Significance
We show that the characteristic variation in the density of grains observed along the inflorescence (spike) of modern cultivated barley (Hordeum vulgare) is the consequence of a perturbed interaction between a microRNA, miR172, and its corresponding binding site in the mRNA of an APELATA2 (AP2)-like transcription factor, HvAP2. Our data indicate that variation in the miR172-driven turnover of HvAP2 regulates the length of a developmental window that is required for elongation of the internodes along the axis of the spike, and this variation results in the striking differences in the size and shape of the barley inflorescence.
Keywords: genome wide association scan, heterochrony
Abstract
Within the cereal grasses, variation in inflorescence architecture results in a conspicuous morphological diversity that in crop species influences the yield of cereal grains. Although significant progress has been made in identifying some of the genes underlying this variation in maize and rice, in the temperate cereals, a group that includes wheat, barley, and rye, only the dosage-dependent and highly pleiotropic Q locus in hexaploid wheat has been molecularly characterized. Here we show that the characteristic variation in the density of grains along the inflorescence, or spike, of modern cultivated barley (Hordeum vulgare) is largely the consequence of a perturbed interaction between microRNA172 and its corresponding binding site in the mRNA of an APELATA2 (AP2)-like transcription factor, HvAP2. We used genome-wide association and biparental mapping to identify HvAP2. By comparing inflorescence development and HvAP2 transcript abundance in an extreme dense-spike mutant and its nearly isogenic WT line, we show that HvAP2 turnover driven by microRNA 172 regulates the length of a critical developmental window that is required for elongation of the inflorescence internodes. Our data indicate that this heterochronic change, an altered timing of developmental events caused by specific temporal variation in the efficiency of HvAP2 turnover, leads to the striking differences in the size and shape of the barley spike.
The cereal grasses are the world’s most economically, sociologically, and ecologically important crops (1). Within and between species, variation in inflorescence architecture results in striking and characteristic morphological diversity. Dissecting the mechanisms and processes underpinning this diversity is central to understanding both its origin and evolution and how it can be harnessed for knowledge-based crop improvement. In maize and rice, a considerable body of work has begun to dissect the genes and gene networks controlling aspects of inflorescence development (e.g., refs. 2–5). However, within the Triticeae cereals, a group that includes some of the world’s most important human foods, only Q, a highly pleiotropic gene that influences a broad range of phenotypic characters including inflorescence compactness, fragility, and free-threshing (6), and genes that determine the development of an extra floret on the lemma (7), additional glumes (8), the number of grains on the inflorescence (9), and floret fertility (10) have been molecularly characterized.
Barley is a diploid inbreeding crop species and a model for other, genetically more complex temperate cereal species such as durum and bread wheats and rye. The barley inflorescence forms a terminal spike that bears reproductive units called “spikelets.” Three spikelets initiate at each alternating node of the spike’s central stem or rachis. Each spikelet forms a single floret that can develop into a kernel of grain. Several loci have been shown to affect spike density in barley (11), including dense spike-ar (dsp.ar), recently mapped to the centromere of chromosome 7H (12) and ZEOCRITON1 (Zeo1), Zeo2, and Zeo3, all of which map to an overlapping interval on the long arm of chromosome 2H (13); however, to date no dense-spike locus in barley has been studied at the molecular level.
Here, using a genome-wide association scan (GWAS) combined with analyses of nearly isogenic lines and biparental genetic linkage mapping, we show that a genetic region on barley chromosome 2HL is a major determinant of spike density in cultivated barley. We identify the gene that from classical genetic studies is termed ZEOCRITON (Zeo) as a promising target for understanding the molecular processes that influence this important agronomic and developmental trait. In particular, we exploited an allelic series of Zeo mutants in a near-isogenic background with the cultivar (cv.) Bowman (hereafter referred to as “WT”) as the recurrent parent (13) to demonstrate that dense spikes in barley are caused largely by polymorphisms in the microRNA172 (miR172)-binding site of the barley ortholog of an APELATA2 transcription factor (HvAP2). We observe that single-nucleotide changes affect the extent of miR172-directed cleavage of HvAP2 transcripts compared with its nearly isogenic WT. By conducting a comparative, fine-scale morphological study of an extremely dense Zeo1 mutant and its WT, we provide insight into the roles of HvAP2 in determining spike density by regulating the timing of specific developmental events.
Results
Genetic Mapping of Grain Density Along the Barley Inflorescence.
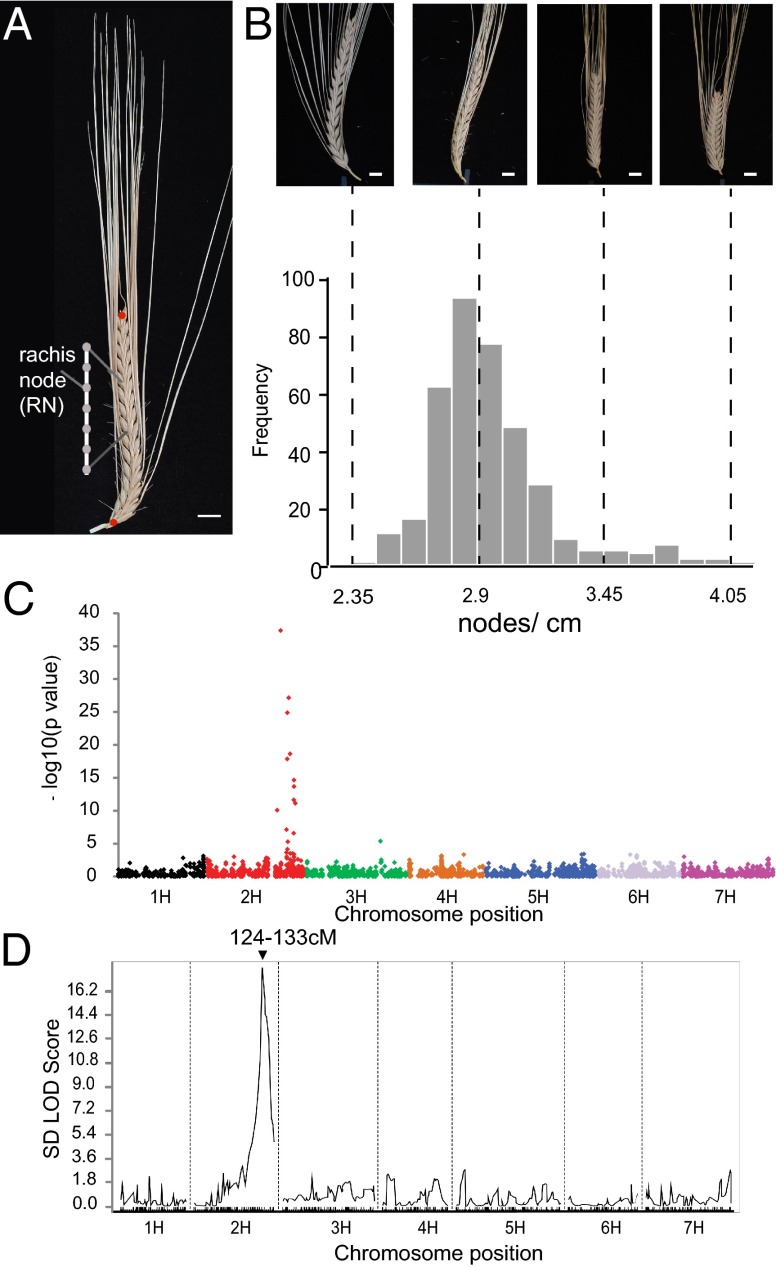
We initially explored the genetic basis of variation in the density of grains along the barley inflorescence using a GWAS. We digitally phenotyped mature spikes from 401 two-row spring-type cultivars (Dataset S1) that we previously had genotyped with a 9K Illumina iSelect single nucleotide polymorphism (SNP) array (14) and recorded variation in two architectural traits: rachis length (RL, the length of the central axis of the inflorescence) and rachis node number (RNN, the number of grain-bearing nodes along the rachis). We derived an integrated spike-density phenotype from the ratio RNN/RL (Fig. 1 A and B). A GWAS for spike density identified a highly significant association peak [−log10(p value) >37] centered around 125 cM on barley chromosome 2H, with a smaller effect on chromosome 3H near the location of the DENSO dwarfing gene (110–115 cM) (Fig. 1C). The former location was supported by analysis of a recombinant inbred line population derived from a cross between the cv. Golden Promise (dense spike) and Morex (normal spike) (GPMx; F11 generation, 160 individuals). We developed a genetic linkage map of this population using the same SNP platform used for GWAS. Quantitative trait loci (QTL) analysis of spike density resolved a single major QTL [logarithm (base 10) of odds (LOD) ≥ 18] in the same region of chromosome 2H (Fig. 1D). This position is consistent with that of previously identified QTL for the orthologous phenotype “rachis internode length” (15, 16).
Fig. 1.
Genome-wide QTL mapping of spike density in a population of 401 two-row spring cultivars. (A) Photograph of a barley spike from the accession “Neruda.” Rachis length, RL, is the distance between the red circles, and a rachis node, RN, is indicated on the diagram inset. Spike density was calculated as the ratio RNN/RL. (B) (Upper) Photographs of selected cultivars highlighting variation in spike density. Cultivars from left to right are Maypole, Appaloosa, Golden Promise, and Forum. (Lower) Histogram of spike density genotypic means in nodes/cm. Dashed lines correspond to the spike density represented by the cultivars in the upper panel. (C) GWAS of spike density across barley’s seven chromosomes (14) resolved two associations, the larger [−log10(p value) >37] around 125 cM on chromosome 2H (indicated in red) and the smaller on chromosome 3H (indicated in green). (D) QTL scan of spike density in the Golden Promise (dense spike) × Morex (normal spike) recombinant inbred line population revealed a single peak (LOD ≥18) overlying the same region on chromosome 2H. (Scale bars, 1 cm in A and B.)
We then surveyed a collection of genetically characterized mutation-containing nearly isogenic introgression lines (known as the “Bowman Nearly Isogenic Lines”) (12) for both phenotypic descriptors related to spike density and for evidence of introgression segments encompassing the spike density QTL. Our attention focused quickly on three semidominant dense-spike loci, Zeo1, -2, and -3. Compared with WT, Zeo1.b spikes have a spike density almost twice that of the recurrent parent Bowman (Fig. S1 A and B and Dataset S1). We developed two F2 populations segregating for the very dense Zeo1.b allele and mapped the spike density phenotype to the same genetic interval defined by GWAS and QTL analysis in Golden Promise x Morex (GPMx). We observed that Golden Promise (a two-rowed cultivar) has about the same rachis length as the six-rowed parent Morex (P = 0.12; t test) but has significantly more nodes (P = 0.016; t test), so that spike density is higher in Golden Promise than in Morex (P = 0.021; t test). No interaction was detected with the closely linked Vrs1 locus (position 85–86 cM) (Fig. 1D). By integrating genetic data from these and additional populations (Fig. S1C and Dataset S1), we resolved the location of Zeo1.b to a region containing 29 barley gene models (according to ref. 17). Two of these, MLOC_81350 and MLOC_43830, were of immediate interest because they encode plant-specific transcription factors containing an AP2 domain, proteins known to regulate developmental processes including cereal spike architecture (18, 19).
Mutations in HvAP2 Contribute to Variation in Rachis Internode Elongation.
To investigate whether either of these genes was responsible for the Zeo phenotype, we extended our collection of natural and induced mutants using broader genetic and phenotypic descriptors [i.e., PYRAMIDATUM (Pyr) and ERECTOIDES (Ert)] on the assumption that colocating and phenotypically similar mutants may represent an allelic series of mutations in ZEO (Dataset S1). We PCR-amplified and sequenced the entire coding sequence, 5′ and 3′ UTRs of MLOC_81350 and MLOC_43830 in 67 lines that included mutants, their WT donors, and recurrent parents. No sequence variation could be associated with the Zeo phenotype in MLOC_81350; however, 31 accessions contained putatively compromised alleles of MLOC_43830 (Dataset S1).
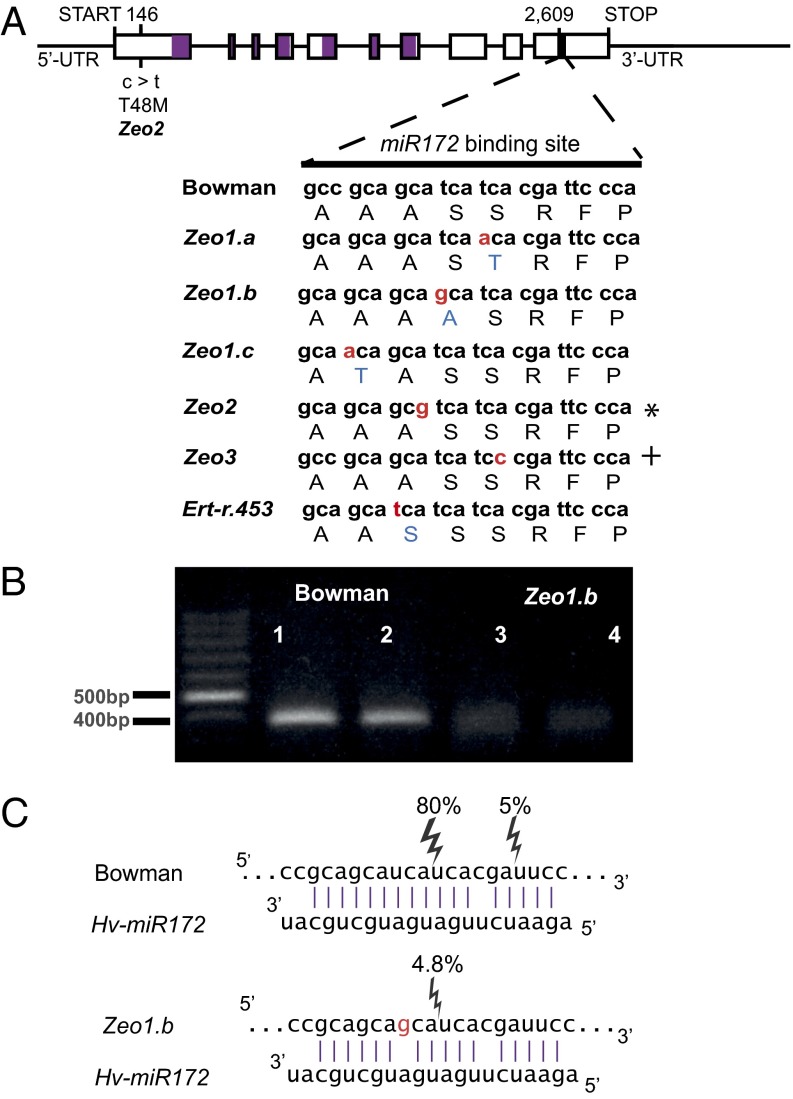
Because some of these dense-spike lines were natural variants, we explored whether these alleles also were represented in the GWAS population. We sequenced MLOC_43830 in 20 lines with the greatest spike density (spike density > 3.3 rachis nodes/cm) and 26 randomly chosen lines. Aligning all sequences (including mutants) revealed a total of 33 SNPs that resolved into 16 haplotype groups, eight of which were phenotypically categorized as containing weak (haplotypes 2, 6, 7, 11, 15, and 16) or strong (haplotypes 1 and 4) Zeo alleles (Dataset S1). We used diagnostic SNPs in MLOC_43830 to refine the genetic position of Zeo1.b in biparental populations (Fig. S1C) and observed complete linkage with the phenotype. Together these data are consistent with MLOC_43830, a transcription factor containing two AP2 DNA-binding domains and an miR172-binding site in its last (10th) exon (Fig. 2A), being ZEOCRITON. We subsequently refer to the gene as HvAP2.
Fig. 2.
Molecular characterization of dense-spike HvAP2 alleles. (A) Genomic structure of the HvAP2 gene and polymorphisms of HvAP2 alleles. The 5′ and 3′ UTRs, exons (black rectangles), and introns (black lines connecting the exons) are illustrated. The positions of mutations in the genomic DNA sequence and the predicted effect on HvAP2 amino acid sequence are indicated. The sequences encoding conserved APETALA2 (AP2) domains are indicated within the exons by purple and are located between nucleotides 334–1006 and 1157–1687. A black rectangle shows the location of a putative miRNA-binding site. SNPs identified by Nair et al. (43) are indicated by an asterisk for cly1.b and by a plus sign for cly1.c. (B) Gel image of amplified miR172-directed cleavage products from two independent amplifications of cv. WT and the Zeo1.b mutant using modified 5′ RACE. WT yielded a clear product of the predicted size of the miR172-derived cleavage product (381 bp), whereas Zeo1.b shows hardly any product. (C) Predicted base-pairing between WT and Zeo1.b HvAP2 mRNAs and Hv-miR172. Cloning and sequencing amplicons generated in B allowed identification of cleavage sites. Percentages of each cleavage fragment compared with total sequenced are indicated. Eighty-five per cent of WT clones showed cleavage of HvAP2 mRNA between bases 10 and 11 of the Hv-miR172 sequence, whereas the Zeo1.b allele showed only 4.8% cleavage at this site. Nucleotide positions of HvAP2 are relative to the start codon.
All eight HvAP2 haplotypes associated with increased spike density contained mutations within the miR172-binding site, suggesting that these polymorphisms may have a role in determining the spike density phenotype (Fig. 2A and Dataset S1). Four haplotypes (1, 2, 4, and 11) contained different nonsynonymous amino acid changes in the miR172-binding site: Zeo1.a (BW937), Zeo1.b (BW938), the cv. Optic mutant (renamed Zeo1.c), and Ert-r.453. The Zeo1.a allele also was detected in the ERECTOIDES line Ert-r.329, whereas Zeo1.b was shared with Ert-r.67, Ert-r.52 (BW322), and Oregon Wolfe Barley dominant. Two HvAP2 haplotypes (15 and 16), previously designated Zeo2.ax and Zeo2.c and shared by 11 dense-spike lines in the GWAS population, are defined by a nonsynonymous mutation in the first exon of HvAP2 and a synonymous mutation within the miR172-binding site. We collectively refer to these haplotypes as Zeo2. The final haplotype groups (6 and 7), which we term Zeo3, contain a synonymous mutation in the miR172-binding site. This mutation also was present in lines previously designated dense spike.av (dsp.av), Zeo, Zeo3, Pyramidatum1 (Pyr1), and Pyr3 as well as in four dense-spike lines in the GWAS population (Dataset S1). We then used Zeo2- and Zeo3-specific SNPs to genotype the GWAS population. Reanalyzing spike density, we observed that the Zeo2 allele induced larger effects on spike density (+0.44 rachis nodes/cm) than did Zeo3 (+0.33 rachis nodes/cm) (Dataset S1). GWAS conducted with the Zeo2 and Zeo3 lines removed from the population revealed minor unlinked but significant effects (0.15 rachis nodes/cm), suggesting that variations in genes other than ZEO also affect spike density, as observed previously (12, 20).
Reduced miRNA-Guided Cleavage Results in Heterochrony and Shorter Rachis Internodes.
Because base-pairing between an miRNA and its target mRNA is crucial for miRNA-directed cleavage (21), we predicted that mismatches in Zeo alleles of HvAP2 would lower miR172-binding affinity and decrease cleavage. Modified 5′-RACE on spike RNA revealed that canonical cleavage of HvAP2 transcripts at Hv-miR172 nucleotide position 10/11 was detected readily in WT but was almost eliminated in Zeo1.b, which effectively represents a miRNA-resistant HvAP2 allele (Fig. 3 B and C and Dataset S1). Furthermore, the semidominant nature of Zeo1.b suggests that spike density reflects a direct read-out of the extent of HvAP2 transcript cleavage. Zeo1.a and Zeo1.b alleles are phenotypically more severe than Zeo2 or Zeo3. Because the latter contain only synonymous mutations in the miR172 site, this additional severity may derive from the nonsynonymous amino acid changes in Zeo1.a and Zeo1.b.
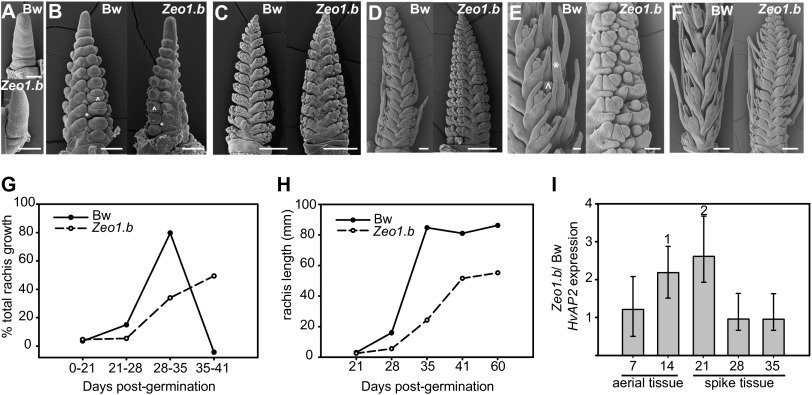
Fig. 3.
Spike development and HvAP2 gene expression in WT and Zeo1.b (BW938). (A–F) Scanning electron micrographs of early spike development in WT (Bw) and Zeo1.b. (A) Elongated vegetative domes form on top of Bw and Zeo1.b shoot apices at 12 dpg. (B) Reproductive apex, at 18 dpg with separated central and lateral spikelet primordia. Central spikelets show the developing outer glumes (*), and lemma initials ( ) are visible surrounding the central floret. (C) Spikelet primordia enlarge, and floral organ initials begin to form at 22 dpg. (D) Bw spikes are clearly advanced compared with Zeo1.b. Rachis length has increased fivefold in Bw but only 1.5-fold in Zeo1.b (Dataset S1). (E) Closer inspection shows that Bw spikelets exhibit awn initiation (*) and floral organ differentiation (
) are visible surrounding the central floret. (C) Spikelet primordia enlarge, and floral organ initials begin to form at 22 dpg. (D) Bw spikes are clearly advanced compared with Zeo1.b. Rachis length has increased fivefold in Bw but only 1.5-fold in Zeo1.b (Dataset S1). (E) Closer inspection shows that Bw spikelets exhibit awn initiation (*) and floral organ differentiation ( ) at 26 dpg; these features are absent in Zeo1.b spikelets, which more closely resemble Bw spikes at 22 dpg. (F) By 31 dpg, awn initials finally have developed on Zeo1.b spikes along with differentiating floral organs. Zeo1.b spikes also elongate but fail to catch up with Bw spikes, which are rapidly approaching maturity at 31 dpg. (G) Growth kinetics of rachis elongation in Bw and Zeo1.b plotted as the percentage of total growth between each time interval. Bw shows clear a growth spurt at 35 dpg, whereas Zeo1.b shows a later and muted growth peak (n = 7–16). (H) Rachis length over time in Bw and Zeo1.b showing the delay in rachis elongation at 21 dpg and a shorter final rachis length in the mature Zeo1.b rachis at 60 dpg (n = 7–16). (I) Bar graph of the ratio of HvAP2 mRNA levels in Zeo1.b/ Bw as detected by RT-qPCR on three or four biological replicates (in triplicate) and calculated by the Pairwise Fixed Reallocation Randomization Test performed by REST 2009 software (Qiagen). Samples harvested at 7 dpg (vegetative) and 14 dpg were from total seedlings. Samples at 21, 28, and 35 dpg consist of spike material only. Error bars represent 68% confidence intervals (one SE). Significant increases in levels of HvAP2 transcript were detected in Zeo1.b versus WT for dpg 14 plants and dpg 21 spikes (1,2P < 0.05). (Scale bars, 100 μm in A; 200 μm in B–E; 500 μm in F.)
) at 26 dpg; these features are absent in Zeo1.b spikelets, which more closely resemble Bw spikes at 22 dpg. (F) By 31 dpg, awn initials finally have developed on Zeo1.b spikes along with differentiating floral organs. Zeo1.b spikes also elongate but fail to catch up with Bw spikes, which are rapidly approaching maturity at 31 dpg. (G) Growth kinetics of rachis elongation in Bw and Zeo1.b plotted as the percentage of total growth between each time interval. Bw shows clear a growth spurt at 35 dpg, whereas Zeo1.b shows a later and muted growth peak (n = 7–16). (H) Rachis length over time in Bw and Zeo1.b showing the delay in rachis elongation at 21 dpg and a shorter final rachis length in the mature Zeo1.b rachis at 60 dpg (n = 7–16). (I) Bar graph of the ratio of HvAP2 mRNA levels in Zeo1.b/ Bw as detected by RT-qPCR on three or four biological replicates (in triplicate) and calculated by the Pairwise Fixed Reallocation Randomization Test performed by REST 2009 software (Qiagen). Samples harvested at 7 dpg (vegetative) and 14 dpg were from total seedlings. Samples at 21, 28, and 35 dpg consist of spike material only. Error bars represent 68% confidence intervals (one SE). Significant increases in levels of HvAP2 transcript were detected in Zeo1.b versus WT for dpg 14 plants and dpg 21 spikes (1,2P < 0.05). (Scale bars, 100 μm in A; 200 μm in B–E; 500 μm in F.)
Across species, the miR172 gene family encodes a set of highly conserved miRNAs which antagonize AP2 function, a key regulatory event that promotes the vegetative-to-reproductive transition and regulates floral patterning (22). To investigate the role that miR172 regulation of HvAP2 plays in controlling spike density, we conducted a comparative fine-scale morphological examination of spike development paired with quantitative measurements of the abundance of the HvAP2 transcript (Fig. 3). Early spike development appeared similar in the two genotypes (Fig. 3 A and B and Dataset S1). Between 22 and 26 d postgermination (dpg), WT spikelets began to differentiate as the rachis elongated by fivefold; however, at the same time Zeo1.b spikes extended only 1.5-fold (Fig. 3 C and D). Moreover, hallmark morphological events observed in the WT, such as the differentiation of awn initials and anthers, were missing in Zeo1.b spikelets (Fig. 3E), which instead resembled earlier growth stages. These results suggest that the switch to spikelet differentiation is stalled in Zeo1.b and that the duration of spikelet initiation is prolonged, consistent with a significant increase in spikelet node numbers on Zeo1.b spikes (P < 0.0001) (Dataset S1). Awn initials eventually form in Zeo1.b but only about 5 d later, when spikes have grown to the length at which awn initiation occurs in WT (6.47 ± 0.05 mm) (Fig. 3F and Dataset S1); this delay suggests that differentiation events are linked to spike size. Synchronicity between spikelet differentiation and internode elongation in Zeo1.b, previously observed across barley (23), indicates a level of genetic interdependency between these traits during maturation. Over a longer term, Zeo1.b spike elongation continues along a slower, shallower rate. Plotting rachis growth as a proportion of final length resolves a single steep peak or growth spurt in WT and a shorter, rounded curve peaking 1 wk later in Zeo1.b (Fig. 3G), reflecting delayed and muted growth to a final length of 46.4 ± 0.49 mm, 65% of the final length of WT (Fig. 3H and Dataset S1).
We tested whether intact HvAP2 mRNA levels were altered because of miR172 resistance in Zeo1.b and if this resistance correlated with observed differences in spike development. Elevated HvAP2 mRNA abundance per total RNA was detected throughout the early phases of Zeo1.b reproductive development, corresponding precisely with delayed maturation; however, HvAP2 transcripts ultimately decline to levels comparable with WT as differences in spike growth become manifest (Fig. 3I). These data suggest that miR172 acts early to limit HvAP2 function but does not restrict HvAP2 later in spike development, hinting that the reduced rachis internode elongation observed in mature Zeo1.b spikes is a consequence of early developmental events.
Identification of Other AP2 Family Members in Barley and Wheat.
Reduced internode growth and corresponding increases in spike density are associated with yield increases in rice (24) and in hexaploid wheat (3). In the latter, the gene responsible, Q, also encodes an AP2-like transcription factor. To examine possible phenotypic relationships within this family, we analyzed AP2-like proteins across grasses using a phylogenetic approach (Fig. S2). In barley the HvAP2 gene family consists of four members (17), three of which can be assigned positions on chromosomes 1HS (HvAP2-1L), 7HL (HvAP2-2L), and 2HL (HvAP2) (Fig. S2) (17). HvAP2-Q is not yet assigned, but Q in wheat is located on chromosome 5AL (25). We were unable to identify a wheat homeologue for each barley gene but did identify a wheat cDNA clone, AK331198.1, that showed 79.7% similarity at the protein level with ZEO1. AK331198.1 is homologous to the genomic sequence from three wheat homeologues, TaAP2-A, TaAP2-B, and TaAP2-D, and is a candidate ortholog of HvAP2 (Fig. S2). In wheat, two loci associated with spike density, COMPACTUM (C) and SOFT OUTER GLUME (SOG), map to the group 2 chromosomes (26). We explored whether mutations in the miR172-binding site of TaAP2 were responsible for the COMPACTUM phenotype. We designed genome-specific primers (27) which enabled us to amplify and sequence all three TaAP2 homeologues from 10 dense-spike wheat accessions (Dataset S1) that contain a dominant COMPACTUM allele. Compared with Chinese Spring, a wheat cultivar that does not have a dense spike, no mutations were observed in any of the homeologues, consistent with previous observations that C is not orthologous to ZEO (26).
Discussion
Our analysis highlighted two major aspects of barley spike development regulated by HvAP2. First, HvAP2 influences the kinetics and extent of rachis growth and delays spikelet maturation at the awn initiation stage. miR172-directed cleavage is lost in Zeo1.b, and this loss appears to correspond with elevated levels of HvAP2 mRNA at the initiation of the transition from juvenile to mature spikelet. These data suggest that miR172 acts early in spike development to limit the abundance of the HvAP2 transcript, corresponding to the observed maturation delay in Zeo1.b, but does not restrict HvAP2 mRNA levels at later time points when differences in rachis elongation in Zeo1.b are manifest. One caveat to this interpretation is that miR172 regulates HvAP2 function primarily via transcript stability, as shown for AP2-like targets in multiple species (28–30). However, miR172 also controls AP2 function at the translational level in Arabidopsis and maize (31–34), suggesting that the mode of miR172 action may depend on the target, tissue, or developmental stage. As in other AP2-like genes, HvAP2 was strongly expressed in juvenile tissue (Fig. 3), agreeing with reports that miR172 expression levels are low early in development and increase toward the reproductive transition (28, 35). Although not as highly expressed, HvAP2 was detected early in spike growth and showed increasing levels in 1.5- to 2-wk-old and 2- to 3-wk-old plants (inflorescence, 5 mm long and 10–12 mm long, respectively) (17).
The antagonism between miR172 and AP2-like genes is deeply conserved across plants (36) and plays pivotal roles in developmental timing. AP2-like targets of miR172 in grasses include INDETERMINATE SPIKELET1 (ids1) and SISTER OF INDETERMINATE SPIKELET 1 (sid1) in maize and SUPERNUMERARY BRACT (SNB) in rice, which control the transition when spikelet meristems commit to floral meristem fate. In general, increasing levels of miR172 promote the development of adult features, acting in opposition to miR156, which is abundant early in the lifecycle and acts to promote juvenile characteristics such as shoot and tiller production (37). Overexpression of miR172 in Arabidopsis, maize, or rice leads to artificially low AP2-like levels, shortened juvenile phases, and precocious reproduction (30). Extending the time before this switch leads to indeterminacy and spikelet branching (29, 38–40). Therefore, we propose that miR172 regulation of HvAP2 in barley controls the length of a critical developmental window early in spike development during the switch from spikelet formation to awn initiation and internode elongation.
Floral organ differentiation and internode elongation occur concurrently in barley, and this link is influenced by neither environmental conditions nor genotype (23, 41), suggesting a highly canalized, genetically buffered transition that may be regulated by miR172 fine-tuning of HvAP2. Although the roles of HvAP2 in developmental timing and internode elongation may be mechanistically separate, our data indicate that this developmental stage is extended in plants containing a Zeo allele, and the resulting excessive HvAP2 mRNA represses rachis internode growth and increases spike density. Variation in spike density therefore could reflect differences in both the timing of the transition and the rate of internode growth, examples of classic heterochronic mechanisms (42).
We conclude that the interaction between miR172 and HvAP2 is a central regulator of spike density in widely grown barley cultivars and induced mutants. Interestingly, Nair et al. (43) previously reported two HvAP2 alleles with SNPs in the miR172-binding sequence: cleistogamous1.b (cly1.b) containing the Zeo2 SNP, and cly1.c containing the Zeo3 SNP. cly1.b alleles were identified in North American and European two-row accessions, and cly1.c alleles were identified in Spanish, Italian, and Finnish six-row types, suggesting that Zeo is neither two-row nor United Kingdom specific. In addition to an association with denser spikes, cultivars with cly1.b or cly1.c showed a failure of lodicules to enlarge at anthesis, leading to florets remaining closed at flowering (i.e., cleistogamy) (43). Screening all Zeo HvAP2 mutants also revealed a failure of lodicule expansion Fig. S3), suggesting that HvAP2 may control both phenotypes pleiotropically. Because spike development is strikingly similar across the Triticeae, it is of interest to explore the morphological and developmental outcomes of interactions between HvAP2 orthologs and miR172 in related species. Homologs of HvAP2 are known in rice and maize, and recently a putative ortholog in wheat, TaAP2 (Fig S2), has been shown to be cleaved by miR172 (27). A dominant compact spike locus, COMPACTUM, is located on wheat chromosome 2D, but previous studies have indicated that this locus is not orthologous to ZEO (26). In agreement, we failed to identify mutations in the miR172-binding site from a range of COMPACTUM wheat cultivars. In barley, ZEO alleles induce a delay in the switch from spikelet initiation to differentiation and internode elongation, eventually reflected in reduced rachis length and high spike density. Our data indicate that miR172 regulation of HvAP2 is critical during a specific developmental window that, when perturbed, induces heterochronic variation that strikingly alters barley inflorescence morphology.
Methods
Plant Material.
All plant materials used in this study are identified in Dataset S1. Barley accessions originated from stocks at The James Hutton Institute, except for 36Ab51 which was provided by Harold Bockelman from the National Small Grains Collection, Aberdeen, ID (www.ars.usda.gov/pwa/aberdeen). GPMx was developed from a cross between cv. Golden Promise (dense spike) and cv. Morex (normal spike) and was advanced to F10 by single-seed descent. Two F2 populations were generated by crossing BW938 (Zeo1.b) to the barley cultivars Morex and Barke. Each population revealed 1:2:1 segregation [BW938 × Barke, (χ2 = 2, n = 182) = 4.17 (P < 0.05) and BW938 × Morex (χ2 = 2, n = 145) = 3.41 (P < 0.05)]. Wheat accessions were provided by Mike Ambrose from the Biotechnology and Biological Sciences Research Council Small Grain Cereal Collection, Swindon, United Kingdom.
GWAS of RL, RNN. and Spike Density.
We selected 401 two-rowed United Kingdom spring barley accessions to create a diverse germplasm stock with minimal population stratification. These accessions were planted in a Latinized row and column design with five replicates. The tallest three spikes per plant were photographed at harvest. Grain number and rachis length per spike were recorded using ImageJ (44). To calculate spike density, the rachis RNNs of three spikes per plant were divided by the RL (Fig. 1A), and genotypic means were calculated using the Restricted Maximum Likelihood directive in Genstat v.14. In the model, rows and columns within replications and test entries were fitted as random effects. We considered 5,251 SNP markers with less than 10% missing data and minimum allele frequency >5% for GWAS analyses. Within a mixed linear model framework, the Eigenstrat relationship model with principal component analysis scores as random terms was used for GWAS as implemented in Genstat v.14 (VSN International).
KASPar Marker Mapping.
Twenty KASP By Design (KBioscience) assays (Dataset S1) were developed using markers surrounding Zeo1 (12) and were used to map the phenotype in BW938 × Morex (145 F2 lines) and BW938 × Barke (182 F2 lines). Reactions were carried out using an 8-µL reaction mix containing 20 ng DNA, 2× KASPar v 4.0 Reagent (KBS-1016) and 0.11 µL KASP By Design Nonvalidated SNP assay (KBS-1013). PCR was performed on StepOnePlus using the following program: pre-PCR read for 2 min at 20 °C; 15 min at 94 °C min; 10 cycles (94 °C for 20 s; 62 °C for 1 min, decreasing by 0.7 °C per cycle); 32 cycles (94 °C for 20 s, 55 °C for 1 min); post-PCR read for 2 min at 20 °C. The analysis performed using default parameters on the StepOnePlus AntMap version 1.2 (45) was used for all linkage analysis. Default settings were used, except that the locus-ordering criterion was changed to minimize the sum of adjacent recombination fractions (SARF).
Resequencing and Mapping of HvAP2.
MLOC_81350 and MLOC_43830 were sequenced in the lines detailed in Dataset S1 exactly as described in Houston et al. (8). Sequences were aligned using Geneious version 6.1.2 (Biomatters). Details of sequences, polymorphisms and KASP primer sequences are given in Dataset S1. To increase the resolution of the association-mapping peak, KASP By Design assays were designed to specific SNPs identified in Zeo2 and Zeo3 and were used to genotype all 401 lines. The COMPACTUM wheat accessions and genome-specific primer sequences used for PCR sequencing are listed in Dataset S1.
miRNA Cleavage Analysis Using Modified 5′ RACE.
Actively elongating spikes were harvested from WT and Zeo1.b at 60% of rachis final length (5.5 cm in WT and 3.0 cm in Zeo1.b). Tissue was collected, RNA was extracted, and integrity was confirmed using a Bioanalyzer 2100 (Agilent Technologies). Cleavage analysis of the HvAP2 transcript was determined using the GeneRacer kit (Invitrogen) according to the manufacturer’s instructions except that the phosphatase and decapping steps were eliminated before ligation of the 5′ RNA oligo adaptor to select for cleaved transcripts. cDNA synthesis was carried out by SuperScript III using oligo dT and random hexamer primers. PCR and Nested PCR on cDNA with a 5′ adaptor-specific and an HvAP2-specific 3′ UTR reverse primer amplified HvAP2 cleavage products. Amplicons were sliced from the gel; in the case of Zeo1.b, where no discrete band was present, a gel slice of the analogous size range was taken. Samples were purified and cloned into the pCR4-TOPO vector. Inserts from 20 independent clones from WT and 21 independent clones from Zeo1.b were sequenced.
Quantitative RT-PCR.
Tissues were harvested at the same time of day from plants by immediate freezing in liquid nitrogen. Three or four biological replicates per developmental time point and per genotype were harvested. Details of RNA extraction, integrity determination, and cDNA synthesis are described in SI Text. Quantitative PCR (qPCr) was performed using TaqMan chemistry on the Applied Biosystems StepOnePlus machine. Intron-spanning primers amplified HvAP2 cDNA amplicons of 88 bp between the eighth and ninth exon (upstream from the miRNA172 cleavage site in exon 10). Three stably expressed genes—26S, HvACTIN2, and HvPPA2—were examined as reference genes for each sample. All primers are listed in Dataset S1. Detailed descriptions of qPCR reaction set-ups, run conditions, probe and primer efficiencies, and reference gene calibration are given in SI Text.
Scanning Electron Microscopy.
Spikes were harvested from WT and Zeo1.b plants at 12, 18, 22, 26, 31, and 36 dpg. Tissue was fixed and processed for scanning electron microscopy as described previously (8).
Analysis of the Barley AP2 Transcription Factor Family and Its Homologs.
We identified other members of the barley AP2 family by conducting a BLASTn search against published full-length cDNA datasets for barley (17) and identifying the corresponding amino acid sequence in the National Center for Biotechnology Information (NCBI) database. To identify homologs of these AP2 transcription factors in species closely related to barley, we used the NCBI database to carry out a BLASTx search. The only exception was wheat, for which we also subsequently queried the Unité de Recherche Génomique Info sequence repository. MEGA version 5 (46) was used to perform protein sequence alignments with ClustalW, and the neighbor-joining method was used to construct a phylogeny.
Supplementary Material
Acknowledgments
This work was supported by funding from the Scottish Government Rural and Environment Science and Analytical Services Division Programme 5, European Research Area in Plant Genomics Grant ERAPGFP/06.046A, and Biotechnology and Biological Sciences Research Council Sustainable Bioenergy Centre Programme SG Grant 0921. S.M.M. was supported by a Royal Society of Edinburgh Personal Research Fellowship and the College of Life Sciences, University of Dundee. N.C.C. was funded by the Australian Government Grains Research and Development Corporation, the Australian Research Council, and the government of South Australia. M.H. and C.D. received support from the Carlsberg Foundation and Deutsche Forschungsgemeinschaft Research Fellowship DO1482. We also acknowledge the support of the Scottish Food Security Alliance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.L. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database, www.ncbi.nlm.nih.gov/genbank/ (accession nos. KC898651–KC898818).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311681110/-/DCSupplemental.
References
- 1. Crop Prospects and Food Situation (FAO) No 2 July 2013 pp 1–34 (available for download at www.fao.org/docrep/018/aq114e/aq114e.pdf)
- 2.Sreenivasulu N, Schnurbusch T. A genetic playground for enhancing grain number in cereals. Trends Plant Sci. 2012;17(2):91–101. doi: 10.1016/j.tplants.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka W, Pautler M, Jackson D, Hirano HY. Grass meristems II: Inflorescence architecture, flower development and meristem fate. Plant Cell Physiol. 2013;54(3):313–324. doi: 10.1093/pcp/pct016. [DOI] [PubMed] [Google Scholar]
- 4.Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ. The control of spikelet meristem identity by the branched silkless1 gene in maize. Science. 2002;298(5596):1238–1241. doi: 10.1126/science.1076920. [DOI] [PubMed] [Google Scholar]
- 5.Vollbrecht E, Springer PS, Goh L, Buckler ES, 4th, Martienssen R. Architecture of floral branch systems in maize and related grasses. Nature. 2005;436(7054):1119–1126. doi: 10.1038/nature03892. [DOI] [PubMed] [Google Scholar]
- 6.Simons KJ, et al. Molecular characterization of the major wheat domestication gene Q. Genetics. 2006;172(1):547–555. doi: 10.1534/genetics.105.044727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller KJ, et al. The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature. 1995;374(6524):727–730. doi: 10.1038/374727a0. [DOI] [PubMed] [Google Scholar]
- 8.Houston K, et al. Analysis of the barley bract suppression gene Trd1. Theor Appl Genet. 2012;125(1):33–45. doi: 10.1007/s00122-012-1814-x. [DOI] [PubMed] [Google Scholar]
- 9.Komatsuda T, et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA. 2007;104(4):1424–1429. doi: 10.1073/pnas.0608580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsay L, et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat Genet. 2011;43(2):169–172. doi: 10.1038/ng.745. [DOI] [PubMed] [Google Scholar]
- 11.Franckowiak JD, Lundqvist U. Descriptions of barley genetic stocks for 2011. Barley Genet Newsl. 2011;41:12–53. [Google Scholar]
- 12.Shahinnia F, et al. High resolution mapping of Dense spike-ar (dsp.ar) to the genetic centromere of barley chromosome 7H. Theor Appl Genet. 2012;124(2):373–384. doi: 10.1007/s00122-011-1712-7. [DOI] [PubMed] [Google Scholar]
- 13.Druka A, et al. Genetic dissection of barley morphology and development. Plant Physiol. 2011;155(2):617–627. doi: 10.1104/pp.110.166249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comadran J, et al. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet. 2012;44(12):1388–1392. doi: 10.1038/ng.2447. [DOI] [PubMed] [Google Scholar]
- 15.Turuspekov Y, et al. Identification and mapping of a QTL for rachis internode length associated with cleistogamy in barley. Plant Breed. 2005;124(6):542–545. [Google Scholar]
- 16.Chen A, Baumann U, Fincher GB, Collins NC. Flt-2L, a locus in barley controlling flowering time, spike density, and plant height. Funct Integr Genomics. 2009;9(2):243–254. doi: 10.1007/s10142-009-0114-2. [DOI] [PubMed] [Google Scholar]
- 17.Mayer KF, et al. International Barley Genome Sequencing Consortium A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491(7426):711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 18.Qi W, et al. Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 2011;157(1):216–228. doi: 10.1104/pp.111.179945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang F, et al. Mutations in an AP2 transcription factor-like gene affect internode length and leaf shape in maize. PLoS ONE. 2012;7(5):e37040. doi: 10.1371/journal.pone.0037040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borje Larsson HE. Genetic analysis of laxatum barley mutants. Heriditas. 1985;103:255–267. [Google Scholar]
- 21.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Q-H, Helliwell CA. Regulation of flowering time and floral patterning by miR172. J Exp Bot. 2011;62(2):487–495. doi: 10.1093/jxb/erq295. [DOI] [PubMed] [Google Scholar]
- 23.Nicholls PB, May LH. Studies on the growth of the barley apex. II. On the initiation of internode elongation in the inflorescence. Aust J Biol Sci. 1964;17:619–630. [Google Scholar]
- 24.Huang X, et al. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet. 2009;41(4):494–497. doi: 10.1038/ng.352. [DOI] [PubMed] [Google Scholar]
- 25.Faris JD, Fellers JP, Brooks SA, Gill BS. A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics. 2003;164(1):311–321. doi: 10.1093/genetics/164.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson EB, Nalam VJ, Zemetra RS, Riera-Lizarazu O. Mapping the compactum locus in wheat (Triticum aestivum L.) and its relationship to other spike morphology genes of the Triticeae. Euphytica. 2008;163:193–201. [Google Scholar]
- 27.Ning S, et al. Structure, transcription and post-transcriptional regulation of the bread wheat orthologs of the barley cleistogamy gene Cly1. Theor Appl Genet. 2013;126(5):1273–1283. doi: 10.1007/s00122-013-2052-6. [DOI] [PubMed] [Google Scholar]
- 28.Jung J-H, et al. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007;19(9):2736–2748. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chuck G, Meeley R, Hake S. Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development. 2008;135(18):3013–3019. doi: 10.1242/dev.024273. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Q-H, Upadhyaya NM, Gubler F, Helliwell CA. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa) BMC Plant Biol. 2009;9:149. doi: 10.1186/1471-2229-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15(11):2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303(5666):2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA. 2005;102(26):9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwab R, et al. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8(4):517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Wu G, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138(4):750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 37.Poethig RS. Small RNAs and developmental timing in plants. Curr Opin Genet Dev. 2009;19(4):374–378. doi: 10.1016/j.gde.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuck G, Meeley RB, Hake S. The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 1998;12(8):1145–1154. doi: 10.1101/gad.12.8.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee DY, Lee J, Moon S, Park SY, An G. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 2007;49(1):64–78. doi: 10.1111/j.1365-313X.2006.02941.x. [DOI] [PubMed] [Google Scholar]
- 40.Lee DY, An G. Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 2012;69(3):445–461. doi: 10.1111/j.1365-313X.2011.04804.x. [DOI] [PubMed] [Google Scholar]
- 41.Paleg LG, Aspinall D. Effects of Daylength and Light Intensity on Growth of Barley II. Influence of Incandescent Light on Apical Development. Bot Gaz. 1964;125(3):149–155. [Google Scholar]
- 42.Smith KK. Time’s arrow: Heterochrony and the evolution of development. Int J Dev Biol. 2003;47(7-8):613–621. [PubMed] [Google Scholar]
- 43.Nair SK, et al. Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc Natl Acad Sci USA. 2010;107(1):490–495. doi: 10.1073/pnas.0909097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwata H, Ninomiya S. AntMap: Constructing genetic linkage maps using an ant colony optimization algorithm. Breed Sci. 2006;56(4):371–377. [Google Scholar]
- 46.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.