Significance
Chronic exposure to adverse social environments is associated with increased risk of disease, and stress-related increases in the expression of proinflammatory genes appear to contribute to these effects. The present study identifies a biological mechanism of such effects in the ability of the sympathetic nervous system to up-regulate bone marrow production of immature, proinflammatory monocytes. These effects are mediated by β-adrenergic receptors and the myelopoietic growth factor GM-CSF, and suggest new targets for interventions to protect health in the context of chronic social stress.
Keywords: social genomics, immunology
Abstract
Across a variety of adverse life circumstances, such as social isolation and low socioeconomic status, mammalian immune cells have been found to show a conserved transcriptional response to adversity (CTRA) involving increased expression of proinflammatory genes. The present study examines whether such effects might stem in part from the selective up-regulation of a subpopulation of immature proinflammatory monocytes (Ly-6chigh in mice, CD16− in humans) within the circulating leukocyte pool. Transcriptome representation analyses showed relative expansion of the immature proinflammatory monocyte transcriptome in peripheral blood mononuclear cells from people subject to chronic social stress (low socioeconomic status) and mice subject to repeated social defeat. Cellular dissection of the mouse peripheral blood mononuclear cell transcriptome confirmed these results, and promoter-based bioinformatic analyses indicated increased activity of transcription factors involved in early myeloid lineage differentiation and proinflammatory effector function (PU.1, NF-κB, EGR1, MZF1, NRF2). Analysis of bone marrow hematopoiesis confirmed increased myelopoietic output of Ly-6chigh monocytes and Ly-6cintermediate granulocytes in mice subject to repeated social defeat, and these effects were blocked by pharmacologic antagonists of β-adrenoreceptors and the myelopoietic growth factor GM-CSF. These results suggest that sympathetic nervous system-induced up-regulation of myelopoiesis mediates the proinflammatory component of the leukocyte CTRA dynamic and may contribute to the increased risk of inflammation-related disease associated with adverse social conditions.
Across a variety of adverse life circumstances, such as social isolation (1, 2), imminent bereavement (3), low socioeconomic status (SES) (4, 5), early life social deprivation (6), late-life social adversity (7), traumatic stress (8), diagnosis with a life-threatening illness (9), and experimentally imposed social stress (7, 10–13), the pool of circulating leukocytes has been found to show a conserved transcriptional response to adversity (CTRA) involving increased expression of genes involved in inflammation (e.g., IL1B, IL6, IL8, TNF) and decreased expression of genes involved in innate antiviral responses (IFNB, IFIs, MX, OAS) (13–17).†Bioinformatic analyses indicate that the proinflammatory component of the leukocyte CTRA originates predominately from monocytes (2, 9, 13). The biological mechanism of this monocyte-related proinflammatory transcriptional skew is unknown, but is important to define because chronic inflammation contributes to many of the diseases that drive contemporary morbidity and mortality (18) and disproportionately impact socially disadvantaged groups (e.g., those of low SES) (19, 20).
Adverse social conditions may up-regulate inflammatory gene expression via stress/threat-related neural and endocrine signals that induce per-cell changes in gene transcription (7) or compartmental redeployment of a fixed population of monocytes (21, 22). Neural and endocrine dynamics could also conceivably expand the total body pool of monocytes via increased hematopoietic output of myeloid lineage immune cells (myelopoiesis) (23). Hematopoiesis and monocyte differentiation occur in a bone marrow microenvironment that receives direct innervation from the sympathetic nervous system (SNS) (24), and SNS activation can alter hematopoietic stem cell mobilization (25–27) and myelopoietic responses to systemic infection (28, 29). It is not known whether social processes can influence monocyte differentiation, or what role the SNS might play in such dynamics.
A proinflammatory dynamic similar to the human leukocyte CTRA has been observed in monocytes from mice exposed to repeated social defeat (RSD; i.e., repeated territorial intrusion by an aggressive conspecific) (30). In addition to enhancing general inflammatory responses (31–36), and antigen-presenting cell (APC) stimulation of adaptive immune responses (34, 37, 38), RSD can functionally desensitize the monocyte glucocorticoid receptor (GR) (39–43). In the present studies, we used genomics-based strategies to dissect the cellular mechanisms of the leukocyte CTRA in observational studies of human social adversity and the RSD experimental mouse model to determine whether chronic social stress might induce a proinflammatory skew in the circulating leukocyte transcriptome through selective expansion of a specific subpopulation of immature, proinflammatory monocytes (Ly-6chigh in mice, CD16− in humans) (23, 44–46). Low SES was selected as a model of human social adversity due to its chronic and pervasive effects, as well its previously documented association with elevated SNS activity (47–49) and up-regulated proinflammatory gene expression (6).
Results
Mouse Social Stress and Monocyte Transcriptome Representation.
To determine whether chronic social stress might increase representation of the monocyte transcriptome within the circulating leukocyte pool, we assessed the effects of six daily cycles of 2-h RSD (32–34) on the prevalence and transcriptome differentiation state of mouse CD11b+ monocytes. Relative to home cage control (HCC) conditions, RSD induced an average fourfold increase in the frequency of monocytes in circulating blood [HCC: 2.2 ± 0.4% of circulating peripheral blood mononuclear cells (PBMCs); RSD: 9.8 ± 1.3%; difference, P < 0.001] and spleen (HCC: 6.0 ± 0.6%; RSD: 22.7 ± 1.8%; difference, P = 0.018), and a twofold increase in bone marrow monocytes (HCC: 5.9 ± 0.7%; RSD: 14.0 ± 3.0%; difference, P < 0.001).
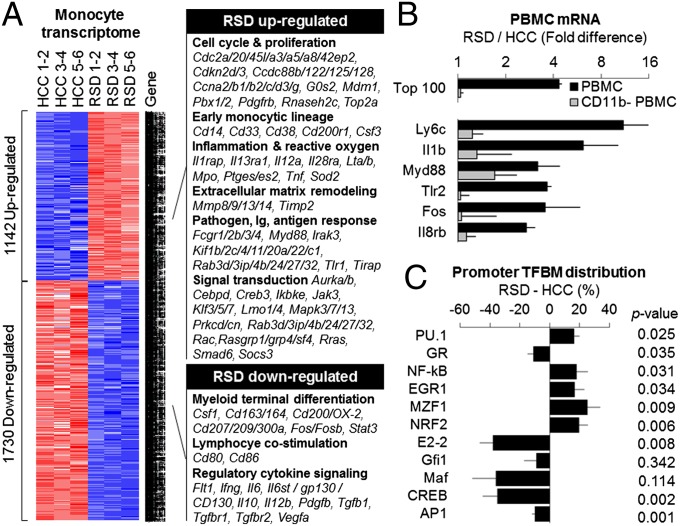
Genome-wide transcriptional profiling of isolated spleen monocytes identified 2,976 transcripts showing ≥50% difference in average expression level in RSD animals vs. HCC, including up-regulation of 1,142 transcripts and down-regulation of 1,730 transcripts (Fig. 1A and Dataset S1). Differentially expressed transcripts derived from 2,013 distinct named genes (6.6% of the mouse genome; 1,142 up-regulated, 871 down-regulated). As shown in Fig. 1A, RSD up-regulated genes involved in cell growth and differentiation (cell cycle progression, proliferation, apoptosis, growth-factor signaling, and immature monocytic lineage), myeloid lineage effector functions (cytokines and their receptors; prostaglandin synthases; reactive oxygen species-related molecules; pathogen pattern recognition receptors and associated signaling molecules; Ig receptors; and mediators of extracellular matrix remodeling), APC function (endocytosis and exocytosis, lipid metabolism, and cytoskeletal remodeling), and signal transduction and transcription control (particularly, MAP kinase signaling). RSD down-regulated genes involved in myeloid lineage terminal differentiation, lymphocyte costimulation, and signaling by regulatory cytokines and their receptors.
Fig. 1.
Differential gene expression in mouse CD11b+ monocytes and PBMCs. (A) Genome-wide transcriptional profiling of (unstimulated) adherent CD11b+ spleen monocytes harvested after 6 d of RSD or HCC conditions identified 2,976 transcripts showing ≥50% difference in mean abundance across groups (n = 6 animals per condition pooled into three groups of two for microarray assay; results representative of two independent experiments). Red, overexpressed in RSD; blue, overexpressed in HCC (underexpressed in RSD). (B) Effects of RSD on gene expression in total PBMCs (black bars) and monocyte-depleted PBMCs (CD11b−, gray bars). Data represent mean fold difference (±SE) in expression of 100 genes showing greatest RSD-induced up-regulation in total PBMCs (RSD transcriptome shift; Upper) or six representative proinflammatory genes (Lower); n = 3 animals per condition in one experiment representative of three independent experiments. (C) Promoter-based bioinformatic analysis of myeloid lineage transcription factors in genes up-regulated in RSD vs. HCC monocytes (A). Data represent mean fold difference (±SE) in prevalence of transcription factor binding motifs, averaged over nine parametric combinations of promoter length and motif detection stringency. P values, two-tailed difference from null difference of 0%.
To determine whether increased monocyte prevalence could account for the proinflammatory CTRA shift in the overall mouse PBMC transcriptome, we conducted genome-wide transcriptional profiling of total PBMCs and monocyte-depleted PBMC populations. RSD up-regulated proinflammatory gene expression in total PBMC (Fig. 1B; +343.8 ± 26.1% difference in average expression of six representative proinflammatory genes, P < 0.001). Both the general transcriptomic effects of RSD and its up-regulation of specific proinflammatory genes were largely abrogated by monocyte depletion (85.5 ± 10.7% reduction in top 100 RSD up-regulated genes, P < 0.001; 91.2 ± 26.1% reduction in six proinflammatory genes, P = 0.013). Monocyte depletion also abrogated bioinformatic indications of RSD-induced activation of the proinflammatory transcription factors NF-κB (total PBMC: +73.7 ± 19.6%, P = 0.015; monocyte-depleted PBMC: −31.9 ± 18.0%, P = 0.951) and AP-1 (total PBMC: +12.3 ± 4.3%, P = 0.025; monocyte-depleted PBMC: −7.0 ± 4.6%, P = 0.857).
Regulation of the Mouse Monocyte Transcriptome.
Promoter-based bioinformatic analysis of genes up-regulated by RSD indicated increased activity of the PU.1 transcription factor involved in early myeloid lineage commitment and decreased activity of transcription factors involved in terminal differentiation of myeloid cells to a mature macrophage fate (cMaf, MafB, CREB, AP-1) or dendritic cell fate (E2-2, Gfi1) (23, 50–52) (Fig. 1C). These results raised the possibility that increased myelopoietic production of immature monocytes might contribute to their relative up-regulation within the total PBMC transcriptome. Consistent with previous results from the RSD paradigm (41–43), promoter-based bioinformatics also indicated increased activity of transcription factors mediating inflammatory/antimicrobial effector functions (NF-κB, EGR1, MZF1, NRF2), and decreased activity of the GR (Fig. 1C). Reduced GR-mediated gene transcription was not attributable to reduced expression of the GR-encoding Nr3c1 gene, which showed an average 3% difference in expression across five Nr3c1 probe sets on the microarray; P = 0.457. This general pattern of results suggested that the monocyte pool itself may show selective expansion of the immature proinflammatory Ly-6chigh subset (23, 44–46).
Mouse Monocyte Subset Differentiation.
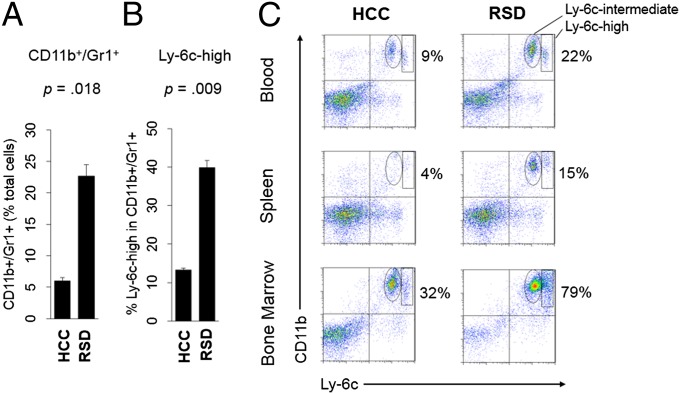
To directly assess whether RSD up-regulated the immature/proinflammatory monocyte transcriptome (45), we used a bioinformatic transcriptome representation analysis (TRA) to decompose the complex gene expression profile of the total splenic monocyte transcriptome into subcomponents originating from distinct Ly-6chigh and Ly-6clow monocyte subsets (Dataset S2). Results indicated an average 22.7% (±3.1%) expansion of the Ly-6chigh monocyte transcriptome in RSD animals vs. controls (P < 0.0001). Flow cytometry confirmed increased Ly-6chigh monocytes in the blood, spleen, and bone marrow (Fig. 2). RSD also increased Ly-6cintermediate cells (granulocytes) in each compartment (Fig. 2C), suggesting a potential effect on myeloid differentiation more broadly.
Fig. 2.
Monocyte subset prevalence in bone marrow and peripheral compartments. (A) Average prevalence (±SE) of CD11b+/Gr1+ monocytes within total spleen cells (n = 6 per condition, representative of three independent experiments). (B) Average prevalence (±SE) of Ly-6chigh cells within splenic CD11b+/Gr1+ monocyte pool (samples from A). (C) Flow cytometric quantification of Ly-6cintermediate and Ly-6chigh (horizontal axis) CD11b+ (vertical axis) mononuclear cells in spleen, blood, and bone marrow from RSD and HCC mice. Percentages represent total CD11b+/Ly-6c+ myeloid cells (monocytes + granulocytes).
Mouse Myeloid Lineage Differentiation.
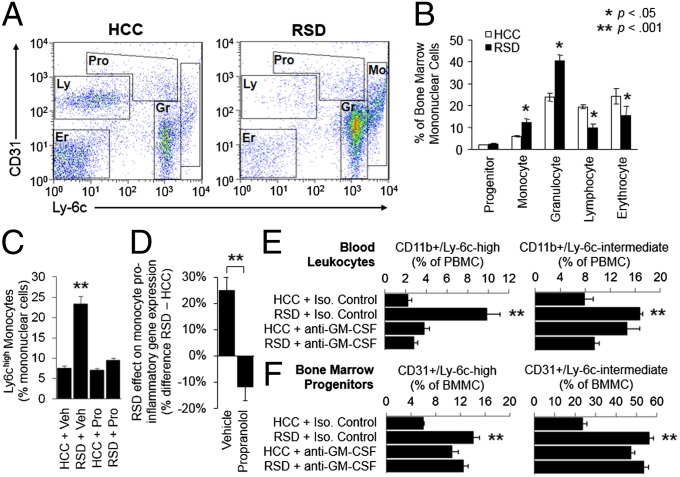
To more directly assess the role of myelopoiesis in RSD up-regulation of Ly-6c+ monocytes and granulocytes, we examined bone marrow prevalence of multilineage progenitor cells (CD31high/Ly-6chigh) and lineage-committed erythroid (CD31−/Ly-6c−), lymphoid (CD31+/Ly-6c−), granulocytic (CD31+/Ly-6c+), and monocytic progenitor cells (CD31+/Ly-6chigh; Fig. 3 A and B) (53, 54). RSD up-regulated both myeloid lineages, approximately doubling monocyte progenitors (P = 0.0074) and increasing granulocyte progenitors by 70% (P = 0.020). Multilineage progenitor-cell prevalence did not change significantly (P = 0.140), and erythroid and lymphoid progenitors both showed relative decreases (P = 0.019 and P = 0.002, respectively).
Fig. 3.
Role of lineage differentiation and β-adrenergic/GM-CSF signaling. (A) Flow cytometric assessment of mixed-lineage progenitors (Pro), and erythroid (Er), lymphoid (Ly), granulocytic (Gr), and monocytic (Mo) lineage cells in bone marrow. (B) Average prevalence (±SE) of each progenitor cell type (n = 3–7 per condition, representative of three independent experiments). *P < 0.05; **P < 0.001. (C) Effect of the β-adrenergic antagonist propranolol on RSD-induced expansion of splenic Ly-6chigh monocytes. Data represent mean prevalence (±SE) of Ly-6chigh/CD11b+ cells as a percent of total splenocytes (n = 9 per condition; Pro, propranolol; Veh, vehicle). (D) Effect of propranolol on RSD-induced up-regulation of 17 proinflammatory gene transcripts in peripheral blood monocytes (Cxcl1, Cxcl2, Fos, Fosb, Fosl1, Il1a, Il1b, Il6, Jun, Junb, Jund1, Ly6c, Myd88, Ptgs1, Ptgs2, Tlr2, Tnf). Data represent mean percent difference in gene expression (RSD – HCC, ±SE) in vehicle-treated vs. propranolol-treated mice (n = 3 per condition; representative of results from four independent experiments). RSD up-regulated proinflammatory gene expression by an average 25.1% in vehicle-treated animals (P < 0.001) vs. an average −11.7% difference in propranolol-treated animals (P = 0.017). (E) Effect of GM-CSF–neutralizing antibody MP1-22E9 (or isotype control antibody) on average prevalence (±SE) of peripheral blood Ly-6chigh monocytes and Ly-6cintermediate granulocytes (as determined by flow cytometry in n = 8 animals per condition pooled across three independent experiments). (F) Parallel assessment of bone marrow CD31+/Ly-6chigh monocyte progenitors and CD31+/Ly-6cintermediate granulocyte progenitors.
Consistent with previous indications that β-adrenergic signaling can promote myelopoiesis (28, 29), pretreatment of animals with the β-antagonist propranolol abrogated both RSD-induced expansion of peripheral Ly-6chigh monocytes (Fig. 3C and Fig. S1; RSD × antagonist interaction, P < 0.001) and RSD-induced up-regulation of proinflammatory gene expression in peripheral blood monocytes (Fig. 3D; RSD × antagonist interaction, P = 0.001). Moreover, among the 17 genes selected a priori as representative inflammatory markers, those showing the greatest magnitude of RSD-induced up-regulation in monocytes from vehicle-treated mice also showed the most pronounced inhibition of RSD response in propranolol-treated mice (r = 0.74, P < 0.001; Fig. 3D).
To define the specific myelopoietic growth factor mediating these effects, we examined the role of the general myeloid lineage differentiation factor GM-CSF and the monocyte-specific differentiation factor M-CSF. Consistent with the fact that RSD up-regulated both myeloid cell lineages (Figs. 2C and 3 A–C), expression of GM-CSF (Csf3) and its receptor (Csf3r) were up-regulated in monocytes from RSD animals (Csf3: +80.6 ± 20.3%, P < 0.001; Csf3r: +38.0 ± 10.6%, P < 0.001). In contrast, M-CSF expression was significantly down-regulated (Csf1: −39.9 ± 10.4%, P < 0.001). A mechanistic role for GM-CSF was confirmed by studies showing that administration of a neutralizing antibody to GM-CSF efficiently blocked RSD-induced expansion of both peripheral blood Ly-6chigh monocytes and Ly-6cintermediate granulocytes (both RSD × antagonist interactions, P < 0.001; Fig. 3E and Fig. S2). GM-CSF blockade also inhibited RSD-induced up-regulation of bone marrow monocyte and granulocyte progenitor cell populations (both P < 0.001; Fig. 3F). These effects were specific to GM-CSF because M-CSF blockade with the antagonist GW2580 failed to inhibit RSD up-regulation of blood monocytes (P = 0.898) or bone marrow monocytic progenitors (P = 0.134).
Human Socioeconomic Status and Monocyte Transcriptome.
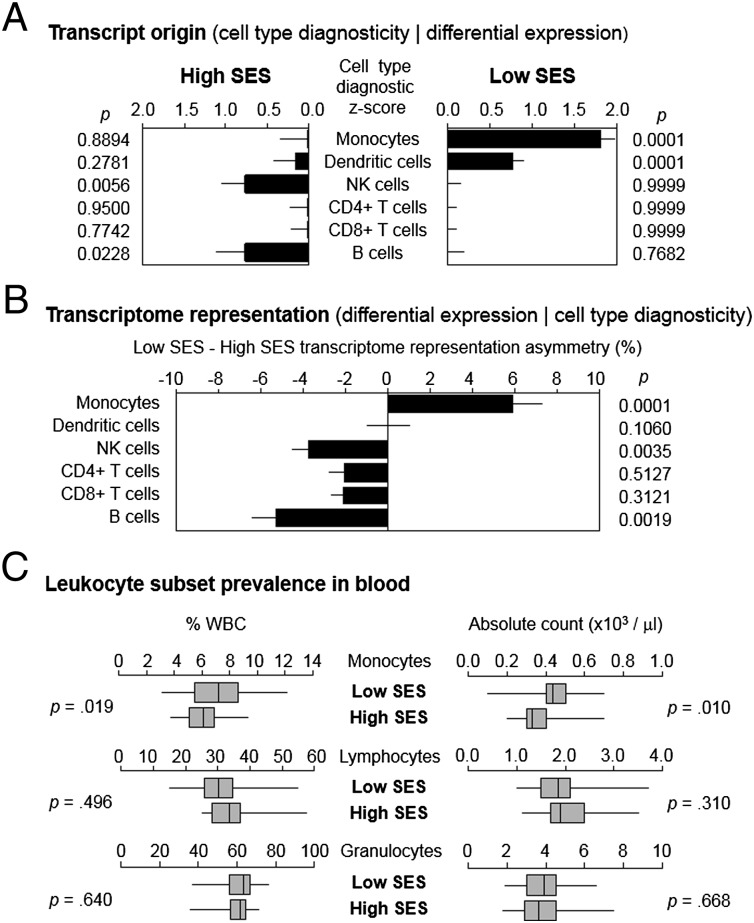
To determine whether monocyte population dynamics similar to those observed in mouse RSD might contribute to the leukocyte CTRA in humans subject to chronic stress, we used TRA to decompose the complex gene expression profile of circulating PBMCs into subcomponents originating from monocytes, plasmacytoid dendritic cells, NK cells, B cells, CD4+ T cells, and CD8+ T cells (2, 55). Analyses of archival PBMC transcriptome profiles from 60 healthy young adults identified 387 gene transcripts showing ≥20% differential expression between individuals with low vs. high SES [as assessed by 5-y occupational status, and controlling for age, sex, race, body mass index, and smoking as previously described (6); genes listed in Dataset S3]. Differentially expressed genes exemplified the general leukocyte CTRA profile (17) in showing increased expression of proinflammatory genes (e.g., IL1A, IL1B, IL8, COX2/PTGS2, TNF) and decreased expression of innate antiviral response genes (e.g., IFI27, IFIT1, IFITM1, ISG15) in low-SES individuals relative to high-SES individuals. Some 32.3% of genes identified as differentially expressed in PBMCs from low-SES humans were also differentially expressed in monocytes from RSD mice (significantly exceeding the <0.1% overlap expected by chance, P < 0.001). As in previous studies of other types of human social adversity, transcript origin analysis (2) identified monocytes and plasmacytoid dendritic cells as the predominate cellular sources of transcripts up-regulated in PBMCs from low-SES individuals (Fig. 4A). Transcript origin analysis cannot distinguish differential cell prevalence from per-cell changes in gene expression, so we conducted a more focused TRA analysis specifically assessing cell prevalence. Results indicated a greater prevalence of the monocyte transcriptome (but not the plasmacytoid dendritic cell transcriptome) in PBMC from low- vs. high-SES individuals (Fig. 4B). Automated complete blood count confirmed greater numbers and percentages of monocytes (but not lymphocytes or granulocytes) in circulating blood from low-SES individuals (Fig. 4C).
Fig. 4.
Socioeconomic status and human leukocyte subsets. (A) Transcript origin analysis assessing predominate cellular sources of 387 gene transcripts found to be differentially expressed by ≥20% in genome-wide transcriptional profiling of PBMCs from 30 low-SES healthy young adults and 30 matched high-SES individuals. (B) Transcriptome representation analysis assessing prevalence of transcriptomes corresponding to major leukocyte subsets in total PBMC RNA samples. (C) Relative percent and absolute frequency of monocytes, lymphocytes, and granulocytes in circulating blood samples.
Human Monocyte Subsets.
To determine whether chronic social adversity might be associated with up-regulated expression of the immature/proinflammatory monocyte transcriptome in humans, we used TRA to assess prevalence of the CD16− monocyte transcriptome (homologous to the mouse Ly-6chigh monocyte subset) (23, 44–46) in PBMCs from individuals with high vs. low SES (Dataset S4). Low-SES individuals show elevated circulating catecholamines (47–49), and promoter-based bioinformatic analysis of genes up-regulated in PBMCs from the present low-SES sample (Dataset S3) indicated increased activity of the β-adrenergic transcription factor cAMP response element-binding protein (CREB; +69.1 ± 8.0%, P < 0.001). Consistent with potential β-adrenergic up-regulation of proinflammatory monocytes, TRA indicated an average 8.4% (±0.9%) expansion of the CD16− monocyte transcriptome in PBMCs from low-SES individuals (P < 0.001). Expression of the GM-CSF receptor (CSF3R) was also up-regulated by an average of 21.8% (±9.1%) in PBMC from low-SES individuals (P = 0.026, controlling for age, sex, race, body mass index, and smoking, as previously described) (6).
Discussion
These studies identify β-adrenergic up-regulation of myelopoiesis as one molecular mechanism by which chronic stress may induce the proinflammatory CTRA gene expression dynamic previously observed in the circulating leukocyte pool of people confronting a diverse array of adverse life circumstances (1–9, 11). In the present study, healthy young adults exposed to the chronic stress of low SES showed selective expansion of both the total monocyte transcriptome and the immature CD16− monocyte transcriptome within the PBMC pool. Parallel expansion of the Ly-6chigh monocyte population was observed in mice subject to a very different type of chronic social stress in RSD, which nevertheless shares with human low SES the involvement of elevated SNS catecholamine signaling (47–49, 56). Bioinformatic analysis of differentially expressed genes suggested increased myelopoiesis as a potential mechanism for the selective up-regulation of immature proinflammatory monocytes. Direct analyses of myelopoiesis confirmed that result and implicated a β-adrenoreceptor–mediated pathway involving the myelopoietic growth factor GM-CSF in driving RSD-induced expansion of the mouse Ly-6chigh monocyte subpopulation. These results suggest that SNS/β-adrenergic up-regulation of myelopoiesis may play a key role in mediating the proinflammatory component of the leukocyte CTRA dynamic (13, 17) and could potentially contribute to the increased risk of inflammatory disease observed in individuals confronting chronic social adversity (19, 20).
The present findings are consistent with previous data showing that myeloid lineage immune cells and their innate effector molecules, such as proinflammatory cytokines and type I interferons, are regulated by β-adrenergic signaling (7, 57–62) and adverse socioenvironmental conditions (2, 7, 10, 13, 17, 63). These results are also consistent with previous findings that nonsocial activators of β-adrenergic signaling, such as burn-induced sepsis, can enhance monocyte differentiation (28, 29). SNS induction of a “primed” or hyperreactive bias in the circulating leukocyte pool may have evolved to help the immune system anticipate wound-related bacterial infections historically associated with adverse social conditions (17, 18). However, in contemporary social environments, such dynamics would needlessly undermine antiviral responses and promote chronic inflammation (17, 18). In addition to identifying one general physiological mechanism of such effects, the present data imply specific molecular strategies (e.g., β-blockade) and cellular biomarkers (e.g., CD16− monocytes) that could potentially be harnessed in future studies to develop health-protective interventions.
Results from the mouse RSD model demonstrate a causal pathway by which chronic social stress can remodel the circulating monocyte pool via a β-adrenergic up-regulation of myelopoiesis. The study showing parallel observations in human low SES is broadly consistent with these experimental results, but is nevertheless subject to some interpretive limitations. The human low-SES data come from an observational study in a single cohort and will require replication in future studies. The associations observed there do not demonstrate a causal effect of SES and do not indicate that observed effects are specific to low SES or mediated solely by SNS activity. Experimental studies will be required to establish a causal effect of SES on human gene expression and define its social, psychological, and neural/endocrine mechanisms [e.g., using randomization to remedial interventions such as those that inhibit CTRA response to other types of adversity (9, 64, 65) or experimental primate models of social stress that more closely mimic human low social status (12, 13)]. Although parallel effects on gene expression and monocyte phenotype emerged in both human low SES and mouse RSD, these two contexts also differ in significant ways, and mouse RSD paradigm should not be interpreted as a direct analog of human low SES. We assessed CTRA gene expression and monocyte up-regulation in low SES to verify the human social epidemiologic rationale for experimental analysis of myelopoietic dynamics, and we tested for β-adrenergic transcription control and expansion of the immature proinflammatory CD16− monocyte subpopulation in low SES to determine whether there is any reason to believe that the myelopoietic dynamics emerging from the mouse experimental model might potentially be relevant to a (clearly distinct) human social stressor that also involves elevated catecholamine signaling (47–49) and a proinflammatory shift in PBMC gene expression (6).
Although mouse RSD and human low SES differ in many respects, these two chronic socials stressors share some general features in common, including periodic exposure to violence, persistent threat of physical harm, and chronically elevated catecholamine levels (47–49, 56). The present results show that both of these chronic social stressors are also associated with similar increases in proinflammatory gene expression and monocyte differentiation. Approximately one-third of the total PBMC gene expression differences observed in human low SES also occurred in mouse RSD, including inflammation- and activation-related genes such as CD163, EGR1, FOSB, G0S2, IRS2, MMP9, SOD2, and TNF. This substantial overlap is especially remarkable in light of the fact that (i) the generally homologous human CD16− and mouse Ly-6chigh monocyte subsets nevertheless show some marked species-specific divergence in expression of specific genes (including converse expression of CD36, which is also observed here) (45), (ii) the present human and mouse transcriptome effects were assessed in distinct cell populations (i.e., PBMCs for humans vs. isolated monocytes for mice, the latter of which would have more favorable signal-to-noise ratio), and (iii) the human data come from a cross-sectional observational study subject to a variety of extraneous influences, whereas the mouse data come from a well-controlled experimental manipulation within a fixed genetic background. The collective effects of these biological and methodological differences likely underlie the sevenfold greater number of differentially expressed genes identified in the mouse RSD model.
These data clarify the basis for the proinflammatory component of the leukocyte CTRA (13, 17), but the type I IFN- and antibody-related components of the CTRA are not explained by the present monocyte dynamics and remain to be addressed in future studies. The present transcript origin analyses implicate plasmacytoid dendritic cells (pDCs) in the observed suppression of type I IFN-related genes, but TRA showed no significant reduction in pDC numbers. The IFN component of the CTRA may instead involve β-adrenergic inhibition of IFN gene expression as previously observed in vitro (57). However, this study’s definition of a β-adrenergic/myelopoietic pathway by which chronic stress can remodel the circulating leukocyte pool does clarify the proinflammatory component of the CTRA (17) and expands the range of physiological mechanisms by which adverse social environments may increase the risk of inflammation-related disease (19, 20).
Methods
Mouse RSD and β-Adrenergic Signaling.
Young adult male C57BL/6 mice were subject to six daily cycles of 2-h exposure to an aggressive male intruder mouse (RSD) or HCC conditions as described (39). Where indicated, mice were injected with 10 mg/kg propranolol (Sigma) or equivalent volume of vehicle 30 min before each RSD cycle; injected with 300 μg of anti–GM-CSF neutralizing antibody (nAb) MP1-22E9 or an equivalent volume of control IgG2a κ Ab (eBioscience) at 48-h intervals starting 1 d before RSD; and administered 160 mg/kg of the CSF1R antagonist GW2580 (R. I. Chemical, Inc.) or vehicle daily by oral gavage beginning 1 d before RSD. Immunomagnetically isolated CD11b+ monocytes (Miltenyi Biotec) were subject to genome-wide transcriptional profiling using Affymetrix MOE430 2.0 high-density oligonucleotide arrays as described (7) [Gene Expression Omnibus (GEO) accession no. GSE28830]. Genes showing ≥50% difference in average (quantile-normalized, log2-transformed) expression were subject to TRA as described below to quantify Ly-6clow and Ly-6chigh monocyte transcriptomes and subject to Transcription Element Listening System (TELiS) promoter bioinformatics to assess transcription factor activity (7). Results were confirmed by flow cytometry for CD11b/Gr1/Ly-6c in splenocytes, PBMCs, and bone marrow mononuclear cells (BMMCs), and flow cytometry for CD31/Ly-6c lineage-specific progenitor/precursor populations in BMMCs. Additional confirmatory transcriptome profiling studies used Illumina MouseRef-8 BeadArrays to survey gene expression in total PBMCs, PBMC-derived CD11b+ monocytes, and CD11b-depleted PBMCs from RSD vs. HCC mice (GEO accession nos. GSE47153 and GSE47154). See SI Methods for detailed methods. Procedures were approved by The Ohio State University Institutional Animal Care and Use Committee.
Human SES and Proinflammatory Monocytes.
PBMCs from 30 low-SES and 30 high-SES young adults were subject to automated complete blood count with differential and parallel genome-wide transcriptional profiling as previously published (6) (GEO accession no. GSE15180) and described in SI Methods. Genes showing ≥20% difference in average expression across low- vs. high-SES individuals (classified by 5-y occupational status) were identified in general linear models controlling for age, sex, race, body mass index, and smoking (6); cellular origin of these genes was analyzed by transcript origin analysis (2); activity of CREB transcription factors was assessed by TELiS promoter-based bioinformatics (7); and TRA was conducted as described below to quantify prevalence of major leukocyte subsets and CD14+/CD16− proinflammatory monocytes. All studies were approved by the University of British Columbia Research Ethics Board, and all subjects gave written informed consent (6).
Transcriptome Representation Analysis.
We assessed a composite index of cell type-diagnostic transcripts (55) to quantify the relative prevalence of discrete cellular transcriptomes within heterogeneous RNA pools (e.g., human monocytes vs. other leukocyte subsets in PBMCs; mouse Ly-6chigh vs. Ly-6clow subsets in total monocytes; human CD14+/CD16+ and CD14+/CD16− subsets in PBMCs). Cell type-diagnostic transcripts were defined by average expression ≥6 SD above the mean level of all other candidate cells (2) in reference gene expression profiles derived from isolated cell types [mouse Ly-6chigh/Ly-6clow monocytes from GEO accession no. GSE17256 (45); human leukocyte subsets from GSE1133 (66); and human CD16−/CD16+ monocytes from GSE18565 (45)] and tested for difference in average expression across groups by paired t test (methods are detailed in SI Methods).
Supplementary Material
Acknowledgments
We thank the Genome Quebec Innovation Centre, the University of California, Los Angeles (UCLA) DNA Microarray Core, and the UCLA Neuroscience Genomics Core for performing microarray assays. This research was supported by National Institutes of Health Grants HD058502, MH46801, MH093473, DE014320, CA116778, AG033590, and AG107265; the Mind, Body, Brain, and Health Initiative of the John D. and Catherine T. MacArthur Foundation; the British Colombia Ministry of Child and Family Development via the Human Early Learning Partnership; and the Allergy, Genes, and Environment Research Network.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE28830, GSE47153, and GSE47154).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310655110/-/DCSupplemental.
†Social epidemiologists use “adversity” to denote health risk factors defined by objective external conditions of one’s life (e.g., low SES, bereavement, caregiving for a dying spouse) as opposed to internal subjective reactions to those conditions (e.g., stress, depression) or physiological responses (e.g., allostatic load). This presentation does not require any distinction between chronic stress and adversity, and we use the terms interchangeably.
References
- 1.Cole SW, et al. Social regulation of gene expression in human leukocytes. Genome Biol. 2007;8(9):R189. doi: 10.1186/gb-2007-8-9-r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci USA. 2011;108(7):3080–3085. doi: 10.1073/pnas.1014218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller GE, et al. A functional genomic fingerprint of chronic stress in humans: Blunted glucocorticoid and increased NF-kappaB signaling. Biol Psychiatry. 2008;64(4):266–272. doi: 10.1016/j.biopsych.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen E, et al. Genome-wide transcriptional profiling linked to social class in asthma. Thorax. 2009;64(1):38–43. doi: 10.1136/thx.2007.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen E, Miller GE, Kobor MS, Cole SW. Maternal warmth buffers the effects of low early-life socioeconomic status on pro-inflammatory signaling in adulthood. Mol Psychiatry. 2011;16(7):729–737. doi: 10.1038/mp.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller GE, et al. Low early-life social class leaves a biological residue manifested by decreased glucocorticoid and increased proinflammatory signaling. Proc Natl Acad Sci USA. 2009;106(34):14716–14721. doi: 10.1073/pnas.0902971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole SW, et al. Computational identification of gene-social environment interaction at the human IL6 locus. Proc Natl Acad Sci USA. 2010;107(12):5681–5686. doi: 10.1073/pnas.0911515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Donovan A, et al. Transcriptional control of monocyte gene expression in post-traumatic stress disorder. Dis Markers. 2011;30(2-3):123–132. doi: 10.3233/DMA-2011-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoni MH, et al. Transcriptional modulation of human leukocytes by cognitive-behavioral stress management in women undergoing treatment for breast cancer. Biol Psychiatry. 2012;71:366–372. doi: 10.1016/j.biopsych.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sloan EK, et al. Social stress enhances sympathetic innervation of primate lymph nodes: Mechanisms and implications for viral pathogenesis. J Neurosci. 2007;27(33):8857–8865. doi: 10.1523/JNEUROSCI.1247-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sloan EK, et al. The sympathetic nervous system induces a metastatic switch in primary breast cancer. Cancer Res. 2010;70(18):7042–7052. doi: 10.1158/0008-5472.CAN-10-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tung J, et al. Social environment is associated with gene regulatory variation in the rhesus macaque immune system. Proc Natl Acad Sci USA. 2012;109(17):6490–6495. doi: 10.1073/pnas.1202734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole SW, et al. Transcriptional modulation of the developing immune system by early life social adversity. Proc Natl Acad Sci USA. 2012;109(50):20578–20583. doi: 10.1073/pnas.1218253109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole SW. Social regulation of human gene expression. Curr Dir Psychol Sci. 2009;18(3):132–137. doi: 10.1111/j.1467-8721.2009.01623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller G, Chen E, Cole SW. Health psychology: Developing biologically plausible models linking the social world and physical health. Annu Rev Psychol. 2009;60:501–524. doi: 10.1146/annurev.psych.60.110707.163551. [DOI] [PubMed] [Google Scholar]
- 16.Cole SW. Elevating the perspective on human stress genomics. Psychoneuroendocrinology. 2010;35(7):955–962. doi: 10.1016/j.psyneuen.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irwin MR, Cole SW. Reciprocal regulation of the neural and innate immune systems. Nat Rev Immunol. 2011;11(9):625–632. doi: 10.1038/nri3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finch CE. The Biology of Human Longevity: Inflammation, Nutrition, and Aging in the Evolution of Life Spans. Burlington, MA: Academic; 2007. [Google Scholar]
- 19.Berkman LF, Kawachi I. Social Epidemiology. New York: Oxford Univ Press; 2000. [Google Scholar]
- 20.Banks J, Marmot M, Oldfield Z, Smith JP. Disease and disadvantage in the United States and in England. JAMA. 2006;295(17):2037–2045. doi: 10.1001/jama.295.17.2037. [DOI] [PubMed] [Google Scholar]
- 21.Engler H, Bailey MT, Engler A, Sheridan JF. Effects of repeated social stress on leukocyte distribution in bone marrow, peripheral blood and spleen. J Neuroimmunol. 2004;148(1-2):106–115. doi: 10.1016/j.jneuroim.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Dhabhar FS, Malarkey WB, Neri E, McEwen BS. Stress-induced redistribution of immune cells—from barracks to boulevards to battlefields: A tale of three hormones—Curt Richter Award winner. Psychoneuroendocrinology. 2012;37(9):1345–1368. doi: 10.1016/j.psyneuen.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327(5966):656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elenkov IJ, Wilder RL, Chrousos GP, Vizi ES. The sympathetic nerve—an integrative interface between two supersystems: The brain and the immune system. Pharmacol Rev. 2000;52(4):595–638. [PubMed] [Google Scholar]
- 25.Katayama Y, et al. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124(2):407–421. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 26.Méndez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452(7186):442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 27.Beiermeister KA, et al. Hematopoietic progenitor cell mobilization is mediated through beta-2 and beta-3 receptors after injury. J Trauma. 2010;69(2):338–343. doi: 10.1097/TA.0b013e3181e5d35e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Y, et al. Norepinephrine modulates myelopoiesis after experimental thermal injury with sepsis. Ann Surg. 2001;233(2):266–275. doi: 10.1097/00000658-200102000-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen MJ, et al. Bone marrow norepinephrine mediates development of functionally different macrophages after thermal injury and sepsis. Ann Surg. 2004;240(1):132–141. doi: 10.1097/01.sla.0000130724.84914.d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avitsur R, Powell N, Padgett DA, Sheridan JF. Social interactions, stress, and immunity. Immunol Allergy Clin North Am. 2009;29(2):285–293. doi: 10.1016/j.iac.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Stark JL, Avitsur R, Hunzeker J, Padgett DA, Sheridan JF. Interleukin-6 and the development of social disruption-induced glucocorticoid resistance. J Neuroimmunol. 2002;124(1-2):9–15. doi: 10.1016/s0165-5728(02)00004-8. [DOI] [PubMed] [Google Scholar]
- 32.Avitsur R, Kavelaars A, Heijnen C, Sheridan JF. Social stress and the regulation of tumor necrosis factor-alpha secretion. Brain Behav Immun. 2005;19(4):311–317. doi: 10.1016/j.bbi.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Bailey MT, Engler H, Powell ND, Padgett DA, Sheridan JF. Repeated social defeat increases the bactericidal activity of splenic macrophages through a Toll-like receptor-dependent pathway. Am J Physiol Regul Integr Comp Physiol. 2007;293(3):R1180–R1190. doi: 10.1152/ajpregu.00307.2007. [DOI] [PubMed] [Google Scholar]
- 34.Powell ND, et al. Repeated social defeat activates dendritic cells and enhances Toll-like receptor dependent cytokine secretion. Brain Behav Immun. 2009;23:225–231. doi: 10.1016/j.bbi.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong-Newsom P, Powell ND, Bailey MT, Padgett DA, Sheridan JF. Repeated social stress enhances the innate immune response to a primary HSV-1 infection in the cornea and trigeminal ganglia of Balb/c mice. Brain Behav Immun. 2010;24(2):273–280. doi: 10.1016/j.bbi.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wohleb ES, et al. β-Adrenergic receptor antagonism prevents anxiety-like behavior and microglial reactivity induced by repeated social defeat. J Neurosci. 2011;31(17):6277–6288. doi: 10.1523/JNEUROSCI.0450-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mays JW, et al. Influenza virus-specific immunological memory is enhanced by repeated social defeat. J Immunol. 2010;184(4):2014–2025. doi: 10.4049/jimmunol.0900183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powell ND, Mays JW, Bailey MT, Hanke ML, Sheridan JF. Immunogenic dendritic cells primed by social defeat enhance adaptive immunity to influenza A virus. Brain Behav Immun. 2011;25(1):46–52. doi: 10.1016/j.bbi.2010.07.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stark JL, et al. Social stress induces glucocorticoid resistance in macrophages. Am J Physiol Regul Integr Comp Physiol. 2001;280(6):R1799–R1805. doi: 10.1152/ajpregu.2001.280.6.R1799. [DOI] [PubMed] [Google Scholar]
- 40.Avitsur R, Stark JL, Dhabhar FS, Padgett DA, Sheridan JF. Social disruption-induced glucocorticoid resistance: Kinetics and site specificity. J Neuroimmunol. 2002;124(1-2):54–61. doi: 10.1016/s0165-5728(02)00010-3. [DOI] [PubMed] [Google Scholar]
- 41.Quan N, et al. Molecular mechanisms of glucocorticoid resistance in splenocytes of socially stressed male mice. J Neuroimmunol. 2003;137(1-2):51–58. doi: 10.1016/s0165-5728(03)00042-0. [DOI] [PubMed] [Google Scholar]
- 42.Engler H, et al. Interleukin-1 receptor type 1-deficient mice fail to develop social stress-associated glucocorticoid resistance in the spleen. Psychoneuroendocrinology. 2008;33:108–117. doi: 10.1016/j.psyneuen.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey MT, et al. Social stress enhances allergen-induced airway inflammation in mice and inhibits corticosteroid responsiveness of cytokine production. J Immunol. 2009;182(12):7888–7896. doi: 10.4049/jimmunol.0800891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5(12):953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 45.Ingersoll MA, et al. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood. 2010;115(3):e10–e19. doi: 10.1182/blood-2009-07-235028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robbins CS, Swirski FK. The multiple roles of monocyte subsets in steady state and inflammation. Cell Mol Life Sci. 2010;67(16):2685–2693. doi: 10.1007/s00018-010-0375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans GW, English K. The environment of poverty: Multiple stressor exposure, psychophysiological stress, and socioemotional adjustment. Child Dev. 2002;73(4):1238–1248. doi: 10.1111/1467-8624.00469. [DOI] [PubMed] [Google Scholar]
- 48.Cohen S, Doyle WJ, Baum A. Socioeconomic status is associated with stress hormones. Psychosom Med. 2006;68(3):414–420. doi: 10.1097/01.psy.0000221236.37158.b9. [DOI] [PubMed] [Google Scholar]
- 49.Janicki-Deverts D, et al. Socioeconomic status is related to urinary catecholamines in the Coronary Artery Risk Development in Young Adults (CARDIA) study. Psychosom Med. 2007;69:514–520. doi: 10.1097/PSY.0b013e3180f60645. [DOI] [PubMed] [Google Scholar]
- 50.Valledor AF, Borràs FE, Cullell-Young M, Celada A. Transcription factors that regulate monocyte/macrophage differentiation. J Leukoc Biol. 1998;63(4):405–417. doi: 10.1002/jlb.63.4.405. [DOI] [PubMed] [Google Scholar]
- 51.Saeki K, Saeki K, Yuo A. Distinct involvement of cAMP-response element-dependent transcriptions in functional and morphological maturation during retinoid-mediated human myeloid differentiation. J Leukoc Biol. 2003;73(5):673–681. doi: 10.1189/jlb.1002512. [DOI] [PubMed] [Google Scholar]
- 52.Sawka-Verhelle D, et al. PE-1/METS, an antiproliferative Ets repressor factor, is induced by CREB-1/CREM-1 during macrophage differentiation. J Biol Chem. 2004;279(17):17772–17784. doi: 10.1074/jbc.M311991200. [DOI] [PubMed] [Google Scholar]
- 53.Leenen PJ, Slieker WA, Melis M, Van Ewijk W. Murine macrophage precursor characterization. I. Production, phenotype and differentiation of macrophage precursor hybrids. Eur J Immunol. 1990;20(1):15–25. doi: 10.1002/eji.1830200104. [DOI] [PubMed] [Google Scholar]
- 54.Nikolic T, de Bruijn MF, Lutz MB, Leenen PJ. Developmental stages of myeloid dendritic cells in mouse bone marrow. Int Immunol. 2003;15(4):515–524. doi: 10.1093/intimm/dxg050. [DOI] [PubMed] [Google Scholar]
- 55.Abbas AR, et al. Immune response in silico (IRIS): Immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6(4):319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 56.Hanke ML, Powell ND, Stiner LM, Bailey MT, Sheridan JF. Beta adrenergic blockade decreases the immunomodulatory effects of social disruption stress. Brain Behav Immun. 2012;26(7):1150–1159. doi: 10.1016/j.bbi.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collado-Hidalgo A, Sung C, Cole S. Adrenergic inhibition of innate anti-viral response: PKA blockade of type I interferon gene transcription mediates catecholamine support for HIV-1 replication. Brain Behav Immun. 2006;20(6):552–563. doi: 10.1016/j.bbi.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 58.Mayer B, et al. Functional improvement in heart failure patients treated with beta-blockers is associated with a decline of cytokine levels. Int J Cardiol. 2005;103(2):182–186. doi: 10.1016/j.ijcard.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 59.Tatli E, Kurum T. A controlled study of the effects of carvedilol on clinical events, left ventricular function and proinflammatory cytokines levels in patients with dilated cardiomyopathy. Can J Cardiol. 2005;21(4):344–348. [PubMed] [Google Scholar]
- 60.Gage JR, et al. Beta blocker and angiotensin-converting enzyme inhibitor therapy is associated with decreased Th1/Th2 cytokine ratios and inflammatory cytokine production in patients with chronic heart failure. Neuroimmunomodulation. 2004;11(3):173–180. doi: 10.1159/000076766. [DOI] [PubMed] [Google Scholar]
- 61.Matsumura T, et al. Effects of carvedilol on plasma levels of interleukin-6 and tumor necrosis factor-alpha in nine patients with dilated cardiomyopathy. J Cardiol. 2002;39(5):253–257. [PubMed] [Google Scholar]
- 62.Ohtsuka T, et al. Effect of beta-blockers on circulating levels of inflammatory and anti-inflammatory cytokines in patients with dilated cardiomyopathy. J Am Coll Cardiol. 2001;37(2):412–417. doi: 10.1016/s0735-1097(00)01121-9. [DOI] [PubMed] [Google Scholar]
- 63.Gidron Y, Armon T, Gilutz H, Huleihel M. Psychological factors correlate meaningfully with percent-monocytes among acute coronary syndrome patients. Brain Behav Immun. 2003;17(4):310–315. doi: 10.1016/s0889-1591(03)00061-8. [DOI] [PubMed] [Google Scholar]
- 64.Creswell JD, et al. Mindfulness-based stress reduction training reduces loneliness and pro-inflammatory gene expression in older adults: A small randomized controlled trial. Brain Behav Immun. 2012;26(7):1095–1101. doi: 10.1016/j.bbi.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Black DS, et al. Yogic meditation reverses NF-κB and IRF-related transcriptome dynamics in leukocytes of family dementia caregivers in a randomized controlled trial. Psychoneuroendocrinology. 2013;38(3):348–355. doi: 10.1016/j.psyneuen.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.