Significance
The Hedgehog (Hh) signaling pathway plays key roles during embryonic development and remains active in adults. Mutations in the genes encoding the Hh signaling pathway proteins lead to developmental disorders and cancer. The glycosaminoglycan (GAG) chains of proteoglycans at the cell surface shape Hh gradients and signal transduction. We determined the crystal structures of Hh proteins with two different GAG chains, heparin and chondroitin sulfate. The GAG-binding site we identified in the Hh protein is previously not identified and the majority of Hh residues forming this GAG-binding site have been previously implicated in developmental diseases. Analysis of the crystal packing, combined with biophysical experiments, revealed GAG-dependent Hh multimerization and suggests a unique mechanism of Hh signaling regulation.
Abstract
Hedgehog (Hh) morphogens play fundamental roles during embryogenesis and adulthood, in health and disease. Multiple cell surface receptors regulate the Hh signaling pathway. Among these, the glycosaminoglycan (GAG) chains of proteoglycans shape Hh gradients and signal transduction. We have determined crystal structures of Sonic Hh complexes with two GAGs, heparin and chondroitin sulfate. The interaction determinants, confirmed by site-directed mutagenesis and binding studies, reveal a previously not identified Hh site for GAG binding, common to all Hh proteins. The majority of Hh residues forming this GAG-binding site have been previously implicated in developmental diseases. Crystal packing analysis, combined with analytical ultracentrifugation of Sonic Hh–GAG complexes, suggests a potential mechanism for GAG-dependent Hh multimerization. Taken together, these results provide a direct mechanistic explanation of the observed correlation between disease and impaired Hh gradient formation. Moreover, GAG binding partially overlaps with the site of Hh interactions with an array of protein partners including Patched, hedgehog interacting protein, and the interference hedgehog protein family, suggesting a unique mechanism of Hh signaling modulation.
Hedgehog (Hh) signaling is a key mediator of embryonic development (1). Mutations in Hh proteins lead to developmental defects, whereas ectopic activation of Hh signaling is oncogenic (2, 3). The mature Hh morphogen is derived from a protein precursor by autocatalytic cleavage and lipid modification, to generate an amino-terminal signaling domain (HhN), modified by palmitoyl and cholesteryl adducts (4). Hh release from secreting cells requires different membrane proteins, e.g., Dispatched and heparan sulfate proteoglycans (HSPGs) (4, 5). HhN appears to be multivalent and part of a lipoprotein particle (6).
Multiple cell surface molecules control Hh activities. Patched (Ptc1) and Smoothened (Smo) are the core components of Hh signal transduction. In the absence of Hh, Ptc1 suppresses the signaling activity of Smo by preventing its ability to activate the Ci/Gli transcription activators (7). Additional extracellular modulators fine tune Hh signaling responses, including the interference hedgehog protein family (Ihog in fly and Cdo and Boc in human), the vertebrate-specific growth arrest-specific protein 1 (Gas1) and hedgehog-interacting protein (Hhip) (reviewed in ref. 8). HSPGs form an additional group of extracellular Hh modulators. They are composed of a protein core to which linear glycosaminoglycan (GAG) chains [e.g., heparan sulfate (HS) or chondroitin sulfate (CS)] are linked and can act as positive or negative Hh regulators (9). Alongside HSPGs, CS proteoglycans (CSPGs) are key players in development and are required for endochronal bone formation, an Indian Hh (Ihh)-dependent process in the developing growth plate (10). Mutations in genes encoding HSPG biosynthesis enzymes resemble hh mutant phenotypes (11, 12).
HhN directly binds to the different types of GAGs (10, 13). The Cardin–Weintraub sequence (CW), a positively charged region (residues 33–38 in mouse Shh), has been identified as a GAG-binding site by molecular modeling (14) and functional studies confirmed its importance for Hh signaling (15, 16). However, the CW is unable to explain all interactions between Hh and GAGs. In vitro measurements using an alkaline phosphatase assay (17), heparin chromatography (15), and surface plasmon resonance (SPR) (13) have shown that mutations in the CW reduce, but do not eliminate, binding to heparin and HS. The CW lies outside the Shh construct, which is sufficient for signaling and binding to Hh receptors (8). We have determined the crystal structures of ShhN in complex with two ubiquitous GAGs, heparin and chondroitin sulfate. Our structural and functional analysis reveals a previously not identified GAG-binding site on Shh and suggests a potential mechanism for GAG-dependent Hh multimerization.
Results
The Shh N-Terminal Core Domain Without the CW Motif Is Sufficient for Heparin and HS Binding.
To measure the affinity of Shh–GAG interactions, SPR experiments were performed with HS and monodisperse 30-mer heparin (which mimics sulfated regions of HS). Two constructs of mouse Shh were tested: the Shh N-terminal signaling domain (ShhNΔ24) and a truncated construct missing the N-terminal CW sequence (ShhNΔ39) (Fig. 1A and Fig. S1). Both constructs lack the residues for lipid attachment. ShhNΔN24 bound heparin (Kd = 0.8 μM) as well as its cognate biological ligand HS (Kd = 14.5 μM) comparably to previously reported binding data (13) (Fig. S2 A and B), whereas ShhNΔ39 bound heparin and HS 2- to 10-fold weaker (Fig. S2 C and D), suggesting that the CW sequence is contributing to GAG binding, but is not the sole GAG interaction site.
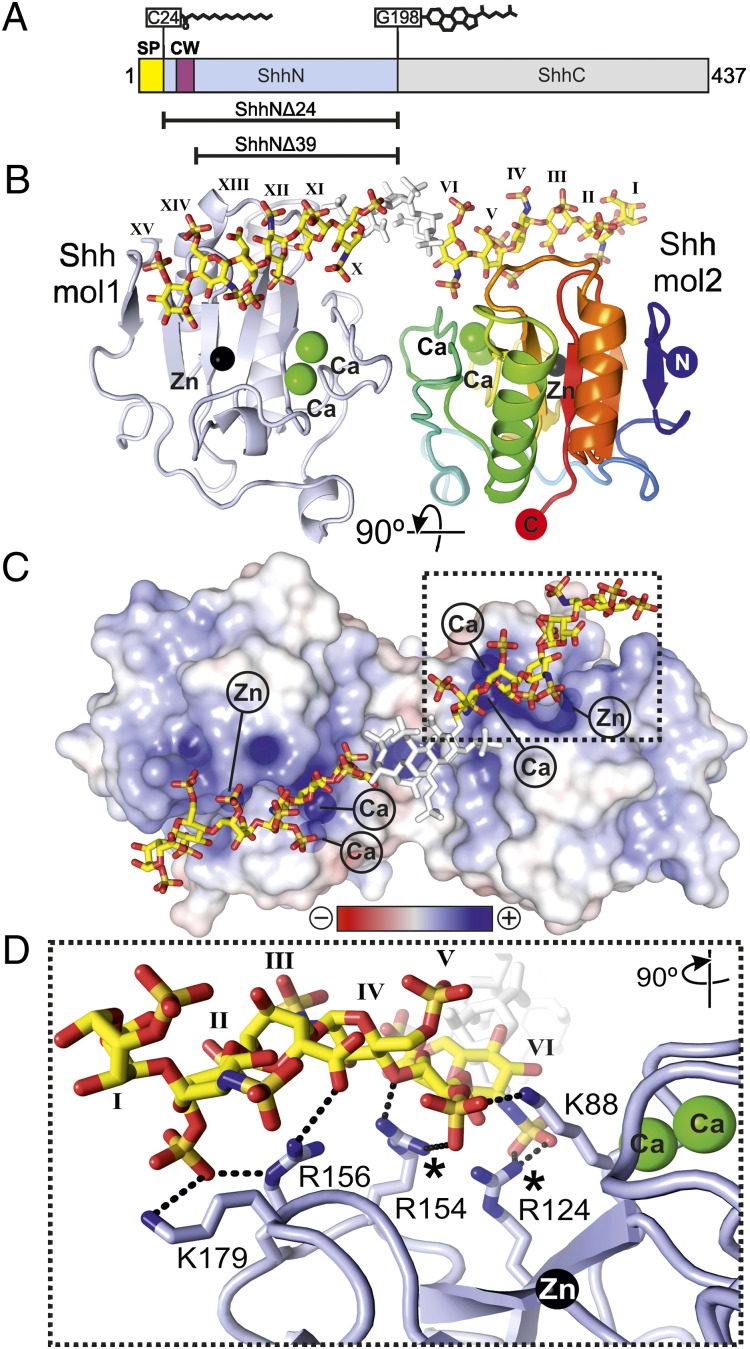
Fig. 1.
Structure of the Shh–heparin complex. (A) Schematic of Shh. CW, Cardin–Weintraub sequence; SP, signal peptide. Lipid modifications are shown. Constructs used are indicated. (B) Ribbon representation of the ShhNΔ39–heparin complex. One molecule is shown in blue, the other in rainbow coloring. Calciums (green), zinc (black), and heparin (yellow, carbon; red, oxygen; blue, nitrogen; and orange, sulfur) are depicted. The three disordered sugar residues (VII, VIII, and IX) are shown in white. Sugar residues are marked in roman numerals (even, N,O6-disulfoglucosamine; odd, O2-sulfoiduronic acid). (C) Electrostatic potential from red (−8 kbT/ec) to blue (+8 kbT/ec). (D) Close-up view of the Shh–heparin-binding site showing the marked region in C. Color coding is as in B. Shh molecule 2 has a similar interaction pattern and architecture due to the pseudosymmetric heparin-binding site (Fig. S3). Equivalent Shh residues of Ihh disease mutations (27, 28) are marked.
Structure Determination of the Shh–Heparin Complex.
The complex of ShhNΔ39 with HPLC-purified 18-mer heparin was solved in space group P21212 and refined to 2.35-Å resolution (Table S1). The asymmetric unit comprises two Shh molecules. A region of additional electron density was immediately detectable and assigned as the heparin chain occupying two distinct orientations (Fig. S3). One heparin chain lay across the Shh dimer, running from a nonreducing end (oxygen O4) to a reducing end (oxygen O1), and the other occupied the same space, but ran in the opposite direction (from O1 to O4) (Fig. S3 B and C). Such an arrangement has been observed before when crystals include highly symmetric polymers in complex with proteins (18) and is potentially due to the regular sulfation pattern of heparin and to the symmetry of the Shh dimer.
The heparin carbohydrate backbone adopts a left-handed helical conformation with 4 monosaccharides per turn, in agreement with previous structural studies (19). Whereas 12 monosaccharide residues could be built into the GAG chain density unambiguously, the central segment of the heparin chain could not be resolved, due to artifactual density arising from the proximity of a crystallographic twofold axis and the lack of heparin contacts with Shh. Three sugar residues would be required to span this segment; therefore we estimate that a heparin chain of 15 monosaccharides is necessary to span the Shh dimer.
Structure of the Shh–Heparin Complex.
The Shh–heparin structure consists of two Shh molecules, a zinc and two calcium ions per Shh molecule, and a heparin chain comprising residues 1–15 (with residues 7–9 omitted) (Fig. 1 and Figs. S4 and S5). The two Shh molecules are essentially identical [root-mean-square deviation (rmsd) of 0.23 Å for 151 equivalent Cα atoms] and show little structural disparity to other Hh structures (e.g., a rmsd of 0.44 Å for 151 equivalent Cα atoms to the first published Shh structure (20) and a rmsd of 0.49 Å over 151 equivalent Cα atoms to the Shh homolog Desert Hh (21). The two Shh molecules are related by a noncrystallographic pseudo–two-fold axis running parallel to the crystallographic axis a. The dimer interface comprises five hydrogen bonds and 46 nonbonded contacts (Fig. S4) with a total buried surface area of 762 Å2, which seems to be lower than observed for physiologically relevant dimer interfaces. Instead, this arrangement appears to be held together by the heparin chain (Fig. 1B). The Shh dimer forms a continuous stretch of positively charged residues, which provides a platform for the interactions with heparin (Fig. 1C). The zinc and calcium ions contribute to the positively charged character of the heparin binding site, albeit without forming direct electrostatic interactions (the closest zinc ion is some 12 Å away from the sulfate groups of N,O6-disulfoglucosamine (SGN) residue II and O2-sulfoiduronic acid (IDS) residue V; the calcium ions reside some 8 Å away from sulfate groups of residues IDS-V and SGN-VI) (Fig. 1B). The metal ions likely shield the negatively charged patches on the Shh surface, and therefore seem to be important for heparin binding.
Next, we mutated four of the five positively charged Shh residues interacting with heparin in our Shh–heparin structure to glutamates (ShhNΔ24–K88E/R124E/R154E/R156E and ShhNΔ39–K88E/R124E/R154E/R156E) and compared the binding to wild-type ShhN using heparin affinity chromatography (Fig. S6). We observed that wild-type ShhNΔ24 bound strongest to the heparin column, whereas ShhNΔ24–K88E/R124E/R154E/R156E and wild-type ShhNΔ39 showed reduced binding (Fig. S6). ShhNΔ39–K88E/R124E/R154E/R156E did not bind at all, validating the observed Shh–heparin interface. This is in agreement with a two-site model for Shh–GAG interactions, where both the CW motif and our identified Shh core GAG-binding site contribute to GAG interactions.
Structure of the Shh–Chondroitin-4-Sulfate (C4S) Complex.
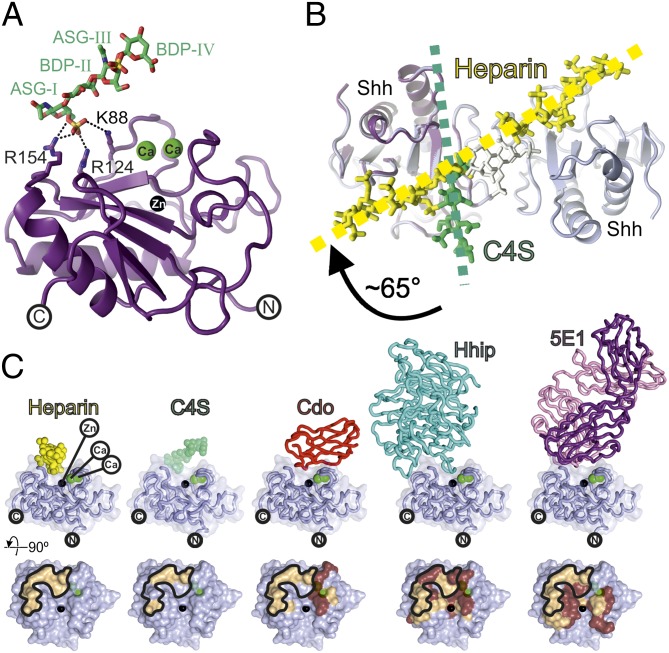
CS is a binding partner for Hh morphogens in vitro and in vivo and direct interactions of CS with purified Ihh and Shh proteins have been observed, albeit showing lower binding affinities compared with heparin or HS (10, 13, 15). We solved the crystal structure of ShhNΔ39 in complex with C4S to 1.74-Å resolution (Table S1 and Fig. S7). The ShhNΔ39–C4S complex reveals one Shh molecule bound to a C4S tetrasaccharide [two N-acetylgalactosamine-4-sulfate (ASG) and two glucuronic acid (BDP) residues] containing two sulfate groups (one on each of the ASG residues) (Fig. 2A). In common with the Shh–heparin complex, the C4S molecule runs head to tail throughout the crystal. The major interaction between C4S and Shh is mediated via hydrogen bonds between the sulfate group of ASG residue I and Shh residues K88, R124, and R154 (Fig. 2A), which form part of the Shh core GAG-binding site identified in our ShhNΔ39–heparin complex. Unlike the Shh–heparin complex, however, no C4S-dependent dimerization of Shh is observed and the C4S tetrasaccharide binds to Shh at a different angle compared with heparin in the Shh–heparin complex (Fig. 2B). This arrangement may be a consequence of the lower sulfation and chain length of the C4S to interact with the basic stripe of the Shh dimer observed in the Shh–heparin structure.
Fig. 2.
Structure of the Shh–C4S complex and analysis of the ShhN core GAG-binding site. (A) Ribbon representation of the ShhNΔ39–C4S complex [Shh, violet; calciums, green; zinc, black; C4S, atomic coloring (green, carbon; red, oxygen; blue, nitrogen; orange, sulfur)]. (B) Comparison of the Shh–C4S and Shh–heparin structures. View is as in Fig. 1C. The Shh–C4S complex is superimposed onto the Shh–heparin complex (Shh, light blue; heparin, yellow). Directions of the GAG chains are shown. (C, Upper) Ribbon representation of Shh (light blue) complexes with (from Left to Right) heparin, C4S, Cdo [Protein Data Bank (PDB) ID 3D1M], Hhip (PDB ID 2WFX), and the antigen-binding fragment of the antibody 5E1 (PDB ID 3MXW). (Lower) Binding footprints mapped on Shh corresponding to Upper. Hydrophilic interactions, beige; hydrophobic interactions, brown. The heparin binding footprint on Shh is marked.
The Shh Core GAG-Binding Site Is a Hotspot for Hh Activity and Receptor Binding.
Our structural analysis reveals that the unique Shh core GAG-binding site is located at a “hotspot” for Hh interactions with its receptors. Hh receptors Cdo/Boc (22), Gas1 (22), Hhip (21, 23), and the monoclonal antibody 5E1 (24) (which blocks Shh binding to Ptc1) as well as the GAGs heparin and CS4 (from this study) all use an overlapping interface on Hh involving the Hh metal binding sites (Fig. 2C). Binding of both positive and negative regulators to the same site on Hh can be explained by considering the Hh morphogen as part of a multimeric assembly where multiple binding sites are displayed and available for interaction with numerous regulators, thus permitting different regulators to act simultaneously (8). In this model, the strength a regulator is capable of exerting on the Hh signal can be determined by its affinity for the Hh morphogen and its local concentration under physiological conditions.
Various mutations identified in human Hh proteins map onto the Shh core GAG-binding site. The human Shh mutation K178S (equivalent to mouse Shh K179, Fig. 1D) reduces Hh signaling activity in C3H10T1/2 cells (25), and proliferation and invasion of PANC1 cells (26). Additionally, a Shh construct harboring three point mutations (two in the CW, the other K178S) was unable to efficiently bind HS and was found to be defective in forming HS-dependent Shh oligomers (26). Shh mutations K179A and R154A also have reduced affinity for the Hh receptor Ptc1 (25). Ihh carrying a R158C (equivalent to mouse Shh residue R154, Fig. 1D) results in an altered signaling capacity and range (27) and reduced gene transcription activity and Ptc1 binding (25). A heterozygous Ihh mutation of this residue leads to the developmental disorder bractydactyly type A1 (BDA1) (27). A heterozygous mutation of Ihh-R128 (equivalent to R124 in mouse Shh, Fig. 1D) also has a BDA1 phenotype (28). A pentameric alanine mutant form of mouse Shh (K88/R124/D153/R154/R156) showed reduced GAG-binding ability and exhibited a severely restricted ability to form Hh oligomers (29). Because the Shh core GAG-binding site resides at a hotspot essential for Hh receptor interactions, it is difficult to dissect the effect that a given mutation at this site will have on each individual receptor. Furthermore, other mechanisms including Ptc binding have been shown to alter the signaling range of Hh morphogens (30). Nonetheless, the signaling range of the Hh morphogen is also shaped by Hh–proteoglycan interactions. The observation that some mutants that map to our identified GAG-binding site affect Hh signaling range, indicates that at least part of the complex signaling output is mediated by interactions between this site and the GAG chains of proteoglycans.
Conservation of the Hh Core GAG-Binding Site in Drosophila Hh Signaling.
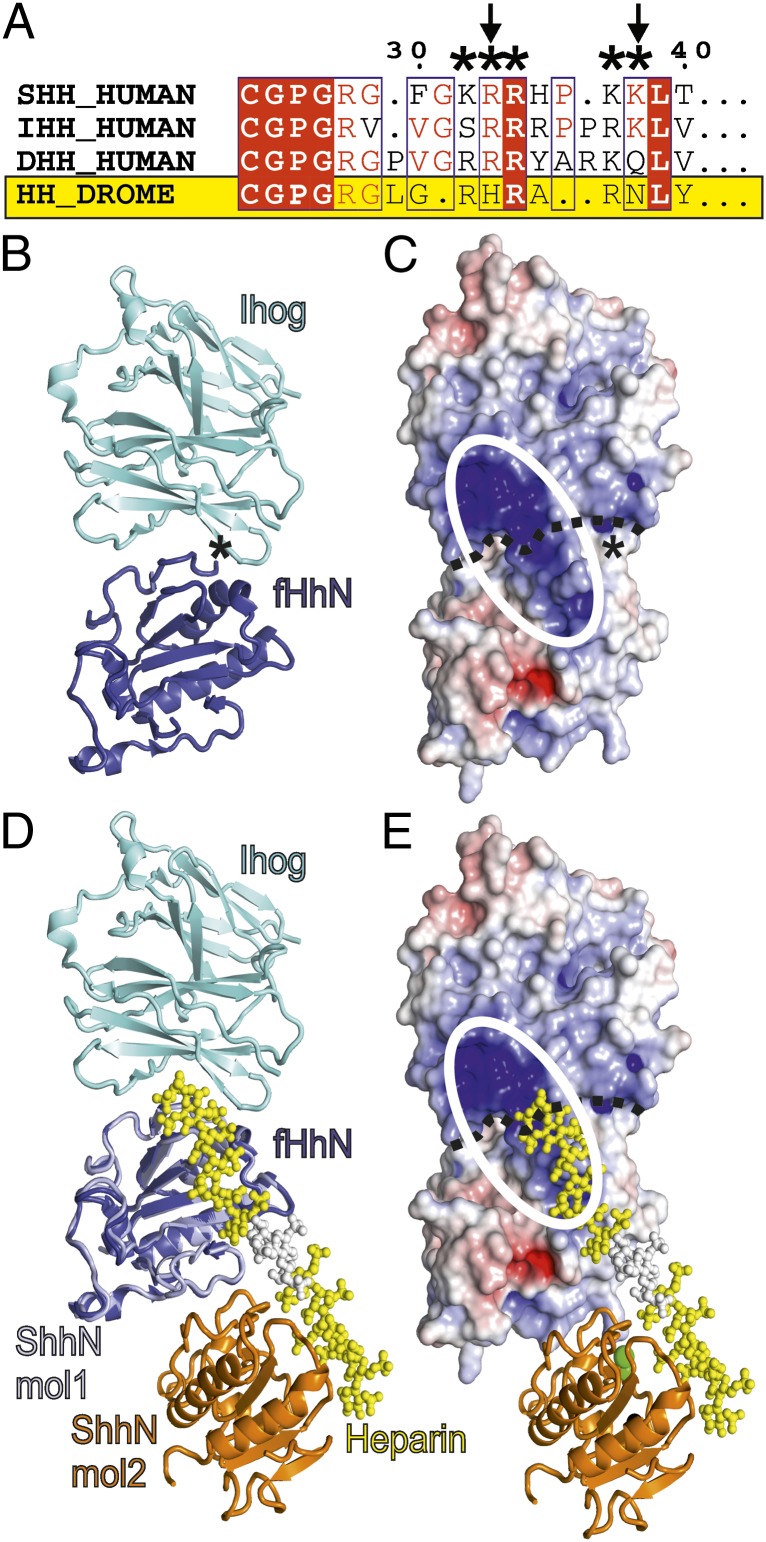
Despite the importance of HS as a key modulator of Drosophila Hh signaling, the CW is only partially conserved in fly (Fig. 3A). As defined bioinformatically (14), the CW consensus sequence is xBBBxxBBx, where B represents the basic residues R or K, and x is a hydrophobic residue. In the Drosophila CW, the second and fifth residues of the consensus sequence are replaced by histidine and asparagine, respectively. The loss of two of the five basic CW residues can be expected to lower the GAG-binding affinity of Drosophila Hh (fHh). In contrast, the Hh core GAG-binding site identified in this study is conserved across all known Hh orthologs, including fly (Fig. S1). This strongly supports its significance for Hh function.
Fig. 3.
Implications of the Shh–GAG-binding site for fly Hh signaling. (A) Sequence alignment of the Shh CW. The CW is marked (*) and is only partially conserved in fly (residue exchanges are indicated with arrows). DROME, Drosophila melanogaster. (B–E) Comparison of the core GAG-binding site in mouse and fly. (B and C) The fHh–Ihog complex (PDB ID 2IBG) (fHh, blue; Ihog, cyan). In C, the surface is color coded according to electrostatic potential from red (−8 kbT/ec) to blue (+8 kbT/ec). The continuous basic surface stretch formed by both fHh and Ihog is marked as a white ellipse. The asterisk marks the fHh N terminus. (D and E) Superposition of the ShhNΔ39–heparin complex onto fHh (Shh, lightblue/orange; heparin, yellow/white) in the same orientation as in B and C. Extension of the heparin chain would result in covering the positively charged patch on Ihog.
The previously determined crystal structure of fHh in complex with its receptor Ihog revealed a continuous strip of positive charge formed between the two proteins (31) (Fig. 3 B and C). Although heparin was not visible in the fHh–Ihog crystal structure, the complex could only be obtained in solution and crystallized in the presence of heparin. McLellan et al. (31) suggested that HS might bind this strip of positive charge, thereby bridging the two molecules, stabilizing the complex, and fulfilling its role as an essential cofactor for this interaction. To validate their findings, McLellan et al. (31) mutated several putative HS-binding residues in fHh (K105, R147, R213, and R239) to glutamate, of which the corresponding mouse Shh residues are K88, R124, R154, and R156. This mutant showed no binding to heparin and much-reduced ability to bind to Ihog overexpressing cells. Intriguingly, the putative HS-binding residues on fHh are located at equivalent positions to the Shh core GAG-binding site reported here (Fig. 3 D and E and Fig. S1), revealing a high degree of functional conservation from fly to human. Further, the location of the potential CW motif at the N terminus of fHh is distant from the positively charged patch important for Ihog binding and is therefore unlikely to directly contribute to the fHh–Ihog interaction—the distance between Tyr-100 (the most N-terminal residue visible in the fHh crystal structure) and Arg-213 (at the core binding site) is 24 Å. These two regions are on opposite sides of the fHh molecule (Fig. 3 B and C).
The Shh–Heparin Structure Suggests a Potential Mechanism for GAG-Dependent Hh Multimerization.
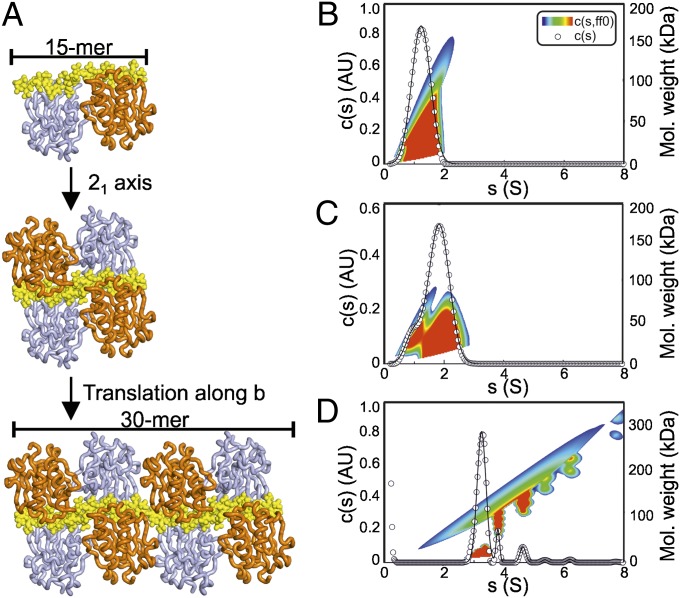
Previous studies have assigned a key role for HSPGs in Hh multimerization (32, 33) and multiple studies have reported reduced oligomerization and signaling range of Hh variants with mutations within the GAG-binding site reported here (25, 26, 29). The arrangement of the Shh–GAG complexes in the crystal suggests a potential mechanism for GAG-dependent Hh multimerization. In the crystal of the Shh–heparin complex, continuous electron density assigned to the heparin molecule runs throughout the asymmetric unit with the heparin molecules arranged head to tail, parallel to the crystallographic axis b (Fig. 4A). The 21 symmetry of the crystallographic b axis generates a sandwich arrangement of two Shh dimers on a single heparin chain (Fig. 4A, second panel from Top), and translation along the crystallographic b axis results in a continuous stretch of a heparin chain, with Shh molecules bound like “beads on a string” (Fig. 4A, Bottom). HS chains of length over 100 sugar moieties exist at the cell surface, and the binding of multiple Hh copies to a single HS molecule suggests an appealing mechanism for GAG-dependent Hh multimerization.
Fig. 4.
Implications for Shh multimerization. (A) Crystal packing analysis of the ShhNΔ39–heparin structure. Heparin lies parallel to the crystallographic b axis, which has 21 symmetry (Top). This operation results in sandwich arrangement of two Shh dimers packed around the heparin chain (Middle). Extension along the b axis reveals a structure comprising Shh molecules attached to heparin like beads on a string (Bottom). (B–D) AUC sedimentation velocity experiments of ShhNΔ39 with heparin. (B) ShhNΔ39 shows a monodisperse peak at 1.2 S corresponding to monomeric ShhNΔ39 (rmsd 0.0322). (C) ShhNΔ39 mixed with a 6-mer heparin shows reduction in the monomeric peak and the appearance of a major peak at 1.8 S indicating a 1:1 Shh:heparin complex (rmsd 0.0183). The minor species observed was residual Shh (at ∼1.2 S). (D) ShhNΔ39 mixed with a 30-mer heparin reveals multiple peaks corresponding to discrete Shh–heparin complexes (rmsd 0.0112). AU, arbitrary unit.
To test whether such an assembly can exist in solution, we used analytical ultracentrifugation (AUC) to analyze Shh–heparin complex formation. Sedimentation velocity AUC revealed that ShhNΔ39 in the absence of heparin is monodisperse and monomeric in solution (Fig. 4B). Addition of an equimolar amount of a six-residue heparin molecule resulted in an increase in the sedimentation coefficient from 1.2 S to 1.8 S, which is consistent with a 1:1 ShhNΔ39:heparin complex (Fig. 4C). We then tested a 30-mer heparin in the same experimental setting. Six clearly resolved species were observed (Fig. 4D), representing discrete populations of Shh–heparin complexes. It was not possible unambiguously to assign each peak as a particular assembly, but it is likely that each peak comprises a 30-mer heparin molecule bound to an increasing number of ShhNΔ39 molecules. To evaluate how discrete the sedimenting species are, we applied a 2D analysis [plotted here as c(s,Mw)] (Fig. 4 B–D). For smaller s values, it is difficult to assign a definite stoichiometry because of a greater range of possible conformers; however, the highest s value peak of the ShhNΔ39–30-mer heparin complex (∼6.2 S corresponding to Mw of ∼200–230 kDa, Fig. 4D) likely corresponds to 7 and 8 Shh bound to one heparin chain. This is in agreement with our ShhNΔ39–heparin structure, where a maximum of 8 Shh molecules are bound to a 30-mer heparin (Fig. 4A, Lower) in a tightly packed crystal environment.
Discussion
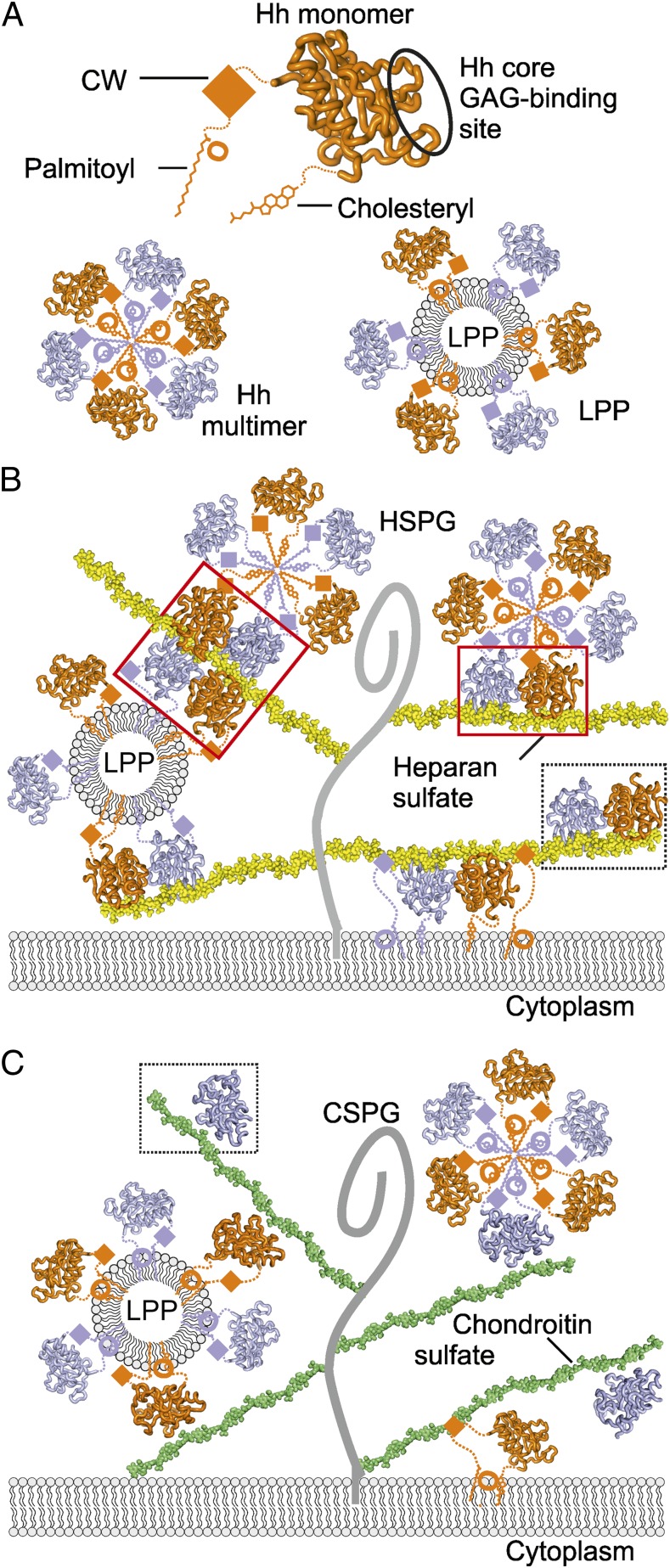
We have identified a unique, evolutionarily conserved GAG-binding site on the Hh morphogen core (Fig. 5A). The identification and characterization of this site elucidates unresolved observations in the control of Hh signaling. Although Hh oligomerization is important for Hh gradient formation (6, 33), analysis of apo-Hh crystal structures found no evidence for a conserved mode of Hh oligomerization (34) raising the question of what might promote it. Our structural and biophysical data address this question and suggest a mode of GAG-dependent Hh oligomerization, in which Hh proteins bind along highly sulfated GAG chains. Indeed, Hh dimers assemble on the heparin chain (which mimics sulfated regions of HS), harboring three sulfate groups per disaccharide (Figs. 1 and 5B), whereas the less sulfated C4S chain (which has only one sulfate group per disaccharide) binds to a single Hh monomer per tetrasaccharide in the crystal structure (Figs. 2A and 5C). This shows that distinctive GAG sulfation patterns differently interact with Hh and may control their oligomeric state via the unique GAG-binding site. This previously not identified site together with the GAG-binding CW motif suggests a two-site model for Hh–GAG interactions.
Fig. 5.
Model of Hh–GAG interactions at the cell surface. (A) The Hh monomer (orange) is covalently linked to palmitoyl and cholesteryl moieties at the N- and C-termini, respectively. CW is located between the palmitoyl and Hh core domain. Our identified Hh core GAG-binding site is circled. Hh monomers assemble into homomultimers by burying lipid moieties within the hydrophobic core (Lower Left) or as part of lipoprotein particles (LPP) (Lower Right). (B) Different monomeric and oligomeric forms of Hh bind to HS chains (yellow) linked to the stalk regions of HSPGs (gray curve). Both CW and the Hh core GAG-binding site mediate Hh binding to HS. High degree of HS sulfation mediates HS-dependent Hh oligomerization. HS-induced tetramer and dimer are shown in red boxes, the Shh–heparin complex observed in the crystallographic asymmetric unit, in a dashed box. (C) Similarly to HS, CS chains (green) bind to multiple forms of Hh (monomers or lipid-induced oligomers). However, in contrast to HS, the less sulfated CS provides a platform for a lower degree of Hh oligomerization on the cell surface. A Shh–C4S complex as observed in the crystal structure is framed within a dashed box.
The Shh constructs used in this study lack lipid modifications, previously implicated in Hh multimerization, whereby oligomerization may be achieved by a sphere of lipid-modified Hh molecules, centered on a hydrophobic core to form a micelle-like structure, either a Hh homomultimer or as part of a bigger lipoprotein particle (6, 35) (Fig. 5A). In this arrangement, Hh lipid modification sites would be positioned distantly from the Hh core GAG-binding site (Fig. 5A), still allowing interactions between lipid layer-attached Hh and GAG chains, whereas the CW would be in close proximity to the hydrophobic core (within seven residues of the N-terminal palymitoylation site) and might be less accessible (Fig. 4A). Thus, a combination of Hh-linked lipids and two distinctive GAG-binding sites suggests that Hh could be incorporated into supramolecular complexes such as lipoprotein particles that further interact with proteoglycans and form Hh morphogen gradients in the extracellular matrix (Fig. 5 B and C).
In a cellular context, the degree of GAG sulfation and distribution of the sulfate groups are important for the interactions with protein partners and their biological consequences (36). Our structural analyses directly illustrate this, as the different relative orientations of the GAG chains in the Shh–heparin and Shh–C4S complexes (Fig. 2B) appear to be controlled by the sulfation pattern. Interestingly, HS showed a different effect compared with CS in the release and processing of the active Shh morphogen in a cellular assay (37), and a mouse model for undersulfated CSPGs indicated that the degree of CS sulfation is crucial for Ihh function in the developing growth plate (10). Now, our data suggest a mechanism in which HSPGs together with CSPGs control Hh morphogen gradients by providing both low- and high-affinity binding sites for multiple Hh forms.
The modulation of the Hh pathway by small molecules is a proven strategy for the treatment of cancer and developmental disorders, and for stem cell therapies (3). As our identified Hh GAG-binding site resides at a Hh receptor-binding region, our Shh–GAG structures might form a platform for the rational design of GAG mimetics as a unique class of Hh pathway modulators. Similar strategies focused on other morphogen signaling systems, for example, the signaling modulator PI-88, a HS mimetic, can specifically modulate the interactions of fibroblast growth factor (FGF) and its receptor (FGFR) (38). FGF–FGFR interactions are heparin dependent and require simultaneous binding of FGF and heparin to form a ternary signaling complex (39, 40), analogous to the fHh–Ihog–heparin complex (31). Furthermore, parallels can be drawn between our findings that the long heparin chains multimerize Shh molecules and previous reports on heparin-dependent supramolecular clustering of FGF–FGFR complexes (41).
Other morphogens, such as Wnts, are lipid modified, form extracellular gradients by associating with lipoprotein particles (6), and are presented to their receptor Frizzled in a HS-dependent manner (42). We note that a recent Wnt8 structure reveals a prominent basic patch (formed by R63, R288, R289, K292, and R293) proximal to a predicted binding site for Lrp5/6 coreceptors (43). Taken together, our studies suggest that binding hotspots that accommodate GAGs as well as protein agonists and antagonists play an important role in fine tuning morphogen signaling.
Materials and Methods
Structure Determination of Shh–GAG Complexes.
ShhNΔ24 (residues 25–194), containing the CW sequence, and ShhNΔ39 (residues 40–194), lacking the CW sequence, were cloned and expressed in Escherichia coli as described previously (21). ShhNΔ39 at 9 mg/mL was crystallized in the presence of 2 mM 18-mer heparin and the ShhNΔ39–C4S complex in the presence of 10 mM chondroitin 4-sulfate and 5 mM CaCl2. Structures were solved by molecular replacement (Table S1).
Shh–GAG-Binding Studies.
Heparin affinity chromatography was performed to test binding to different Shh constructs. SPR-based experiments were performed using a Biacore T100 machine. Analytical ultracentrifugation sedimentation velocity experiments were carried out in a Beckman Optima XL-I analytical ultracentrifuge using a scanning absorbance of 280 nm and interference optics. Further details for all these methods are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank T. Walter and K. Harlos for help with crystallization; B. Bishop for laboratory support; the staff of Diamond Light Source Beamline I03 for assistance with data collection; and the Biophysical Instrument Facility, Department of Biochemistry, and the Wellcome Trust Centre for Human Genetics. The work was funded by Wellcome Trust and Cancer Research UK (C.S.). T.M. was supported by Cancer Research UK. D.M.W. was a Medical Research Council Student. R.J.C.G. was a Royal Society University Research Fellow. C.S. is a Cancer Research UK Senior Research Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.J.L. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4C4N and 4C4M).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310097110/-/DCSupplemental.
References
- 1.Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001;15(23):3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Belloni E, et al. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat Genet. 1996;14(3):353–356. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- 3.Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5(12):1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 4.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12(6):393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 5.Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol. 2009;1(3):a002493. doi: 10.1101/cshperspect.a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panáková D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435(7038):58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- 7.Rohatgi R, Scott MP. Patching the gaps in Hedgehog signalling. Nat Cell Biol. 2007;9(9):1005–1009. doi: 10.1038/ncb435. [DOI] [PubMed] [Google Scholar]
- 8.Beachy PA, Hymowitz SG, Lazarus RA, Leahy DJ, Siebold C. Interactions between Hedgehog proteins and their binding partners come into view. Genes Dev. 2010;24(18):2001–2012. doi: 10.1101/gad.1951710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams EH, et al. Dally-like core protein and its mammalian homologues mediate stimulatory and inhibitory effects on Hedgehog signal response. Proc Natl Acad Sci USA. 2010;107(13):5869–5874. doi: 10.1073/pnas.1001777107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortes M, Baria AT, Schwartz NB. Sulfation of chondroitin sulfate proteoglycans is necessary for proper Indian hedgehog signaling in the developing growth plate. Development. 2009;136(10):1697–1706. doi: 10.1242/dev.030742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han C, et al. Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development. 2004;131(7):1563–1575. doi: 10.1242/dev.01051. [DOI] [PubMed] [Google Scholar]
- 12.Bellaiche Y, The I, Perrimon N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature. 1998;394(6688):85–88. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, McLellan JS, Ayala AM, Leahy DJ, Linhardt RJ. Kinetic and structural studies on interactions between heparin or heparan sulfate and proteins of the hedgehog signaling pathway. Biochemistry. 2007;46(13):3933–3941. doi: 10.1021/bi6025424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardin AD, Weintraub HJ. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9(1):21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 15.Rubin JB, Choi Y, Segal RA. Cerebellar proteoglycans regulate sonic hedgehog responses during development. Development. 2002;129(9):2223–2232. doi: 10.1242/dev.129.9.2223. [DOI] [PubMed] [Google Scholar]
- 16.Farshi P, et al. Dual roles of the Cardin-Weintraub motif in multimeric Sonic hedgehog. J Biol Chem. 2011;286(26):23608–23619. doi: 10.1074/jbc.M110.206474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan JA, et al. Proteoglycan interactions with Sonic Hedgehog specify mitogenic responses. Nat Neurosci. 2009;12(4):409–417. doi: 10.1038/nn.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker S, Groner B, Müller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394(6689):145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 19.Mulloy B, Forster MJ. Conformation and dynamics of heparin and heparan sulfate. Glycobiology. 2000;10(11):1147–1156. doi: 10.1093/glycob/10.11.1147. [DOI] [PubMed] [Google Scholar]
- 20.Hall TM, Porter JA, Beachy PA, Leahy DJ. A potential catalytic site revealed by the 1.7-A crystal structure of the amino-terminal signalling domain of Sonic hedgehog. Nature. 1995;378(6553):212–216. doi: 10.1038/378212a0. [DOI] [PubMed] [Google Scholar]
- 21.Bishop B, et al. Structural insights into hedgehog ligand sequestration by the human hedgehog-interacting protein HHIP. Nat Struct Mol Biol. 2009;16(7):698–703. doi: 10.1038/nsmb.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLellan JS, et al. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature. 2008;455(7215):979–983. doi: 10.1038/nature07358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosanac I, et al. The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nat Struct Mol Biol. 2009;16(7):691–697. doi: 10.1038/nsmb.1632. [DOI] [PubMed] [Google Scholar]
- 24.Maun HR, et al. Hedgehog pathway antagonist 5E1 binds hedgehog at the pseudo-active site. J Biol Chem. 2010;285(34):26570–26580. doi: 10.1074/jbc.M110.112284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuse N, et al. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc Natl Acad Sci USA. 1999;96(20):10992–10999. doi: 10.1073/pnas.96.20.10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang SC, Mulloy B, Magee AI, Couchman JR. Two distinct sites in sonic Hedgehog combine for heparan sulfate interactions and cell signaling functions. J Biol Chem. 2011;286(52):44391–44402. doi: 10.1074/jbc.M111.285361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stattin EL, Lindén B, Lönnerholm T, Schuster J, Dahl N. Brachydactyly type A1 associated with unusual radiological findings and a novel Arg158Cys mutation in the Indian hedgehog (IHH) gene. Eur J Med Genet. 2009;52(5):297–302. doi: 10.1016/j.ejmg.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Byrnes AM, et al. Brachydactyly A-1 mutations restricted to the central region of the N-terminal active fragment of Indian Hedgehog. Eur J Hum Genet. 2009;17(9):1112–1120. doi: 10.1038/ejhg.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohlig S, et al. Sonic hedgehog shedding results in functional activation of the solubilized protein. Dev Cell. 2011;20(6):764–774. doi: 10.1016/j.devcel.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Casali A, Struhl G. Reading the Hedgehog morphogen gradient by measuring the ratio of bound to unbound Patched protein. Nature. 2004;431(7004):76–80. doi: 10.1038/nature02835. [DOI] [PubMed] [Google Scholar]
- 31.McLellan JS, et al. Structure of a heparin-dependent complex of Hedgehog and Ihog. Proc Natl Acad Sci USA. 2006;103(46):17208–17213. doi: 10.1073/pnas.0606738103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eugster C, Panáková D, Mahmoud A, Eaton S. Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev Cell. 2007;13(1):57–71. doi: 10.1016/j.devcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 33.Vyas N, et al. Nanoscale organization of hedgehog is essential for long-range signaling. Cell. 2008;133(7):1214–1227. doi: 10.1016/j.cell.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 34.Kavran JM, Ward MD, Oladosu OO, Mulepati S, Leahy DJ. All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. J Biol Chem. 2010;285(32):24584–24590. doi: 10.1074/jbc.M110.131680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goetz JA, Singh S, Suber LM, Kull FJ, Robbins DJ. A highly conserved amino-terminal region of sonic hedgehog is required for the formation of its freely diffusible multimeric form. J Biol Chem. 2006;281(7):4087–4093. doi: 10.1074/jbc.M511427200. [DOI] [PubMed] [Google Scholar]
- 36.Coles CH, et al. Proteoglycan-specific molecular switch for RPTPσ clustering and neuronal extension. Science. 2011;332(6028):484–488. doi: 10.1126/science.1200840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dierker T, Dreier R, Petersen A, Bordych C, Grobe K. Heparan sulfate-modulated, metalloprotease-mediated sonic hedgehog release from producing cells. J Biol Chem. 2009;284(12):8013–8022. doi: 10.1074/jbc.M806838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cochran S, et al. Probing the interactions of phosphosulfomannans with angiogenic growth factors by surface plasmon resonance. J Med Chem. 2003;46(21):4601–4608. doi: 10.1021/jm030180y. [DOI] [PubMed] [Google Scholar]
- 39.Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407(6807):1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 40.Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98(5):641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- 41.Harmer NJ, et al. Multimers of the fibroblast growth factor (FGF)-FGF receptor-saccharide complex are formed on long oligomers of heparin. Biochem J. 2006;393(Pt 3):741–748. doi: 10.1042/BJ20050985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ai X, et al. QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol. 2003;162(2):341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337(6090):59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.