Abstract
Summary: We have developed a new web application for peptide matching using Apache Lucene-based search engine. The Peptide Match service is designed to quickly retrieve all occurrences of a given query peptide from UniProt Knowledgebase (UniProtKB) with isoforms. The matched proteins are shown in summary tables with rich annotations, including matched sequence region(s) and links to corresponding proteins in a number of proteomic/peptide spectral databases. The results are grouped by taxonomy and can be browsed by organism, taxonomic group or taxonomy tree. The service supports queries where isobaric leucine and isoleucine are treated equivalent, and an option for searching UniRef100 representative sequences, as well as dynamic queries to major proteomic databases. In addition to the web interface, we also provide RESTful web services. The underlying data are updated every 4 weeks in accordance with the UniProt releases.
Availability: http://proteininformationresource.org/peptide.shtml
Contact: chenc@udel.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Locating occurrences of a specific peptide in a protein sequence database is important for protein identification in proteomics studies as well as for sequence-based protein retrieval. The Protein Information Resource (Wu et al., 2003) has hosted a peptide match service for >10 years. It is one of the most popular tools on the Protein Information Resource website.
Although exact peptide matching is algorithmically simple, quickly locating a peptide in a large protein sequence database has become challenging, as the number of protein sequences exponentially increases. So far, to our knowledge, no other peptide match services provide all of the key characteristics in our new service, including search speed, support for any query peptide of ≥3 amino acids long, coverage of all UniProtKB sequences including isoforms (UniProt Consortium, 2013) and frequent update. Other major peptide match methods are UniPept (Mesuere et al., 2012), which indexes in silico digested tryptic peptides, and Pep2Pro (Askenazi et al., 2010), which uses indexes based on 6-mers. A detailed comparison of these and our method is provided in the Supplementary Material (Supplementary Table S1). Another related work is SANS (Koskinen and Holm, 2012), which uses suffix arrays to effectively identify protein sequence similarities in the range of 50–100% identity.
As brute-force peptide matching is slow, we need effective ways to index protein sequences, analogous to indexing in relational databases. The recent advance in search engine technology, such as the Apache LuceneTM (McCandless et al., 2010) information retrieval software library, provides a scalable and high-performance index structure as well as powerful and efficient search algorithms to build a fast search engine for our Peptide Match service. Here we present our new peptide matching application, with an enhanced web user interface.
2 METHODS
The source protein sequence database is UniProtKB (including isoform sequences), currently containing >40 million protein sequences. The index engine is built based on Apache Lucene. We index the original protein sequences as well as leucine and isoleucine equivalent sequences. We include accession number, protein name, organism, taxonomic group and lineage from UniProtKB records to annotate the matched proteins. We also provide protein cross-references to proteomic/peptide spectral databases if the matched proteins are present in those databases. We store the corresponding UniProtKB accession numbers that are mapped to NIST Peptide Libraries (Stein and Rudnick, 2009), PeptideAtlas (Desiere et al., 2006), PRIDE (Vizcaíno et al., 2013) and Immune Epitope Database (IEDB) (Vita et al., 2009) using our ID mapping (Huang et al., 2011) services. These informational fields are stored, and key fields are indexed to support field-specific search and fast retrieval.
The protein sequence is analysed by Lucene NGramTokenFilter, which generates a series of 3-gram terms from the original sequence, and therefore, allows searching for a tri-peptide. These 3-gram terms filter search space analogous to other approaches: trypic peptides in UniPept, 6-mers in Pep2Pro and suffixes in SANS. Internally, Lucene index maintains an inverted table for a term and its corresponding protein accession numbers, and stores the term frequency and position occurrence. The Lucene’s Term Query method can only find the exact match with the same length term from the indexes. For example, the Term Query for 3-gram terms would match ‘MKE’ but not ‘MKEV’. We, therefore, use Lucene’s Phrase Query to search for overlapping consecutive 3-gram terms derived form the query peptide. We also use Lucene’s Boolean Query to combine the Phrase Query of sequence field and Term Query of other informational fields if the peptide search is restricted by a specific set of organisms, taxonomic groups or lineage.
To allow users to determine whether the given query peptide has previously been observed in mass spectrometry (MS)-based proteomics experiments, we provide a dynamic search of the query peptide against four major proteomic/peptide spectral databases: gpmDB (Craig et al., 2004), NIST Peptide Libraries, PeptideAtlas and PRIDE from our web application using the search engines of those individual databases. The current implementation of Peptide Match service uses Apache Lucene 3.5 and Apache SolrTM 3.5 for indexing and searching. The web application is developed in Java Server Pages/Servlets running on Apache Tomcat 6.
3 RESULTS
The search engine is fast and accurate. A typical peptide match from our web interface takes only a second, including time spent rendering the results. On the search time itself, our test results showed an average of 0.1 s from a random sample of ∼11 million peptides of 10-mers derived from the UniProtKB Human complete proteome. For UniProtKB release 2013_07 containing ∼40 million protein sequences with annotations (24 GB), the index size is 98 GB and the indexing time is 5 h on a Linux server with Intel® Xeon® 2.00 Ghz CPUs and 128 GB RAM.
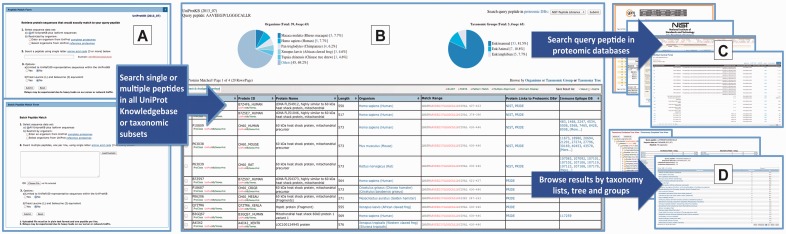
The Peptide Match web interface provides multiple functionalities, both for input query and output navigation (Fig. 1). A user can input a query peptide and specify whether to search the entire UniProtKB or limit the search to sequences from one or more selected organisms (Fig. 1A). For the latter, a user can either enter an organism name/taxonomic ID of a UniProt complete proteome or select a set of organisms from the UniProt reference proteomes. Additional options are provided to (i) restrict the matches in UniRef100 (Suzek et al., 2007) representative sequences to reduce redundancy and (ii) equate isobaric leucine and isoleucine to support MS-based proteomics. The match results are summarized by organism and taxonomic group as pie charts and listed as tables. The matched proteins are shown in paginated tables (Fig. 1B), along with annotations, matched region(s) in the protein sequence and hypertext links to corresponding databases. The match results can be sorted by different fields and browsed by organisms, taxonomic group or taxonomy tree (Fig. 1D). As an example (Fig. 1), peptide ‘AAVEEGIVLGGGCALLR’ matches 65 proteins in UniProtKB, among which 53 are from mammals, and 10, 5, 4 have links to corresponding proteins in PRIDE, NIST Peptide Libraries and IEDB, respectively. For direct proteomic evidence of the query peptide, the dynamic search using search engines of the proteomic/peptide spectral databases further displayed the peptide with links to the source MS data (Fig. 1C).
Fig. 1.
The search interfaces and functionalities of the Peptide Match server. (A) search interfaces; (B) matched result summary; (C) dynamic proteomic databases search results; (D) taxonomic views
For a given peptide, the matching protein entries from UniProtKB database can be directly accessed using a simple URL (e.g. http://proteininformationresource.org/peptide/SVQYDDVPEYK), and used for other servers as in PeptideAtlas. We also provide a web interface for batch submission of peptides to our service as well as RESTful web services. The underlying data of our new Peptide Match server are updated every 4 weeks in accordance with the UniProt releases. The Peptide Match services will be forthcoming from the UniProt website (www.uniprot.org).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Drs J. Garavelli, J. Wyffels, P. McGarvey, N. Edwards, P. Rudnick and X. Yang for comments and suggestions.
Funding: NIH (1U41HG006104-03) and Institutional Resources at University of Delaware.
Conflict of Interest: none declared.
REFERENCES
- Askenazi M, et al. The complete peptide dictionary-a meta-proteomics resource. Proteomics. 2010;23:4306–4310. doi: 10.1002/pmic.201000270. [DOI] [PubMed] [Google Scholar]
- Craig R, et al. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- Desiere F, et al. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, et al. A comprehensive protein-centric ID mapping service for molecular data integration. Bioinformatics. 2011;27:1190–1191. doi: 10.1093/bioinformatics/btr101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen JP, Holm L. SANS: high-throughput retrieval of protein sequences allowing 50% mismatches. Bioinformatics. 2012;28:i438–i443. doi: 10.1093/bioinformatics/bts417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCandless M, et al. Lucene in Action. 2nd edn. Greenwich, CT, USA: Manning Publications; 2010. [Google Scholar]
- Mesuere B, et al. Unipept: tryptic peptide-based biodiversity analysis of metaproteome samples. J. Proteome Res. 2012;11:5773–5780. doi: 10.1021/pr300576s. [DOI] [PubMed] [Google Scholar]
- Stein,S. and Rudnick,P. (eds.) (2009) NIST Peptide Tandem Mass Spectral Libraries. Human Peptide Mass Spectral Reference Data, H. sapiens, ion trap, Official Build Date: Feb. 4, 2009. NIST, Gaithers-burg, MD, 20899.
- Suzek BE, et al. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, et al. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R, et al. The immune epitope database 2.0. Nucleic Acids Res. 2009;38:D854–D862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, et al. The protein information resource. Nucleic Acids Res. 2003;31:345–347. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.