Abstract
Dopamine D4 receptor (DRD4) gene variation has been associated with biased attention for contextually relevant information (e.g. images of cigarettes among smokers). No research has examined whether DRD4 variation is associated with biased attention for contextually cued emotion stimuli, an important putative intermediate phenotype for a number of pathologies (e.g. depression and anxiety). We conducted two studies examining the relationship between the DRD4 variable number tandem repeats polymorphism and attention bias for facial expressions of emotion following a mood-state manipulation in healthy young adult samples. Study 1 demonstrated that long (i.e. seven or greater tandem repeats) DRD4 allele carriers vs. short DRD4 homozygotes had increased attention for sad facial stimuli, but only after a sad mood provocation. Study 2 demonstrated an association between the long DRD4 allele and attention for negative stimuli (sad and fear expressions) following a sad mood provocation. These studies are the first to demonstrate an association between the long DRD4 allele and biased attention for contextually cued emotion stimuli, an important cognitive mechanism thought to increase risk for affective psychopathology. Implications of these studies for vulnerability and plasticity models of psychiatric genetics are discussed.
Keywords: Attention, dopamine, DRD4, mood
Introduction
The dopamine system is hypothesized to play a crucial role in regulating attentional processes. In particular, the variable number tandem repeats (VNTR) polymorphism in exon III of the dopamine D4 receptor (DRD4) gene is associated with dysregulation and modulation of the attentional system. For example, presence of an allele with seven or more repeats (a ‘long’ allele) is associated with attention-deficit hyperactivity disorder (ADHD; Faraone et al. 2005; Kuntsi et al. 2006) and difficulties with sustained attention in infants and young children (Auerbach et al. 2001; Schmidt et al. 2001) compared to individuals with two alleles with fewer than seven repeats (‘short’ alleles). The long allele is also associated with retrospective reports of increased difficulties with attention and increased dysphoria in childhood compared to individuals without the long allele (Levitan et al. 2003).
The DRD4 also appears to influence attention for salient cues. For instance, the DRD4 long allele is associated with greater attention to smoking cues in current smokers (Hutchison et al. 2002) and increased selective attention for smoking-related stimuli in exsmokers (Munafò & Johnstone, 2008). Functional neuroimaging data suggest that the DRD4 long allele is associated with increased neural response in the superior frontal gyrus and insula to smoking cues in smokers (McClernon et al. 2007). That is, smokers with the long DRD4 allele demonstrated a greater neural response to smoking cues compared to individuals homozygous for the short allele. This suggests that the DRD4 VNTR polymorphism moderates the relationship between smoking status and attention to smoking-relevant information.
Although these studies indicate that DRD4 variation is associated with attention for contextually relevant information among smokers, biases for contextually relevant information also have an important role in other neuropsychiatric disorders, such as depression (Leppänen, 2006), anxiety (Bar-Haim et al. 2007) and eating disorders (Lee & Shafran, 2004). Attention for disorder-relevant information is hypothesized to contribute to maintenance of these conditions through continued attentional focus on, and elaboration of, the disorder-relevant material. Additionally, these biases often appear to be context specific for each of the disorders. For example, individuals with depression demonstrate biased attention for sad faces but not for angry faces (Gotlib et al. 2004) and individuals with social anxiety display biases for socially threatening information (Schultz & Heimberg, 2008). Due to the hypothesized importance of biased attention in the aetiology and maintenance of psychological disorders, and the apparent context specificity of these biases, attention for contextually relevant stimuli may represent a promising intermediate phenotype for the expression of vulnerability for a number of psychiatric conditions.
In the current studies, we sought to examine whether DRD4 variation might contribute to biased attention for context-relevant stimuli. We manipulated context by measuring attention bias for emotional faces in both a naturally euthymic mood and an experimentally provoked sad mood. Emotional facial expressions represent powerful, interpersonally relevant, ecologically valid stimuli (Ekman, 1993) and may be interpreted as contextually relevant, depending on the mood of the individual. We examined attention bias in psychologically healthy individuals to better study the impact of DRD4 variation on attention bias independent from the effects of psychopathology. In each study, we experimentally provoked a sad mood among participants and subsequently measured biased attention for sad and happy stimuli in study 1 and sad and fearful stimuli in study 2. Consistent with previous research, we hypothesized that the presence of the long DRD4 allele would be associated with increased attentional bias for contextually relevant information. That is, the long DRD4 allele would be associated with increased attentional bias for sad stimuli in the context of a sad mood provocation, but not in the context of a euthymic mood.
Study 1
Method
Participants
Participants were 186 healthy young adults recruited at the University of Texas at Austin. Interviewers used structured clinical interviews to determine the presence of current or past psychopathology. Exclusion criteria included presence of any current or past mood, anxiety, substance use, psychotic or eating disorder. In addition, all participants had Beck Depression Inventory II (BDI-II; Beck et al. 1996) scores of ≤12 and were unmedicated at the time of testing. Genotype data were unavailable for six participants, leaving a final sample size of 180. Participants partially fulfilled a research requirement for an introductory psychology course by completing this study. The Internal Review Board at the University of Texas at Austin approved all procedures.
Assessments
Structured clinical interview for DSM-IV
To assess exclusion criteria, the patient version of the Structured Clinical Interview for DSM-IV (SCID; First et al. 1995) was administered at the time of study participation. Twenty percent of all interviews were rated by an independent assessor. Agreement between study and independent assessor was perfect for diagnoses for mood, anxiety, psychotic and eating disorders. Agreement for substance dependence (κ = 0.66) and alcohol abuse (κ = 0.79) was acceptable.
BDI-II
The BDI-II (Beck et al. 1996) is a widely used 21-item, self-report questionnaire that assesses symptoms of depression. The BDI-II has demonstrated adequate internal consistency, test–retest reliability and construct validity (Dozois et al. 1998). A score of ≤12 is considered non-depressed (Dozois et al. 1998).
Current mood
Current sad and happy mood were each measured independently with a single-item scale ranging from 1 (‘not at all sad’ or ‘not at all happy’) to 9 (‘very sad’ or ‘very happy’). These manipulation check measures were intended to be brief and face valid assessments of mood. This approach is commonly used in mood provocation studies (van der Does, 2002a). Current mood was measured before and after each experimental mood condition.
Mood conditions
Participants completed the dot-probe task (described below) in both neutral (euthymic) and sad mood conditions. Consistent with previous work using a sad mood provocation (Segal et al. 1999, 2006; van der Does, 2002a), in the sad mood condition participants listened to sad music while imagining a time in their life when they were very sad. The sad music (Samuel Barber’s Adagio for Strings) has been effectively used to induce a sad mood in previous mood provocation research (Hunt & Forand, 2005). This type of sad mood provocation is effective in eliciting a temporary sad mood (van der Does, 2002b).
In the neutral (euthymic) mood condition, the participants listened to neutral music while thinking of their day in as much detail as possible. They were instructed to think about the details of their day rather than their feelings throughout the day. The neutral music (Wolfgang Amadeus Mozart’s Concerto no. 17 in G Major) was shown in pilot testing to neither increase nor decrease a sad or happy mood. Instructions for both conditions were presented via a computer monitor and the prompt (to imagine a sad time in their life or to think of the details of their day) remained on the screen for the duration of the music (approximately 7 min).
Genotyping
Genomic DNA was collected and isolated from buccal swabs using published procedures (Freeman et al. 1997; Lench et al. 1988). The 48-bp VNTR in exon 3 of the DRD4 gene was assayed using previously reported methods (McGeary et al. 2006, 2007). In short, polymerase chain reactions (PCRs) contained 1 µl genomic DNA (≤20 ng), 10% DMSO (Hybra-Max® grade; Sigma, USA), 1.8 mM MgCl2, 180 µm deoxynucleotides, with 7′-deaza-2′-deoxyGTP (Roche Applied Science, USA) substituted for one half of the dGTP, forward and reverse primers (DRD4, 480 nm, obtained from IDT, USA) and 1 U AmpliTaq Gold® polymerase (ABI, USA), in a total volume of 20 µl. Amplification was performed using touchdown PCR. A 95 °C incubation for 10 min is followed by two cycles of 95 °C for 30 s, 65 °C for 30 s and 72 °C for 60 s. The annealing temperature is decreased every two cycles from 65 to 57 °C in 2 °C increments (10 cycles total) and a final 30 cycles of 95 °C for 30 s, 65 °C for 30 s and 72 °C for 60 s and a final 30-min incubation at 72 °C. After amplification, 2 µl PCR product is combined with 20 µl Hi-Di formamide and 0.5 µl Genescan 500 ROX Size Standard (ABI, USA). Then, 0.5 µl of this mixture undergoes electrophoresis with an ABI 3130xl DNA sequencer using unmodified protocols supplied by the company. Ten percent of samples were randomly selected to be re-run and all calls were consistent between these runs. Of 186 samples, 180 (96.8%) were genotyped successfully. Allele sizes are scored by two investigators independently and inconsistencies are reviewed and rerun when necessary. The primer sequences were forward, 5′-AGGACCCTCATGGCCTTG-3′ (fluorescently labelled) and reverse, 5′-GCGACTACGTGGTCTACTCG-3′ (Lichter et al. 1993). The distribution of DRD4 genotypes can be seen in Table 1. Results of an exact test for Hardy–Weinberg proportions using Markov chain-Monte Carlo implementation (Guo & Thompson, 1992) indicate that our observed genotype frequencies do not differ from the Hardy–Weinberg equilibrium (HWE; p = 0.6779). For statistical analyses, participants were grouped by number of repeats in the VNTR by conventional methods with DRD4 long composed of those with at least one copy of the seven or greater repeats and those in the DRD4 short group being those who had neither copy being greater than six repeats (McGeary, 2009).
Table 1.
DRD4 genotype distribution
| Study 1 (n = 180) |
Study 2 (n = 163) |
|||
|---|---|---|---|---|
|
DRD4 genotypea |
n | % | n | % |
| 2/2 | 3 | 1.67 | 7 | 4.29 |
| 2/3 | 2 | 1.11 | 0 | 0.00 |
| 2/4 | 21 | 11.67 | 22 | 13.50 |
| 2/6 | 1 | 0.56 | 0 | 0.00 |
| 2/7 | 4 | 2.22 | 2 | 1.23 |
| 3/3 | 0 | 0.00 | 2 | 1.23 |
| 3/4 | 9 | 5.00 | 2 | 1.23 |
| 3/5 | 1 | 0.56 | 0 | 0.00 |
| 3/7 | 2 | 1.11 | 3 | 1.84 |
| 4/4 | 72 | 40.00 | 78 | 47.85 |
| 4/5 | 6 | 3.33 | 3 | 1.84 |
| 4/6 | 2 | 1.11 | 2 | 1.23 |
| 4/7 | 42 | 23.33 | 28 | 17.18 |
| 4/8 | 3 | 1.67 | 4 | 2.45 |
| 5/7 | 2 | 1.11 | 3 | 1.84 |
| 5/8 | 1 | 0.56 | 0 | 0.00 |
| 6/7 | 2 | 1.11 | 1 | 0.61 |
| 7/7 | 6 | 3.33 | 4 | 2.45 |
| 7/8 | 1 | 0.56 | 2 | 1.23 |
For statistical analyses DRD4 alleles were classified as ‘short’ ( < 7 repeats) or ‘long’ ( ≥ 7 repeats).
Percentages do not add up to 100 due to rounding.
Dot-probe task
Each trial began with a white fixation cross on a black background in the middle of the screen for 500 ms, followed by a pair of faces presented for 1000 ms. Face pairs consisted of one emotional (happy or sad) face and one neutral face of the same actor. Stimuli appeared in the left and right halves of the screen. Following the offset of the images, a small white asterisk probe on a black background appeared in the location of one of the faces and remained on the screen until the participant responded with a key press on a standard keyboard. The computer recorded the latency and accuracy of each response. Each type of face stimulus (emotional or neutral) and the probe appeared in the left and right position with equal frequency.
Task stimuli were images of faces from the Pictures of Facial Affect (Ekman & Friesen, 1976) photo set. Human faces were selected because they have been used successfully in this paradigm in previous research (Gotlib et al. 2004). Eight faces (four female, four male) were selected from happy and sad emotional expressions and then paired with the neutral emotional expression of the same actor. The block of 16 face pairs was presented seven times for a total of 112 trials. Each block of 16 face pairs was fully randomized for each participant.
Participants sat approximately 60 cm from a 17-inch computer monitor. Each stimulus was approximately 10.5×17 cm (10°×16.1° visual angle) when presented on the screen and the face pairs were spaced approximately 20 cm (18.9° visual angle) apart when measured from the centre. Participants were told that their goal was to locate the position of the asterisk (‘left’ or ‘right’) as quickly and accurately as possible. They used their left index finger to press the ‘D’ key when the asterisk appeared in the location of the left image and their right index finger to press the ‘K’ key when the asterisk appeared in the location of the right image. Participants completed 10 practice trials using neutral–neutral face pairs and repeated the practice until they responded accurately to at least eight of the 10 practice trials.
Consistent with previous research (Gotlib et al. 2004), attentional bias scores were calculated for each participant using the following equation:
| (1) |
where R = right position, L = left position, p = probe and e = emotional word stimulus. Therefore, RpLe indicates the mean response latency when the probe is in the right position and the emotional word stimulus is in the left position, and so on. Positive bias scores indicate a bias toward the emotional stimuli while negative bias scores indicate a bias away from the emotional stimuli.
Procedure
Mass pre-testing identified participants who scored <4 on the short-form of the Beck Depression Inventory (Beck et al. 1974). Upon arrival at the laboratory, trained interviewers administered the SCID to confirm the absence of any current or past psychopathology. Participants meeting diagnostic criteria for a current or past mood, anxiety, psychotic, substance use or eating disorder were excluded, as were participants currently taking a psychotropic medication. Participants were then randomized by computer to receive either the neutral or sad mood condition and subsequently completed the dot-probe task. Participants then returned 24–96 h later to receive the other mood condition and again complete the dotprobe task. As a manipulation check, current mood was assessed immediately before and after each mood condition with the single item scales.
Results
Sample characteristics
Descriptive statistics for the sample are presented in Table 2 by DRD4 status. There were no significant differences between allele groups for age (F1,179 <1, p = 0.35), gender (, p = 0.48), race, ( = 0.03, p = 0.86) or depressive symptoms (F1,179 <1, p = 0.87).
Table 2.
Demographic characteristics
| Demographic |
DRD4 status |
|||
|---|---|---|---|---|
| Study 1 |
Study 2 |
|||
| Short homozygote |
Long carrier | Short homozygote |
Long carrier | |
| n | 117 | 63 | 116 | 47 |
| Age (yr) | 18.9 (1.8) | 18.7 (0.9) | 19.1 (1.2) | 18.9 (0.9) |
| Gender (female/male) | 51%/49% | 56%/44% | 54%/46% | 64%/36% |
| Race (Caucasian/’other’) | 59%/41% | 60%/40% | 67%/33% | 74%/26% |
| Depressive symptoms (BDI-IIa) | 3.4 (3.2) | 3.5 (3.2) | 3.1 (2.8) | 3.2 (3.0) |
BDI-II, Beck Depression Inventory – II.
Data reduction
We deleted trials with incorrect responses (0.6% of all trials) and did not include them in analyses. Consistent with prior research (Gotlib et al. 2004), we deleted reaction times that were faster than 150 ms or slower than 1000 ms (1.1%). Together, these procedures resulted in the exclusion of <1.8% of the data.
Manipulation check
The sad mood provocation successfully increased sad mood, t17 9 = 14.45, p <0.001, Cohen’s d =1.42 and decreased happy mood, t179 = 13.79, p <0.001, Cohen’s d = 1.18. Participants experienced a mean increase of 2.6 points (s.d. = 2.4) for sadness and a mean decrease of 2.1 points (s.d. = 2.1) for happiness on the respective 9-point scales. The neutral mood condition did not change sad (t179 = 1.02, p = 0.31) or happy (t179 <1, p = 0.99) mood. There were no effects of DRD4 genotype on change in sad (F1,179 <1, p = 0.40) or happy (F1,179 <1, p = 0.79) mood after the neutral condition. Similarly, there was no genotype effect for change in sad mood after the sad mood provocation, F1,179 <1, p = 0.42. However, there was a non-significant DRD4 effect for change in happy mood following the sad provocation, F1,179 = 3.50, p = 0.063, with DRD4 long allele carriers showing a less pronounced decrease in happy mood (mean = 1.75, s.d. = 2.29) than short allele homozygotes (mean = 2.35, s.d. = 1.93).
DRD4 variation and attention bias
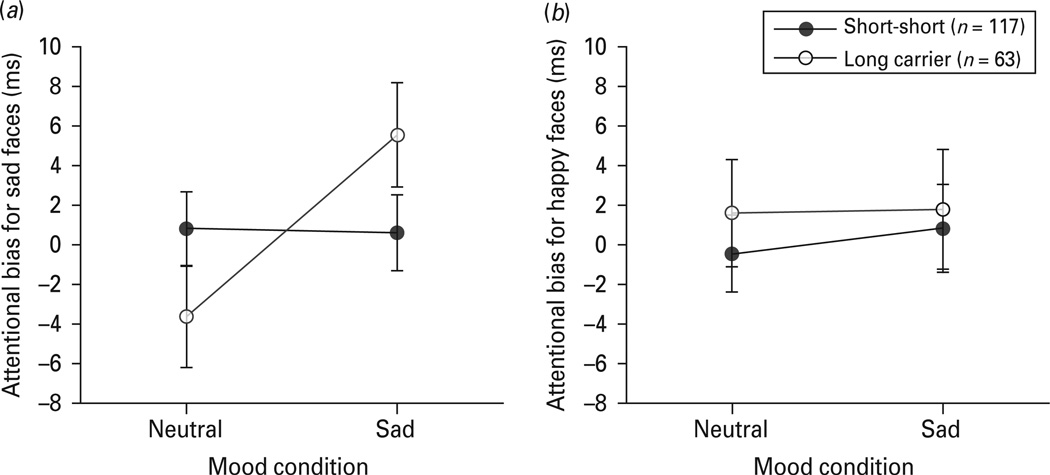
A 2 (DRD4 genotype: short-short, long-carrier) × 2 (mood condition: neutral, sad) repeated measures analysis of variance (ANOVA) was conducted with change in attention bias score from neutral to sad mood as the within-subjects factor. For attention bias for sad stimuli, there was no significant main effect for DRD4 genotype, F1,78 < 1, p = 0.92, but there was a significant main effect for mood condition, F1,78 = 4.14, p = 0.043, partial χ2 = 0.023. More importantly, the DRD4 genotype × mood condition interaction was also statistically significant, F1,78 = 4.47, p = 0.036, partial χ2 = 0.025. The DRD4 genotype × mood condition interaction on attention bias for sad stimuli remained significant when self-reported race was entered as a covariate, F1,177 = 4.43, p = 0.037, partial χ2 = 0.024. Mood condition order did not significantly interact with attention for sad stimuli, F1,178 <1, p = 0.42. Posthoc analyses indicated a significant increase in bias for sad faces (mean = 9.2 ms, s.d.= 32.6) after the sad mood provocation relative to neutral mood in DRD4 long allele carriers, t62= 2.25, p = 0.028, Cohen’s d = 0.41. Bias for sad faces did not change significantly in short allele homozygotes, t116 < 1, p = 0.62. There were no significant differences for attention bias for sad stimuli between genotype groups in either the neutral mood (t178 = 1.42, p = 0.16) or sad mood (t178 = 1.52, p = 0.13) conditions. These results can be seen in Fig. 1.
Fig. 1.
Study 1 attention bias scores (ms) for sad (a) and happy (b) stimuli as a function of DRD4 allele grouping and mood condition. Error bars represent S.E.M.
For attention bias for happy stimuli, an identical repeated measures ANOVA revealed no significant main effects for DRD4 genotype, F1,178 < 1, p = 0.55, mood condition, F1,178 < 1, p = 0.77 and no significant interaction, F1,178 < 1, p = 0.82. Attentional bias for happy facial expression was not influenced by DRD4 status or the mood manipulation.
Discussion
The findings from study 1 provide initial evidence that DRD4 long allele carriers demonstrate a significant shift in attention towards sad stimuli after a sad mood provocation relative to a neutral mood condition. In contrast, participants homozygous for the short allele did not demonstrate a change in attention bias for sad stimuli. Furthermore, the bias demonstrated by long allele carriers was specific to sad stimuli. Neither allele group demonstrated a change in bias for happy stimuli. This is the first evidence that DRD4 genotype influences selective attention for contextually cued emotional information in a carefully selected psychiatrically healthy sample.
Nonetheless, it is important to replicate and extend these findings for two reasons. First, initial behavioural genetics findings are often not replicated (Ioannidis et al. 2001). Although the sample size of our study 1 considerably exceeded the sample sizes of previous studies examining the association between DRD4 variation and smoking cues in smokers and exsmokers (Hutchison et al. 2002; Munafò & Johnstone, 2008), we considered it prudent to replicate the results to increase confidence that the findings are valid. Second, we added face stimuli expressing fear to the dot-probe task to assess the specificity of the bias exhibited by DRD4 long allele carriers. We wanted to determine if the attentional bias would be specific to sad faces or if it would generalize to other negative emotions (i.e. fear). Therefore, we conducted a second study with this modification to replicate and extend our initial findings.
Study 2
For study 2, we recruited a convenient sample of nondepressed young adults who provided buccal cells for genetic analyses and completed the dot-probe task before and after a sad mood provocation. Fearful faces, in addition to sad faces, were presented in the dotprobe task. On the basis of findings from study 1, we expected long DRD4 allele carriers to demonstrate increased bias for sad faces after the sad mood provocation. Addition of the fearful faces to the dot-probe task allowed us to test the specificity of this bias; however, given the lack of previous research, we did not make a specific hypothesis regarding the association between the long allele and attention bias for fearful faces.
Method
Participants
Participants were 166 non-depressed young adults recruited from introductory psychology classes at the University of Texas at Austin. All participants had BDI-II scores of ≤12, reported no history of depression and were unmedicated at the time of testing. Genotype data were unavailable for three participants, leaving a final sample of 163. Participants partially fulfilled a research requirement by completing this study. The Internal Review Board at the University of Texas at Austin approved all procedures.
Assessments
Inventory to diagnose depression – lifetime version
The Inventory to Diagnose Depression – Lifetime Version (IDD-L; Zimmerman et al. 1986) is a 22-item self-report inventory designed to assess the presence and severity of the diagnostic criteria for a major depressive episode. The IDD-L has good agreement with diagnostic interview on presence of a previous major depressive episode (Goldston et al. 1990).
Current mood
Due to the lack of differences in mood change, as measured with two separate sad and happy mood scales, current mood was measured with a single item incorporating both sad and happy mood ranging from 1 (very sad) to 9 (very happy).
Sad mood provocation
Participants completed only a sad mood provocation. The procedure for the provocation was identical to that used in study 1.
Genotyping
Genotyping methods were identical to study 1. In study 2, 163 of 166 samples (98.2%) were genotyped successfully. The distribution of DRD4 genotypes can be seen in Table 1. Using the same methods as study 1, results of an exact test for Hardy–Weinberg proportions indicate that our observed genotype frequencies differ significantly from HWE (p = 0.002316).
Dot-probe task
The dot-probe task was identical to the one used in study 1 with a few exceptions. In study 2, 12 faces (six female, six male) were selected from each emotional category (sad and fearful) and then paired with the neutral expression of the same actor. The block of 24 face pairs was presented three times for a total of 72 trials.
Procedure
Mass pre-testing identified participants who scored <4 on the short-form of the Beck Depression Inventory (Beck et al. 1974). Upon arrival to the laboratory, depression severity was re-assessed using the BDI-II and history of depression was assessed with the IDD-L. Participants with BDI-II scores > 12, a history of depression according to the IDD-L or currently taking psychotropic medications were excluded from the study. Participants who qualified completed a demographics form and provided buccal cells via a cheek swab and mouthwash for genotyping. Next, participants completed the dot-probe task. They then completed a sad mood provocation and again completed the dot-probe task. Current mood was also assessed immediately before and after the mood provocation with the single item scale. In contrast to study 1, all assessments were administered in one laboratory session.
Results
Sample characteristics
Descriptive statistics for the sample are presented in Table 2 by DRD4 status. There were no significant differences between allele groups for age, F1,161 = 1.0, p = 0.32, gender, , p = 0.27, race, , p = 0.39 or depressive symptoms, F1,62<1, p = 0.93.
Data reduction
As in study 1, we deleted trials with incorrect responses and did not include them in analyses (0.5% of all trials). Furthermore, we deleted reaction times that were faster than 150 ms or slower than 1000 ms (0.5%). Together, these procedures resulted in the exclusion of < 1.1% of the data.
Manipulation check
A paired samples t test indicated a significant increase in sadness following the sad mood provocation, t162 = 15.54, p < 0.001, Cohen’s d = 1.25, with participants experiencing a mean change of 1.88 points (s.d. = 1.54) on the 9-point scale. Mean pre-provocation mood was 6.25 (‘somewhat happy’) and mean post-provocation mood was 4.37 (‘somewhat sad’). There was no effect of DRD4 genotype on change in mood, F1,162 = 1.75, p = 0.19.
DRD4 variation and attention bias
Due to the lack of an a priori hypothesis regarding differences between the sad and fear stimuli, we conducted a 2 (DRD4 genotype: short-short, long-carrier) × 2 (time: pre-mood provocation, post-mood provocation) × 2 (stimulus type: sad, fearful) mixed plot ANOVA with attention bias score pre- and post-mood provocation as the within-subjects factor. There were no significant main effects for DRD4 genotype, F1,161 = 3.0, p = 0.08, time, F1,61 = 2.06, p = 0.15 or stimulus type, F1,161 = 2.07, p = 0.15. However, there was a significant DRD4 genotype × time interaction, F1,61 = 9.66, p = 0.002, partial n2 = 0.057. The DRD4 genotype × time interaction on attention bias remained significant when self-reported race was entered as a covariate, F1,60 = 8.89, p = 0.003, partial n2 = 0.053. The DRD4 genotype × stimulus type interaction and the three way interaction were not significant, F’s1,161 < 1, p = n.s.
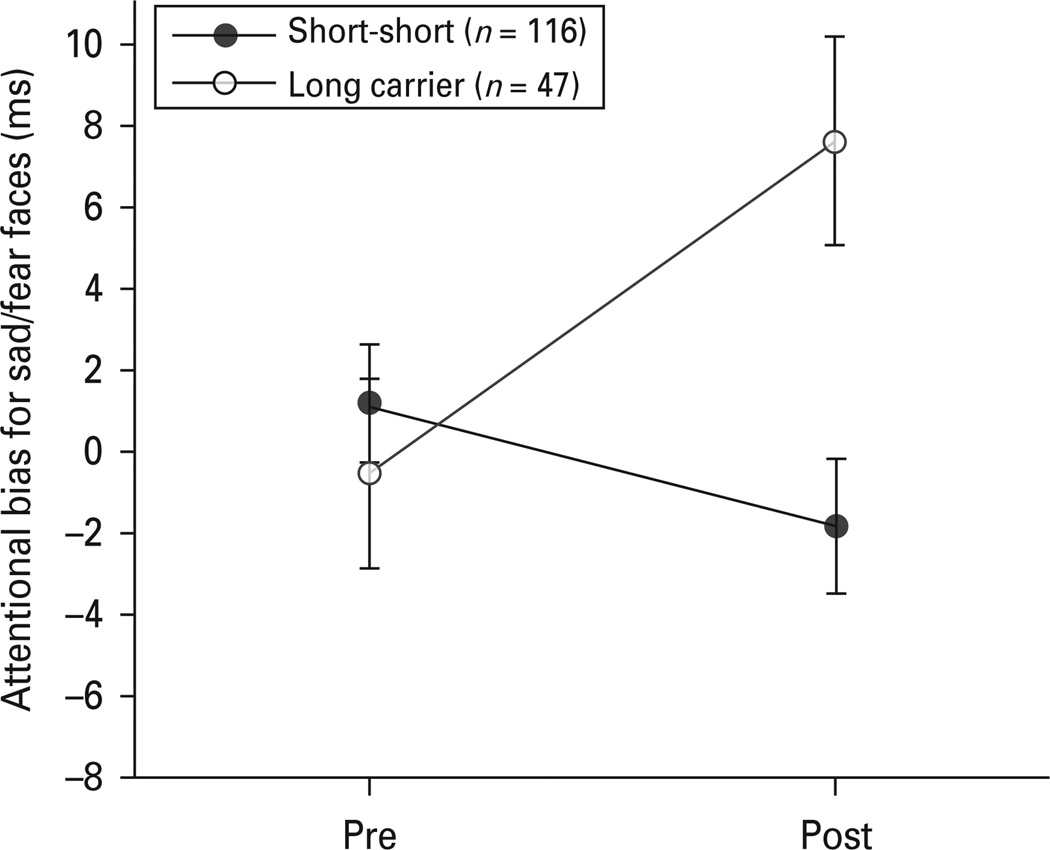
Due to the lack of interaction between stimulus type and our other variables, we collapsed the sad and fear stimuli into a single negative stimuli category to investigate the genotype by time interaction. This interaction was driven by an increase in bias for negative stimuli among long allele carriers following the sad mood provocation, t93 = 2.73, p = 0.008, Cohen’s d = 0.36. There was no change in bias for negative stimuli among short allele homozygotes, t231 = 1.53, p = 0.13. There was no significant difference between genotype groups in attention bias for negative stimuli prior to the sad mood provocation, t324<1, p = 0.55. However, there was a significant difference between genotype groups in attention bias for negative stimuli following the sad mood provocation, t324 = 3.19, p = 0.002, Cohen’s d = 0.39. These results can be seen in Fig. 2.
Fig. 2.
Study 2 attention bias scores (ms) pre and post sad mood provocation for negative stimuli as a function of DRD4 allele grouping. Error bars represent s.e.m.
General discussion
Results from these two studies suggest that the long DRD4 allele is associated with a shift in attention towards negative information during a sad mood in psychiatrically healthy young adults. Study 1 showed that long allele carriers demonstrated increased attentional bias for sad facial stimuli after a sad mood provocation compared to a neutral mood condition. Study 2 replicated and extended these findings. In study 2, long allele carriers demonstrated increased attentional bias for both sad and fearful stimuli after a sad mood provocation. These studies are the first to demonstrate an association between DRD4 variation and selective attention for contextually cued emotion stimuli in a psychiatrically healthy sample.
Our results are consistent with the suggestion that the DRD4 behaves as a ‘plasticity’ gene rather than a vulnerability gene (Belsky et al. 2009). That is, rather than a particular allele simply conferring risk in the context of exposure to adverse environmental stimuli, the ‘risk’ allele may lead to increased susceptibility to environmental influence in general. For example, the DRD4 long allele has been associated with increased vulnerability to ADHD in the context of prenatal smoking exposure, but also with reduced vulnerability in the absence of prenatal smoking exposure (Pluess et al. 2006). The current studies, along with those examining attention to smoking cues in smokers and ex-smokers (Hutchison et al. 2002; Munafo` & Johnstone, 2008), suggest that one potential mechanism through which the long allele increases susceptibility to environmental stimuli may be increased salience of contextually relevant information. Future studies examining selective attention for emotional information after a positive mood provocation would help to further clarify the potential plastic relationship between DRD4 variation and attentional bias for enhanced stimulus salience.
Biased attention for disorder-relevant stimuli has been associated with a broad range of psychiatric conditions, including problem drinking, anxiety, depression and paranoia (Ceballos et al. 2009; Mogg & Bradley, 2005; Moritz & Laudan, 2007). Our results suggest that psychiatrically healthy individuals also display biased attention for contextually relevant emotional stimuli and that DRD4 variation contributes to this process. Thus, contextually relevant attention bias represents a potential intermediate phenotype that is observable in the context of ‘normal’ negative mood in addition to the pathological mood states observed in depression, anxiety and other psychiatric disorders.
Our results also have potential implications for treatment of psychiatric problems. Recent research has shown that attention training with disorder-relevant stimuli can reduce symptoms of anxiety, depression and problem drinking (Amir et al. 2009; Fadardi & Cox, 2009; Schmidt et al. 2009; Wells & Beevers, 2010). If attention to contextually relevant stimuli is found to be a reliable intermediate phenotype of the DRD4, intervention programmes could be refined and more efficiently targeted at those most likely to benefit, such as DRD4 long allele carriers, thereby increasing the potency of such treatments.
Findings from the present study should be interpreted with some limitations in mind. First, our analyses were limited to one candidate gene, but several genes likely play a role in attention for emotional stimuli (Beevers et al. 2009). In addition, the possibility that the DRD4 is in linkage disequilibrium with another functional genetic marker should be considered as a possible explanation for the observed effects. Population stratification is a potential concern in any genetic association study (Hutchison et al. 2004). This confound is unlikely as allele frequencies did not significantly differ across race. Furthermore, we included self-reported race as a covariate in our primary analyses in each study and the results maintained statistical significance with a similar magnitude of effect size. This approach has been suggested to account for much of the effect of stratification and thus may play an important role in the control for population structure (Liu et al. 2006; Sinha et al. 2006; Tang et al. 2005). Future studies could include ancestry informative markers to even more rigorously control for potential population stratification (Kosoy et al. 2009). Finally, genotype frequency in study 2 differed significantly from HWE. As such, results from study 2 should be interpreted cautiously as the genetotypic distribution of this sample may not represent a nonrandom sample of the population. However, we note that, despite the study 2 sample differing significantly from HWE, the results are consistent with those in study 1, which did not differ from HWE.
Despite these limitations, we believe that these studies make an important and interesting contribution to understanding the role of the VNTR polymorphism of the DRD4 gene. Of particular importance is the implication for the attentional plasticity associated with the DRD4. These studies, together with previous work, suggest that the attentional system of DRD4 long allele carriers is more susceptible to environmental influence. Our results are the first to demonstrate this association using emotion stimuli in psychiatrically healthy samples. Future research should continue to investigate potential plasticity effects of candidate genes on intermediate phenotypes and endeavour to determine whether individual differences in these phenotypes confer susceptibility or resilience to future psychopathology dependent on environmental context.
Acknowledgements
This research was supported in part by the Mike Hogg Endowed Fellowship from the University of Texas at Austin to Tony Wells, a grant (R01MH076897) from the National Institute of Mental Health (NIMH) to Christopher Beevers, and a shared equipment grant (S10RR023457) from the National Center for Research Resources (NCRR) and US Department of Veteran Affairs (VA) shared equipment program to John McGeary. We thank the research assistants of the Mood Disorders Laboratory at the University of Texas at Austin for their help with data collection. Tony T. Wells is now at the Department of Psychology, Oklahoma State University.
Footnotes
Statement of Interest
None.
References
- Amir N, Beard C, Taylor CT, Klumpp H, et al. Attention training in individuals with generalized social phobia: a randomized controlled trial. Journal of Consulting and Clinical Psychology. 2009;77:961–973. doi: 10.1037/a0016685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach JG, Benjamin J, Faroy M, Geller V, et al. DRD4 related to infant attention and information processing: a developmental link to ADHD? Psychiatric Genetics. 2001;11:31–35. doi: 10.1097/00041444-200103000-00006. [DOI] [PubMed] [Google Scholar]
- Bar-Haim Y, Lamy D, Pergamin L, Bakermans-Kranenburg MJ, et al. Threat-related attentional bias in anxious and nonanxious individuals: a meta-analytic study. Psychological Bulletin. 2007;133:1–24. doi: 10.1037/0033-2909.133.1.1. [DOI] [PubMed] [Google Scholar]
- Beck AT, Rial WY, Rickels K. Short form of depression inventory: cross-validation. Psychological Reports. 1974;34:1184–1186. [PubMed] [Google Scholar]
- Beck AT, Steer RA, Brown GK. Beck Depression Inventory-II (BDI-II) San Antonio, TX: Psychological Corporation; 1996. [Google Scholar]
- Beevers CG, Wells TT, Ellis AJ, McGeary JE. Association of the serotonin transporter gene promoter region (5-HTTLPR) polymorphism with biased attention for emotional stimuli. Journal of Abnormal Psychology. 2009;118:670–681. doi: 10.1037/a0016198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky J, Jonassaint C, Pluess M, Stanton M, et al. Vulnerability genes or plasticity genes? Molecular Psychiatry. 2009;14:746–754. doi: 10.1038/mp.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos NA, Komogortsev OV, Marc T. Ocular imaging of attentional bias among college students: automatic and controlled processing of alcohol-related scenes. Journal of Studies on Alcohol and Drugs. 2009;70:652–659. doi: 10.15288/jsad.2009.70.652. [DOI] [PubMed] [Google Scholar]
- Dozois DJ, Dobson KS, Ahnberg JL. A psychometric evaluation of the Beck Depression Inventory–II. Psychological Assessment. 1998;10:83–89. [Google Scholar]
- Ekman P. Facial expression and emotion. American Psychologist. 1993;48:384–392. doi: 10.1037//0003-066x.48.4.384. [DOI] [PubMed] [Google Scholar]
- Ekman P, Friesen WV. Pictures of Facial Affect. Palo Alto, CA: Consulting Psychologists Press; 1976. [Google Scholar]
- Fadardi JS, Cox WM. Reversing the sequence: reducing alcohol consumption by overcoming alcohol attentional bias. Drug and Alcohol Dependence. 2009;101:137–145. doi: 10.1016/j.drugalcdep.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Perlis RH, Doyle AE, Smoller JW, et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biological Psychiatry. 2005;57:1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV Axis I Disorders: Patient Edition (SCIDI/P). Version 2.0. New York: Biometric Research, New York State Psychiatric Institute; 1995. [Google Scholar]
- Freeman B, Powell J, Ball D, Hill L, et al. DNA by mail: an inexpensive and noninvasive method for collecting DNA samples from widely dispersed populations. Behavior Genetics. 1997;27:251–257. doi: 10.1023/a:1025614231190. [DOI] [PubMed] [Google Scholar]
- Goldston DB, O’Hara MW, Schartz HA. Reliability, validity, and preliminary normative data for the Inventory to Diagnose Depression in a college population. Psychological Assessment. 1990;2:212–215. [Google Scholar]
- Gotlib IH, Krasnoperova E, Yue DN, Joormann J. Attentional biases for negative interpersonal stimuli in clinical depression. Journal of Abnormal Psychology. 2004;113:127–135. doi: 10.1037/0021-843X.113.1.121. [DOI] [PubMed] [Google Scholar]
- Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Hunt M, Forand N. Cognitive vulnerability to depression in never depressed subjects. Cognition and Emotion. 2005;19:763–770. [Google Scholar]
- Hutchison KE, LaChance H, Niaura R, Bryan A, et al. The DRD4 VNTR polymorphism influences reactivity to smoking cues. Journal of Abnormal Psychology. 2002;111:134–143. doi: 10.1037//0021-843x.111.1.134. [DOI] [PubMed] [Google Scholar]
- Hutchison KE, Stallings M, McGeary J, Bryan A. Population stratification in the candidate gene study: fatal threat or red herring? Psychological Bulletin. 2004;130:66–79. doi: 10.1037/0033-2909.130.1.66. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nature Genetics. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- Kosoy R, Nassir R, Tian C, White PA, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Human Mutation. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntsi J, McLoughlin G, Asherson P. Attention deficit hyperactivity disorder. Neuro Molecular Medicine. 2006;8:461–484. doi: 10.1385/NMM:8:4:461. [DOI] [PubMed] [Google Scholar]
- Lee M, Shafran R. Information processing biases in eating disorders. Clinical Psychology Review. 2004;24:215–238. doi: 10.1016/j.cpr.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Lench N, Stanier P, Williamson R. Simple non-invasive method to obtain DNA for gene analysis. Lancet. 1988;331:1356–1358. doi: 10.1016/s0140-6736(88)92178-2. [DOI] [PubMed] [Google Scholar]
- Leppa¨nen JM. Emotional information processing in mood disorders: a review of behavioral and neuroimaging findings. Current Opinion in Psychiatry. 2006;19:34–39. doi: 10.1097/01.yco.0000191500.46411.00. [DOI] [PubMed] [Google Scholar]
- Levitan RD, Masellis M, Lam RW, Muglia P, et al. Childhood inattention and dysphoria and adult obesity associated with the dopamine D4 receptor gene in overeating women with seasonal affective disorder. Neuropsychopharmacology. 2003;29:179–186. doi: 10.1038/sj.npp.1300314. [DOI] [PubMed] [Google Scholar]
- Lichter JB, Barr CL, Kennedy JL, van Tol HH, et al. A hypervariable segment in the human dopamine receptor D4 (DRD4) gene. Human Molecular Genetics. 1993;2:767–773. doi: 10.1093/hmg/2.6.767. [DOI] [PubMed] [Google Scholar]
- Liu X, Paterson A, John E, Knight J, et al. The role of self-defined race/ethnicity in population structure control. Annals of Human Genetics. 2006;70:496–505. doi: 10.1111/j.1469-1809.2005.00255.x. [DOI] [PubMed] [Google Scholar]
- McClernon FJ, Hutchison KE, Rose JE, Kozink RV. DRD4 VNTR polymorphism is associated with transient fMRI-BOLD responses to smoking cues. Psychopharmacology. 2007;194:433–441. doi: 10.1007/s00213-007-0860-6. [DOI] [PubMed] [Google Scholar]
- McGeary J. The DRD4 exon 3 VNTR polymorphism and addiction-related phenotypes: a review. Pharmacology Biochemistry and Behavior. 2009;93:222–229. doi: 10.1016/j.pbb.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeary JE, Esposito-Smythers C, Spirito A, Monti PM. Associations of the dopamine D4 receptor gene VNTR polymorphism with drug use in adolescent psychiatric inpatients. Pharmacology Biochemistry and Behavior. 2007;86:401–406. doi: 10.1016/j.pbb.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeary JE, Monti PM, Rohsenow DJ, Tidey J, et al. Genetic moderators of naltrexone’s effects on alcohol cue reactivity. Alcoholism: Clinical and Experimental Research. 2006;30:1288–1296. doi: 10.1111/j.1530-0277.2006.00156.x. [DOI] [PubMed] [Google Scholar]
- Mogg K, Bradley BP. Attentional bias in generalized anxiety disorder vs. depressive disorder. Cognitive Therapy and Research. 2005;29:29–45. [Google Scholar]
- Moritz S, Laudan A. Attention bias for paranoiarelevant visual stimuli in schizophrenia. Cognitive Neuropsychiatry. 2007;12:381–390. doi: 10.1080/13546800601119982. [DOI] [PubMed] [Google Scholar]
- Munafo` MR, Johnstone EC. Smoking status moderates the association of the dopamine D4 receptor (DRD4) gene VNTR polymorphism with selective processing of smoking-related cues. Addiction Biology. 2008;13:435–439. doi: 10.1111/j.1369-1600.2008.00098.x. [DOI] [PubMed] [Google Scholar]
- Pluess M, Belsky J, Neuman RJ. Prenatal smoking and attention-deficit/hyperactivity disorder: DRD4–7R as a plasticity gene. Journal of Abnormal Psychology. 2006;86:103–126. [Google Scholar]
- Schmidt LA, Fox NA, Perez-Edgar K, Hu S, et al. Association of DRD4 with attention problems in normal childhood development. Psychiatric Genetics. 2001;11:25–29. doi: 10.1097/00041444-200103000-00005. [DOI] [PubMed] [Google Scholar]
- Schmidt NB, Richey JA, Buckner JD, Timpano KR. Attention training for generalized social anxiety disorder. Journal of Abnormal Psychology. 2009;118:5–14. doi: 10.1037/a0013643. [DOI] [PubMed] [Google Scholar]
- Schultz LT, Heimberg RG. Attentional focus in social anxiety disorder: potential for interactive processes. Clinical Psychology Review. 2008;28:1206–1221. doi: 10.1016/j.cpr.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Segal ZV, Gemar M, Williams S. Differential cognitive response to a mood challenge following successful cognitive therapy or pharmacotherapy for unipolar depression. Journal of Abnormal Psychology. 1999;108:3–10. doi: 10.1037//0021-843x.108.1.3. [DOI] [PubMed] [Google Scholar]
- Segal ZV, Kennedy S, Gemar M, Hood K, et al. Cognitive reactivity to sad mood provocation and the prediction of depressive relapse. Archives of General Psychiatry. 2006;63:749–755. doi: 10.1001/archpsyc.63.7.749. [DOI] [PubMed] [Google Scholar]
- Sinha M, Larkin EK, Elston RC, Redline S. Selfreported race and genetic admixture. New England Journal of Medicine. 2006;354:421–422. doi: 10.1056/NEJMc052515. [DOI] [PubMed] [Google Scholar]
- Tang H, Quertermous T, Rodriguez B, Kardia SLR, et al. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. American Journal of Human Genetics. 2005;76:268–275. doi: 10.1086/427888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Does W. Cognitive reactivity to sad mood: structure and validity of a new measure. Behaviour Research and Therapy. 2002a;40:105–119. doi: 10.1016/s0005-7967(00)00111-x. [DOI] [PubMed] [Google Scholar]
- van der Does W. Different types of experimentally induced sad mood? Behavior Therapy. 2002b;33:551–561. [Google Scholar]
- Wells TT, Beevers CG. Biased attention and dysphoria: manipulating selective attention reduces subsequent depressive symptoms. Cognition and Emotion. 2010;24:719–728. [Google Scholar]
- Zimmerman M, Coryell W, Corenthal C, Wilson S. A self-report scale to diagnose major depressive disorder. Archives of General Psychiatry. 1986;43:1076–1081. doi: 10.1001/archpsyc.1986.01800110062008. [DOI] [PubMed] [Google Scholar]