechnology company that receives funding from the National Institutes of Health to develop gene editing in commercial livestock.
This article is a PNAS Direct Submission.
See Commentary on page 16295.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310478110/-/DCSupplemental.
References
- 1.Fahrenkrug SC, et al. Precision genetics for complex objectives in animal agriculture. J Anim Sci. 2010;88(7):2530–2539. doi: 10.2527/jas.2010-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan W, Carlson DF, Walton MW, Fahrenkrug SC, Hackett PB. Precision editing of large animal genomes. In: Friedmann T, Dunlap JC, Goodwin SF, editors. Advances in Genetics. Vol 80. New York: Academic Press; 2012. pp. 37–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammer RE, et al. Production of transgenic rabbits, sheep and pigs by microinjection. Nature. 1985;315(6021):680–683. doi: 10.1038/315680a0. [DOI] [PubMed] [Google Scholar]
- 4.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23(8):967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 5.Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science. 2011;333(6051):1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 6.Jinek M, et al. RNA-programmed genome editing in human cells. eLIFE. 2013 doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tesson L, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29(8):695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 11.Huang P, et al. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29(8):699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 12.Lei Y, et al. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) Proc Natl Acad Sci USA. 2012;109(43):17484–17489. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30(5):390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, et al. TALENs enable efficient plant genome engineering. Plant Physiol. 2012;161(1):20–27. doi: 10.1104/pp.112.205179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlson DF, et al. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109(43):17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 18.Mussolino C, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39(21):9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li T, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39(14):6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bedell VM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491(7422):114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wefers B, et al. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc Natl Acad Sci USA. 2013;110(10):3782–3787. doi: 10.1073/pnas.1218721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kambadur R, Sharma M, Smith TPL, Bass JJ. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997;7(9):910–916. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- 24.Grobet L, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17(1):71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 25.McPherron AC, Lee S-J. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997;94(23):12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graf B, Senn M. Behavioural and physiological responses of calves to dehorning by heat cauterisation with or without local anaesthesia. Appl Anim Behav Sci. 1999;62:153–171. [Google Scholar]
- 27.Long CR, Gregory KE. Inheritance of the horned, scurred, and polled condition in cattle. J Hered. 1978;69(6):395–400. [Google Scholar]
- 28.Medugorac I, et al. Bovine polledness—an autosomal dominant trait with allelic heterogeneity. PLoS ONE. 2012;7(6):e39477. doi: 10.1371/journal.pone.0039477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen F, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8(9):753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer M, Ortiz O, Hrabé de Angelis M, Wurst W, Kühn R. Modeling disease mutations by gene targeting in one-cell mouse embryos. Proc Natl Acad Sci USA. 2012;109(24):9354–9359. doi: 10.1073/pnas.1121203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beumer KJ, Trautman JK, Mukherjee K, Carroll D. Donor DNA utilization during gene targeting with zinc-finger nucleases. G3 (Bethesda) 2013;3(4):657–664. doi: 10.1534/g3.112.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boch J, et al. Breaking the code of DNA-binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 33.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326(5959):1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 34.Carter DB, et al. Phenotyping of transgenic cloned piglets. Cloning Stem Cells. 2002;4(2):131–145. doi: 10.1089/153623002320253319. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro IM, Lubennikova EI. Population kinetics of cells in tissue culture incubated at low temperature. Exp Cell Res. 1968;49(2):305–316. doi: 10.1016/0014-4827(68)90182-1. [DOI] [PubMed] [Google Scholar]
- 36.Clark KJ, Carlson DF, Fahrenkrug SC. Pigs taking wing with transposons and recombinases. Genome Biol. 2007;8(Suppl 1):S13. doi: 10.1186/gb-2007-8-s1-s13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrels W, et al. Germline transgenic pigs by Sleeping Beauty transposition in porcine zygotes and targeted integration in the pig genome. PLoS ONE. 2011;6(8):e23573. doi: 10.1371/journal.pone.0023573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, et al. TALEN-mediated editing of the mouse Y chromosome. Nat Biotech. 2013;31(6):530–532. doi: 10.1038/nbt.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palgrave CJ, et al. Species-specific variation in RELA underlies differences in NF-κB activity: A potential role in African swine fever pathogenesis. J Virol. 2011;85(12):6008–6014. doi: 10.1128/JVI.00331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson T, et al. Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol Reprod. 2001;64(4):1225–1235. doi: 10.1095/biolreprod64.4.1225. [DOI] [PubMed] [Google Scholar]
- 41.Freking BA, et al. Identification of the single base change causing the callipyge muscle hypertrophy phenotype, the only known example of polar overdominance in mammals. Genome Res. 2002;12(10):1496–1506. doi: 10.1101/gr.571002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317(6034):230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 43.Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: A general strategy for targeting mutations to non-selectable genes. Nature. 1988;336(6197):348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 44.Hayes BJ, Lewin HA, Goddard ME. The future of livestock breeding: Genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends Genet. 2013;29(4):206–214. doi: 10.1016/j.tig.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 45. Zeng W, et al. (2012) Viral transduction of male germline stem cells results in transgene transmission after germ cell transplantation in pigs. Biol Reprod 88(1):27, 1–9. [DOI] [PMC free article] [PubMed]
- 46.Honaramooz A, et al. Fertility and germline transmission of donor haplotype following germ cell transplantation in immunocompetent goats. Biol Reprod. 2003;69(4):1260–1264. doi: 10.1095/biolreprod.103.018788. [DOI] [PubMed] [Google Scholar]
- 47.Rilianawati R, Speed R, Taggart M, Cooke HJ. Spermatogenesis in testes of Dazl null mice after transplantation of wild-type germ cells. Reproduction. 2003;126(5):599–604. doi: 10.1530/rep.0.1260599. [DOI] [PubMed] [Google Scholar]
- 48.Richardson TE, Chapman KM, Tenenhaus Dann C, Hammer RE, Hamra FK. Sterile testis complementation with spermatogonial lines restores fertility to DAZL-deficient rats and maximizes donor germline transmission. PLoS ONE. 2009;4(7):e6308. doi: 10.1371/journal.pone.0006308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481(7381):295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guschin D, et al. (2010) A rapid and general assay for monitoring endogenous gene modification. Engineered Zinc Finger Proteins: Methods and Protocols, Methods in Molecular Biology, eds Mackay JP, Segal DJ (Humana Press, Totowa, NJ) Vol 649, pp 247–256. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.