Abstract
Motivation: Second-generation sequencing technologies produce high coverage of the genome by short reads at a low cost, which has prompted development of new assembly methods. In particular, multiple algorithms based on de Bruijn graphs have been shown to be effective for the assembly problem. In this article, we describe a new hybrid approach that has the computational efficiency of de Bruijn graph methods and the flexibility of overlap-based assembly strategies, and which allows variable read lengths while tolerating a significant level of sequencing error. Our method transforms large numbers of paired-end reads into a much smaller number of longer ‘super-reads’. The use of super-reads allows us to assemble combinations of Illumina reads of differing lengths together with longer reads from 454 and Sanger sequencing technologies, making it one of the few assemblers capable of handling such mixtures. We call our system the Maryland Super-Read Celera Assembler (abbreviated MaSuRCA and pronounced ‘mazurka’).
Results: We evaluate the performance of MaSuRCA against two of the most widely used assemblers for Illumina data, Allpaths-LG and SOAPdenovo2, on two datasets from organisms for which high-quality assemblies are available: the bacterium Rhodobacter sphaeroides and chromosome 16 of the mouse genome. We show that MaSuRCA performs on par or better than Allpaths-LG and significantly better than SOAPdenovo on these data, when evaluated against the finished sequence. We then show that MaSuRCA can significantly improve its assemblies when the original data are augmented with long reads.
Availability: MaSuRCA is available as open-source code at ftp://ftp.genome.umd.edu/pub/MaSuRCA/. Previous (pre-publication) releases have been publicly available for over a year.
Contact: alekseyz@ipst.umd.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Following the creation of draft versions of the human genome in 2001, many small and large genomes were sequenced using first-generation (i.e. Sanger) sequencing technology, with read lengths exceeding 800 bp. More recently, a variety of types of second-generation sequencing (SGS) technologies have appeared with read lengths ranging from 50–400 bp. The lowest-cost sequencers today produce 100-bp reads at a cost many thousands of times lower than Sanger sequencing. New assembly methods have been developed in response to the challenge of short-read assembly, and they have steadily improved in recent years. Despite this progress, though, the problem of determining the sequence of a genome is far from a solved problem. Virtually all assemblies published today are ‘draft’ genomes with varying levels of quality, containing many gaps and assembly errors that present significant problems for scientists who rely on these genomes for downstream analysis. This article reports progress in assembling genomes facilitated by a new approach to genome assembly. First, we briefly describe the two general approaches that have been used for assembly of whole-genome shotgun sequencing data.
Overlap–layout–consensus (OLC) assembly. Briefly, the OLC paradigm first attempts to compute all pairwise overlaps between reads, using sequence similarity to determine overlap. Then an OLC algorithm creates a layout, which is an alignment of all overlapping reads. From this layout, the algorithm extracts a consensus sequence by scanning the multiread alignment, column by column. Most assemblers for Sanger sequencing data, including Celera Assembler (Miller et al., 2008; Myers et al., 2000), PCAP (Huang, 2003), Arachne (Batzoglou et al., 2002) and Phusion (Mullikin and Ning, 2003), are based on the OLC approach.
Two of the main benefits of the OLC approach are flexibility with respect to read lengths and robustness to sequencing errors. To improve the likelihood that apparent overlaps are real (and not repeat-induced), OLC algorithms typically require them to exceed some minimum length, e.g. the Celera Assembler requires overlaps of 40 bp or longer, allowing for a small error rate (1–2%) in the overlapping region.
To compensate for shorter read length and lack of uniformity in coverage, SGS de novo assembly projects typically generate 100 times as many reads as Sanger-sequencing projects; e.g. the original human (Lander et al., 2001; Venter et al., 2001) and mouse (Mouse Genome Sequencing Consortium et al., 2002) projects generated ∼35 million reads each, whereas recent human sequencing projects (Li et al., 2010) generated 3–4 billion reads. The de Bruijn graph approach avoids the pairwise overlap computation entirely, which is one reason why it has become the leading method for SGS assembly.
The de Bruijn graph approach. The de Bruijn graph assembly algorithm was pioneered by Pevzner et al. (Idury and Waterman, 1995; Pevzner, 1989), who first implemented these ideas in the Euler assembler (Pevzner et al., 2001). Although Euler was designed for Sanger reads, the same general framework has been adopted recently by programs for assembling SGS data, and for Illumina read data in particular. Recently developed assemblers that use the de Bruijn strategy include Allpaths-LG (Gnerre et al., 2010), SOAPdenovo (Li et al., 2008), Velvet (Zerbino and Birney, 2008), EULER-SR (Chaisson and Pevzner, 2008) and ABySS (Simpson et al., 2009).
This approach begins by creating a de Bruijn graph from the read data, as follows. For a fixed value k, every substring of length k (a k-mer) from every read is assigned to a directed edge in a graph connecting nodes A and B. Nodes A and B correspond to the first and last k-1 nucleotides of the original k-mer. Any path through the graph that visits every edge exactly once, formally known as an Eulerian path, forms a draft assembly of the reads. In practice, these graphs are complex with many intersecting cycles, and many alternative Eulerian paths, and therefore creating the graph is merely the first small step in creating a good draft assembly. Complete assembly requires incorporating mate pair information into the graph and attempting to disentangle the many complex cycles created by repetitive sequences. Because the k-mers are shorter than reads, the graph contains less information than the reads, so the reads need to be retained for later use in disambiguating paths in the graph.
The main benefit of this approach is its computational efficiency, which it gains from the fact that the immense number of overlaps is not computed. The main drawbacks are loss of k-mer adjacency information in the graph and spurious branching caused by errors in the data.
Super-reads, a new alternative. In this article, we propose a third paradigm for assembly of short-read data, based on the creation of what we call super-reads. The aim is to create a set of super-reads that contains all of the sequence information present in the original reads despite the fact that there are far fewer super-reads than original reads. For the ideal error-free case, see the Theorem below.
The basic concept of super-reads is to extend each original read forwards and backwards, base by base, as long as the extension is unique. The concept can be explained as follows. We create a k-mer count look-up table (using an efficient hash table) to determine quickly how many times each k-mer occurs in our reads. Given a k-mer found at the end of a read, there are four possible k-mers that could be the next k-mer in a genome’s sequence: these are the strings formed by appending A, C, G or T to the last k-1 bases in the read. Our algorithm looks up, which of these k-mers occur in the table. If only one of the four possible k-mers occurs, we say the read has a unique following k-mer and we append that base to the read. We continue until the read can no longer be extended uniquely; i.e. there is more than one possible continuing base, or we have reached a dead end and no base is permissible. We perform this extension on both the 3′ and 5′ ends of the read. The new longer string is called a super-read. Many reads extend to the same super-read as shown in Figure 1. Notice that if two reads have an interior difference by even one base—as, for example, would occur if they derived from two non-identical repeats or from two divergent haplotypes—then they will generate distinct super-reads. Of course super-reads can easily be computed using a de Bruijn graph. The point is that once the super-reads are created, they–together with mate pairs that connect super-reads–collectively replace the de Bruijn graph. Incorporation of mate-pair information is carried out using the OLC assembly step described below. The two most important properties of the super-read data computation are as follows:
each of the original reads is contained in a super-read (so no information has been lost); and
many of the original reads yield the same super-read, so using super-reads leads to vastly reduced dataset.
Fig. 1.

Reads 1, 2 and 3 yield the same super-read. Reads are depicted by the black solid lines. Dashed lines represent the k-mer extensions starting from k-mers in the reads 1, 2 and 3. The super-read is depicted by the thick solid line. All reads that extend to the same super-read are replaced by that super-read
Hundreds of times fewer super-reads than reads. MaSuRCA uses a modified version of the CABOG assembler (Miller et al., 2008), for the overlap-based assembly following super-read construction. In creating its fundamental unit of unitigs, CABOG uses only ‘maximal’ reads, i.e. reads that are not proper substrings of other larger reads. In principle, this could cause assembly errors but in practice they seem to be rare. Because of this practice, we carry out one extra step: the only super-reads we use are maximal super-reads, i.e. those that are not exact substrings of another super-read. We then assemble the maximal super-reads along with other available data including mate pairs with the modified CABOG assembler. The ‘other data’ include jumping libraries and possibly 454 read data and Sanger read data and mate pairs.
We observe that the coverage of the genome by maximal super-reads typically varies from ∼2–3×, independent of whether the raw read coverage is 50, 100 or even higher. Note that each heterozygous single nucleotide polymorphism increases the number of super-reads. For a haploid genome, super-reads will tend towards 2× coverage, whereas for highly heterozygous diploid genomes the super-read coverage may be closer to 4×. In the two example genomes described in the Results section, Rhodobacter sphaeroides and Mus musculus, the reads outnumber the maximal super-reads by factors of ∼400 and 300, respectively. The N50 lengths for the super-reads themselves are 3314 and 2241 bp, respectively. MaSuRCA automatically chooses the k-mer size for creating super-reads, and in these two cases, k is 33 and 69, respectively.
The following Theorem lays the theoretical foundation of equivalence of assemblies made from the original reads and the super-reads for the case of perfect error-free reads.
The super-reads theorem for the ideal case of perfect (error-free) reads. To understand the underpinnings of the super-reads approach, we consider the simplest case. Here we ignore mate pairs. The above construction of super-reads is based on a fixed k-mer size, so for clarity we can call them k-super-reads. The genome is a collection of strings (chromosomes) and loops (plasmids or organelles) with a four-letter alphabet. To avoid end effects, we assume that all DNA in the genome is circular, as is often the case for bacteria, but we shall still speak of their ‘substrings’. A string (read or super-read) is called perfect if it is identical to a substring of the genome. Such a substring of the genome together with its coordinates is called a placement. A string may have multiple placements. We say that a set of strings R is k-perfect with respect to a genome G if (i) every base of the genome G is covered by some placement, and (ii) adjacent placements overlap by at least k bases.
When a set of reads is k-perfect, we can distill the information in the reads by the usually much smaller set of k-super-reads. The following result says that k-super-reads contain all of the information in the reads.
Theorem —
Assume a set of reads is k-perfect for some genome G. Then the corresponding set of k-super-reads has the same property.
In other words, the set of super-reads contains all of the information in the reads, and they have introduced no errors. The proof follows from the construction of super-reads. Because each read is contained in a super-read, no data are lost. At the same time, if the original read data are inadequate for deducing what the genome is, then so are the super-read data. If both flanks of some copy of the repeat were not covered in the read set, there would be no maximal k-super-read that could be placed at that copy of the repeat.
In practice reads are not perfect, and because the super-reads can only represent the information in the original reads, there will always be some super-reads that contain errors that were in the original reads. The task of the assembly algorithm used downstream of super-reads is to detect and correct most of the errors and create a mostly correct assembly. Assemblers have long been designed to do exactly that, because reads used in assembly projects were never assumed to be perfect. The MaSuRCA assembler benefits from the advanced assembly techniques in the CABOG assembler for creating contigs and scaffolds from super-reads.
2 RESULTS
Here we report on the application of the MaSuRCA assembler version 2.0 to the assembly of two organisms: the bacterium R.sphaeroides str. 2.4.1 (Rhodobacter) and chromosome 16 of M.musculus lineage B6 (mouse). Note that MaSuRCA 2.0 is a new release of a system that was formerly known as MSR-CA.
Choice of genomes. These genomes were chosen for several reasons. First, they represent two widely different challenges: a small clonal genome and a much larger diploid one. Second, the finished sequence is available for both genomes, which allows us to evaluate correctness of the assemblies. Third, one of the assemblers with which we compared MaSuRCA, Allpaths-LG, requires that data are generated according to a preset recipe, which was followed for both the Rhodobacter and mouse datasets. Fourth, for both of these genomes, up to 9× coverage by long (Sanger) reads is available. This allows us to show how the MaSuRCA assembler can benefit from combining Illumina data with additional relatively low (1–4×) genome coverage by long reads (LR). We also compare the performance of MaSuRCA with the performance of CABOG only for the 9× Sanger data on the bacterial dataset. Although we used Sanger sequencing technology for LR, similar improvements in assembly results can be achieved using the latest Roche 454 sequencing technology. We do not require the LR to have mate pairs, and we did not use mate-pair information for the Sanger reads in our experiments on the mouse chromosome assembly.
We also chose the R.sphaeroides bacterium because it has a high GC content, ∼68%, which makes it challenging to sequence. Illumina technology is much less effective in high GC-content regions. In particular, even though we used 45× overall genome coverage, short (100–200 bp) windows whose GC content is above 80% or below 20% may be covered by only one or two reads. This makes assembly of high- or low-GC genomes from Illumina data particularly difficult, and frequently results in fractured assemblies.
Both datasets that we chose had Illumina data generated by the Broad Institute sequencing center and had high-quality libraries with tightly controlled fragment lengths, as shown in the Supplementary Material. Such low deviations from the target library size may not be typical for all sequencing centers and genome projects. The MaSuRCA assembler uses a modified version of the CABOG assembler for contiging and scaffolding, and in practice it will produce good assemblies with libraries whose standard deviations are up to 20% of the library mean.
One of the popular genomes used in evaluations of genome assemblers is Escherichia coli (Simpson et al., 2009). There is ample sequencing data available for E.coli genome, and the finished sequence is also available. However, we decided against using this genome for our evaluations because E.coli is relatively easy to assemble, because of its low repeat content, and thus does not provide a stern test of an assembly algorithm: most assemblers do reasonably well on this genome. We also decided against evaluations using artificially generated ‘faux’ data. We do not know of any working and published technique that comes close to simulating the whole spectrum of errors and biases that are present in real-life sequencing data. Thus an assembler that performs well on simulated data may perform poorly on the real data. See, e.g. Tables 1 and 2 in Luo et al. (2012), where the SOAPdenovo2 assembler performed better than Allpaths-LG on the faux Assemblathon 1 data, but was much worse on the real data for both bacterial datasets.
Table 1.
Comparison of the assemblies of R.sphaeroides
| Assembler/ data type | Quast NGA50 contig (kb) | Quast misas- semblies in contigs | NGA50 scaffold (Mb) | Scaffold misas- semblies/ Mb |
|---|---|---|---|---|
| Allpaths-LG | 41.5 | 15 | 3.2 | 0.2 |
| SOAPdenovo2 | 17.5 | 5 | 0.067 | 0.9 |
| MaSuRCA | 41.4 | 13 | 3.1 | 0.7 |
| Assemblies including some Long Read (LR) data | ||||
| CABOG LR only | 52.7 | 12 | 1.5 | 0.4 |
| MaSuRCA + 1× LR | 63.9 | 21 | 3.2 | 0.7 |
| MaSuRCA + 2× LR | 87.2 | 17 | 3.2 | 0.2 |
| MaSuRCA + 4× LR | 228.4 | 20 | 3.2 | 0.2 |
Note: The best value for each column is shown in boldface. All assemblies used Illumina data. The size of the finished reference sequence for this genome was 4 603 067 bp and the largest chromosome is about 3.2 Mb. All assemblies had <1 misassembly/1 Mb of scaffold sequence.
Table 2.
Comparison of the assemblies of mouse chromosome 16 using Illumina-only data (top three rows) and MaSuRCA using a mixture of Illumina data and long Sanger reads (bottom)
| Assembler | Quast contig NGA50 | Quast contig misas- semblies | NGA50 scaffold (Kb) | Scaffold misas- semblies/ MB |
|---|---|---|---|---|
| Allpaths-LG | 28 | 175 | 261 | 0.03 |
| SOAPdenovo2 | 8 | 369 | 1828 | 0.17 |
| MaSuRCA | 56 | 283 | 3445 | 0.19 |
| Assemblies including some Long Read (LR) data | ||||
| MaSuRCA + 1× LR | 70 | 256 | 4472 | 0.04 |
| MaSuRCA + 2× LR | 82 | 248 | 3704 | 0.21 |
| MaSuRCA + 4× LR | 102 | 246 | 4511 | 0.21 |
Note: The best value for each column is shown in boldface. The total size of the finished sequence of this chromosome was 98 319 150 bp. All assemblies generated accurate scaffolds, but the number of the contig misassemblies differs significantly.
Choice of assembly programs for comparison. We chose to compare the performance of MaSuRCA with the two most popular large-scale genome assemblers: Allpaths-LG and SOAPdenovo. We decided against doing more comparisons to avoid repetition of the results of studies done in the recent GAGE evaluations. One can judge the relative performance of other popular assemblers from that project (Salzberg et al., 2012, http://gage.cbcb.umd.edu). The original GAGE assembly comparison (Salzberg et al., 2012) compared the following assembly programs:
ABySS (Simpson et al., 2009)
ALLPATHS-LG (Gnerre et al., 2011)
Bambus2 (Koren et al., 2011)
CABOG (Miller et al., 2008)
MSR-CA (now renamed MaSuRCA 1.0)
SGA (Simpson and Durbin, 2012)
SOAPdenovo (Luo et al., 2012)
Velvet (Zerbino and Birney, 2008)
The best performers in GAGE were AllPaths-LG and SOAPdenovo. Hence, we have included those two programs for comparison with MaSuRCA 2.0. For a more recent comparison one should see the GAGE-B competition (Magoc et al., 2013). It reports on assemblies of 12 bacterial genomes by ABySS, CABOG, MaSuRCA 1.8.3, Mira v3.4.0 (Chevreux et al., 2004), SOAPdenovo, SPAdes v2.3.0 (Bankevich et al., 2012) and Velvet. MaSuRCA 1.8.3 produced the best assemblies for the majority of the 12 species. SPAdes did well, especially on 250-bp reads, but is not designed for larger genomes. Allpaths-LG was not used in that competition because it requires two libraries, whereas this test had only one library per species.
Evaluating the assemblies. We evaluated the performance of the assemblers using two separate techniques. We evaluated the contigs using the recently published Quast 2.1 software (Gurevich et al., 2013). In the tables below, we report the contig sizes in terms of NGA50 reported by Quast. The NGA50 size is defined as the value N such that 50% of the finished sequence is contained in contigs whose alignments to the finished sequence are of size N or larger. Note that NGA50 differs from N50 in that N50 is defined by the total size of the assembled contigs, whereas NGA50 is defined by the actual size of the genome itself. If the assembly size is close to the true genome size, then N50 and NGA50 are roughly equivalent.
Quast does not report the scaffold statistics in the way we would prefer to look at them. In evaluating the scaffolding, we look for the correct order and orientation of the contigs and contiguity of the coverage allowing for reasonable (shorter than a longest mate pair) gaps. Thus we evaluated the scaffolds separately by mapping them to the finished sequence using Nucmer (Kurtz et al., 2004). We then clustered the matches of each scaffold to the finished sequence based on proximity of the matches in terms of finished sequence coordinates. Within each cluster of the matches of the scaffold to the finished sequence, we required that the matches are in the same order in terms of finished sequence and scaffold match coordinates (no rearrangements), same orientation and the distance between the consecutive matches is smaller than 40 kb (the size of the longest library) for the mouse genome and 3.5 kb for the bacteria. Then we counted the number of clusters and the number of the scaffolds. The number of scaffold misassemblies is the difference between these two numbers. We defined NGA50 for the set of scaffolds as the value N such that 50% of the finished sequence is spanned by clusters where the span of each individual cluster is of size N or larger.
Bacteria genome assembly. For the first comparison, we chose two Illumina datasets: (i) a paired-end library (i.e. PE), in which reads were generated from both ends of 180-bp DNA fragments (SRA accession SRR081522), and (ii) a ‘jumping’ library in which paired ends were sequenced from 3600-bp fragments, (SRA accession SRR034528). We randomly down-sampled both libraries to 45× genome coverage. For LR we used 59 211 Sanger reads, with average length 772 bp, from the National Center for Biotechnology Information (NCBI) Trace Archive entry for R.sphaeroides str. 2.4.1 (Choudhary et al., 2007). The Sanger reads provided ∼10× genome coverage. For our experiments, we used randomly down-sampled LR datasets of 1×, 2× and 4× coverage. The data are summarized in Supplementary Table S1.
The parameters used for the Allpaths-LG, MaSuRCA and SOAPdenovo2 assemblies are described in the Supplementary Material. Because the creation of super-reads is critical in the MaSuRCA design, we first present the analysis of the number and correctness of the super-reads.
For this dataset, MaSuRCA reduced the original 2 050 868 paired-end reads to 5168 super-reads, a reduction by a factor of almost 400. In addition to those, the MaSuRCA submits to the modified Celera Assembler 18 970 linking mates (PE pairs where the two reads ended up in two different super-reads, see Methods). Note that all linking mates were contained in super-reads. Thus we transformed 2 050 868 original reads into a total of 24 138 (=18 970 + 5168) reads with a 77-fold reduction in the number of reads. The N50 size of the super-reads was 3314 bp, the minimum size was 33 bp, and the longest super-read was 13 283 bp. The total amount of sequence in super-reads was 9 138 989 bp, ∼2× coverage of the genome.
To determine how well the (maximal) super-reads agreed with the genome, we mapped the super-reads to the finished sequence by Nucmer (Kurtz et al., 2004) using a k-mer seed size of 15 (parameters: -l 15 -c 32 –maxmatch). The total number of bases in the super-reads that matched the finished sequence was 9 106 770 in 4845 super-reads. A total of 99.2% of the matching bases were in at least one of 4673 super-reads that matched with at least 99% identity over at least 99% of their length. The remaining 323 super-reads that did not match the finished sequence contained 32 219 bp of sequence, and their maximum size was 150 bp. We examined the reads that were used to produce the non-matching super-reads and could not find a match to the genome of length ≥32 bp in any of these reads. It is likely that these reads primarily contained adapter sequences with errors or other contaminants
Table 1 shows the comparison of the performance of the MaSuRCA assembler with the others on the R.sphaeroides dataset. The MaSuRCA assembler using only Illumina data performs on par with Allpaths-LG, with nearly identical NGA50 sizes, two fewer contig errors and two more scaffold errors. All scaffold errors were in small scaffolds whose sizes were well below the N50 scaffold size and this did not influence the NGA50 scaffold size. Moreover, the performance of MaSuRCA on Illumina data alone is comparable with performance of CABOG on only the Sanger (long-read) data.
Although SOAPdenovo2 had the smallest number of contig errors (5), its contigs were significantly smaller than those produced by the other assemblers. As we introduced additional coverage by LR into the mix, the assemblies produced by MaSuRCA assembler become superior in contiguity to all other assemblers (bottom three rows of Table 2). In particular, the contig N50 value increased from 41.4 to 52.7 kb with just 1× Sanger data, and to 228 kb with deeper 4× Sanger data. We note that neither SOAPdenovo2 nor Allpaths-LG allows for mixed datasets of this type.
Mouse genome assembly. To save time and to allow for more detailed examination of the results, we created a restricted dataset for a single chromosome of the mouse genome, chromosome 16 (Mmu16). We downloaded the same data for the mouse genome as was used in the evaluation of Allpaths-LG (Gnerre et al., 2011), which are available from the NCBI SRA under the study Mouse_B6_Genome_on_Illumina. These sequences were generated from mouse strain C57BL/6J, the same strain used for the finished mouse sequence (Mouse Genome Sequencing Consortium et al., 2002).
We mapped the reads to the finished sequence for the entire mouse genome using Bowtie2 (Langmead and Salzberg, 2012), allowing up to five best hits of identical quality for each read. We then extracted the reads whose best hit either for the read or for its mate was in chromosome 16. We also downloaded the original Sanger reads from NCBI Trace Archive (Mouse genome Sequencing Consortium et al., 2002), and mapped them against the finished sequence. MaSuRCA does not require the LR to be mated, and we excluded mate-pair information for these reads during assembly. Supplementary Table S2 lists the mouse datasets used in our experiments.
From the paired-end dataset containing 50 million reads, the super-reads module of MaSuRCA produced 297 279 super-reads containing 210 839 005 bp, with an N50 size of 2241 bases. The reads outnumber the (maximal) super-reads by a factor of over 300. The original ∼45× coverage by the 101-bp paired-end reads reduced to just over 2× coverage by super-reads. In addition, the super-reads module output 940 390 linking mates from the PE library; these are paired reads that link together two super-reads. Thus we reduced 50 M reads to ∼1.24 M super-reads and linking mates, a 40-fold reduction. After mapping the super-read sequences to the finished sequence using Nucmer, we found that 209 017 737 bp in 284 179 super-reads matched Mmu16. Of these matching bases, 98% were contained in at least one of the 258 927 super-reads that had at least 99% identity to MMu16 over at least 99% of the super-read’s length.
Results for the mouse assemblies are provided in Table 2. Not unexpectedly, the MMu16 dataset was more challenging than the bacterial genome. For assembly with Illumina-only data, the NGA50 contig size for MaSuRCA assembly was twice as big compared with the Allpaths-LG assembly, whereas the number of errors was 62% larger. SOAPdenovo2 produced small contigs with a large number of errors. The MaSuRCA assembler produced the largest scaffolds, with NGA50 more than an order of magnitude larger than the Allpaths-LG scaffolds and almost twice bigger than the SOAPdenovo2 scaffolds.
MaSuRCA produced progressively larger and more accurate contigs as LR were added into the mix. Additional 4× LR coverage almost doubled the N50 contig size while reducing the number of contig misassemblies by 13%. The LR data did not have any mate pairs by design; thus we did not expect a significant improvement in scaffolding, however, the scaffolds improved as well. We note that for each run the same set of super-reads, jumping library reads and linking mates went into the CABOG assembler; the only difference between runs was in the number of LR. As we introduced more LR, the number of assembly errors decreased, whereas the contig N50 size increased significantly.
3 METHODS
The key idea driving the development of the MaSuRCA assembler is to reduce the complexity of the data by transforming the high coverage (typically 50–100× or deeper) of the paired-end reads into 2–3× coverage by fairly long (maximal) super-reads. The reduced data could then be efficiently assembled by a modified Celera Assembler. Here we want to describe some of the modules in MaSuRCA. In this section, we simply say super-read for maximal super-reads, as non-maximal super-reads are not used in assembly.
QuorUM error correction. We use the QuorUM error corrector due to its stability and high performance (Marçais et al., 2013). One may substitute another error corrector, such as Quake (Kelley et al., 2010), as long as the output is processed in such a way that read names are preserved and the mates are reported together.
Creation of k-unitigs. Extending each read base by base is computationally inefficient. Therefore, we use error-corrected reads to construct k-unitigs as follows. We create a k-mer count look-up table, using a hash constructed by the Jellyfish program (Marçais and Kingsford, 2011), which allows us to determine quickly how many times each k-mer occurs in the reads. Given any k-mer, there are four possible k-mers that may follow it, one for each possible extension by A,C,G or T. We look up how often each of these occurs, and if we find only one of these four extensions, we say that our original k-mer has a unique following k-mer. We similarly check whether there is a unique preceding k-mer, analogously defined.
We call a k-mer simple if it has a unique preceding k-mer and a unique following k-mer. A k-unitig is a string of maximal length such that every k-mer in it is simple except for the first and the last. There are alternative names for quite similar concepts, such as ‘unipaths’ in Allpaths-LG (Gnerre et al., 2011). By construction, no k-mer can belong to more than one k-unitig. (But note that a k-unitig itself can occur at multiple sites in the genome, as will happen for exact repeats.) Hence, if a k-mer from a k-unitig U is encountered in the genome, then the whole sequence of the k-unitig U must appear at that location in the genome. We will use this property of k-unitigs when creating super-reads.
Super-reads from paired-end reads. Because no k-mer can belong to more than one k-unitig, if a read has a k-mer that occurs in a k-unitig, the read and the k-unitig can be aligned to one another. In the simplest case, when a read is a substring of a k-unitig, then that k-unitig is the read’s super-read. In other cases, we use individual reads to merge the k-unitigs that overlap them into a single longer super-read. Figure 2 shows an example of this process, in which a read R is extended to a super-read that consists of two k-unitigs that overlap by k-1 bases. K-mers M1 and M2 belong to R and also to k-unitigs K1 and K2, which may extend beyond the ends of R. Two reads that differ even at only one base will map and be extended by different sets of k-unitigs. The k-unitigs by construction can be connected by the exact k-1 end sequence overlaps. We call them k-unitig overlaps.
Fig. 2.
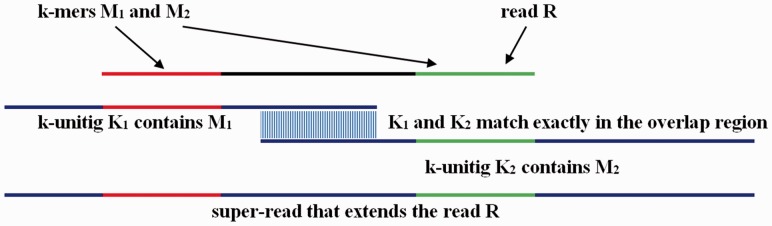
An example of a read whose super-read has two k-unitigs. Read R contains k-mers M1 and M2 on its ends. M1 and M2 each belong to k-unitigs K1 and K2, respectively. K-unitigs K1 and K2 are shown in blue, and the matching k-mers M1 and M2 are shown in red and green. K1 and K2 overlap by k-1 bases. We extend read R on both ends producing a super-read, also depicted in blue. A super-read can consist of one k-unitig or can contain many k-unitigs
If the reads are paired, we examine each pair of reads (we also call them mated reads or mate pairs) and map each read to the k-unitigs, and then look for a unique path of k-unitigs connected by k-unitig overlaps that connects the two reads. If we find such a path, then we extend both paired-end reads to a new super-read, created by merging the k-unitigs on this unique path. Often this process of creating a super-read from a pair fails, e.g. when there is a repeated sequence or a gap in coverage between the mates. In this case, we form a super-read for each of the mates separately, and the mate pair is submitted to the assembler as linking mate pair along with the corresponding super-reads.
Jumping library filter. Although not required for MaSuRCA, long ‘jumping’ libraries (i.e. in which each pair of reads are several kilobases apart) are often part of genome sequencing projects, where they provide valuable long-range connectivity data for the scaffolding. However, these libraries sometimes contain reads that are chimeric, i.e. they derive from two distant parts of the genome, or they may be mis-oriented because of problems in library construction. In particular, some jumping libraries use a circularization protocol, in which the DNA fragment is circularized to bring together its ends. The resulting read pairs face outward, meaning that the opposite ends of the original DNA fragment occur on the 3′ ends of the two reads. Misoriented mates from these libraries are ‘innies’ that originated from one ‘side’ of the original circle that did not contain a junction site. These ‘innies’ should be treated as regular short insert (300–400 bp) paired-end reads. Because these kinds of errors create many problems in the scaffolding phase, it is imperative that most of them be removed before giving the data to the assembly program.
We use QuORUM error correction and the super-reads procedures to perform library cleaning. First, during error correction we create a k-mer database from the paired-end reads only, excluding the jumping libraries. Note that circularized reads include a linker sequence, and the concatenation of the linker and the source DNA represents sequence that should not appear in the genome. If one of the reads in a jumping library mate pair contains a junction site (identified by the linker), then k-mers that span the junction site in that read will not be found in the k-mer database. The error corrector then trims the read at the junction site.
We then create super-reads from the jumping library reads. Here we introduce a modification to the algorithm that accepts any path of overlapping k-unitigs in building a super-read. By doing aggressive joining, we are able to identify most of the non-junction ‘innie’ pairs, because they end up in the same super-read. Next we look for redundant jumping library pairs, where the same DNA fragment was amplified before circularization, producing two or more pairs of reads that represent the same fragment. Because the assembly process assumes that each mate pair is an independent sample from the genome, such redundant pairs can lead to assembly errors later. We examine the positions of reads in the resulting super-reads and look for redundant pairs, where both sets of forward and reverse reads end up in the same one or two super-reads with the same offset. We reduce such pairs to a single copy.
Contiging and scaffolding with the CABOG assembler. We keep track of the number of reads that generated each maximal super-read and the positions of those reads in the maximal super-read; thus allowing us to precisely report positions of all reads in the assembly. After creating the super-reads, we assemble the data with a modified version of the CABOG assembler (Miller et al., 2008, 2010), updated to allow long super-reads as well as short reads in the same assembly. We supply the following four types of data to CABOG:
super-reads
linking mates
cleaned and de-duplicated jumping library mate pairs (if available)
other available LR
We note that the modified version of CABOG 6.1 used in MaSuRCA is not capable of supporting the long high-error-rate reads generated by the PacBio technology.
CABOG uses read coverage statistics (Myers et al., 2000) to distinguish between unique and repetitive regions of the assembly. Each super-read typically represents multiple reads, and we keep track of how many reads belong to each super-read and the position of these reads. We modified CABOG to incorporate these data, using counts of all the original reads in its computation of coverage statistics. This major modification allowed us to use CABOG to assemble super-read data together with Illumina, 454 and Sanger reads.
Gap filling. The final major step in the MaSuRCA assembler is gap filling. This step is aimed at filling gaps in scaffolds that are relatively short and do not contain complicated repetitive structures. Such gaps may occur because the assembler software over-trimmed the reads in the error correction step, or because it misestimated the coverage statistics for some contigs or because of other reasons. Our gap-filling technique is again centered on our super-reads algorithm.
For each gap in each scaffold, we create faux 100-bp paired-end reads from the ends of the contigs surrounding the gap. We call these contig-end reads. From each pair of contig-end reads, we then use the 21mers whose counts are globally below a threshold of 1000 (to avoid highly repetitive sequences) to pull in the original uncorrected reads where either the read or its mate contains one of those k-mers. From this set, we create a local bin of reads corresponding to the gap in question. We then create k-unitigs from the reads in the bin for k ranging from 17 to 85, and see if we can create a unique super-read that contains both contig-end reads. If we are able to achieve that for some value of k, then the resulting super-read is used as a local patch of sequence to fill the gap. We found that, depending on the genome, 10–20% of the gaps in the scaffolds can be filled using this approach.
4 DISCUSSION
We began this project when we were faced with the prospect of assembling a 20+ Gbp pine tree genome with perhaps 15+ billion Illumina reads. That was far larger than anything that had been assembled. Along the way, we have found our philosophy of reducing Illumina reads to super-reads is useful. We discuss possible shortcomings and problems of our approach, as well as data problems that can result in a poor assembly if the user does not address them.
We have mentioned the GAGE B study of bacterial genomes, in which MaSuRCA was declared highly effective. At the end of this section, we list larger genomes that have been assembled by MaSuRCA and are publicly available.
Overall evaluation. In Tables 1 and 2, the Quast NGA50 contig size and the NGA50 scaffold size can be viewed loosely as N50 sizes after the contigs and scaffolds have been broken at each major misassembly. When comparing two assemblies, if after breaking at errors the N50 contig or scaffold size is doubled in one assembly compared with the other while introducing fewer than twice as many errors, we believe the doubling is justified.
In Table 1 on Illumina data only, we view Allpaths as doing slightly better than MaSuRCA on scaffolds and both did significantly better than SOAPdenovo. Note that the scaffold NGA50 for Allpaths and MaSuRCA are about three-fouth of the size of the genome, indicating that unlike SOAPdenovo, they both got the biggest chromosome in a correct scaffold. The fact that MaSuRCA has long scaffolds ∼3 Mb after breaking at errors, and the errors occur at roughly ∼1 Mb in spacing indicates that the errors lie near the ends of the big scaffold or in small scaffolds (in this case they all were in small scaffolds), so that the overall size of the scaffolds is not severely impacted by breaking at errors. When significant amounts of long read data are available, MaSuRCA makes use of that resource and does better. Its contig sizes rise dramatically and the scaffold error rates drop.
For R.sphaeroides the high GC content of the genome results in greater variability in the read coverage because of biases present in Illumina sequencing technology. This case shows that good assemblies are still possible even for high (or low) GC genomes.
Table 2 is a better test of scaffolding, as the scaffolds are not approaching the size of the genome. MaSuRCA’s scaffolds are roughly 13 times larger than Allpaths’ while introducing only about 6 times as many errors. Errors seem inevitable unless contigs and scaffolds are built conservatively and remain small. SOAPdenovo’s assembly suffers from small contigs. Again, adding LR improves the assembly significantly.
It is clear that even if assembler A is significantly better than assembler B on a collection of genome datasets, A may do worse than B on some datasets (Magoc et al., 2013). Here we have chosen datasets for which the PE mate pairs overlap each other, as is required by Allpaths. Our limited experience MaSuRCA assemblies are better if multiple PE libraries are used, varying the fragment length. Generally a jumping library should also be available such as one built from 3 kb (or longer) fragments.
Error correction. Error correction greatly simplifies the de Bruijn graph and typically results in larger k-unitigs and thus larger super-reads. Our algorithm works best on error-corrected reads, but is not tied to a particular error correction technique. In the MaSuRCA software package, we use the QuORUM error correction algorithm (Marcais et al., in preparation). However, one can substitute other techniques, such as Quake (Kelley et al., 2010) or Hammer (Medvedev et al., 2011).
Data problems. A variety of problems with the input reads and libraries can reduce the quality of an assembly. One of the most common issues is mislabeled or poorly size-selected fragment libraries. For example, we have encountered jumping libraries identified as 8 Kb (made from 8 Kb fragments), but later found that the sequences include a mixture of pairs created from 2 to 8 Kb fragments. Similar problems often arise with long-distance paired reads. Various explanations have been offered for this type of error, but regardless of the source, the misidentified mate pairs create difficulties when the assembler tries to place them 8 Kb apart in the assembly. An examination of the assembly may reveal the problem, at which point it can be corrected and the assembly can be restarted. We have observed libraries that were designed to be longer than 5 Kb but were entirely comprised of 2 Kb fragments. Another problem that arises with current technology is that the forward reads might be of excellent quality, but their mates (which are created in a separate run) are of far lower quality. We encountered one dataset where some of the libraries had so many errors that the assembly was better when made without those libraries. For example, when using 454 paired-end data, if the wrong linker sequence is provided to the assembler, the assembly will be severely fragmented. In general, severe fragmentation of an assembly is an indication of some kind of data error, which in turn requires a form of ‘data debugging’ to fix the errors and restart the assembly. No list of possible data errors will be complete.
A data diagnostic, U/k. Before running an assembler, one should evaluate the quality of the input data with any tools available. One strategy that we have found useful is to count the number of unique k-mers in the reads. Given a project with deep coverage, e.g. 30× or higher, any k-mers that occurs just once in the set of reads almost certainly contains at least one error. [This is the insight used by the Quake error corrector (Kelley et al., 2010)]. We can compare the number of unique k-mers in forward and reverse reads as a means of evaluating the quality of the reverse reads.
We can also use k-mer counts to estimate the real error rate in the read data, as follows. A sequencing error in the middle of a read is likely to result in k unique k-mers, because every k-mer containing the error will be unique. If the average number of unique k-mers per read is U, then U/k is a lower bound estimate of the average number of sequencing errors per read in the data. This estimate ignores the fact that an error near the end of a read will result in fewer erroneous k-mers, and it does not take into account cases when there are two or more errors per k-mer. The U/k value should be used as a minimum fitness criterion for the input read data: if the estimated number of errors is >2–3 for 100-bp reads, then it is likely that there was a problem in the sequencing run (current Illumina technology usual has an error rate below 1%). It may be more effective to ignore or redo a run with a high error rate than to use it for assembly.
Polymorphic genomes. Differences between the two copies of homologous chromosomes in a diploid genome can increase the number of super-reads ∼2-fold. This does not usually constitute a problem as long as subsequent assembly steps handle the polymorphic super-reads. The Celera Assembler (CABOG) will attempt to combine polymorphic regions that differ by up to 6%. If the haplotype divergence rate is higher, it will result in a fragmented assembly, where many scaffolds will terminate in regions of haplotype difference. This occurs because, even though the mate pairs may suggest that two scaffolds representing two haplotypes should be merged, the contigs within those scaffolds will not align sufficiently well, and therefore the scaffolder will not make the merge. In this case, the assembly can be post-processed to split the haplotypes and create scaffolds representing both heterozygous chromosomes.
Available genomes assembled by MaSuRCA. MaSuRCA has been used to assemble de novo a variety of genomes, sometimes improving on published genomes using added data, sometimes creating the first publicly available draft genome for the species. Below is a partial list of genomes that were recently assembled with MaSuRCA, including the types of read data used for each project:
Loblolly pine, Pinus taeda, a 22 Gbp genome, draft assembly using Illumina data only, in collaboration with the Pinerefseq consortium.
Indian cow, Bos indicus, 454/Illumina mixed data, in collaboration with USDA/ARS.
Rhesus macaque, Macaca mulatta, Sanger/Illumina mixed data, in collaboration with University of Nebraska.
Water buffalo, Bubalus bubalus, 454/Illumina mixed data, in collaboration with USDA-ARS and CASPUR, Italy.
Domestic cat, Felis felis, Sanger/454/Illumina mixed data, in collaboration with Washington University.
Philippine tarsier, Tarsier syrichta, Sanger/Illumina mixed data, in collaboration with Washington University.
Fire ant, Wasmannia auropunctata, 454/Illumina mixed data, in collaboration with OIST, Japan.
Stalk-eyed fly, Teleopsis dalmanni, 454/Illumina mixed data, in collaboration with University of Maryland.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kristian Stevens (University of California-Davis) for his helpful comments. We also thank one of the referees for noticing an important error in the original statement of the theorem.
Funding: National Research Initiative competitive grants 2009-35205-05209 and 2008-04049 from the United States Department of Agriculture National Institute of Food and Agriculture (in part); National Institutes of Health grants R01-HG002945 and R01-HG006677.
Conflict of Interest: none declared.
REFERENCES
- Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzoglou S, et al. ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary M, et al. Genome analyses of three strains of Rhodobacter sphaeroides: evidence of rapid evolution of chromosome II. J. Bacteriol. 2007;189:1914–1921. doi: 10.1128/JB.01498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl Acad. Sci. USA. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, et al. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, et al. PCAP: a whole-genome assembly program. Genome Res. 2003;13:2164–2170. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idury RM, Waterman MS. A new algorithm for DNA sequence assembly. J. Comput. Biol. 1995;2:291–306. doi: 10.1089/cmb.1995.2.291. [DOI] [PubMed] [Google Scholar]
- Kelley DR, et al. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 2010;11:R116. doi: 10.1186/gb-2010-11-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, et al. Bambus 2: scaffolding metagenomes. Bioinformatics. 2011;27:2964–2971. doi: 10.1093/bioinformatics/btr520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, et al. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T, et al. GAGE-B: an evaluation of genome assemblers for bacterial organisms. Bioinformatics. 2013;29:1718–1725. doi: 10.1093/bioinformatics/btt273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, et al. 2013. QuoUM: an error corrector for Illumina reads. arXiv.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev P, et al. Error correction of high-throughput sequencing datasets with non-uniform coverage. Bioinformatics. 2011;27:i137–i141. doi: 10.1093/bioinformatics/btr208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, et al. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, et al. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Mullikin JC, Ning Z. The Phusion assembler. Genome Res. 2003;13:81–90. doi: 10.1101/gr.731003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers G, et al. A whole genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- Pevzner PA. 1-Tuple DNA sequencing: computer analysis. J. Biomol. Struct. Dyn. 1989;7:63–73. doi: 10.1080/07391102.1989.10507752. [DOI] [PubMed] [Google Scholar]
- Pevzner PA, et al. An Eulerian path approach to DNA fragment assembly. Proc. Natl Acad. Sci. USA. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL, et al. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res. 2012;22:557–567. doi: 10.1101/gr.131383.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


