Abstract
Nucleus-encoded ribonucleases and RNA-binding proteins influence chloroplast gene expression through their roles in RNA maturation and stability. One mechanism for mRNA 5′ end maturation posits that sequence-specific pentatricopeptide repeat (PPR) proteins define termini by blocking the 5′→3′ exonucleolytic activity of ribonuclease J (RNase J). To test this hypothesis in vivo, virus-induced gene silencing was used to reduce the expression of three PPR proteins and RNase J, both individually and jointly, in Nicotiana benthamiana. In accordance with the stability-conferring function of the PPR proteins PPR10, HCF152 and MRL1, accumulation of the cognate RNA species atpH, petB and rbcL was reduced when the PPR-encoding genes were silenced. In contrast, RNase J reduction alone or combined with PPR deficiency resulted in reduced abundance of polycistronic precursor transcripts and mature counterparts, which were replaced by intermediately sized species with heterogeneous 5′ ends. We conclude that RNase J deficiency can partially mask the absence of PPR proteins, and that RNase J is capable of processing chloroplast mRNAs up to PPR protein-binding sites. These findings support the hypothesis that RNase J is the major ribonuclease responsible for maturing chloroplast mRNA 5′ termini, with RNA-binding proteins acting as barriers to its activity.
INTRODUCTION
Chloroplasts arose from the phagocytotic uptake of a photosynthetic bacterium by a primitive heterotrophic eukaryote. Following endosymbiosis, the bacterium surrendered its autonomy through massive gene transfer to the host nucleus (1). Although the chloroplast retains its own transcription and translation apparatus, gene regulation is predominately conferred by nucleus-encoded proteins targeted to the chloroplast (2). The majority of chloroplast-encoded genes are arranged into operons or clusters, and the resulting polycistronic transcripts are frequently processed into mono- and oligocistronic RNAs. These processing events involve a mosaic of trans-factors, namely endo- and exoribonucleases of mainly prokaryotic ancestry together with RNA-binding proteins of more recent origins (3). Here, the cooperative functions of ribonuclease J (RNase J) and the pentatricopeptide repeat (PPR) family of RNA-binding proteins were investigated.
RNase J was discovered in Bacillus subtilis, where it was demonstrated to be an essential protein harboring both endonuclease and 5′→3′ exonuclease activities (4,5). It is found in Archaea, Gram-positive bacteria, cyanobacteria and chloroplasts but is absent from Escherichia coli. Nuclear RNase J-encoding genes are ubiquitous in plants, and Arabidopsis RNase J is targeted to the chloroplast, possesses dual ribonuclease activities (6) and is essential for embryo development (7). Tobacco (Nicotiana benthamiana) plants made deficient in RNase J by virus-induced gene silencing (VIGS) exhibit a remarkable accumulation of antisense RNAs (asRNAs), which duplex with sense transcripts and impair their translation (6). These asRNA substrates of RNase J are hypothesized to originate from inefficient chloroplast transcriptional termination. RNase J, therefore, participates in a chloroplast RNA quality-control mechanism by eliminating read-through transcription products.
Recent studies have also implicated RNase J in a chloroplast mRNA maturation pathway directed by PPR proteins. PPR proteins are characterized by helical repeat structures formed from tandem arrays of a degenerate 35 amino acid motif and are prevalent among land plants but absent from bacteria (8). Many are organelle targeted, bind RNA in a gene-specific manner through an amino acid code (9,10) and confer transcript stability by obstructing ribonucleolytic digestion and defining mature mRNA termini (11). In the case of polycistronic mRNAs, PPR proteins bind overlapping transcripts relieved from precursors by multiple endonucleolytic cleavages within intergenic regions and block exonucleolytic processing from both the 3′ and 5′ directions (12,13). PPR10 and HCF152 bind the atpI-atpH and psbH-petB intergenic regions, respectively, and shield transcript 5′ and 3′ termini mapping to those regions (13,14). PPR proteins are also known to define monocistronic transcript ends (15). MRL1, for instance, binds a sequence in the 5′ untranslated region (UTR) of the primary rbcL transcript, leading to the accumulation of a shorter processed form (16). The enzyme responsible for maturing PPR protein-defined 5′ ends has yet to be determined. As a commercial 5′→3′ exonuclease can carry out this function in vitro (17), RNase J is a prime candidate for this role in vivo as the only identified chloroplast 5′→3′ exoribonuclease, as previously speculated (13).
To evaluate the involvement of RNase J in PPR protein-mediated 5′ mRNA maturation, genes encoding PPR10, HCF152, MRL1 and RNase J were silenced, independently and in pairwise fashion, by VIGS in N. benthamiana. Deficiency of single PPR proteins resulted in diminished accumulation of processed cognate RNAs, as expected, an effect that could be partially masked by deficiency of RNase J. Based on these results, we conclude that chloroplast RNase J trims mRNA 5′ termini to mature forms defined by bound PPR proteins.
MATERIALS AND METHODS
Plant material
Nicotiana benthamiana plants were grown in a greenhouse at 25°C under a 14-h light/10-h dark regime.
Design of VIGS target regions
Arabidopsis genes for PPR10 (At2g18940), HCF152 (At3g09650) and MRL1 (At4g34830) were submitted as BLAST queries of the Nicotiana tabacum Unigene sequence set of the Sol Genomics Network (18). The largest open reading frame of each Unigene result was translated and aligned with the corresponding Arabidopsis PPR protein sequence using ClustalW2 (19), verifying that the orthologous N. tabacum genes had been identified. For VIGS target regions, two unique stretches of 250–350 nt for each N. tabacum sequence were selected, which returned only a single hit on submission to siRNA Scan (http://bioinfo2.noble.org/RNAiScan.htm). All primer sequences are given in Supplementary Table S1.
Plasmid construction and VIGS infection
One or two target regions for PPR10, HCF152, MRL1 and RNJ were PCR-amplified from N. benthamiana DNA. The amplicons were cloned into pCR®8/GW (Invitrogen) as both single inserts and RNase J-PPR protein tandem inserts. The VIGS targets were then inserted into pYL170/GW (20) and transformed into Agrobacterium tumefaciens GV3101 by electroporation. As a control, the empty vector pYL170, along with the expression component of the bipartite VIGS system, pTRV1, were introduced into Agrobacterium. VIGS was carried out as previously described (6). Given that genes silenced with either of two targeting regions consistently yielded indistinguishable molecular phenotypes results from only one VIGS targeting region per gene or gene combination are presented here.
RNA extraction, 5′ rapid amplification of cDNA ends and quantitative RT-PCR
Leaf tissue was harvested 18 days after VIGS infections, and RNA was isolated with TRI reagent (Sigma-Aldrich) per the manufacturer’s instructions. Three micrograms of RNA were treated with 2 U DNase I (Ambion) for 30 min at 37°C. The reaction was terminated by phenol:chloroform extraction, followed by ethanol precipitation. Treatment with tobacco acid pyrophosphatase (TAP) (Epicentre; 0.5 U per 3 μg RNA) was carried out at 37°C for 1 h with 40 U RNaseOUT (Invitrogen). The RNA was re-purified and incubated with 1.7 µl of RNA adapter (10 µM; see Supplementary Table S1) at 65°C for 5 min, briefly chilled on ice and then added to the ligation mixture [1 mM ATP, 4 U RNaseOUT, 1× ligase buffer, 0.5 U T4 RNA ligase (New England Biolabs)]. SuperScript III (Invitrogen) was used for first-strand synthesis, primed with random hexamers, in accordance with the manufacturer’s instructions. Reverse transcription products were amplified by PCR with 1 U of GoTaq (Promega) using primers specific for the RNA adapter and the gene of interest. The following conditions were used: 2 min at 95°C; 30 s at 95°C, 30 s at 54°C, 70 s at 72°C, repeated for 30 cycles; and 2 min at 72°C. Rapid amplification of cDNA ends (RACE) products were separated in 1.5–3% agarose gels, and DNA fragments were gel purified and ligated into pGEM-T Easy (Promega) for sequencing. For quantitative RT-PCR (qRT-PCR), strand-specific cDNA synthesis was performed using the 3′ qPCR gene-specific primers (Supplementary Table S1), and PCR was conducted as previously described (21) using the same reference genes, 18S rRNA and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Standard errors were calculated from three biological replicates.
RNA gel blot analysis
Total RNA (0.5 µg/lane) was fractionated in 1.2% formaldehyde-agarose gels and transferred to Amersham Hybond-N+ membranes (GE Healthcare). Strand- and gene-specific probe templates were PCR-amplified from N. benthamiana DNA (see Supplementary Table S1) and gel purified. RNA probes were generated by T7 RNA polymerase (Promega), in accordance with the manufacturer’s protocol, with 40 µCi of α32P-uridine-5′-triphosphate (UTP). RNA gel blots were prehybridized as previously described (6), hybridized with riboprobes for 14 h at 68°C, washed twice in 1× SSC, 0.6% SDS for 10 min at 68°C and twice with 0.1× SSC, 0.6% SDS for 45 min at 68°C. DNA-probed RNA gel blots were performed as previously described (22).
RESULTS
RNase J processes atpH 5′ ends defined by PPR10
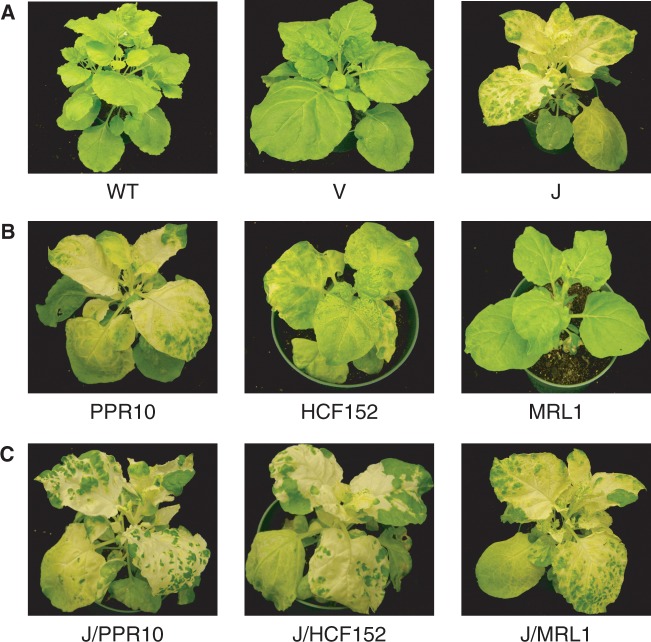
To study the function of RNase J in vivo, expression of the enzyme was repressed in N. benthamiana by VIGS using a bipartite tobacco rattle virus system (23). As we previously reported (6), infected plants deficient for RNase J exhibited a chlorotic phenotype (Figure 1A), indicative of impaired photosynthetic capabilities. Indistinguishable physiological and molecular phenotypes were seen when VIGS targeted either of two unique regions within the RNase J-encoding transcript, indicating minimal off-target silencing. Efforts to use immunoblot analysis to quantify RNase J deficiency were unsuccessful; however, our earlier work showed that VIGS reduces the transcript level for RNase J.
Figure 1.
VIGS-mediated deficiency of RNase J and PPR proteins in N. benthamiana. All plants are shown 18 days post-infiltration. (A) WT, wild-type; V, plants infiltrated with the empty pYL170 vector as a negative control; J, RNase J-silenced plants. (B) Plants targeted for PPR proteins PPR10, HCF152 and MRL1. (C) VIGS of both RNase J and the indicated PPR protein.
Although RNase J deficiency is known to impair chloroplast function by allowing the deleterious accumulation of double-stranded RNA (6), defects in processing of functional RNAs could also arise, given that RNase J has such a role in bacteria (5,24). Indeed, RNase J-deficient tissues in both Arabidopsis and N. benthamiana, accumulate abnormally sized sense mRNA species as judged by RNA gel blot, consistent with a role of RNase J in chloroplast RNA maturation (6). Here, we used RACE to precisely map the mRNA 5′ ends in RNase J-depleted tissues of N. benthamiana.
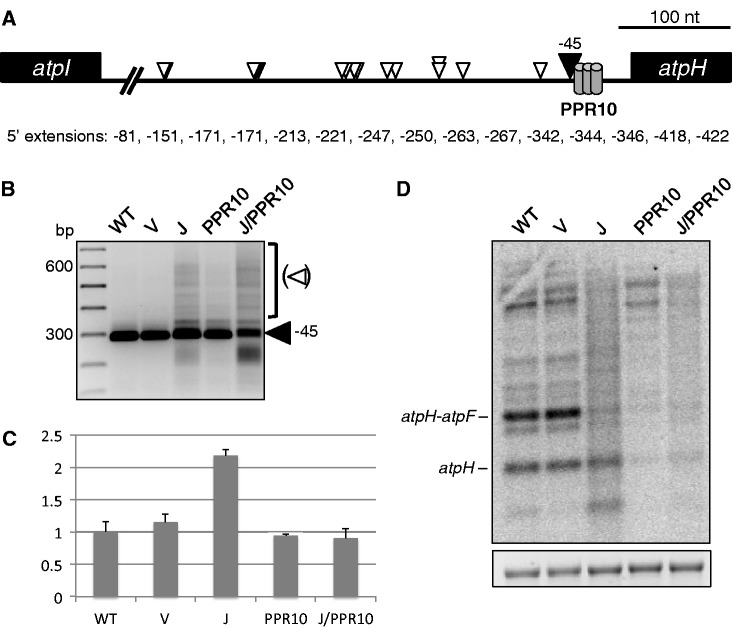
We initially chose atpH for analysis because its 5′ end can be correctly generated in vitro by a combination of PPR10 and a commercial 5′→3′ exonuclease (17). As shown in Figure 2A and B, 5′ end analysis revealed a single major band for the wild-type (WT) and the vector control (V) tissues corresponding to the previously described (13) processed terminus at position −45 (filled arrowhead), which abuts the PPR10 binding site. In contrast, RT-PCR amplifications of 5′ ends from RNase J-deficient material revealed additional longer and a few shorter termini. Sequence analysis of the longer 5′ RACE products showed that extended species map between the mature WT 5′ end of atpH to >400 nt upstream, as displayed in Figure 2A. Local secondary structure predictions suggest that nearly all of the sequenced 5′ extensions terminate in regions of single-stranded character (Supplementary Figure S1). These findings indicate RNase J plays a critical role in the 5′ maturation of atpH mRNA, perhaps by resecting unstructured intergenic regions following endonucleolytic cleavage by itself or another enzyme.
Figure 2.
Analysis of atpH by 5′ RACE, qRT-PCR and RNA gel blot. (A) 15 sequenced atpH 5′ termini from RNase J-VIGS plants. The numbers shown correspond to nucleotide distances upstream of the atpH translation initiation codon. PPR10, conserved minimal PPR10 binding site (17). (B) Gel separation of atpH 5′ RACE products. The filled arrowhead indicates the WT atpH 5′ end, whereas the open arrowheads designate atpH 5′ extensions that accumulate in RNase J-deficient tissue, as presented in (A). Left lane, 100 bp DNA ladder. WT, wild-type; V, vector control; J, RNase J-VIGS. (C) Relative amounts of atpH-containing fragments measured by qRT-PCR. (D) Strand-specific RNA gel blot analysis of atpH. An ethidium bromide-stained image of 28S rRNA is shown below to reflect loading.
qRT-PCR and RNA gel blots were used for additional assessments of sense strand transcript accumulation. The qRT-PCR data (Figure 2C) revealed a 2-fold increase of sense strand atpH-containing RNAs in RNase J-VIGS material compared with control tissues. RNA gel blot analysis of the RNase J-deficient tissues (Figure 2D), using a strand-specific probe, revealed a background smear along with a decrease in the abundance of discrete species. One interpretation is that qRT-PCR measurements reflect a modest increase of atpH mRNAs with heterogeneous 5′ ends. The results also indicate that RNase J normally limits abundance of atpH mRNA, in the short segment of the coding region defined by the qRT-PCR primer set.
To test whether accurate RNase J-mediated atpH mRNA maturation requires the cognate PPR protein, PPR10, VIGS was used to create PPR10 deficiency with or without concomitant RNase J silencing. Targeting of PPR10 (Figure 1B) resulted in a chlorotic phenotype, reminiscent of the maize ppr10 null mutant phenotype (13). To verify that PPR10 in tobacco and maize has orthologous functions, RNA analysis was performed. Figure 2C shows that PPR10 deficiency does not cause a general loss of atpH-containing transcripts, whereas Figure 2D clearly shows that the levels of transcripts with atpH at their 5′ ends is strongly reduced, in particular the atpH monocistronic mRNA and the atpH-atpF dicistronic species (atpF is immediately downstream of atpH). Furthermore, because this reduction is not accompanied by an increase higher molecular weight bands, we conclude that the forms with reduced abundance are unstable. PPR10 deficiency was also associated with a shift in migration of the major polycistronic species, which may suggest that PPR10 influences RNA processing at secondary sites. The 5′ RACE (Figure 2B) showed that although the WT 5′ end could still be detected, as expected from the partial deficiency that VIGS creates, some longer 5′ ends were also present, suggesting that the role of PPR10 in defining 5′ ends had been compromised. Overall, our results agree with findings from maize that PPR10 is necessary for the accumulation of processed atpH transcripts (13).
Simultaneous VIGS silencing was used to investigate the relationship between RNase J and PPR10 during the maturation of atpH 5′ ends, leading to the same whole-plant phenotype that was observed following silencing of only RNase J (Figure 1). Figure 2B shows that based on 5′ RACE, dual RNase J/PPR10 deficiency resulted in similar extended atpH 5′ ends as observed in RNase J-depleted plants. However, an effect of PPR10 deficiency was also evident, as atpH-containing transcripts were reduced to control levels as judged by qRT-PCR analysis (Figure 2C). Apart from transcript abundance, the overall similarity of molecular phenotypes between RNase J/PPR10- and RNase J-deficient plants suggests that the reduction of RNase J masks the compromised protective role of PPR10. Therefore, RNase J is involved in the 5′ maturation of atpH transcripts bound by PPR10 in vivo.
RNase J and HCF152 are required for the maturation of petB 5′ termini
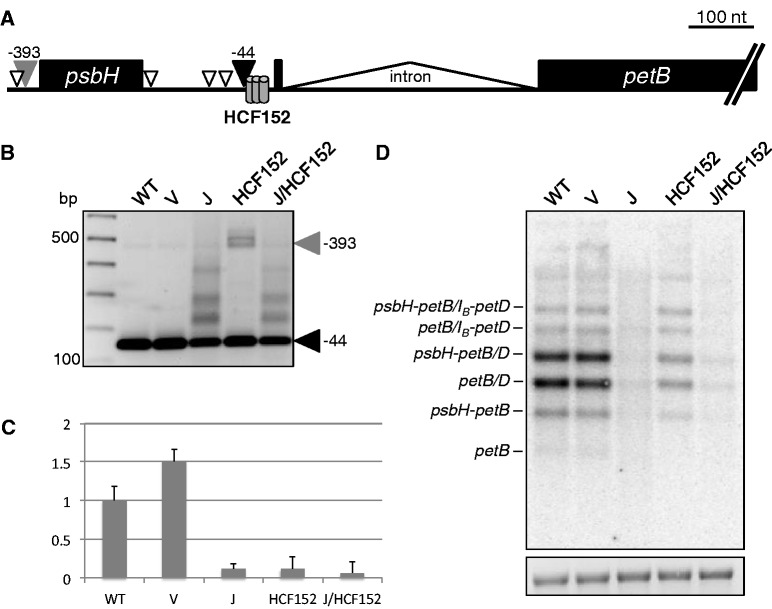
To see whether the paradigm verified for RNase J and PPR10 holds true in the context of another gene cluster, we analyzed the 5′ end of petB, a transcript derived from the psbB operon, which is protected by the PPR protein HCF152 (15). We used the same VIGS strategy as for PPR10, and the expected plant phenotypes were obtained as shown in Figure 1—Arabidopsis HCF152 null mutants are chlorotic and require sucrose for growth (14). Figure 3A and B show that heterogeneous and extended petB 5′ termini, compared with the WT -44 position, accumulate in tissue with reduced RNase J. This contrasts with the HCF152-depleted tissue, where the major species not found in WT (gray arrowhead) corresponds to the psbH 5′ end (−393), indicative of transcripts that have not undergone intercistronic processing. The presence of residual WT 5′ ends is consistent with the incomplete silencing typical of VIGS and is in agreement with results from the weak mutant allele hcf152-2 of Arabidopsis (14). RNA from RNase J/HCF152-depleted material exhibited a 5′ end pattern most similar to that seen with RNase J deficiency alone, suggesting that as for PPR10, RNase J deficiency can at least partially mask HCF152 deficiency.
Figure 3.
Analysis of petB mRNA. (A) Schematic of the major bands detected by 5′ RACE. The filled gray and black arrowheads correspond to the similarly-labeled bands in (B). Open arrowheads represent estimated 5′ ends from products of intermediate sizes. HCF152, conserved minimal HCF152 binding site (15). (B) 5′ RACE products from RT priming within the petB exon. The filled black arrowhead marks the mature WT petB 5′ end (25) and the filled gray arrowhead marks products from a precursor transcript that terminates upstream of psbH, corresponding to the psbH-petB dicistron. Numbers at right are nucleotide positions relative to the petB translation initiation codon. (C) Relative amounts of petB-containing RNA fragments measured by qRT-PCR. (D) Strand-specific RNA gel blot for the petB exon. IB indicates unspliced petB species. An ethidium bromide-stained image of 28S rRNA is shown below to reflect loading.
Additional assessment of petB mRNAs was performed using qRT-PCR and strand-specific RNA gel blot analysis. As shown in Figure 3C, RNase J deficiency was linked to a 5-fold reduction of petB-containing RNAs, accompanied by a diffuse RNA gel blot signal (Figure 3D). Reduced RNA accumulation is likely to be at least in part a secondary consequence of diminished PEP activity that is associated with the chloroplast translational defects in RNase J-depleted chloroplasts: petB transcription is mainly dependent on the upstream psbB PEP promoter, in contrast to atpH whose transcription is increased in the absence of PEP (26). HCF152-deficient plants also exhibited reduced accumulation of petB-containing transcripts, including the ones that had been cleaved within the psbH-petB intergenic region and especially those containing spliced petB. The persistence of higher molecular weight petB RNAs in HCF152-VIGS tissue suggests a comparative instability of the spliced forms, a phenomenon first noted in Arabidopsis hcf152 T-DNA mutants (14). Joint reduction of RNase J and HCF152 led to similar petB mRNA defects as were observed for RNase J depletion, where an accumulation of 5′-extended petB species (Figure 3B) was accompanied by a general decrease in petB RNA abundance (Figure 3C and D). These results suggest RNase J and HCF152 operate on the same petB transcripts and are both required for proper 5′ maturation and stability.
To see whether there was a differential effect of RNase J and/or HCF152 deficiency on unspliced petB species, we used 5′ RACE initiated by reverse priming within the petB intron and RNA gel blots (Supplementary Figure S2). Although the major unspliced petB RNAs accumulated in HCF152-VIGS material at similar levels as in WT tissue, as judged by gel blot, few petB 5′ ends at position −44 were detected by RACE, with the products being dominated by 5′ ends upstream of psbH and species mapping slightly downstream of the −44 site. In the cases of RNase J deficiency, 5′ extensions were also mapped between −44 and psbH. The effects of HCF152 deficiency thus differed significantly for spliced versus unspliced RNAs, where the −44 5′ end was readily found for spliced species but barely detectable for unspliced RNAs. One interpretation is that spliced RNAs have a higher affinity for residual HCF152. The small size of the first exon would be consistent with an effect of the intron on RNA structure in the HCF152 binding region (see Figure 3A). Although HCF152 binding to different regions surrounding, the petB 5′ end has been assessed (15,27), to our knowledge, a direct comparison of spliced and unspliced substrates has not been conducted.
RNase J and MRL1 are necessary for the processing of rbcL 5′ ends
We chose the monocistronic rbcL mRNA as a third model to investigate RNase J involvement in 5′ end processing, as a complement to the polycistronic transcript derivatives atpH and petB. Two forms of rbcL mRNA accumulate in land plants: a primary transcript and a shorter form generated by ribonucleolytic processing (28). These can be differentiated before 5′ RACE by treatment of RNA with TAP because primary transcripts can only be amplified following TAP treatment. As shown in Figure 4A and B, WT and vector control plants possess both primary (−182) and processed (−59) forms of rbcL. In the +TAP lanes, the product representing the primary form predominates owing to its higher abundance, whereas in −TAP lanes, only the processed form is represented. Depletion of RNase J led to a strong reduction of primary rbcL transcripts, suggesting that PEP-mediated transcription of rbcL had diminished, and to the appearance of 5′ extended species relative to the −59 form. The species mapping between positions −59 and −182 represent incomplete maturation owing to depletion of RNase J. The −59 form, but not the primary transcript, is found in tobacco where PEP has been genetically eliminated, reflecting the presence of additional promoters upstream (30), which also accounts for the accumulation of novel longer transcripts on the gel blot for RNase J-depleted material.
Figure 4.
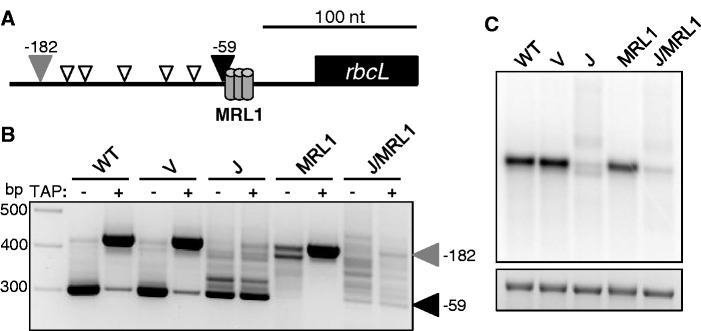
rbcL 5′ end analysis. (A) Graphic representation of rbcL 5′ ends detected by RACE. The filled gray and black arrowheads correspond to the labeled bands in (B). Open arrowheads represent transcript ends of estimated sizes. MRL1, predicted MRL1 binding site (15). (B) Gel separation of 5′ RACE products. +, TAP treatment prior to RACE; −, samples not treated with TAP. The filled black arrowhead marks the processed rbcL 5′ end (29) and the filled gray arrowhead designates the primary transcript. Values at right designate nucleotide positions relative to the rbcL translation initiation codon. (C) Strand-specific RNA gel blot probed for rbcL. An ethidium bromide-stained image of 28S rRNA is shown below to reflect loading.
We next partially silenced expression of the PPR protein MRL1, which specifically protects the processed rbcL transcript. VIGS material was indistinguishable from the WT in terms of plant growth phenotype (Figure 1B), consistent with results from Arabidopsis (16). This is because the accumulation of primary rbcL transcripts is unaffected by MRL1 deficiency (Figure 4B), while the processed form is essentially undetectable. Because of their small size difference, the two isoforms of rbcL mRNA did not always resolve in RNA gel blot analysis. When RNase J was silenced along with MRL1, plants accumulated a ladder of rbcL 5′ ends that differed little between +TAP and −TAP samples, as was the case when only RNase J was silenced. The additive effect of reducing both MRL1 and RNase J expression was a near-complete loss of −59 rbcL termini, as well as disappearance of a discrete species seen by gel-blot analysis, that migrates slightly faster than the WT transcript (Figure 4C). It is possible that this species is defined by a secondary MRL1 binding site.
To cross-check our conclusions for rbcL, we examined psbB 5′ ends. Like rbcL, psbB mRNA features closely spaced primary and processed 5′ ends, and its primary transcript is also PEP-dependent. As shown in Supplementary Figure S3, RNase J depletion leads to near-absence of the primary transcript and a ladder of 5′ ends ranging from shorter than the processed form to well upstream. RNase J, therefore, appears to be required for generating fully processed 5′ ends from both rbcL and psbB primary transcripts, an event originally assumed to be catalyzed by a single endonucleolytic cleavage (25). The possible trans-factor that protects the processed psbB 5′ end has not been identified: mutants deficient for the Arabidopsis ortholog of Mbb1, which protects psbB in Chlamydomonas (31), have RNA defects only for psbH (32).
RNase J processes psbA mRNA derived from read-through transcription of an upstream tRNA gene
Another monocistronic transcript investigated by 5′ RACE was psbA. Unlike rbcL, psbA has a single predominant form, assumed to be entirely derived from transcription initiation at its well-studied PEP promoter at −85 [(33); Figure 5A]. As shown in Figure 5B, RACE analysis of TAP-treated RNA from WT and vector control plants revealed only the expected primary transcript. When processed species were specifically amplified (minus-TAP lanes), smaller products were observed, the major ones being 50–75 bp shorter than the primary transcript. Indeed, minor 5′ ends at approximately those locations were previously noted during mapping of Arabidopsis psbA transcripts (35), and shorter fragments were also detected during RNA decay assays in isolated spinach chloroplasts (36).
Figure 5.
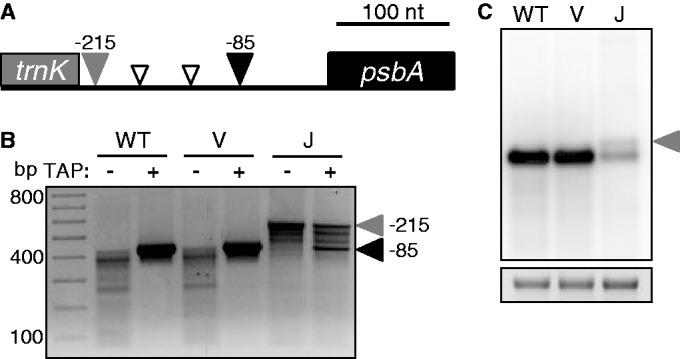
psbA 5′ end analysis. (A) Schematic of major psbA 5′ RACE products. The gray rectangle represents the 3′ end of trnK exon 2. The filled gray and black arrowheads correspond to the 5′ ends marked in (B). Open arrowheads represent 5′ ends of estimated sizes. (B) 5′ RACE products for psbA. Primary transcript initiating at the PEP promoter (filled black arrowhead) (34); read-through transcript from the upstream trnK gene (filled gray arrowhead). Positions shown are relative to the psbA translation initiation codon. (C) Strand-specific RNA gel blot analysis of psbA. The filled gray arrowhead designates psbA transcripts arising as byproducts of trnK 3′ end maturation. An ethidium bromide-stained image of 28S rRNA is shown below to represent loading.
When RNase J was depleted, none of the putative WT processed forms were detected, and only a trace amount of the primary transcript was found, which appeared to be replaced by a product 130 bp longer (Figure 5B). Loss of the primary transcript is consistent with reduced PEP activity. Similarly, RNA gel blot analysis showed relatively little of the WT-sized band, along with a somewhat less abundant and slightly larger species (Figure 5C). The lengthiest of the 5′-extended species (filled gray arrowheads in Figure 5A and B) was sequenced and found to match precisely the 3′ end of mature tRNALys, which is encoded immediately upstream and whose 3′ maturation is catalyzed by the endonuclease RNase Z (37,38). The fact that amplification of these extended transcripts is independent of TAP treatment confirms their identity as processing products rather than derivatives of a second psbA promoter. To estimate the frequency of read-through transcription, a run-on transcription assay was performed, where lysed chloroplasts were labeled for 5 min in the presence of α-32P-UTP to extend previously initiated transcripts (39). This analysis showed that transcription activity within the psbA coding region was ∼12 times higher than the activity upstream of the PEP promoter in WT chloroplasts (Supplementary Figure S4). Assuming similar stabilities of transcripts generated through either means, one can estimate that 9–10% of psbA transcripts result from read-through transcription, and there is no reason to believe that those transcripts are not functional.
DISCUSSION
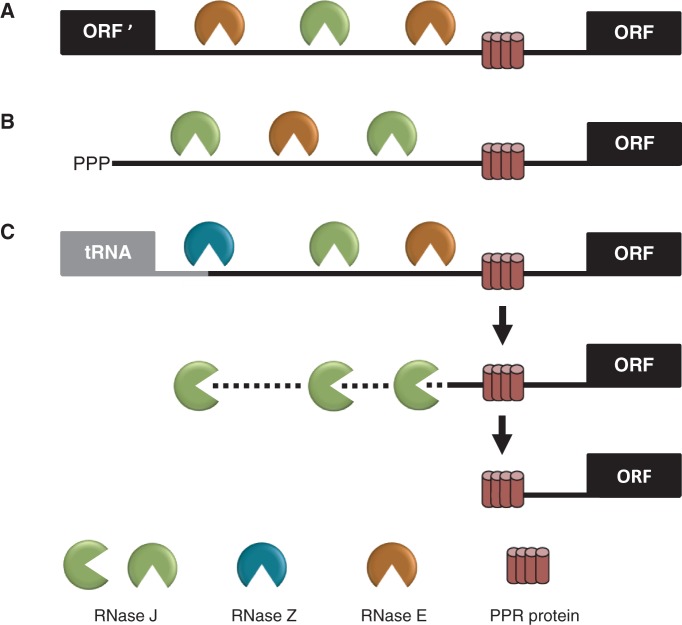
The results presented here provide evidence that chloroplast RNase J plays a major role in the 5′ maturation of mRNA. This could occur by 5′→3′ exonuclease activity following RNase J-catalyzed endonucleolytic cleavage, or RNase J could act mainly in a 5′→3′ mode following endonuclease cleavage by other enzymes such as RNase E or RNase Z. Figure 6, which builds on a previous model (13), illustrates the various modes of RNase J participation in chloroplast mRNA 5′ end maturation that are consistent with our data, depending on whether the substrates are derived from intercistronic cleavage (A), nearby transcription initiation (B) or 3′ processing of an upstream tRNA (C). Figure 6 posits that RNase J acts in partnership with other endoribonucleases, either RNase E or RNase Z. Evidence for RNase Z participation comes from results for psbA (Figure 5), whereas results for atpH, where RNase J deficiency leads to accumulation of RNAs with a range of 5′ ends mainly in predicted single-stranded regions, implicate the activity of a second endoribonuclease (Supplementary Figure S1). Concomitant silencing of RNases J and E could in principle test this hypothesis.
Figure 6.
Model for the involvement of RNase J in the processing of chloroplast mRNA 5′ ends defined by PPR proteins. Maturation of a generic mRNA (ORF) 5′ end is shown here. Precursor transcripts originate from polycistronic (A) and monocistronic (B) transcriptional units, as well as read-through transcripts from upstream genes such as tRNAs (C). Processing is initiated by endonucleolytic cleavages by RNase J or RNase E within unstructured intergenic regions, or in the case of tRNAs by RNase Z. The resultant 5′ ends are subsequently trimmed to their mature forms by the exonuclease activity of RNase J to PPR protein-bound sites.
Although the role of RNase J in 5′ end maturation is readily observed following VIGS, the associated reduction of PEP activity must be taken into account. PEP cannot be produced without chloroplast translation, and RNase J deficiency globally increases antisense RNA accumulation, which in turn impedes translation (6). Some RNAs we examined are more dependent on PEP than others, leading to varying effects on their accumulation as measured by qRT-PCR or RNA gel blot. Unlike PEP, NEP does not depend on chloroplast translation, as it lacks chloroplast-encoded subunits, and its activity generally increases in the absence of competition from PEP (40). It is thus unsurprising that one transcript more prevalent in RNase J-deficient material was atpH, which is more strongly transcribed when PEP is absent (26). As another example, atpB is transcribed by both NEP and PEP in WT tissue (41), and in plants, depleted of RNase J, the NEP-transcribed atpB persisted while PEP-initiated forms were not detected (Supplementary Figure S5).
Mechanism of 5′ maturation by RNase J: exonucleolytic processing of endonucleolytic products
A central premise of the model presented in Figure 6 is that RNase J behaves as a 5′→3′ exonuclease while trimming the 5′ ends of mRNA precisely to the borders of bound PPR proteins. A recent study revealed that recombinant PPR10 and a commercial 5′→3′ exonuclease were sufficient to generate atpH 5′ ends that match the processed PPR10-dependent termini found in vivo (17). Fine mapping has further shown that the majority of processed chloroplast RNAs in land plants may be generated by an analogous mechanism (15). In Chlamydomonas chloroplasts, RNA-binding proteins also shield mRNA from 5′→3′ exonucleolytic degradation (31,42), suggesting the mechanism of 5′ stability elements functioning as roadblocks against 5′→3′ exonucleases is well conserved. The present work in N. benthamiana shows that RNase J is required for the maturation of several mRNA 5′ ends in vivo (Figures 2–4), which likewise map precisely to the 5′ borders of predicted PPR protein-binding sites (Supplementary Figure S6).
The 5′→3′ exonuclease activity of B.subtilis RNase J1 is currently regarded as the enzyme’s predominant mode of action in vivo (43). B. subtilis possesses an RNA processing system related to that of the plant PPR protein-mediated maturation, as 5′ stability elements can protect kilobases of downstream RNA (5). However, Arabidopsis RNase J has a relatively robust endonuclease activity in vitro (6). This may suggest that in higher plants, the two primary endonucleases that initiate mRNA processing are RNase J and RNase E, with RNase Z contributing where tRNAs are present immediately upstream of protein-coding genes. Arabidopsis RNase E null mutants accumulate a few lengthy precursor mRNAs, but processed forms are still detected, and the abundance of many mRNAs is unperturbed (44,45). These findings support the notion that RNase E is not the sole endoribonuclease that processes chloroplast mRNAs. Although it is tempting to speculate that the 5′-extended mRNA species that accumulate on depletion of RNase J arise directly from RNase E cleavages, we cannot rule out that some proportion of them arise due to stalled exonucleolytic processing by residual RNase J in VIGS tissue or from activity of yet another enzyme.
Arguing against the hypothesis that RNase J carries out rate-limiting endonuclease cleavages that initiate mRNA processing is the fact that lengthy mRNA precursors were not found to accumulate in RNase J-VIGS tissue (Figures 2D, 3D, 4C and 5C; Supplementary Figures S2C and S3C). As VIGS can be assumed to only partially deplete plants of RNase J, it is plausible that residual RNase J was sufficient to mediate near-normal endonucleolytic cleavage. In this respect, recent structural analyses of B. subtilis RNase J1 (46) and Thermus thermophilus RNase J (47) suggest that complete or partial subunit dissociation is necessary for the positioning of an internal RNA segment across the enzyme’s catalytic cleft for endonucleolytic cleavage. If this is also the case for the chloroplast enzyme, diluting the concentration of RNase J by VIGS would promote greater endonuclease activity, potentially allowing proper initiation of mRNA processing even while its exonucleolytic activity was impaired.
A prevalent mechanism of chloroplast mRNA 5′ end maturation involves RNase J and PPR proteins
RNase J is involved in the maturation of mRNAs with diverse origins: atpH and petB are derived from larger transcriptional units, rbcL and psbB are primary transcripts with processed isoforms, and a previously unrecognized form of psbA results from 3′ maturation of an upstream tRNA. In addition to its role in processing sense transcripts, RNase J degrades transcriptional read-through products that are prolific in chloroplasts (6), suggesting that the enzyme operates on a global scale and with little sequence specificity. Gene-specific RNA-binding factors, such as PPR proteins, are therefore critical in stabilizing functional mRNAs. By precisely designating the ends of chloroplast RNAs that are destined for maturation, PPR proteins serve as molecular barriers against the non-specific degradation by RNase J, permitting compatibility of the enzyme’s roles in both asRNA cleanup and mRNA processing (15). Stem-loops may also serve as barriers to RNase J and define 5′ ends, although we did not demonstrate such a mechanism in this study.
Beyond the PPR proteins investigated here (PPR10, HCF152 and MRL1), many other PPR, PPR-like and sequence-specific proteins bind mRNA termini and confer stability. In Chlamydomonas, MCA1 and MCD1 protect petA and petD mRNAs from 5′→3′ exonucleolytic decay (42,48). Another chloroplast RNA-binding protein, HCF107, serves to define psbH mRNA 5′ termini by resisting 5′→3′ exonuclease degradation in Arabidopsis and Chlamydomonas (31,49,50). Recent deep sequencing of small RNAs (sRNAs) has greatly augmented the ability to identify transcripts that undergo protein-mediated processing of the type studied here because PPR or analogous proteins leave in vivo sRNA footprints (15,51). Not only do sRNAs validate the known protective functions of PPR10 and the other PPR proteins studied here but conserved sRNAs also map to the 5′ ends of psbB and psbA (Supplementary Figure S6). This suggests the involvement of as-yet uncharacterized RNA-binding proteins in 5′ end maturation of these transcripts.
The psbA case is intriguing because this mRNA has been presumed to arise uniquely from transcription initiation and thus harbor a protective triphosphate at its 5′ end; an assumption supported by the TAP-dependence of RT-PCR amplification shown in Figure 5B, and earlier data showing that the 5′ end could be labeled by a capping enzyme (52). Our data, however, suggest that some portion of psbA mRNA results from the processing of trnK read-through transcripts, and this RNA would lack a 5′ triphosphate. As all the −85 5′ ends are TAP dependent, RNase J may convert the downstream byproduct of RNase Z-catalyzed trnK 3′ maturation to the slightly shorter RNAs seen in minus-TAP lanes (Figure 5B). If one accepts, however, that the sRNA footprint results from protection of the abundant psbA mRNA 5′ end by an RNA-binding protein, then the transcripts produced by RNase J should likewise be protected. Isolation and analysis of this predicted psbA binding factor may help to resolve this ambiguity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including (53,54).
FUNDING
Binational Scientific Foundation award [2009253]; and Binational Agriculture Research and Development Foundation award [IS 4152-08]. Funding for open access charge: BSF grant listed in Funding.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Germain A, Hotto AM, Barkan A, Stern DB. RNA processing and decay in plastids. Wiley Interdiscip Rev RNA. 2013;4:295–316. doi: 10.1002/wrna.1161. [DOI] [PubMed] [Google Scholar]
- 4.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 6.Sharwood RE, Halpert M, Luro S, Schuster G, Stern DB. Chloroplast RNase J compensates for inefficient transcription termination by removal of antisense RNA. RNA. 2011;17:2165–2176. doi: 10.1261/rna.028043.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 2004;135:1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yagi Y, Hayashi S, Kobayashi K, Hirayama T, Nakamura T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One. 2013;8:e57286. doi: 10.1371/journal.pone.0057286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barkan A. Studying the structure and processing of chloroplast transcripts. Methods Mol. Biol. 2011;774:183–197. doi: 10.1007/978-1-61779-234-2_12. [DOI] [PubMed] [Google Scholar]
- 12.Choquet Y. 5′ and 3′ ends of chloroplast transcripts can both be stabilised by protein ‘caps': a new model for polycistronic RNA maturation. EMBO J. 2009;28:1989–1990. doi: 10.1038/emboj.2009.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhelyazkova P, Hammani K, Rojas M, Voelker R, Vargas-Suarez M, Borner T, Barkan A. Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 2012;40:3092–3105. doi: 10.1093/nar/gkr1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman F-A, Vallon O. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell. 2010;22:234–248. doi: 10.1105/tpc.109.066266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 20.Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP. Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol. 2006;142:21–27. doi: 10.1104/pp.106.084624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hotto AM, Huston ZE, Stern DB. Overexpression of a natural chloroplast-encoded antisense RNA in tobacco destabilizes 5S rRNA and retards plant growth. BMC Plant Biol. 2010;10:213. doi: 10.1186/1471-2229-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchive C, Yehudai-Resheff S, Germain A, Fei Z, Jiang X, Judkins J, Wu H, Fernie AR, Fait A, Stern DB. Abnormal physiological and molecular mutant phenotypes link chloroplast polynucleotide phosphorylase to the phosphorus deprivation response in Arabidopsis. Plant Physiol. 2009;151:905–924. doi: 10.1104/pp.109.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratcliff F, Martin-Hernandez AM, Baulcombe DC. Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001;25:237–245. doi: 10.1046/j.0960-7412.2000.00942.x. [DOI] [PubMed] [Google Scholar]
- 24.Madhugiri R, Evguenieva-Hackenberg E. RNase J is involved in the 5′-end maturation of 16S rRNA and 23S rRNA in Sinorhizobium meliloti. FEBS Lett. 2009;583:2339–2342. doi: 10.1016/j.febslet.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 25.Westhoff P, Herrmann RG. Complex RNA maturation in chloroplasts: the psbB operon from spinach. Eur. J. Biochem. 1988;171:551–564. doi: 10.1111/j.1432-1033.1988.tb13824.x. [DOI] [PubMed] [Google Scholar]
- 26.Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM. Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 2002;31:171–188. doi: 10.1046/j.1365-313x.2002.01349.x. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura T, Meierhoff K, Westhoff P, Schuster G. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 2003;270:4070–4081. doi: 10.1046/j.1432-1033.2003.03796.x. [DOI] [PubMed] [Google Scholar]
- 28.Mullet J, Orozco E, Chua NH. Multiple transcripts for higher plant rbcL and atpB genes and localization of the transcription initiation sites of the rbcL gene. Plant Mol. Biol. 1985;4:39–54. doi: 10.1007/BF02498714. [DOI] [PubMed] [Google Scholar]
- 29.Serino G, Maliga P. RNA polymerase subunits encoded by the plastid rpo genes are not shared with the nucleus-encoded plastid enzyme. Plant Physiol. 1998;117:1165–1170. doi: 10.1104/pp.117.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allison LA, Simon LD, Maliga P. Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 1996;15:2802–2809. [PMC free article] [PubMed] [Google Scholar]
- 31.Vaistij FE, Goldschmidt-Clermont M, Wostrikoff K, Rochaix JD. Stability determinants in the chloroplast psbB/T/H mRNAs of Chlamydomonas reinhardtii. Plant J. 2000;21:469–482. doi: 10.1046/j.1365-313x.2000.00700.x. [DOI] [PubMed] [Google Scholar]
- 32.Felder S, Meierhoff K, Sane AP, Meurer J, Driemel C, Plucken H, Klaff P, Stein B, Bechtold N, Westhoff P. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell. 2001;13:2127–2141. doi: 10.1105/TPC.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruissem W, Zurawski G. Analysis of promoter regions for the spinach chloroplast rbcL, atpB and psbA genes. EMBO J. 1985;4:3375–3383. doi: 10.1002/j.1460-2075.1985.tb04093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi K, Shiina T, Ishii N, Iwai K, Ishizaki Y, Morikawa K, Toyoshima Y. A role of the -35 element in the initiation of transcription at psbA promoter in tobacco plastids. Plant Cell Physiol. 2003;44:334–341. doi: 10.1093/pcp/pcg041. [DOI] [PubMed] [Google Scholar]
- 35.Shen Y, Danon A, Christopher DA. RNA binding-proteins interact specifically with the Arabidopsis chloroplast psbA mRNA 5′ untranslated region in a redox-dependent manner. Plant Cell Physiol. 2001;42:1071–1078. doi: 10.1093/pcp/pce142. [DOI] [PubMed] [Google Scholar]
- 36.Klaff P. mRNA decay in spinach chloroplasts: psbA mRNA degradation is initiated by endonucleolytic cleavages within the coding region. Nucleic Acids Res. 1995;23:4885–4892. doi: 10.1093/nar/23.23.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiffer S, Rosch S, Marchfelder A. Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J. 2002;21:2769–2777. doi: 10.1093/emboj/21.11.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugita M, Shinozaki K, Sugiura M. Tobacco chloroplast tRNA(UUU) gene contains a 2.5-kilobase-pair intron: An open reading frame and a conserved boundary sequence in the intron. Proc. Natl Acad. Sci. USA. 1985;82:3557–3561. doi: 10.1073/pnas.82.11.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng XW, Stern DB, Tonkyn JC, Gruissem W. Plastid run-on transcription: Application to determine the transcriptional regulation of spinach plastid genes. J. Biol. Chem. 1987;262:9641–9648. [PubMed] [Google Scholar]
- 40.Emanuel C, Weihe A, Graner A, Hess WR, Borner T. Chloroplast development affects expression of phage-type RNA polymerases in barley leaves. Plant J. 2004;38:460–472. doi: 10.1111/j.0960-7412.2004.02060.x. [DOI] [PubMed] [Google Scholar]
- 41.Schweer J, Loschelder H, Link G. A promoter switch that can rescue a plant sigma factor mutant. FEBS Lett. 2006;580:6617–6622. doi: 10.1016/j.febslet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 42.Loiselay C, Gumpel NJ, Girard-Bascou J, Watson AT, Purton S, Wollman FA, Choquet Y. Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 2008;28:5529–5542. doi: 10.1128/MCB.02056-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Condon C. What is the role of RNase J in mRNA turnover? RNA Biol. 2010;7 doi: 10.4161/rna.7.3.11913. [DOI] [PubMed] [Google Scholar]
- 44.Walter M, Piepenburg K, Schöttler MA, Petersen K, Kahlau S, Tiller N, Drechsel O, Weingartner M, Kudla J, Bock R. Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J. 2010;64:851–863. doi: 10.1111/j.1365-313X.2010.04377.x. [DOI] [PubMed] [Google Scholar]
- 45.Stoppel R, Manavski N, Schein A, Schuster G, Teubner M, Schmitz-Linneweber C, Meurer J. RHON1 is a novel ribonucleic acid-binding protein that supports RNase E function in the Arabidopsis chloroplast. Nucleic Acids Res. 2012;40:8593–8606. doi: 10.1093/nar/gks613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman JA, Hewitt L, Rodrigues C, Solovyova A, Harwood CR, Lewis RJ. Unusual, Dual Endo- and Exonuclease Activity in the Degradosome Explained by Crystal Structure Analysis of RNase J1. Structure. 2011;19:1241–1251. doi: 10.1016/j.str.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 47.Dorleans A, Li de la Sierra-Gallay I, Piton J, Zig L, Gilet L, Putzer H, Condon C. Molecular basis for the recognition and cleavage of RNA by the bifunctional 5′-3′ exo/endoribonuclease RNase J. Structure. 2011;19:1252–1261. doi: 10.1016/j.str.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 48.Drager RG, Girard-Bascou J, Choquet Y, Kindle KL, Stern DB. In vivo evidence for 5′-3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J. 1998;13:85–96. doi: 10.1046/j.1365-313x.1998.00016.x. [DOI] [PubMed] [Google Scholar]
- 49.Hammani K, Cook WB, Barkan A. RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl Acad. Sci. USA. 2012;109:5651–5656. doi: 10.1073/pnas.1200318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaistij FE, Boudreau E, Lemaire SD, Goldschmidt-Clermont M, Rochaix JD. Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 2000;97:14813–14818. doi: 10.1073/pnas.97.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruwe H, Schmitz-Linneweber C. Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic Acids Res. 2012;40:3106–3116. doi: 10.1093/nar/gkr1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boyer SK, Mullet JE. Characterization of P. sativum chloroplast psbA transcripts produced in vivo, in vitro and in E. coli. Plant Mol. Biol. 1986;6:229–243. doi: 10.1007/BF00015229. [DOI] [PubMed] [Google Scholar]
- 53.Zhelyazkova P, Sharma CM, Forstner KU, Liere K, Vogel J, Borner T. The Primary Transcriptome of Barley Chloroplasts: Numerous Noncoding RNAs and the Dominating Role of the Plastid-Encoded RNA Polymerase. Plant Cell. 2012;24:123–136. doi: 10.1105/tpc.111.089441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka M, Obokata J, Chunwongse J, Shinozaki K, Sugiura M. Rapid splicing and stepwise processing of a transcript from the psbB operon in tobacco chloroplasts: Determination of the intron sites in petB and petD. MGG. 1987;209:427–431. doi: 10.1007/BF00331145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.