Abstract
The role of the mRNA-binding protein human antigen R (HuR) in stabilization and translation of AU-rich elements (ARE) containing mRNAs is well established. However, the trafficking of HuR and bound mRNA cargo, which comprises a fundamental requirement for the aforementioned HuR functions is only poorly understood. By administering different cytoskeletal inhibitors, we found that the protein kinase Cδ (PKCδ)-triggered accumulation of cytoplasmic HuR by Angiotensin II (AngII) is an actin-myosin driven process functionally relevant for stabilization of ARE-bearing mRNAs. Furthermore, we show that the AngII-induced recruitment of HuR and its bound mRNA from ribonucleoprotein particles to free and cytoskeleton bound polysomes strongly depended on an intact actomyosin cytoskeleton. In addition, HuR allocation to free and cytoskeletal bound polysomes is highly sensitive toward RNase and PPtase and structurally depends on serine 318 (S318) located within the C-terminal RNA recognition motif (RRM3). Conversely, the trafficking of the phosphomimetic HuRS318D, mimicking HuR phosphorylation at S318 by the PKCδ remained PPtase resistant. Co-immunoprecipitation experiments with truncated HuR proteins revealed that the stimulus-induced association of HuR with myosin IIA is strictly RNA dependent and mediated via the RRM3. Our data implicate a microfilament dependent transport of HuR, which is relevant for stimulus-induced targeting of ARE-bearing mRNAs from translational inactive ribonucleoprotein particles to polysomes.
INTRODUCTION
The embryonic lethal abnormal vision (ELAV)-like protein human antigen R (HuR) is a ubiquitously expressed member of a highly conserved RNA-binding protein family functionally involved in the stabilization of AU-rich element (ARE)-bearing mRNAs (1–3). In addition to its originally described role as a mRNA stability factor, HuR functions have expanded to many other aspects of mRNA processing including splicing, polyadenylation, translation and modulation of miRNA repression (4–6). HuR is implicated in key functions of cells including proliferation, differentiation, apoptosis and migration. Consequently, perturbations of physiologic HuR regulation play a causative role in many pathologic states (7–9). Although HuR is most abundantly localized within the cell nucleus, its posttranscriptional impact on target mRNA is tightly linked to the translocation of HuR from the nucleus to the cytoplasm and backwards, which is structurally related to a HuR nucleo-cytoplasmic shuttling (HNS) domain residing in the basic hinge region of the protein (3,10). Published data implicate that phosphorylation of HuR by different protein kinases including checkpoint kinase 2 (11), cyclin-dependent kinase 1 (12), protein kinase Cα (PKCα) (13), PKCβ (14) and PKCδ (15,16) can alter subcellular localization of HuR and posttranscriptional effects by HuR. Mapping of potential PKC phosphorylation sites in HuR revealed several serines targeted by PKC (13). Importantly, we found that PKCδ through tandem phosphorylation of different HuR domains coordinates target mRNA binding within the nucleus and export of HuR to the cytoplasm (16). Based on previously published work, HuR distribution between the nucleus and the cytoplasm is mediated by different transport receptors including transportin 1 and 2, importin α1 and chromosome maintenance region 1 /exportin 1 (17–20). However, the mechanisms underlying the cytoplasmic trafficking of HuR and its cargo mRNA are far less understood.
The intracellular mRNA transport is mediated by microfilaments together with their corresponding motor proteins including kinesins, dyneins and myosins (21–24). In contrast to microtubule-dependent transport, which is assumed as the major trafficking system for mRNA in oocytes and axons, the actin-myosin system seems more relevant in the short-distance mRNA transport in non-neuronal cells (22). Importantly, the contact of mRNA with the cytoskeleton in most cases is indirect and mediated by RNA-binding proteins that target specific cis-regulatory elements in the 3′UTR of mRNA as has been convincingly demonstrated for the interaction of the Drosophila mRNA-binding protein Staufen with oskar mRNA (25). Results from several studies suggest an involvement of neuronal ELAV proteins in the microtubule-mediated mRNA transport in axons (26,27). In contrast, a comparable role of HuR in cytoskeleton-guided mRNA transport has not been shown so far. Here, we aimed to analyze the functional role of cytoskeletal elements in the AngII-induced nucleo-cytoplasmic HuR redistribution in human renal mesangial cells (HMC). We unveiled that the actin–myosin cytoskeleton is essential for the nuclear export and transport of HuR to the translationally active polysomes. We furthermore demonstrate that myosin IIA is able to associate with HuR only if phosphorylated at S318 via the C-terminal RRM3. Our data suggest that dynamic interactions with the cytoskeleton play a critical role in the spatio-temporal control of mRNA transport and translation by the ubiquitous mRNA-binding protein HuR.
MATERIALS AND METHODS
Reagents
Latrunculin-A and colchicine were obtained from Calbiochem (Schwalbach, Germany). Actinomycin D (from Streptomyces species), human angiotensin II, blebbistatin, 4′, 6′-diamidino-2-phenylindole (DAPI), phalloidin-rhodamine, MG-132, the anti-Flag antibody and a secondary rhodamin coupled anti-mouse antibody were purchased from Sigma-Aldrich (Deisenhofen, Germany). Ribonucleotides and modifying enzymes were from Life Technologies (Karlsruhe, Germany). Antibodies raised against HuR, histone deacetylase-1 (HDAC1), c-myc, anti-rabbit and anti-mouse horseradish peroxidase linked immunoglobulin G (IgG) were purchased from Santa Cruz Biotechnology (Heidelberg, Germany). Antibodies raised against green fluorescent protein (GFP), α-actinin, myosin IIA, phospho-p38 MAPK and p38 MAPK were obtained from Cell Signaling (Frankfurt am Main, Germany). The secondary Alexa-Fluoro 488-coupled and Cy3 antibodies were from Molecular Probes (Karlsruhe, Germany). Protein G sepharose, the ECL system and Hyperfilm were from GE Healthcare (München, Germany). All cell culture media and supplements were purchased from Life Technologies (Karlsruhe, Germany).
Cell culture
Human primary mesangial cells (HMC) were isolated from collagenase IV-treated human glomeruli and cultivated as described previously (28). Serum-free preincubations were performed in Dulbecco’s modified Eagle’s medium supplemented with 0.1 mg of fatty acid-free bovine serum albumin/ml. For experiments, HMC were grown to 80% confluency before stimulated.
RNA interference
Gene silencing was performed by using the FlexiTube small interfering RNA (siRNA) for human non-muscle myosin IIA heavy chain 9 (siRNA-MYH9) from Qiagen (Hilden, Germany), HuR-specific siRNAs from Santa Cruz Biotechnology and a control siRNA from Dharmacon (Fermentas, St. Leon-Rot, Germany). siRNA transfection of subconfluent HMC was performed by using the Oligofectamine reagent (Invitrogen, Karlsruhe, Germany) according to the manufacturer’s instructions.
Separation of polysomes from translational inactive ribonucleoprotein particles
Trypsinized cells were collected by centrifugation for 5 min at 5000g at 4°C before cell pellets were mixed with two volumes of ice-cold lysis buffer [20 mM HEPES, 250 mM sucrose, 250 mM KCl, 5 mM MgCl2, 2 mM dithiothreitol, 1 mg/ml heparine (pH 7.5)] and homogenized by freezing and thawing in liquid nitrogen followed by centrifugation at 4000g for 10 min at 4°C. Supernatants were supplemented with Triton X-100 (final 1%) and 40 U/ml RNaseOUT (Invitrogen, Karlsruhe, Germany) and equal volumes loaded (0.5 ml) onto a sucrose cushion (1 M) and polysomes isolated by centrifugation at 100 000g for 2 h at 4°C. Polysomal pellets were dissolved in polysomal lysis buffer [10 mM Tris–HCl, 100 mM NaCl, 10 mM EDTA, 1% SDS (pH 7.4)], whereas the supernatants were further centrifuged at 300 000g for 3 h at 4°C for isolation of postpolysomal messenger ribonucleoproteins particles (mRNP) fractions, which were also dissolved in polysomal lysis buffer. To control the relative purity of the separated fractions, aliquots from both fractions were monitored by optical density measurement (A254).
Polysomal fractionation
The isolation of different polysomal fractions was performed by using a protocol published by Hovland et al. (29). Thereby, cells were homogenized in 1 ml of ice-cold lysis buffer I [10 mM Tris–HCl (pH 7.6), 0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 0.5 mM CaCl2, 0.5% NP40] and cell lysates sedimented by centrifugation at 1000g for 5 min with supernatants contain free polysomal fractions. The remaining pellet including cell nuclei, insolube cell membranes and cytoskeletal proteins was washed with lysis buffer I before cytoskeletal-bound polysomes (CBP) were isolated by the addition of lysis buffer II [10 mM Tris–HCl (pH 7.6), 0.25 M sucrose, 130 mM KCl, 5 mM MgCl2, 0.5 mM CaCl2, 0.05% NP40]. Lysates were again centrifugated at 2000g for 5 min to separate CBP accumulated in the supernatants, from membrane-bound polysomes (MBP), which were enriched in the pellets after being resuspended in buffer III containing 10 mM Tris–HCl (pH 7.6), 0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 0.5 mM CaCl2, 0.5% NP40 and 0.5% deoxycholate. After incubation for 10 min on ice, MBPs were collected by a final centrifugation step at 3000 × g. The activity of lactate dehydrogenase, which is preferentially detected in the fraction of free polysomes, was quantified by using the CytoTox96 Nonradioactive Cytotoxicity Assay (Promega, Mannheim, Germany) and normalized to protein concentrations of corresponding samples.
PPtase and RNase treatments of polysomal fractions
To assess the possible role of RNA and phosphorylation in polysomal HuR distribution, a portion (0.3 ml) of the cell homogenates, which had been lyzed in buffer I, before centrifugation were incubated with 1U protein phosphatase 1 (Roche, Mannheim, Germany) for 30 min at 30°C before the polysomal fractionation was continued. Correspondingly, another portion of the cell homogenate was treated with 10 µg RNase A/ 25U T1 for 10 min at 37°C and, subsequently, polysomal fractions were prepared as described earlier in the text.
Construction of eukaryotic expression plasmids
The plasmid pEGFP-PKCδ containing a C-terminal fusion of full-length human PKCδ was generated according to a strategy described previously (30).
Flag-tagged recombinant wild-type HuR (pCMV-Flag-HuRwt) was generated by subcloning the pQE30-His-wild-type HuR plasmid DNA into the eukaryotic pCMV-Flag-N3 expression vector as described previously (15). The plasmid pCMV-Flag-Ser318DHuR, which encompasses a cDNA encoding a phosphomimetic HuR mutant with a serine to aspartate substitution at amino acid position 318, was generated as described previously (31), and pCMV-Flag-Ser318AHuR was generated as described previously (16). Plasmids bearing wild-type or, alternatively, different c-myc-tagged HuR deletions were constructed by subcloning a cDNA fragment containing the coding region of HuR plus the myc-tag into HindIII/EcoRI cut pcDNA3.1 vector (Invitrogen) by using the plasmid pTet-Myc-HuR as a template (32) and are further described in Supplementary Methods. Point mutations and proper in-frame insertion of cDNAs was proven by DNA sequencing. HMC were transiently transfected with different c-myc-tagged cDNA constructs by using the Lipofectamine reagent (Invitrogen). Transfection of pCMV-Flag constructs was done using Effectene (Qiagen, Hilden, Germany) by following the manufacturer’s protocol.
Immunoprecipitation-qRT-PCR analysis
HuR-bound mRNA was isolated by Immunoprecipitation (IP)-qRT-PCR analysis (pull-down RT assay) as reported previously (15).
RNA isolation and RT-PCR analysis
Total RNA was extracted from whole-cell lysates by using the Tri-reagent (Sigma-Aldrich). First strand cDNA was synthesized using random hexamer primer (Fermentas) and Superscript reverse transcriptase (Life Technologies). Primers used for PCR are listed in the Supplementary Methods section. Semiquantitative PCRs were performed with 28 cycles (30 s at 95°C, 30 s at 60°C and 60 s at 72°C).
Western blot analysis
Nuclear and cytoplasmic extracts from HMC were prepared following to a protocol described by Schreiber et al. (33). To confirm an equal loading of nuclear or cytoplasmic proteins, the blots were additionally probed with either an antibody raised against HDAC1 or with a β-actin specific antibody. Total protein lysates were prepared by standard procedures. Proteins were detected by using specific primary and the appropriate secondary antibodies and visualized with chemiluminescence using an ECL system from GE Healthcare.
IP
IP was performed as described previously (16) and is described in more detail in the Supplementary Methods section.
Blot overlay assay
Blot overlay was performed by using a modified protocol from Côté and Richard (34). For blot overlay assay experiments, 300 µg of total cell lysates derived from untreated HMC were subjected to myosin II IP as described. Subsequently, the myosin IIA-precipitated proteins were separated by SDS–PAGE and transferred to polyvinylidene difluoride membranes. After blocking, the membrane was incubated for 16 h at 4°C with total cell lysates (0.2 mg/ml) derived from vehicle- or AngII-treated HMC overexpressing either c-myc tagged HuRwt (c-myc-HuRwt) or c-myc-tagged HuRΔHinge (c-myc-HuRΔHinge) proteins. After several washing steps with TBST, myosin-bound c-myc fusion proteins were detected by immunoblotting by using an anti-c-myc specific antibody.
Fluorescence microscopy
Cells were grown on coverslips at 60–80% confluence and rendered serum-free for 16 h before stimulated for the indicated periods. Immediately following treatment, cells were rinsed once with phosphate buffered saline (PBS) and subsequently fixed for 20 min at RT with 4% paraformaldehyde in PBS, before cells were blocked and permeabilized in 2% BSA plus 0.1% Triton X-100 (PBS) at RT. Subsequently, the fixed cells were stained with the indicated primary antibodies for 30 min at RT, before cells were incubated with a secondary Alexa Fluor 488-or rhodamine-coupled antibody for 1 h at RT. F-actin was visualized with rhodamine-conjugated phalloidin, and cell nuclei were counterstained with DAPI before mounted on slides by using Fluoromount-G (Southern Biotech, Eching, Germany).
Cells were imaged with a inverted confocal microscope (LSM 510) from Zeiss (Göttingen, Germany) by using a Plan Apochromat 40×/1.4 oil-immersion objective and 488, 543 and 633 nm laser lines, respectively. Image acquisition was performed with the ZEN2009 Light Edition software (Carl Zeiss, Inc).
Statistical analysis
Results are depicted as mean values ± standard deviation (SD). The data are presented as x-fold induction compared with untreated conditions (*) or to stimulated values (#). Statistical analysis was performed using Student’s t-test and analysis of variance with the Bonferroni multicomparison test. P < 0.05 (*), (#) or P < 0.01 (**), (##) or P < 0.005 (***), (###) were considered significant.
RESULTS
The stimulus-dependent increase in cytoplasmic HuR by AngII depends on the actin-myosin cytoskeleton
To address the functional role of the cytoskeleton in HuR shuttling induced by the vasoactive hormone AngII in renal HMC (15,16), we tested the effects of two well-established cytoskeleton inhibitors specifically interfering with the integrity of microfilaments (latrunculin A) or microtubules (colchicine). Western blot analysis revealed a strong increase in cytoplasmic HuR abundance on stimulation with AngII (0.1 µM) (Figure 1A, left panel). Interestingly, a short preincubation of cells for 30 min with latrunculin A (0.1 µM), which prevents F-actin polymerization, completely inhibited cytoplasmic HuR accumulation by AngII, whereas colchicine (0.1 mg/ml) had no effects on cytoplasmic HuR levels (Figure 1A, left panel). In contrast, the nuclear and total content of HuR remained unchanged, independent of which inhibitor was applied (Supplementary Figure S1). As actin-dependent transport of mRNPs in many cases depends on the molecular motor protein myosin (22), we furthermore tested whether inhibition of myosin activity could similarly impair the cytoplasmic HuR accumulation by AngII. For this purpose, we tested the effects of blebbistatin (5 µM), a small molecule inhibitor with a reported high selectivity toward the non-muscle myosin IIA ATPase (35). To achieve a maximal inhibitory effect on enzymatic activity, blebbistatin instead of 30 min, was preincubated for 4 and 16 h, respectively. Thereby, we observed significant inhibitory effects on AngII-induced HuR by blebbistatin at both incubation periods but most clearly seen at 16 h (Figure 1A, right panel). Interestingly, in contrast to its inhibitory effect on the AngII-induced cytoplasmic HuR accumulation, blebbistatin increased the content of cytoplasmic HuR when given alone, and this effect was most clearly seen at 16 h (Figure 1A, right panel). We assume that the stimulatory effect on HuR shuttling by blebbistatin is independent of myosin inhibition and likely due to a drug-dependent induction of stress responses as indicated by the rapid increase in p38 MAPK phosphorylation (Supplementary Figure S2A). Importantly in HMC, the p38 MAPK pathway is functionally irrelevant for cytoplasmic HuR accumulation by AngII (15). In addition, colchicine that had no inhibitory effect on the AngII-induced nucleo-cytoplasmic HuR shuttling (Figure 1A), clearly reduced the cytoplasmic HuR accumulation by blebbistatin (Supplementaryl Figure S2B), thus indicating that the blebbistatin-triggered HuR redistribution depends on a microtubuli-dependent transport mechanism. To verify the results from pharmacological myosin inhibition, we additionally tested the impact of myosin by siRNA depletion. We decided to deplete the heavy chain 9 of non-muscle myosin IIA (MYH9), as it is known to mediate many cellular processes by myosin IIA (36). In agreement with the long-term inhibition of cytoplasmic HuR accumulation, we observed a total blockade in cytoplasmic HuR accumulation (Figure 1B). In contrast, the blebbistatin-induced cytoplasmic HuR accumulation observed at 16 h was not affected by myosin IIA depletion (Supplementary Figure S2C), which further substantiates our finding that the stimulatory activity of HuR export by blebbistatin is independent of myosin IIA. As phosphorylation and subsequent export of nuclear HuR to the cytoplasm in response to the octapeptide AngII depends on a prior PKCδ import to the nucleus (15), we furthermore tested whether the inhibitory effects of latrunculin A on cytoplasmic HuR accumulation is possibly indirect and due to an inhibition of PKCδ translocation. For this purpose, we monitored the cytoplasmic HuR accumulation after ectopical expression of PKCδ (16). Notably, HMC displayed a clear increase in cytoplasmic HuR content 24 h after transient transfection of GFP-PKCδ, whereas cells that were co-treated in the presence of latrunculin A showed a strong reduction in cytoplasmic HuR levels. As expected, colchicine had no effect on cytoplasmic HuR levels induced by PKCδ overexpression (Figure 1C).
Figure 1.
AngII-induced and PKCδ-mimicked cytoplasmic HuR accumulation depend on the actin-myosin cytoskeleton. (A, left panel). Cells were serum starved for 16 h before stimulated for 2 h with vehicle (−) or AngII (0.1 µM) without or with either latrunculin A (Lat. A) or colchicine (Colch.) as indicated. Both inhibitors were preincubated for 30 min before the addition of AngII. Protein lysates (20 µg) of cytoplasmic fractions were subjected to SDS–PAGE and successively immunoblotted with anti-HuR and anti-β-actin antibodies. (A, right panel). Western blot analysis demonstrating a time-dependent inhibitory effect of blebbistatin on cytoplasmic HuR levels. HMC were treated for the indicated time points with blebbistatin (Blebbistat.) before cells were additionally stimulated for 2 h with vehicle (−) or AngII as indicated. Data summerize densitometric analysis of cytoplasmic HuR relative to β-actin levels and represent means ± SD. (n = 3). **P ≤ 0.01, ***P ≤ 0.005 compared with control. ##P ≤ 0.01 and ###P ≤ 0.005 versus AngII-treated conditions. (B). Serum starved HMC were transfected for 72 h with duplex siRNA of human myosin heavy chain 9 of myosin IIA (siRNA-MYH9) or with control siRNA (siRNA-control). After transfection, cells were serum starved for 16 h before treated for further 2 h with vehicle or with AngII (0.1 µM). Thereafter, cells were lyzed for either total or cytoplasmic and nuclear protein extracts. The levels of myosin IIA and HuR protein in total cell lysates and cytoplasmic extracts were monitored by western blot analysis with corresponding antibodies. Nuclear fractions are characterized by the high contents of HDAC1, which was not detectable in cytoplasmic cell lysates. Cytoplasmic extracts are characterized by positive β-actin staining. (C). HMC cells were transfected with a GFP-tagged PKCδ expression plasmid (PKCδ-GFP), or with the corresponding empty vector (mock transfected). 72 h after transfection, cells were starved for 16 h and preincubated for additional 30 min with latrunculin A or with colchicine as indicated. Thereafter, duplicates were extracted for either total, nuclear or cytoplasmic extracts, respectively. The contents of GFP-tagged PKCδ and HuR in the different fractions were monitored by Western blot analysis using corresponding antibodies. To correct for variations in protein loading, the blots were stripped and additionally monitored for β-actin (cytoplasmic) or HDAC1 (nuclear) contents. The data shown are from a single experiment representative for two independent experiments giving similar results.
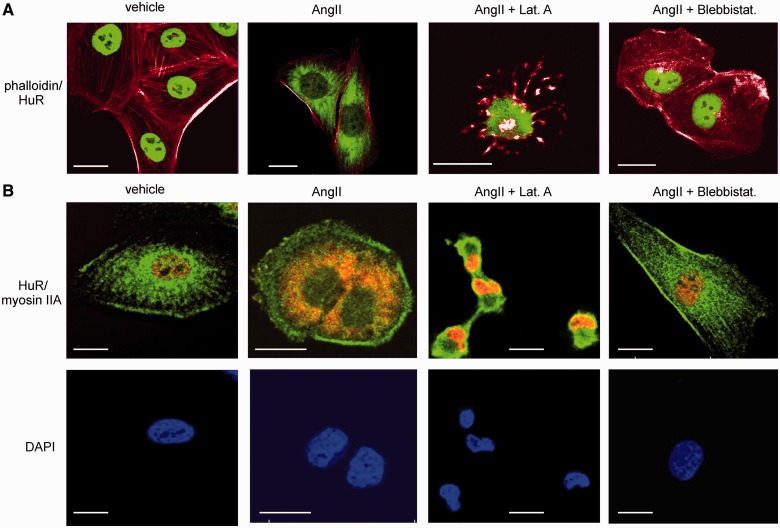
Concordant with the data from Western blotting, the visualization of intracellular HuR distribution by confocal microscopy shows an intense staining of HuR exclusively in the nuclei of untreated HMC but a strong extension of HuR staining to cytoplasmic regions in cells that were exposed to AngII for 2 h (Figure 2A and B). Notably, in contrast to the results from western blot analysis, the nuclear HuR signal was strongly diminished on AngII treatment (Figure 2A and B). Concomitant with the strong disruption of microfilaments, the AngII-induced increase in cytosolic HuR was totally impaired if cells were pretreated with latrunculin (Figure 2). The strong shrinkage that was observed in many cells and nuclei after administration of latrunculin is expected and results from the impaired tension of the cytoskeleton after disruption of F-actin filaments (Figure 2A). Similar to latrunculin A, blebbistatin impaired cytoplasmic HuR abundance by AngII when used for longer preincubations (Figure 2). Importantly, none of these inhibitors had any effect on the high basal HuR abundance in cell nuclei (Supplementary Figure S3).
Figure 2.
Disruption of the actin-myosin cytoskeleton impairs nucleo-cytoplasmic HuR shuttling by AngII. (A). Confocal microscopy was applied to visualize changes in the actin–myosin cytoskeleton by chemical inhibitors and nucleo-cytoplasmic redistribution of HuR. Serum starved HMC were treated for 2 h with vehicle or AngII (0.1 µM) in the absence or presence of either latrunculin A (0.1 µM) or blebbistatin (5 µM), which were preincubated for 30 min (Lat. A) or 4 h (Blebbistat.), respectively, before the addition of AngII. After fixation and permeabilization, cells were stained with an anti-HuR antibody and subsequently with the Alexa-Fluoro 488 coupled (green) secondary antibody. F-actin was visualized with rhodamine-conjugated phalloidin with merged pictures showing HuR plus F-actin staining. Cell nuclei were visualized by staining with DAPI (4′,6′-DAPI). Bars, 20 µm. (B). Cells were treated as in (A), but simultaneously immunostained for myosin IIA (green), HuR (red) using either Alexa-Fluoro 488 or Cy3 coupled secondary antibodies. Additionally, cells were stained with DAPI (blue). Bars, 25 µm. Data shown are from a single experiment representative of two repeats giving similar results.
Modulatory effects on HuR-triggered mRNA expression by cytoskeletal inhibitors
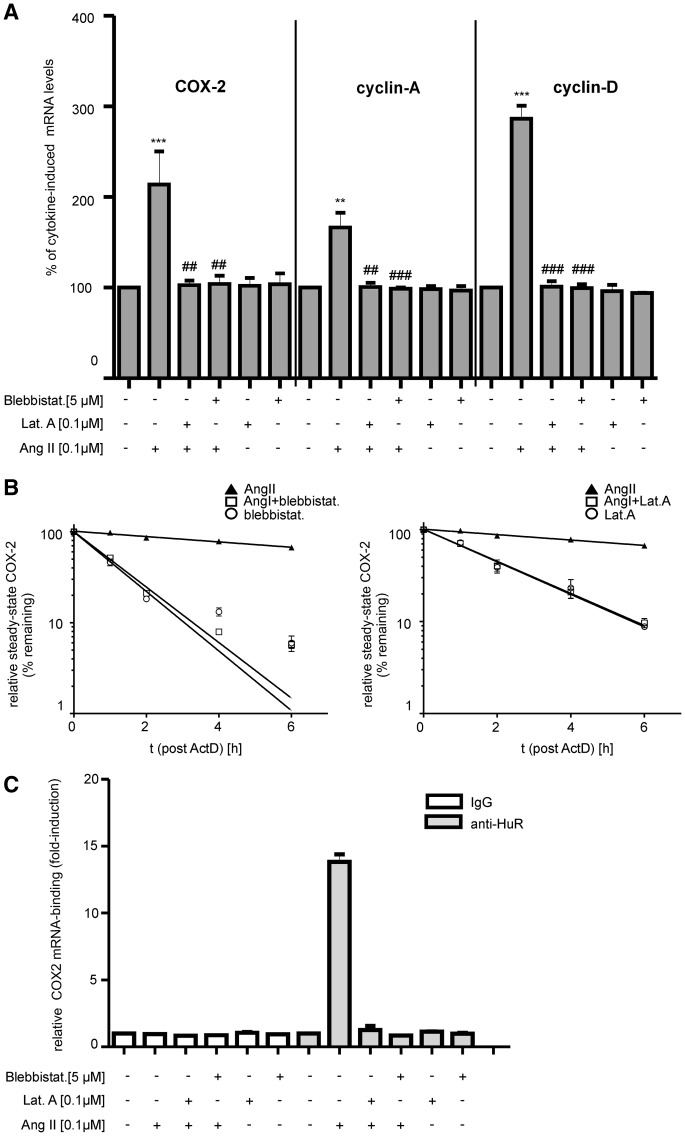
Next, we tested whether an inhibition of cytoplasmic HuR by microfilament inhibitors is functionally relevant for the expression of the HuR target mRNAs COX-2, cyclin A and cyclin D1, which is similarly increased by AngII via HuR-dependent stabilization of corresponding mRNAs (16). To induce COX-2 mRNA, cells were pretreated with TNFα and IL-1β (both at 2 nM) for 16 h before AngII was administered for further 2 h. In full accordance with our previous findings (16), AngII caused a substantial increase in cytokine-induced levels of COX-2, cyclin A and cyclin D1 encoding mRNAs (Figure 3A). The increased abundance of these mRNAs by AngII was totally blocked when cells were treated in the presence of latrunculin A or blebbistatin (Figure 3A). Furthermore, monitoring the time-course of mRNA decay with actinomycin D, demonstrated that the stimulatory effects of AngII on the cytokine-induced COX-2 mRNA were strongly antagonized by both inhibitors (Figure 3B). In particular, AngII caused a substantial raise in COX-2 mRNA stability, increasing the half-life from almost 3 to >8 h when compared with GAPDH mRNA levels (15). Importantly, the AngII-evoked increase in COX-2 mRNA half-life significantly dropped from ≥8 to ≤2 h if blebbistatin was additionally applied to the cells (Figure 3B, left panel). Correspondingly, a strong reduction in COX-2 mRNA half-life (from ≥8 h to ≤2.5 h) was observed if cells were treated in the presence of latrunculin A (Figure 3B, right panel). In contrast, the half-life of COX-2 mRNA was below 3 h when cells were treated with either of the mentioned inhibitors alone (Figure 3B, open circles). Concomitant with their strong suppressive effects on COX-2 mRNA-stability, both compounds totally impaired HuR binding to COX-2 mRNA as analyzed by pull-down RT-PCR assay (Figure 3C).
Figure 3.
Microfilament inhibitors suppress the AngII-mediated increase in COX-2 expression by preventing HuR binding to the corresponding mRNA. (A). HMC cells were treated for 2 h with either vehicle (−) or with AngII, or together with different microfilament inhibitors as indicated. Inhibitors were preincubated for 30 min (Lat.A) or 4 h (Blebbistat.) before the addition of AngII. To achieve transcriptional induction of different HuR target genes, cells were stimulated for 16 h with a cytokine mix containing IL-1β and TNFα (both at 2 nM) before AngII treatment. Steady-state mRNA levels of COX-2, cyclin A and cyclin D1 were quantified by qRT-PCR using β-actin as a normalization control. Results are depicted as relative mRNA levels (%) in comparison with cytokine-stimulated conditions (vehicle), which were set to 100% and are means ± SD (n=3). **P ≤ 0.01, ***P ≤ 0.005 compared with cytokine-induced conditions ##P ≤ 0.01 and ###P ≤ 0.005, compared with cytokine plus AngII-treated conditions. (B). Reduction of AngII-mediated stabilization of cytokine-triggered COX-2 mRNA by blebbistatin (left panel) or latrunculin A (right panel). Quiescent HMC were treated for 20 h with a cytokine mixture containing IL-1β and TNFα (both at 2 nM) before the administration of Act D, which was added 30 min before AngII (filled triangles). Importantly, the indicated cytoskeletal inhibitors were given 4.5 h before the addition of AngII and remained for the indicated times with vehicle (open circles) or with 0.1 µM AngII (open squares) before cells were harvested and extracted for total cellular RNA. Steady-state mRNA levels of COX-2 were determined by qRT-PCR using β-actin mRNA as a normalization control. Graphs show means ± SD (n = 3) and depict the percentage of remaining COX-2 mRNA levels compared with the content of steady-state COX-2 mRNA measured immediately before the addition of Act D (0 h). (C). HMC were treated similarly as described in (A) before total cell lysates were assayed by IP using an anti-HuR antibody (gray bars), or the same amount of isotypic IgG (open bars). HuR-bound COX-2 mRNA was determined by pulldown RT assays. Data shown are means of two independent experiments and are depicted as fold induction versus cytokine-treated cells.
AngII-induced association of HuR with free and cytoskeletal bound polysomes is impaired after microfilament inhibition
An increase in cytoplasmic HuR in many cases is functionally equivalent with a recruitment of HuR-bound target mRNAs to the translation apparatus. Three major sites of translation can be distinguished including free cytoplasmic polysomes (FP), CBP and, finally, polysomes associated with the endoplasmatic reticulum (MBP) (21). Here, we explored whether microfilament disruption would also affect the stimulus-induced HuR translocation to distinct polysomal aggregations by using polysomal fractionation. Western blotting revealed that the AngII-induced accumulation of HuR to FP and CBP, the latter fraction of which is characterized by high α-actinin levels, is totally impaired in the presence of latrunculin A or blebbistatin (Figure 4A). In contrast, the content of β-actin mRNA, which characteristically can be found in both polysome fractions, remained unchanged independent of which inhibitor was applied (Figure 4A, upper panel). In addition to the lack of α-actinin, the high activity of the cytosolic marker enzyme lactate dehydrogenase within free polysomal fractions confirmed an adequate separation of this fraction (Supplementary Figure S4A). In contrast, HuR was not detectable in the fraction of MBP (Figure 4A), which is characterized by the strong expression of the acetylcholine receptor α (AchRα) (29). In addition, we tested whether the effects on polysomal HuR distribution by both microfilament inhibitors are functionally redundant with changes in the polysomal loading of COX-2 mRNA. Again, to achieve transcriptional induction of COX-2, cells were stimulated for 16 h with a cytokine mix before AngII was added for further 2 h. Concomitant with the observed changes in polysomal HuR distribution, COX-2 mRNA was exclusively detected in fractions of FP and CBP from cytokine-treated cells costimulated with AngII (Figure 4B). By contrast, polysomal COX-2 mRNA was neither detectable in control cells that had only been treated with a cytokine mix nor in cells that were costimulated with latrunculin or blebbistatin, thus indicating that the increase in the polysomal COX-2 mRNA content is mainly triggered by AngII (Figure 4B). Conversely, the amount of nonpolysomal COX-2 mRNA, which is restricted to the nucleus, was highest in cytokine-treated cells either treated in the absence or presence of latrunculin or blebbistatin, respectively (Supplementary Figure S4B).
Figure 4.
HuR recruitment to free and cytoskeleton-bound polysomes by AngII is impaired after microfilament disruption and depends on HuR phosphorylation. (A). HMC were treated for 2 h either with vehicle (−) or with AngII (0.1 µM) in the absence or presence of either latrunculin A (0.1 µM) or blebbistatin (5 µM). Latrunculin was preincubated for 30 min and blebbistatin for 4 h before the addition of AngII. HMC were subsequently subjected to sequential detergent and salt extraction to release FP, CBP and MBP as described in ‘Materials and Methods’ section. Similar amounts of proteins corresponding to the different polysomal fractions were subjected to SDS–PAGE and successively immunoblotted with antibodies against HuR or α-actinin, the latter of which was used as a marker for cytoskeletal cell fractions. Additionally, RT-PCR products corresponding to α-actin and AchRα mRNAs were used as specific markers of cytoplasmic (β-actin) or membrane bound proteins (AchRα). Data are representative for two independent experiments giving similar results. (A, right upper panels). Before the isolation of polysomal subfractions, cell lysates were additionally treated with protein phosphatase 1 (+PPtase) or, alternatively, with RNaseA/T1 (+RNase) as described in ‘Materials and Methods’ section. To validate the accuracy of the treatment, a portion from PPtase-treated cell extracts was subjected to HuR-IP, and phosphorylated HuR levels were monitored by western blot analysis using a p-Ser-PKC substrate antibody (right panel). Accordingly, one portion of the RNase-treated cell homogenates was subjected to total RNA isolation and the presence of β-actin mRNA was subsequently analyzed by RT-PCR (right panel). (B). HMC were preincubated for 16 h with a cytokine mix consisting of IL-1β and TNFα (both at 2 nM) before addition of AngII (+AngII) for 2 h in the absence (−) or presence of the indicated cytoskeletal inhibitors, which were given 4 h before the administration of AngII. Subsequently, different polysomal fractions were separated as described in (A) before RNA from these fractions was harvested and steady-state levels of COX-2 mRNA measured by qRT-PCR. Values show stimulus-dependent changes in COX-2 mRNA contents related to total cytokine-induced COX-2 mRNA levels measured in duplicates. (C). HMC were transfected either with pCMV-Flag-HuRwt (left panel) or with pCMV-Flag-S318DHuR (middle) or, alternatively, with pCMV-Flag-S318AHuR (right panel) for 24 h before treated without (−) or with AngII (0.1 µM) for 2 h. Subsequently, cells were lyzed and before polysomal fractionation, similar portions of lysates were treated with either protein phosphatase 1 (+PPtase) or RNase (+RNase). Thereafter, different polysomal fractions (FP, CBP, MBP) were isolated and analyzed for HuR enrichment by Western blot analysis.
HuR recruitment to free and cytoskeleton bound polysomes by AngII is phosphorylation and RNA dependent
As HuR phosphorylation at S318 by PKCδ is implicated in the AngII-induced and constitutive binding and stabilization of ARE mRNAs by HuR (16,31), we assessed whether the AngII-induced HuR recruitment to polysomes depends on phosphorylation events and/or on the presence of RNA. To this end, polysomal fractions were isolated from alkaline phosphatase or RNase A/T1-treated cell lysates from AngII-stimulated HMC. To confirm the efficiency of phosphatase treatment, the phosphorylation status of immunoprecipitated HuR was monitored by a phospho-Ser-PKC substrate-specific antibody (Figure 4A, right panel, +PPtase). Accordingly, RNase A/T1 effects were confirmed by semiquantitative PCR and agarose gel electrophoresis (Figure 4A, right panel, +RNase). Interestingly, the high content of HuR in FP and CBP triggered by AngII totally disappeared if polysome subfractions were isolated from phosphatase (+PPtase) or RNase treated (+RNase) cell lysates (Figure 4A, middle panel). Based on the observation that treatment of cell extracts with either RNase or PPtase eliminated HuR from all polysomal fractions, we reasoned that probably proteasomal degradation could be responsible for the disappearance of HuR, as ubiquitin-mediated proteolysis of HuR resambles an additional paradigm of HuR regulation (37,38). To address this issue, before separating polysomal fractions, cell lysates were pretreated with MG-132, a broad spectrum proteasome inhibitor before either RNase or PPtase was applied to cell extracts. Interestingly, preincubation of cell lysates with MG-132 restored a HuR accumulation to the fraction of FP even if RNase or PPtase was applied (Supplementary Figure S4C). By contrast, the co-sedimentation of HuR with CBPs was still fully impaired by RNase and PPtase (Supplementary Figure S4C). These data indicate that RNase or PPtase treatment leads to the dissociation of HuR from CBP and FP to soluble cytoplasmic fractions where the HuR protein is a target of rapid proteasomal degradation. The fact that MG-132 did only rescue HuR if dissociated from FP strongly indicates that the FP fraction additionally contains soluble cytoplasmic proteins. To further verify the assumption that HuR distribution to both polysomal fractions may critically depend on the presence of RNA and on the phosphorylation status of HuR, we used a phosphomimetic Ser 318 to Asp mutated Flag-tagged HuR protein (pCMV-FlagS318DHuR), which in contrast to wild-type HuR (pCMV-FlagHuRwt) displays a high constitutive RNA binding to several ARE mRNAs (31). HMC transfected with Flag-tagged HuRwt showed the same dramatic changes in HuR distribution by AngII as endogenous HuR (Figure 4C). Importantly, a similar stimulus-dependent accumulation of Flag-tagged HuR in polysomes was observed in pCMV-FlagS318DHuR transfectants, and, notably, the co-sedimentation of phosphomimetic HuR protein with both polysomal fractions was insensitive toward phosphatase treatment but still RNase sensitive (Figure 4C). Conversely, a phosphorylation deficient HuR chimera (pCMV-Flag-S318AHuR) was refractory to AngII-mediated recruitment to polysomes (Figure 4C). Similar to endogenous HuR, none of the HuR fusion proteins were detected in MBP subfractions (Figure 4C). These data clearly implicate that the RNA-bound state of HuR, which is triggered by PKCδ-dependent phosphorylation at S318, is critical for HuR recruitment to free and cytoskeleton-bound polysomes.
AngII-induced HuR association with polysomes depends on the actin-myosin cytoskeleton
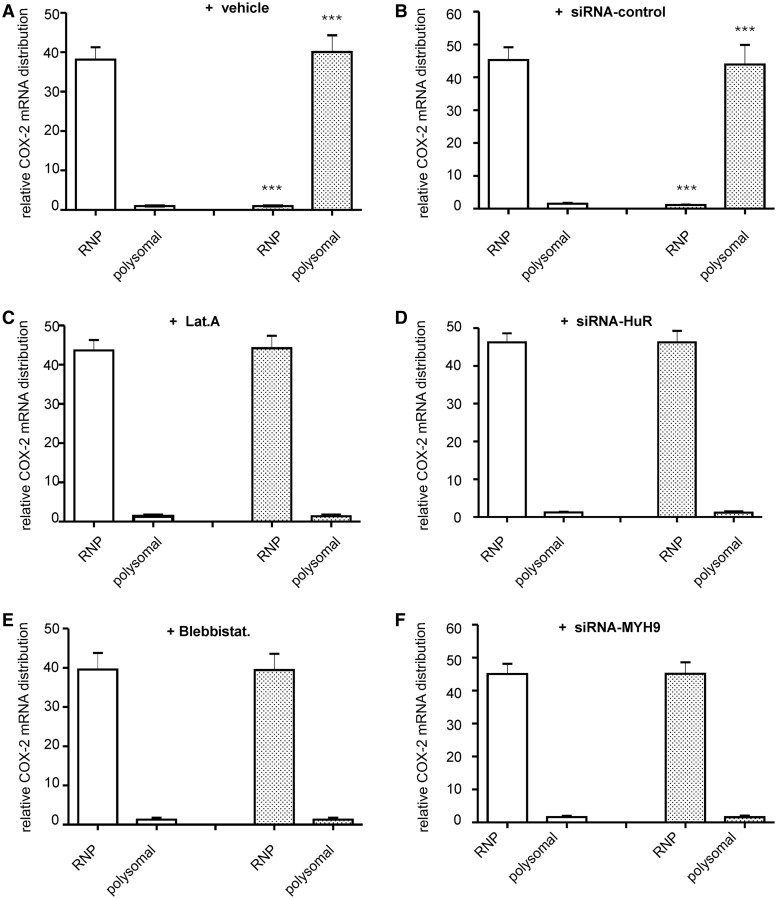
The redistribution of target mRNA from non-polysomal mRNPs to translational active polysomes is a non-random path, which reflects a general increase in translational activity of an mRNA. To examine whether the temporal shift in HuR and its bound target mRNA from mRNPs to translationally active polysomes is affected by disrupting the actin-myosin cytoskeleton, we used sequential ultracentrifugation on sucrose cushion, which enables fractionation of free mRNPs devoid of polysomes from polyribosomes. Western blot analysis of both fractions identified that in unstimulated HMC, HuR is exclusively detectable in the fraction of non-polysomal RNPs (RNPs), whereas stimulation of cells with AngII-induced co-sedimentation of HuR with polysomal fractions concomitant with a reduction of HuR in the corresponding RNP fraction (Figure 5A, left panel). As a control, disruption of polysome assembly after addition of EDTA to cell lysates, resulted in an almost total loss of HuR in the polysomal fraction (Figure 5A, right panel). Consistently, the AngII-induced shift in HuR attachment from RNPs to polysomes was prevented when cells were stimulated in the presence of latrunculin or blebbistatin (Figure 5A, left panel). Similar inhibitory effects were observed after transfection of MYH9-specific siRNAs (siRNA-MYH9) (Figure 5B, left panel). A similar stimulus-dependent shift in HuR distribution was observed with Flag-tagged wild-type HuR (Figure 5B, right panel). In addition to the changes in HuR distribution, AngII treatment caused a strong relocalization of cytokine-induced COX-2 mRNA from non-polysomal RNPs to polysomes (Figure 6A), which was totally impaired by either latrunculin (Figure 6C) or blebbistatin (Figure 6E). As depicted in Figure 6D and F, similar inhibitory effects were achieved after depletion of HuR (Supplementary Figure S5) or myosinIIA (Figure 1B) by specific siRNAs.
Figure 5.
AngII-induced HuR redistribution between non-polysomal RNPs and polysomes depends on the actin–myosin based cytoskeleton. (A). Serum-deprived HMC were treated for 2 h with vehicle (−) or with AngII in the absence or presence of latrunculin (Lat. A) or blebbistatin (Blebbistat.), which were applied for 30 min (Lat. A) or 4 h (Blebbistat.) before AngII stimulation. Thereafter, cells were lyzed for separation of total polysomes from post-polysomal RNPs (RNPs) by ultracentrifugation and HuR distribution in both fractions determined by immunoblotting. (A, left panel). Before ultracentrifugation, cells were either lyzed in the absence (−) or in the presence of EDTA (A, right panel). (B). HMC were either transfected with control siRNA (siRNA-control) or with siRNA of myosin heavy chain 9 (siRNA-MYH9). Seventy-two hours after transfection, cells were serum starved and stimulated for further 2 h with vehicle (−) or AngII (+). The efficiency of silencing of myosin IIA is proven by western blot analysis. In parallel, cells were extracted for total polysomes and non-polysomal RNP particles, before HuR distribution in the different fractions was assessed by Western blotting. The blots shown are from a single experiment representative of two repeats with similar results. (right panel). Stimulation with AngII for 2 h induces a total redistribution of Flag-tagged HuR from RNPs to polysomes.
Figure 6.
AngII-induced COX-2 mRNA redistribution between RNPs and polysomes depends on the actin-myosin based cytoskeleton. Effects of microfilament inhbitors (C and E) or siRNA transfections (B, D and F) on the AngII-induced RNP-polysomal COX-2 mRNA redistribution. Polysomes were separated from free RNPs as described in ‘Materials and Methods’ section, before RNA from both fractions was isolated and levels of COX-2 mRNA were measured by qRT-PCR. (A, C and E). Serum starved HMC were preincubated for 16 h with a cytokine-mix of IL-1β and TNFα (both at 2 nM) before stimulated for further 2 h in the presence (filled bars) or absence (open bars) of AngII without (+ vehicle) or with the indicated inhibitors, which were given 4 h before administration of AngII. (B, D and F). HMC were either treated with control siRNA (siRNA-control), siRNA specific for HuR (siRNA-HuR) or myosin heavy chain 9 (siRNA-MYH9) for 72 h before cells were stimulated for 16 h with the cytokine-mix in the presence (filled bars) or absence (open bars) of AngII, which was added 2 h before cell lysis. Results show the stimulus dependent changes in COX-2 mRNA content within the indicated fractions relative to total cellular COX-2 mRNA contents. Data are means ± SD of three independent experiments and depicted as relative COX-2 mRNA distribution., ***P < 0.005 versus control conditions.
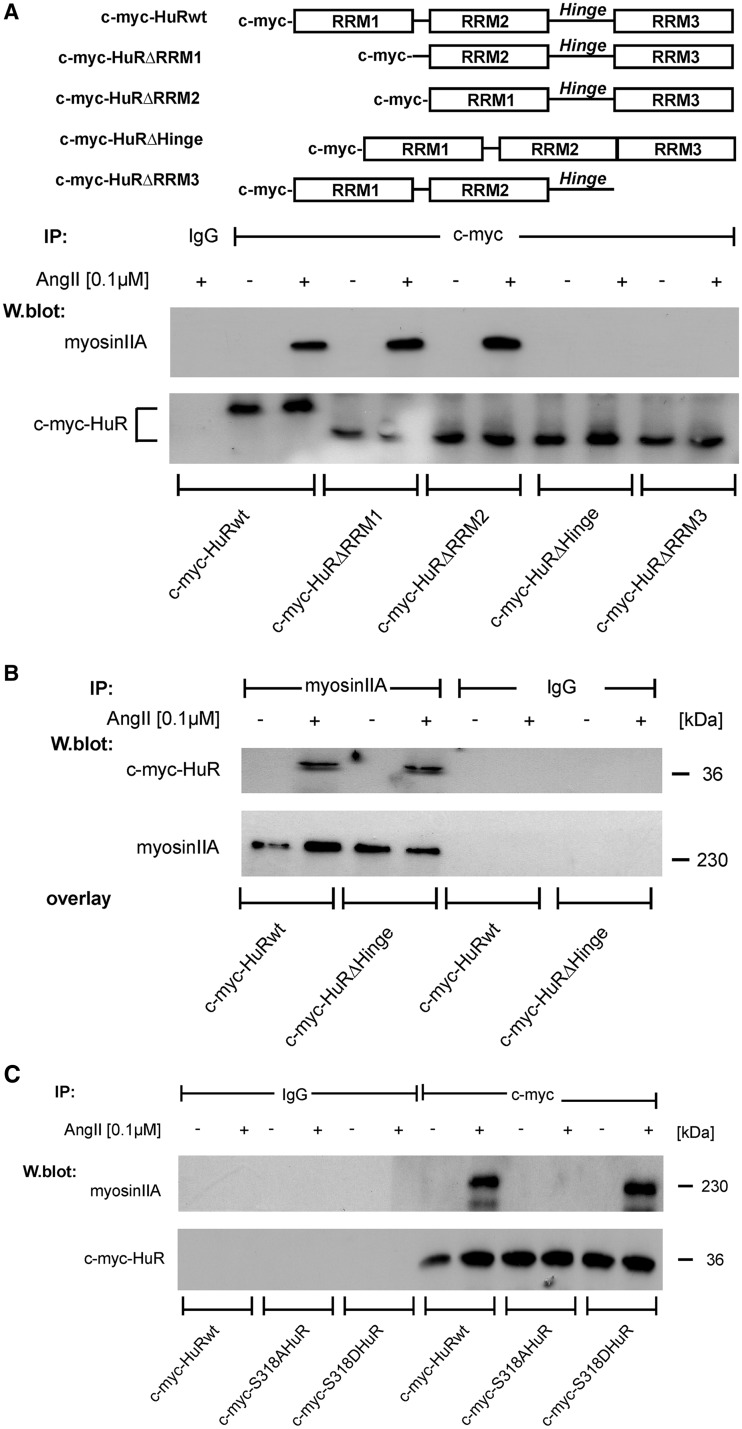
AngII induces a physical interaction of HuR with myosin IIA, which depends on the C-terminal RRM 3 and on HuR phosphorylation at S318
In a next approach, we determined whether AngII may induce a physical interaction of HuR with the motor protein myosin IIA. For this purpose, we used pull-down assays with cell lysates from HMC expressing different c-myc-HuR constructs bearing wild-type HuR, or different truncations in HuR as summerized in Figure 7A. IP of c-myc-HuR fusion proteins with an anti-c-myc antibody specifically coprecipitated a protein of ∼230 kDa corresponding to the molecular weight of myosin IIA in cell lysates from AngII-stimulated HMC (Figure 7A). In contrast, myosin IIA was not precipitated if lysates from untreated cells were used for IP indicating a stimulus-dependent HuR interaction with myosin IIA (Figure 7A). Structurally, HuR and its neuronal relatives contain three highly conserved RRMs and a less conserved hinge region, which bears the HNS relevant for nucleo-cytoplasmic HuR shuttling and which furthermore separates the two N-terminal RRMs (RRM1 and 2) from the C-terminal RRM3 (10,39). To further delineate which of these HuR domains is implicated in myosin binding, we compared the potential binding of a series of c-myc-tagged HuR truncations (32) with endogenous myosin in the cell lysates of different c-myc-HuR transfectants shown in Figure 7A. Importantly, myosin binding to c-myc-tagged HuR was totally lost in cells expressing a RRM3 truncated HuR mutant or c-myc-HuRΔHinge (Figure 7A). As the loss in myosin binding observed with the HuRΔHinge mutant may simply result from an impaired nucleo-cytoplasmic HuR transport, we performed a blot overlay assay as an alternative approach, which independent of the subcellular HuR distribution, allows detection of interactions between ectopically expressed HuR proteins and endogenous myosin IIA. Thereby, myosin was immunoprecipitated from total lysates of untreated HMC, and the bound proteins were resolved by denaturating SDS–PAGE. As shown in Figure 7B, we observed a strong immunopositive myosin IIA band when the membranes were incubated with the cell lysates from AngII-treated transfectants independent of whether cells were transfected with a wild-type or mutated HuR construct (c-myc-HuRΔHinge) with a truncated HNS. These results clarly indicate that the hinge region of HuR in contrast to the RRM3 is irrelevant for myosin IIA binding.
Figure 7.
AngII promotes a physical interaction of HuR with non-muscle myosin IIA, which structurally depends on phosphorylated S318 within the RRM3. (A, upper panel). Schematic representation of c-myc-tagged wild-type HuR and different deletion mutants bearing truncations in the specific HuR domains. (A, lower panel). HMC were transfected with the indicated c-myc-HuR coding plasmids. Seventy-two hours after transfection, cells were treated without (−) or with AngII (+) for 2 h. Thereafter, c-myc-tagged HuR proteins were immunoprecipitated with an anti-c-myc antibody (IP: c-myc), or isotype specific control (IP: IgG). HuR-bound myosin IIA was detected by Western blot analysis with an anti-myosin IIA specific antibody. Equal pull-down (input levels) of c-myc was ascertained by reincubating the blot with the same antibody used for IP. (B). Blot-overlay assay demonstrating that the hinge region of HuR is not relevant for AngII-induced myosin IIA binding. Pull-down of myosin IIA from vehicle (−) or AngII treated HMC was performed as described in ‘Materials and Methods’ section. Blots were probed with equal amounts of total cell lysates cells overexpressing c-myc-HuR wild-type or c-myc-HuRΔHinge. Myosin IIA-HuR interaction was assessed by reincubating the blot with an anti-c-myc antibody. Equal pull-down of myosin is ascertained by reincubating the blot with the same antibody used for the myosin IIA co-IP. (C). HMC were transfected with the indicated c-myc-point mutated HuR and IPs were performed similar as described for (A). Equal pull-down (input levels) of c-myc was ascertained by reincubating the blot with the same antibody used for IP. (D, left panel). HuR binding to myosin IIA is sensitive toward RNase treatment. HMC overexpressing c-myc-HuRwild-type or c-myc-HuRΔHinge were treated for 2 h with AngII (0.1 µM) before total cell lysates from transfectants were immunoprecipitated with an anti-c-myc antibody or with control IgG. Before the IP reaction, the cell homogenates were subjected to RNase treatment and subsequently immunoblotted with an anti-myosin IIA specific antibody. Equal pull-down of c-myc is ascertained by reincubating the blot with anti-c-myc antiserum. (right panel). To test for AngII-dependent association of myosinIIA with endogenous HuR, 200 µg of total cell extracts from unstimulated (−) or AngII-stimulated (+) HMC were subjected to (IP) as described in ‘Materials and Methods’ section. The data shown are from a single experiment representative of two repeats with similar results.
Next, we tested the impact of HuR phosphorylation at Ser 318, which is critical for AngII-induced HuR binding to the AREs of target mRNA (16). Interestingly, AngII-induced myosin binding to the phosphorylation deficient HuR bearing a serine-to-alanine substitution at position 318 (c-myc-S318AHuR) was totally impaired (Figure 7C), and, conversely, a stimulus-dependent myosinIIA binding was observed with a phosphomimetic S318 to Asp HuR protein (c-myc-S318DHuR). According to the critical impact of Ser 318 for mRNA binding, the affinity of wild-type HuR to myosin IIA was completely abrogated if cell lysates had been treated with RNase A, indicating that the AngII-induced HuR interaction with myosin IIA critically depends on RNA bound status of HuR (Figure 7D, left panel). Similar to the findings with ectopic HuR proteins, an AngII-induced interaction of myosin was also observed for endogenous HuR (Figure 7D, right panel).
DISCUSSION
According to a validated model of cytoplasmic mRNA trafficking, translationally silent mRNAs are assembled with various trans-acting RNA-binding proteins into an export competent RNP, which is ready for transit through the nuclear pore complex. Thereby, RNPs, once they have passed the nucleus, can associate with cytoskeletal elements and guide cytoplasmic RNA to its specific destination (22,23,40). However, the mechanisms that specifically control stimulus dependent temporal changes in the docking and release of RNPs from the cytoskeleton are unknown. A microtubule dependent transport of neuronal ELAV proteins is necessary for an association with mRNA to small mRNP granules and its subsequent targeting to the translation apparatus (26). A microtubule dependent trafficking is furthermore implied in the fast and long-distance mRNA transport in neurons and oocytes (24,41–43). In contrast to these reports, a microtubule-mediated transport seems irrelevant for HuR. Here, we have identified the involvement of an actin-myosin IIA-based mechanism as indicated by the strong inhibitory effects by latrunculin A and blebbistatin (Figure 1A) and by experiments with myosin IIA specific siRNA (Figure 1B). The reduction in cytoplasmic HuR in AngII plus latrunculin treated cells suggests that the lack of cytoplasmic HuR is a direct consequence of abrogated nuclear HuR export to the cytoplasm after microfilament disruption and implies a microfilament-mediated cross-talk between both compartments. A cytoskeleton-dependent communication between the nucleus and the cytoplasm has been demonstrated by various studies (44–47). It is noteworthy that the actin polymerizing protein profilin is critical for a Ran-mediated export in Drosophila (48), which is also implicated in nucleo-cyoplasmic HuR shuttling (47).
Given the fact that HuR co-sedimented exclusively with the free and cytoskeleton bound polysomes after cells were stimulated with AngII (Figure 4A and C) indicates that AngII-activated HuR is preferentially transported to translationally active cell compartments. Further studies are needed to elucidate the general impact of a specific external stimulus to the allocation of HuR to a specific polysomal subfraction. Functionally, the AngII-induced accumulation of HuR in specific subpolysomal fractions is consistent with a stimulus-dependent appearance of COX-2 mRNA within the same fractions (Figure 4B).
The finding that the AngII-induced HuR association to polysomes is totally lost if cell lysates were treated with phosphatase or RNase implicates that both RNA binding and phosphorylation of HuR are indispensable for the recruitment of HuR to polysomes (Figure 4A, right panel). This notion is further confirmed by the observation that the co-sedimentation of Flag-tagged phosphomimetic S318D HuR with cytoskeleton bound and free polysomes remained unaffected by phosphatase treatment but still sensitive toward RNase (Figure 4C). As S318 is a functional PKCδ phosphorylation site, which is critical for AngII-induced HuR binding to specific target mRNAs (16), results with a corresponding phosphorylation deficient HuR mutation strongly suggest that HuR binding to target mRNA is critical for its redistribution from RNPs to translational active polysomes. Similar to HuR recruitment to polysomes, myosin IIA association with HuR is a phosphorylation and RNA-dependent event. Accordingly, AngII-induced physical interaction of myosin II with HuR was totally impaired in HuR bearing a serine-to-alanine substitution at position 318 in the RRM3 (Figure 7C). In addition, mapping of putative myosin-binding domains revealed that the C-terminal RRM3 is critical for myosin IIA binding (Figure 7A and B). The fact that phosphorylation at S318 in the RRM3 is critical for AngII-induced RNA-binding of HuR (16) as well as for myosin binding clearly indicates that the RNA-bound status of HuR is indispensable for its association with nonmuscle myosin. Further studies are needed to elucidate whether HuR does directly interact with myosin IIA or, whether alternatively, the interaction is indirect and bridged by either an ancillary RNA-binding protein and/or is directly mediated through HuR-bound mRNA. Consequently, our data propose an active role of phosphorylated HuR in the anchorage and active transport of ARE-bearing mRNA species to microfilaments through stimulus-induced myosin binding. Collectively, our study implicates that mRNA binding, nuclear export and subsequent cytoplasmic trafficking of HuR and its cargo mRNA comprise an intimately linked series of events that are initiated and coordinated via nuclear HuR phosphorylation at S318 (Figure 8). The discovery of an actin-myosin involvement in HuR-directed mRNA trafficking furthermore emphasizes the pharmacological potential of latrunculin and its semisynthetic derivatives to interfere with posttranscriptional gene regulation by the multifunctional protein HuR.
Figure 8.
Schematic summary of regulation of HuR trafficking by the actin-myosin cytoskeleton. AngII triggers nuclear translocation of the PKCδ, which in turn phosphorylates nuclear HuR at RRM3, thereby increasing HuR binding to target mRNAs. The HuR-bound mRNA assembles to a motile transport RNP particle and is exported to the cytoplasm by a mechanism, which essentially requires the actin-myosin-based cytoskeleton. HuR and bound cargo-mRNA is directed to specific cytoplasmic destinations via a myosin-driven transport along filamentous (F) actin. Importantly, the AngII-induced recruitment of HuR from translation inactive RNPs to FP and CBP occurs via an interaction of HuR with myosin IIA, which is highly sensitive toward RNase and PPtase treatment. AngII = angiotensin II, CBP = cytoskeleton bound polysomes, FP = free polysomes, PKCδ = protein kinase Cδ, PPtase = protein phosphatase, RNP = ribonucleoprotein, RRM = RNA recognition motif, 3′UTR = 3′untranslated region.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation (DFG) [EB 257/5-1, GRK1172, SFB 815]; the state of Hesse (Onkogene Signaltransduktion [L-4-518/55.004]); and the Excellence Cluster ‘Cardiopulmonary System (ECCPS)’ EXC 147/2. Funding for open access charge: DFG [EB 257/5-1].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Ann-Bin Shyu (University of Texas Medical School, Houston) for providing the Myc-tagged HuR plasmids and to Heinfried Radeke (Goethe University of Frankfurt, Germany) for providing primary human mesangial cells (HMC).
REFERENCES
- 1.Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl Acad. Sci. USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu H, Zhou HL, Hasman RA, Lou H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J. Biol. Chem. 2007;282:2203–2210. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]
- 5.Izquierdo JM. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J. Biol. Chem. 2008;283:19077–19084. doi: 10.1074/jbc.M800017200. [DOI] [PubMed] [Google Scholar]
- 6.Meisner NC, Filipowicz W. Properties of the regulatory RNA-binding protein HuR and its role in controlling miRNA repression. Adv. Exp. Med. Biol. 2011;700:106–123. doi: 10.1007/978-1-4419-7823-3_10. [DOI] [PubMed] [Google Scholar]
- 7.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khabar KS. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell. Mol. Life Sci. 2010;67:2937–2955. doi: 10.1007/s00018-010-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascale A, Govoni S. The complex world of post-transcriptional mechanisms: is their deregulation a common link for diseases? Focus on ELAV-like RNA-binding proteins. Cell. Mol. Life Sci. 2012;69:501–517. doi: 10.1007/s00018-011-0810-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan XC, Steitz JA. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl Acad. Sci. USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HH, Abdelmohsen K, Lal A, Pullmann R, Jr, Yang X, Galban S, Srikantan S, Martindale J, Blethrow J, Shokat KM, et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008;22:1804–1815. doi: 10.1101/gad.1645808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doller A, Huwiler A, Müller R, Radeke HH, Pfeilschifter J, Eberhardt W. Protein kinase Cα-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amadio M, Bucolo C, Leggio GM, Drago F, Govoni S, Pascale A. The PKCβ/HuR/VEGF pathway in diabetic retinopathy. Biochem. Pharmacol. 2010;80:1230–1237. doi: 10.1016/j.bcp.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 15.Doller A, Akool el-S, Huwiler A, Müller R, Radeke HH, Pfeilschifter J, Eberhardt W. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cδ elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol. Cell. Biol. 2008;28:2608–2625. doi: 10.1128/MCB.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doller A, Schlepckow K, Schwalbe H, Pfeilschifter J, Eberhardt W. Tandem phosphorylation of serines 221 and 318 by protein kinase Cδ coordinates mRNA-binding and nucleocytoplasmic shuttling of HuR. Mol. Cell. Biol. 2010;30:1397–1410. doi: 10.1128/MCB.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallouzi IE, Brennan CM, Steitz JA. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA. 2001;7:1348–1361. doi: 10.1017/s1355838201016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rebane A, Aab A, Steitz JA. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA. 2004;10:590–599. doi: 10.1261/rna.5224304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Güttinger S, Mühlhäusser P, Koller-Eichhorn R, Brennecke J, Kutay U. Transportin2 functions as importin and mediates nuclear import of HuR. Proc. Natl Acad. Sci. USA. 2004;101:2918–2923. doi: 10.1073/pnas.0400342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Yang X, Kawai T, López de Silanes I, Mazan-Mamczarz K, Chen P, Chook YM, Quensel C, Köhler M, Gorospe M. AMP-activated protein kinase-regulated phosphorylation and acetylation of importin α1: involvement in the nuclear import of RNA-binding protein HuR. J. Biol. Chem. 2004;279:48376–48388. doi: 10.1074/jbc.M409014200. [DOI] [PubMed] [Google Scholar]
- 21.Hovland R, Hesketh JE, Pryme IF. The compartmentalization of protein synthesis: importance of cytoskeleton and role in mRNA targeting. Int. J. Biochem. Cell Biol. 1996;28:1089–1105. doi: 10.1016/1357-2725(96)00059-3. [DOI] [PubMed] [Google Scholar]
- 22.Jansen RP. RNA-cytoskeletal associations. FASEB J. 1999;13:455–466. [PubMed] [Google Scholar]
- 23.Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brendza RP, Serbus LR, Duffy JB, Saxton WM. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science. 2000;289:2120–2122. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antic D, Keene JD. Messenger ribonucleoprotein complexes containing human ELAV proteins: interactions with cytoskeleton and translational apparatus. J. Cell Sci. 1998;111:183–197. doi: 10.1242/jcs.111.2.183. [DOI] [PubMed] [Google Scholar]
- 27.Fujiwara Y, Kasashima K, Saito K, Fukuda M, Fukao F, Sasano Y, Inoue K, Fujiwara T, Sakamoto H. Microtubule association of a neuronal RNA-binding protein HuD through its binding to the light chain of MAP1B. Biochimie. 2011;93:817–822. doi: 10.1016/j.biochi.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Radeke HH, Meier B, Topley N, Flöge J, Habermehl GG, Resch K. Interleukin 1-β and tumor necrosis factor-α induce oxygen radical production in mesangial cells. Kidney Int. 1990;37:767–775. doi: 10.1038/ki.1990.44. [DOI] [PubMed] [Google Scholar]
- 29.Hovland R, Campbell G, Pryme I, Hesketh J. The mRNAs for cyclin A, c-myc and ribosomal proteins L4 and S6 are associated with cytoskeletal-bound polysomes in HepG2 cells. Biochem. J. 1995;310:193–196. doi: 10.1042/bj3100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaefer M, Albrecht N, Hofmann T, Gudermann T, Schultz G. Diffusion-limited translocation mechanism of protein kinase C isotypes. FASEB J. 2001;15:1634–1636. doi: 10.1096/fj.00-0824fje. [DOI] [PubMed] [Google Scholar]
- 31.Doller A, Winkler C, Azrilian I, Schulz S, Hartmann S, Pfeilschifter J, Eberhardt W. High-constitutive HuR phosphorylation at Ser 318 by PKCδ propagates tumor relevant functions in colon carcinoma cells. Carcinogenesis. 2011;32:676–685. doi: 10.1093/carcin/bgr024. [DOI] [PubMed] [Google Scholar]
- 32.Chen CY, Xu N, Shyu AB. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol. Cell. Biol. 2002;22:7268–7278. doi: 10.1128/MCB.22.20.7268-7278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Côté J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 35.Kovács M, Tóth J, Hetényi C, Málnási-Csizmadia A, Sellers JR. Mechanism of blebbistatin inhibition of myosin IIA. J. Biol. Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- 36.Mermall V, Post PL, Mooseker MS. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science. 1998;279:527–533. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- 37.Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, Kuwano Y, Galban S, Becker KG, Kamara D, de Cabo R, et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009;28:1271–1282. doi: 10.1038/emboj.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou HL, Geng C, Luo G, Lou H. The p97-UBXD8 complex destabilizes mRNA by promoting release of ubiquitinated HuR from mRNP. Genes Dev. 2013;27:1046–1058. doi: 10.1101/gad.215681.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Good PJ. A conserved family of elav-like genes in vertebrates. Proc. Natl Acad. Sci. USA. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daneholt B. Assembly and transport of a premessenger RNP particle. Proc. Natl Acad. Sci. USA. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilhelm JE, Vale RD. RNA on the move: the mRNA localization pathway. J. Cell Biol. 1993;123:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem. Sci. 2006;31: 687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Sotelo-Silveira JR, Calliari A, Kun A, Koenig E, Sotelo JR. RNA trafficking in axons. Traffic. 2006;7:508–515. doi: 10.1111/j.1600-0854.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 44.Schindler M, Jiang LW. Nuclear actin and myosin as control elements in nucleocytoplasmic transport. J. Cell Biol. 1986;102:859–862. doi: 10.1083/jcb.102.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S, Buder K, Burkhardt C, Schlott B, Görlach M, Grosse F. Nuclear DNA helicase II/RNA helicase A binds to filamentous actin. J. Biol. Chem. 2002;277:843–853. doi: 10.1074/jbc.M109393200. [DOI] [PubMed] [Google Scholar]
- 46.Zhang S, Köhler C, Hemmerich P, Grosse F. Nuclear DNA helicase II (RNA helicase A) binds to an F-actin containing shell that surrounds the nucleolus. Exp. Cell Res. 2004;293:248–258. doi: 10.1016/j.yexcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 47.Tretyakova I, Zolotukhin AS, Tan W, Bear J, Propst F, Ruthel G, Felber BK. Nuclear export factor family protein participates in cytoplasmic mRNA trafficking. J. Biol. Chem. 2005;280:31981–31990. doi: 10.1074/jbc.M502736200. [DOI] [PubMed] [Google Scholar]
- 48.Minakhina S, Myers R, Druzhinina M, Steward R. Crosstalk between the actin cytoskeleton and Ran-mediated nuclear transport. BMC Cell Biol. 2005;6:32. doi: 10.1186/1471-2121-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brennan CM, Gallouzi IE, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 2000;151:1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.