Abstract
Newcastle disease (ND) is one of the most lethal diseases of poultry worldwide. It is caused by an avian paramyxovirus 1 that has high genomic diversity. In the framework of an international surveillance program launched in 2007, several thousand samples from domestic and wild birds in Africa were collected and analyzed. ND viruses (NDV) were detected and isolated in apparently healthy fowls and wild birds. However, two thirds of the isolates collected in this study were classified as virulent strains of NDV based on the molecular analysis of the fusion protein and experimental in vivo challenges with two representative isolates. Phylogenetic analysis based on the F and HN genes showed that isolates recovered from poultry in Mali and Ethiopia form new groups, herein proposed as genotypes XIV and sub-genotype VIf with reference to the new nomenclature described by Diel’s group. In Madagascar, the circulation of NDV strains of genotype XI, originally reported elsewhere, is also confirmed. Full genome sequencing of five African isolates was generated and an extensive phylogeny reconstruction was carried out based on the nucleotide sequences. The evolutionary distances between groups and the specific amino acid signatures of each cluster allowed us to refine the genotype nomenclature.
Introduction
Newcastle disease (ND) is one of the most severe infectious diseases of birds, particularly poultry, and has been the cause of major economic losses worldwide [1]. It is one of the 14 avian diseases notifiable to the World Organization for Animal Health (Office International des Epizooties, OIE) [2]. The cause of ND, Newcastle disease virus (NDV) or avian paramyxovirus type 1 (APMV-1), belongs to the Avulavirus genus, Paramyxoviridae family, and has a negative-sense single-stranded RNA genome of about 15.2 kilobases. The genome encodes eight proteins, nucleocapsid (NP), phosphoprotein (P), matrix (M), fusion (F), hemagglutinin-neuraminidase (HN), a large RNA-directed RNA polymerase (L), and two additional nonstructural proteins, V and W, generated by RNA editing during P gene transcription [3], [4]. NDV can be categorized into highly pathogenic (velogenic), intermediate (mesogenic), and nonpathogenic (lentogenic) strains based on pathogenicity in chickens [5]. Although V, HN, NP, P and L proteins play a role in virulence [6], [7], [8], [9], the most important molecular determinant of virulence appears linked to the amino acid motif present at the protease cleavage site of the F0 precursor of the fusion protein [10]. In virulent isolates, this motif is constituted of basic amino acids, and rapid typing of this region by RT-PCR and sequencing is a good indicator of the NDV pathotype. However, other viral factors affect the virulence of isolates, so pathogenicity should be confirmed by in vivo tests, including the intracerebral pathogenicity index (ICPI) in 1-day-old chickens, the mean death time (MDT) of specific-pathogen-free hen’s embryos after inoculation, and the intravenous pathogenicity index (IVPI) in 6-week-old chickens [2].
NDV strains are divided into two clades (class I and class II) according to the genome size and the sequence of the F and L genes [4]. Restriction enzyme site mapping of the F protein gene and phylogenetic analysis of the partial nucleotide sequence of the F gene have been used to classify NDV of class II [11], [12]. However, there is no consensus on NDV classification and taxonomy, since some authors use the classification of the group of Lomniczi and Ballagi-Pordany [11], [12] based on “genotypes” whereas others use the “lineage” classification of Aldous et al [1]. Both cover distinct isolate clusters but are based on the same genomic information. According to the evolutionary distances, Miller et al [13] showed inconsistencies between the two nomenclatures (for example lineage 3 is not monophyletic and contains genotypes III, IV, V, and VIII: detailed discrepancies between the two nomenclatures can be found in Table S1). Calling for objective criteria to unify the NDV nomenclature, those authors favored the use of genotypes. In the first genotype classification [12], genotype I contained mostly avirulent strains whereas genotypes II, III, and IV were involved in the first panzootic that started in 1920 and vanished around 1950. Genotypes V, VIa, and VIII were responsible for the second panzootic between the 1960s and the 1970s. Sub-genotypes VIb, VIc, and VId caused the third panzootic that emerged from pigeons during the 1980s, and sub-genotypes VIIa, VIIb, VIIc, and VIId appeared in the 1980s and the 1990s in the Far East, Europe, and South Africa [14], [15]. Genotype VII has been the predominant genotype circulating throughout the world, particularly in Asia and Africa, and it was recently reported in South America [16], [17], [18]. South African, European, American, and Asian isolates were previously typed as genotypes VIb, VIIb, and VIII [12], [16], [17], [19], [20]. In Uganda, strains from an undetermined genotype but close to genotype VI were also isolated [21]. In many parts of Africa, ND is considered endemic but only few data are available on virus isolates circulating there. In West Africa, new sub-genotypes VIIf, VIIg, and VIIh were recently described [22], [23]. Based on phylogenetic analyses of a partial F coding sequence of NDV isolates recovered from apparently healthy poultry in Mali in 2007 and 2008, we previously proposed a new sub-genotype, VIIi [24]. In Madagascar, other original strains were also isolated, sequenced, and ascribed to a new genotype, genotype XI [24], [25]. Six new genotypes were subsequently proposed by others, with objective criteria, to unify the genotype nomenclature of NDV [26], [27]. In this work, new African NDV isolates are described, and their sequences and others publicly available were included in an extensive phylogeny reconstruction based on various methods. It is shown that the “genotype” nomenclature is better adapted to the resulting genetic discrimination of NDV isolates. In addition, 10 sub-genotypes are defined. According to these results and previous publications, a rooted classification with 14 distinct genotypes is now proposed.
Materials and Methods
Ethics Statement
All animal experiments (ICPI tests) were conducted according to internationally approved OIE standards, under authorizations set forth by the director of the veterinary services of Côtes d’Armor on behalf of the prefect of Côtes d’Armor (N°22–18) and by the director of the veterinary services of Hérault on behalf of the prefect of Hérault (N° 34–114). The experiments were approved by the Regional Committee for Ethics in animal experiments (Comité d’éthique ComEth Afssa/ENVA/UPEC) under number 14/06/2011-4. Certificates are available from the authors upon request.
Samples
Samples were collected in the framework of the Gripavi project (http://gripavi.cirad.fr/en/) launched in 2007 and terminated in 2011 by CIRAD in collaboration with the Laboratoire Central Vétérinaire of (LCV) Bamako (Mali), Centre National d’Elevage et de Recherche Veterinaire (CNERV) of Nouakchott (Mauritania), the Département de Recherche Zootechnique et Vétérinaire du Centre National de la Recherche Appliquée au Développement Rural (FOFIFA-DRZV) in Madagascar, the National Animal Health Diagnostic and Investigation Center (NADHIC) of Sebeta in Ethiopia, and with the support of the French Ministry of Foreign Affairs. Cloacal and tracheal swabs were collected from 9,609 domestic and 10,343 wild birds in different areas of Mali, Mauritania, Madagascar, and Ethiopia from 2007 to the end of 2011. The samples were dipped in a transport medium consisting of isotonic phosphate-buffered saline, pH 7.0–7.4, with antimicrobial additives (penicillin 10,000 U/mL, streptomycin 10 mg/mL, amphotericin B 25 µg/mL, and gentamycin 250 µg/mL) supplemented with 20% glycerol and stored in liquid nitrogen containers. They were shipped in dry ice and kept in the laboratory at −80°C until processing. All samples were manipulated in a bio-safety level 3 laboratory.
RNA Extraction and Molecular Detection
Viral RNA was extracted from samples by a high throughput automated workstation Biomek FXP (Beckman) using the Nucleospin RNA virus kit (Macherey Nagel). The viral RNA was resuspended in nuclease-free water and stored at −80°C. NDV was detected on the F gene by real-time one step RT-PCR (rRT-PCR) using Stratagene Machine Mx3000 or 3005. The forward primer was F+4839 5′-TCCGGAGGATACAAGGGTCT [28] and the reverse primer was F2AS 5′-TGGCAGCATTCTGGTTGGCT [29]. RT-PCR was carried out in a 25 µl reaction mixture with the Brilliant SYBR Green QRT-PCR II Master mix Kit (Stratagene) according to the manufacturer’s instructions. The conditions for reverse transcription were 50°C for 30 min and 95°C for 15 min. PCR consisted of 40 cycles of 95°C for 30 sec, 60°C for 1 min, and 72°C for 30 sec, followed by the final dissociation stage: 1 min 95°C, 30 sec 55°C, 30 sec 95°C.
Virus Isolation and Pathogenicity Tests
All samples positive on rRT-PCR were inoculated into the allantoic cavities of 9- to 11-day-old embryonated fowl eggs from a commercial specific-pathogen-free flock. After one to three passages, allantoic liquid from dead eggs was tested by NDV rRT-PCR, and NDV-positive samples were then stored at −80°C and used as working stock for sequence analysis.
For the determination of pathogenicity, the mean death time for chick embryo (MDTE) was calculated. Then, as recommended by the latest version of the OIE Manual of Standards for Diagnostic Tests and Vaccines, for three strains (2008/Mali/ML007, 2009/Mali/ML008, 2007/Mali/ML029), the allantoic liquid was also inoculated in Specific Pathogen Free (SPF) one-day-old chicks to determine the intracerebral pathogenicity index (ICPI) [2].
Amplification for Sequencing
Viral RNA was extracted from NDV-positive allantoic liquid using the Nucleospin RNA virus kit (Macherey Nagel) according to the manufacturer’s instructions. RNA was quantified by UV absorbance using a Nanodrop spectrophotometer (Thermo scientific). Complementary DNA was synthesized using 20 µl of eluted RNA with oligo pd(N)6 primer and the (M-MuLV) reverse transcriptase of the First-Strand cDNA Synthesis Kit (Ge Healthcare). Conventional PCR was carried out with 20 ng of cDNA using Taq DNA polymerase (Qiagen) according to the manufacturer’s instructions. Five distinct PCR runs allowed the amplification of the complete sequence of the F and HN genes. The primers used for amplification of F and HN genes are shown in Table 1. The PCR products were separated by gel electrophoresis using 2% w/v agarose gel in Tris-acetate buffer, stained with ethidium bromide, and visualized under ultraviolet light. The product was excised and purified using QiaQuick gel extraction Kit (Qiagen).
Table 1. Primers used for complete sequencing of the F and HN genes.
| Primer | Sequence 5′-3′ | Directiona | Nucleotide positionon AY741404 | Gene targetb | PCR product size | Reference |
| MFS1 | GACCGCTGACCACGAGGTTA | F | 4306 | M | 766 | [30] |
| F3AS | TGCATCTTCCCAACTGCCAC | R | 5072 | F | c | |
| F+4952 | GCAGCCGCAGCTCTAATAC | F | 4952 | F | 747 | c |
| MFS3 | GGCAATAACTGAGCCTTTGAG | R | 5699 | F | c | |
| F+886 | AATAATATGCGTGCCACCTA | F | 5429 | F | 1040 | c |
| HN49rev | GCGCCATGTRTTCTTTGC | R | 6469 | HN | c | |
| P6A | ATCAGATGAGAGCCACTACA | F | 6177 | F | 1120 | [31] |
| HN886rev | ACTCCTGGGTAATTTGCCAC | R | 7297 | HN | c | |
| 3HNOV | GTCTTGCAGTGTGAGTGCAAC | F | 7119 | HN | 1271 | [30] |
| P7B | TCTGCCCTTTCAGGACCGGA | R | 8390 | L | [31] |
F, forward; R, reverse.
M, F, HN, and L matrix, fusion protein, hemagglutinin-neuraminidase, and polymerase genes, respectively.
primers designed on conserved sequences based on the alignment of the complete sequences of 219 F genes and 74 HN genes published in GenBank.
For F and HN genes, DNA was sequenced by the GATC-Biotech company using the same primers described for PCR reactions. For the full genome sequencing, genomic virus RNA was extracted from 200 µL of allantoic fluid using the QIAamp viral Rneasy Mini kit (QIAGEN, Hilden, Germany). RNAs were reverse transcribed using Superscript II (Invitrogen) with hexanucleotides. Twenty-six overlapping PCR products were obtained with Platinum Taq DNA polymerase (Platinum® TaqDNA polymerase High Fidelity, Invitrogen). All these primers were defined in the laboratory (primers available upon request). The DNA sequences were determined by sequencing, in both senses, using a Big dye Terminator v3.1 (Applied Biosystem) cycle sequencing kit according to the manufacturer’s instructions.
Sequence Alignments, Phylogenetic Analyses, and Evolutionary Distances
The forward and reverse sequences were aligned using Vector NTI software version 10.3 (Invitrogen, Europe). The validated open reading frames of the different genes were aligned with others retrieved in GenBank using the CLUSTAL W program of the Mega5 software suite. The sequence of a class I NDV isolate was used as an outgroup. Full length genomes, including the genomes from the Malian and Madagascar strains, were aligned using Muscle, which is a faster and more reliable method than CLUSTAL W for larger sets of long sequences [32]. From the full genomes, individual and concatenated genes were generated using Mega5 software [33]. Putative recombination events in sequence alignment of available NDV genomes and the new Malian and Madagascar genomes studied here were investigated by RDP suite (versions RDP3 Alpha 44 and RDP4 Beta 4.16 [34]. This software uses multiple recombination signal detection methods like CHIMAERA, Maximum chi2, RDP, RECSCAN, and Geneconv. All recombined sequences were discarded before analysis. Phylogenetic reconstructions were first carried out by the Maximum Likelihood method implemented in Treefinder [35] (version of March 2011). The data sets consisted of the full genome sequences (13740 nt, n = 110), the six individual genes from these full genomes, the complete F genes (1653 nt, n = 741), the complete HN genes (1713 nt, n = 323), and the partial coding sequences (445 and 372 nt) of the F gene (for n = 796 and 1921, respectively). All these multiple sequence alignments are provided in Table S7. The best models of nucleotide substitution for each data set were selected from the uncorrected and corrected Akaike Information Criterion, the Hannan and Quinn performance-based decision theory [36], and Bayesian Information Criterion [37] of Treefinder version March 2010. A General Time Reversible (GTR) model with a discrete gamma distribution (+G) with 5 classes, allowing for invariant sites (+I), was the consensus substitution model proposed for the different data and was used for all Maximum Likelihood analyses. Bayesian inference was also performed with a GTR model using MrBayes_3.2.2 [38,(http://sourceforge.net/p/mrbayes/code/HEAD/tree/release/installers/)]. All Bayesian reconstructions were initially set at 100,000,000 trees with a sample frequency on the chains of 1/1,000 (targeted sample size = 100,000 trees). All priors were set by default, except the evolutionary model and the branch length. For the latter, an inverse gamma Dirichlet prior was selected to avoid overestimation of branch lengths by MrBayes [39], [40]. For all reconstructions, two runs were launched in parallel with two chains (one heated, one cold). A convergence rule between the two runs was set at a standard deviation of split frequencies lower than 0.01, which then stops the reconstruction. Alternatively, when the standard deviation of split frequencies followed a stationary fluctuation above 0.01 for several consecutive days, very long reconstructions were manually stopped. To assess convergence, the expected sampling sizes (ESS) for posterior probabilities and the Potential Scale Reduction Factor (PSRF) were checked for all reconstructions: validation criteria for all parameters were average ESS>200 and PSRF within [0.99 and 1.01] (as the chains converge in the runs, the variance between the runs becomes more similar and the PSRF approaches 1.0). The numbers of trees used for the generation of the consensus tree and values for the two convergence parameters are indicated in the results. Final trees were laid out using Figtree software, version 1.4.0. Topologies of the Maximum Likelihood and Bayesian consensus trees obtained for the different genes and the full genomes (n = 110 sequences in the different data sets) were compared by Treefinder using the Shimodaira and Hasegawa test [41]. The best representation was then selected. For the F gene, comparisons were performed using different phylogenetic methods, including Maximum Likelihood, Neighbor Joining, and Maximum Parsimony methods from Mega5 software [33] and the Bayesian approach for phylogenetic reconstruction. Branch support values were obtained using nonparametric bootstrapping with 1,000 resamplings for the first three phylogenetic methods, and the posterior probabilities for the Bayesian approach were estimated on 16,806 samples with a Burn-in phase for the first 25% of tree samples. The best tree for the F gene was determined by calculating the minimum branch length distance (K tree score) between the phylogenetic trees by the Ktreedist program [42]. The complete F gene data set was also used to calculate the mean evolutionary distances within and between clusters. The pairwise distance matrix was generated by Treefinder and analyzed in Excel. In addition to the evolutionary distance, nucleic and amino acid signatures specific for the different clusters were sought in the multiple alignments of the F gene and protein. New genotypes and sub-genotypes were assigned according to the criteria described by Diel et al [26] with the following modifications:
new genotypes and sub-genotypes were assigned on at least three independent isolates without a direct epidemiologic link in the phylogenetic trees generated for the complete F gene sequence and confirmed by at least two HN gene sequences, using both the Maximum Likelihood and Bayesian methods and the optimal nucleotide model (GTR +G +I, Г5), as determined by Treefinder.
the mean distance between genotypes and sub-genotypes, as determined by the distance matrix from the complete F gene sequence generated by the Maximum Composite Likelihood model of Treefinder, was higher than or equal to 0.100 and 0.03, respectively.
sub-genotypes were included into a monophyletic genotype and were identified by unique amino or nucleic acid signatures, as described in the results section.
Results
Detection and Initial Characterization of NDV Isolates in African Samples
Using real-time RT-PCR detection, we found 421 samples positive for NDV among 9,609 domestic samples (4.38%, 95% CI: 4–4.8%) and 211 positive samples among 10,343 collected from wild birds (2.04%, 95% CI: 1.8–2.3). Nine viruses were isolated from domestic birds in Mali, ten in Madagascar, and five in Ethiopia. However, note that eight of ten isolates from Madagascar were obtained from farms where clinical signs invoking Newcastle disease were obvious. This was not the case for the two other isolates from Madagascar and all the others from Mali and Ethiopia. In addition, two viruses were isolated from healthy wild birds in Madagascar. Unfortunately, no isolates were obtained from Mauritanian and Malian wild birds. However, 14 cleavage sites could be sequenced (Table 2). All isolates from Mali had a virulent cleavage site with at least four basic amino acids (sequences of domestic birds with an accession number in Table 2). Interestingly, the 2007/Mali/ML029 and 2007/Mali/ML031 isolates have five basic amino acids on their cleavage site and a V118 associated with the cleavage site (112RRRKR116↓FV), a motif described only rarely, and just recently in the neighboring country, Burkina Faso [22]. The second velogenic cleavage site (112RRQKR116↓FI) found in all other sequences from Malian domestic bird isolates was also encountered in some wild birds collected in Mali and Mauritania. This F cleavage site is identical to that of most NDV strains isolated in different parts of Asia and Africa [17], [19], [20], [21], [43]. The rest of the wild bird sequences from Mali and Mauritania shared a lentogenic F cleavage site (112GKQGR116↓LIA/V). All the strains isolated from domestic birds of Madagascar had five arginines (R) in the cleavage site 112GRRRRR116↓F and clustered in the same genotype XI as previously described [25]. The lentogenic cleavage site found in two wild birds of Madagascar was identical to the one in Mali and Mauritania (Table 2). The five Ethiopian isolates showed three velogenic cleavage sites: 112RRQKR116FV, 112RRRKR116FV, and 112RRHKR116↓FV. To our knowledge, a histidine in position 114 of a cleavage site of F protein has never been described before.
Table 2. Characteristics of viruses analyzed in this study.
| Group | Sequence reference | Collecting year | Origin | Host | Cleavage site(aa 112–118) |
| domestic birds | 2007/Mali/ML029/07 | 2007 | Mopti market/Mali | Gallus gallus | RRRKR↓FV |
| 2007/Mali/ML038/07 | 2007 | Mopti market/Mali | Guinea fowl | RRQKR↓FI | |
| 2007/Mali/ML031/2007 | 2007 | Mopti market/Mali | Gallus gallus | RRRKR↓FV | |
| 2008/Mali/ML007/08 | 2008 | Sikasso/Mali | Gallus gallus | RRQKR↓FI | |
| 2008/Mali/ML225/08 | 2008 | Mopti/Gninitogo/Mali | Gallus gallus | RRQKR↓FI | |
| 2008/Mali/ML230/2008 | 2008 | Mopti/Gninitogo/Mali | Gallus gallus | RRQKR↓FI | |
| 2010/Mali/ML57071T | 2010 | Mali | Gallus gallus | RRQKR↓FI | |
| 2010/Mali/ML57072T | 2010 | Mali | Gallus gallus | RRQKR↓FI | |
| 2009/Mali/ML008* | 2009 | Mopti market/Mali | Gallus gallus | RRQKR↓FI | |
| 2009/Madagascar/MGBBS | 2009 | Tsarahonenana/Antananarivo/Madagascar | Gallus gallus | RRRRR↓FV | |
| 2010/Madagascar/MGF003C | 2010 | Amparihitsokatra/East_lakeside/Alaotra_region/Madagascar | Gallus gallus | RRRRR↓FV | |
| 2011/Madagascar/MGF015C | 2011 | Ambohimandroso/West lakeside/Alaotra_region/Madagascar | Gallus gallus | RRRRR↓FV | |
| 2010/Madagascar/MGF082T | 2010 | Amparafaravola/West_lakeside/Alaotra_region/Madagascar | Gallus gallus | RRRRR↓FV | |
| 2011/Madagascar/MGF120T | 2011 | Anororo/West_lakeside/Alaotra_region/Madagascar | Gallus gallus | RRRRR↓FV | |
| 2011/Madagascar/MGF166 | 2011 | Anororo/West_lakeside/Alaotra_region/Madagascar | Gallus gallus | RRRRR↓FV | |
| 2011/Madagascar/MGF192C | 2011 | Bejofo/East_lakeside/Alaotra_region/Madagascar | Gallus gallus | RRRRR↓FV | |
| 2009/Madagascar/MGMNJ | 2009 | Manjakandriana/Antananarivo/Madagascar | Gallus gallus | RRRRR↓FV | |
| 2011/Madagascar/MGS1130T | 2011 | Anororo/West_lakeside/Alaotra_region/Madagascar | Gallus gallus | RRRRR↓FV | |
| 2011/Madagascar/MGS1595T | 2011 | Imerimandroso/East_lakeside/Alaotra_region/Madagascar | Gallus gallus | RRRRR↓FV | |
| 2011/Ethiopia/ETH10065 | 2011 | Dhera market/Ethiopia | Gallus gallus | RRQKR↓FV | |
| 2011/Ethiopia/ETH10073 | 2011 | Dire market/Ethiopia | Gallus gallus | RRHKR↓FV | |
| 2011/Ethiopia/ETH8755 | 2011 | Maliyu/Ethiopia | Gallus gallus | RRQKR↓FV | |
| 2011/Ethiopia/ETHAN01 | 2011 | Arsinegele/Ethiopia | Gallus gallus | RRQKR↓FV | |
| 2011/Ethiopia/ETHMG1C | 2011 | Migra/Ethiopia | Gallus gallus | RRRKR↓FV | |
| Wild birds | 2009/Madagascar/MGA057C | 2009 | Alaotra_lake/Madagascar | Anas Melleri | GKQGR↓LI |
| 2009/Madagascar/MGA184C | 2009 | Alaotra_lake/Madagascar | Anas erythrorhyncha | GKQGR↓LI | |
| 2008/Mali/DvML160_08 | 2008 | Lake Fati/Mali | Dendrocygna viduata | RRQKR↓FI | |
| 2008/Mali/NnML495_08 | 2008 | Lake Horo/Mali | Nycticorax nycticorax | RRQKR↓FI | |
| 2008/Mali/PpML283_08 | 2008 | Lake Horo/Mali | Porphyrio porphyrio | GKQGR↓LI | |
| 2008/Mali/CaMT197_08 | 2008 | Banc d’Arguin/Mauritania | Calidris alpina | RRQKR↓FI | |
| 2008/Mali/CpML476_08 | 2008 | Lake Fati/Mali | Charadrius pecuarius | GKQGR↓LI | |
| 2008/Mauritania/LgMTLG199_08 | 2008 | Banc d’Arguin/Mauritania | Larus genei | RRQKR↓FI | |
| 2008/Mauritania/LgMTLG223_08 | 2008 | Banc d’Arguin/Mauritania | Larus genei | GKQGR↓LI | |
| 2008/Mauritania/LgMTLG228_08 | 2008 | Banc d’Arguin/Mauritania | Larus genei | GKQGR↓LI | |
| 2008/Mauritania/LgMTLG232_08 | 2008 | Banc d’Arguin/Mauritania | Larus genei | GKQGR↓LI | |
| 2008/Mauritania/LgMTLG241_08 | 2008 | Banc d’Arguin/Mauritania | Larus genei | RRQKR↓FI | |
| 2008/Mauritania/LgMTLG243_08 | 2008 | Banc d’Arguin/Mauritania | Larus genei | RRQKR↓FI | |
| 2008/Mauritania/LgMTLG254_08 | 2008 | Banc d’Arguin/Mauritania | Larus genei | RRQKR↓FI | |
| 2008/Mauritania/LgMTLG258_08 | 2008 | Banc d’Arguin/Mauritania | Larus genei | RRQKR↓FI | |
| 2008/Mauritania/LgMTLG261_08 | 2008 | Banc d’Arguin/Mauritania | Larus genei | RRQKR↓FI |
- *full genome sequence.
The virulence of the 2007/Mali/ML029 isolate was verified by determining the MDTE and ICPI. The minimum dilution killing all inoculated eggs was 10−8 and the MDTE for this dilution was 48 and 60 hours in two independent tests, thus confirming the velogenic nature of this strain (mesogenic strain killed eggs after 60 hours). The ICPIs of 2007/Mali/ML029, 2008/Mali/ML007, and 2009/Mali/ML008 were all 1.8, close to the maximal score of 2. The ICPI of Madagascar isolates was previously established at 1.9 [25]. These results are in line with the virulent F cleavages site motifs described above.
Genotyping of African NDV Isolates using Complete Sequences
To determine the genotype of the isolates, the F and HN genes of the African isolates were sequenced. The analyses of the total F and HN genes showed that 2007/Mali/ML031 and 2008/Mali/ML230 were 100% identical to 2007/Mali/ML029 and 2008/Mali/ML225, respectively (data not shown). For this reason, phylogenetic analyses were carried out on only seven of the Malian isolates (2007/Mali/ML029, 2007/Mali/ML038, 2008/Mali/ML225, 2008/Mali/ML007, 2009/Mali/ML008, 2010/Mali/ML57071T, and 2010/Mali/ML57072T). The degree of nucleic acid variation between these seven strains was 0.1–2.0% for the F gene (0.9–6.5% for the protein) and 2.0–2.3% for the HN gene (8.1–8.6% for the protein). Similarly, the divergences among the five Ethiopian strains were 0.2–1.0% and 0.1–0.7% for the F and HN genes, respectively (1.3–3.4% and 0.9–3.9% for the F and HN proteins, respectively). For the eight Madagascar isolates from domestic birds, the divergences ranged from 0–0.6% for the F gene (0–3.3% for the protein).
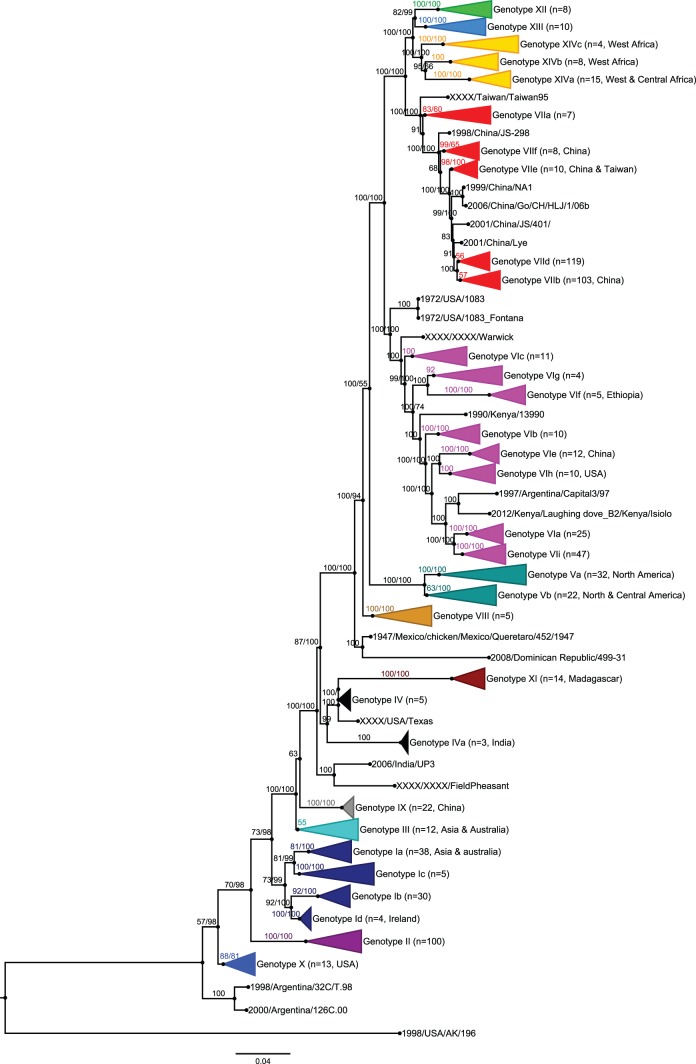
Phylogenetic analyses were performed by comparing the complete sequences of F and HN genes by the Maximum Likelihood and Bayesian inference methods with the best model of substitution proposed by Treefinder (GTR +G +I, Г5). The trees generated by the two methods were very close, as shown by the Shimodaira and Hasegawa test (Table S2). This was also the case for all individual genes and the full genome. However, the clustering of some strains was sometimes better with the Bayesian inference (data not shown). Bayesian trees were thus preferred for final representation. Since the topology of the two Bayesian trees rendered by the F and HN genes was identical, only the tree of the F gene is shown in Figure 1. The bootstrap values for 1000 replicates for the two genes are reported on this figure. Because sub-lineages 3a, 3b, 3c, and 3d described by Aldous et al. [1] were not found monophyletic in our reconstructions, a division into clusters gIII, gIV, gV, and gVIII and the use of nomenclature based on genotypes from Ballagi-Pordany et al. [12] and Lomniczi et al. [11] were preferred. For more clarity on NDV nomenclatures, the reader can see correspondences between various nomenclatures in Table S1. According to the criteria described in Material and Methods for the definition of genotype and sub-genotype and in Figure 1, 14 genotypes and 23 sub-genotypes are proposed here. The Malian isolates are all clustered with other isolates from West Africa [22], [23] into genotype XIV. This cluster is clearly separated from genotype VII in terms of genetic distances (0.139 versus 0.038, which is the average intra-genotype distance, see Table 3).The Malian isolates and others from Niger, Burkina Faso, Cameroon, Nigeria, Mauritania, and Ivory Coast also form three distinct sub-genotypes, here proposed as sub-genotypes XIVa, XIVb, and XIVc. The isolates from Madagascar domestic birds, all responsible for clinical outbreaks, were confirmed as members of genotype XI (Figure 1). Genotype VI contains highly diverse strains from pigeons and wild birds collected throughout the world. It is suspected that pigeon type-PMV1 emerged as a result of multiple transmissions of viruses from chickens to pigeons, but this hypothesis is currently not confirmed. Ujvári et al [44] revealed the existence of four sub-genotypes of VIb/1 PPMV1 and a new clade designated VIb/2 with recent isolates from Croatia and even more recent ones from Russia [45]. According to Diel’s criteria [26] and our analyses, four new sub-genotypes VI were defined. The Ethiopian isolates constitute the new sub-genotype VIf. Sub-genotype VIb/2 [45] including Russian strains was renamed sub-genotype VIg. Sub-genotypes VIh and VIi contain only isolates from the USA and isolates from pigeons and wild bird strains from the USA and Europe, respectively.
Figure 1. Phylogenetic analysis of 741 complete F nucleic acid sequences of NDV.
Trees were constructed using Bayesian inference with 16,806,000 iterations and 1/1000 tree sampled in the chain. Minimal average ESS for all parameters was 1681, and PSRF were between 0.99996 and 1.00084. A class 1 virus sequence was introduced as an outgroup. Consensus tree posterior probabilities are indicated on the branch (first number). A Bayesian inference based on 323 complete HN nucleic acid sequences was also done (all details in Figure S3A, Figure S5J and Figure S9R). Since the F and HN trees were superimposable, only the F tree is represented; where informative, branch support values for HN are indicated after the F posterior probabilities. Sequences from this study are grouped in genotypes Ib (2 strains), VIf (5 strains), XI (10 strains), XIVa (1 strains), XIVb (5 strains), and XIVc (1 strain). The complete list of the 741 sequences, the corresponding multiple sequence alignments, and the tree in Newick and in Figtree format can be found in Table S8, Table S7, Figure S5G, and Figure S9O, respectively.
Table 3. Estimates of evolutionary distances between genotypes.
| I | II | III | IV | IX | V | VI | VII | VIII | X | XI | XII | XIII | XIV | XV | |
| I | 0.052 | ||||||||||||||
| II | 0.140 | 0.018 | |||||||||||||
| III | 0.115 | 0.156 | 0.031 | ||||||||||||
| IV | 0.133 | 0.174 | 0.105 | 0.046 | |||||||||||
| IX | 0.117 | 0.158 | 0.089 | 0.100 | 0.004 | ||||||||||
| V | 0.237 | 0.277 | 0.209 | 0.188 | 0.204 | 0.068 | |||||||||
| VI | 0.226 | 0.266 | 0.199 | 0.177 | 0.193 | 0.195 | 0.077 | ||||||||
| VII | 0.225 | 0.263 | 0.197 | 0.176 | 0.192 | 0.194 | 0.158 | 0.036 | |||||||
| VIII | 0.173 | 0.213 | 0.145 | 0.124 | 0.140 | 0.153 | 0.142 | 0.141 | 0.041 | ||||||
| X | 0.124 | 0.129 | 0.140 | 0.158 | 0.142 | 0.262 | 0.252 | 0.250 | 0.198 | 0.021 | |||||
| XI | 0.235 | 0.275 | 0.207 | 0.160 | 0.202 | 0.289 | 0.279 | 0.278 | 0.225 | 0.260 | 0.015 | ||||
| XII | 0.232 | 0.272 | 0.205 | 0.184 | 0.200 | 0.201 | 0.165 | 0.128 | 0.148 | 0.258 | 0.285 | 0.021 | |||
| XIII | 0.227 | 0.266 | 0.199 | 0.178 | 0.194 | 0.196 | 0.160 | 0.122 | 0.143 | 0.252 | 0.280 | 0.114 | 0.061 | ||
| XIV | 0.244 | 0.283 | 0.216 | 0.195 | 0.211 | 0.213 | 0.177 | 0.139 | 0.160 | 0.269 | 0.296 | 0.134 | 0.128 | 0.086 | |
| XV | 0.135 | 0.175 | 0.108 | 0.086 | 0.102 | 0.129 | 0.118 | 0.117 | 0.064 | 0.161 | 0.188 | 0.124 | 0.119 | 0.136 | 0.000 |
Average intergenotype distance: 0.190.
Average intragenotype distance: 0.038.
The two Madagascar isolates from wild birds were classified as sub-genotype Ib. Beyond the new sequences of African isolates provided in this study, we confirm most of the genotypes/sub-genotypes proposed by Diel et al. [26], except sub-genotype VIIg and genotype XV, both including only recombined strains that were excluded from our data set to avoid any side-effect on the reconstruction. Relative to Diel et al [26], we propose the creation of sub-genotypes Ic, Id, IVa, VIf, XIVa, XIVb, and XIVc. Sub-genotype Ic (n = 5) is constituted by different strains from Europe, Asia and America. Sub-genotype Id (n = 4) is constituted by strains from Ireland only, and the genetic distance between this cluster and Ia, Ib sub-genotypes is 0.054 and 0.052, respectively. Our phylogenetic reconstructions show a cluster constituted by strains only from India (n = 3), with a common ancestor branched to genotype IV. These three viruses are distant enough (0.089) from genotype IV to propose a new sub-genotype IVa (Table 4). Genotype VI was previously classified into eight sub-genotypes by Wang et al, 2006 [46]. However, we confirm the existence of only four of these sub-genotypes, VIa, VIb, VIc, and VIe, in agreement with others [26]. Our five Ethiopian isolates are clustered in a distinct group branched to genotype VI. Its genetic distance from the other sub-genotypes VI (≥0.128), compared to the mean evolutionary distances between VIa, VIb, VIc, and VIe (<0.11, Table 4), supports the creation of a new sub-genotype, VIf. This sub-genotype was previously assigned to a cluster of Korean NDV strains described by Kwon et al. [47]. However, the phylogenetic tree was generated on a few isolates with shorter F gene sequences (319 nt) and by Neighbor Joining method. According to our reconstruction based on 796 sequences of 445 nucleotides (nt 55–500), the Korean isolates [47] clearly fall into sub-genotype VIc (Figure S3B, Figure S5K and Figure S9S). We also propose new sub-genotypes VIg, VIh, and VIi based on the phylogenetic sub-genotype distances observed between them (>0.03). Sub-genotype VIg was previously described by Wang et al, 2006 [46] to classify Chinese strains which, however, are included in VIc in our reconstructions. Genotype XIV, previously classified as sub-lineage 5f, 5g, and 5h by Snoeck et al. [22] and as a new lineage 7 by Cattoli et al [23], contains viruses obtained mainly in West and Central Africa between 2006 and 2008. The five strains from Mali isolated during this study are part of this cluster containing other strains from West Africa, previously described [23]. The genetic distance between this genotype and the two other close genotypes, XII and XIII, is very similar (0.134 to 0.128, Table 3), supporting the creation of a new genotype. In addition, three branches were identified within this genotype, and the genetic distance between these branches (>0.13) supports the creation of three new sub-genotypes: XIVa (former lineages 5g or 7b) that includes our strain 2008/Mali/ML007 and others from Burkina Faso, Niger, Nigeria, and Cameroon; XIVb (former lineage 7a) containing five of our Malian strains and others from Mauritania and Ivory Coast; and XIVc (former lineages 5f or 7d) containing our strain 2007/Mali/ML029 and another one from Niger.
Table 4. Estimates of evolutionary distances between sub-genotypes.
| Ia | Ib | Ic | Id | IVa | IV | Va | Vb | VIa | VIb | VIc | VIe | VIf | VIg | VIh | VIi | VIIa | VIIb | VIId | VIIe | VIIf | XIVa | XIVb | XIVc | |
| Ia | 0.016 | |||||||||||||||||||||||
| Ib | 0.078 | 0.019 | ||||||||||||||||||||||
| Ic | 0.070 | 0.094 | 0.053 | |||||||||||||||||||||
| Id | 0.054 | 0.052 | 0.069 | 0.008 | ||||||||||||||||||||
| IVa | 0.005 | 0.089 | ||||||||||||||||||||||
| IV | 0.014 | |||||||||||||||||||||||
| Va | 0.031 | |||||||||||||||||||||||
| Vb | 0.104 | 0.042 | ||||||||||||||||||||||
| VIa | 0.021 | |||||||||||||||||||||||
| VIb | 0.075 | 0.016 | ||||||||||||||||||||||
| VIc | 0.105 | 0.076 | 0.043 | |||||||||||||||||||||
| VIe | 0.089 | 0.070 | 0.100 | 0.014 | ||||||||||||||||||||
| VIf | 0.158 | 0.128 | 0.138 | 0.152 | 0.023 | |||||||||||||||||||
| VIg | 0.125 | 0.095 | 0.104 | 0.119 | 0.132 | 0.052 | ||||||||||||||||||
| VIh | 0.081 | 0.062 | 0.092 | 0.063 | 0.145 | 0.111 | 0.031 | |||||||||||||||||
| VIi | 0.057 | 0.074 | 0.104 | 0.087 | 0.157 | 0.123 | 0.080 | 0.026 | ||||||||||||||||
| VIIa | 0.052 | |||||||||||||||||||||||
| VIIb | 0.086 | 0.023 | ||||||||||||||||||||||
| VIId | 0.081 | 0.037 | 0.022 | |||||||||||||||||||||
| VIIe | 0.075 | 0.041 | 0.036 | 0.018 | ||||||||||||||||||||
| VIIf | 0.072 | 0.054 | 0.049 | 0.044 | 0.020 | |||||||||||||||||||
| XIVa | 0.021 | |||||||||||||||||||||||
| XIVb | 0.120 | 0.033 | ||||||||||||||||||||||
| XIVc | 0.142 | 0.137 | 0.062 |
Average intra-subgenotype distance: 0.03.
Genotypes XII and XIII were proposed recently by others [26] and are fully confirmed by our reconstruction including new sequences. Genotype XII contains virulent viruses recently isolated from poultry in South America and from geese in China [26]. Genotype XIII, previously classified as genotype VII [46], comprises virulent viruses that were isolated in Pakistan, Russia, Burundi, India, and Sweden between 1997 and 2008. Genotype VII, which comprises the largest number of NDV isolates and the highest diversity, is sub-divided into four sub-genotypes named VIIb, VIId, VIIe, and VIIf, as previously described [26]. Genotypes VII, XII, XIII, and XIV share a common ancestor. The recently described genotype XI found in Madagascar [25] was also consistently branched as a separate group from genotype IV.
Concerning sub-genotype IIa, we support on the basis of genetic distance from genotype II (0.13, Table 3) the proposal of Diel et al. [26] to introduce a new genotype X. Furthermore, the distinction between these two groups is reinforced by the exclusive combination of amino acid signatures (Table 5).
Table 5. List of specific signatures on the F complete gene for genotypes and subsequent sub-genotype discrimination.
| Genotype | Exclusive signatures (amino acids/nucleic acids) Underline:specific signature to this cluster | Sub-genotype | Exclusive signatures (amino acids/nucleic acids) Underline: specific signature to this cluster |
| Ia | – | ||
| T1104 T1503A1539 | |||
| Ib | – | ||
| C528 A753 A1029 C1542 | |||
| I | V17 Q422 | Ic | V20 |
| – | C1332 T1377 | ||
| Id | – | ||
| G132 C609 A1293 | |||
| V19 N30 L69 K232 I386 N403 K421 I509 | |||
| II | – | ||
| III | T16 I17 V26 S139 Q195 T288 | ||
| – | |||
| IV | T22 R115 | IVa | I50 S132 N380 |
| – | – | ||
| Va | R46 | ||
| V | A106 K421 S476 N494 A508 I517 | – | |
| Vb | K312 V516 | ||
| – | |||
| VIa | I50 S132 N380 | ||
| – | |||
| VIb | T19 S132 S406 | ||
| – | |||
| – | VIc | P13 A132 S514 | |
| VI | – | VIe | P13 I50 S132 S515 |
| VIf | S132 P31 E342 | ||
| – | |||
| VIg | S132 H272 G304 I537 | ||
| VI h | T9 E111 S132 S406 V432 | ||
| VI i | I50 S132 N380K494 | ||
| A108 | |||
| VII a | V255 F314 | ||
| VIIb | V52 R71 I255 Y314 R480 | ||
| T1044 G1608 | |||
| S176 | VIId | V52 I255 Y314 | |
| VII | – | T1044 G1608 | |
| VIIe | V52 I255 Y314 R480 | ||
| G1608 | |||
| VIIf | A29 I255 Y314 R480 | ||
| – | |||
| VIII | T11 T107 A 203 S514 V516 | ||
| – | |||
| IX | R27 N30 A106 R115 Q195 V339 I386 G497 I509 | ||
| – | |||
| X | G4 K113 G115 G124 Q422 Q451 | ||
| – | |||
| XI | L18 S211 S272 M289 L384 R394 H411 S442 I522 | ||
| – | |||
| XII | L10 I43 T549 | ||
| – | |||
| XIII | A192 T447 A696 C1033 T1368 | ||
| XIVa | S12 P15 Y27 M28 | ||
| – | |||
| XIVb | S170 N550 | ||
| XIV | I44 K51 | – | |
| XIVc | A220A465 L510 | ||
| – |
The second column indicates the combination of specific amino (first line) and nucleic (second line) signatures that discriminate the genotypes. The fourth column shows the same for the sub-genotype discrimination and is applicable once the genotype has been assigned. Underlined amino or nucleic acids represent specific unique signatures.
Finally, with only two sequences from Dominican Republic and Mexico in our phylogenetic reconstructions, but showing a long branch between old and recent genotypes, the criteria were not fully met to assign a new genotype. However, very recent work adding more sequences from those countries supports the existence of a XVth genotype [27].
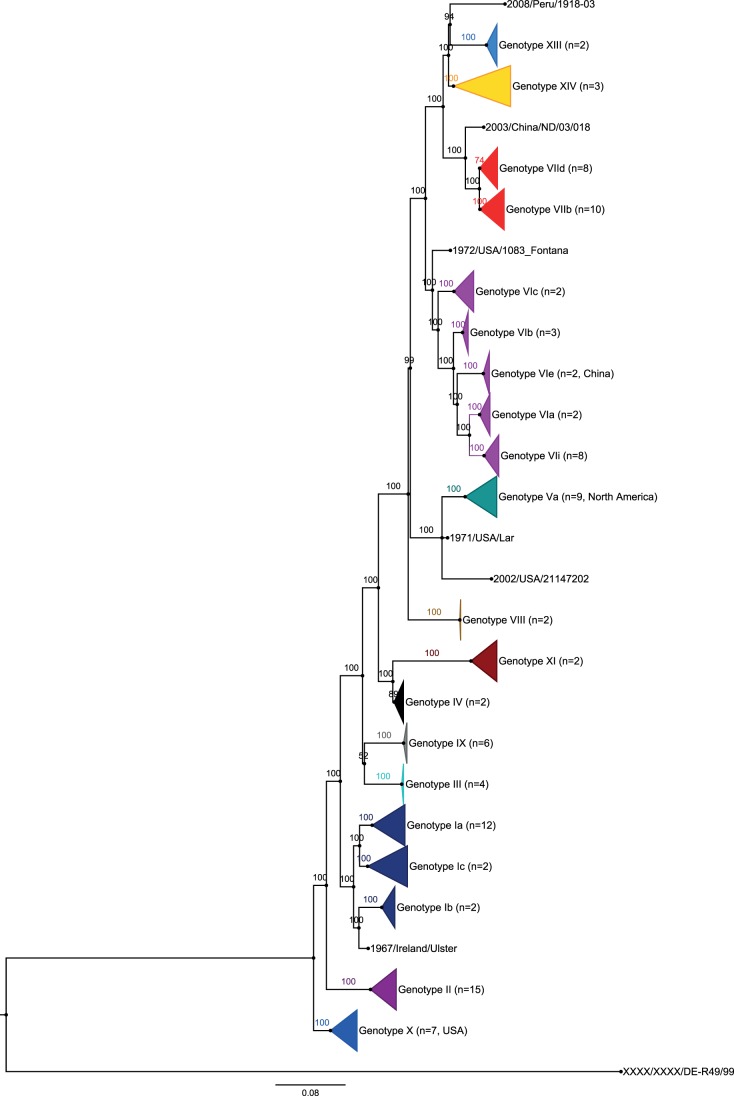
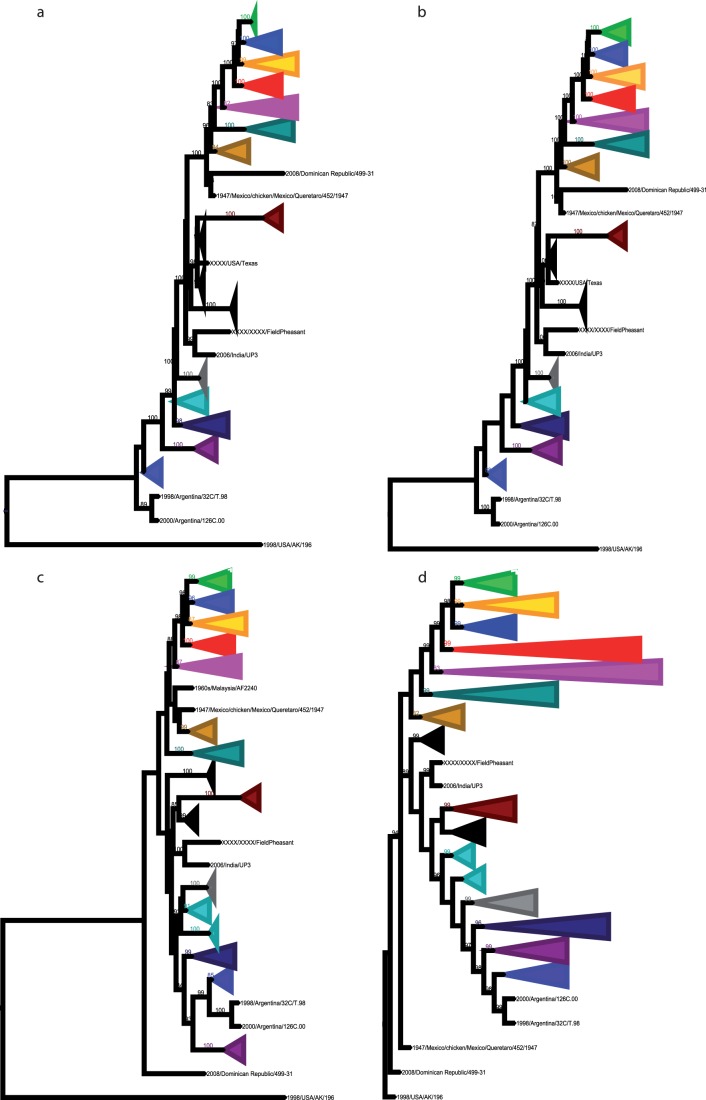
These results are confirmed by the identification of specific amino or nucleic acid signatures on the full F gene for all genotypes and sub-genotypes (Table 5). They are also confirmed by phylogenetic analyses based on the HN gene (Figure S3A), the N, P, M, and L genes (data not shown), and the full genomes (Figure 2). With the “Test topologies” option of Treefinder (Table S2), the most probable tree for representing the different sets of nucleic acid sequences was the one obtained with the full genome by Maximum Likelihood and Bayesian inference (tree in Figure 2). This tree branches the different genotypes in the following order from the root of the phylogenetic tree: X, II, I, III, (IV, XI), VIII, V, VI, and (VII, (XIII, XIV). The complete F tree confirms this classification, adding genotype IX between III and (IV, XI) and genotype XII with XIII and XIV. The complete HN tree switches genotypes III and IX but conserves the rest. Recombination analysis performed on all the sequences used in this study failed to identify any recombination events in our Malian, Ethiopian, and Madagascar isolates that could have interfered with the phylogenetic analyses. All our analyses were conducted using the Maximum Likelihood (ML) or Bayesian methods. To confirm our findings with these methods, a comparison with other methods was carried out on the F gene. The methods included Maximum Parsimony (MP) and Neighbor Joining (NJ) (JTT matrix with 5 categories for the gamma distribution) using Mega. The different representations are shown in Figure 3. The robustness of the trees is estimated by comparing bootstrap values for ML, NJ, and MP and the posterior probabilities of MrBayes for the main clusters (Figure 3)]. In addition, Ktreedist was used to compare the branch lengths between MrBayes, ML, and NJ (not applicable for MP). The best K-score was obtained with the Bayesian inference (Table S4).
Figure 2. Phylogenetic analysis of 110 complete genome sequences.
Trees were constructed using Bayesian inference with 2,960,000 iterations and 1/1000 tree sampled in the chain Minimal average ESS for all parameters was 365, and PSRF were between 0.99978 and 1.0059. A class I virus was used as an outgroup. Sequences from this study are three Malian strains, one in each of sub-genotypes XIVa, b, and c. The complete list of the 110 sequences, the corresponding multiple sequence alignments, and the tree in Newick and Figtree format can be found in Table S8, Table S7, Figure S5HandFigure S9P, respectively.
Figure 3. Comparison of tree topology after phylogenetic reconstruction on the 741 complete F gene sequences using four phylogenetic methods: (a) Maximum Likelihood (ML), (b) Bayesian (MrBayes), (c) Maximum Parsimony (MP), and (d) Neighbor Joining (NJ).
Branch support values correspond to 1000 bootstrap replicates for ML, MP, and NJ, and posterior probabilities estimated for 10,000 samples of the Markov chain for MrBayes. The area of the triangle is proportional to the number of isolates within the genotype.
Confirmation of the New Nomenclature using Shorter Sequences
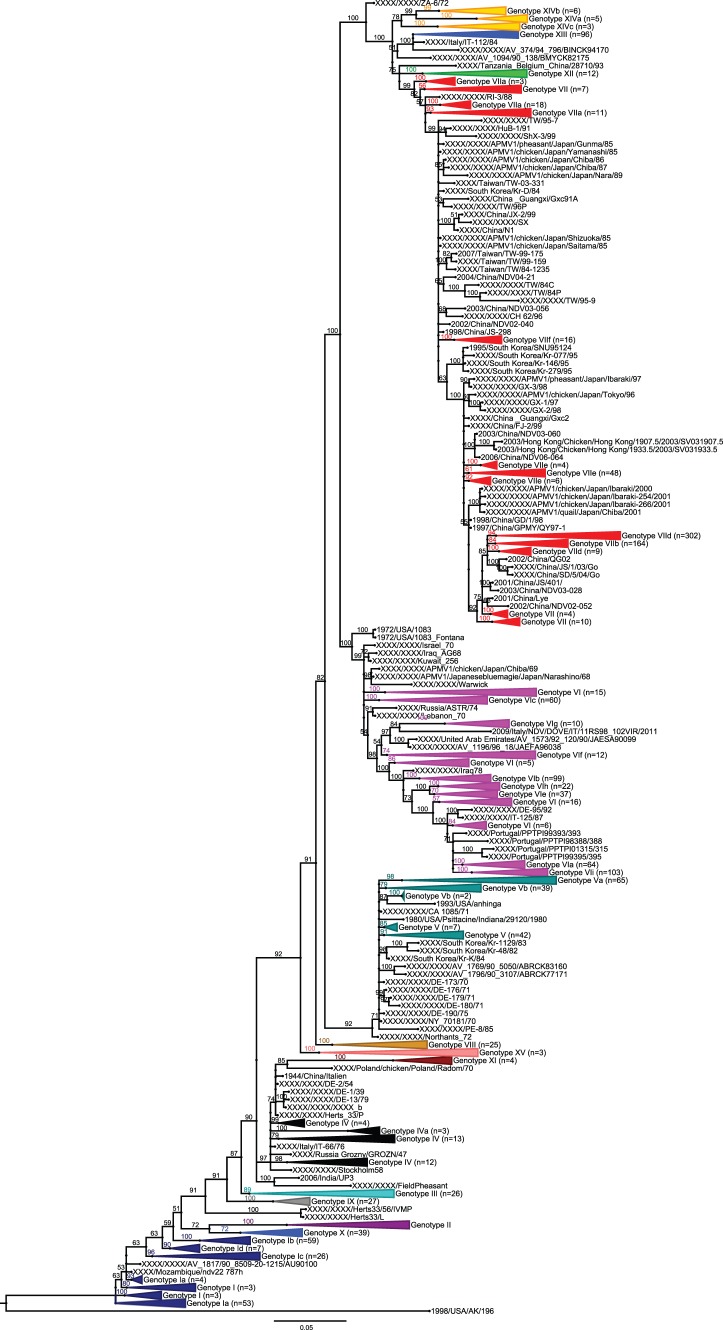
Since partial F gene sequences are often used in the literature, we made a Bayesian phylogenetic reconstruction with 1921 sequences of 372 bases retrieved in GenBank. The genotypes were generally discriminated as with the full F gene except for genotype I, which cannot be maintained as monophylogenetic because its sub-genotypes are spread around genotypes II and X (Figure 4). At the sub-genotype level, the partial sequence does not allow precise delineation and so it is complicated to identify strains that are not close enough to the standard sequence of a given sub-genotype. The reconstruction with partial F gene is therefore less accurate than with the full F gene as shown by the inability to achieve the convergence rule and validation criteria with MrBayes (standard deviation of split frequencies lower than 0.01 or minimal average ESS <200 and PSRF within [0.99 and 1.01], see Figure 4 caption). It suggests that a 372-base-long segment should be the minimal genetic information required for correct genotyping but should be used with caution for further sub-clustering.
Figure 4. Phylogenetic analysis of 1921 partial F gene sequences based on nucleotides from positions 1 to 372.
Trees were constructed using Bayesian inference with 47,408,000 iterations and 1/1000 tree sampled in the chain. Minimal average ESS for all parameters was 88, and PSRF were between 1.00003 and 1.01335. A class I virus was used as an outgroup. Sequences from this study are 5 Malian strains (1 in sub-genotype XIVa, 3 in XIVb, and 1 in XIVc), 5 Ethiopian strains (VIf), and 4 Madagascar strains (XI). The complete list of the 1921 sequences, the corresponding multiple sequence alignments, and the tree in Newick and Figtree format can be found in Table S8, Table S7, Figure S5I, and Figure S9Q, respectively.
Discussion
In this study, a comprehensive phylogenetic reconstruction of NDV strains was carried out using robust inference methods based on Maximum Likelihood and MrBayes. We included sequences in public databases representing all genotypes described so far and new isolates obtained and sequenced in this study in the framework of an international survey on wild and domestic birds in Africa. In this active surveillance survey, the prevalence of NDV in domestic birds was three times lower than that found in a study carried out in Burkina Faso [48]. In that study, the Burkina samples were also tested for the presence of influenza A viruses; the prevalence was estimated to be 3.2%. This prevalence was also three times higher than ours (data not shown). It is unknown whether this reflects a difference in sensitivity of detection methods used by the investigating groups or in the virus prevalence in different African countries. The presence of NDV in apparently healthy poultry in Africa is not surprising with regard to low or moderate virulent strains. However, most of our isolates have a cleavage site of virulent strains. In addition, some representative isolates were confirmed to be highly virulent in experimental challenges under controlled conditions. It remains unclear whether the apparent absence of signs in poultry infected by virulent strains results from the incubation time before disease onset or an incomplete vaccine protection (no information available about the pre-immune status). In line with the second assumption, the fact that viruses currently involved in Mali and Ethiopia belong to new genotypes (XIV and VIf, respectively) could question the efficacy of vaccines used in these countries, which all derived a long time ago from genotype II.
The data sets used in this study and the comparison of different methods for tree reconstruction allowed us to refine the NDV classification. The clustering of the different genotypes as well as the analyses of the minimal distances between genotypes and the identification of specific signatures for each sub-genotype show that the nomenclature proposed by Ballagi-Pordany [12] and Lomniczi et al [11] is more appropriate to describe the phylogenetic relationships between the isolates than the lineage nomenclature proposed by Aldous et al [1] and adopted by others [11], [12], [13], [26].
Most NDV phylogenetic reconstructions are based on the most variable part of the F gene (nucleotide 47 to 420). However, recombination occurs in NDV genomes and thus the use of only a partial sequence of the F gene for genotyping can lead to incomplete or false conclusions [49]. For instance, Qin et al. observed that strain SRZ03 isolated in China was the result of a recombination inside the F open reading frame between a genotype II and a genotype VII strain [49]. Moreover, Chong et al. showed incongruence in genotyping for 6 out of 54 complete genomes of NDV because of recombination events [50]. Here, to avoid any mistake due to recombination events, an analysis was carried out to show the absence of recombinations. We found evidence of recombination in 47 strains in the public database (Table S6). More than 45% of the recombinations of F gene involved the most variable part of the gene, thus including the region habitually used for NDV phylogeny. In this study, all phylogeny is based on the analysis of the complete sequence of the F and HN genes and confirmed by full genomes and N, P, M, and L genes (data not shown). In addition, different methods have been used for NDV phylogenetic analysis, like Neighbor Joining (NJ) [4], [11], [12], [13], [49], Maximum Likelihood (ML) [1], [50], [51], or Maximum Parsimony (MP) [44]. Using different algorithms for phylogenetic reconstructions may have an impact on the interpretation of the trees and thus on NDV classification. To strengthen our observations and interpretations, we compared the performances of four main methods (NJ, ML, MP, and Bayesian) for phylogenetic tree reconstruction on the complete F gene sequence. The robustness of the trees was assessed by comparison of the bootstrap values for NJ, MP, and ML and posterior probabilities of the Bayesian approach [38]. A K-score analysis was also done to compare MrB, ML, and NJ using the Ktreedist program [42]. From these comparisons, the performance of MrBayes was slightly better than ML, and both were clearly higher than NJ and MP. In our previous publication describing new genotype XI in Madagascar [25], we discussed some clustering incongruence between phylogenetic trees based on full genome and on the 374 nt fragment of the F gene. At that time, our analyses were carried out using nucleotide sequences and NJ method. The robustness of the results found in the current study in comparison with our previous ones reinforces the concept that phylogenetic analyses based on nucleic acid sequences and Maximum Likelihood or Bayesian approaches are more trustworthy.
Our results confirm most of the genotypes/sub-genotypes proposed by Diel et al, [26], except sub-genotype VIIg and genotype XV, which we do not recommend for validation at the moment. Both include only recombined strains and there is no clear indication that all strains in each of these clusters are phylogenetically related (e.g. arising from a common ancestor). Indeed, there is no consistency in the position of recombination events in the F gene and in the strains that are involved in these events, for both genotypes VIIg and XV, making it difficult to assume a monophyletic lineage for each of these clusters. We describe then the existence of 14 genotypes and propose 10 new sub-genotypes: Ic, Id, IVa, VIf, VIg, Vh, VIi, XIVa, XIVb, and XIVc. Furthermore, all the clusters have exclusive single or combined amino/nucleic acid signatures. Genotype VI was previously classified into eight sub-genotypes by Wang et al [46]. We confirm only four of these sub-genotypes (VIa, b, c, and e) but add four additional clusters consisting of Ethiopian strains grouped in a new sub-genotype VIf, Russian strains grouped in sub-genotype VIg (previously described as VIb/2), and North American strains clustered in sub-genotype VIh and sub-genotype VIi.
All seven strains isolated from Mali in this study clustered in genotype XIV with other strains previously described by Snoeck et al. [22] and Cattoli et al. [23]. All strains in genotype XIV have a cleavage site characteristic of virulent strains. It contains at least three basic amino acid residues (arginine or lysine) at the cleavage site of F protein (positions 113 to 116) in addition to a phenylalanine residue at position 117 [2]. The three distinct branches within this genotype with a high degree of genetic variability between them and the specific amino acid signatures allow sub-clustering into XIVa, b, and c. The analysis of recent western African isolates suggests that these viruses have evolved from a common ancestor with genotype VII, which contains a majority of strains from Asia [22], [23]. The ancestor seems to have generated two distinct lineages: on the one hand, genotype VII and on the other hand, genotypes XII, XIII, and XIV. The fact that only African strains are found in genotype XIV is in favor of the hypothesis that a unique variant was introduced some time ago, possibly from Asia, and that it subsequently evolved into different sub-genotypes.
In the framework of this study, we have also detected eight new virulent NDV strains in Madagascar, all of them clustered in genotype XI previously described by our group [25]. Moreover, we consolidate the identity of genotype XI by nine exclusive amino acid signatures. The presence of only genotype XI strains in NDV outbreaks in domestic birds and the PCR detection of such strains in healthy wild birds (data not shown) suggest large circulation of this particularly virulent genotype between domestic and wild bird compartments in Madagascar.
For NDV tree reconstruction and genotyping, we suggest using a Bayesian or Maximum Likelihood inference on the complete F gene, as recombinations may occur. In this study, the genotypes were generally well discriminated with the full F gene. Consistent reconstruction cannot be performed with full confidence with sequences of only 375 bases of F gene, as is usually done. However, phylogenetic trees based on 375-nt can at least discriminate major genotype clusters, even if discrimination between genotypes I and II and certain sub-genotypes may become less reliable. In large epidemiological surveys, like the one that allowed us to identify new (sub-) genotypes in West Africa, Ethiopia, and Madagascar [this study, 25], it is not always possible to generate the full-length sequence of the F gene from PCR positive swabs, in particular when all attempts to isolate the virus failed. In these situations, a phylogeny based on partial F gene sequences may help but the limitations of such an approach should be kept in mind.
Supporting Information
Table of correspondence between the nomenclature by Aldous et al. [1] or Lomniczi et al. [11] and the new nomenclature proposed for all genotypes based on our results and those of Diel et al. [26] and Courtney et al. [27] .
(DOCX)
Comparison of tree topologies generated on the individual gene and the full genome sequence data set (110 sequences in the different data sets) by Maximum Likelihood and Bayesian inference. Comparison was made by Treefinder using the Shimodaira and Hasegawa test [41].
(XLSX)
Supplementary tree representations. Figure A: Phylogenetic analysis of 323 complete HN nucleic acid sequences of NDV. Trees were constructed using Bayesian inference with 2,648,000 iterations 1/1000 tree sampled in the chain. Minimal average ESS for all parameters was 400, and PSRF were between 0.99976 and 1.00196. A class 1 virus sequence was introduced as an outgroup. Consensus tree posterior probabilities are indicated on the branch. Sequences from this study are grouped in genotypes VIf (5 Ethiopian strains), XI (8 Madagascar strains), XIVa (1 Malian strain), XIVb (5 Malian strains), and XIVc (1 Malian strain). The complete list of the 323 sequences, the corresponding multiple sequence alignments, and the tree in Newick and Figtree format can be found in Table S8, Table S7, Figure S5J. and Figure S9R, respectively. Figure B: Phylogenic inference of 496 partial F gene sequences (445 nt) using MrBayes. Trees were constructed using Bayesian inference with 6,013,000 iterations and 1/1000 tree sampled in the chain. Minimal average ESS for all parameters was 614, and PSRF were between 0.99989 and 1.00057. A class I virus was used as an outgroup. Sequences from our study are 5 Malian strains (1 in sub-genotype XIVa, 3 in XIVb, and 1 in XIVc), 5 Ethiopian strains (VIf), and 12 Madagascar strains (2 and 10 in genotypes Ib and XI, respectively). The complete list of the 1921 sequences, the corresponding multiple sequence alignments, and the tree in Newick and Figtree format can be found in Table S8, Table S7, Figure S5I,and Figure S9Q, respectively.
(ZIP)
Comparison of trees generated by MrBayes, Maximum Likelihood, and Neighbor Joining on 741 F gene sequences. The comparison was made by calculating the minimum branch length distance (K tree score) between the phylogenetic trees using the Ktreedist program [42]. The minimal score represents the best tree.
(XLSX)
Trees generated by MrBayes in Newick format for 741 F gene sequences (Figure G), 110 full genome (Figure H), 1921 F partial sequences –372 nt (Figure I), 796 F partial sequences –445 nt (Figure N) and 323 HN sequences (Figure J). Trees generated in Newick format on F gene sequences by Maximum Likelihood (Figure K), Neighbor Joining (Figure L) and Maximum Parsimony (Figure M).
(ZIP)
Detection of recombination events in the different sequence data sets by RDP suite.
(XLSX)
Multiple sequence alignments in Fasta format of 741 F gene sequences, 110 full genome, 1921 F partial sequences –372 nt, 796 F partial sequences – 445 nt and 323 HN sequences.
(ZIP)
List of sequences included in the different data sets (full genome, complete F gene, partial F gene –372 nt, partial F gene –445 nt and complete HN gene).
(XLSX)
Trees generated by MrBayes in Figtree format for 741 F gene sequences (Figure O), 110 full genome (Figure P), 1921 F partial sequences –372 nt (Figure Q), 323 HN sequences (Figure R) and 796 F partial sequences –445 nt (Figure S).
(ZIP)
Acknowledgments
This work was supported by the high performance cluster of the UMR AGAP CIRAD.
Funding Statement
This study was mainly funded by the French Ministry of Foreign Affairs (MAE) via the Fonds de Solidarité Prioritaire project [GRIPAVI 2006-26] and partly by the EU network of excellence project [EPIZONE (016236, 01/06/2006–31/05/2011)]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Aldous EW, Mynn JK, Banks J, Alexander DJ (2003) A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol 32: 239–256. [DOI] [PubMed] [Google Scholar]
- 2.Office-International-des-Epizooties (2008 and 2009) Newcastle disease. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals.
- 3. Steward M, Vipond IB, Millar NS, Emmerson PT (1993) RNA editing in Newcastle disease virus. J Gen Virol. 74: 2539–2547. [DOI] [PubMed] [Google Scholar]
- 4. Czegledi A, Ujvari D, Somogyi E, Wehmann E, Werner O, et al. (2006) Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Research 120: 36–48. [DOI] [PubMed] [Google Scholar]
- 5. Alexander DJ (2000) Newcastle disease and other avian paramyxoviruses. Rev Sci Tech 19: 443–462. [DOI] [PubMed] [Google Scholar]
- 6. de Leeuw OS, Koch G, Hartog L, Ravenshorst N, Peeters BP (2005) Virulence of Newcastle disease virus is determined by the cleavage site of the fusion protein and by both the stem region and globular head of the haemagglutinin-neuraminidase protein. J Gen Virol 86: 1759–1769. [DOI] [PubMed] [Google Scholar]
- 7. Huang Z, Krishnamurthy S, Panda A, Samal SK (2003) Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J Virol 77: 8676–8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dortmans JC, Koch G, Rottier PJ, Peeters BP (2011) Virulence of newcastle disease virus: what is known so far? Vet Res 42: 122, 1–11. [DOI] [PMC free article] [PubMed]
- 9. Rout SN, Samal SK (2008) The large polymerase protein is associated with the virulence of Newcastle disease virus. J Virol 82: 7828–7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glickman RL, Syddall RJ, Iorio RM, Sheehan JP, Bratt MA (1988) Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. J Virol 62: 354–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lomniczi B, Wehmann E, Herczeg J, Ballagi-Pordany A, Kaleta EF, et al. (1998) Newcastle disease outbreaks in recent years in western Europe were caused by an old (VI) and a novel genotype (VII). Arch Virol 143: 49–64. [DOI] [PubMed] [Google Scholar]
- 12. Ballagi-Pordany A, Wehmann E, Herczeg J, Belak S, Lomniczi B (1996) Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Arch Virol 141: 243–261. [DOI] [PubMed] [Google Scholar]
- 13. Miller PJ, Decanini EL, Afonso CL (2010) Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect Genet Evol 10: 26–35. [DOI] [PubMed] [Google Scholar]
- 14. Herczeg J, Wehmann E, Bragg RR, Travassos Dias PM, Hadjiev G, et al. (1999) Two novel genetic groups (VIIb and VIII) responsible for recent Newcastle disease outbreaks in Southern Africa, one (VIIb) of which reached Southern Europe. Arch Virol 144: 2087–2099. [DOI] [PubMed] [Google Scholar]
- 15. Yu L, Wang Z, Jiang Y, Chang L, Kwang J (2001) Characterization of newly emerging Newcastle disease virus isolates from the People’s Republic of China and Taiwan. J Clin Microbiol 39: 3512–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu H, Wang Z, Wu Y, Zheng D, Sun C, et al. (2007) Molecular epidemiological analysis of Newcastle disease virus isolated in China in 2005. Journal of Virological Methods 140: 206–211. [DOI] [PubMed] [Google Scholar]
- 17. Lien Y-Y, Lee J-W, Su H-Y, Tsai H-J, Tsai M-C, et al. (2007) Phylogenetic characterization of Newcastle disease viruses isolated in Taiwan during 2003–2006. Veterinary Microbiology 123: 194–202. [DOI] [PubMed] [Google Scholar]
- 18.Perozo F, Marcano R, Afonso CL (2102) Biological and phylogenetic characterization of a genotype VII Newcastle disease virus from Venezuela: efficacy of field vaccination. [DOI] [PMC free article] [PubMed]
- 19. Abolnik C, Horner RF, Maharaj R, Viljoen GJ (2004) Characterization of a pigeon paramyxovirus (PPMV-1) isolated from chickens in South Africa. Onderstepoort J Vet Res 71: 157–160. [DOI] [PubMed] [Google Scholar]
- 20. Abolnik C, Gerdes GH, Kitching J, Swanepoel S, Romito M, et al. (2008) Characterization of pigeon paramyxoviruses (Newcastle disease virus) isolated in South Africa from 2001 to 2006. Onderstepoort J Vet Res 75: 147–152. [DOI] [PubMed] [Google Scholar]
- 21.Otim MO, Christensen H, Jorgensen PH, Handberg KJ, Bisgaard M (2004) Molecular Characterization and Phylogenetic Study of Newcastle Disease Virus Isolates from Recent Outbreaks in Eastern Uganda. 2802–2805. [DOI] [PMC free article] [PubMed]
- 22. Snoeck CJ, Ducatez MF, Owoade AA, Faleke OO, Alkali BR, et al. (2009) Newcastle disease virus in West Africa: new virulent strains identified in non-commercial farms. Arch Virol 154: 47–54. [DOI] [PubMed] [Google Scholar]
- 23. Cattoli G, Fusaro A, Monne I, Molia S, Le Menach A, et al. (2009) Emergence of a new genetic lineage of Newcastle disease virus in West and Central Africa–implications for diagnosis and control. Vet Microbiol 142: 168–176. [DOI] [PubMed] [Google Scholar]
- 24. Servan de Almeida R, Maminiaina OF, Gil P, Hammoumi S, Molia S, et al. (2009) Africa, a reservoir of new virulent strains of Newcastle disease virus? Vaccine 27: 3127–3129. [DOI] [PubMed] [Google Scholar]
- 25. Maminiaina OF, Gil P, Briand FX, Albina E, Keita D, et al. Newcastle Disease Virus in Madagascar: Identification of an Original Genotype Possibly Deriving from a Died Out Ancestor of Genotype IV. PLoS One 5: e13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diel DG, da Silva LH, Liu H, Wang Z, Miller PJ, et al. (2012) Genetic diversity of avian paramyxovirus type 1: Proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect Genet Evol 12: 1770–1779. [DOI] [PubMed] [Google Scholar]
- 27.Courtney SC, Susta L, Gomez D, Hines NL, Pedersen JC, et al.. (2012) Highly Divergent Virulent Isolates of Newcastle Disease Virus from the Dominican Republic are Members of a New Genotype that May have Evolved Unnoticed for Over Two Decades. J Clin Microbiol Nov 28. [DOI] [PMC free article] [PubMed]
- 28. Wise MG, Suarez DL, Seal BS, Pedersen JC, Senne DA, et al. (2004) Development of a real-time reverse-transcription PCR for detection of newcastle disease virus RNA in clinical samples. J Clin Microbiol 42: 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pham HM, Konnai S, Usui T, Chang KS, Murata S, et al. (2005) Rapid detection and differentiation of Newcastle disease virus by real-time PCR with melting-curve analysis. Arch Virol 150: 2429–2438. [DOI] [PubMed] [Google Scholar]
- 30. Fuller CM, Collins MS, Easton AJ, Alexander DJ (2007) Partial characterisation of five cloned viruses differing in pathogenicity, obtained from single isolate of pigeon paramyxovirus type 1 (PPMV-1) following passage in fowls’ eggs. Arch Virol 152: 1575–1582. [DOI] [PubMed] [Google Scholar]
- 31. Zou J, Shan S, Yao N, Gong Z (2005) Complete genome sequence and biological characterizations of a novel goose paramyxovirus-SF02 isolated in China. Virus Genes 30: 13–21. [DOI] [PubMed] [Google Scholar]
- 32. Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin DP (2009) Recombination detection and analysis using RDP3. Methods Mol Biol 537: 185–205. [DOI] [PubMed] [Google Scholar]
- 35. Jobb G, von Haeseler A, Strimmer K (2004) TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36. Hannan EJ, Quinn B (1979) The determination of the order of an autoregression. J Roy Statist Soc B 41: 190–191. [Google Scholar]
- 37. Schwartz G (1978) Estimating the Dimension of a Model. Ann Statist 6: 461–464. [Google Scholar]
- 38. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, Rannala B, Yang Z (2012) Robustness of compound Dirichlet priors for Bayesian inference of branch lengths. Syst Biol: 779–784. [DOI] [PubMed]
- 40.Rannala B, Zhu T, Yang Z (2012) Tail paradox, partial identifiability, and influential priors in Bayesian branch length inference. Mol Biol Evol: 325–335. doi: 10.1093/molbev/msr210. [DOI] [PubMed]
- 41. Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16: 1114–1116. [Google Scholar]
- 42. Soria-Carrasco V, Talavera G, Igea J, Castresana J (2007) The K tree score: quantification of differences in the relative branch length and topology of phylogenetic trees. Bioinformatics 23: 2954–2956. [DOI] [PubMed] [Google Scholar]
- 43. Mohamed MH, Kumar S, Paldurai A, Megahed MM, Ghanem IA, et al. (2009) Complete genome sequence of a virulent Newcastle disease virus isolated from an outbreak in chickens in Egypt. Virus Genes 32: 234–237. [DOI] [PubMed] [Google Scholar]
- 44. Ujvári D, Wehmann E, Kaleta EF, Werner O, Savić V, et al. (2003) Phylognetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columbia livia) and suggests multiple species transmission. Virus research 96: 63–73. [DOI] [PubMed] [Google Scholar]
- 45. Pchelkina IP, Manin TB, Kolosov SN, Starov SK, Andriyasov AV, et al. (2013) Characteristics of Pigeon Paramyxovirus Serotype-1 Isolates (PPMV-1) from the Russian federation from 2011 to 2009. Avian diseases 57: 2–7. [DOI] [PubMed] [Google Scholar]
- 46. Wang Z, Liu H, Hu J, Bao J, Zheng D, et al. (2006) Genotyping of Newcastle disease viruses isolated from 2002 to 2004 in China. Ann. N.Y.Acad.Sci. 1081: 228–239. [DOI] [PubMed] [Google Scholar]
- 47. Kwon HJ, Cho SH, Ahn YJ, Seo SH, Choi KS, et al. (2003) Molecular epidemiology of Newcsatle disease in Republic of Korea. Vet microbial 95(1–2): 39–48. [DOI] [PubMed] [Google Scholar]
- 48. Tarnagda Z, Yougbare I, Kam A, Tahita MC, Ouedraogo JB (2011) Prevalence of infectious bronchitis and Newcastle disease virus among domestic and wild birds in H5N1 outbreaks areas. J Infect Dev Ctries 5: 565–70. [DOI] [PubMed] [Google Scholar]
- 49. Qin Z, Sun L, Ma B, Cui Z, Zhu Y, et al. (2008) F gene recombination between genotype II and VII Newcastle disease virus. Virus Res 131: 299–303. [DOI] [PubMed] [Google Scholar]
- 50. Chong YL, Padhi A, Hudson PJ, Poss M (2010) The Effect of Vaccination on the Evolution and Population Dynamics of Avian Paramyxovirus-1. PLoS Pathog 6: e1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Khan TA, Rue CA, Rehmani SF, Ahmed A, Wasilenko JL, et al. (2010) Phylogenetic and biological characterization of Newcastle disease virus isolates from Pakistan. J Clin Microbiol 48: 1892–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table of correspondence between the nomenclature by Aldous et al. [1] or Lomniczi et al. [11] and the new nomenclature proposed for all genotypes based on our results and those of Diel et al. [26] and Courtney et al. [27] .
(DOCX)
Comparison of tree topologies generated on the individual gene and the full genome sequence data set (110 sequences in the different data sets) by Maximum Likelihood and Bayesian inference. Comparison was made by Treefinder using the Shimodaira and Hasegawa test [41].
(XLSX)
Supplementary tree representations. Figure A: Phylogenetic analysis of 323 complete HN nucleic acid sequences of NDV. Trees were constructed using Bayesian inference with 2,648,000 iterations 1/1000 tree sampled in the chain. Minimal average ESS for all parameters was 400, and PSRF were between 0.99976 and 1.00196. A class 1 virus sequence was introduced as an outgroup. Consensus tree posterior probabilities are indicated on the branch. Sequences from this study are grouped in genotypes VIf (5 Ethiopian strains), XI (8 Madagascar strains), XIVa (1 Malian strain), XIVb (5 Malian strains), and XIVc (1 Malian strain). The complete list of the 323 sequences, the corresponding multiple sequence alignments, and the tree in Newick and Figtree format can be found in Table S8, Table S7, Figure S5J. and Figure S9R, respectively. Figure B: Phylogenic inference of 496 partial F gene sequences (445 nt) using MrBayes. Trees were constructed using Bayesian inference with 6,013,000 iterations and 1/1000 tree sampled in the chain. Minimal average ESS for all parameters was 614, and PSRF were between 0.99989 and 1.00057. A class I virus was used as an outgroup. Sequences from our study are 5 Malian strains (1 in sub-genotype XIVa, 3 in XIVb, and 1 in XIVc), 5 Ethiopian strains (VIf), and 12 Madagascar strains (2 and 10 in genotypes Ib and XI, respectively). The complete list of the 1921 sequences, the corresponding multiple sequence alignments, and the tree in Newick and Figtree format can be found in Table S8, Table S7, Figure S5I,and Figure S9Q, respectively.
(ZIP)
Comparison of trees generated by MrBayes, Maximum Likelihood, and Neighbor Joining on 741 F gene sequences. The comparison was made by calculating the minimum branch length distance (K tree score) between the phylogenetic trees using the Ktreedist program [42]. The minimal score represents the best tree.
(XLSX)
Trees generated by MrBayes in Newick format for 741 F gene sequences (Figure G), 110 full genome (Figure H), 1921 F partial sequences –372 nt (Figure I), 796 F partial sequences –445 nt (Figure N) and 323 HN sequences (Figure J). Trees generated in Newick format on F gene sequences by Maximum Likelihood (Figure K), Neighbor Joining (Figure L) and Maximum Parsimony (Figure M).
(ZIP)
Detection of recombination events in the different sequence data sets by RDP suite.
(XLSX)
Multiple sequence alignments in Fasta format of 741 F gene sequences, 110 full genome, 1921 F partial sequences –372 nt, 796 F partial sequences – 445 nt and 323 HN sequences.
(ZIP)
List of sequences included in the different data sets (full genome, complete F gene, partial F gene –372 nt, partial F gene –445 nt and complete HN gene).
(XLSX)
Trees generated by MrBayes in Figtree format for 741 F gene sequences (Figure O), 110 full genome (Figure P), 1921 F partial sequences –372 nt (Figure Q), 323 HN sequences (Figure R) and 796 F partial sequences –445 nt (Figure S).
(ZIP)