Abstract
Eugenol is an aromatic component of clove oil that has therapeutic potential as an antifungal drug, although its mode of action and precise cellular target(s) remain ambiguous. To address this knowledge gap, a chemical-genetic profile analysis of eugenol was done using ∼4700 haploid Saccharomyces cerevisiae gene deletion mutants to reveal 21 deletion mutants with the greatest degree of susceptibility. Cellular roles of deleted genes in the most susceptible mutants indicate that the main targets for eugenol include pathways involved in biosynthesis and transport of aromatic and branched-chain amino acids. Follow-up analyses showed inhibitory effects of eugenol on amino acid permeases in the yeast cytoplasmic membrane. Furthermore, phenotypic suppression analysis revealed that eugenol interferes with two permeases, Tat1p and Gap1p, which are both involved in dual transport of aromatic and branched-chain amino acids through the yeast cytoplasmic membrane. Perturbation of cytoplasmic permeases represents a novel antifungal target and may explain previous observations that exposure to eugenol results in leakage of cell contents. Eugenol exposure may also contribute to amino acid starvation and thus holds promise as an anticancer therapeutic drug. Finally, this study provides further evidence of the usefulness of the yeast Gene Deletion Array approach in uncovering the mode of action of natural health products.
Introduction
Infectious diseases are responsible for more than 17 million deaths each year worldwide [1]. Among other things, this is due to a lack of resources to combat diseases in impoverished regions and an increasing incidence of microbial resistance to existing antibiotics [2]. Phytomedicines are broadly accessible and could also provide a solution to the problem of drug resistance, as plant secondary metabolites may inhibit microbial growth by different mechanisms than the presently used antibiotics [3]. Syzygium aromaticum is a well-known aromatic plant species that is widely cultivated in Asian and African countries [4]. It is reported that the buds of S. aromaticum (cloves) are used in folk medicine as diuretic, odontalgic, stomachic, tonicardiac, and as a condiment with carminative and stimulant activity [5]. Clove essential oils have been described as having useful antiseptic and analgesic effects and are frequently used in dental medicine [6] and for sedating fish for research purposes. Several studies have also shown that clove oil has strong antibacterial, antifungal, antiviral and antioxidant activities. Many of these activities are believed to be due to the main biologically active phenolic component of clove essential oils, eugenol [6]–[10].
Despite many reports on the antimicrobial properties of the essential oils, including eugenol, found in most Syzygium species (e.g.s [6], [10]), the antifungal mode of action of these compounds was only investigated by in vitro pharmacological assays, and the precise cellular target of eugenol in fungi remains unclear. The leading view is that phenolic compounds such as eugenol disrupt the cytoplasmic membrane and result in cell leakage [10]–[12], and that a subsequent Ca2+ influx may serve a protective function against eugenol [13]. Considering that eugenol-rich essential oils are gaining increasing importance for their pronounced antimicrobial activity, the objective of our present research was to evaluate the antifungal activity and mechanism of action of eugenol using cell-based phenotypic screens in the yeast Saccharomyces cerevisiae.
Materials and Methods
Growth Media and Compounds
Standard rich (YPD) and synthetic complete (SC) media were used for the experiments [14]. S. cerevisiae cells were grown at 30°C for 1–2 days. The YPD medium containing Geneticin (G418; 200 µg/ml) was used for the maintenance of deletion strains carrying the G418R selectable marker. G418 and eugenol were purchased from Sigma-Aldrich (Oakville, ON, Canada). Ethanolic root extracts of Echinacea purpurea were used as a positive control in membrane disruption assays and prepared as described previously [15].
Antifungal Activity
S. cerevisiae (S288C) was used in antifungal activity assays. Minimum Inhibitory Concentration (MIC) for eugenol was measured using the broth microdilution assay protocol [16]. A three-fold serial dilution of eugenol (concentration range of 21.4 to 1.7×10−7 mg/ml) was added to sterile 96-well microtitre plates. Plates were incubated at 30°C for 1–2 days. Inhibition of growth was visually compared with control wells containing no eugenol.
Gene Deletion Array (GDA) Analysis
For high throughput phenotypic screenings, approximately 4700 MATa haploid gene deletion strains of S. cerevisiae derived from BY4741 (MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0) were maintained in an ordered array of approximately 384 individual strains in each of 16 plates [17]. YPD agar plates without (control), and with a subinhibitory concentration of eugenol (0.18 mg/ml, experimental), were inoculated by hand-pinning sets of 384 mutant strains per plate using a floating pin replicator as previously described [18]. After 1–2 days incubation at 30°C, digital images of the plates were captured and analyzed using Growth Detector software [19]. The relative size of colonies was used as a measure for growth differences under experimental and control conditions. Each experiment was repeated three times. Colonies that demonstrated 70% or more reduction in size in at least two replicates were classified as supersensitive (i.e. highly susceptible mutants). Deletion strains for genes associated with multidrug resistance were omitted from the list of supersensitives as they provide little information about the specific mode of activity of antifungal. Gene ontology annotation analysis was done with online software (gprofiler, http://biit.cs.ut.ee/gprofiler/; Profcom, http://webclu.bio.wzw.tum.de/profcom/; GeneMANIA, http://www.genemania.org/) and Saccharomyces Genome Database [20] was used for functional profiling of highly susceptible mutants in our large-scale experiment.
Spot Test Analysis
Sensitivity of selected mutant strains identified in the GDA screens were confirmed by spot test analyses. Yeast cells were grown in YPD liquid medium to mid-log phase and 10-fold serially diluted. From each dilution, 15 µl was spotted on medium containing subinhibitory concentrations of eugenol (0.18 mg/ml), or without eugenol (control). The growth patterns were compared after 2 days at 30°C. Each experiment was repeated a minimum of three times.
Liposome Leakage Assay
Carboxyfluorescein (CF, Life Technologies Inc., Burlington ON, Canada) was encapsulated in large unilamellar vesicles (LUVs) made of total yeast lipids (Avanti Polar Lipids, Alabaster AL, USA) in the quenched state for use in this experiment as described previously [21]. A black opaque microplate containing 190 µl of the optimal dilution of the LUVs suspension in each well had its fluorescent emissions measured with a FLUOstar microplate reader (OPTIMA BMG LABTECH Inc., Durham NC, USA) prior to addition of any of the test compounds to establish the fluorescence at time zero (F0). A row in the same plate was reserved for 190 µl per well of the HEPES buffer as a control to test for compound autofluorescence. Eugenol was two-fold serially diluted across a clear 96-well microplate in HEPES buffer and 10 µl from each well of the clear microplate was transferred to the well of a black opaque 96-well microplate. The final concentrations of the eugenol across the black opaque microplate were 1.6 to 0.003 mg/ml. Similarly, a dilution of Echinacea purpurea ethanol extract was used as a positive lysis control and the carrier solvent was used as the negative control. Following 1 h incubation at room temperature in the dark, the fluorescence units of each well was measured to determine Ft. Immediately after, 20 µl of a 10% solution of Triton X-100 was added to each well causing 100% release and dequenching of CF. The microplate was read again after ∼10 min to determine the final fluorescence measurement F100 (100% fluorescence after addition of Triton X-100). The percentage of leakage for each point was calculated as: % Leakage = {(Ft – F0)/(F100 – F0)}×100.
β-galactosidase Expression Assay
This assay used an inducible β-galactosidase reporter gene in p416 as described previously [22]. Briefly, cells of BY4741 harboring p416GAL1-lacZ were incubated in SC-ura medium containing 2% galactose with subinhibitory amounts (0.21 and 0.27 mg/ml) of eugenol and without eugenol (negative control). Cycloheximide (1 µg/ml, Sigma) was used as a positive control for translation inhibition. After 20 h at 30°C, yeast cells were harvested by centrifugation, cell density was measured at OD600 and β-galactosidase activity was measured as described in Miller et al. [23].
Auxotroph Supplement Assay
All of the haploid gene deletion strains of S. cerevisiae derived from BY4741 (MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0) that were available in our GDA library and involved in tryptophan, phenylalanine, tyrosine or isoleucine biosynthesis pathways were grown in YPD liquid medium to mid-log phase and 10-fold serially diluted. From each dilution, 15 µl of each strain was spotted onto experimental (containing a subinhibitory 0.18 mg/ml concentration of eugenol) and control (no eugenol) plates of synthetic medium supplemented with four targeted amino acids (tryptophan, phenylalanine, tyrosine, isoleucine) together with uracil, leucine, histidine, methionine (that were essential for BY4741 background strain growth). The growth patterns were compared after 2 days at 30°C.
Phenotypic Suppression Analysis
Overexpression constructs of four permeases that are either general (GAP1) or specific (TAT1, TAT2 and BAP2) transporters for aromatic and branched-chain amino acids were obtained from the yeast gene overexpression array [24]. Suppression analysis was performed as described by Alamgir et al. [25]. Overexpression plasmids were transformed into the aro1Δ strain by the lithium acetate method [26]. Transformants were grown overnight in SC-ura medium, adjusted to OD600 ∼1.0 and diluted 1∶300 before 100 µl aliquots of the diluted broth cultures were added to each well of a sterile microtiter plate. Sensitivity to 0.18 mg/ml eugenol by the transformants containing the overexpression constructs was compared to that with a control plasmid after 2 days growth at 30°C by measuring the optical density (OD600) of cells in each well with a FLUOstar microplate reader (OPTIMA BMG LABTECH Inc., Durham NC, USA).
Statistical Analyses
Statistical significance in the data sets was assessed by Student T-test using Microsoft Excel 2007 (Microsoft Corporation, USA). The difference was considered to be statistically significant when P-value≤0.05.
Results and Discussion
Antifungal Activity of Eugenol
We determined that the minimum inhibitory concentration (MIC100) for eugenol was in the range of 0.27–0.32 mg/ml. This MIC100 was defined as the lowest concentration that resulted in complete inhibition of visible growth of S. cerevisiae strain S288C after 2 days using a broth microdilution assay [16].
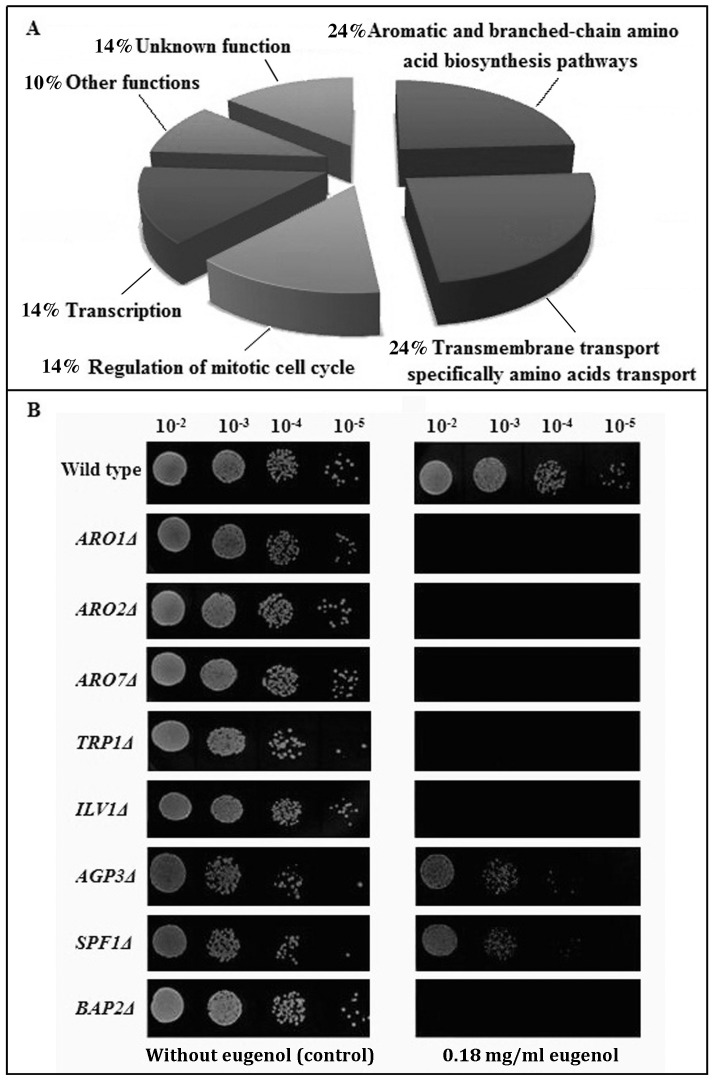
GDA shows Eugenol Interacts with Aromatic and Branched-chain Amino Acid Pathway(s)
A range of subinhibitory concentrations (0.16–0.21 mg/ml) was initially tested using a representative plate of yeast deletion strains to determine that a concentration of 0.18 mg/ml gave approximately 1% supersensitives. The entire haploid yeast gene deletion array (yGDA) was then replica plated onto medium with this subinhibitory concentration of eugenol and subsequent colony size measurements revealed that 21 deletion mutants were super-sensitive to eugenol (Table 1). The deleted genes are not normally required for growth under laboratory conditions, and slow growth by these supersensitive deletion strains is likely a result of a chemical-genetic interaction. The genes deleted in supersensitive strains were clustered based on the cellular processes in which they participate (Figure 1A). Of the 21 most sensitive gene deletions, 48% have known roles in aromatic and branched-chain amino acid biosynthesis and transport pathways (P-value: 8.6×10−5). These genes include ARO1, ARO2, ARO7, TRP1, ILV1 which are involved in aromatic and branched-chain amino acid biosynthesis pathways (24%) and BAP2, AGP3, SLM4, SPF1, WSC4 which are linked to transmembrane transport and/or linked to amino acid transport (24%) through the cytoplasmic membrane. Specifically, BAP2 and AGP3 encode amino acid permeases [27], [28]. Slm4p is a component of the GSE complex that is required for proper sorting of amino acid permease Gap1p from the late endosome to the plasma membrane [29]. Spf1p, a P-type ATPase that may provide an electrochemical gradient across the plasma membrane needed for active amino acid transport, has physical interactions with amino acid permeases such as Bap2p and Gap1p, as well as Dip5p and Can1p that are dicarboxylic amino acid and arginine permeases, respectively [30]. Finally, Wsc4p interacts genetically with amino acid transporter, Tat1p, and is both co-expressed and co-localized with Gap1p [31].
Table 1. List of eugenol-sensitive gene deletion mutants from GDA analysis that showed greater than 70% reduction in colony size.
| Gene function | Systematic Name | Standard Name | Average % colony size reduction |
| Aromatic and branched-chain amino acid biosynthesis pathways | |||
| YDR127W | ARO1 | 95.1 | |
| YGL148W | ARO2 | 97.6 | |
| YPR060C | ARO7 | 91.6 | |
| YDR007W | TRP1 | 93.1 | |
| YER086W | ILV1 | 96.8 | |
| Transmembrane transport specifically amino acids transport | |||
| YBR068C | BAP2 | 94.3 | |
| YFL055W | AGP3 | 71.2 | |
| YBR077C | SLM4 | 90.7 | |
| YEL031W | SPF1 | 70.7 | |
| YHL028W | WSC4 | 77.1 | |
| Regulation of mitotic cell cycle | |||
| YOR026W | BUB3 | 86.3 | |
| YNL273W | TOF1 | 75.5 | |
| YGL173C | KEM1 | 84.8 | |
| Transcription | |||
| YBR245C | ISW1 | 91.6 | |
| YHL025W | SNF6 | 76.1 | |
| YPL042C | SSN3 | 72.0 | |
| Other functions | |||
| YDR312W | SSF2 | 82.1 | |
| YMR202W | ERG2 | 95.3 | |
| Unknown function | |||
| YDR442W | YDR442W | 88.5 | |
| YMR326C | YMR326C | 78.9 | |
| YPR087W | VPS69 | 78.2 | |
Figure 1. Nearly half of eugenol-sensitive strains have deletions in genes involved in aromatic and branched-chain amino acid synthesis or uptake.
(A) The haploid non-essential yeast gene deletion array was subjected to a subinhibitory concentration of eugenol. Colony size reduction was used to detect sensitivity. The mutants most sensitive to eugenol were clustered according to the cellular processes in which their deleted genes participated. (B) Eugenol-sensitive strains identified by GDA were verified by drop out plates. Wild type and eight randomly selected gene deletion mutant strains that were eugenol-sensitive based on GDA analysis were 10-fold serially diluted and spotted on solid YPD medium with a subinhibitory concentration (0.18 mg/ml) of eugenol or without eugenol (control). The plates were incubated at 30°C for 1–2 days and then photographed. All deletion mutants selected exhibited increased sensitivity to eugenol, providing verification of the GDA analysis.
Among the next largest clusters, approximately 14%, 14% and 10% of sensitive strains are associated with regulation of mitotic cell cycle, transcription, and other functions, respectively (excluding genes with unknown functions). These smaller clusters in the profile could represent additional target sites (side effects) of eugenol on yeast cells.
Spot Test Analysis Verifies GDA
To investigate the accuracy of our large-scale approach for identifying eugenol-sensitive mutants, eight deletion strains that were supersensitive to eugenol based on the GDA assay were randomly selected and subjected to spot test analyses (Figure 1B). These spot test assays confirmed that deletion of these genes confers increased sensitivity to eugenol, and verified the large-scale screen based on the GDA approach.
Hypotheses on the Antifungal Mode of Action of Eugenol
Existing reports on the antimicrobial activity propose that phenolic compounds, such as eugenol and its analogs, compromise the structural and functional integrity of the cytoplasmic membrane and thus cause cell leakage [10]–[12]. To test this possibility, we examined the effect of eugenol on a model of the S. cerevisiae cytoplasmic membrane by monitoring the release of carboxyfluorescein (CF) from large unilamellar vesicles (LUVs). As seen in Figure 2A, only ∼12.5% leakage occurred at the eugenol MIC100 (0.27 mg/ml), and liposomes were only slightly destabilized even at concentrations of 1.6 mg/ml (6 times the eugenol MIC100). In contrast, Echinacea purpurea extract (positive control) caused 100% leakage at a concentration that corresponds to 0.04 times the yeast MIC100 value. This indicates that eugenol’s antifungal mode of action is not likely related to membrane disruption per se as suggested previously [11].
Figure 2. Eugenol does not induce leakage of liposomes made of total yeast lipids or inhibit protein synthesis.
(A) Release of carboxyfluorescein from large unilamellar vesicles (LUVs) over a series of eugenol concentrations (0.003 to 1.6 mg/ml, bottom axis) was compared to 100% release from liposome exposed to Triton-X 100. Echinacea purpurea extract was used as a positive lysis control over a concentration gradient of 0.01 to 5% (top axis). Data correspond to the mean % leakage values (±SD) of three independent experiments. (B) Yeast exposed to subinhibitory (0.21 mg/ml) or inhibitory (0.27 mg/ml) concentrations of eugenol do not have significantly decreased β-galactosidase activity in comparison to the untreated control. In contrast, the inhibitor of protein translation, cycloheximide, significantly reduced β-galactosidase activity in the assay. These observations indicate that eugenol does not reduce efficiency of translation in yeast as would be expected for compounds that perturb the intracellular pool of amino acids. The values are expressed as mean ±SD of triplicates, difference between treatment and untreated control are indicated as p<0.05 (*) and p<0.01 (**).
According to our GDA analysis, we can hypothesize two additional antifungal modes of action: (i) Eugenol may interfere with factors that are involved in aromatic and branched-chain amino acid biosynthesis pathways, and thus is expected to reduce the internal pool of these amino acids and, as a consequence, efficiency of translation in yeast cells. (ii) Eugenol may interfere with transporters, particularly aromatic and branched-chain amino acids permeases, in the cytoplasmic membrane of yeast cells. We therefore designed secondary assays to test these two hypotheses.
Eugenol does not Significantly Reduce Translation Efficiency
According to the first hypothesis, the efficiency of translation in yeast cells would decrease in the presence of eugenol relative to the untreated control. To investigate this possibility, we used an inducible β-galactosidase reporter construct and found that the addition of subinhibitory (0.21 mg/ml) or inhibitory (0.27 mg/ml) concentrations of eugenol to yeast cells did not significantly decrease β-galactosidase activity in comparison to the untreated control (P≥0.1, Figure 2B). In contrast, 1 µg/ml of cycloheximide, a known inhibitor of protein translation, significantly reduced β-galactosidase activity in the assay. These observations do not support the first hypothesis, that eugenol interferes with factors involved in aromatic and branched-chain amino acid biosynthesis, since any perturbation by eugenol of the amino acid pool would likely reduce the efficiency of translation in yeast.
Eugenol Blocks Uptake of Aromatic and Branched-chain Amino Acids
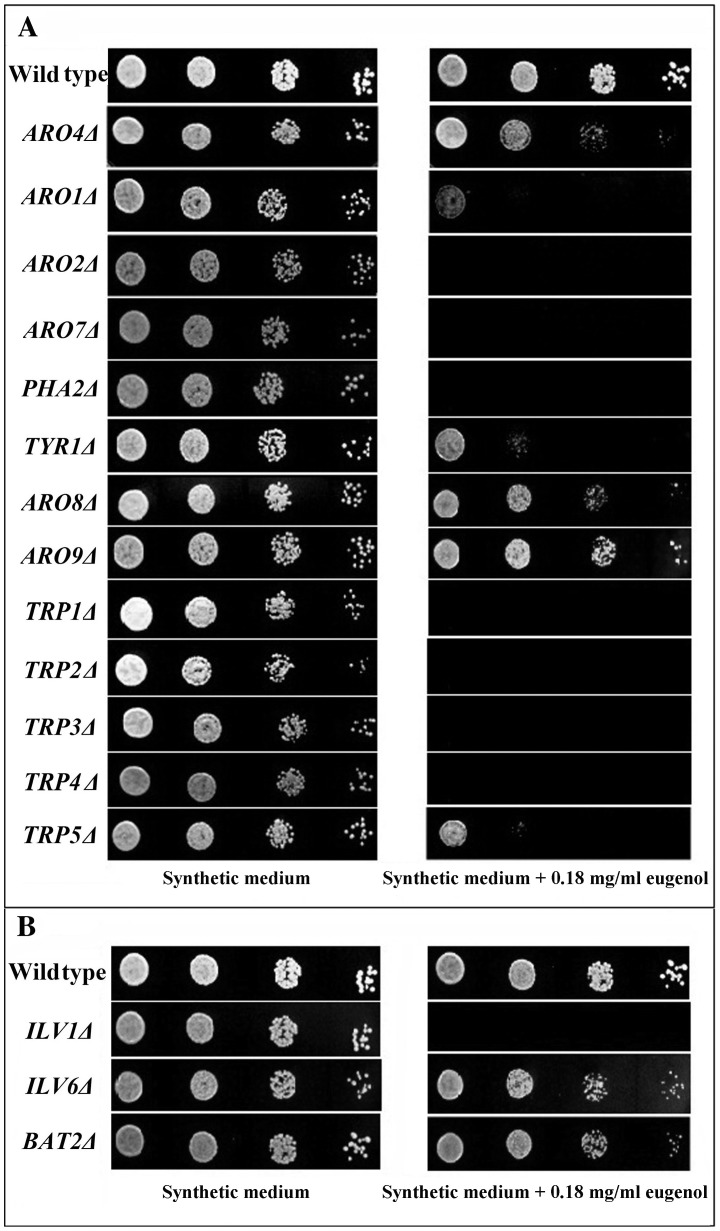
To test our second hypothesis, that eugenol interferes with specific amino acid transporters, we used an auxotroph supplement assay. The basis of this assay is as follows. A yeast strain that carries a mutation in a gene encoding an enzyme in a tryptophan, phenylalanine, tyrosine and/or isoleucine biosynthesis pathway will only be able to grow if these amino acids are provided in the culture medium. In contrast, auxotrophic strains are expected to grow poorly on synthetic medium supplemented with these amino acids if eugenol is present and interferes with aromatic and branched-chain amino acids permeases. Using the auxotroph supplement assay we examined the 13 strains shown in Figure 3A that carry mutations in aromatic amino acid biosynthesis genes. Of these 13 strains, only three (aro4Δ, aro8Δ and aro9Δ) grew well on a synthetic medium supplemented with aromatic amino acids that contained a subinhibitory concentration of eugenol (0.18 mg/ml). The remaining ten strains grew well on aromatic amino acid-supplemented medium, but grew poorly or not at all when eugenol was included in the medium. It has been reported that neither of the ARO8 or ARO9 single mutants display any nutritional requirements on minimal ammonia medium whereas the ARO8 and ARO9 double mutant is auxotrophic for both phenylalanine and tyrosine [32]. Similarly, there is no evidence that the ARO4 single mutant displays an auxotrophic phenotype for aromatic amino acids [20]. In addition, as seen in Figure 3B, among the three strains with mutations in the branched-chain amino acid (isoleucine) biosynthetic pathway (ilv1Δ, ilv6Δ and bat2Δ were available in our GDA library), only the ilv1Δ strain could not grow on synthetic medium supplemented with isoleucine in the presence of 0.18 mg/ml eugenol. Kispal et al. [33] reported that on glucose-containing media, a single deletion of either of the two BAT genes (BAT1 or BAT2) does not impair cell growth, but deletion of both genes results in branched-chain amino acid auxotrophy and severe growth retardation. In addition, there is no evidence that mutations in the ILV6 gene results in an auxotrophic phenotype for isoleucine [20]. These reports are consistent with our results and noticeably confirm our second hypothesis that eugenol interferes with transporters responsible for uptake of aromatic and branched-chain amino acids across the yeast cytoplasmic membrane. This putative interference with amino acid uptake, in itself, would not appear to fully explain the antifungal mode of action of eugenol since growth by amino acid prototrophs is nevertheless inhibited by eugenol.
Figure 3. Auxotroph supplement assay shows that eugenol inhibits the functions of aromatic and branched-chain amino acid transporters.
Strains of S. cerevisiae from GDA library were selected with gene deletions in the tryptophan, phenylalanine, tyrosine (A) or isoleucine (B) biosynthesis pathways. Cultures were 10-fold serially diluted and spotted on synthetic medium supplemented with tryptophan, phenylalanine, tyrosine and isoleucine, either containing a subinhibitory concentration of eugenol (0.18 mg/ml) or without eugenol (control). The plates were incubated at 30°C for 1–2 days and then photographed.
Phenotypic Suppression Assay
To further test whether eugenol interferes with aromatic and branched-chain amino acid permeases, we expressed BAP2, GAP1, TAT1 and TAT2 (which encode general or specific transporters for aromatic and branched-chain amino acids in the cytoplasmic membrane of yeast) from high copy number plasmids in the aro1Δ strain. As indicated in Figure 4, the growth defect of the aro1Δ strain grown in SC-ura medium containing 0.18 mg/ml eugenol was partially compensated by the overexpression of GAP1 (P<0.01) and TAT1 (P<0.05) in comparison to the aro1Δ strain with a control plasmid. However, no significant differences in growth were evident when the aro1Δ strain carried either BAP2 or TAT2 overexpression constructs compared to that with the control plasmid (P>0.1). Therefore, we propose that eugenol specifically interferes with permeases with a dual transport function for both aromatic and branched-chain amino acids (i.e. Tat1p and Gap1p), rather than Bap2p and Tat2p which are high-affinity transporters for branched-chain and aromatic amino acids, respectively [20], [34], [35]. Furthermore, the inferred eugenol-specific interaction with Tat1p rather than Tat2p provides additional evidence for substrate specificity of these two permeases as reported by Regenberg et al. [28].
Figure 4. Phenotypic suppression assay shows that Tat1p and Gap1p permeases are targets of eugenol in the yeast cytoplasmic membrane.
Overexpression constructs of four permeases, BAP2, GAP1, TAT1 and TAT2 were transformed separately into the aro1Δ strain. Transformants were grown overnight in SC-ura medium, diluted 1∶300 and then added to each well of a sterile microtiter plate with or without 0.18 mg/ml eugenol. Growth of the aro1Δ transformants containing the overexpression constructs were compared to one with a control plasmid by measuring the optical density of cells in each well (OD600). The values are expressed as mean (±SD, n = 3) and significant differences between treatment and plasmid control are indicated as P<0.05 (*) and P<0.01 (**) based on Student T-test. Inset: eugenol is structurally similar to aromatic amino acids and is synthesized in plants via the phenylpropanoid pathway from phenylalanine.
Eugenol is a member of the phenylpropanoid class of plant secondary metabolites. Aromatic amino acids, specifically phenylalanine, are precursors of eugenol in the phenylpropanoid biosynthesis pathway [36]. As shown in the inset of Figure 4, the molecular structure of eugenol is very similar to these aromatic amino acid precursors and because of these structural similarities eugenol may interfere with active sites of both Tat1p and Gap1p permeases. While our results indicate that eugenol does not likely perturb the phospholipid bilayer directly, binding of eugenol to these membrane-bound permeases could alter their permeability or cause conformational changes in the targeted permeases that disrupt the yeast cytoplasmic membrane to account for release of intracellular components as reported previously [10]–[12]. Indeed, current models recognize that there is a tight association between proteins, including Gap1p in yeast, and lipids in the cell membrane [37]. However, this model seems incongruent with the partial rescue of aro1Δ strain from eugenol sensitivity that is observed when Gap1p and Tat1p are overexpressed; in this case, an increased abundance of eugenol targets may be expected to increase cytoplasmic leakage and thus sensitivity to eugenol. Further investigations should be carried out to test whether or not there is a direct effect by eugenol on the conformation of Tat1p and Gap1p permeases. Another possible explanation is that Gap1p and Tat1p serve other important functions in the cell. For example, in addition to amino acid transport, Gap1p plays a role in amino acid sensing in a protein kinase A (PKA)-mediated protein phosphorylation cascade [38]. Perturbation of these additional functions may result in cell death and subsequent cytoplasmic leakage.
Approximately 45% of the genes in yeast are homologous to mammalian genes (BLAST e-value<10−10) [39], supporting the view that chemical-genetic profiles obtained from yeast can reflect disease processes in human cells [40]. It should be noted in this context, that amino acid starvation is an effective strategy for cancer therapy. It has been demonstrated that murine and human melanoma cells are induced to undergo apoptosis by phenylalanine and tyrosine starvation [41]. Tsukahara et al. showed that the novel anticancer chemical E7070 inhibits leucine and uracil transporters in the fission yeast, Schizosaccharomyces pombe, and may also target mammalian transporters [42]. It has also been reported that inhibitors of the mammalian amino acid permease, LAT1, that preferentially transports branched-chain and aromatic amino acids through the plasma membrane, may be an effective therapeutic option for human astrocytic tumors [43]. Based on these considerations, amino acid permease inhibitors such as eugenol may hold chemotherapeutic promise for human cancers. However, detailed studies are required to profile the genome-wide effects of eugenol in human cell lines to exploit its therapeutic potential more effectively.
The interesting mode of action of eugenol identified herein for the first time is notable as being distinct from those of commercially available antifungals such as azoles and amphotericin B. Of concern, cross-resistance to amphotericin B and azole antifungals can result from a single mutation in genes involved in ergosterol biosynthesis [44]. Given their different targets in the cell, permeases vs ergosterol, eugenol would likely be useful against increasingly common clinical isolates of pathogenic fungi that are cross-resistant to amphotericin B and azole drugs.
Acknowledgments
We would like to thank I. Cruz for valuable technical advice.
Funding Statement
Funding was from Natural Sciences and Engineering Research Council of Canada to AG and MLS; Ministry of Science, Research and Technology, Iran to ED. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hagan P, Projan S, Rosamond J (2002) Infectious diseases: back to the future? Trends in Microbiology 10: sl–s2. [DOI] [PubMed] [Google Scholar]
- 2. Loeffler J, Stevens DA (2003) Antifungal drug resistance. Clinical Infectious Diseases 36: S31–S41. [DOI] [PubMed] [Google Scholar]
- 3. Eloff JN (1998) Which extractant should be used for the screening and isolation of antimicrobial components from plants? Journal of Ethnopharmacology 60: 1–8. [DOI] [PubMed] [Google Scholar]
- 4. Schmid R (1972) A resolution of the Eugenia-Syzygium controversy (Myrtaceae). American Journal of Botany 59: 423–436. [Google Scholar]
- 5.Boulos L (1983) In Medicinal plants of North Africa, Reference publications, Algonac, Michigan 48001, 286 p. [Google Scholar]
- 6. Chaieb K, Hajlaoui H, Zmantar T, Kahla-Nakbi AB, Mahmoud R, et al. (2007) The chemical composition and biological activity of clove essential oil, Eugenia caryophyllata (Syzigium aromaticum L. Myrtaceae): a short review. Phytotherapy Research 21: 501–506. [DOI] [PubMed] [Google Scholar]
- 7. Gayoso CW, Lima EO, Oliveira VT, Pereira FO, Souza EL, et al. (2005) Sensitivity of fungi isolated from onychomycosis to Eugenia cariophyllata essential oil and eugenol. Fitoterapia 76: 247–249. [DOI] [PubMed] [Google Scholar]
- 8. Lopez P, Sanchez C, Batlle R, Nerın C (2005) Solid- and vapour-phase antimicrobial activities of six essential oils: susceptibility of selected food borne bacterial and fungal strains. Journal of Agricultural and Food Chemistry 53: 6939–6946. [DOI] [PubMed] [Google Scholar]
- 9. Jirovetz L, Buchbauer G, Stoilova I, Stoyanova A, Krastanov A, et al. (2006) Chemical composition and antioxidant properties of clove leaf essential oil. Journal of Agricultural and Food Chemistry 54: 6303–6307. [DOI] [PubMed] [Google Scholar]
- 10. Pinto E, Vale-Silva L, Carlos C, Salgueiro L (2009) Antifungal activity of the clove essential oil from Syzygium aromaticum on Candida, Aspergillus and dermatophyte species. Journal of Medical Microbiology 58: 1454–1462. [DOI] [PubMed] [Google Scholar]
- 11. Carrasco H, Raimindi M, Svetaz L, Liberto MD, Rodriguez MV, et al. (2012) Antifungal activity of eugenol analogues. Influence of different substituents and studies on mechanism of action. Molecules 17: 1002–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zore GB, Thakre AD, Jadhav S, Karuppayil SM (2011) Terpenoids inhibit Candida albicans growth by affecting membrane integrity and arrest of cell cycle. Phytomedicine 18: 1181–1190. [DOI] [PubMed] [Google Scholar]
- 13. Roberts SK, McAinsh M, Widdicks L (2012) Cch1p mediates Ca2+ influx to protect Saccharomyces cerevisiae against eugenol toxicity. PLoS One 7: e43989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherman F, Fink GR, Lawrence CW (1986) Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor Laboratory press, New York, NY.
- 15. Mir-Rashed N, Cruz I, Jessulat M, Dumontier M, Chesnais C, et al. (2010) Disruption of fungal cell wall by antifungal Echinacea extracts. Medical Mycology 48: 949–58. [DOI] [PubMed] [Google Scholar]
- 16.CLSI (2008) Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. Approved standard M27- A3, 3rded, Clinical and Laboratory Standards Institute, Wayne, PA.
- 17. Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, et al. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- 18. Galván IJ, Mir-Rashed N, Jessulat M, Atanya M, Golshani A, et al. (2008) Antifungal and antioxidant activities of the phytomedicine pipsissewa, Chimaphila umbellata . Phytochemistry 69: 738–746. [DOI] [PubMed] [Google Scholar]
- 19. Memarian N, Jessulat M, Alirezaie J, Mir-Rashed N, Xu J, et al. (2007) Colony size measurement of the yeast gene deletion strains for functional genomics. BMC Bioinformatics 8: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong EL, Balakrishnan R, Christie KR, Costanzo MC, Dwight SS, et al. (2006) Saccharomyces Genome Database. http://www.yeastgenome.org/(accessed August 2012).
- 21. Trombetta D, Castelli F, Sarpietro MG, Venuti V, Cristani M, et al. (2005) Mechanisms of antibacterial action of three monoterpenes. Antimicrobial Agents and Chemotherapy 49: 2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alamgir M, Eroukova V, Jessulat M, Xu J, Golshani A (2008) Chemical-genetic profile analysis in yeast suggests that a previously uncharacterized open reading frame, YBR261C, affects protein synthesis. BMC Genomics 9: 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory, New York. 352–355.
- 24. Sopko R, Huang D, Preston N, Chua G, Papp B, et al. (2006) Mapping pathways and phenotypes by systematic gene overexpression. Molecular Cell 21: 319–330. [DOI] [PubMed] [Google Scholar]
- 25. Alamgir M, Erukova V, Jessulat M, Azizi A, Golshani A (2010) Chemical-genetic profile analysis of five inhibitory compounds in yeast. BMC Chemical Biology 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods in Enzymology 350: 87–96. [DOI] [PubMed] [Google Scholar]
- 27. Schreve JL, Garrett JM (2004) Yeast Agp2p and Agp3p function as amino acid permeases in poor nutrient conditions. Biochemical and Biophysical Research Communication 313: 745–751. [DOI] [PubMed] [Google Scholar]
- 28. Regenberg B, Düring-Olsen L, Kielland-Brandt MC, Holmberg S (1999) Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae . Current Genetics 36: 317–328. [DOI] [PubMed] [Google Scholar]
- 29. Gao M, Kaiser CA (2006) A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nature Cell Biology 8: 657–667. [DOI] [PubMed] [Google Scholar]
- 30. Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, et al. (2008) An in vivo map of the yeast protein interactome. Science 320: 1465–1470. [DOI] [PubMed] [Google Scholar]
- 31. Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, et al. (2009) Combinatorial effects of environmental parameters on transcriptional regulation in Saccharomyces cerevisiae: a quantitative analysis of a compendium of chemostat-based transcriptome data. BMC Genomics 10: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Urrestarazu A, Vissers S, Iraqui I, Grenson M (1998) Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Molecular Genetics and Genomics 257: 230–237. [DOI] [PubMed] [Google Scholar]
- 33. Kispal G, Steiner H, Court DA, Rolinski B, Lill R (1996) Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. Journal of Biological Chemistry 271: 24458–24464. [DOI] [PubMed] [Google Scholar]
- 34. Grauslund M, Didion T, Kielland-Brandt MC, Andersen HA (1995) BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae . Biochimica et Biophysica Acta 1269: 275–280. [DOI] [PubMed] [Google Scholar]
- 35. Schmidt A, Hall MN, Koller A (1994) Two FK506 resistance conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino-acid permeases mediating tyrosine and tryptophan uptake. Molecular and Cellular Biology 14: 6597–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Louie GV, Baiga TJ, Bowman ME, Koeduka T, Taylor JH, et al. (2007) Structure and reaction mechanism of basil eugenol synthase. PLoS ONE 2: e993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lauwers E, Grossmann G, André B (2007) Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Molecular Biology of the Cell 18: 3068–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Donaton MCV, Holsbeeks I, Lagatie O, Zeebroeck GV, Crauwels M, et al. (2003) The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae . Molecular Microbiology 50: 911–929. [DOI] [PubMed] [Google Scholar]
- 39. Hughes TR (2002) Yeast and drug discovery. Functional and integrative genomics 2: 199–211. [DOI] [PubMed] [Google Scholar]
- 40. Dixon SJ, Stockwell BR (2009) Identifying druggable disease-modifying gene products. Current Opinion in Chemical Biology 13: 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fu YM, Yu ZX, Playo BA, Ferrans VJ, Meadows GG (1999) Focal adhesion kinase-dependent apoptosis of melanoma induced by tyrosine and phenylalanine deficiency. Cancer Research 59: 758–765. [PubMed] [Google Scholar]
- 42. Tsukahara K, Watanabe T, Hata-sugi N, Yoshimatsu K, Okayama H, et al. (2001) Anticancer agent E7070 inhibits amino acid and uracil transport in fission yeast. Molecular Pharmacology 6: 1254–1259. [DOI] [PubMed] [Google Scholar]
- 43. Nawashiro H, Otani N, Shinomiya N, Fukui S, Ooigawa H, et al. (2006) L-type amino acid transporter 1 as a potential molecular target in human astrocytic tumors. International Journal of Cancer 119: 484–492. [DOI] [PubMed] [Google Scholar]
- 44. Esponel-Ingroff A (2008) Mechanisms of resistance to antifungal agents: Yeasts and filamentous fungi. Revista Iberoamericana Micologia 25: 101–106. [DOI] [PubMed] [Google Scholar]