Abstract
Progesterone receptors (PR) are transcription factors relevant to breast cancer biology. Herein, we describe an N-terminal common docking (CD) domain in PR-B, a motif first described in mitogen-activated protein kinases. Binding studies revealed PR-B interacts with dual-specificity phosphatase 6 (DUSP6) via the CD domain. Mutation of the PR-B CD domain (mCD) attenuated cell cycle progression and expression of PR-B target genes (including STAT5A and Wnt1); mCD PR-B failed to undergo phosphorylation on Ser81, a ck2-dependent site required for expression of these genes. PR-B Ser81 phosphorylation was dependent on binding with DUSP6 and required for recruitment of a transcriptional complex consisting of PR-B, DUSP6 and ck2 to an enhancer region upstream of the Wnt1 promoter. STAT5 was present at this site in the absence or presence of progestin. Furthermore, phospho-Ser81 PR-B was recruited to the STAT5A gene upon progestin treatment, suggestive of a feed-forward mechanism. Inhibition of JAK/STAT-signaling blocked progestin-induced STAT5A and Wnt1 expression. Our studies show that DUSP6 serves as a scaffold for ck2-dependent PR-B Ser81 phosphorylation and subsequent PR-B-specific gene selection in coordination with STAT5. Coregulation of select target genes by PR-B and STAT5 is likely a global mechanism required for growth promoting programs relevant to mammary stem cell biology and cancer.
INTRODUCTION
Progesterone is an ovarian steroid hormone essential for breast development and implicated in breast cancer progression. Progesterone receptors (PR) exist primarily as two coexpressed isoforms, PR-A and PR-B (1,2), encoded by the same gene downstream of distinct promoters (3). PR-B, the full-length receptor, contains 164 amino acids at the N-terminus, not present in PR-A, termed the B-upstream segment (BUS) (4). Both receptors contain the same DNA-binding domain (DBD), a hinge region (H) and two activator function (AF) domains; PR-B contains a third AF domain in the BUS (5). Unliganded PR rapidly shuttles between the cytoplasm and the nucleus. After ligand binding, however, PR undergoes dimerization and is retained in the nucleus. Nuclear PR, together with coactivators and corepressors, activates or represses transcription of PR target genes, either directly through DNA binding to progesterone response elements (PREs) or indirectly through tethering interactions with other transcription factors (AP1, SP1, STATs) (6–9).
PR-mediated regulation of gene expression is controlled by many posttranslational modifications to the receptor, primarily on N-terminal serine (phosphorylation) and lysine (ubiquitination, sumoylation and acetylation) residues (10–15). These modifications significantly alter receptor stability, localization, transcriptional activity and target gene selectivity. PR is phosphorylated on serines (Sers) 294, 345 and 400 by mitogen-activated protein kinase (MAPK) and cyclin-dependent kinase 2 (cdk2). PR-B is also phosphorylated on Ser81 by ck2 [formerly casein kinase II; (16–19)], a ubiquitously expressed, constitutively active kinase that is overexpressed in every cancer examined thus far, including breast cancer (16,20). ck2-dependent PR-B phosphorylation of Ser81 regulates a specific subset of PR-B target genes involved in breast cancer cell growth and pro-survival, including BIRC3, HSD11β2 and HbEGF (19). In addition, ck2 is recruited along with Ser81-phosphorylated PR-B to enhancer sites of a subset of progesterone-responsive target genes (19). Notably, these studies have shown that ck2-dependent phosphorylation of PR-B Ser81 is unique to PR-B and thereby a primary determinant of PR isoform-specific activity. However, the molecular interactions necessary to support PR-B Ser81 phosphorylation have yet to be understood.
Posttranslational modifications to PR regulate the receptor’s interactions with other proteins (21). PR protein interaction domains include the estrogen receptor (ER) interaction domains [ERIDs (22)] and a poly-proline-rich (p-Pro) domain that is required for interaction between PR and the SH3-domain of c-Src (23). PR interacts with many other proteins via unknown domains (i.e. MEK1, FGFR2, STAT5) (21,24–26). In a recent in silico analysis of the PR amino acid sequence aimed at identifying protein interaction domains, we identified a putative common docking (CD) domain in the N-terminal BUS region of full-length PR-B (21), a region that is not present in other PR isoforms. CD domains are commonly found in members of the MAPK family, where they mediate interactions between MAPKs and their upstream activators (MEKs), negative regulators (MAPK phosphatases [MKPs] or dual-specificity phosphatases [DUSPs]) and downstream targets (27,28). CD domains are characterized by clusters of negatively charged amino acids (aspartic or glutamic acid) that form electrostatic interactions with a positively charged ‘D domain’ in their respective binding partner [MKKs, DUSPs, MAPK substrates; reviewed in (29)]. The PR-B CD domain is an identical match to the CD domain of the MAPK family member, Erk2. This identical match suggests that PR-B interacts with the same D domain-containing proteins as Erk2. However, the function of this domain, unique to the PR-B isoform, has not yet been determined.
We predict that both PR-B Ser81 phosphorylation and PR-B CD domain interactions may be involved in breast cancer progression. Indeed, PR has been implicated in breast cancer progression in recent clinical studies of hormone-replacement therapy. These studies found that women taking estrogen and progesterone had more and larger breast tumors than women taking estrogen alone (30,31). In addition, we recently identified a phosphorylated PR-B gene signature associated with decreased survival in women with luminal (ER+ and/or PR+) breast cancer whose disease stopped responding to anti-ER therapy with tamoxifen (13). Mitogenic protein kinases—such as MAPK, c-Src, cdk2 and ck2—are commonly upregulated in cancer (17,32–34) and represent likely pathway components in PR-mediated gene expression in breast cancer. Understanding how PR interacts with these protein kinases and their regulatory protein partners is critical to understanding breast tumor etiology and developing better treatments. Herein, we sought to identify proteins that interact with PR-B via a novel N-terminal CD domain and how protein–protein interactions through this domain alter PR-B phosphorylation and transcription factor function in breast cancer models.
MATERIALS AND METHODS
Cell lines and constructs
The estrogen-independent ER/PR-positive T47Dco (T47D) variant cell line has been previously described (35), and is the parent cell line from which all T47D-variants used herein were created. T47D-Y (PR negative), T47D-YB (stably expressing wt PR-B) and T47D-YA (stably expressing wt PR-A) cells were characterized by Sartorius et al. (36). T47D-S79/81A PR-B cells have been previously described (19). T47D-mCD PR-B cells were created by stable expression of pSG5-mCD PR-B and pSV-neo in T47D-Y cells using FuGene-HD (Roche). Individual colonies were selected in 500 µg/ml G418 and maintained in 200 µg/ml G418 after initial selection in cMEM (as described in the Supplementary Materials and Methods). All cells were maintained as previously described or described in the Supplementary Materials and Methods (19). Creation of the pSG5-mCD PR-B, CD-PR-A and mCD-PR-A plasmids was performed as described in the Supplemental Materials and Methods. The DUSP6 construct [pcDNA3.1(-)Myc-His] was a gift from Stefanie Dimmeler, University of Frankfurt.
Gene expression profiling
T47D cells stably expressing pSG5 empty vector, wt PR-B or mCD PR-B were serum starved in modified iMEM (Gibco) for 1 day and treated with R5020 (10 nM) or vehicle control for 6 h. Total RNA was extracted using a RNeasy kit (QIAgen) with on-column DNase I treatment (QIAgen). Triplicate RNA samples were labeled and hybridized to the Illumina HT-12v4 bead chip platform according to the manufacturer’s protocols. Chip scanning within Genome Studio software produced raw expression values that were analyzed within R software using the Bioconductor (37) package called lumi, where raw intensities were log2 transformed and quantile normalized. Differentially expressed genes were analyzed using the limma package, where empirical Bayes was used to better estimate the variance of the genes. Reported gene expression data contain log2 normalized intensities and biological comparisons (e.g. R5020/vehicle) contain log2 fold change with the Benjamini and Hochberg (BH)-adjusted P-value (38). Heat maps were generated through unsupervised hierarchical clustering of probes by the heatmap.2 function in the gplots R software package. Clustering was performed using Euclidean distance and complete linkage. Rows were scaled to have mean zero and standard deviation (SD) equal to one. Gene expression data are available in the NCBI Gene Expression Omnibus database (accession number: GSE46850.
Pathway and gene set analysis
Ingenuity Pathway Analysis (IPA) software (Ingenuity® Systems, www.ingenuity.com) was used to compare biological functions or network pathways in cells expressing wt or mCD PR-B after progestin treatment (wt + R5020/wt – R5020 or mCD + R5020/mCD – R5020). Default IPA settings for Core analyses were used to evaluate upregulated genes (fold change >1.5, P < 0.05). IPA Comparison analyses were used to reveal whether cells expressing wt or mCD PR-B regulated functionally distinct pathways. Analyses were scored based on significance (the BH-adjusted P-value, corrected for multiple hypothesis testing) and the threshold for a gene list to be significantly involved in a particular biological function was P < 0.05 [i.e. –log10(BH-adjusted P-value) >1.30].
Gene set enrichment (GSEA) and Leading Edge analysis (39,40) was performed using the javaGSEA desktop software; all five gene set collections (c1–c5) from the Molecular Signatures Database (MSigDB) version 3.1 were queried. Data set files were developed based on normalized Illumina expression intensities from cells that constitutively express PR. Specifically, the log2 fold change values were compared for two phenotypes in our ligand-dependent analysis: (wt +R5020/–R5020) versus (mCD +R5020/–R5020). GSEA was executed using the default settings, except the permutation type was set to Gene_set with 1000 permutations, and the metric for ranking genes was set to Diff_of_Classes because normalized expression data were log2 transformed. Each MSigDB collection was analyzed individually in multiple GSEA runs.
Immunoblotting
Immunoblotting was performed as previously described (19) and in the Supplementary Materials and Methods.
Coimmunoprecipitation experiments
For coimmunoprecipitation (CoIP) experiments, cell lysates were collected in RIPA (supplemented as described in the Supplementary Materials and Methods) and incubated on ice for 60 min. Cell lysates containing equivalent protein concentrations (1000 µg) were incubated overnight at 4°C with 2 µg appropriate antibody or control IgG. Protein G agarose (Roche Diagnostics) was added for the final 1 h of incubation time. Immune complexes were washed three times with supplemented RIPA buffer, resuspended in Laemmli sample buffer containing dithiothreitol and β-mercaptoethanol, boiled for 5 min and subjected to western blotting analysis.
Transient transfections
Transient transfections were performed as previously described (19) and in the Supplementary Materials and Methods.
H2O2 assays
After T47D-wt PR-B cells were starved overnight in serum-fee iMEM, they were pretreated with 1 mM H2O2 for 20 min, followed by 30 min with 10 nM R5020. Protein lysates were isolated and analyzed as described above.
Luciferase transcription assays
Luciferase assays were performed as previously described (41) using the Dual Luciferase Reporter Assay (Promega). Relative luciferase units were normalized to Renilla ± SD.
siRNA
ON-TARGETplus SMARTpool designed to target human DUSP6 (DUSP6) and nonsilencing siRNA controls were purchased from Dharmacon. For siRNA experiments, T47D-YB cells were plated, and 24 h later they were transfected with 50 nM nonsilencing or DUSP6 siRNA. After 72 h, cells were treated with EtOH or 10 nM R5020 for 60 min. Protein lysates were isolated and analyzed as described in the Supplementary Materials and Methods.
Reagents
Cells were treated with the following reagents (when applicable): R5020 (10 nM; Sigma), AG490 (50 µM; CalBioChem) and H2O2 (1 mM; from Cell Biolabs OxiSelect™ Intracellular ROS Assay Kit).
Cell cycle analysis/flow cytometry
Flow cytometry for cell cycle analysis was performed as previously described (42).
Real-Time Quantitative polymerase chain reaction
Real-time quantitative polymerase chain reaction (RT-qPCR) was performed as described previously (19) and in the Supplementary Materials and Methods. Primer sets used for qPCR are listed in Supplementary Table S2. Relevant genomic sequence information and enhancer positioning for Wnt1 genomic sequence is based on GR37 Release 57 (March 2010).
ChIP assays
ChIP was performed using the ChIP-IT Express Kit (Active Motif) according to the manufacturer’s instructions using sonication as the method for chromatin shearing. Lysates were immunoprecipitated (IP) overnight (18 h) with the following antibodies: PR (ThermoScientific #MS-298-P), STAT5 (Santa Cruz Biotechnology sc-1081 and sc-836), ck2α (Santa Cruz sc-12738), DUSP6 (Santa Cruz Biotechnology sc-100374) or an equal amount of mouse or rabbit IgG. Resulting DNA was analyzed using qPCR as described above, and data are represented as a percentage of input DNA. In silico analysis using MatInspector (Genomatix) identified potential PRE-binding sites using the following consensus sequence: RGNACANRNTGTNCY. Primer sets used for ChIP-qPCR are listed in Supplementary Table S2.
CEAS
Web-based CEAS analysis (http://ceas.cbi.pku.edu.cn/index.html) was performed on a publicly available PR ChIP–Chip data set (http://cistrome.org/NR_Cistrome/Cistrome.html). TRANSFAC and JASPAR motifs were used to determine putative transcription factor binding sites.
Statistics
Statistical significance for all experiments was determined using an unpaired Student’s t-test, unless otherwise specified.
RESULTS
PR-B CD domain is required for progestin-induced S-phase entry
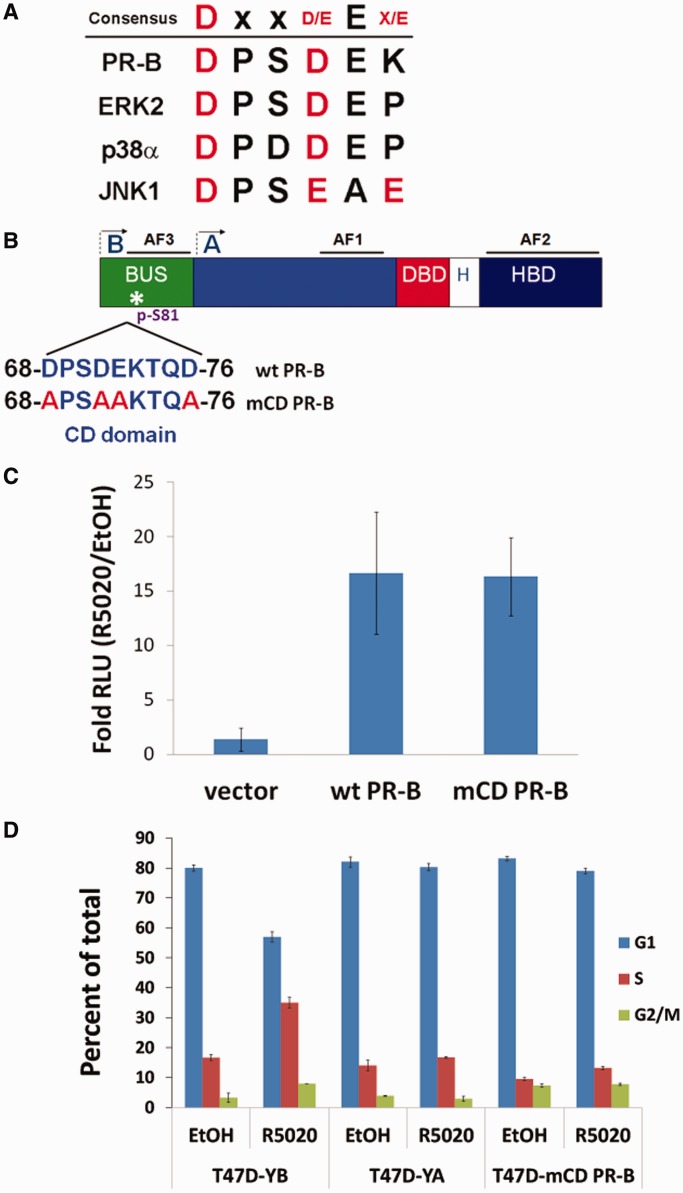
We previously identified a putative CD domain (Figure 1A) (21) located in the N-terminal BUS region of full-length PR-B, a region that is absent from other PR isoforms (Figure 1B). To study the importance of this newly identified CD domain in modulating PR-B-specific functions, we mutated the critical negatively charged amino acids (D-aspartic acid, E-glutamic acid) to alanines (A), creating an mCD PR-B mutant (Figure 1B). Similar mutational strategies have been used to study CD domains present in Erks and other MAPKs (27,43). We then determined whether mCD PR-B was able to bind DNA and activate PRE-dependent transcription in luciferase reporter gene assays. PRE-luciferase expression levels were increased at similar levels in HeLa cells transiently expressing wt or mCD PR-B following treatment with vehicle (ethanol) or 10 nM R5020 (synthetic progesterone) (Figure 1C). These data suggest that the PR-B CD domain is not required for intrinsic PR-B transcriptional activity.
Figure 1.
CD domain in PR-B is required for progestin-induced S-phase entry. (A) Table comparing the CD domain core amino acid sequences in PR-B and selected MAPKs. (B) Schematic representing functional domains in wt PR-B. The full-length receptor has an N-terminal region unique to PR-B, termed the BUS. Both PR-B and the shorter form, PR-A, contain two activating function (AF 1 and 2) domains, a DNA-binding domain (DBD), hinge region (H) and a hormone-binding domain (HBD). PR-B contains an additional AF3, located in the BUS. Arrows mark the translational start sites for PR-B and PR-A. The CD domain is located between amino acids 68–76, immediately upstream of a ck2-dependent phosphorylation site, Ser81. Negatively charged amino acids were mutated to alanines (in red) to create the mCD PR-B mutant. (C) Reporter gene assays measuring transcriptional activity of PR constructs. HeLa cells were transiently transfected with plasmids expressing wt PR-B, mCD PR-B or vector, as well as a firefly PRE-luciferase reporter construct and Renilla expression control. Luciferase assays were performed as described in the ‘Materials and Methods’ section. Fold relative luciferase units (PRE-luciferase over Renilla luciferase controls) of R5020/EtOH-treated cells are plotted. Error bars represent ± SD of three independent experiments. (D) PR-B CD domain required for progestin-induced S-phase entry. T47D cells stably expressing wt PR-B (T47D-YB), PR-A (T47D-YA) or mCD PR-B (T47D-mCD PR-B) were starved for 18 h in serum-free media, followed by treatment with 10 nM R5020 or vehicle for 18 h. Single cells were analyzed by flow cytometry.
Progesterone or synthetic progestins (R5020) induce S-phase entry in breast cancer cells expressing PR-B (44–46), but not PR-A (47). Experimental isolation of PR isoform-specific actions is complicated by the fact that estradiol is generally required for robust PR expression in steroid hormone receptor-positive breast cancer models. Not only is estrogen itself a potent mitogen, but it tightly controls PR isoform expression (48). To overcome this barrier, we used the ER-positive T47Dco cell line, which expresses abundant PR-A and PR-B in the absence of added estrogen (35). A naturally occurring PR-negative variant of T47Dco cells, termed T47D-Y, was used to create stable cell lines constitutively expressing either wt PR-B (T47D-YB) or wt PR-A (T47D-YA) (49). We then tested the contribution of the PR-B CD domain to cell cycle progression by stably expressing mCD PR-B (T47D-mCD PR-B) in T47D-Y cells and analyzed progestin-induced S-phase entry. As predicted, there was an increase in progestin-induced S-phase entry in T47D-YB cells but not in T47D-YA cells (Figure 1D). Interestingly, cells stably expressing mCD PR-B also failed to enter S-phase upon treatment with R5020; these cells resembled PR-A expressing cells (Figure 1D). These data suggest that the PR-B CD domain is essential for proliferative signaling in breast cancer cells, as measured by progestin-induced S-phase entry.
PR-B CD domain regulates select PR-B target genes
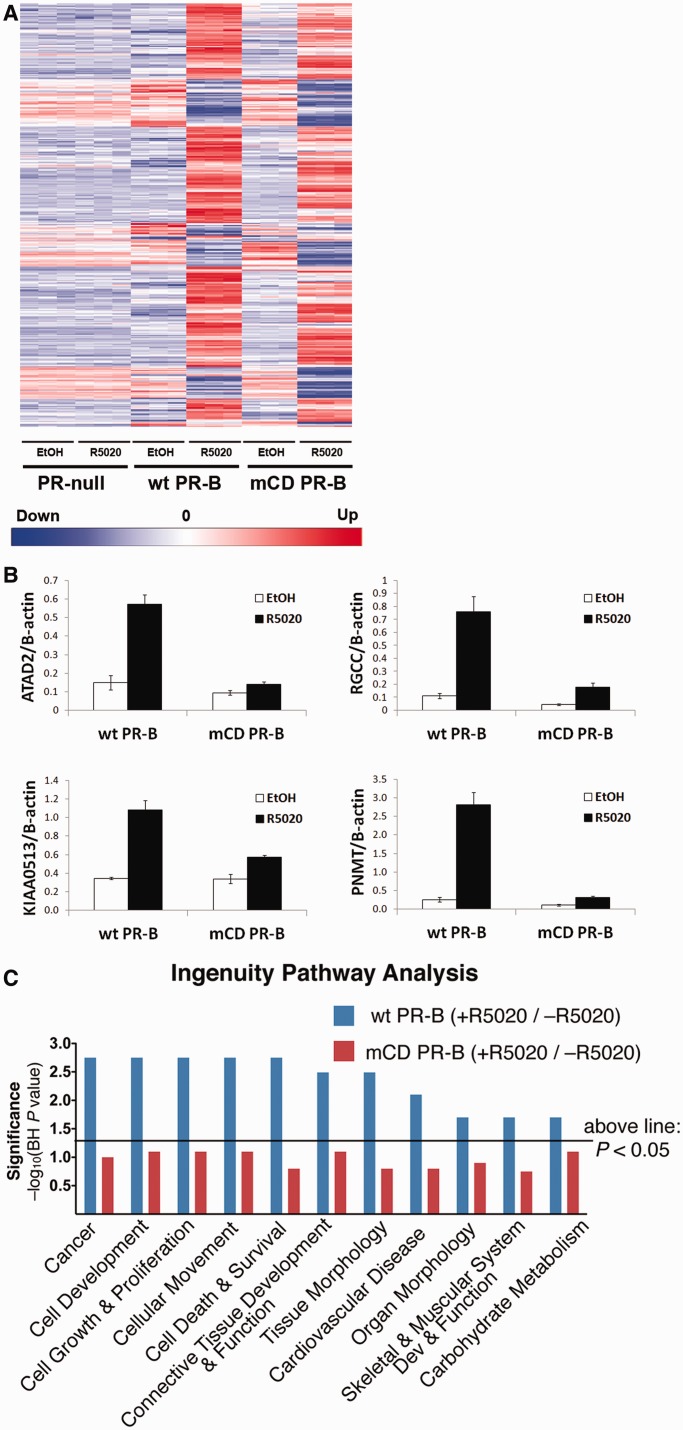
Although mutation of the PR-B CD domain did not appreciably alter the absolute levels of PR-B transcriptional activity, posttranslational modifications can dramatically alter PR target gene selectivity, directing PR to specific enhancer or promoter regions in chromatin (50–52). This is an important feature of steroid hormone receptor action that is missed in reporter assay systems such as luciferase. To determine whether the PR-B CD domain functions in the regulation of endogenous PR target genes, we performed global gene expression analyses using Illumina HT-12v4 whole genome bead arrays. Triplicate gene expression analyses were performed on T47D-Y (PR-null), T47D-YB and T47D-mCD PR-B cells after 6 h of treatment with R5020 or vehicle (ethanol). Transcriptional differences between cells expressing wt and mCD PR-B are evident in the heat map of significantly upregulated or downregulated (≥1.5-fold) genes (Figure 2A). Differential regulation of numerous genes that require an intact CD domain for ligand-induced expression was validated by RT-qPCR (Figure 2B). Using IPA software, we compared the wt and mCD PR-B gene sets to a large database of genes that have been manually assigned to molecularly defined pathways, biological functions or disease states. Interestingly, genes that were specifically upregulated (≥1.5-fold) in cells expressing wt but not mCD PR-B (P < 0.05, BH adjusted) were identified as significantly involved in pathways regulating cell proliferation, survival and cancer (Figure 2C; line represents P = 0.05 significance). Importantly, the individual genes validated by RT-qPCR (Figure 2B) were included in the CD-regulated gene set assigned to these IPA-defined pathways. These data suggest that the CD domain in PR-B is critical for PR’s ligand-dependent contributions to cell growth and survival pathways, and verify that the CD domain regulates a biologically significant subset of PR-B target genes.
Figure 2.
Gene expression analysis in mCD PR-B cells. (A) Heat map highlighting transcriptional differences between cells stably expressing wt PR-B, mCD PR-B or PR-null. The indicated T47D cells were treated with R5020 or vehicle. Genes differentially expressed >1.5 fold (BH-adjusted P < 0.01) are displayed for each treatment group. The experiment was performed in triplicate. (B) Validation of CD-regulated genes. T47D-Y cells stably expressing either wt or mCD PR-B were starved for 18 h in serum-free media, followed by treatment with 10 nM R5020 or EtOH for 6 h. mRNA levels for selected genes were analyzed by qPCR. Error bars represent ± SD. (C) IPA comparing >1.5-fold upregulated genes from wt PR-B and mCD PR-B expressing cells. Line represents P = 0.05 significance.
PR-B CD domain is required for PR-B Ser81 phosphorylation in response to ligand
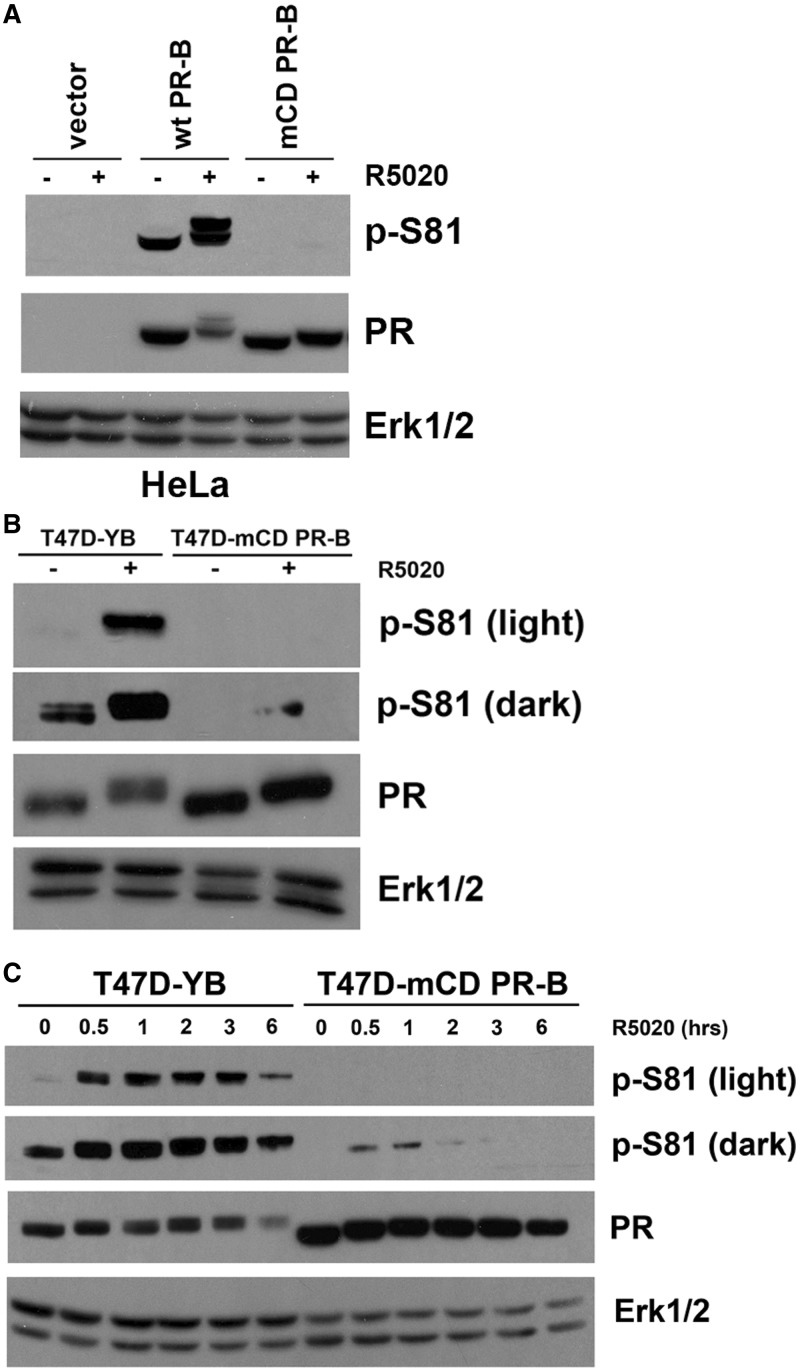
PR phosphorylation occurs on multiple sites and is a key determinant of receptor localization, ubiquitin-dependent turnover, tethering interactions and hormone responsiveness at selected PR target genes [reviewed in (53)]. We therefore screened for differences in basal and regulated phosphorylation of wt and mCD PR-B using phospho-specific PR antibodies. Notably, Ser81, a basally phosphorylated site that is further upregulated by ck2 in response to ligand-binding (19), failed to undergo basal (absence of ligand) phosphorylation in HeLa cells transiently expressing mCD PR-B (Figure 3A) or T47D cells stably expressing mCD PR-B (Figure 3B). Similarly, ligand-induced PR-B Ser81 phosphorylation was greatly diminished in cells expressing mCD PR-B relative to wt PR-B (Figure 3A and B). A time course of PR-B Ser81 phosphorylation in response to R5020 treatment verified that mCD PR-B is persistently weakly phosphorylated relative to wt PR-B (Figure 3C). Additionally, we analyzed PR phosphorylation on proline-directed sites (Sers 294, 345 and 400) in HeLa cells transiently transfected with either wt or mCD PR-B and then treated with R5020. Despite relatively equal levels of total wt or mCD PR-B expression, PR phosphorylation occurred with faster kinetics in cells expressing mCD PR-B relative to cells expressing wt PR-B. After 60 min, however, similar levels of phosphorylation were achieved in both groups (Supplementary Figure S1). These data suggest that mutation of PR-B’s CD domain dramatically alters PR phosphorylation in response to ligand. Namely, Ser81 fails to be persistently phosphorylated, while a number of proline-directed sites (Sers 294, 345 and 400) appear to exhibit transient or temporary hyper-phosphorylation that is not persistently maintained.
Figure 3.
PR-B phosphorylation is altered by mutation to the CD of PR-B. (A) PR-B Ser81 phosphorylation is reduced in cells expressing mCD PR-B. HeLa cells were transiently transfected with wt PR-B, mCD PR-B or vector only. Twenty-four hours after transfection, cells were starved for 18 h in serum-free media and then treated with 10 nM R5020 or EtOH for 60 min. Lysates were analyzed via western blotting using p-S81, PR-B and Erk1/2 antibodies. (B) T47D cells stably expressing wt PR-B or mCD PR-B were starved and treated as in (A). Lysates were analyzed as in (A). A light and dark exposure of the western blotting film is shown for blotting with the p-Ser81 antibody. (C) Time course of Ser81 phosphorylation in cells expressing wt and mCD PR-B. T47D cells stably expressing wt PR-B or mCD PR-B were starved for 18 h in serum-free media, followed by treatment with 10 nM R5020 for 0–6 h. Lysates were analyzed via western blotting as in (A).
PR-B CD domain interacts with DUSP6
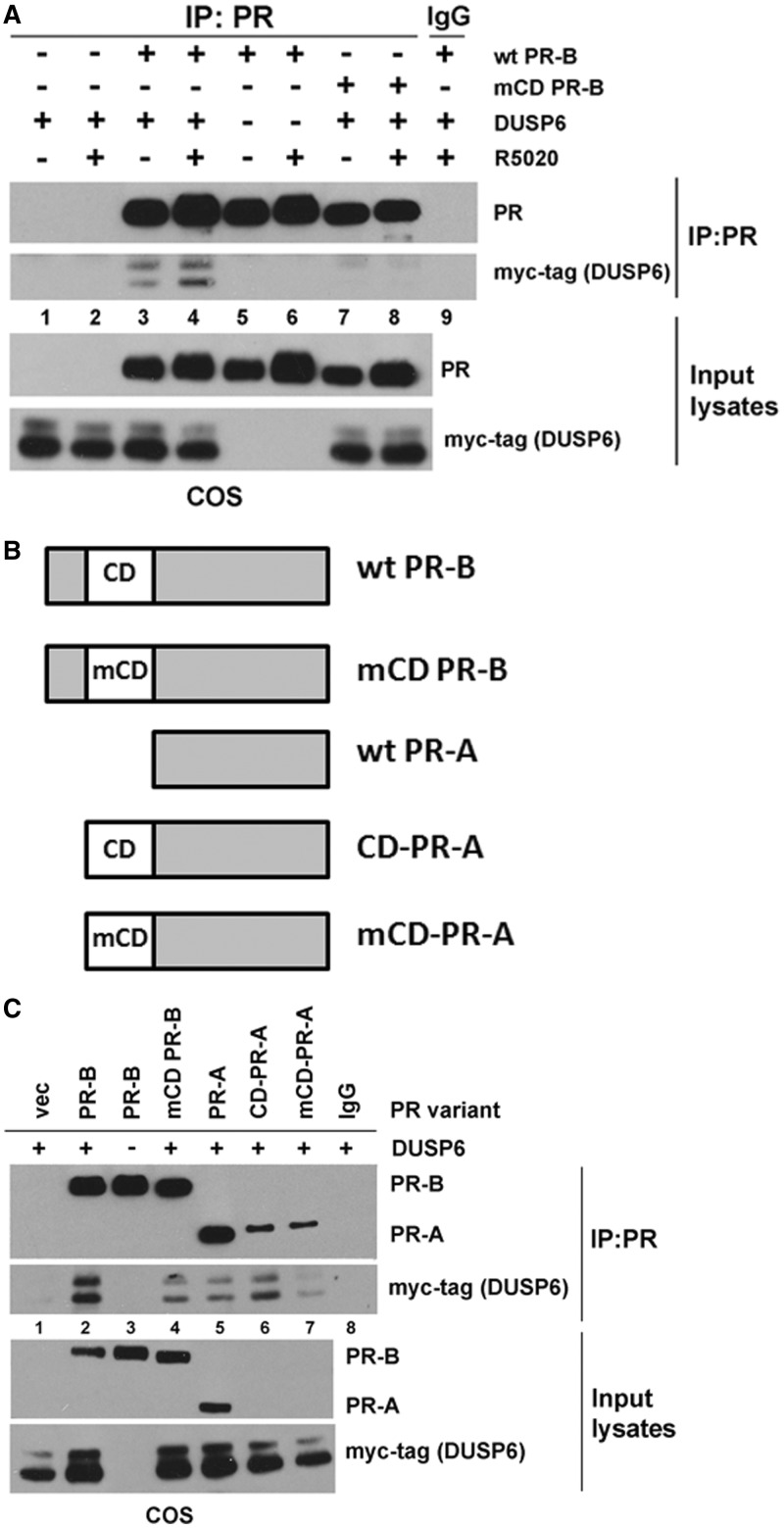
The fact that mCD PR-B lacks Ser81 phosphorylation suggests that the CD domain may facilitate a specific interaction(s) between PR-B and one or more factors that are required for this phosphorylation event. An interaction between ck2 and DUSP6 [also called MAPK phosphatase 3 (MKP3)] has previously been reported (54). To test whether PR-B also interacts with DUSP6, COS cells were transiently cotransfected with constructs encoding wt or mCD PR-B and DUSP6 (myc-tagged) or vector-only controls. In cells transfected with wt PR-B and DUSP6, myc-tagged DUSP6 clearly coimmunoprecipitated with wt PR-B in a largely ligand-independent manner (Figure 4A; lanes 3–4). In contrast, myc-tagged DUSP6 weakly copurified with mCD PR-B under the same experimental conditions (Figure 4A; lanes 7–8), indicating that the CD domain is mediating an interaction between PR-B and DUSP6.
Figure 4.
PR-B interacts with DUSP6 through the CD domain. (A) CD-domain dependent interaction between PR-B and DUSP6. COS cells were transiently transfected with wt PR-B, mCD PR-B or vector alone, as well as myc-tagged DUSP6 (or vector). Twenty-four hours after transfection, cells were starved for 18 h in serum-free media and then treated with 10 nM R5020 or EtOH for 60 min. Lysates were IP'd with PR antibody or mouse IgG (control). IP lysates or input lysates (25 µg not subjected to immunoprecipitation) were analyzed via western blotting using PR-B or myc-tag (to detect myc-tagged DUSP6) antibodies. Myc-tagged DUSP6 resolves as a doublet by western blotting due to alternate translational start sites. (B) Schematic representing wt and mutant PR-B constructs. The CD domain in wt PR-B (top) was mutated to create mCD PR-B (as described above). PR-A lacks a BUS region where the CD domain is located. A synthetic wt or mutant CD domain was fused to the N-terminus of wt PR-A to create CD-PR-A or mCD-PR-A, respectively. (C) CD domain-mediated interaction between PR-B and DUSP6. COS cells were transiently transfected with the indicated PR constructs or vector alone, as well as myc-tagged DUSP6 (or vector). CoIP and western blotting were performed as described in (A).
To determine the specificity of PR-B’s interaction with DUSP6, we engineered two additional PR-specific mutants (Figure 4B) in which a wt or mutant PR CD domain was fused to the N-terminus of PR-A (termed CD-PR-A and mCD-PR-A, respectively). COS cells were transiently transfected with control (wt PR-B) and mutant PR constructs, as well as myc-tagged DUSP6. Reproducibly, wt PR-B and DUSP6 exhibited robust interaction as measured by CoIP, and mCD PR-B again displayed greatly reduced interaction with DUSP6 (Figure 4C; lanes 2 and 4). Wt PR-A coimmunoprecipitated with low levels of myc-tagged DUSP6, similar to levels observed for mCD PR-B (Figure 4C; lanes 5 and 4). Transient expression of CD-PR-A and mCD-PR-A fusion proteins remained poor relative to wt PR-A (Figure 4C; lanes 6–7 of Input lysates). However, wt PR-A and both PR-A fusion proteins were visible in western blots of immunoprecipitates (Figure 4C; lanes 5–7 of IP:PR blot). Despite relatively poor expression, the CD-PR-A fusion receptor coimmunoprecipitated with myc-tagged DUSP6 at greater levels than wt PR-A (Figure 4C; lanes 6 and 5). However, the mCD PR-A fusion receptor completely reversed this effect (Figure 4C; lane 7), returning DUSP6 CoIP levels to approximately those seen with wt PR-A alone (no CD domain) and mCD PR-B. An additional region(s) common to both PR-B and PR-A (i.e. outside the BUS) is likely capable of weak interaction with DUSP6 (lanes 5 and 7). These data indicate that the PR-B CD domain is primarily responsible for mediating the interaction between wt PR-B and DUSP6.
DUSP6 is required for ck2-dependent PR-B Ser81 phosphorylation
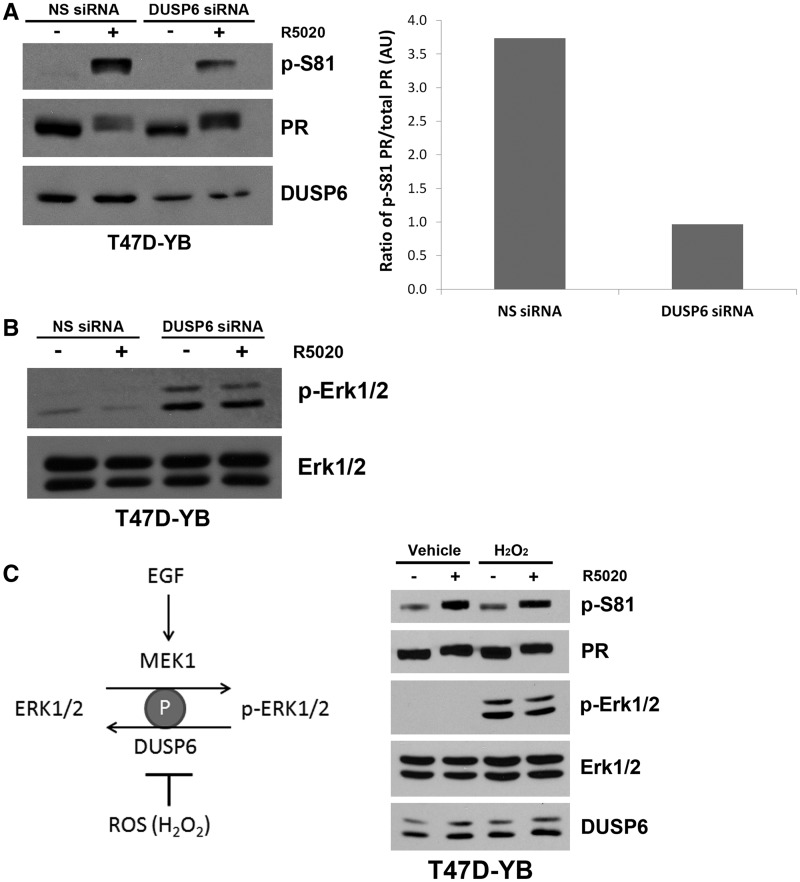
We next sought to determine how PR-B’s interaction with DUSP6 is related to PR-B Ser81 phosphorylation. We previously identified PR-B Ser81 as a ck2-dependent site regulated in response to treatment of breast cancer cells with progestin, and during the S-phase of the cell cycle in the absence of progestin (19). If DUSP6 primarily functions to recruit ck2 for PR-B Ser81 phosphorylation, then loss of DUSP6 should block this phosphorylation event. To test this hypothesis, a DUSP6-specific siRNA was used to knock down DUSP6 protein expression in breast cancer cells before analysis of progestin-induced PR-B Ser81 phosphorylation. Although DUSP6 knockdown efficiency remained weak (∼50% as determined by densitometry), T47D-YB cells transfected with DUSP6 siRNA consistently exhibited decreased PR-B Ser81 phosphorylation relative to cells transfected with nonsilencing control siRNA (Figure 5A, left panel); a 50% decrease in DUSP6 protein levels resulted in at least 75% reduction of PR-B Ser81 phosphorylation (Figure 5A, right panel). As a control for functional DUSP6 knockdown, we measured Erk1/2 phosphorylation under similar conditions because DUSP6 phosphatase activity is a negative regulator of Erk1/2 phosphorylation. As expected, T47D-YB cells transfected with DUSP6 siRNA contained elevated levels of Erk1/2 phosphorylation relative to controls, indicating effective DUSP6 knockdown (Figure 5B). To confirm these results using independent methods, we chemically modulated DUSP6 phosphatase activity. Reactive oxygen species (ROS), produced as a result of treatment of cells with agents such as H2O2, block MKP enzyme activity (55,56), thereby resulting in high levels of Erk1/2 phosphorylation (Figure 5C, left panel). T47D-YB cells treated with either 1 mM H2O2 or vehicle alone, followed by R5020, exhibited similar levels of PR-B Ser81 phosphorylation (Figure 5C, right panel) despite effective DUSP6 enzyme (i.e. phosphatase) inhibition, as measured by increased phospho-Erk1/2. DUSP6 protein levels remained unchanged in the presence of high ROS (Figure 5C, right panel). Phosphorylation on other selected PR sites (Ser345 and Ser400) was relatively insensitive to H2O2 treatment (Supplementary Figure S2). These data suggest that DUSP6 enzyme activity is not required for PR-B Ser81 phosphorylation, as phospho-Ser81 levels remained unchanged even under conditions where DUSP6 phosphatase activity was greatly diminished. Cumulatively, these data suggest that the DUSP6 protein, but not its phosphatase activity, is required for efficient PR-B Ser81 phosphorylation, indicating that DUSP6 serves as a scaffolding protein that supports ck2-dependent PR-B Ser81 phosphorylation.
Figure 5.
PR-B’s interaction with DUSP6 is required for PR-B Ser81 phosphorylation. (A) DUSP6 is necessary for ligand-induced PR-B Ser81 phosphorylation. Left: T47D cells stably expressing wt PR-B (T47D-YB) were transfected with 50 nM nonsilencing (NS) or DUSP6 siRNA. Seventy-two hours after transfection, cells were treated with 10 nM R5020 or EtOH for 60 min. Lysates were analyzed via western blotting using p-S81, PR-B and DUSP6 antibodies. Right: Densitometry was used to determine the ratio of Ser81 phosphorylated PR-B in R5020-treated cells to total PR-B in cells transfected with NS or DUSP6 siRNA. (B) Knockdown of DUSP6 increased MAPK activation. T47D-YB cells were transfected and treated as in (A), and lysates were analyzed via western blotting using p-Erk1/2 and Erk1/2 antibodies. (C) DUSP6 phosphatase activity is not needed to mediate ligand-induced PR-B Ser81 phosphorylation. Left: Schematic showing how ROS production (increased through treatment with H2O2) decreases DUSP6 phosphatase activity, which subsequently leads to an increase in Erk1/2 activity and phosphorylation. Right: T47D-YB cells were pretreated with 1 mM H2O2 for 20 min, followed by 10 nM R5020 (or EtOH) for 30 min. Lysates were analyzed via western blotting using p-S81, PR, p-Erk1/2, Erk1/2 and DUSP6 antibodies.
PR-B Ser81 phosphorylation is required for STAT5A and Wnt1 expression
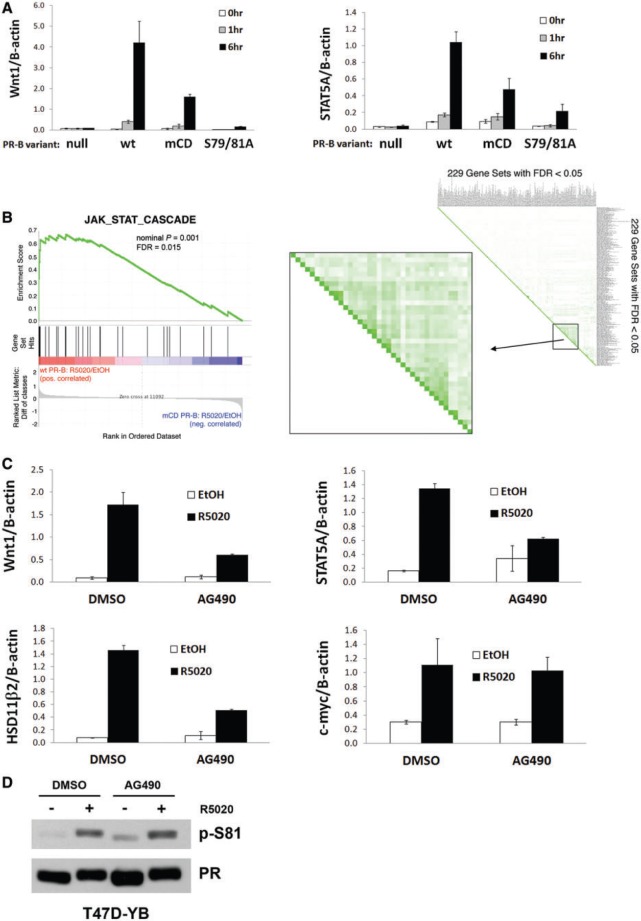
To link CD domain-dependent regulation of PR-B Ser81 to gene expression, we examined known PR isoform-specific target genes for sensitivity to disruption of the CD domain. STAT5A (signal transducer and activator of transcription 5A) (25,57) and Wnt1 (wingless-type MMTV integration site family, member 1) (41,58–60) are primarily regulated by PR-B in response to progestin (25,41). To study the effects of PR-B Ser81 on STAT5A and Wnt1 gene expression, we used a previously well-characterized PR-B phospho-mutant that cannot be phosphorylated on Ser81, S79/81A PR-B (Ser81 and nearby Ser79 were mutated to ensure that this PR mutant remained fully resistant to phosphorylation on Ser81) (19). T47D cells expressing empty vector control (PR-null) or wt, mCD or S79/81A PR-B were treated with progestin (0, 1 and 6 h), and mRNA was harvested for RT-qPCR analyses. After progestin treatment, cells stably expressing wt PR-B robustly activated STAT5A and Wnt1 relative to PR-null cells, whereas cells expressing S79/81A PR-B exhibited greatly diminished STAT5A and Wnt1 mRNA relative to cells expressing wt PR-B (Figure 6A). Interestingly, cells expressing mCD PR-B displayed an intermediate (reduced expression) phenotype, consistent with our finding that this mutant is only weakly phosphorylated on Ser81 over time (Figure 3B). STAT5B expression remained unaffected in cells expressing wt or mutant receptors (Supplementary Figure S3A). Notably, cells expressing the S79/81A phospho-mutant PR-B were phenotypically identical to cells expressing wt PR-A with regard to STAT5A and Wnt1 gene expression (Supplementary Figure S3B), confirming that these are PR-B–specific target genes. Additionally, tissue factor (TF) mRNA expression, which is independent of PR-B Ser81 (19), was similar in cells expressing either wt or S79/81A PR-B (Supplementary Figure S4A). Taken together, these data suggest that PR-B Ser81 phosphorylation—which is dependent on a CD domain-mediated scaffolding interaction between PR-B, DUSP6 and ck2—is required for expression of select PR-B target genes (STAT5A and Wnt1) known to be critical mediators of mammary gland development, stem cell self-renewal and breast cancer cell proliferation.
Figure 6.
Endogenous phospho-Ser81 PR-B-target genes are JAK/STAT-dependent. (A) Select genes regulated by PR-B Ser81 phosphorylation. T47D-Y cells stably expressing either wt PR-B, mCD PR-B, S79/81A PR-B or unmodified (PR-null) cells were starved for 18 h in serum-free media, followed by treatment with 10 nM R5020 or EtOH for 0–6 h. mRNA levels were analyzed by qPCR. Error bars represent ±SD. (B) GSEA comparing gene expression data sets obtained using the Illumina Microarray described in Figure 3. Left: wt PR-B R5020/EtOH compared with mCD PR-B R5020/EtOH is shown as compared with the c5 Gene Ontology database, JAK_STAT_Cascade gene set. Right: Leading Edge analysis within GSEA (c2 Curated Gene Set) included 229 gene sets significantly enriched (FDR < 0.05) in wt PR-B cells, as compared with mCD PR-B. A subset of 38 gene sets (boxed region) contained substantial overlap (green shading) and were significantly involved in interferon signaling pathways. See Supplementary Table S1 for gene set names and statistics. (C) Ser81-dependent PR-B target genes are JAK/STAT-dependent. T47D-Y cells stably expressing wt PR-B (T47D-YB) were starved for 18 h in serum-free media. Cells were then pretreated (60 min) with 50 μM AG490, followed by 10 nM R5020 for 6 h. mRNA levels were analyzed by qPCR. Error bars represent ±SD. (D) PR-B Ser81 phosphorylation is unaffected by JAK/STAT-inhibition. T47D-YB cells were starved for 18 h in serum-free media. Cells were then pretreated (60 min) with 50 uM AG490, followed by 10 nM R5020 for 30 min. Lysates were analyzed via western blotting using p-S81 and total PR antibodies.
PR-B CD domain is required for JAK/STAT-dependent transcriptional responses
GSEA is a powerful computational tool that can be used to determine whether publicly available gene sets (via the MSigDB) are significantly enriched in gene expression data sets (39,40). GSEA can identify gene sets that are significantly regulated in a particular microarray sample group (e.g. wt PR-B or mCD PR-B). Using GSEA, we compared ligand-regulated wt and mCD PR-B expression data sets (wt PR-B R5020/EtOH versus mCD PR-B R5020/EtOH) and identified enriched gene sets from the c5 Gene Ontology collection. Interestingly, genes from the JAK/STAT signaling pathway (JAK_STAT_Cascade gene set) were significantly enriched in cells expressing wt PR-B relative to cells expressing mCD PR-B (Figure 6B, left panel), suggesting that mCD PR-B loses the ability to regulate genes in the JAK/STAT pathway. Leading edge analysis is a deeper examination used to compare only the significant gene sets (identified through GSEA) to each other to identify the following: (i) the individual genes that are highly associated in a particular sample group and (ii) the core gene sets that contain the majority of those highly associated genes. These methods help identify the pathways (and the core subset of genes within these pathways) that are significantly associated with a sample group. We performed leading edge analysis on gene sets that were significantly enriched (229 c2 Curated gene sets with FDR < 0.05) in cells expressing wt PR-B relative to mCD PR-B. Our analyses identified 38 gene sets that contained substantial overlap (Figure 6B, right panel; Supplementary Table S1) and revealed a substantial overlap of gene sets involved in interferon signaling, which is primarily mediated via the actions of STAT proteins. Cumulatively, these data suggest that the CD domain in PR-B is critical for progestin-dependent regulation of interferon/JAK/STAT-related signaling pathways.
Interestingly, female STAT5 and PR-B knockout mice exhibit similar developmental blocks in mammary gland alveologenesis (61). Furthermore, numerous studies have implicated the JAK/STAT pathway in PR target gene regulation (25,57). Notably, progestin-induced expression of selected PR-B target genes, including HSD11β2 and STAT5A, was significantly blocked by the JAK/STAT inhibitor, AG490 (62). Similar to STAT5A (Figure 6A), we previously showed that progestin-induced expression of HSD11β2 requires phospho-Ser81 PR-B (19). To test the requirement for JAK/STAT signaling in PR-B Ser81-dependent regulation of Wnt1 expression, T47D cells stably expressing wt PR-B were pretreated (1 h) with AG490 (50 µM) followed by progestin (10 nM R5020 for 6 h) or respective vehicle controls. AG490 significantly diminished progestin-induced Wnt1 mRNA expression (Figure 6C); HSD11β2 and STAT5A were included as additional Ser81-dependent PR-B target genes and positive controls (62). In contrast, c-myc, a well-characterized PR target gene whose regulation is independent of both PR-B Ser81 (19) and JAK/STAT (62) remained unaffected by AG490 inhibition of JAK/STAT signaling (Figure 6C). Importantly, progestin-induced PR-B Ser81 phosphorylation was unaffected by pretreatment of cells with AG490 (Figure 6D). Thus, in addition to PR-B Ser81 phosphorylation, JAK/STAT signaling is also required for progestin-induced expression of a subset of PR-B target genes (STAT5A, Wnt1, HSD11β2).
To determine the role of STAT5 in coregulating phospho-Ser81 PR-B-dependent transcription, we analyzed a publicly available (http://cistrome.org/Cistrome/Cistrome_Project.html) PR chromatin immunoprecipitation ChIP–chip data set for the presence of STAT5 binding sites within or nearby PR binding sites. These ChIP–chip data were created using T47D breast cancer cells treated with vehicle or estrogen (to increase endogenous PR expression) followed by progesterone treatment for 45 min. Interestingly, CEAS [cis-regulatory element annotation system; (63)] analysis revealed a 1.8-fold enrichment of STAT5 DNA sequence binding motifs within PR binding sites as compared with random genome sampling (P = 8.22 × 10−11). These data suggest that STAT5 may function as a pioneer factor by ‘opening’ sites in chromatin for subsequent transcriptional activation by PR-B, DUSP6 and ck2.
PR-B Ser81 phosphorylation is required for binding to an enhancer region upstream of the Wnt1 promoter
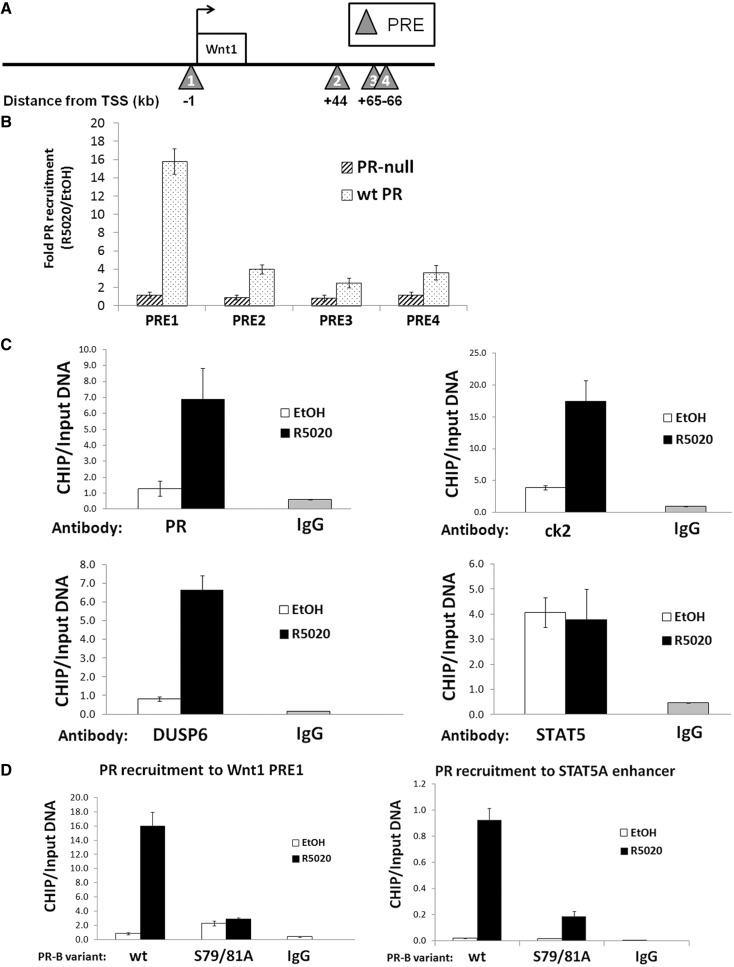
Wnt1 is a potent breast oncogene and stem cell regulatory factor/morphogen. We previously demonstrated the existence of a PR-driven autocrine loop in which frizzled receptors, activated by progestin-induced Wnt1, transactivated epidermal growth factor receptor (EGFR), leading to increased cyclin D1 expression and breast cancer cell proliferation (41). Importantly, knockdown of Wnt1 blocked progestin-induced breast cancer cell growth in soft agar. To further understand how phospho-Ser81 PR-B regulates Wnt1 expression in collaboration with STATs, we performed an in silico analysis of Wnt1 promoter and enhancer regions. We identified four putative full-length PRE binding regions (Figure 7A; triangles), including a site located in the proximal enhancer region (∼1 kb upstream from the transcriptional start site [TSS]; PRE1) and three sites located downstream of the TSS (+44, 65 and 66 kb from the TSS; PREs 2–4, respectively). To determine whether PR-B directly regulates Wnt1, we performed ChIP assays to detect relative PR-B recruitment to sites within the Wnt1 enhancer region. After cross-linking and sonication was performed, lysates from vehicle- (EtOH) or R5020-treated T47D-YB cells were subjected to ChIP using PR-specific antibodies. PR-null (T47D-Y) cells served as a negative control. In the presence of ligand, we detected robust recruitment (∼16-fold) of wt PR-B to PRE1, while moderate levels of PR-B recruitment (∼2–4-fold with ligand) were detected at the other PREs located downstream of the Wnt1 TSS (PREs 2, 3 and 4; Figure 7B). We next tested whether DUSP6 and ck2 were present in the transcription complexes at PRE1, the same region where wt PR-B was detected. Both DUSP6 and ck2 proteins were IP'd from similarly prepared lysates of ligand-treated T47D-YB cells; PR-B was also IP'd as a positive control (Figure 7C). In addition, STAT5-specific antibodies revealed that STAT5 was present at this Wnt1 regulatory site, although protein recruitment was not ligand-dependent (Figure 7C, bottom right). Cumulatively, these data demonstrate direct regulation of the Wnt1 enhancer at region PRE1 by PR-B, DUSP6 and ck2. Although these molecules were recruited on progestin treatment, STAT5 was pre-associated with this site.
Figure 7.
PR-B regulates Wnt1 transcription through binding to Wnt1 enhancer regions. (A) Schematic of the Wnt1 promoter/enhancer regions. PRE sites (triangles) are located upstream and downstream of the Wnt1 transcription start site (TSS; denoted with an arrow). Distance of the regulatory regions from the TSS is listed in kilobases (kb). (B) PR-B is recruited to Wnt1 PREs. T47D-Y cells stably expressing wt PR-B or unmodified cells (PR-null) were serum-starved for 18 h. Cells were then treated with 10 nM R5020 or EtOH for 60 min. Fixed lysates were subjected to ChIP with antibodies against PR-B or species-specific IgG (control), and qPCR was performed on the isolated DNA using primers designed to amplify the respective regulatory region (PREs 1–4). Recruitment of PR-B to PREs is shown as fold change between R5020- or EtOH-treated cells (R5020/EtOH). Error bars represent ±SD of triplicate experiments. (C) PR-B Ser81 phosphorylation-scaffolding complex is recruited to Wnt1 enhancer. T47D-Y cells stably expressing wt PR-B were serum-starved for 18 h. Cells were then treated with 10 nM R5020 or EtOH for 60 min. Fixed lysates were subjected to ChIP with antibodies against PR-B, ck2, DUSP6, STAT5 or species-specific IgG (controls), and qPCR was performed on the isolated DNA using primers designed to amplify Wnt1 PRE1. ChIP experiments were performed in triplicate. A representative experiment is shown here; error bars represent ±SD of technical replicates. (D) PR-B Ser81 phosphorylation is necessary for PR-B recruitment to Ser81-regulated genes. T47D-Y cells stably expressing wt PR-B or S79/81A PR-B were serum-starved for 18 h. Cells were then treated with 10 nM R5020 or EtOH for 60 min. Fixed lysates were subjected to ChIP with antibodies against PR or species-specific IgG (controls), and qPCR was performed on the isolated DNA using primers designed to amplify Wnt1 PRE1 or a STAT5A-enhancer site. ChIP experiments were performed in triplicate. A representative experiment is shown here; error bars represent ±SD of technical replicates.
PR-B Ser81 phosphorylation mediates recruitment of PR-B to Wnt1 and STAT5A enhancer regions
Finally, we examined PR-B recruitment to Wnt PRE1 in cells expressing either wt or S79/81A PR-B. In contrast to efficient recruitment of wt PR-B to the Wnt1 enhancer region, S79/81A PR-B failed to be recruited to PRE1 at the Wnt1 enhancer (Figure 7D, left panel). Similarly, ChIP assays revealed that wt but not S79/81A PR-B was recruited to a regulatory region within the STAT5A gene (Figure 7D, right panel). Importantly, both wt and S79/81A PR-B showed similar levels of recruitment to PRE regions associated with expression of TF [Supplementary Figure S4B and (19)], indicating that the lack of S79/81A PR-B recruitment to Wnt1 and STAT5A enhancer regions is not due to an inherent nuclear localization or DNA-binding defect of phospho-mutant S79/81A PR-B. These data suggest a possible feed-forward mechanism involving phospho-Ser81 PR and STAT5. Cumulatively, these results provide a basis for understanding mechanisms of PR-B isoform-specific target gene selection and suggest that phosphorylation of Ser81 is required for PR-B recruitment to selected PREs located within JAK/STAT-regulated genes.
DISCUSSION
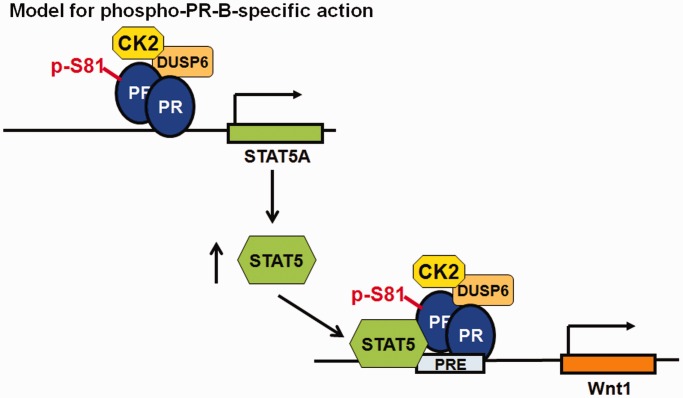
Our studies identify a novel protein interaction domain (CD domain) in the PR-B N-terminus that mediates interaction between PR-B, DUSP6 and ck2, and is required for progestin-induced S-phase cell cycle entry of breast cancer cells. DUSP6 binding to PR-B is required for ck2-dependent PR-B Ser81 phosphorylation and subsequent regulation of Wnt1 and STAT5A, two PR-B-specific target genes known to be critically involved in mammary stem cell maintenance and breast cancer cell proliferation. DUSP6 appears to play a novel scaffolding role in this complex, as DUSP6 knockdown but not inhibition of phosphatase activity blocked PR-B Ser81 phosphorylation. Cumulatively, these data support a model where DUSP6 binds the PR-B CD domain and scaffolds activated ck2 to mediate PR-B Ser81 phosphorylation (54). In addition, we defined Wnt1 and STAT5A as members of the same gene set regulated by phospho-Ser81 PR-B and JAK/STAT signaling. This finding complements previous studies showing that PR-B Ser81 phosphorylation is required for progestin-induced expression of HSD11β2 and BIRC3 (19) and that both genes are sensitive to JAK/STAT pathway inhibition (62). Notably, we found that PR-B, STAT5, DUSP6 and ck2 are recruited to the same hormone-responsive PRE-containing region of the Wnt1 enhancer. Taken together, our data demonstrate a role for PR-B Ser81 phosphorylation in target gene selection of JAK/STAT-dependent genes that are seminal to mammary gland biology (Figure 8). Furthermore, our data demonstrate a functional linkage between PR-B and STAT5 that provides a paradigm for coordinate activation of proliferative gene programs in mammary gland development and, pathologically, in ER+/PR+ breast cancer.
Figure 8.
Model of phospho-PR-B–specific action. PR-B CD domain-dependent recruitment of DUSP6 and ck2 is required for PR-B phosphorylation on Ser81. This complex is required for PR-B–dependent expression of STAT5A, whose protein product in turn then complexes with Ser81-phosphorylated PR-B on a specific-subset of PR-target genes, such as Wnt1. JAK/STAT-dependent regulation of phospho-Ser81 PR-B target genes regulates critical genes involved in mammary gland development, mammary stem cell maintenance/expansion and early events in breast cancer progression.
CD domains and nuclear receptors
The CD domain of PR-B is located in the N-terminal BUS region of full-length PR-B, a region that is absent from other PR isoforms (21). An in silico analysis of the most closely related, steroid hormone-activated nuclear receptors (i.e. ER, glucocorticoid receptor and androgen receptor) revealed the presence of weak potential candidate CD domain-like regions. However, none contained the conserved Erk2 amino acid sequence similarity found in the PR-B CD domain. A similar domain was identified in PPARγ, a more distant member of the nuclear receptor family, which mediates an interaction between PPARγ and MEK1 (64). Interestingly, deletion of the CD-like domain in PPARγ did not completely abrogate MEK1 interaction, suggesting that other areas of PPARγ also make contact with MEK1 or proteins that bind MEK1 (64). Similarly, in our study, mutation of PR-B’s CD domain did not completely disrupt DUSP6 binding. Additionally, mutation of PR-B’s CD domain did not alter MEK1 binding (data now shown), indicating that the CD domain in PR-B is not required for PR-MEK1 complex formation (21). Stable PR binding to components of MAPK pathways (MEKs or DUSPs) occurs via multiple domains (CD domain, p-Pro, ERIDs) in PR that are required for robust progestin-induced MAPK activity. Moreover, both c-Src and ER are also found in protein complexes with components of the MAPK pathway (22,23,65). PR interaction with multiple signaling molecules illustrates that these pathways are fully integrated, and this integrated circuitry provides a basis for the highly selective context-dependent regulation of PR target genes. Notably, both the CD domain and p-Pro region occur in human but not mouse PR-B. This fact may explain why mouse models have not strongly implicated PR-B as a breast oncogene; PR-B is not well expressed in virgin mice relative to PR-A, and mouse PR-B lacks key domains required for linkage of mitogenic signaling pathways to PR-B–specific gene expression.
Regulation of ck2-dependent PR-B Ser81 phosphorylation
ck2 is a ubiquitously expressed kinase with >300 substrates (16). Unlike traditional growth factor-activated protein kinases that require upstream inputs for full activation, ck2 is constitutively active. Although ck2 regulation is poorly understood, evidence suggests that ck2 activation is modulated primarily through subcellular localization, substrate distribution/complex formation, ck2 holoenzyme formation, small molecule interactions or autophosphorylation (66). In a previous report, we show that PR-B Ser81 is phosphorylated by ck2 in the presence of progestin, but in the absence of progestin PR-B Ser81 is mainly phosphorylated by ck2 during the S-phase of the cell cycle, when ck2 is nuclear and exposed to PR-B (19). Herein, we discovered that DUSP6 binding via the PR-B CD domain provides a mechanism for robust ck2-dependent phosphorylation of PR-B Ser81. Perhaps PR-B must be bound to DUSP6 to ‘accept’ Ser81 phosphorylation by ck2, either because of proximity restrictions whereby DUSP6 recruits ck2 into close proximity with PR-B Ser81 or because of substrate conformation changes in which DUSP6 binding to PR-B induces conformational changes that permit ck2-dependent Ser81 phosphorylation. Importantly, we observed constitutive PR-B binding with DUSP6 in the absence or presence of progestins. However, transcriptional complexes containing PR-B, DUSP6 and ck2 are clearly recruited to the Wnt1 enhancer (PRE1) in a progestin-dependent manner. Whether these protein–protein and protein–DNA interactions are regulated by additional factors (e.g. growth factors or STAT5) or circumstances (e.g. cell cycle) are questions requiring further study. Notably, ck2 is upregulated in many human cancers, including breast cancer (16,20). Preliminary data obtained from a small subset of PR-positive breast tumors [described in (13)] demonstrated that roughly half contained phospho-Ser81 PR-B (A. Daniel and C. Lange, unpublished observations). These findings suggest that PR-B Ser81 phosphorylation is clinically relevant, and underscore the importance of further study of PR-B phosphorylation and associated isoform-specific target gene expression in human breast tumors.
DUSP6 may function as a scaffolding protein to promote cancer growth
DUSP6 is a potent phosphatase responsible for reversing Erk1/2 phosphorylation and thus is a negative regulator of MAPK activity. Because of its role as a negative regulator of MAPK signaling, the central dogma has been that DUSP6 functions as a tumor suppressor in cancer, even though DUSP6 overexpression was often predictive of poor clinical outcomes [reviewed in (67,68)]. Recent data, however, have implicated DUSP6 overexpression as a negative prognostic marker in cancer development, progression and survival in many different types of primary cancers and cancer cell lines, including thyroid (69), lung (70), myeloma (71), melanoma (72,73), breast (74,75), colon (76), cervical (77), pancreatic (78) and glioblastoma (79). These paradoxical data suggest that DUSP6 may have multiple biological functions, independent of its long-studied role in attenuating MAPK activation. Herein, our data support a novel mechanism in which DUSP6 functions as a scaffold for assembly of transcriptional coactivators that drive tumor growth. We found that DUSP6 phosphatase activity was not required for PR-B Ser81 phosphorylation, but rather, DUSP6 acted as a scaffold protein to bridge the interaction between PR-B and ck2, thereby bringing ck2 into close proximity with its substrate (i.e. PR-B Ser81) (19,54). This scaffolding action represents a unique role for DUSP6. Indeed, DUSP6’s scaffolding function, rather than its phosphatase activity, may provide a molecular explanation for the growing body of data linking DUSP6 overexpression to poor clinical outcomes in many different types of cancer, including breast (74,75). In addition, classically defined roles of kinases and phosphatases clearly have broader scopes of action, such as bridging pathways previously thought to be unrelated (i.e. PR-B regulation by ck2) and direct participation in gene regulation as part of transcription complexes (19,21,80).
Functional linkage of STAT5 and PR-B signaling
Whole genome (cistrome) analyses have identified ‘pioneer factors’ for nuclear receptors. Pioneer factors are specialized subsets of transcriptional coregulators that bind to transcriptional enhancers, making them competent for subsequent transcriptional activation by transcription factors, such as steroid hormone receptors [reviewed in (81,82)]. For example, multiple pioneer factors (FOXA1, PBX-1, GREB1, AP2-γ) have been identified for modulation of ER binding, and similar factors have been identified for other nuclear receptors (81,83–86). Importantly, pioneer factors bind DNA before activation of transcription and function to ‘open’ sites in chromatin for subsequent transcriptional activation. We found that JAK/STAT pathway inhibition blocked the expression of multiple phospho-Ser81 PR-B target genes. We also identified STAT5 DNA sequence binding motifs within PR binding sites and STAT5 protein associated with PR-B target genes. Thus, it is tempting to speculate that STAT5 may act as a pioneer factor for phospho-Ser81 PR-B binding by ‘opening’ the enhancer regions of phospho-PR-B target genes. Interestingly, we also observed that phospho-Ser81 PR-B was required for STAT5A mRNA expression, suggestive of a feed-forward regulatory loop wherein many PR-B and STAT5 genes are coordinately regulated.
Recent reports provide some insight into how PR-B/DUSP6/ck2-containing protein complexes and STAT5A coordinate gene expression during breast cancer development and early breast cancer progression. STAT5A and Wnt1 have recently been implicated in PR control of mammary stem cell maintenance and mammary gland biology (60,87). Like PR-B, STAT5A is required for mammary gland development, and both STAT5A and PR-B knockout mice have similar defects in mammary gland development (87). Progesterone is a known activator of STAT5A mRNA and protein expression (25,57); however, the mechanism by which progesterone induces STAT5A expression is not well understood. Similarly, Wnts are important mediators of progesterone action in the normal (58) and pregnant mammary gland (59). Previous work published from our lab showed that phospho-PR-B-dependent upregulation of Wnt1 is required for breast cancer cell soft-agar growth in response to progestins (41). Wnts have recently been shown to be critical paracrine mediators of progesterone-induced expansion of mammary stem cells (60), and Wnt loss as a result of early parity has been linked to protection from breast cancer (88). Deregulation of the Wnt/β-catenin signaling pathway has been found in many human cancers, including breast cancer (89). Interestingly, unlike most other cancers, direct mutations of positive and negative regulators of the Wnt/β-catenin signaling pathway are rarely seen in breast cancer, despite the clear upregulation of downstream pathway endpoints, such as β-catenin stabilization and nuclear accumulation (90,91). Notably, progesterone/PR-B is a direct activator of this pathway (58,60). Potential involvement of these key mediators (Wnts, STAT5A) of mammary gland biology in progestin-induced breast cancer development or early tumor progression underscores the need to understand precisely how PR-B regulates these genes.
PR-B Ser81 phosphorylation is a major determinant of PR isoform specificity
We showed previously that phosphorylation of Ser81 is a significant determinant of PR isoform specificity; mutation of this residue in PR-B (or mutation of CD) confers PR-A–specific behavior with regard to target gene expression (19) and cell cycle entry (Figure 2C). Coordinate regulation of PR-B–specific target genes by phospho-Ser81 PR-B and STAT5 may explain the requirement for both factors during the same stage of mammary gland development. Notably, paracrine factors (i.e. Wnts, RANKL) derived from PR-B-positive progenitors or luminal precursor cells are believed to induce self-renewal of PR-null stem cells (60,92). PR-A and PR-B are usually coexpressed in the same tissues; cells that express only a single PR isoform are rare (93–95). A 1:1 ratio of PR-A to PR-B seen in normal tissues is often altered in malignant breast tissues, suggesting that balanced isoform action is crucial to normal adult mammary gland biology (93,96). PR-A, but not PR-B, gene silencing via promoter methylation was significantly associated with tamoxifen-resistant breast cancer (97). Understanding the critical differences between PR-A and PR-B-dependent gene regulation as linked to DUSP6-dependent PR-B Ser81 phosphorylation by ck2 and JAK/STAT signaling may allow for highly selective isoform-specific therapies. Restoration of the balance between PR isoform actions may provide an innovative and complementary approach to existing endocrine therapies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [T32 CA009138 to C.H., 1K99CA166643-01 to C.H., R01 CA159712 to C.L.]; Department of Defense Breast Cancer Research Program [BC085608 to C.H., BC093529 to T.K.]. Funding for open access charge: NIH K99.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Andrea R. Daniel (Minnesota) for her critical contributions to the development and execution of this project, and Michael Freeman (Minnesota) for critical review/editorial assistance. The authors acknowledge the following Masonic Cancer Center Core facilities: Minnesota Supercomputing Institute for administration and access to Ingenuity Pathway Analysis and the University of Minnesota Genomics Center (UMGC) for microarray execution.
REFERENCES
- 1.Kraus WL, Montano MM, Katzenellenbogen BS. Cloning of the rat progesterone receptor gene 5′-region and identification of two functionally distinct promoters. Mol. Endocrinol. 1993;7:1603–1616. doi: 10.1210/mend.7.12.8145766. [DOI] [PubMed] [Google Scholar]
- 2.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill KK, Roemer SC, Churchill ME, Edwards DP. Structural and functional analysis of domains of the progesterone receptor. Mol. Cell. Endocrinol. 2012;348:418–429. doi: 10.1016/j.mce.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sartorius CA, Melville MY, Hovland AR, Tung L, Takimoto GS, Horwitz KB. A third transactivation function (AF3) of human progesterone receptors located in the unique N-terminal segment of the B-isoform. Mol. Endocrinol. 1994;8:1347–1360. doi: 10.1210/mend.8.10.7854352. [DOI] [PubMed] [Google Scholar]
- 6.Owen GI, Richer JK, Tung L, Takimoto G, Horwitz KB. Progesterone regulates transcription of the p21(WAF1) cyclin- dependent kinase inhibitor gene through Sp1 and CBP/p300. J. Biol Chem. 1998;273:10696–10701. doi: 10.1074/jbc.273.17.10696. [DOI] [PubMed] [Google Scholar]
- 7.Stoecklin E, Wissler M, Schaetzle D, Pfitzner E, Groner B. Interactions in the transcriptional regulation exerted by Stat5 and by members of the steroid hormone receptor family. J. Steroid Biochem. Mol. Biol. 1999;69:195–204. doi: 10.1016/s0960-0760(99)00052-7. [DOI] [PubMed] [Google Scholar]
- 8.Cicatiello L, Addeo R, Sasso A, Altucci L, Petrizzi VB, Borgo R, Cancemi M, Caporali S, Caristi S, Scafoglio C, et al. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol. Cell. Biol. 2004;24:7260–7274. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocrine Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 10.Daniel AR, Faivre EJ, Lange CA. phosphorylation-dependent antagonism of sumoylation de-represses progesterone receptor action in breast cancer cells. Mol. Endocrinol. 2007;21:2890–2906. doi: 10.1210/me.2007-0248. [DOI] [PubMed] [Google Scholar]
- 11.Lange CA, Shen T, Horwitz KB. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc. Natl Acad. Sci. USA. 2000;97:1032–1037. doi: 10.1073/pnas.97.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weigel NL, Bai W, Zhang Y, Beck CA, Edwards DP, Poletti A. Phosphorylation and progesterone receptor function. J. Steroid. Biochem. Mol. Biol. 1995;53:509–514. doi: 10.1016/0960-0760(95)00098-k. [DOI] [PubMed] [Google Scholar]
- 13.Knutson TP, Daniel AR, Fan D, Silverstein KA, Covington KR, Fuqua SA, Lange CA. Phosphorylated and sumoylation-deficient progesterone receptors drive proliferative gene signatures during breast cancer progression. Breast Cancer Res. 2012;14:R95. doi: 10.1186/bcr3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel AR, Gaviglio AL, Czaplicki LM, Hillard CJ, Housa D, Lange CA. The progesterone receptor hinge region regulates the kinetics of transcriptional responses through acetylation, phosphorylation, and nuclear retention. Mol. Endocrinol. 2011;24:2126–2138. doi: 10.1210/me.2010-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierson-Mullany LK, Lange CA. Phosphorylation of progesterone receptor serine 400 mediates ligand-independent transcriptional activity in response to activatoin of cyclin-dependent protein kinase2. Mol. Cell. Biol. 2004;24:10542–10557. doi: 10.1128/MCB.24.24.10542-10557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 17.Tawfic S, Yu S, Wang H, Faust R, Davis A, Ahmed K. Protein kinase CK2 signal in neoplasia. Histol. Histopathol. 2001;16:573–582. doi: 10.14670/HH-16.573. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Beck CA, Poletti A, Edwards DP, Weigel NL. Identification of phosphorylation sites unique to the B form of human progesterone receptor. In vitro phosphorylation by casein kinase II. J. Biol. Chem. 1994;269:31034–31040. [PubMed] [Google Scholar]
- 19.Hagan CR, Regan TM, Dressing GE, Lange CA. ck2-dependent phosphorylation of progesterone receptors (PR) on Ser81 regulates pr-b isoform-specific target gene expression in breast cancer cells. Mol. Cell. Biol. 2011;31:2439–2452. doi: 10.1128/MCB.01246-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerra B, Issinger OG. Protein kinase CK2 in human diseases. Curr. Med. Chem. 2008;15:1870–1886. doi: 10.2174/092986708785132933. [DOI] [PubMed] [Google Scholar]
- 21.Hagan CR, Faivre EJ, Lange CA. Scaffolding actions of membrane-associated progesterone receptors. Steroids. 2009;74:568–572. doi: 10.1016/j.steroids.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballare C, Uhrig M, Bechtold T, Sancho E, Di Domenico M, Migliaccio A, Auricchio F, Beato M. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol. Cell. Biol. 2003;23:1994–2008. doi: 10.1128/MCB.23.6.1994-2008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boonyaratanakornkit V, Scott MP, Ribon V, Sherman L, Anderson SM, Maller JL, Miller WT, Edwards DP. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol. Cell. 2001;8:269–280. doi: 10.1016/s1097-2765(01)00304-5. [DOI] [PubMed] [Google Scholar]
- 24.Cerliani JP, Guillardoy T, Giulianelli S, Vaque JP, Gutkind JS, Vanzulli SI, Martins R, Zeitlin E, Lamb CA, Lanari C. Interaction between FGFR-2, STAT5, and progesterone receptors in breast cancer. Cancer Res. 2011;71:3720–3731. doi: 10.1158/0008-5472.CAN-10-3074. [DOI] [PubMed] [Google Scholar]
- 25.Richer JK, Lange CA, Manning NG, Owen G, Powell R, Horwitz KB. Convergence of progesterone with growth factor and cytokine signaling in breast cancer. Progesterone receptors regulate signal transducers and activators of transcription expression and activity. J. Biol. Chem. 1998;273:31317–31326. doi: 10.1074/jbc.273.47.31317. [DOI] [PubMed] [Google Scholar]
- 26.Wardell SE, Boonyaratanakornkit V, Adelman JS, Aronheim A, Edwards DP. Jun dimerization protein 2 functions as a progesterone receptor N-terminal domain coactivator. Mol Cell Biol. 2002;22:5451–5466. doi: 10.1128/MCB.22.15.5451-5466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 28.Tanoue T, Nishida E. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol. Ther. 2002;93:193–202. doi: 10.1016/s0163-7258(02)00188-2. [DOI] [PubMed] [Google Scholar]
- 29.Enslen H, Davis RJ. Regulation of MAP kinases by docking domains. Biol. Cell. 2001;93:5–14. doi: 10.1016/s0248-4900(01)01156-x. [DOI] [PubMed] [Google Scholar]
- 30.Beral V. Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet. 2003;362:419–427. doi: 10.1016/s0140-6736(03)14065-2. [DOI] [PubMed] [Google Scholar]
- 31.Anderson GL, Limacher M, Assaf AR, Bassford T, Beresford SA, Black H, Bonds D, Brunner R, Brzyski R, Caan B, et al. Effects of conjugated equine estrogen in postmenopausal women with hysterectomy: the Women's Health Initiative randomized controlled trial. JAMA. 2004;291:1701–1712. doi: 10.1001/jama.291.14.1701. [DOI] [PubMed] [Google Scholar]
- 32.Gregory CW, Fei X, Ponguta LA, He B, Bill HM, French FS, Wilson EM. Epidermal growth factor increases coactivation of the androgen receptor in recurrent prostate cancer. J. Biol. Chem. 2004;279:7119–7130. doi: 10.1074/jbc.M307649200. [DOI] [PubMed] [Google Scholar]
- 33.Wilson GR, Cramer A, Welman A, Knox F, Swindell R, Kawakatsu H, Clarke RB, Dive C, Bundred NJ. Activated c-SRC in ductal carcinoma in situ correlates with high tumour grade, high proliferation and HER2 positivity. Br. J. Cancer. 2006;95:1410–1414. doi: 10.1038/sj.bjc.6603444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steeg PS, Zhou Q. Cyclins and breast cancer. Breast Cancer Res. Treat. 1998;52:17–28. doi: 10.1023/a:1006102916060. [DOI] [PubMed] [Google Scholar]
- 35.Horwitz KB, Mockus MB, Lessey BA. Variant T47D human breast cancer cells with high progesterone-receptor levels despite estrogen and antiestrogen resistance. Cell. 1982;28:633–642. doi: 10.1016/0092-8674(82)90218-5. [DOI] [PubMed] [Google Scholar]
- 36.Sartorius CA, Groshong SD, Miller A, Powell RL, Tung L, Takimoto G, Horwitz K. New T47D breast cancer cell lines for the independent study of progesterone B- and A-receptors; only antiprogestin-occupied B-receptors are switched to transcriptional agonists by cAMP. Cancer Res. 1994;54:3868–3877. [PubMed] [Google Scholar]
- 37.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 39.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 40.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faivre EJ, Lange CA. Progesterone receptors upregulate Wnt-1 to induce epidermal growth factor receptor transactivation and c-Src-dependent sustained activation of Erk1/2 mitogen-activated protein kinase in breast cancer cells. Mol. Cell. Biol. 2007;27:466–480. doi: 10.1128/MCB.01539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faivre EJ, Daniel AR, Hillard CJ, Lange CA. Progesterone receptor rapid signaling mediates serine 345 phosphorylation and tethering to specificity protein 1 transcription factors. Mol. Endocrinol. 2008;22:823–837. doi: 10.1210/me.2007-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanoue T, Maeda R, Adachi M, Nishida E. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 2001;20:466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Groshong SD, Owen GI, Grimison B, Schauer IE, Todd MC, Langan TA, Sclafani RA, Lange CA, Horwitz KB. Biphasic regulation of breast cancer cell growth by progesterone: role of the cyclin-dependent kinase inhibitors, p21 and p27(Kip1) Mol. Endocrinol. 1997;11:1593–1607. doi: 10.1210/mend.11.11.0006. [DOI] [PubMed] [Google Scholar]
- 45.Musgrove EA, Lee CS, Sutherland RL. Progestins both stimulate and inhibit breast cancer cell cycle progression while increasing expression of transforming growth factor alpha, epidermal growth factor receptor, c-fos, and c-myc genes. Mol. Cell. Biol. 1991;11:5032–5043. doi: 10.1128/mcb.11.10.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skildum A, Faivre E, Lange CA. Progesterone receptors induce cell cycle progression via activation of mitogen activated protein kinases. Mol. Endocrinol. 2005;19:327–339. doi: 10.1210/me.2004-0306. [DOI] [PubMed] [Google Scholar]
- 47.Boonyaratanakornkit V, Bi Y, Cheskis B, Mancini M, Rudd M, Buser A, Rosen J, Mukherjee A, Lyndon J, Edwards D. Keystone Symoposia on Molecular and Cellular Biology, Nuclear Receptors: Orphan Brothers and Steroid Sisters. Keystone Symposia, Whistler, British Columbia; 2008. p. 109. [Google Scholar]
- 48.Horwitz KB, Koseki Y, McGuire WL. Estrogen control of progesterone receptor in human breast cancer: role of estradiol and antiestrogen. Endocrinology. 1978;103:1742–1751. doi: 10.1210/endo-103-5-1742. [DOI] [PubMed] [Google Scholar]
- 49.Sartorius CA, Groshong SD, Miller LA, Powell RL, Tung L, Takimoto GS, Horwitz KB. New T47D breast cancer cell lines for the independent study of progesterone B- and A-receptors: only antiprogestin-occupied B-receptors are switched to transcriptional agonists by cAMP. Cancer Res. 1994;54:3868–3877. [PubMed] [Google Scholar]
- 50.Hagan CR, Daniel AR, Dressing GE, Lange CA. Role of phosphorylation in progesterone receptor signaling and specificity. Mol. Cell. Endocrinol. 2012;357:43–49. doi: 10.1016/j.mce.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daniel AR, Hagan CR, Lange CA. Progesterone receptor action: defining a role in breast cancer. Expert Rev. Endocrinol. Metab. 2011;6:359–369. doi: 10.1586/eem.11.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daniel AR, Knutson TP, Lange CA. Signaling inputs to progesterone receptor gene regulation and promoter selectivity. Mol. Cell. Endocrinol. 2009;308:47–52. doi: 10.1016/j.mce.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagan CR, Daniel AR, Dressing GE, Lange CA. Role of phosphorylation in progesterone receptor signaling and specificity. Mol. Cell. Endocrinol. 2012;357:43–49. doi: 10.1016/j.mce.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castelli M, Camps M, Gillieron C, Leroy D, Arkinstall S, Rommel C, Nichols A. MAP kinase phosphatase 3 (MKP3) interacts with and is phosphorylated by protein kinase CK2alpha. J. Biol. Chem. 2004;279:44731–44739. doi: 10.1074/jbc.M407669200. [DOI] [PubMed] [Google Scholar]
- 55.Cui Y, Parra I, Zhang M, Hilsenbeck SG, Tsimelzon A, Furukawa T, Horii A, Zhang ZY, Nicholson RI, Fuqua SA. Elevated expression of mitogen-activated protein kinase phosphatase 3 in breast tumors: a mechanism of tamoxifen resistance. Cancer Res. 2006;66:5950–5959. doi: 10.1158/0008-5472.CAN-05-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levinthal DJ, Defranco DB. Reversible oxidation of ERK-directed protein phosphatases drives oxidative toxicity in neurons. J. Biol. Chem. 2005;280:5875–5883. doi: 10.1074/jbc.M410771200. [DOI] [PubMed] [Google Scholar]
- 57.Santos SJ, Haslam SZ, Conrad SE. Estrogen and progesterone are critical regulators of Stat5a expression in the mouse mammary gland. Endocrinology. 2008;149:329–338. doi: 10.1210/en.2007-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brisken C, Heineman A, Chavarria T, Elenbaas B, Tan J, Dey SK, McMahon JA, McMahon AP, Weinberg RA. Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev. 2000;14:650–654. [PMC free article] [PubMed] [Google Scholar]
- 59.Gavin BJ, McMahon AP. Differential regulation of the Wnt gene family during pregnancy and lactation suggests a role in postnatal development of the mammary gland. Mol. Cell. Biol. 1992;12:2418–2423. doi: 10.1128/mcb.12.5.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joshi PA, Jackson HW, Beristain AG, Di Grappa MA, Mote PA, Clarke CL, Stingl J, Waterhouse PD, Khokha R. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 61.Santos SJ, Haslam SZ, Conrad SE. Signal transducer and activator of transcription 5a mediates mammary ductal branching and proliferation in the nulliparous mouse. Endocrinology. 2010;151:2876–2885. doi: 10.1210/en.2009-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subtil-Rodriguez A, Millan-Arino L, Quiles I, Ballare C, Beato M, Jordan A. Progesterone induction of the 11beta-hydroxysteroid dehydrogenase type 2 promoter in breast cancer cells involves coordinated recruitment of STAT5A and progesterone receptor to a distal enhancer and polymerase tracking. Mol. Cell. Biol. 2008;28:3830–3849. doi: 10.1128/MCB.01217-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–W554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burgermeister E, Chuderland D, Hanoch T, Meyer M, Liscovitch M, Seger R. Interaction with MEK causes nuclear export and downregulation of peroxisome proliferator-activated receptor gamma. Mol. Cell. Biol. 2007;27:803–817. doi: 10.1128/MCB.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Migliaccio A, Piccolo D, Castoria G, Di Domenico M, Bilancio A, Lombardi M, Gong W, Beato M, Auricchio F. Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. EMBO J. 1998;17:2008–2018. doi: 10.1093/emboj/17.7.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olsten ME, Litchfield DW. Order or chaos? An evaluation of the regulation of protein kinase CK2. Biochem. Cell Biol. 2004;82:681–693. doi: 10.1139/o04-116. [DOI] [PubMed] [Google Scholar]
- 67.Caunt CJ, Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs): shaping the outcome of MAP kinase signalling. FEBS J. 2013;280:489–504. doi: 10.1111/j.1742-4658.2012.08716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008;27:253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- 69.Degl'Innocenti D, Romeo P, Tarantino E, Sensi M, Cassinelli G, Catalano V, Lanzi C, Perrone F, Pilotti S, Seregni E, et al. DUSP6/MKP3 is overexpressed in papillary and poorly differentiated thyroid carcinoma and contributes to neoplastic properties of thyroid cancer cells. Endocr. Relat. Cancer. 2013;20:23–37. doi: 10.1530/ERC-12-0078. [DOI] [PubMed] [Google Scholar]
- 70.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, Cheng CL, Wang CH, Terng HJ, Kao SF, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N. Engl. J. Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 71.Croonquist PA, Linden MA, Zhao F, Van Ness BG. Gene profiling of a myeloma cell line reveals similarities and unique signatures among IL-6 response, N-ras-activating mutations, and coculture with bone marrow stromal cells. Blood. 2003;102:2581–2592. doi: 10.1182/blood-2003-04-1227. [DOI] [PubMed] [Google Scholar]
- 72.Bloethner S, Chen B, Hemminki K, Muller-Berghaus J, Ugurel S, Schadendorf D, Kumar R. Effect of common B-RAF and N-RAS mutations on global gene expression in melanoma cell lines. Carcinogenesis. 2005;26:1224–1232. doi: 10.1093/carcin/bgi066. [DOI] [PubMed] [Google Scholar]
- 73.Li W, Song L, Ritchie AM, Melton DW. Increased levels of DUSP6 phosphatase stimulate tumourigenesis in a molecularly distinct melanoma subtype. Pigment Cell Melanoma Res. 2012;25:188–199. doi: 10.1111/j.1755-148X.2011.00949.x. [DOI] [PubMed] [Google Scholar]
- 74.Lucci MA, Orlandi R, Triulzi T, Tagliabue E, Balsari A, Villa-Moruzzi E. Expression profile of tyrosine phosphatases in HER2 breast cancer cells and tumors. Cell. Oncol. 2010;32:361–372. doi: 10.3233/CLO-2010-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lonne GK, Masoumi KC, Lennartsson J, Larsson C. Protein kinase Cdelta supports survival of MDA-MB-231 breast cancer cells by suppressing the ERK1/2 pathway. J. Biol. Chem. 2009;284:33456–33465. doi: 10.1074/jbc.M109.036186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Quyun C, Ye Z, Lin SC, Lin B. Recent patents and advances in genomic biomarker discovery for colorectal cancers. Recent Pat. DNA Gene Seq. 2010;4:86–93. doi: 10.2174/187221510793205764. [DOI] [PubMed] [Google Scholar]
- 77.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat. Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 78.Furukawa T, Fujisaki R, Yoshida Y, Kanai N, Sunamura M, Abe T, Takeda K, Matsuno S, Horii A. Distinct progression pathways involving the dysfunction of DUSP6/MKP-3 in pancreatic intraepithelial neoplasia and intraductal papillary-mucinous neoplasms of the pancreas. Mod Pathol. 2005;18:1034–1042. doi: 10.1038/modpathol.3800383. [DOI] [PubMed] [Google Scholar]
- 79.Messina S, Frati L, Leonetti C, Zuchegna C, Di Zazzo E, Calogero A, Porcellini A. Dual-specificity phosphatase DUSP6 has tumor-promoting properties in human glioblastomas. Oncogene. 2011;30:3813–3820. doi: 10.1038/onc.2011.99. [DOI] [PubMed] [Google Scholar]
- 80.Madak-Erdogan Z, Lupien M, Stossi F, Brown M, Katzenellenbogen BS. Genomic collaboration of estrogen receptor alpha and extracellular signal-regulated kinase 2 in regulating gene and proliferation programs. Mol. Cell. Biol. 2011;31:226–236. doi: 10.1128/MCB.00821-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nat. Rev. Cancer. 2012;12:381–385. doi: 10.1038/nrc3263. [DOI] [PubMed] [Google Scholar]
- 82.Magnani L, Eeckhoute J, Lupien M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet. 2011;27:465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 83.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 84.Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERalpha signaling underlying progression in breast cancer. PLoS Genet. 2011;7:e1002368. doi: 10.1371/journal.pgen.1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mohammed H, D'Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, Rueda OM, Holmes KA, Theodorou V, Robinson JL, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tan SK, Lin ZH, Chang CW, Varang V, Chng KR, Pan YF, Yong EL, Sung WK, Cheung E. AP-2gamma regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J. 2011;30:2569–2581. doi: 10.1038/emboj.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Santos SJ, Haslam SZ, Conrad SE. Signal transducer and activator of transcription 5a mediates mammary ductal branching and proliferation in the nulliparous mouse. Endocrinology. 2010;151:2876–2885. doi: 10.1210/en.2009-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meier-Abt F, Milani E, Roloff T, Brinkhaus H, Duss S, Meyer DS, Klebba I, Balwierz PJ, van Nimwegen E, Bentires-Alj M. Parity induces differentiation and reduces Wnt/Notch signaling ratio and proliferation potential of basal stem/progenitor cells isolated from mouse mammary epithelium. Breast Cancer Res. 2013;15:R36. doi: 10.1186/bcr3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Howe LR, Brown AM. Wnt signaling and breast cancer. Cancer Biol. Ther. 2004;3:36–41. doi: 10.4161/cbt.3.1.561. [DOI] [PubMed] [Google Scholar]
- 90.Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, Pestell RG, Hung MC. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc. Natl Acad. Sci. USA. 2000;97:4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Collu GM, Meurette O, Brennan K. Is there more to Wnt signalling in breast cancer than stabilisation of beta-catenin? Breast Cancer Res. 2009;11:105. doi: 10.1186/bcr2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tanos T, Sflomos G, Echeverria PC, Ayyanan A, Gutierrez M, Delaloye JF, Raffoul W, Fiche M, Dougall W, Schneider P, et al. Progesterone/RANKL Is a Major Regulatory Axis in the Human Breast. Science translational medicine. 2013;5:182ra155. doi: 10.1126/scitranslmed.3005654. [DOI] [PubMed] [Google Scholar]
- 93.Mote PA, Bartow S, Tran N, Clarke CL. Loss of co-ordinate expression of progesterone receptors A and B is an early event in breast carcinogenesis. Breast Cancer Res Treat. 2002;72:163–172. doi: 10.1023/a:1014820500738. [DOI] [PubMed] [Google Scholar]
- 94.Mote PA, Balleine RL, McGowan EM, Clarke CL. Colocalization of progesterone receptors A and B by dual immunofluorescent histochemistry in human endometrium during the menstrual cycle. J Clin Endocrinol Metab. 1999;84:2963–2971. doi: 10.1210/jcem.84.8.5928. [DOI] [PubMed] [Google Scholar]
- 95.Mote PA, Graham JD, Clarke CL. Progesterone receptor isoforms in normal and malignant breast. Ernst Schering Found Symp Proc. 2007:77–107. [PubMed] [Google Scholar]
- 96.Graham JD, Yeates C, Balleine RL, Harvey SS, Milliken JS, Bilous AM, Clarke CL. Characterization of progesterone receptor A and B expression in human breast cancer. Cancer Res. 1995;55:5063–5068. [PubMed] [Google Scholar]
- 97.Pathiraja TN, Shetty PB, Jelinek J, He R, Hartmaier R, Margossian AL, Hilsenbeck SG, Issa JP, Oesterreich S. Progesterone receptor isoform-specific promoter methylation: association of PRA promoter methylation with worse outcome in breast cancer patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:4177–4186. doi: 10.1158/1078-0432.CCR-10-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.