Abstract
Idiopathic clubfoot is a common orthopedic birth defect that affects approximately 135,000 newborns worldwide. It is characterized by equinus, varus and adductus deformities of the ankle and foot. While numerous studies suggest a multifactorial etiology, the specific genetic and environmental components have yet to be delineated. Maternal smoking during pregnancy is the only common environmental factor consistently shown to increase the risk for clubfoot. Moreover, a positive family history of clubfoot, in conjunction with maternal smoking, increases the risk twenty-fold. These findings suggest that genetic variation in smoking metabolism (xenobiotic) genes may increase susceptibility to clubfoot. Based on this reasoning, we interrogated eight candidate genes (CYP1A1, CYP1A2, CYP1B1, CYP2A6, EPHX1, NAT2, GSTM1 and GSTT1), chosen based on their involvement in xenobiotic metabolism. Twenty-two SNPs and two null alleles in these genes were genotyped in a dataset composed of nonHispanic white and Hispanic multiplex and simplex families. Only rs1048943/CYP1A1 had significantly altered transmission in the aggregate and multiplex NHW datasets (p=0.003 and p=0.009, respectively). Perturbation of CYP1A1 can cause an increase in harmful, adduct forming metabolic intermediates. A significant interaction between EPHX1 and NAT2 was also found (p=0.007). Importantly, for CYP1A2, significant maternal (p=0.03; RR=1.24; 95% CI: 1.04–1.44) and fetal (p=0.01; RR=1.33; 95% CI: 1.13–1.54) genotypic effects were identified, suggesting that both maternal and fetal genotypes can negatively impact limb development. No association was found between maternal smoking status and variation in xenobiotic metabolism genes. Together, these results suggest that xenobiotic metabolism genes are unlikely to play a major role in clubfoot, however, perturbation of this pathway may still play a contributory role.
Keywords: clubfoot, smoking, tobacco, genetics, xenobiotic genes, CYP
INTRODUCTION
Idiopathic talipes equinovarus, or isolated (non-syndromic) clubfoot is a common birth defect that has been recognized and described for centuries. Equinus, varus and adduction of the foot and ankle are the abnormal findings that comprise a clubfoot. In 50–75% of cases, the clubfoot is isolated, with no known cause(s) [Bakalis et al. 2002; Gurnett et al. 2008]. The birth prevalence of clubfoot is approximately 1/1,000 live births, although it can vary approximately 10-fold among different populations. The highest prevalence of clubfoot is in Polynesian populations (6.8/1,000 live births), while the lowest prevalence is observed in Orientals (0.57/1,000 live births) [Ching et al. 1969].
Although clubfoot is a common and well-studied birth defect, its cause(s) and risk factors have not yet been identified. Many etiologies of nonsyndromic clubfoot have been hypothesized and include vascular obstruction, abnormal muscle development, intrauterine growth restriction and neurological abnormalities [Dunn 1972; Hootnick et al. 1982; Reimann 1967; Turco 1981]. Evidence for a genetic etiology comes from twin and family studies. For example, the risk of clubfoot significantly increases as the number of affected relatives increases [Cardy et al. 2007; Cartlidge 1984; Engell et al. 2006; Harper 2004; Idelberger 1939; Lochmiller et al. 1998; Wynne-Davies 1964; Wynne-Davies 1965].
A number of additional environmental factors have been studied, but most have not been known to be consistently to be associated with clubfoot [Alderman et al. 1991; Byron-Scott et al. 2005; Moorthi et al. 2005; Wynne-Davies 1964]. While early amniocentesis (10–13 weeks) has been shown to increase the risk of clubfoot, maternal smoking is the only other environmental factor that has consistently been shown to be associated with clubfoot [Alderman et al. 1991; CEMAT 1998; Centini et al. 2003; Delisle and Wilson 1999; Skelly et al. 2002].
In the United States for 2007, the overall rate of cigarette smoking during pregnancy was 13.2%; for nonHispanic white (NHW) women the rate was more than six times then that of Hispanic women (18.1% versus 2.8%) [Heron et al.]. The relative risk for clubfoot when a mother smokes during pregnancy ranges from 1.34 (95% CI: 1.04–1.72) - 2.6 (95% CI: 1.6–4.0) suggesting an association between clubfoot and smoking (refs). The combination of smoking and a positive history of clubfoot increases the risk for clubfoot twenty-fold (Odds Ratio (OR) = 20.3 95% CI: 7.90–52.17) [Honein et al. 2000]. Therefore, certain genotypes (maternal and fetal) may confer an increased risk, which may be amplified by maternal smoking.
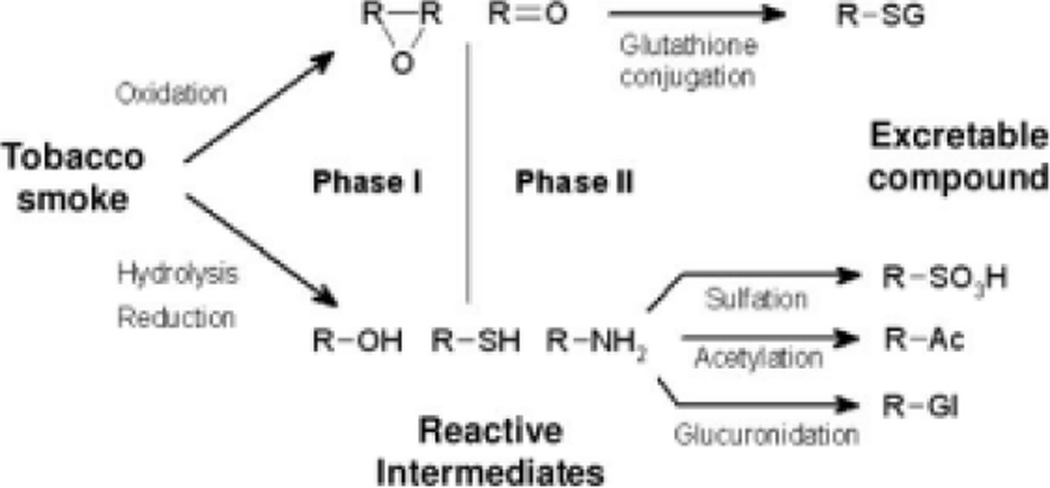
Cigarette smoke consists of more than 4,000 chemical compounds, including dioxins, dioxin-like compounds and other arylhydrocarbon receptor (AhR) agonists [Hecht 1999; Kitamura and Kasai 2007]. The main toxins in cigarette smoke are polycyclic aromatic hydrocarbons (PAHs) [Hecht 1999; Hoffmann and Hoffmann 1997]. Metabolism of tobacco smoke is accomplished through the xenobiotic metabolism pathway (Fig. 1 need to put in this fig which was 5) [Meyer 1996]. This pathway involves biotransformation of a xenobiotic compound by functionalization and/or conjugation reactions into polar, water-soluble metabolites that can be excreted [Meyer 1996; Voet 2004]. This pathway consists of phase I and phase II. Phase I is characterized by the functionalization reactions and utilizes enzymes capable of dehydrogenation/hydrogenation, oxidation, hydrolysis, reduction and mono-oxygenation [Meyer 1996]. Phase II consists of the conjugation reactions and utilizes enzymes capable of glucuronidation, sulphation, acetylation, GSH-conjugation and methylation.
Figure 1.
Xenobiotic metabolism pathway. Adapted from Tim Vickers, http://en.wikipedia.org/wiki/File:Xenobiotic_metabolism.png.
Biotransformation through the xenobiotic metabolism pathway can detoxify a compound or it can create a more toxic/reactive intermediate metabolite. The damaging PAHs and reactive metabolic intermediates from tobacco smoke cross the placenta and form adducts in maternal and fetal tissues [Czekaj et al. 2005; Gladen et al. 2000; Pasanen 1999; Shugart and Matsunami 1985]. Adducts are molecules that can covalently bind DNA or proteins and could therefore interfere with the normal developmental process by altering transcription and replication or damaging protein structure or function [Lopachin and Decaprio 2005; Meyer and Bechtold 1996; Uchida and Stadtman 2000]. For example, PAH-DNA adducts and dioxins from cigarette smoke are mutagenesis, carcinogenesis and teratogenesis [Kitamura and Kasai 2007; Nock et al. 2007].
Given the importance of the xenobiotic metabolism pathway in the clearance of harmful metabolic intermediates and the impact these intermediates may have on a developing fetus, the enzymes in this pathway may offer insight into the observed interaction between maternal smoking and family history on the risk for clubfoot. Variation within key genes involved in xenobiotic metabolism may lead to perturbation of the metabolism pathways and increase DNA or protein adduct forming compounds in fetal tissues which in turn could affect fetal development.
While many genes play a role in this pathway, only a few genes are known to specifically metabolize tobacco smoke. CYP1A1, CYP1A2 and CYP1B1 ? CYP2A6? are members of the cytochrome P450 (CYP450) superfamily and all play a role in phase I metabolism of compounds found in tobacco smoke, such as PAHs and dioxins [2004; Meyer 1996; Pavek and Dvorak 2008; Rodriguez-Antona and Ingelman-Sundberg 2006]. CYP1A1, CYP1A2, CYP1B1 and CYP2A6 expression has been identified in key organs, including the placenta [Pavek and Dvorak 2008]. Epoxide hydrolases (EH), such as EPHX1, also activate and detoxify PAHs during phase I [Omiecinski et al. 2000]. Specifically, EHs metabolize reactive epoxides to less-harmful dihydrodiol derivatives [Hassett et al. 1994b; Omiecinski et al. 2000]. Additionally, certain variants in EPHX1 decrease its activity by 40%, inhibiting effective biotransformation of exogenous compounds [Hassett et al. 1994b].
Genes involved in phase II metabolism include NAT2, GSTM1 and GSTT1. NAT2 is of particular interest because of its “slow acetylator” phenotype [Meyer and Zanger 1997]. The activity of the slow acetylation phenotype ranges from X-X of normal and is associated with increased in harmful adduct levels [Grant et al. 1990; Hecht et al. 2007; Meyer and Zanger 1997]. Approximately 40–70% of European and Northern American individuals have the “slow acetylator” phenotype [Meyer and Zanger 1997]. A study by Hecht, et al. (2007) suggested that the slow-acetylator phenotype of NAT2 may play a role in the development of clubfoot. GST mu?µ (GSTM) and GST theta?τ (GSTT) are of interest because of their role in detoxification of activated nicotine metabolites and other xenobiotics during phase II [Pasanen 1999]. They are involved in the detoxification of epoxides created by the CYP450s [Bolt and Thier 2006]. Only about 40–60% of individuals in the population express GSTM1 and, for those who do not express the gene, there is an increased susceptibility to DNA-adduct formation and cytogenetic damage [Seidegard and Pero 1985; Seidegard et al. 1986; Wiencke et al. 1990]. GSTT1 also has a null allele that can be found in 10–40% of individuals [Seidegard et al. 1988].
These genes interact in the sense that they all play a role in a pathway responsible for metabolizing cigarette smoke, both through activating and detoxifying reactions. Because the level of genotoxic damage in individuals is the result of complex gene-environment and gene-gene interactions, genetic variants within multiple genes may act additively and alter the overall ability of the pathway to detoxify exogenous compounds or toxic intermediate metabolites [Georgiadis et al. 2004]. Specifically, individuals with polymorphisms that increase the activity in activating reactions and decrease the activity of inactivating reactions are more susceptible to the effects of genotoxic compounds [Georgiadis et al. 2004]. Therefore, a mutation in any one of these genes may have a large impact on the efficiency of biotransformation of toxic compounds and the presence of multiple mutations may increase the susceptibility even further. This study was undertaken to determine whether genetic variation in specific smoking xenobiotic genes involved in both phases I and II of cigarette smoke metabolism is associated with clubfoot in the presence or absence of maternal smoking.
MATERIALS AND METHODS
IRB Approval
This study was reviewed and approved by the Committee for the Protection of Human Subjects at the University of Texas Health Science Center at Houston (HSC-MS-09-0328) and all collaborating centers.
Clubfoot Study Samples and Sample Preparation
Probands and their families were identified at four pediatric orthopedic centers: Shriners Hospital for Children of Houston and Los Angeles, Texas Scottish Rite Hospital for Children of Dallas and University of British Columbia. The diagnosis of clubfoot was based on the presence of adducted forefoot, varus hindfoot and ankle equinus deformities as determined by either examination and/or by review of medical records. All patients with syndromic clubfoot were excluded.
All families were either NHW or Hispanic (Mexican) and ethnicity was self-reported. Family history and exposure information were obtained by interview with the proband’s mother and/or by chart review. Two-generation pedigrees were constructed for all families and pedigrees were extended to include all affected individuals if a positive family history was elicted. Blood or saliva samples were collected on all available family members. DNA was extracted from the blood or saliva using either the Roche DNA Isolation Kit for Mammalian Blood (Roche, Basel, Switzerland) or the Oragene Purifier for saliva (DNA Genotek, INC., Kanata, Ontario, Canada) following the manufacturer’s protocol.
The dataset consisted of 1,776 individuals from 619 families. Families were considered to be multiplex (242; 149 NHW and 92 Hispanic) or simplex (377 simplex; 149 NHW and 226 Hispanic) based on the presence or absence of a family history of clubfoot, respectively.
Gene and SNP Identification and Genotyping
Candidate genes were selected following a thorough literature review. Hundreds of enzymes play a role in xenobiotic metabolism, however only a subset of these enzymes are well-characterized and an even smaller subset have been shown to be specifically involved in the metabolism of tobacco smoke. Only genes known to metabolize compounds in cigarette smoke and that interact in multiple steps of a common general pathway were included in this study need some review refs here.
SNPs in the CYP1A1, CYP1A2, CYP1B1, CYP2A6, EPHX1 genes were identified using NCBI and Ensembl websites. SNPs in the NAT2 gene were identified previously [Hecht et al. 2007]. SNPs were selected based on a standard set of criteria including: heterozygosity >0.3, inter- and intragenic positions, coverage of the gene and tagging ability. SNPs with a higher heterozygosity that caused a missense mutation and/or tagged for multiple SNPs were always used. Table 1 lists the SNPs genotyped in this study. All SNPs were genotyped using TaqMan Genotyping Assays (Applied Biosystems, Foster City, CA) and detected on a 7900 HT Sequence Detection System (Applied Biosystems). All genotype results were entered into a Progeny and checked for incompatibility using Pedcheckref.
Table 1.
Smoking Metabolism Genes: SNP Location, Alleles and Ethnic Frequenciesa
| Xenobiotic phase |
Gene | SNP | Base pair no. | Allelesb | Locationc | Typed | cDNA change |
Protein change |
MAF | HCFe |
|---|---|---|---|---|---|---|---|---|---|---|
| I | CYP1A1 15q22–24 5.99kb | rs2470893 | 72806502 | G/A | U | – | – | – | 0.30 | 0.13 |
| rs1048943 | 72800038 | A/G | E | M | 1384A>G | Ile462Val | 0.04 | 0.35 | ||
| rs1456432 | 72790104 | G/A | D | – | – | – | 0.16 | 0.44 | ||
| CYP1A2 15q22-qter 7.76kb | rs2472299 | 72820453 | G/A | U | – | – | – | 0.28 | 0.27 | |
| rs2470890 | 72834479 | C/T | E7 | S | 1548T>C | Asn516Asn | 0.37 | 0.68 | ||
| rs11854147 | 72839824 | C/T | D | – | – | – | 0.33 | 0.61 | ||
| CYP1B1 2p22-p21 8.55kb | rs4646429 | 38160439 | A/G | U | – | – | – | 0.32 | 0.31 | |
| rs10012 | 38155894 | G/C | E2 | M | 142C>G | Arg48Gly | 0.32 | 0.33 | ||
| rs1056836 | 38151707 | G/C | E3 | M | 1294C>G | Leu432Val | 0.42 | 0.25 | ||
| rs163084 | 38144420 | T/C | D | – | – | – | 0.20 | 0.14 | ||
| CYP2A6 19q13.2 6.90kb | rs4105144 | 46050464 | C/T | U | – | – | – | 0.32 | 0.25 | |
| rs7250713 | 46047035 | C/G | I2 | – | – | – | 0.40 | 0.34 | ||
| rs7246742 | 46037235 | G/T | D | – | – | – | 0.13 | 0.18 | ||
| EPHX1 1q42.1 20.29kb | rs2854450 | 224079200 | C/T | U | – | – | – | 0.20 | 0.18 | |
| rs1051740 | 224086256 | T/C | E3 | M | 337T>C | Tyr113His | 0.30 | 0.41 | ||
| rs2234922 | 224093029 | A/G | E4 | M | 416A>G | His139Arg | 0.17 | 0.08 | ||
| rs360063 | 224102932 | G/A | D | – | – | – | 0.44 | 0.48 | ||
| II | NAT28p23.1-p21.3 9.97kb | rs1041983 | 18302075 | C/T | E2 | S | 282C>T | Tyr94Tyr | 0.33 | 0.31 |
| rs1801280 | 18302134 | T/C | E2 | M | 341T>C | Ile114Thr | 0.44 | 0.32 | ||
| rs1799929 | 18302274 | C/T | E2 | S | 481C>T | Leu161Leu | 0.42 | 0.32 | ||
| rs1799930 | 18302383 | G/A | E2 | M | 590G>A | Arg197Gln | 0.30 | 0.18 | ||
| rs1799931 | 18302650 | G/A | E2 | M | 857G>A | Gly286Glu | 0.04 | 0.13 | ||
| GSTM1 | WT/null | – | Null | – | Null | 0.47 | 0.56 | |||
| GSTT1 | WT/null | – | Null | – | Null | 0.20 | 0.13 |
HCF = Hispanic corresponding frequency to NHW minor allele; MAF = minor allele frequency in non-Hispanic white sample; WT = wild-type.
Ancestral allele/alternate allele.
U = upstream, E = exon, I = intron, D = downstream.
Type of mutation: M = missense; S = synonymous.
Values in bold = HCF significantly different from MAF [p < 0.01].
GSTM1 and GSTT1 were genotyped to identify individuals with wild type or null alleles following the protocol of Arand et al. (1996). The GSTM1 and GSTT1 alleles were amplified simultaneously using Takara Ex Taq Polymerase PCR (Takara Bio USA). The PCR was modified by an addition of 0.5 µL of MgCl per reaction at an annealing temperature of 64°C. This method combines primers for GSTM1, GSTT1 and ALB as an internal positive control into one assay [Arand et al. 1996]. Amplified samples were run on a 2% agarose gel and scored according to the presence of the wild type or null allele for both GSTM1 and GSTT1. Absence of GSTM1 or GSTT1 were scored as null alleles [Arand et al. 1996].
Analysis
Allele frequencies and Hardy-Weinberg equilibrium (HWE) were calculated using SAS (v9.1). SNPs out of HWE (P<0.001) were excluded from the subsequent analyses. Chi-squared (X2) analysis was performed to determine allele frequency differences between the NHW and Hispanic populations. Pair-wise linkage disequilibrium (LD) values (D’ and r2) were calculated using GOLD [Abecasis and Cookson 2000].
Linkage and/or association were tested using multiple analytic methods to extract the greatest amount of information from the data. Parametric and non-parametric linkage analyses were performed using Merlin [Abecasis et al. 2002]. Association was tested for using Pedigree Disequilibrium Test (PDT), genotype-pedigree disequilibrium test (geno-PDT) and Association in the Presence of Linkage (APL) [Martin et al. 2000]. [Martin et al. 2003] [Chung et al. 2006]. Two-SNP intrgenic haplotypes were evaluated using APL. Generalized estimating equations (GEE) as implemented in SAS was used to evaluate gene interactions [Hancock et al. 2007]. and Gene-environment interactions were assessed using FBATI hoffman ref.
Log-linear regression models were used to evaluate the independent effects of maternal and child genotypes [van Den Oord and Vermunt 2000]. Only one triad was selected per family. For each SNP, log-likelihoods were computed for the full models (including both maternal and child genotypes) and compared to the log-likelihoods computed for partial models (including either the maternal genotype or the child genotype only).
Chi-square analysis was used to evaluate the relationship between smoking and clubfoot in the presence of null GSTM1 or GSTT1 genotypes.
Protein function and transcription binding sites analyses
To evaluate the effect of the ancestral and alternate alleles on protein function, analyses of exonic missense mutations were performed using in silico SNPs3D and Polyphen programs [Teng et al. 2008; Yue et al. 2006]. Alibaba2, Patch and Transcription Element Search Software (TESS) were used to assess potential regulatory SNPs [Grabe 2002; Matys et al. 2006; Schug 2003]. The ancestral and alternate allele sequences were obtained from the NCBI Entrez SNP Database (www.ncbi.nlm.nih.gov).
RESULTS
Twenty-two SNPs in six genes, CYP1A1, CYP1A2, CYP1B1, CYP2A6, EPHX1, NAT2, and two null allele systems, GSTM1 and GSTT1, were genotyped in our NHW and Hispanic families (Table 1). All SNPs had call rates of >95%. All SNPs were in HWE in the NHW group. In the Hispanic subset, three SNPS, rs1048943 (p=0.02), rs2470893 (p=0.0004) and rs4105144 (p=0.01), were not in HWE and were excluded from further. Allele frequencies for 16 of the 22 SNPs differed between the NHW and Hispanic groups; therefore the data was stratified by ethnicity. In addition, the data was further stratified by the presence or absence of family history (FH) of clubfoot. There was signficant intragenic LD and the LD patterns were similar between ethnicities (data not shown).
Results of the single SNP analyses are presented in Table 2 and Supplemental Table 1. rs1048943 in CYP1A1 had altered transmission in both the NHW aggregate and multiplex family subset (p=0.003 and p=0.009). SNPs in CYP1A2 (1), CYP1B1 (2), EPHX1 (1) and NAT2 (1) had marginal evidence for altered transmission (Table 2A). None of the two-SNP haplotypes had altered transmission (data not shown).
Table 2.
Single SNP Association Results by Family Historya
| A. NHW | ||||||||||
| All | Multiplex | Simplex | ||||||||
| Gene | dbSNP | PDT | GENO-PDT | APL | PDT | GENO-PDT | APL | PDT | GENO-PDT | APL |
| CYP1A1 | rs1048943 | 0.003 | 0.003 | 0.12 | 0.009 | 0.009 | 0.71 | 0.17 | 0.17 | 0.06 |
| CYP1A2 | rs2472299 | 0.85 | 0.29 | 0.03 | 0.61 | 0.22 | 0.27 | 0.21 | 0.43 | 0.07 |
| CYP1B1 | rs1056836 | 0.53 | 0.76 | 0.14 | 0.92 | 0.93 | 0.81 | 0.13 | 0.14 | 0.05 |
| CYP1B1 | rs163084 | 0.10 | 0.14 | 0.05 | 0.40 | 0.59 | 0.40 | 0.05 | 0.04 | 0.05 |
| EPHX1 | rs2234922 | 0.05 | 0.09 | 0.13 | 0.05 | 0.06 | 0.06 | 0.73 | 0.68 | 0.93 |
| NAT2 | rs1799931 | 0.12 | 0.07 | 0.55 | 0.59 | 0.70 | 0.41 | 0.01 | 0.01 | 0.04 |
| B. Hispanic | ||||||||||
| All | Multiplex | Simplex | ||||||||
| Gene | dbSNP | PDT | GENO-PDT | APL | PDT | GENO-PDT | APL | PDT | GENO-PDT | APL |
| CYP1A1 | rs1456432 | 0.44 | 0.68 | 0.52 | 0.39 | 0.70 | 0.37 | 0.03 | 0.12 | 0.21 |
| CYP2A6 | rs7250713 | 0.37 | 0.17 | 0.74 | 0.88 | 0.39 | 0.01 | 0.25 | 0.20 | 0.34 |
| EPHX1 | rs360063 | 0.62 | 0.13 | 0.28 | 0.42 | 0.08 | 0.04 | 0.03 | 0.15 | 0.01 |
Results for p ≤ 0.05.
For the Hispanic group, no SNPs had altered transmission in the aggregate group (Table 2B). rs7250713 in CYP2A6 (p=0.01) and rs360063 in EPHX1 (p=0.04) had altered transmission in the multiplex family subset, whereas rs1456432/CYP1A1 (p=0.03) and rs360063/EPHX1 (PDT: p=0.03; APL: p=0.01) had altered transmission in the simplex families. None of the two-SNP haplotypes had altered transmission.
Evidence for a gene interaction was seen only in the NHW group between rs105740 in EPHX1 and rs1799929 in NAT2 (p=0.007) (Table 3). Suggestive evidence for interactions was found for SNPs in CYP2A6 and SNPs in CYP1B1, EPHX1 and NAT2. There was minimal evidence for gene interactions in the Hispanic group.
Table 3.
Gene-Gene Interactions
| A. NHW | ||||
| Gene | SNP | Gene | SNP | p value |
| EPHX1 | rs1051740 | NAT2 | rs1799929 | 0.007 |
| NAT2 | rs1801280 | 0.03 | ||
| CYP2A6 | rs7250713 | 0.05 | ||
| rs360063 | CYP2A6 | rs4105144 | 0.04 | |
| rs2234922 | CYP2A6 | rs7246742 | 0.05 | |
| CYP2A6 | rs4105144 | CYP1B1 | rs1056836 | 0.02 |
| NAT2 | rs1799930 | 0.04 | ||
| B. Hispanic | ||||
| Gene | SNP | Gene | SNP | p value |
| CYP1B1 | rs1056836 | NAT2 | rs1799929 | 0.04 |
| rs1801280 | 0.04 | |||
| rs163084 | rs1799929 | 0.05 | ||
| rs1056836 | CYP1A2 | rs2472299 | 0.05 | |
| CYP1A1 | rs1456432 | CYP1A2 | rs2472299 | 0.04 |
Evidence for the independent effects of maternal and fetal genotypes was found for only two SNPs in CYP1A2 (Table 4). For rs11854147, a significant maternal genotypic effect (p= 0.03) was found with a relative risk of 1.24 (95% CI: 1.04–1.44). A significant fetal genotypic effect (p= 0.01), with a relative risk of 1.33 (95% CI: 1.13–1.54), was found for rs2470890‥
Table 4.
Log-Linear Regression Modeling of Maternal and Fetal Genotypesa
| RR (95% CI) | LRT p-value | ||||
|---|---|---|---|---|---|
| Gene | SNP | Fetal | Maternal | Fetal | Maternal |
| CYP1A2 | rs2470890 | 1.33 (1.13–1.54) | 1.23 (1.02–1.45) | 0.01 | 0.06 |
| rs11854147 | 1.19 (0.99–1.39) | 1.24 (1.04–1.44) | 0.99 | 0.03 | |
RR = relative risk; LRT = log likehood ratio test.
Two associated SNPs, rs2472299/CYP1A2 and rs4105144/CYP2A6 are located potential regulatory regions (Table 5). All three DNA-binding site algorithms predicted that the alternate rs2472299 allele eliminates a glucocorticoid receptor (GR) TFBS. In contrast, DNA binding sites for rs4105144 in CYP2A6 were inconsistent although two programs predicted a change between ancestral and alternate alleles.
Table 5.
Predicted Transcription Factor Binding Sites for 5′-Associated SNPsa
| Alibaba2 Alleles |
Patch Alleles |
TESS Alleles |
|||||
|---|---|---|---|---|---|---|---|
| Gene/SNP | Location | Ancestral | Alternate | Ancestral | Alternate | Ancestral | Alternate |
| CYP1A2/ rs2472299 | 8.7 kb upstream | GR | None | GR, AR | None | GR, AR | None |
| CYP2A6/ rs4105144 | 2.3 kb upstream | None | PU.1 | None | None | Bcd, Ft2.2 | LEF |
AR = androgen receptor; Bcd = bicoid; GR = glucocorticoid receptor; LEF = lymphoid enhancer factors.
Forty-seven percent of individuals genotyped were homozygous for the GSTM1 null allele and 18% for GSTT1 null allele, which is consistent with previous reports [Seidegard et al. 1988]. Smoking was reported by 23% of NHW mothers and 4.6% of Hispanic mothers. There was no evidence for an association between in utero exposure to maternal smoking and GSTM1 or GSTT1 maternal or fetal phenotypes. In addition, there was no difference in the percentage of null mothers versus null fathers for either gene for either ethnic group. Lastly, FBAT-I did detect any evidence for an effect of smoking in any of SNPs in the other six genes.
DISCUSSION
An explanation for the development of clubfoot has been sought almost since the beginning of recorded history. The current hypothesis invokes both genes and environmental factors, although the exact roles of each still need to be defined refs. Candidate gene studies have begun to uncover the clubfoot etiologic pathways. For example, apoptotic genes, such as Casp8/10 and Casp3, as well as HOX genes have recently been associated with clubfoot heck[Ester et al. 2007; Ester et al. 2009]. Maternal smoking is the only common environmental exposure that has consistently been shown to increase the risk of clubfoot [Alderman et al. 1991; Honein et al. 2000; Skelly et al. 2002]. Importantly, the risk of clubfoot is significantly increased for women who smoke during pregnancy and have a positive family history [Honein et al. 2000]. These observations led us to hypothesize that genetic variation in smoking metabolism genes increases susceptibility to clubfoot. Therefore, we interrogated eight candidate genes, chosen for their involvement in cigarette smoke metabolism [Ambrosone et al. 1996; Omiecinski et al. 2000; Rodriguez-Antona and Ingelman-Sundberg 2006][Pasanen 1999; Seidegard et al. 1988].
We hypothesized that perturbation of phase I reactions of xenobiotic metabolism, that increase enzyme activity and adduct formation, play a role in the development of clubfoot. Compounds from cigarette smoke form DNA and protein adducts, which can cause mutagenesis and teratogenesis [Detmar et al. 2008; Kitamura and Kasai 2007; Nock et al. 2007]. Additionally, simultaneous increase in phase I activity with a perturbation in phase II activity may also play a role. Because phase I xenobiotic metabolism genes create harmful metabolic intermediates in the normal biotransformation pathway, it may be that an increase in phase I enzyme activity and a decrease in phase II degradation of these intermediates increases the concentration of harmful compounds to damaging levels that can interfere with fetal development. Some of the metabolic intermediates known to be produced by phase I xenobiotic metabolism are active oxygen species and DNA or protein binding adducts [Meyer 1996]. While it has been suggested that harmful oxygen species and adducts can interfere with normal fetal development, the role of these compounds in the pathogenesis of clubfoot is unknown [Izzotti et al. 2003; Pasanen 1999; Pinorini-Godly and Myers 1996]. Our results suggest that an increase in harmful metabolic intermediates could contribute to abnormal foot development or rotation of the foot.
Considering the strength of the association between clubfoot and smoking from previous population-based studies, there was surprisingly minimal evidence to support a role of these eight genes in clubfoot. The strongest evidence for association was for CYP1A1 (rs1048943; p=0.003) in the NHW dataset in the single SNP analysis. The is a missense mutation (1384A>G) in exon 7 that changes an isoleucine to a valine at amino acid position 462 (www.ncbi.nlm.nih.gov/) and confers higher phase I enzymatic activity, which may increase exposure to harmful, adduct forming, metabolic intermediates [Lamba et al. 2002; Schwarz et al. 2005]. Another CYP1A1-related SNP, rs1456432, gave minimal evidence of altered transmission in the Hispanic group. This SNP is located 9.1 kb downstream of CYP1A1 and therefore, may play a role in stabilization of the mRNA [Wickens et al. 2002].
One other significant finding in the NHW group was the interaction between rs1051740 in EPHX1 and rs1799929 in NAT2 (p=0.007) (Table 3). EPHX1 plays major role in hydrolysis of PAH, a phase I process [Hassett et al. 1994a; Omiecinski et al. 2000]. The rs1051740 variant has decreased activity and has been associated with decreased formation of DNA adducts [Georgiadis et al. 2004; Nock et al. 2007]. NAT2 plays an important role in phase II reactions of tobacco smoke metabolism [Ambrosone et al. 1996]. The rs1799929/NAT2 codes for a synonymous amino acid change in exon 2, which could decrease the rate of NAT2 translation [Nielsen et al. 2007]. Importantly, NAT2 is known to have variants with decreased activity, though the in vivo effect of the rs1799929 on enzyme activity is not known [Meyer and Zanger 1997].
Our results also identified other marginal associations that could potentially be important. Many of these SNPs are known or predicted to alter the function of the gene and may impact the efficiency of the xenobiotic metabolism pathway by perturbing the activity of phase I and/or phase II reactions. While these associations are marginal, they suggest that multiple variants are required to perturb different parts of the xenobiotic metabolism pathway, and thereby contribute to clubfoot. Additional studies are needed to further explore these potential interactions and confirm the associations.
One such association involves rs1799931 in NAT2, which had marginal evidence of association in the single SNP analyses only in NHW simplex cases. This SNP encodes a missense mutation that changes a glycine to a glutamine in the mature protein (www.ncbi.nlm.nih.gov/snp/) and is predicted to affect protein stability (SNPs3D.org). In vitro studies showing decreased activity in Chinese hamster ovary (CHO) cells and unstable protein formation in E. coli [Deguchi 1992; Hein et al. 1994; Meyer and Zanger 1997]. The activity of this SNP in vivo is not known. These results support our earlier finding of suggestive evidence for an association with rs1799931 in both NHW and Hispanic simplex families Hecht, et al., 2007. However, in that study, a more significant association was found for the Hispanic simplex families suggesting that this variant may be a greater risk factor in the Hispanic population. The current results suggest that variation in NAT2 may also be an important risk factor for the NHW population.
In addition to the significant interaction between rs1799929/EPHX1 and rs105740/NAT2, a marginal interaction was found between rs1051740 in EPHX1 and rs1801280 in NAT2, the latter of which causes a “slow acetylator” phenotype [Bell et al. 1993; Cascorbi et al. 1995; Meyer and Zanger 1997]. Interestingly, we observed a marginal interaction between rs1056836 in CYP1B1 and the same NAT2 SNPs seen in the interactions with EPHX1 (rs1799929 and rs1801280). EPHX1 and CYP1B1 interact through sequential reactions in phase I metabolism of PAH and NAT2 is a well-characterized phase II enzyme with variants that have been shown to cause an increase in harmful adduct levels [Grant et al. 1990; Meyer and Zanger 1997; Nock et al. 2007]. These results provide additional support for the findings by Hecht, et al., 2007 and provide additional evidence that NAT2 plays a role in clubfoot. Moreover, these results support our hypothesis that perturbation of phase I and phase II enzymatic activity may leads to an increased risk of clubfoot.
Other marginal but potentially interesting interactions involved CYP1B1, CYP2A6 and EPHX1. Because almost all of the marginal gene interactions involved one of these three genes, our results suggest that EPHX1 and CYP2A6 may play a role in clubfoot in the NHW group while CYP1B1 may be important in Hispanics. Additionally, the gene interaction results are intriguing when the functional effect of the SNPs and the possible effect on the metabolic pathway are considered. In general, we found interactions involving between variants that are known to alter the activity of the phase I enzyme or could affect regulation of the gene. Although the these findings need to be further explored, collectively these results support our hypothesis that disruption of normal activity in both phase I and II of xenobotic metabolism can increase the concentration of harmful intermediates and may play a role in clubfoot.
GSTM1 and GSTT1 also play a role in phase II of tobacco smoke metabolism and the null alleles of both genes cause absence of enzymatic activity [Pasanen 1999]. Loss of activity causes increased susceptibility to DNA adducts and could increase harmful metabolic intermediates [Pemble et al. 1994; Seidegard and Pero 1985; Seidegard et al. 1986; Seidegard et al. 1988; Wienckeet et al. 1990]. Interestingly, our results provide no support for a relationship between a homozygous null allele at either locus and an increased risk of clubfoot, even in the presence of smoking. However, these results must be carefully interpreted because heterozygotes cannot be discriminated from homozygous wild type individuals [Pemble et al. 1994; Seidegard et al. 1988]. Our results suggest that GSTM1 and GSTT1 null genotypes, whether they are present in the fetus or the mother, are not associated with an increased risk of clubfoot when the mother smokes during pregnancy.
Because both the maternal and fetal smoking metabolism genes may affect the risk for clubfoot, we evaluated whether each SNP possessed an independent maternal genetic effect or an independent inherited genetic effect in the fetus [van Den Oord and Vermunt 2000]. For CYP1A2, we found evidence for a significant deleterious effect for rs11854147 (p=0.03; RR=1.24; 95% CI: 1.04–1.44) in the mother and for rs2470890 (p=0.01; RR=1.33; 95% CI: 1.13–1.54) when in the fetus. This suggests that variation in CYP1A2 can increase the risk of clubfoot but the maternal or fetal mechanisms differ. Differences in the gene expression, induction and enzyme activity between maternal and fetal tobacco metabolism genes have been reported and these results support evidence that variants in tobacco metabolism genes have different consequences when they are of maternal and/or fetal origin [Legraverend et al. 1984; Pasanen 1999; Pavek and Dvorak 2008].
Interestingly, although an association between xenobiotic metabolism genes and clubfoot was found, we were unable to confirm the association between maternal smoking and clubfoot reported in other studies [Alderman et al. 1991; Dickinson et al. 2008; Honein et al. 2000; Skelly et al. 2002]. In our dataset, only 23% of NHW mothers and 4.6% of Hispanics mothers reported smoking during pregnancy as compared to 18.1% and 2.8%, respectively in the general population. In epidemiological studies that have found an association between maternal smoking and clubfoot, an average of 30% of mothers reported smoking during pregnancy [Dickinson et al. 2008; Honein et al. 2000; Skelly et al. 2002]. ?do any of these studies have a breakdown by ethnicity?? Because smoking does not appear to be as great a risk factor in our population, we would not expect to see strong associations between xenobiotic metabolism genes and clubfoot. Evaluation of the role of these genes in a dataset for which smoking is found to significantly increase the risk of clubfoot may reveal stronger associations between these genes and clubfoot and should be explored in future studies.
If tobacco smoke exposure is not affecting the fetus through the xenobiotic metabolism pathway, there may be other etiologic mechanisms. For example, early amniocentesis also increases the risk of clubfoot suggesting that a teratogenic mechanism may be common to both exposures. The simplest explanation would be vascular insufficiency and hypoxia that would deprive the fetus of blood flow and necessary nutrients. Maternal smoking and nicotine exposure in mice specifically reduces blood flow and increases vascular resistance in the uterus [Albuquerque et al. 2004; Bruner and Forouzan 1991; Clark and Irion 1992; Detmar et al. 2008; Shea and Steiner 2008]. Moreover, mice exposed to PAHs have been shown to have abnormal vasculature in the placenta that significantly reduces arterial surface area and volume of the fetal arterial vasculature [Detmar et al. 2008]. Reduction in vascular efficiency and an increased susceptibility to hypoxia could cause abnormal limb development and has been shown to cause transverse limb defects in prolonged (30–60 minutes) cases of anoxia [Webster and Abela 2007]. Therefore, variable degrees of hypoxia may increase the risk of other limb abnormalities, the mildest of which could be clubfoot.
The results of this study are important but must be carefully interpreted until validation studies can be undertaken. We focused on only eight of the most important tobacco smoke metabolism enzymes. There are likely hundreds of genes involved in the overall biotransformation of the compounds found in tobacco smoke, which were not considered in this study. While the genes in this study are the most likely candidates, we cannot rule out that other genes play a major role or interact with the genes in this study and contribute to the clubfoot phenotype. Our results suggest that there is an association between CYP1A1 and an interaction between EPHX1 and NAT2 xenobiotic metabolism genes and an increased risk for clubfoot. The genes in this study are likely to interact in pathways that affect fetal development. Further studies are needed to better delineate the role of maternal smoking and xenobiotic metabolism genes during pregnancy and the effects on the developing fetus.
Table 6.
Functional Effects for Smoking Metabolism Gene Interactionsa
| A Interactions between Phase I Genes | |||||||
| Gene 1 | SNP 1 | Functional effect | Gene 2 | SNP 2 | Functional effect | p value | Pop. |
| CYP1B1 | rs1056836 | Variant has increased metabolism of BaP and decreased metabolism of BaP-7,8-dihydrodiols; increased risk for adductsb–c | CYP2A6 | rs4105144 | 5′ of gene; may affect TFBSe | 0.02 | NHW |
| CYP1A2 | rs2472299 | 5′ of gene; may affect TFBSe | 0.06 | H | |||
| CYP1A1 | rs1456432 | 3′ of gene;possible enhancer region | CYP1A2 | rs2472299 | 5′ of gene; may affect TFBSe | 0.04 | H |
| EPHX1 | rs360063 | 3′ of gene;possible enhancer region | CYP2A6 | rs4105144 | 5′ of gene; may affect TFBSe | 0.04 | NHW |
| rs2234922 | Variant has decreased activity and decreased adductsb | CYP2A6 | rs7246742 | 3′ of gene; possible enhancer region | 0.06 | NHW | |
| rs1061740 | Variant has decreased activity and decreased adductsb | rs7250713 | Intronic | 0.06 | NHW | ||
| B. Interactions between Phase I and Phase II Genes | |||||||
| Phase I gene | SNP | Functional effect | Phase II gene | SNP 2 | Functional effect | p value | Pop. |
| EPHX1 | rs1061740 | Variant has decreased activity and decreased adductsb | NAT2 | rs1799929 | Activity unknown in vivod | 0.007 | NHW |
| NAT2 | rs1801280 | Decreased activity = “slow acetylator”d | 0.03 | NHW | |||
| CYP2A6 | rs4106144 | 5′ of gene; may effect TFBSe | NAT2 | rs1799930 | Decreased activity in vitrod | NAT2 | NHW |
| CYP1B1 | rs1056836 | Variant has increased metabolism of BaP and decreased metabolism of BaP-7,8-dihydrodiols; increased risk for adductsb–c | NAT2 | rs1799929 | Activity unknown in vivod | 0.04 | H |
| NAT2 | rs1801280 | Decreased activity = “slow acetylator”d | 0.04 | H | |||
| rs163084 | 3′ of gene; possible enhancer region | NAT2 | rs1799929 | Activity unknown in vovod | 0.05 | H | |
Published literature listed in References. TFBS = transcription factor binding site; variant = alternate allele.
AliBaba2,Patch, TESS.
REFERENCES
- Invitrogen, editor. Technical Resource Guide. Second ed. 2004. Drug Metabolizing Enzymes. [Google Scholar]
- Smoking During Pregnancy. Quick Reference: Fact Sheets: March of Dimes. 2008 [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cookson WO. GOLD--graphical overview of linkage disequilibrium. Bioinformatics. 2000;16(2):182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Adam MP, Chueh J, El-Sayed YY, Stenzel A, Vogel H, Weaver DD, Hoyme HE. Vascular-type disruptive defects in fetuses with homozygous alpha-thalassemia: report of two cases and review of the literature. Prenat Diagn. 2005;25(12):1088–1096. doi: 10.1002/pd.1276. [DOI] [PubMed] [Google Scholar]
- Albuquerque CA, Smith KR, Johnson C, Chao R, Harding R. Influence of maternal tobacco smoking during pregnancy on uterine, umbilical and fetal cerebral artery blood flows. Early Hum Dev. 2004;80(1):31–42. doi: 10.1016/j.earlhumdev.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Alderman BW, Takahashi ER, LeMier MK. Risk indicators for talipes equinovarus in Washington State, 1987–1989. Epidemiology. 1991;2(4):289–292. doi: 10.1097/00001648-199107000-00009. [DOI] [PubMed] [Google Scholar]
- Ambrosone CB, Freudenheim JL, Graham S, Marshall JR, Vena JE, Brasure JR, Michalek AM, Laughlin R, Nemoto T, Gillenwater KA, Shields PG. Cigarette smoking, N-acetyltransferase 2 genetic polymorphisms, and breast cancer risk. JAMA. 1996;276(18):1494–1501. [PubMed] [Google Scholar]
- Arand M, Muhlbauer R, Hengstler J, Jager E, Fuchs J, Winkler L, Oesch F. A multiplex polymerase chain reaction protocol for the simultaneous analysis of the glutathione S-transferase GSTM1 and GSTT1 polymorphisms. Anal Biochem. 1996;236(1):184–186. doi: 10.1006/abio.1996.0153. [DOI] [PubMed] [Google Scholar]
- Bakalis S, Sairam S, Homfray T, Harrington K, Nicolaides K, Thilaganathan B. Outcome of antenatally diagnosed talipes equinovarus in an unselected obstetric population. Ultrasound Obstet Gynecol. 2002;20(3):226–229. doi: 10.1046/j.1469-0705.2002.00780.x. [DOI] [PubMed] [Google Scholar]
- Bell DA, Taylor JA, Butler MA, Stephens EA, Wiest J, Brubaker LH, Kadlubar FF, Lucier GW. Genotype/phenotype discordance for human arylamine N-acetyltransferase (NAT2) reveals a new slow-acetylator allele common in African-Americans. Carcinogenesis. 1993;14(8):1689–1692. doi: 10.1093/carcin/14.8.1689. [DOI] [PubMed] [Google Scholar]
- Bolt HM, Thier R. Relevance of the deletion polymorphisms of the glutathione S-transferases GSTT1 and GSTM1 in pharmacology and toxicology. Curr Drug Metab. 2006;7(6):613–628. doi: 10.2174/138920006778017786. [DOI] [PubMed] [Google Scholar]
- Bruner JP, Forouzan I. Smoking and buccally administered nicotine. Acute effect on uterine and umbilical artery Doppler flow velocity waveforms. J Reprod Med. 1991;36(6):435–440. [PubMed] [Google Scholar]
- Byron-Scott R, Sharpe P, Hasler C, Cundy P, Hirte C, Chan A, Scott H, Baghurst P, Haan E. A South Australian population-based study of congenital talipes equinovarus. Paediatr Perinat Epidemiol. 2005;19(3):227–237. doi: 10.1111/j.1365-3016.2005.00647.x. [DOI] [PubMed] [Google Scholar]
- Cardy AH, Barker S, Chesney D, Sharp L, Maffulli N, Miedzybrodzka Z. Pedigree analysis and epidemiological features of idiopathic congenital talipes equinovarus in the United Kingdom: a case-control study. BMC Musculoskelet Disord. 2007;8:62. doi: 10.1186/1471-2474-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartlidge I. Observations on the epidemiology of club foot in Polynesian and Caucasian populations. J Med Genet. 1984;21(4):290–292. doi: 10.1136/jmg.21.4.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascorbi I, Drakoulis N, Brockmoller J, Maurer A, Sperling K, Roots I. Arylamine N-acetyltransferase (NAT2) mutations and their allelic linkage in unrelated Caucasian individuals: correlation with phenotypic activity. Am J Hum Genet. 1995;57(3):581–592. [PMC free article] [PubMed] [Google Scholar]
- CEMAT TCEaM-tATG. Randomised trial to assess safety and fetal outcome of early and midtrimester amniocentesis. Lancet. 1998;351(9098):242–247. [PubMed] [Google Scholar]
- Centini G, Rosignoli L, Kenanidis A, Scarinci R, Petraglia F. A report of early (13 + 0 to 14 + 6 weeks) and mid-trimester amniocenteses: 10 years' experience. J Matern Fetal Neonatal Med. 2003;14(2):113–117. doi: 10.1080/jmf.14.2.113.117. [DOI] [PubMed] [Google Scholar]
- Ching GH, Chung CS, Nemechek RW. Genetic and epidemiological studies of clubfoot in Hawaii: ascertainment and incidence. Am J Hum Genet. 1969;21(6):566–580. [PMC free article] [PubMed] [Google Scholar]
- Chung RH, Hauser ER, Martin ER. The APL test: extension to general nuclear families and haplotypes and examination of its robustness. Hum Hered. 2006;61(4):189–199. doi: 10.1159/000094774. [DOI] [PubMed] [Google Scholar]
- Clark KE, Irion GL. Fetal hemodynamic response to maternal intravenous nicotine administration. Am J Obstet Gynecol. 1992;167(6):1624–1631. doi: 10.1016/0002-9378(92)91752-v. [DOI] [PubMed] [Google Scholar]
- Collins P, Rosano GM, Sarrel PM, Ulrich L, Adamopoulos S, Beale CM, McNeill JG, Poole-Wilson PA. 17 beta-Estradiol attenuates acetylcholine-induced coronary arterial constriction in women but not men with coronary heart disease. Circulation. 1995;92(1):24–30. doi: 10.1161/01.cir.92.1.24. [DOI] [PubMed] [Google Scholar]
- Czekaj P, Wiaderkiewicz A, Florek E, Wiaderkiewicz R. Tobacco smoke-dependent changes in cytochrome P450 1A1, 1A2, and 2E1 protein expressions in fetuses, newborns, pregnant rats, and human placenta. Arch Toxicol. 2005;79(1):13–24. doi: 10.1007/s00204-004-0607-7. [DOI] [PubMed] [Google Scholar]
- Deguchi T. Sequences and expression of alleles of polymorphic arylamine N-acetyltransferase of human liver. J Biol Chem. 1992;267(25):18140–18147. [PubMed] [Google Scholar]
- Delisle MF, Wilson RD. First trimester prenatal diagnosis: amniocentesis. Semin Perinatol. 1999;23(5):414–423. doi: 10.1016/s0146-0005(99)80007-x. [DOI] [PubMed] [Google Scholar]
- Detmar J, Rennie MY, Whiteley KJ, Qu D, Taniuchi Y, Shang X, Casper RF, Adamson SL, Sled JG, Jurisicova A. Fetal growth restriction triggered by polycyclic aromatic hydrocarbons is associated with altered placental vasculature and AhR-dependent changes in cell death. Am J Physiol Endocrinol Metab. 2008;295(2):E519–E530. doi: 10.1152/ajpendo.90436.2008. [DOI] [PubMed] [Google Scholar]
- Dickinson KC, Meyer RE, Kotch J. Maternal smoking and the risk for clubfoot in infants. Birth Defects Res A Clin Mol Teratol. 2008;82(2):86–91. doi: 10.1002/bdra.20417. [DOI] [PubMed] [Google Scholar]
- Dunn PM. Congenital postural deformities: perinatal associations. Proc R Soc Med. 1972;65(8):735–738. [PMC free article] [PubMed] [Google Scholar]
- Engell V, Damborg F, Andersen M, Kyvik KO, Thomsen K. Club foot: a twin study. J Bone Joint Surg Br. 2006;88(3):374–376. doi: 10.1302/0301-620X.88B3.16685. [DOI] [PubMed] [Google Scholar]
- Ester AR, Tyerman G, Wise CA, Blanton SH, Hecht JT. Apoptotic gene analysis in idiopathic talipes equinovarus (clubfoot) Clin Orthop Relat Res. 2007;462:32–37. doi: 10.1097/BLO.0b013e318073c2d9. [DOI] [PubMed] [Google Scholar]
- Ester AR, Weymouth KS, Burt A, Wise CA, Scott A, Gurnett CA, Dobbs MB, Blanton SH, Hecht JT. Altered transmission of HOX and apoptotic SNPs identify a potential common pathway for clubfoot. Am J Med Genet A. 2009;149A(12):2745–2752. doi: 10.1002/ajmg.a.33130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis P, Demopoulos NA, Topinka J, Stephanou G, Stoikidou M, Bekyrou M, Katsouyianni K, Sram R, Autrup H, Kyrtopoulos SA. Impact of phase I or phase II enzyme polymorphisms on lymphocyte DNA adducts in subjects exposed to urban air pollution and environmental tobacco smoke. Toxicol Lett. 2004;149(1–3):269–280. doi: 10.1016/j.toxlet.2003.12.038. [DOI] [PubMed] [Google Scholar]
- Gladen BC, Zadorozhnaja TD, Chislovska N, Hryhorczuk DO, Kennicutt MC, 2nd, Little RE. Polycyclic aromatic hydrocarbons in placenta. Hum Exp Toxicol. 2000;19(11):597–603. doi: 10.1191/096032700671433928. [DOI] [PubMed] [Google Scholar]
- Grabe N. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. 2002;2(1):S1–S15. [PubMed] [Google Scholar]
- Grant DM, Morike K, Eichelbaum M, Meyer UA. Acetylation pharmacogenetics. The slow acetylator phenotype is caused by decreased or absent arylamine N-acetyltransferase in human liver. J Clin Invest. 1990;85(3):968–972. doi: 10.1172/JCI114527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurnett CA, Boehm S, Connolly A, Reimschisel T, Dobbs MB. Impact of congenital talipes equinovarus etiology on treatment outcomes. Dev Med Child Neurol. 2008;50(7):498–502. doi: 10.1111/j.1469-8749.2008.03016.x. [DOI] [PubMed] [Google Scholar]
- Habek D. Effects of smoking and fetal hypokinesia in early pregnancy. Arch Med Res. 2007;38(8):864–867. doi: 10.1016/j.arcmed.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Hancock DB, Martin ER, Li YJ, Scott WK. Methods for interaction analyses using family-based case-control data: conditional logistic regression versus generalized estimating equations. Genet Epidemiol. 2007;31(8):883–893. doi: 10.1002/gepi.20249. [DOI] [PubMed] [Google Scholar]
- Harper P. Practical Genetic Counseling. London: Hodder Arnold; 2004. [Google Scholar]
- Hassett C, Aicher L, Sidhu JS, Omiecinski CJ. Human microsomal epoxide hydrolase: genetic polymorphism and functional expression in vitro of amino acid variants. Hum Mol Genet. 1994a;3(3):421–428. doi: 10.1093/hmg/3.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett C, Robinson KB, Beck NB, Omiecinski CJ. The human microsomal epoxide hydrolase gene (EPHX1): complete nucleotide sequence and structural characterization. Genomics. 1994b;23(2):433–442. doi: 10.1006/geno.1994.1520. [DOI] [PubMed] [Google Scholar]
- Hecht JT, Ester A, Scott A, Wise CA, Iovannisci DM, Lammer EJ, Langlois PH, Blanton SH. NAT2 variation and idiopathic talipes equinovarus (clubfoot) Am J Med Genet A. 2007;143A(19):2285–2291. doi: 10.1002/ajmg.a.31927. [DOI] [PubMed] [Google Scholar]
- Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91(14):1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- Hein DW, Ferguson RJ, Doll MA, Rustan TD, Gray K. Molecular genetics of human polymorphic N-acetyltransferase: enzymatic analysis of 15 recombinant wild-type, mutant, and chimeric NAT2 allozymes. Hum Mol Genet. 1994;3(5):729–734. doi: 10.1093/hmg/3.5.729. [DOI] [PubMed] [Google Scholar]
- Heron M, Sutton PD, Xu J, Ventura SJ, Strobino DM, Guyer B. Annual summary of vital statistics. Pediatrics. 2007;125(1):4–15. doi: 10.1542/peds.2009-2416. [DOI] [PubMed] [Google Scholar]
- Hoffmann D, Hoffmann I. The changing cigarette, 1950–1995. J Toxicol Environ Health. 1997;50(4):307–364. doi: 10.1080/009841097160393. [DOI] [PubMed] [Google Scholar]
- Honein MA, Paulozzi LJ, Moore CA. Family history, maternal smoking, and clubfoot: an indication of a gene-environment interaction. Am J Epidemiol. 2000;152(7):658–665. doi: 10.1093/aje/152.7.658. [DOI] [PubMed] [Google Scholar]
- Hootnick DR, Levinsohn EM, Crider RJ, Packard DS., Jr Congenital arterial malformations associated with clubfoot. A report of two cases. Clin Orthop Relat Res. 1982;(167):160–163. [PubMed] [Google Scholar]
- Idelberger KH. Die Ergenbnisse der Zwillingsforschung beim angeborenen Klumpfuss. Verh Dtsch Orthop Ges. 1939;(33):272–276. [Google Scholar]
- Izzotti A, Balansky RM, Cartiglia C, Camoirano A, Longobardi M, De Flora S. Genomic and transcriptional alterations in mouse fetus liver after transplacental exposure to cigarette smoke. FASEB J. 2003;17(9):1127–1129. doi: 10.1096/fj.02-0967fje. [DOI] [PubMed] [Google Scholar]
- Kitamura M, Kasai A. Cigarette smoke as a trigger for the dioxin receptor-mediated signaling pathway. Cancer Lett. 2007;252(2):184–194. doi: 10.1016/j.canlet.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54(10):1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- Legraverend C, Guenthner TM, Nebert DW. Importance of the route of administration for genetic differences in benzo[a]pyrene-induced in utero toxicity and teratogenicity. Teratology. 1984;29(1):35–47. doi: 10.1002/tera.1420290106. [DOI] [PubMed] [Google Scholar]
- Lochmiller C, Johnston D, Scott A, Risman M, Hecht JT. Genetic epidemiology study of idiopathic talipes equinovarus. Am J Med Genet. 1998;79(2):90–96. [PubMed] [Google Scholar]
- Lopachin RM, Decaprio AP. Protein adduct formation as a molecular mechanism in neurotoxicity. Toxicol Sci. 2005;86(2):214–225. doi: 10.1093/toxsci/kfi197. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Gilbert JR, Pericak-Vance MA, Hauser ER. Genotype-based association test for general pedigrees: the genotype-PDT. Genet Epidemiol. 2003;25(3):203–213. doi: 10.1002/gepi.10258. [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67(1):146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews TJ. Smoking during pregnancy in the 1990s. Natl Vital Stat Rep. 2001;49(7):1–14. [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MJ, Bechtold WE. Protein adduct biomarkers: state of the art. Environ Health Perspect. 1996;104(Suppl 5):879–882. doi: 10.1289/ehp.96104s5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer UA. Overview of enzymes of drug metabolism. J Pharmacokinet Biopharm. 1996;24(5):449–459. doi: 10.1007/BF02353473. [DOI] [PubMed] [Google Scholar]
- Meyer UA, Zanger UM. Molecular mechanisms of genetic polymorphisms of drug metabolism. Annu Rev Pharmacol Toxicol. 1997;37:269–296. doi: 10.1146/annurev.pharmtox.37.1.269. [DOI] [PubMed] [Google Scholar]
- Moorthi RN, Hashmi SS, Langois P, Canfield M, Waller DK, Hecht JT. Idiopathic talipes equinovarus (ITEV) (clubfeet) in Texas. Am J Med Genet A. 2005;132(4):376–380. doi: 10.1002/ajmg.a.30505. [DOI] [PubMed] [Google Scholar]
- Nielsen KB, Sorensen S, Cartegni L, Corydon TJ, Doktor TK, Schroeder LD, Reinert LS, Elpeleg O, Krainer AR, Gregersen N, Kjems J, Andresen BS. Seemingly neutral polymorphic variants may confer immunity to splicing-inactivating mutations: a synonymous SNP in exon 5 of MCAD protects from deleterious mutations in a flanking exonic splicing enhancer. Am J Hum Genet. 2007;80(3):416–432. doi: 10.1086/511992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nock NL, Tang D, Rundle A, Neslund-Dudas C, Savera AT, Bock CH, Monaghan KG, Koprowski A, Mitrache N, Yang JJ, Rybicki BA. Associations between smoking, polymorphisms in polycyclic aromatic hydrocarbon (PAH) metabolism and conjugation genes and PAH-DNA adducts in prostate tumors differ by race. Cancer Epidemiol Biomarkers Prev. 2007;16(6):1236–1245. doi: 10.1158/1055-9965.EPI-06-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omiecinski CJ, Hassett C, Hosagrahara V. Epoxide hydrolase--polymorphism and role in toxicology. Toxicol Lett. 2000;112–113:365–370. doi: 10.1016/s0378-4274(99)00235-0. [DOI] [PubMed] [Google Scholar]
- Pasanen M. The expression and regulation of drug metabolism in human placenta. Adv Drug Deliv Rev. 1999;38(1):81–97. doi: 10.1016/s0169-409x(99)00008-3. [DOI] [PubMed] [Google Scholar]
- Pavek P, Dvorak Z. Xenobiotic-induced transcriptional regulation of xenobiotic metabolizing enzymes of the cytochrome P450 superfamily in human extrahepatic tissues. Curr Drug Metab. 2008;9(2):129–143. doi: 10.2174/138920008783571774. [DOI] [PubMed] [Google Scholar]
- Pemble S, Schroeder KR, Spencer SR, Meyer DJ, Hallier E, Bolt HM, Ketterer B, Taylor JB. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994;300(Pt 1):271–276. doi: 10.1042/bj3000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinorini-Godly MT, Myers SR. HPLC and GC/MS determination of 4-aminobiphenyl haemoglobin adducts in fetuses exposed to the tobacco smoke carcinogen in utero. Toxicology. 1996;107(3):209–217. doi: 10.1016/0300-483x(95)03263-f. [DOI] [PubMed] [Google Scholar]
- Reimann I. Congenital idiopathic club foot with special reference to aetiology, pathogenesis, and possibilities of correction within the first years of life. Vol. 223. Munksgaard: Køobenhavn; 1967. p. p. [Google Scholar]
- Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 pharmacogenetics and cancer. Oncogene. 2006;25(11):1679–1691. doi: 10.1038/sj.onc.1209377. [DOI] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. In: Baxevanis A, editor. Current protocols in bioinformatics. Hobokem, NJ: John Wiley and Sons, Inc.; 2003. pp. 2.6.1–2.6.15. [DOI] [PubMed] [Google Scholar]
- Schwarz D, Kisselev P, Chernogolov A, Schunck WH, Roots I. Human CYP1A1 variants lead to differential eicosapentaenoic acid metabolite patterns. Biochem Biophys Res Commun. 2005;336(3):779–783. doi: 10.1016/j.bbrc.2005.08.172. [DOI] [PubMed] [Google Scholar]
- Seidegard J, Pero RW. The hereditary transmission of high glutathione transferase activity towards trans-stilbene oxide in human mononuclear leukocytes. Hum Genet. 1985;69(1):66–68. doi: 10.1007/BF00295531. [DOI] [PubMed] [Google Scholar]
- Seidegard J, Pero RW, Miller DG, Beattie EJ. A glutathione transferase in human leukocytes as a marker for the susceptibility to lung cancer. Carcinogenesis. 1986;7(5):751–753. doi: 10.1093/carcin/7.5.751. [DOI] [PubMed] [Google Scholar]
- Seidegard J, Vorachek WR, Pero RW, Pearson WR. Hereditary differences in the expression of the human glutathione transferase active on trans-stilbene oxide are due to a gene deletion. Proc Natl Acad Sci U S A. 1988;85(19):7293–7297. doi: 10.1073/pnas.85.19.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea AK, Steiner M. Cigarette smoking during pregnancy. Nicotine Tob Res. 2008;10(2):267–278. doi: 10.1080/14622200701825908. [DOI] [PubMed] [Google Scholar]
- Shugart L, Matsunami R. Adduct formation in hemoglobin of the newborn mouse exposed in utero to benzo[a]pyrene. Toxicology. 1985;37(3–4):241–245. doi: 10.1016/0300-483x(85)90087-3. [DOI] [PubMed] [Google Scholar]
- Skelly AC, Holt VL, Mosca VS, Alderman BW. Talipes equinovarus and maternal smoking: a population-based case-control study in Washington state. Teratology. 2002;66(2):91–100. doi: 10.1002/tera.10071. [DOI] [PubMed] [Google Scholar]
- Teng S, Michonova-Alexova E, Alexov E. Approaches and resources for prediction of the effects of non-synonymous single nucleotide polymorphism on protein function and interactions. Curr Pharm Biotechnol. 2008;9(2):123–133. doi: 10.2174/138920108783955164. [DOI] [PubMed] [Google Scholar]
- Tredwell SJ, Wilson D, Wilmink MA. Review of the effect of early amniocentesis on foot deformity in the neonate. J Pediatr Orthop. 2001;21(5):636–641. [PubMed] [Google Scholar]
- Turco VJ. Clubfoot. Vol. 193. New York: Churchill Livingstone; 1981. p. xiii. p. p. [Google Scholar]
- Uchida K, Stadtman ER. Quantitation of 4-hydroxynonenal protein adducts. Methods Mol Biol. 2000;99:25–34. doi: 10.1385/1-59259-054-3:25. [DOI] [PubMed] [Google Scholar]
- van Den Oord EJ, Vermunt JK. Testing for linkage disequilibrium, maternal effects, and imprinting with (In)complete case-parent triads, by use of the computer program LEM. Am J Hum Genet. 2000;66(1):335–338. doi: 10.1086/302708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet D, Voet JG. Biochemistry. John Wiley & Sons, Inc.; 2004. [DOI] [PubMed] [Google Scholar]
- Webster WS, Abela D. The effect of hypoxia in development. Birth Defects Res C Embryo Today. 2007;81(3):215–228. doi: 10.1002/bdrc.20102. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 2002;18(3):150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wiencke JK, Kelsey KT, Lamela RA, Toscano WA., Jr Human glutathione S-transferase deficiency as a marker of susceptibility to epoxide-induced cytogenetic damage. Cancer Res. 1990;50(5):1585–1590. [PubMed] [Google Scholar]
- Wynne-Davies R. Family Studies and the Cause of Congenital Club Foot. Talipes Equinovarus, Talipes Calcaneo-Valgus and Metatarsus Varus. J Bone Joint Surg Br. 1964;46:445–463. [PubMed] [Google Scholar]
- Wynne-Davies R. Family studies and aetiology of club foot. J Med Genet. 1965;2(4):227–232. doi: 10.1136/jmg.2.4.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H, de Crombrugghe B. Joint formation requires muscle formation and contraction. Dev Cell. 2009;16(5):625–626. doi: 10.1016/j.devcel.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SF, Liu JP, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41(2):89–295. doi: 10.1080/03602530902843483. [DOI] [PubMed] [Google Scholar]