Table 1.
Performance comparison in the LC-MS proteomic and glycomic datasets
| Performance measure | Proteomics |
Glycomics |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Raw | GP | SP | GPSP | GPMP | Raw | GP | SP | GPSP | GPMP | |
| RT | 83.08 (20.84) | 18.02 (22.74) | 19.37 (28.56) | 11.74 (19.61) | 10.70 (20.67) | 103.20 (32.63) | 42.36 (29.76) | 67.45 (42.39) | 44.65 (31.35) | 24.85 (27.21) |
| CV | 1.634 (0.256) | 1.118 (0.372) | 1.150 (0.415) | 1.060 (0.380) | 1.052 (0.380) | 1.194 (0.346) | 0.931 (0.284) | 1.090 (0.399) | 0.978 (0.366) | 0.821 (0.292) |
| Precision | 0.937 (0.026) | 0.983 (0.007) | 0.985 (0.004) | 0.988 (0.004) | 0.990 (0.003) | 0.943 (0.008) | 0.965 (0.005) | 0.967 (0.008) | 0.976 (0.003) | 0.980 (0.002) |
| Recall | 0.638 (0.241) | 0.933 (0.089) | 0.952 (0.043) | 0.962 (0.050) | 0.970 (0.027) | 0.612 (0.215) | 0.819 (0.136) | 0.773 (0.166) | 0.829 (0.167) | 0.907 (0.095) |
Note: Five approaches are compared: no alignment performed (raw), alignment performed using a GP regression, SP alignment performed without using a GP prior (SP), SP alignment performed with a GP prior (GPSP) and multi-profile alignment (G = 4) performed with a GP prior (GPMP). Performance comparison based on RT difference (in seconds) across runs for consensus peaks, CV of the extracted ion chromatograms of consensus peaks, precision and recall. For RT difference and CV, means (standard deviations) are reported based on the 273 and 106 consensus peaks in the proteomic and glycomic datasets, respectively. For precision and recall, means (standard deviations) are reported based on 72 pairs of tolerance parameters of 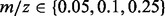 and RT
and RT 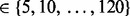 in SIMA.
in SIMA.
