Abstract
Yeast 25S rRNA was reported to contain a single cytosine methylation (m5C). In the present study using a combination of RP-HPLC, mung bean nuclease assay and rRNA mutagenesis, we discovered that instead of one, yeast contains two m5C residues at position 2278 and 2870. Furthermore, we identified and characterized two putative methyltransferases, Rcm1 and Nop2 to be responsible for these two cytosine methylations, respectively. Both proteins are highly conserved, which correlates with the presence of two m5C residues at identical positions in higher eukaryotes, including humans. The human homolog of yeast Nop2, p120 has been discovered to be upregulated in various cancer tissues, whereas the human homolog of Rcm1, NSUN5 is completely deleted in the William's-Beuren Syndrome. The substrates and function of both human homologs remained unknown. In the present study, we also provide insights into the significance of these two m5C residues. The loss of m5C2278 results in anisomycin hypersensitivity, whereas the loss of m5C2870 affects ribosome synthesis and processing. Establishing the locations and enzymes in yeast will not only help identifying the function of their homologs in higher organisms, but will also enable understanding the role of these modifications in ribosome function and architecture.
INTRODUCTION
All cellular RNAs contain various chemical modifications. These modifications provide RNA, the necessary complexity to perform more sophisticated biological processes like gene regulation and catalysis (1). There are >100 structurally distinct ribonucleosides that have been identified in all three domains of life (http://rna-mdb.cas.albany.edu/RNAmods/ (2). Many modified nucleosides are conserved throughout bacteria, archaea and eukaryotes, while some are unique to each branch of life (3). Different enzymes and snoRNP complexes add most of these modifications posttranscriptionally. Methylation of nitrogenous bases and ribose sugars is the most common chemical modification observed in RNA (3). All four nitrogenous bases of RNA have been discovered to undergo methylation either at nitrogen (N) or carbon (C) atoms. Interestingly, the methylation of the cytosine at fifth carbon atom (m5C) is the most frequent base modification found in cellular RNAs.
The m5C was first observed in DNA, and until now, apart from adenine modification in prokaryotes, it is the most common modified base found in the DNA (4,5). The role of m5C in eukaryotic DNA has been extensively analyzed and has been instrumental in understanding the molecular basis of epigenetic gene regulation (6,7). On the other hand, in prokaryotes, especially in bacteria, the methylation of cytosine along with adenine act as molecular tag that helps the restriction endonucleases to differentiate between self and foreign DNA (8).
Widespread analysis of tRNAs in the past, have allowed the identification and precise mapping of m5C residues in tRNAs from all three living domains (9). Interestingly, these analyses have revealed that several archaeal and eukaryotic tRNAs harbors m5C residues in functionally vital regions (10). Most of the eukaryotic tRNAs contain m5C residues in the junction between the variable region and the TΨC-stem, especially at positions 48 and 49. Recent nuclear magnetic resonance and circular dichroism studies have shown that these m5C modifications play a significant role in the structural stability of tRNAs (11,12). A series of studies in Saccharomyces cerevisiae have also demonstrated that the loss of these modifications in tRNAs leads to their rapid degradation (13).
In contrast to tRNAs, the rRNAs have not been subjected to a comprehensive analysis for m5C modifications. Nevertheless, preliminary analyses have revealed that rRNA contains fewer m5C residues as compared with tRNAs (14,15). One m5C modification at position 2278 was mapped in the 25S rRNA of yeast by Planta's lab (16). In contrast to yeast, Escherichia coli was revealed to contain two m5C residues: m5C965 and m5C1407 in 16S and a single m5C residue, m5C1962, in the 23S rRNA (17,18). The human 28S rRNA has also been discovered to contain two m5C residues at position 3782 and 4447 (14,19,20).
The m5C modifications of both RNA and DNA are catalyzed by specific methyltransferases, belonging to class I Methyltransferase (MTases), characterized by a Rossmann-like fold SAM binding domain (21).Both RNA cytosine methyltransferases (RCMTs) and DNA cytosine methyltransferases (DNMTs) share eight signature sequence motifs (22,23). The structural and functional analyses of various RCMTs have revealed that unlike DNMTs, RCMTs make use of a distinct cysteine amino acid in motif VI as the catalytic nucleophile (24–26). The catalytic mechanism of the RCMTs involves an attack by the thiolate of the motif VI cysteine on the carbon atom 6 of the target cytosine base to form a covalent link, thereby activating C-5 for electrophilic substitution by the methyl group of SAM (27). After the addition of the methyl group, motif IV cysteine acts as a general base in the β-elimination of the proton from the methylated cytosine ring, thus releasing the substrate (RNA) (27,28).
The homology search using known amino acid sequences of RCMTs from prokaryotic organisms have led to the identification of three RCMTs, Trm4, Rcm1 (Ynl022c) and Nop2, in S. cerevisiae (22,29,30). Surprisingly, apart from Trm4, the substrates for both Nop2 and Rcm1 remained unknown. Trm4 was identified to catalyze the C-5 methylation of cytosine at several positions of tRNA (30). Nop2 has been shown to be critical for 60S biogenesis, nevertheless the catalytic function of Nop2 remained unclear (31). Rcm1 is the only uncharacterized RCMT in yeast and has not been subjected to any biochemical analysis so far.
Both Nop2 and Rcm1 are highly conserved proteins. The human homolog of yeast Nop2, NSUN1 or p120 has been found to be upregulated in various cancer tissues, whereas the human homolog of Rcm1, NSUN5A, is completely deleted in the Williams–Beuren syndrome, which is a complex developmental disorder with a significant penetration in human population (32–34). Like in yeast, the substrate and function of both human homologs remained to be identified.
In the present study, we analyzed for the first time the m5C modification of 25S rRNA in S. cerevisiae using RP-HPLC analysis. Surprisingly, we discovered that S. cerevisiae contains two m5C residues in the 25S rRNA. In addition, we demonstrated that Rcm1 and Nop2 perform these two modifications, individually. We also analyzed the significance of these modifications in yeast and provide insight into the importance of these two m5C residues in ribosome synthesis and functionality. Together with the identification of substrates in yeast and helices alignment of both human and yeast ribosomal RNA, we could also assign the substrate specificity to the human homologs of yeast Nop2 (NSUN1, p120) and Rcm1 (NSUN5).
MATERIALS AND METHODS
Yeast strains and plasmids
The strains and plasmids used in the present study are listed in Supplementary Table S1. The PCR primers used for the construction of the plasmids are listed in Supplementary Table S2. The rDNA point mutants were constructed as described previously (35). A detailed protocol for construction of all plasmids will be provided on request.
Growth conditions and media
Yeast strains were grown at 30°C in YPD or YPG medium (1% yeast extract, 2% peptone, 2−4% glucose or galactose) or in synthetic dropout medium (0.5% ammonium sulphate, 0.17% yeast nitrogen base, 2−4% glucose). For serial dilution growth assays, yeast cells were grown over night in YPD/YPG medium and diluted to an OD600 of 1 followed by 1:10 serial dilutions. From the diluted cultures, 5 µl were spotted onto YPD plates and incubated at 30 or 19°C. For the antibiotic analysis, 5 µl of a paromomycin solution (200 mg/ml) and anisomycin (20 µg/ml) were spotted on filter discs, which were then placed on YPD plates containing the strains to be tested.
RNA extraction and northern hybridization
For northern blot analysis, RNA was prepared by phenol/chloroform extraction as previously described (36). Northern blotting was performed as described previously (37). For the extraction of 25S rRNA, 60S subunits were isolated using sucrose density centrifugation. The fractions corresponding to 60S subunits were collected with the Density Gradient Fractionation System (Teledyne Isco) and precipitated with 2.5 volume of 100% ethanol at −20°C for 16 h. Precipitated 60S subunits were dissolved in water and 25S rRNA was purified using the RNeasy Kit (QIAGEN).
Reversed phase−high performance liquid chromatography and mung bean nuclease assay
RP-HPLC were performed as described previously (37). Seventy micrograms of 25S rRNA was digested to nucleosides by nuclease P1 and alkaline phosphatase. The hydrolyzate was analyzed by HPLC according to the method described before (38,39). Nucleosides were analyzed by RP-HPLC on a Supelcosil LC-18-S HPLC column (25 cm × 4.6 mm, 5 µm) equipped with a precolumn (4.6 × 20 mm) at 30°C on an Agilent 1200 HPLC system. For mass spectrometry analysis, nucleosides were collected from four HPLC experiments and desalted twice with a Zorbax Eclipse XDB-C18 column (Agilent; 4.6 × 150 mm, 5 µm) using 5 mM ammonium acetate, pH 6.0, with a flow rate of 0.5 ml/min. After buffer evaporation, samples were resolved in water and applied to MALDI mass spectrometry on a VG Tofspec (Fisons Instruments) in the negative ion mode.
The mung bean nuclease protection assay was performed exactly as described previously (37,40). Specific sequence of the 25S rRNA was isolated by hybridization to complementary deoxyoligonucleotides. One thousand picomoles of the synthetic deoxyoligonucleotides complementary to yeast 25S rRNA were incubated with 100 pmoles of total rRNA and 1.5 µl of DMSO in 0.3 volumes of hybridization buffer (250 mM HEPES, 500 mM KCl, pH 7.0). The fragments were digested by mung bean nuclease and 0.02 µg/µl RNase A (Sigma-Aldrich) and were separated on a 13% polyacrylamide gel containing 7 M urea. RNA was extracted from the gel using D-TubeTM Dialyzers according to the manufacturer’s protocol for electroelution (Novagen®)
Polysome profiles
The polysome profiles were performed exactly as described previously (37). The yeast strains were grown in YPD medium (100 ml) at 30°C to early logarithmic phase (OD600 = 1.0). Before cell disruption, the cycloheximide was added to the final concentration of 100 µg/ml. A 100 ml YPD culture (OD600 = 1.0) was harvested by centrifugation at 4°C. Cells were then washed twice with 10 ml of Polysome Buffer A (20 mM HEPES, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EGTA and 1 mM DTT), resuspended in 0.5 ml of buffer A and disrupted by vortexing with an equal volume of glass beads. Equivalent amounts of absorbing material were layered on a 10−50% (w/v) sucrose gradient in buffer A. The gradient was made using Gradient Master 107 (Biocomp). The samples were then centrifuged at 19 000 rpm for 17 h at 4°C in a SW40 rotor using Beckman ultracentrifuge (L-70: Beckman). Gradients were fractionated in an ISCO density gradient fractionar and the absorbance profile at 254 nm was analyzed in ISCO UA-5 absorbance monitor.
Western blot analysis
For western blotting, 30 µg of total protein from each sample were separated with 12% sodium dodecyl sulphate polyacrylamide gel and blotted on a PVDF membrane (Millipore). The membrane was blocked with 5% nonfat dry milk, and the protein was detected with anti-His monoclonal antibody (Roche; 1:1000 dilution) in case of Rcm1 and with anti-Nop2 (EnCor Biotechnology, Florida, USA), followed by anti-mouse IgG-conjugated horseradish peroxidase (Bio-Rad; 1:10 000 dilution).
Protein localization
The plasmid pSH30 encoding GFP-Rcm1 fusion protein was constructed using plasmid pUG35 plasmid (EUROSCARF). Plasmid pSH30 was then transformed in a strain containing a gene encoding for ScNop56-mRFP. Transformants carrying plasmid were grown in synthetic medium lacking uracil at 30°C. GFP-fused Rcm1 was visualized using a Leica TCS SP5. The RFP-fused Nop56 was used as reference for nucleolar localization.
RESULTS
RP-HPLC analysis of the 25S rRNA revealed the presence of two m5C residues
The 25S rRNA of yeast has been predicted to contain one m5C residue at position 2278 (10,16). Surprisingly, the enzyme responsible for this base modification remained unknown.
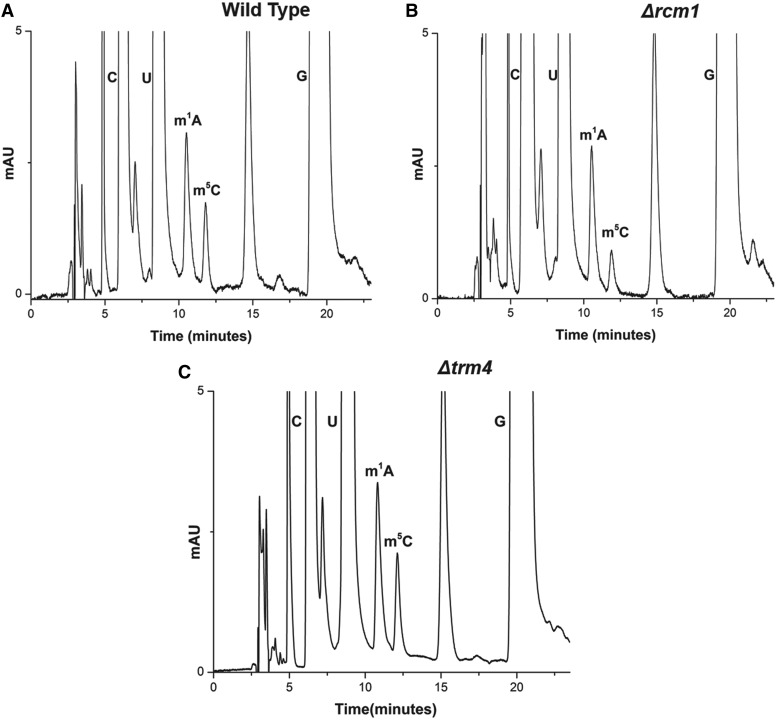
In silico sequence alignments of yeast proteins with already known cytosine methyltransferases have predicted the presence of three m5C methyltransferases in yeast, Nop2, Trm4 and Rcm1. Interestingly, apart from Nop2, both Rcm1 and Trm4 have been found not to be essential for viability. To identify the enzyme responsible for m5C2278 base modification of 25S rRNA, we analyzed the 25S rRNA composition of deletion mutants, Δrcm1 and Δtrm4, using RP-HPLC (Figure 1).
Figure 1.
RP-HPLC analysis of the 25S rRNA of Δrcm1 and Δtrm4. Nucleosides compositions of the 25S rRNAs from the wild type, Δrcm1 and Δtrm4 mutant were analyzed by RP-HPLC. (A) RP-HPLC chromatogram from the wild type, (B) Δrcm1 and (C) Δtrm4 mutant. The peak corresponding to the m5C with a retention time of approximately 12 min reduces to half in the Δrcm1 mutant compared with wild type and Δtrm4.
Surprisingly, in the Δrcm1 mutant, the peak corresponding to m5C reduced to half, suggesting that there are two m5C modifications in the 25S rRNA of S. cerevisiae. This, however, was in contrast to what have been mentioned before by other groups and discussed previously during our analysis of m1A modifications in 25S rRNA (10,37,41). To validate that the m5C peak comprised of only m5C nucleosides, we analyzed the composition of m5C peak with MALDI mass spectrometry as described in ‘Materials and Methods’ section. The mass spectrometry analysis further confirmed that the peak consists of only m5C nucleosides (data not shown).
To further corroborate the presence of two m5C residues, rRNA point mutants were created, where cytosine C2278 of 25S rRNA were exchanged with U and G. For this analysis, we used a plasmid borne copy of 35S rDNA transcribed under the native promoter in a strain where the genomic rDNA was deleted. The mutated 25S rRNA was expressed from plasmids pPK655 and pPK657 corresponding to C2278G, C2278T mutants, respectively. The wild-type 25S rRNA was expressed from the pAV164 plasmid. As evident in the Supplementary Figure S1, RP-HPLC analysis of these strains validated the presence of second m5C modification in the 25S rRNA of S. cerevisiae, as the amount of m5C peak reduced to half, when the cytosine at position 2278 was exchanged with either G or U. Besides, this analysis further confirmed the position of m5C residue at position 2278 in S. cerevisiae. Taken together, our results undoubtedly showed the presence of two m5C residues in the 25S rRNA and more importantly emphasized the involvement of Rcm1 in catalyzing one of the m5C modifications of 25S rRNA.
Localization of the second m5C residue in the 25S rRNA of S. cerevisiae
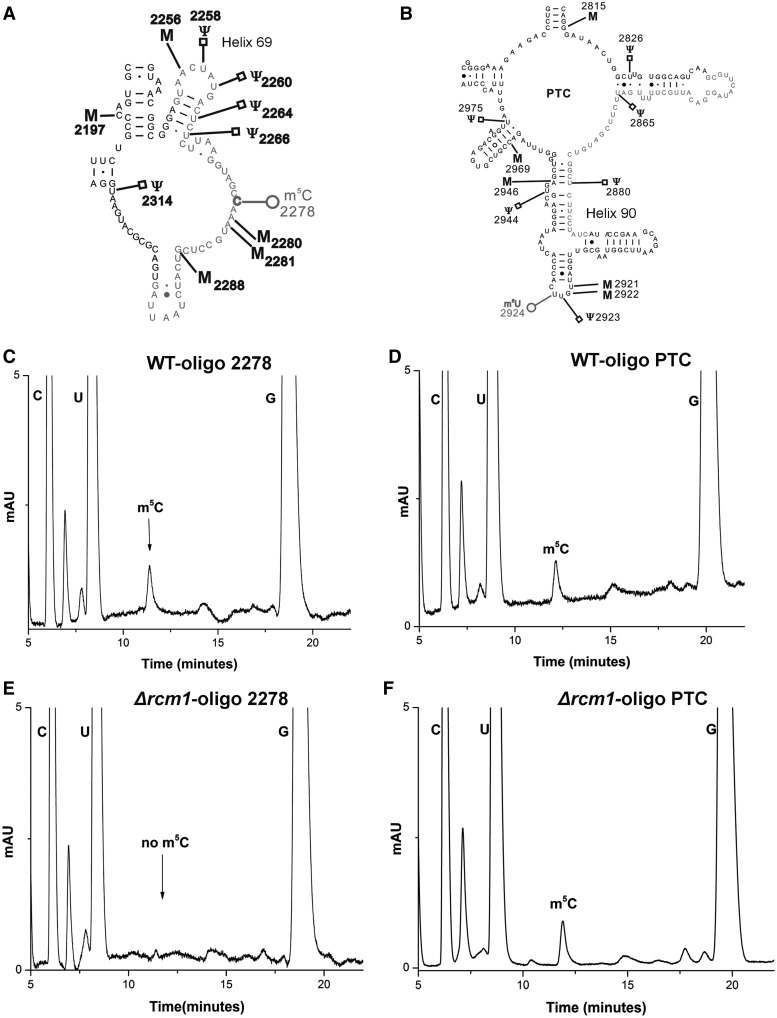
To identify the second m5C modification, we subjected the entire 25S rRNA to mung bean nuclease protection assay. Synthetic oligonucleotides, 50mers in length and complementary to the different regions of 25S rRNA were designed. rRNA (25S) isolated from the wild-type cells was then hybridized to these oligonucleotides in different independent reactions and subjected to the mung bean nuclease digestion. Because mung bean nuclease is a single-strand specific endonuclease, only single-stranded extension from the ends of RNA and DNA were digested. Consequently, desired fragments corresponding to the different regions of 25S rRNA could be protected from the digestion by providing complementary oligonucleotides. These protected RNA–DNA hybrid fragments were then separated from the rest of the debris and from each other using 8M-Urea polyacrylamide gel electrophoresis gels. The RNA fragments were then eluted from the gels using D-TubeTM. Dialyzers by electroelution and their nucleosides composition was next examined using RP-HPLC. Two different fragments derived from region (2253–2300) corresponding to m5C2278, (Figure 2A) and 2839–2890 (Figure 2B) were identified to contain m5C residues (Figure 2C and D).
Figure 2.
Mung bean nuclease protection assay for the identification of new m5C residue in the 25S rRNA of S. cerevisiae. To identify the second m5C modification, whole 25S rRNA was subjected to mung bean nuclease protection assay. Synthetic oligonucleotides complementary to the different regions of 25S rRNA were designed and hybridized to the 25S rRNA of wild type. Two fragments corresponding to domain IV of 25S rRNA (from base 2253 to 2300) marked red (A) and region corresponding to domain V of 25S rRNA (from base 2840 to 2889) also marked in red (B) were discovered to contain m5C residues. (C) RP-HPLC chromatogram of the fragments derived from region displayed in red (A) corresponding to already predicted m5C 2278 residue. (D) RP-HPLC chromatogram of the fragments derived from region marked in red (B). Both fragments contain single m5C residue with the retention time of ∼12 min. To identify the m5C residue missing in Δrcm1, the statuses of these two m5C residues were analyzed in the 25S rRNA of Δrcm1 strain. (E) RP-HPLC chromatogram of fragments corresponding to region 2253–2300 of 25S rRNA marked in red (A). (F) RP-HPLC chromatogram of fragments corresponding to region 2840–2889 of 25S rRNA marked in red (B). Interestingly, m5C residue from the fragments corresponding to m5C2278 was absent, highlighting the involvement of Rcm1 in catalyzing the m5C2278 modification of 25S rRNA.
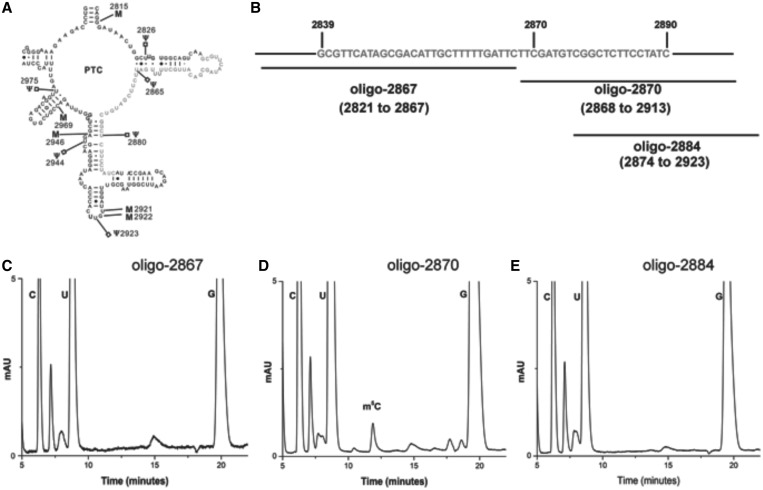
To map the precise location of the m5C residue within the fragment (2839–2890), three overlapping fragments corresponding to the regions displayed in Figure 3B were isolated. The mung bean nuclease assays using oligo-2867, oligo-2870 and oligo-2884 were performed to isolate these fragments. The compositions of these fragments were next analyzed using RP-HPLC (Figure 3C–E). The RP-HPLC chromatograms from these fragments allowed us to map the precise location of the m5C residue to position 2870.
Figure 3.
Mapping of the exact position of m5C residue in the domain V of S. cerevisiae. To identify the precise location of the m5C residue within the fragment corresponding to the PTC region of 25S rRNA, shown in panel (A), fragments marked in panel (B) were isolated using mung bean nuclease protection assay. The nucleosides composition of these fragments was next analyzed using RP-HPLC. (C) RP-HPLC chromatogram of oligo-2867, oligo-2870 (D) and oligo-2884 (E). As evident from the chromatograms, the 25S rRNA fragments corresponding to oligo-2870 contains m5C residues, whereas other two fragments lack m5C residues. Together, this distinctly confirmed that the m5C residue in the PTC region (2839 to 2890) is located at position 2870.
Deletion of Rcm1 specifically abolishes the methylation of m5C 2278 in the 25S rRNA
To determine the nucleotide position at which the modification was absent in the Δrcm1 mutant, a mung bean nuclease protection assay was performed. The fragments corresponding to m5C2278 and m5C2870 as shown in Figure 2A and B from Δrcm1 cells were isolated and the composition of m5C was examined using RP-HPLC. Interestingly, in the Δrcm1 mutant, as evident from RP-HPLC chromatograms shown in Figure 2E and F, the m5C residue from fragment corresponding to m5C2278 was absent. This clearly showed that Rcm1 is specifically involved in the modification of m5C2278 residue.
Rcm1 catalyzes methylation of m5C2278 of 25S rRNA in vivo and the highly conserved cysteine 404 is indispensable for methylation reaction
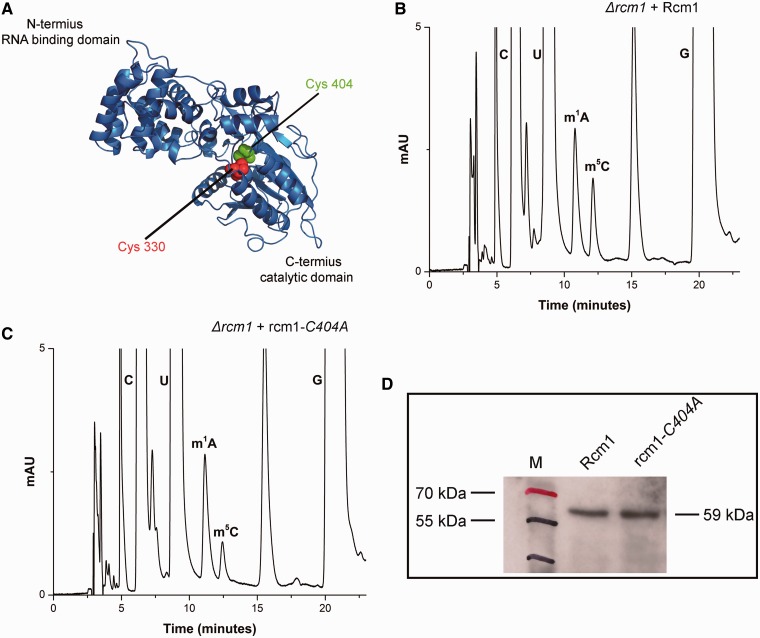
To validate Rcm1 to be an MTase involved in performing m5C modification, the Δrcm1 mutant was complemented with plasmid pSH31 carrying C-terminally heptahistidine tagged Rcm1. The 25S rRNA from this strain was then isolated and composition of its 25S rRNA was analyzed with RP-HPLC. As seen in Figure 4B, the episomally expressed Rcm1 catalyzed the m5C modification in vivo, seen as an increase in the amount of m5C compared with the Δrcm1.
Figure 4.
Rcm1 catalyzes methylation of m5C2278 of 25S rRNA in vivo. (A) In silico 3D structure model of Rcm1. The 3D structure prediction of Rcm1 was performed using a recently described protocol of Kelley and Sternberg, (2009), explained in detail in Supplementary Methods. Rcm1 is predicted to be a Rossmann-like fold methyltransferase. Two highly conserved cysteine residue, Cys330 in motif IV and Cys404 in motif VI, are highlighted as red and green spheres in the cartoon. To substantiate, Rcm1 to be involved in performing m5C modification, plasmid pSH31 carrying C-terminally heptahistidine tagged Rcm1 was transformed into Δrcm1 mutant strain. (B) RP-HPLC chromatogram of 25S rRNA from Δrcm1 strain carrying pSH31 plasmid. As a control, the status of m5C modification of the Δrcm1 strain with mutant rcm1-C404A (C). Like wild-type Rcm1, mutant protein was expressed as C-terminally heptahistidine tagged from plasmid pSH31-b. It became apparent from the chromatograms that the recombinant Rcm1 expressed from pSH31 plasmid was able to methylate the C-5 of C2278 of 25S rRNA, whereas the substitution of catalytically vital cysteine404 to alanine abolished the catalytic potential of Rcm1. (D) Western blot using anti-His antibodies was made to analyze the expression of heptahistidine tagged Rcm1 and mutant rcm1-C404A.
The catalytic mechanism of the RCMTs involves an attack by the thiolate of the motif VI cysteine on the position 6 of the target cytosine base to form a covalent link, thereby activating C-5 for electrophilic substitution by the methyl group of SAM. The motif IV cysteine acts as a general base in the β-elimination of the proton from the methylated cytosine ring. Rcm1 also carries these two highly conserved cysteine residues at position 330 (motif IV) and position 404 (motif VI) shown in Figure 4A.
To corroborate the direct involvement of Rcm1 in performing the m5C2278 modification, we constructed mutant alleles of Rcm1, where both cysteine residues in motif IV and VI were replaced with the alanine. The point mutations were generated on plasmid pSH31 and could be visualized using anti-His antibodies due to their heptahistidine tags. The plasmids pSH31-a (rcm1-C330A) and pSH31-b (rcm1-C404A) carrying the mutant allele of the Rcm1 were then transformed into a Δrcm1 strain. Surprisingly, the transformation with plasmid pSH31-a (rcm1-C330A) turned out to be lethal for the cell (Supplementary Figure S3A). Conversely, the transformation of pSH31-b (rcm1-C404A) caused no growth defect. To examine the functionality of mutant protein (rcm1-C404A), we next analyzed the 25S rRNA composition of its transformants using RP-HPLC. Intriguingly, as seen in Figure 4C, the substitution of cysteine 404 to alanine abolished the catalytic function of Rcm1, as mutant protein failed to perform the C-5 methylation of C2278 residue. Taken together, our results demonstrated the direct involvement of Rcm1 in performing m5C modification and showed that motif VI cysteine is indispensable for C-5 methylation of m5C residues as observed previously for other RCMTs (28).
To further explore the role of cysteine 330 in conferring the lethality and rule out any secondary effect of plasmid on the growth of cell, we transformed the plasmid pSH31-a in the CEN.PK1170-1C strain, where the cytosine 2278 was replaced with the guanine (C2278G). Previous studies with RCMTs have demonstrated that the exchange of motif IV cysteine with alanine abolishes the ß-elimination reaction, responsible for the separation of enzyme from its RNA substrate and consequently results in fixing of the enzyme to its substrate (28). To prove that a covalent fixation of the rcm1-C330A to the 25S rRNA is responsible for the cell death, we transformed the plasmid pSH31a in CEN.PK1170-1C strain, which lacks C2278, the substrate of Rcm1. As expected and shown in Supplementary Figure S3B, the plasmid pSH31a did not exhibit any lethality in CEN.PK1170-1C strain. Moreover western blot analysis confirmed the expression of the mutant protein (Supplementary Figure S3C).
To further support the role of motif IV cysteine residue, we also generated plasmid pSH31c carrying the double mutations, where both conserved cysteines (C330 and C404) were changed to alanine and observed its ability to support viability. Interestingly, the substitution of both cysteines together resulted in no lethality (Supplementary Figure S3D). Taken together, our results clearly showed that the irreversible fixation of rcm1-C330A is apparently responsible for its lethality and together with previous analyses of Trm4 and Nop2, support that cysteine in motif IV is important for the release of methylated RNA, whereas cysteine in motif VI is essential for nucleophillic catalysis (25,28).
Cellular localization of Rcm1
To investigate the cellular localization of Rcm1, a plasmid pSH30 was constructed, where Rcm1 was expressed as Rcm1-GFP fusion protein. The plasmid pSH30 was next transformed into a strain where Nop56, a nucleolar protein, was expressed as a RFP fusion protein. The cells expressing the fusion protein were visualized with the help of a confocal Leica TCS SP5 microscope. As displayed in Supplementary Figure S2, Rcm1 was localized in the nucleolus.
Phenotypic characterization of Rcm1
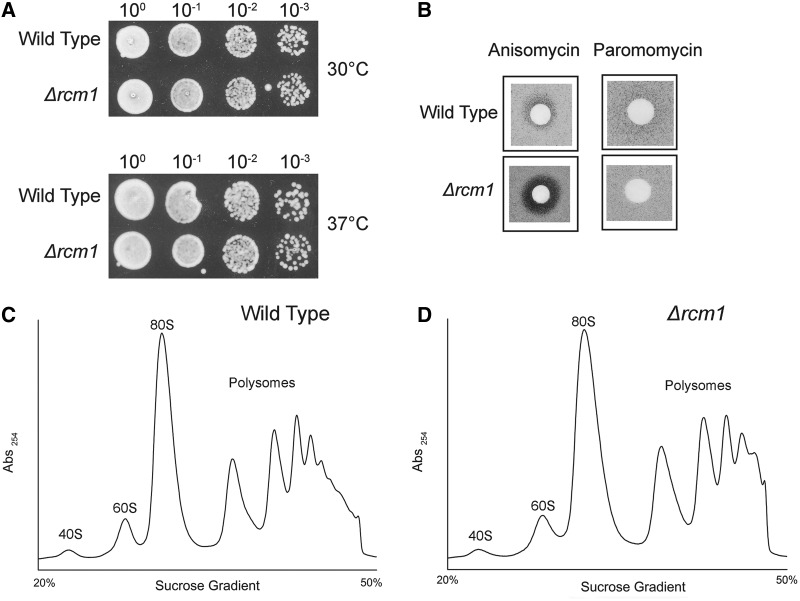
To investigate the impact of loss of Rcm1 and consequently the loss of m5C 2278, we analyzed the growth of Δrcm1 mutant along with rDNA point mutants. Surprisingly, the deletion of rcm1 did not result in any significant growth defect at 30 and 37°C (Figure 5A). Similar to the Δrcm1, the exchange of C2278 to G and T in the 25S rRNA had also no effect on the growth of cells at 30 and 37°C (data not shown).
Figure 5.
Growth analysis and antibiotic sensitivity of the Δrcm1. (A) Ten-fold serial dilutions of the wild-type and Δrcm1 strains were spotted onto solid YPD plates and were incubated at different temperatures. (B) Anisomycin and Paromomycin sensitivity tests were performed by spotting 5 μl of anisomycin (5 µg/ml) and paromomycin solution (200 mg/ml) on filter discs, which were then applied on YPD plates containing the strains indicated. Polysome profile analysis was performed to detect any defects in 60S biogenesis and translation of Δrcm1 compared with isogenic wild type. (C) Polysome profile of the wild type and (D) Polysome profile of Δrcm1. The 59 kDa band corresponds to Rcm1 protein. M stands for protein marker.
We previously showed that the lack of base methylation confers antibiotic hypersensitivity to the cells (37). Moreover the antibiotic analyses are effective tools to detect the changes in conformation of RNA, as the hypersensitivity is often the result of change in RNA structure (42). Interestingly, as seen in Figure 5B, Δrcm1 exhibited hypersensitivity to anisomycin. Anisomycin sensitivity of the Δrcm1 strain would suggest that 25S rRNA experience alterations in the structure owing to lack of m5C2278 modification because the rDNA and Rcm1 point mutants displayed similar hypersensitivity (data not shown).
To investigate any possible role of m5C modification in 60S biogenesis, we analyzed the polysome profile of Δrcm1. As seen in Figure 5C and D, deletion of rcm1 did not disturb the 60S stoichiometry and polysome assembly.
Nop2 catalyzes the methylation of m5C2870 in 25S rRNA
Nop2 has long been predicted to be a RNA cytosine methyltransferase (31). Additionally, previous biochemical and genetic analysis have established Nop2 to be a critical factor for the processing and synthesis of 60S subunits (43). Therefore, we next investigated any possible role of Nop2 in preforming the second m5C2870 base methylation of 25S rRNA.
Nop2 is an essential protein and therefore to validate the involvement of Nop2 as an enzyme responsible for performing m5C modification, we initially attempted to selectively deplete Nop2 using tetracycline aptamer construct and analyzed the effect of the depletion on the m5C modification of 25S rRNA by RP-HPLC (38). Although addition of 250 µM tetracycline arrested growth, we only observed a mild effect of just 10–15% decrease in m5C modification on depletion of Nop2 for 8 h (data not shown). Although only a minor reduction was observed, this was a first hint that Nop2 might be responsible for m5C modification in 25S rRNA.
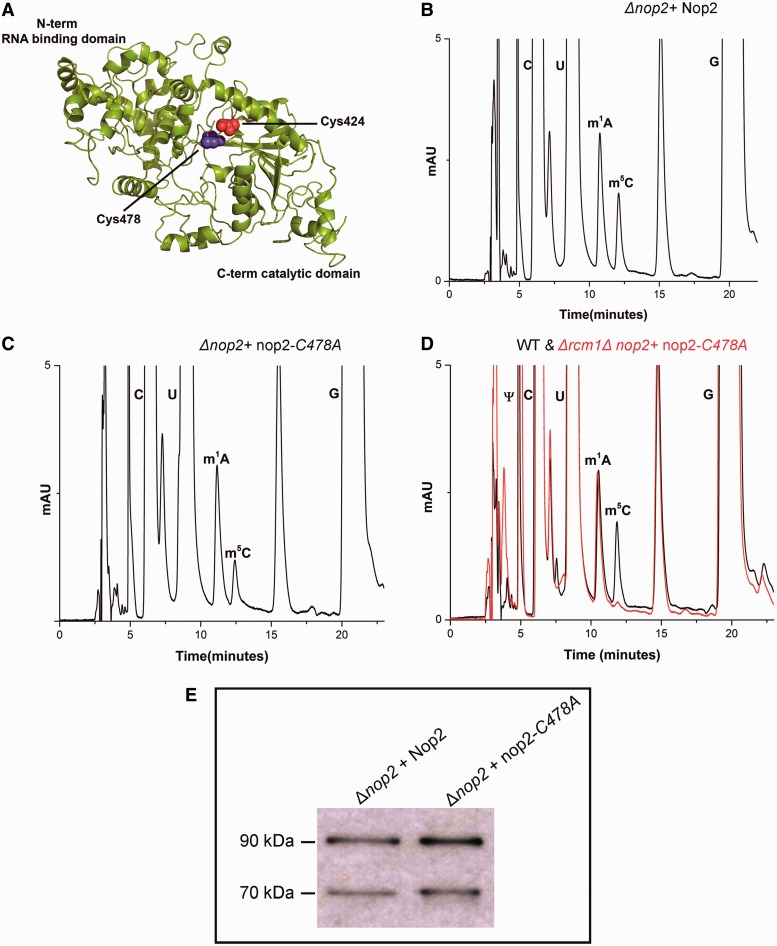
Nop2 contains two highly conserved cysteine residues, Cys424 and Cys478, in motif IV and VI, respectively (Figure 6A). Therefore as performed for Rcm1, we next replaced these conserved cysteine residues of Nop2 in domain IV (Cys424) and domain VI (Cys478) with alanine. These mutations were generated on plasmid pSH17 carrying wild-type Nop2. As observed for Rcm1, the plasmid pSH17-a carrying nop2-C424A (mutation of motif IV cysteine) mutant protein failed to complement the deletion of nop2 (data not shown), as observed previously by Redman's lab (44), whereas pSH17-b carrying nop2-C478A complemented the deletion of NOP2.
Figure 6.
Nop2 catalyzes the C-5 methylation of C2870 in the 25S rRNA of yeast. (A) In silico 3D structure model of Nop2. Like Rcm1, Nop2 is also predicted to be a Rossmann-like fold methyltransferase. Two highly conserved cysteine residues; Cys424 in motif IV and Cys478 in motif VI are highlighted as red and blue spheres in the cartoon. To demonstrate Nop2 to be a RNA cytosine methyltransferase, we generated methyltransferase-dead mutant of nop2, where the catalytically significant cysteine residues Cys424 and Cys478 were exchanged with alanine. All these mutant proteins were expressed from plasmid pSH17-a (C424A) and pSH17-b (C478A) in a Δnop2 strain. Surprisingly, the exchange of Cys424 to alanine failed to complement the deletion of Nop2. (B) RP-HPLC chromatogram of 25S rRNA from Δnop2 carrying wild-type Nop2 expressed from plasmid pSH17. (C) RP-HPLC chromatogram of 25S rRNA from Δnop2 with mutant nop2-C478A expressed from plasmid pSH17-b. Approximately 50% reduction as observed in Δrcm1, in the chromatogram of 25S rRNA of strain carrying mutant nop2-C478A authenticated Nop2 to be an m5C methyltransferase. To further validate that Nop2 is responsible for m5C 2870, we constructed a strain where rcm1 was deleted from Δnop2 + nop2-C478A strain. (D) Overlaid RP-HPLC chromatogram of the nucleosides derived from the 25S rRNA of double mutant Δrcm1Δnop2 + nop2-C478A (red) and isogenic wild type (black). The Δrcm1Δnop2 + nop2-C478A contains no m5C nucleosides, validating the specific involvement of Rcm1 and Nop2 in the m5C modification of 25S rRNA at position 2278 and 2870. (E) To examine the expression of mutant nop2-C478A protein, a western blot using monoclonal antibodies against Nop2 was made. The Nop2 protein was detected at 90 kDa. The lower 70 kDa band corresponds to degradation product.
To analyze the effect of the exchange of C478A on the status of m5C modification of 25S rRNA, the 25S rRNA from the Δnop2 + nop2-C478A mutant were isolated and processed for RP-HPLC analysis. Interestingly, in comparison with wild type, a similar 50% reduction in the amount of m5C nucleosides was detected, as observed previously for the Δrcm1 mutant (Figure 6B and C). This clearly showed that Nop2 is indeed a RCMT and is involved in m5C modification of 25S rRNA. To further substantiate our result, we deleted rcm1 gene from the Δnop2 + nop2-C478A strain and processed the 25S rRNA from this strain for RP-HPLC analysis. As seen in Figure 6D, the peak corresponding to the m5C modification completely disappeared from the 25S rRNA of the double mutant (Δrcm1Δnop2 + nop2-C478A). This clearly confirmed that Nop2 and Rcm1 are RCMTs and are responsible for the methylation of m5C2870 and m5C2278, respectively.
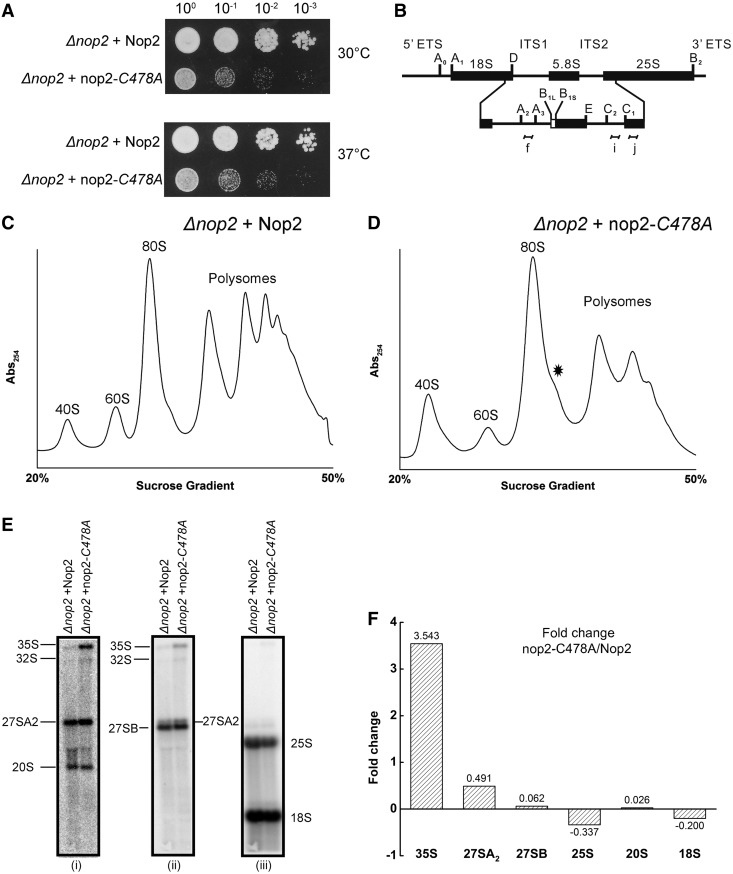
The mutant allele nop2-C478A strongly affects the 60S biogenesis
Interestingly, the growth analysis of the strain carrying mutant nop2-C478A displayed a strong growth phenotype as seen in Figure 7A. The polysome profile from the mutant revealed defect in 60S biogenesis and significant reduction in polysome fractions compared with isogenic wild type (Figure 7C and D). Intriguingly, the northern blot depicted a slower processing of 35S rRNA and 27SA2 pre-rRNA and concomitant decrease in both 25S and 18S rRNA (Figure 7E). The 25S rRNA experienced a stronger reduction as compared with 18S rRNA, which was also supported by the polysome profiles. Generally, the decrease in 60S also leads to the significant accumulation of 43S particles (the translation initiation complex) in the polysome profiles, seen as halfmers. The absence of such significant halfmers from the polysome profiles also implies that in the mutant strain, the biogenesis of 40S is also affected. It is this mutual decrease of both subunits that precludes any halfmer formation. Interestingly, similar phenotypes have been observed on Nop2 depletion (31). Because the mutant allele is expressed in a similar amount to wild type (Figure 6D), comparable defects in processing of pre-rRNA species in the methyltransferase-dead mutant of nop2 and on Nop2 deletion would suggest that these phenotypes are apparently the result of reduced m5C modification. In addition, the change in steady state levels of precursors would also indicate that the modification is performed early during rRNA synthesis and is critical for early rRNA processing.
Figure 7.
Loss of m5C2870 affects ribosome biogenesis and polysome assembly. (A) Ten-fold serial dilutions of the strains were spotted onto solid YPG plates and were incubated at different temperatures. (B) Illustration for the 35S primary transcript. 35S rRNA contains 18S, 5.8S and 25S rRNA sequences separated by ITS1 and ITS2. The processing of the 35S precursor to mature rRNA involves endonucleolytic and exonucleolytic steps at specific sites (C) Polysome profile of Δnop2 + Nop2 and (D) of Δnop2 + nop2-C478A strain. Halfmers formations are indicated by asterisk. (E) Northern blot analysis of the Δnop2 + nop2-C478A mutant. The membrane was hybridized with radioactive labelled probes for ITS1 (i), ITS2 (ii) and to oligonucleotides specific to 18S and 25S rRNA (iii), shown as f, i, d and j in the panel (B), respectively. (F) Quantitative analysis of northern blot. The northern blots were quantified using Image Quant v 5.2 (GE life sciences) and the fold change for various precursors in the mutant strain nop2-C478A were calculated using isogenic wild type as a control.
Analysis of any possible role of Trm112 in catalyzing the m5C modifications
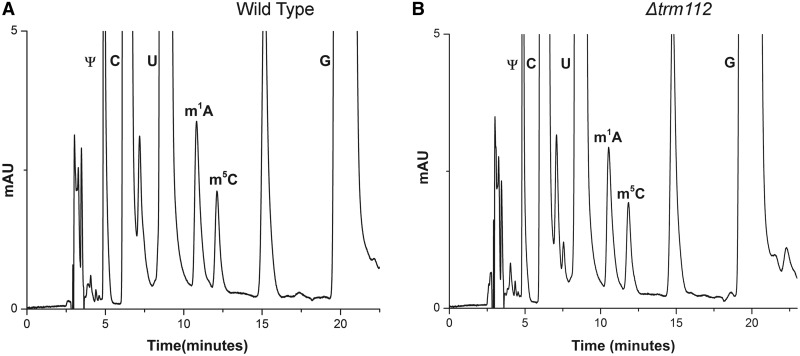
Trm112, a MTase adaptor protein, has been recently observed to play an important role in assisting Bud23 in performing the N-7 methylation of G1575 in 18S rRNA (45,46). These studies have also highlighted a physical interaction of Trm112 with both m5C methyltransferases, Rcm1 and Nop2. This has raised the speculation of Trm112 to be playing a similar role in supporting these two methyltransferases in performing their respective modification. Therefore, to analyze any role of Trm112 in assisting Nop2 and Rcm1 in performing the m5C modification, we analyzed the status of m5C modifications in Δtrm112 mutant by RP-HPLC. As seen in Figure 8, the deletion of trm112 has no significant effect on any of the two cytosines modifications of 25S rRNA. However, as Trm112 physically interacts with Rcm1 and Nop2, it might be possible that Trm112 only partially assists Nop2 and Rcm1 in performing m5C modifications, the effect of which might be difficult to capture. Conversely, as observed for Bud23 and Trm11 mediated methylations, where the modifications disappear completely upon the loss of trm112, and this certainly is not the case for Nop2 and Rcm1. Nevertheless, future work should be directed in analyzing any such subtle effect of Trm112 on cytosine methylations of 25S rRNA.
Figure 8.
Investigation of any role of Trm112 in m5C modifications of 25S rRNA. To analyze any role of Trm112 in assisting Nop2 and Rcm1 in performing the m5C modification, the status of m5C modifications in Δtrm112 mutant were analyzed. RP-HPLC chromatogram of 25S rRNA isolated from (A) wild type and (B) Δtrm112. No significant change in the amount of m5C residues was observed in Δtrm112 mutant compared with the isogenic wild type.
DISCUSSION
The presence of methylated cytosine in DNA of higher eukaryotes is a well-established epigenetic marker. Surprisingly, S. cerevisiae lacks any such modification in the genomic DNA, which is also in consent with the absence of DNA cytosine methyltransferase in yeast. In yeast, only cellular RNAs have been discovered to undergo m5C base modification. The presence of this modification in 25S rRNA was first recognized by Planta's lab in 1973 (15). However, the position and number of the m5C nucleotides in the 25S rRNA of S. cerevisiae remained a mystery for almost three decades.
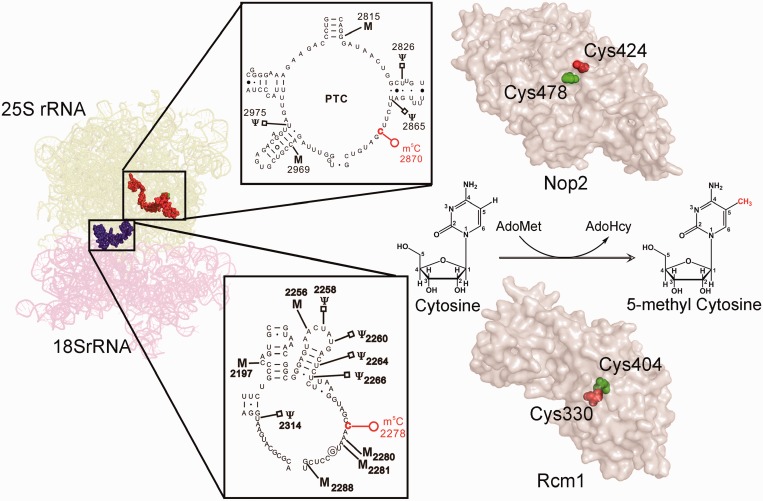
In the present study, we subjected the entire 25S rRNA to a comprehensive m5C analysis, using RP-HPLC and discovered for the first time that S. cerevisiae contain two m5C modifications in the 25S rRNA at position 2278 and 2870 (Figure 9). The m5C2278 is localized in the helix 70, adjacent to helix 69 of 25S rRNA in S. cerevisiae. H70 belongs to the domain IV, which accounts for most of the intersubunit surface of the large subunit of 25S RNA. On the other hand, m5C 2870 is localized in the helix 89 and together with helix 90 and 92 builds the critical A loop of the peptidyl transferase centre. The A loop is responsible for the initial interaction with the incoming peptidyl-tRNAs and to accommodate the tRNAs in the correct conformation for the optimal peptidyl transfer reaction (47).
Figure 9.
Localization of m5C residues and summary of the present study. S. cerevisiae contains two m5C residues, m5C2278 in the helix 70 (shown as blue spheres) and m5C2870 in the helix 89 (shown as red spheres) of 25S rRNA. Nop2 and Rcm1 perform the methylation of these cytosines, individually and the two highly conserved cysteine residues (shown in red and green spheres) are indispensible for the catalytic reaction. The PDB files 3U5B and 3U5E were used for the representation of ribosomal RNA. The cartoon was made by PyMol software (PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.). The pdb files generated by Phyre were used for the surface model of Nop2 and Rcm1.
As far as biological role of m5C modifications is concerned, our results exhibited that the loss m5C2278 in the Helix 70 makes the cells hypersensitive to the anisomycin, whereas the loss of m5C2870 in the helix 89 is vital for the ribosome synthesis and consequently for the translation. As antibiotic sensitivity analyses are significant tools to explore the structural alteration, the increased anisomycin sensitivity would imply that the loss of m5C2278 disturbs the optimal conformation of the helix 70. Interestingly, together with our previous analysis of base modification in helix 65, the present anisomycin hypersensitivity would also implicate that the topology of domain IV of 25S rRNA is crucial for the anisomycin sensitivity.
In the present study, we also identified Rcm1 and Nop2 as the methyltransferases responsible for catalyzing the m5C modifications of 25S rRNA. Rcm1 methylates the cytosine at position 2278, whereas Nop2 catalyzes the methylation of cytosine 2870 (Figure 9). The amino acid sequence analysis of the Rcm1 and Nop2 has a characteristic motif of MTases (Rossmann-like fold protein family). Both Nop2 and Rcm1 share a significant sequence homology at both N and C terminus with the Fmu protein. As observed for Fmu, the N terminal of both Rcm1 and Nop2 contains RNA-binding motifs, supporting their possible role in RNA binding (27). The C terminal of both Rcm1 and Nop2 contains the characteristic MTase (methyltransferase) domain and is apparently involved in the methylation of C-5 of cytosine. In the present study, we demonstrated that the cysteine residue in motif VI, distinctive for RCMTs is crucial for the methylation reactions of Nop2 and Rcm1. The substitution of this cysteine with the alanine residues for both Rcm1 and Nop2 led to the methyltransferase-dead mutant. Conversely, for both Rcm1 and Nop2, the substitution of cysteine in motif IV turned out to be lethal. Intriguingly, as observed previously for Nop2, this lethality of rcm1-C330A mutant protein could be suppressed by together substituting the motif VI cysteine to alanine and by exchanging the cytosine 2278 of 25S rRNA to either guanine or uracil (28,44).
The structural analyses of other RNA cytosine methyltransferase have revealed the distinct catalytic function of both cysteine residues during m5C modification (24,27). The cysteine in motif VI makes a nucleophillic attack on to the C-6 of cytosine and form a covalent adduct with the RNA, whereas the cysteine of motif IV allows the release of substrate. The lethality of both rcm1-C330A and nop2-424 A mutant proteins implies that owing to the replacement of cysteines in motif IV, the mutant proteins fails to detach themselves from their substrate (25S rRNA) and apparently makes a stable protein–rRNA complex. This fixing of the mutant protein then blocks the 25S rRNA processing and eventually results in cell death. This was also supported by the viability of double mutants, where both cysteines (domain IV and VI) were substituted together with alanine, as this prevented the formation of any covalent complex. Together with previous analyses of Trm4 and Nop2, our results further supported that cysteine in motif IV is important for the release of methylated RNA, whereas cysteine in motif VI is essential for nucleophillic catalysis (28). The viability of the methyltransferase-dead mutant of Nop2 exhibited that the catalytic function of Nop2 is not essential for viability. Therefore, the question, what precisely is the essential function of Nop2 that dictates the viability still remains open?
Nop2 and Rcm1 are highly conserved among eukaryotes. Therefore identification of the substrates for these methyltransferases in yeast provides an excellent opportunity to assign the substrates to their homologs in higher organisms, which are otherwise difficult to analyze. Our sequence comparisons for the human homolog of yeast Nop2, p120 (NSUN1), would suggest that NSUN1 is the most suitable candidate for catalyzing m5C4447 residue in the PTC region of human 28S rRNA, as both yeast and human's m5C residue in the helix 89 is located at identical position (Supplementary Figure S4A and B). Similarly, our analysis with Rcm1 demonstrated that NSUN5a is apparently the RCMT responsible for catalyzing the m5C3782 modification of 28S rRNA in human (Supplementary Figure S5). Nevertheless, future biochemical experiments are required to assign the substrates for both NSUN1 and NSUN5. Both NSUN1 and NSUN5A belong to NSUN-related proteins in human (10). There are at least nine different members (NSUN1 to NSUN7, genes NSUN5A, B and C) in human. Interestingly, most of the NSUN proteins carry the putative 5-cytosine methyltransferase domain containing two catalytic cysteines (10). Surprisingly, apart from NSUN2, the human homolog of yeast Trm4, the enzymatic activity of other NSUN members has never been examined (48).
Finally, the present study also emphasized once again the utility of RP-HPLC and mung bean nuclease protection assay in mapping and analyzing the chemical modification of RNA, especially m5C, which is difficult to analyze with other techniques like primer extension. Bisulphite sequencing has been useful for the analysis of m5C residues in DNA and smaller RNA molecules, but for larger RNAs like rRNA, the method lacks reproducibility and need further improvements. Conversely, RP-HPLC allows not only simultaneous qualitative and quantitative analysis of the modified residues but also provide insights into their physical properties. The retention time in an RP-HPLC chromatogram is a convenient measure of the hydrophobicity of the residues. Because hydrophobic interactions play a crucial role in maintaining the RNA tertiary structure, these preliminary hints about the physical properties of the residues can be crucial for understanding the apparent role of these modifications in RNA structure.
In summary, we showed that S. cerevisiae contains two m5C modifications at position 2278 in the helix 70 and at position 2870 in the helix 89. We also showed that Rcm1 and Nop2 methylate these two cytosines, respectively, and the cysteines in motif IV and VI of both proteins are indispensable for the methylation reaction. Furthermore, we also demonstrated that Trm112 plays no role in formation of m5C residues. Determining the substrates of Nop2 and Rcm1 along with the location of these modifications will enable future analyses of these modifications in various organisms to understand their precise role in ribosome structure and function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online, including [48–51].
FUNDING
Funding for open access charge: Deutsche Forschungsgemeinschaft (En134-9).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
S.S. thanks DAAD for the award of PhD scholarship. We thank the members of Entian lab and Prof. Jens Wöhnert for critical and fruitful discussions as well as Prof. Michael Karas for providing us access to mass spectrometry.
REFERENCES
- 1.Yi C, Pan T. Cellular dynamics of RNA modification. Acc. Chem. Res. 2011;44:1380–1388. doi: 10.1021/ar200057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grosjean H, editor. Fine-Tuning of RNA Functions by Modification and Editing. Topics in Current Genetics Series. Vol. 12. Berlin/Heidelberg: Springer; 2005. p. 79. [Google Scholar]
- 3.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways—2012 update. Nucleic Acids Res. 2012;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson TB, Coghill RD. Researches on pyrimidines. c111. the discovery of 5-methyl-cytosine in Tuberculinic acid, the nucleic acid of the Tubercle Bacillus. J. Am. Chem. Soc. 1925;47:2838–2844. [Google Scholar]
- 5.Wyatt GR. Occurrence of 5-methyl-cytosine in nucleic acids. Nature. 1950;166:237–238. doi: 10.1038/166237b0. [DOI] [PubMed] [Google Scholar]
- 6.Allis CD, Jenuwein T, Reinberg D, Caparros ML. Epigenetics. 2007;1:76–80. [Google Scholar]
- 7.Wolffe AP. Epigenetics: regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 8.Wilson GG, Murray NE. Restriction and modification systems. Annu. Rev. Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 9.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basti MM, Stuart JW, Lam AT, Guenther R, Agris PF. Design, biological activity and NMR-solution structure of a DNA analogue of yeast tRNAPhe anticodon domain. Nat. Struct. Biol. 1996;3:38–44. doi: 10.1038/nsb0196-38. [DOI] [PubMed] [Google Scholar]
- 12.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 13.Kadaba S. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maden B. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man: clustering in the conserved core of molecule. J. Mol. Biol. 1988;201:289–314. doi: 10.1016/0022-2836(88)90139-8. [DOI] [PubMed] [Google Scholar]
- 15.Klootwijk J, PLANTA RJ. Analysis of the methylation sites in yeast ribosomal RNA. Eur. J. Biochem. 1973;39:325–333. doi: 10.1111/j.1432-1033.1973.tb03130.x. [DOI] [PubMed] [Google Scholar]
- 16.Veldman GM, Klootwijk J, de Regt VC, Planta RJ, Branlant C, Krol A, Ebel JP. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981;9:6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res. 1993;21:4577–4585. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith JE, Cooperman BS, Mitchell P. Methylation sites in Escherichia coli ribosomal RNA: Localization and identification of four new sites of methylation in 23 S rRNA. Biochemistry. 1992;31:10825–10834. doi: 10.1021/bi00159a025. [DOI] [PubMed] [Google Scholar]
- 19.Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2008;37:e12. doi: 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozbial PZ, Mushegian AR. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 2005;5:19. doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavlopoulou A, Kossida S. Phylogenetic analysis of the eukaryotic RNA (cytosine-5)-methyltransferases. Genomics. 2009;93:350–357. doi: 10.1016/j.ygeno.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Reid R, Greene PJ, Santi DV. Exposition of a family of RNA m5C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999;27:3138–3145. doi: 10.1093/nar/27.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bujnicki JM. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walbott H, Husson C, Auxilien S, Golinelli-Pimpaneau B. Cysteine of sequence motif VI is essential for nucleophilic catalysis by yeast tRNA m5C methyltransferase. RNA. 2007;13:967–973. doi: 10.1261/rna.515707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Santi DV. m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc. Natl Acad. Sci. USA. 2000;97:8263–8265. doi: 10.1073/pnas.97.15.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foster PG, Nunes CR, Greene P, Moustakas D, Stroud RM. The first structure of an RNA m 5 C methyltransferase, Fmu, provides insight into catalytic mechanism and specific binding of RNA substrate. Structure. 2003;11:1609–1620. doi: 10.1016/j.str.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 28.King MY, Redman KL. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry. 2002;41:11218–11225. doi: 10.1021/bi026055q. [DOI] [PubMed] [Google Scholar]
- 29.Reid R, Greene PJ, Santi DV. Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999;27:3138–3145. doi: 10.1093/nar/27.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motorin Y, Grosjean H. Multisite-specific tRNA: m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong B, Brockenbrough JS, Wu P, Aris JP. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato G, Saijo Y, Uchiyama B, Kumano N, Sugawara S, Fujimura S, Sato M, Sagawa M, Ohkuda K, Koike K, et al. Prognostic value of nucleolar protein p120 in patients with resected lung adenocarcinoma. J. Clin. Oncol. 1999;17:2721–2727. doi: 10.1200/JCO.1999.17.9.2721. [DOI] [PubMed] [Google Scholar]
- 33.Sarrió D, Pérez-Mies B, Hardisson D, Moreno-Bueno G, Suárez A, Cano A, Martín-Pérez J, Gamallo C, Palacios J. Cytoplasmic localization of p120ctn and E-cadherin loss characterize lobular breast carcinoma from preinvasive to metastatic lesions. Oncogene. 2004;23:3272–3283. doi: 10.1038/sj.onc.1207439. [DOI] [PubMed] [Google Scholar]
- 34.Doll A, Grzeschik KH. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in williams-beuren syndrome. Cytogenet. Cell Genet. 2001;95:20–27. doi: 10.1159/000057012. [DOI] [PubMed] [Google Scholar]
- 35.Meyer B, Wurm JP, Kotter P, Leisegang MS, Schilling V, Buchhaupt M, Held M, Bahr U, Karas M, Heckel A, et al. The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of 1191 in yeast 18S rRNA. Nucleic Acids Res. 2011;39:1526–1537. doi: 10.1093/nar/gkq931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McEntee CM, Hudson AP. Preparation of RNA from unspheroplasted yeast cells (Saccharomyces cerevisiae) Anal. Biochem. 1989;176:303–306. doi: 10.1016/0003-2697(89)90313-8. [DOI] [PubMed] [Google Scholar]
- 37.Sharma S, Watzinger P, Kötter P, Entian KD. Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2013;41:5428–5443. doi: 10.1093/nar/gkt195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotter P, Weigand JE, Meyer B, Entian KD, Suess B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. 2009;37:e120. doi: 10.1093/nar/gkp578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gehrke CW, Kuo KC. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J. Chromatogr. 1989;471:3–36. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- 40.Andersen TE, Porse BT, Kirpekar F. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004;10:907–913. doi: 10.1261/rna.5259404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peifer C, Sharma S, Watzinger P, Lamberth S, Kotter P, Entian KD. Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA. Nucleic Acids Res. 2013;41:1151–1163. doi: 10.1093/nar/gks1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PLoS One. 2007;2:e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talkish J, Zhang J, Jakovljevic J, Horsey EW, Woolford JL. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:8646–8661. doi: 10.1093/nar/gks609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King M, Ton D, Redman KL. A conserved motif in the yeast nucleolar protein Nop2p contains an essential cysteine residue. Biochem. J. 1999;337(Pt 1):29–35. [PMC free article] [PubMed] [Google Scholar]
- 45.Figaro S, Wacheul L, Schillewaert S, Graille M, Huvelle E, Mongeard R, Zorbas C, Lafontaine DLJ, Heurgue-Hamard V. Trm112 Is Required for Bud23-Mediated Methylation of the 18S rRNA at Position G1575. Mol. Cell. Biol. 2012;32:2254–2267. doi: 10.1128/MCB.06623-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sardana R, Johnson AW. The methyltransferase adaptor protein Trm112 is involved in biogenesis of both ribosomal subunits. Mol. Biol. Cell. 2012;23:4313–4322. doi: 10.1091/mbc.E12-05-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 48.Auxilien S, Guérineau V, Szweykowska-Kulińska Z, Golinelli-Pimpaneau B. The human tRNA m (5) C methyltransferase Misu is multisite-specific. RNA Biol. 2012;9:1331–1338. doi: 10.4161/rna.22180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 50.Chernoff YO, Vincent A, Liebman SW. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13:906. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelley LA, Sternberg M. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.