Abstract
Although the nucleotides in tRNA required for aminoacylation are conserved in evolution, bacterial aminoacyl-tRNA synthetases (aaRSs) are unable to acylate eukaryotic tRNA. The cross-species barrier may be due to the absence of eukaryote-specific domains from bacterial aaRSs. Here we show that while E. coli CysRS cannot acylate human tRNACys, the fusion of a eukaryote-specific domain of human CysRS overcomes the cross-species barrier in human tRNACys. In addition to enabling recognition of the sequence differences in the tertiary core of tRNACys, the fused eukaryotic domain redirects the specificity of E. coli CysRS from the A37 present in bacterial tRNACys to the G37 in mammals. Further experiments show that the accuracy of codon recognition on the ribosome was also highly sensitive to the A37-to-G37 transition in tRNACys. These results raise the possibility of the development of tRNA nucleotide determinants for aminoacylation being interdependent with those for ribosome decoding.
Introduction
The accuracy of translating genetic information into protein sequences is important for organismal growth and viability, and depending on the complexity of each organism, it is tightly controlled1. In E. coli, the average protein has ~300 amino acids2, demanding an error frequency of less than 3 × 10−3 per amino acid, which is supported by experimental estimations3. In humans, the average protein has ~500 amino acids4 with many involved in larger complexes, suggesting a need for higher accuracy than that required for E. coli. This demand is likely true for most eukaryotes, where proteins are typically synthesized as longer chains with more complexity. Indeed, the accuracy of protein synthesis in the unicellular eukaryote Saccharomyces cerevisiae is estimated to be ~3-fold higher than that in E. coli5.
The accuracy of protein synthesis is jointly determined by the aaRS-catalyzed tRNA aminoacylation and by the ribosome-catalyzed tRNA decoding. Each aaRS synthesizes an aminoacyl-tRNA (aa-tRNA), which enters the ribosome at specific codons for peptide bond formation. In the progressive organismal evolution into higher complexity, how tRNAs, aaRSs, and the ribosome adapted to one another is an important question. Relevant to this question is the frequently observed cross-species barriers to aminoacylation, which block bacterial aaRSs from acylating their eukaryotic tRNAs, despite conservation of the “identity” nucleotides (e.g., the anticodon sequences). The existence of barriers suggests the emergence of eukaryote-specific tRNA determinants possibly to meet the demand for higher accuracy of protein synthesis. Notably, eukaryotic aaRSs are more elaborate than their bacterial counterparts6, possessing extension domains in addition to the core domains for aminoacylation. While the extension domains are not essential for aminoacylation, some can enhance aminoacylation specificity over that of bacterial enzymes7. Additionally, eukaryotic ribosomes have expanded from their bacterial counterparts to include new proteins and RNA segments that are implicated in cellular control processes unique to eukaryotes. The expansion of both aaRSs and ribosomes in eukaryotes raises the possibility for the development of eukaryote-specific tRNA determinants interdependent on aminoacylation and translation. However, while eukaryotic tRNA nucleotides that act as cross-species barriers have been studied in some cases (e.g. tRNACys, tRNATyr, and tRNALys)8–10, whether they function to adapt with the expansion of aaRSs and ribosomes has not been tested. Also, while replacement of a bacteria-specific peptide in E. coli TyrRS (eTyrRS) with a eukaryote-specific peptide from human TyrRS (hTyrRS) overcomes a cross-species barrier11, whether this barrier is linked to the demand for higher accuracy of protein synthesis in eukaryotes is unknown.
A cross-species barrier exists in aminoacylation of tRNACys. The major determinants for CysRS reside in the U73 nucleotide at the 3' end of tRNACys and in the GCA anticodon7,12,13, both of which are strictly conserved in bacteria, eukarya, and even in archaea14,15. However, eCysRS and several other bacterial CysRS, while able to acylate each other’s tRNA, are unable to efficiently acylate tRNACys in humans8, indicating the presence of a eukaryote-specific barrier in htRNACys. The barrier8 is localized to the tRNA tertiary core and to nucleotide 37 in htRNACys. In the tertiary core, while htRNACys uses the normal Levitt base pair G15-C48 to join the D and variable loops16, etRNACys uses a rare G15-G48 base pair17,18, which is recognized by eCysRS in an indirect readout mechanism that monitors the backbone structure of the base pair18 (Supplementary Fig. S1a,b). Structural probing shows that the shape of the core in etRNACys as defined by the unusual G15-G48 is similar to the shape of the core in other bacterial tRNACys using different 15–48 base pairs19. At nucleotide 37, the base is conserved as an A in bacteria and in yeast, but is conserved as a G in higher eukaryotes. In the tRNA-bound co-crystal structure18, eCysRS only makes contact with the ribose of A37 (Supplementary Fig. S1c), indicating another example of indirect readout. However, because the tertiary core and nucleotide 37 are physically separated, whether their indirect readout mechanisms are coupled or independent of each other is unknown.
While eCysRS is unable to cross-acylate htRNACys, hCysRS efficiently cross-acylates its bacterial tRNA8. The human enzyme shares high homology with eCysRS throughout the core domains18, but contains three eukaryote-specific extensions, of which only the C-terminal extension (CTE) is broadly conserved in eukaryotes. The CTE enhances aminoacylation specificity of hCysRS relative to eCysRS7, suggesting that it may mediate the transition of the translational apparatus from bacteria to humans.
Here we present a comprehensive analysis of the human CTE and its role in the development of eukaryote-specific tRNA determinants interdependent on aminoacylation and ribosome decoding. The analysis consists of the construction of a CTE-fused eCysRS, demonstrating that the fusion overcomes the barrier at both the tertiary core and at nucleotide 37, and the analysis of eukaryote-specific nucleotides for tRNA selectivity on the ribosome, demonstrating that G37 in htRNACys imparts increased selectivity relative to A37 in etRNACys at both the initial selection and proofreading stages. We also show that G37 has become a eukaryote-specific major determinant of aminoacylation, illustrating a principle of interdependent development of G37 required for both aminoacylation and ribosome decoding. We suggest that this interdependency can increase the accuracy of protein synthesis in eukaryotes relative to bacteria, and that it has implications for the maintenance of the genetic code stability in eukaryotes.
Results
Fusion of eCysRS overcomes the cross-species barrier
A fusion protein “eCysRS+CTE” was created, in which the sequence of eCysRS was extended at the C-terminus with the sequence of the human CTE. This fusion markedly improved aminoacylation of htRNACys by eCysRS, elevating the catalytic efficiency kcat/Km from ~600-fold below to ~30-fold below that observed with hCysRS (Fig. 1a,b; Supplementary Table S1). The improvement of activity by the fusion was even more pronounced in single turnover assays, where the effect of the fusion on the aminoacylation rate constant (kchem) and on the tRNA affinity constant (Kd) was significant. Analysis of kchem/Kd showed that the fusion increased aminoacylation of htRNACys by eCysRS from ~700-fold to ~10-fold below that observed with hCysRS (Fig. 1c). In both assays, the fusion protein acted similarly to hCysRS by retaining full activity to aminoacylate etRNACys.
Figure 1.
Aminoacylation of tRNACys with cysteine. (a) Sequences of transcripts of etRNACys and htRNACys, showing the conserved U73 and GCA anticodon for aminoacylation and the non-conserved determinants at position 15–48 in the tertiary core and at base 37 in the anticodon loop. (b) Steady-state activity (kcat/Km) of aminoacylation of etRNACys and htRNACys by CysRS enzymes. (c) Single turnover activity (kchem/Kd) of aminoacylation of etRNACys and htRNACys by CysRS enzymes. Data are shown in a logarithmic scale and are the average of 3 determinations. Error bars represent standard deviations.
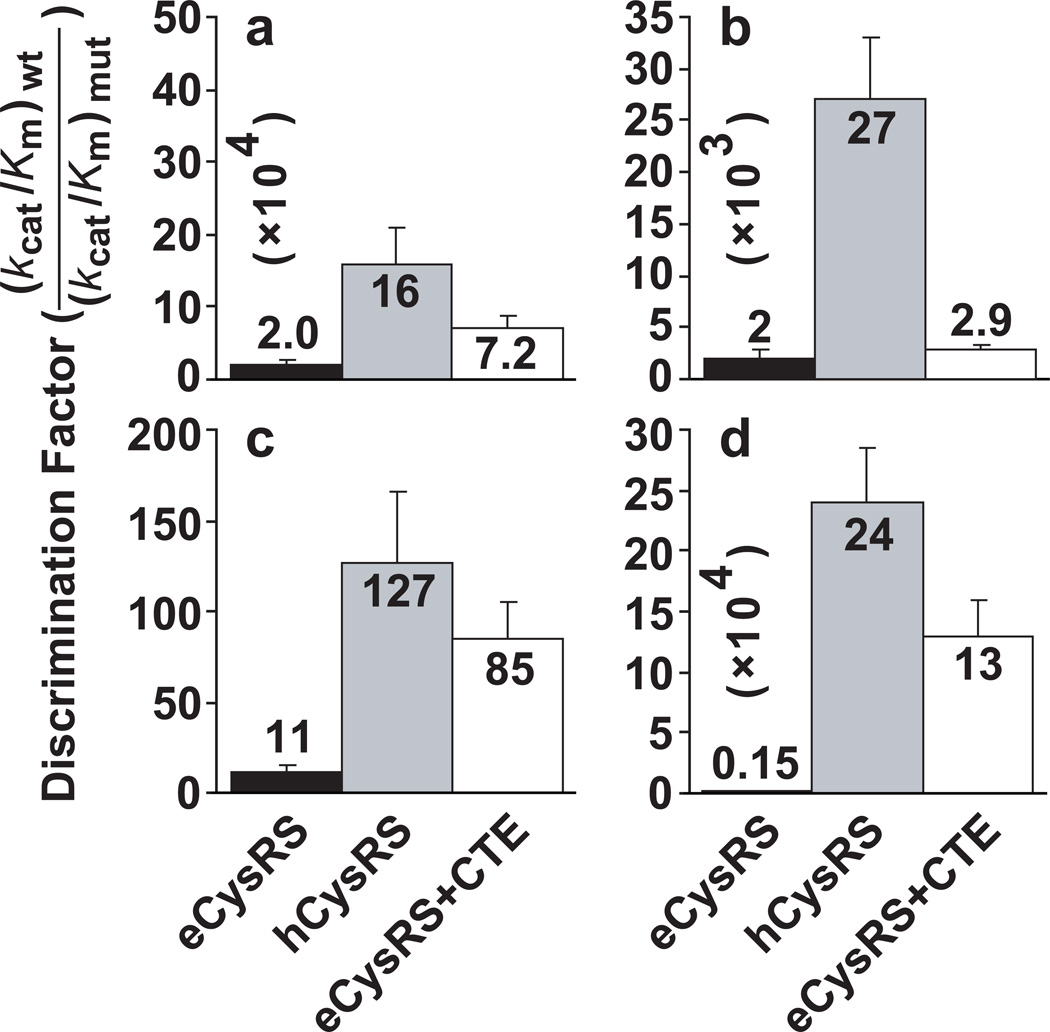
The fusion protein provided a simpler model than the full-length hCysRS to address how the CTE removes the cross-species barrier for eCysRS. Using htRNACys as the substrate, we tested if the fusion protein recognized the eukaryote-specific nucleotides in a way that paralleled hCysRS. We showed that the determinants in htRNACys that are conserved in etRNACys lack such differentiation among the three enzymes. For example, the conserved U73 is the most important nucleotide for tRNA aminoacylation with cysteine7, 8, whose substitutions are typically discriminated by CysRSs by 104–105-fold based on the kcat/Km ratio for wild-type (wt) over mutant tRNAs. Indeed, eCysRS, hCysRS, and the fusion protein all strongly discriminated against the U73A mutation, showing discrimination factors of 2.0, 16, and 7.2 × 104, respectively (Fig. 2a, Supplementary Table S2). The degrees of discrimination by the three enzymes is similar, confirming that U73 is not unique to htRNACys. G34 of the conserved anticodon is the second most important nucleotide for aminoacylation7, whose mutations are typically discriminated by 102–103-fold. Analysis of the G34C mutation showed similar discrimination by eCysRS and by the fusion protein (2.0 and 2.9 × 103, respectively) but much greater discrimination by hCysRS (27 × 103) (Fig. 2b). The weak discrimination by the bacterial and fusion enzymes indicates that they recognize G34 similarly and that G34 is not a eukaryote-specific nucleotide.
Figure 2.
Discrimination against mutations in htRNACys. (a) Discrimination against the U73A mutation. (b) Discrimination against the G34C mutation. (c) Discrimination against the G15-G48 mutation. (d) Discrimination against the G37A mutation. Data were obtained by analysis of the steady-state kcat/Km. Each point was the average of 3 determinations and error bars represented standard deviations.
Discrimination of the tertiary core is typically by 10–100-fold7,20,21. Alteration of the normal G15-C48 in the human core to G15-G48 was discriminated by 11-fold by eCysRS, 127-fold by hCysRS, and 85-fold by the fusion protein (Fig. 2c). The close similarity between hCysRS and the fusion enzyme indicates that the latter had captured the ability of hCysRS to more strongly recognize the G15-C48 base pair than eCysRS. Similarly, discrimination of the G37A mutation in the anticodon loop of htRNACys was 0.15 × 104-fold for eCysRS but much stronger, 24 and 13 × 104-fold, for hCysRS and the fusion enzyme (Fig. 2d), indicating that the human and fusion enzymes behaved similarly with an ~100-fold higher discrimination relative to eCysRS. Together these results showed that the fusion enabled eCysRS to recognize the human-specific tertiary core and G37 of htRNACys. Additionally, mutational analysis of G37 revealed that the level of discrimination against the G37A mutation by hCysRS and the fusion protein is as strong as that against the U73A mutation (16 × 104-fold, Fig. 2a, d), suggesting that G37 has emerged as a eukaryote-specific key determinant equally important as the conserved U73 for aminoacylation of htRNACys.
Coordination of the core and G37 in the barrier
Additional analysis (Supplementary Table S3, Supplementary Fig. S2) revealed that the recognition of the core and G37 in htRNACys is coordinated by the CTE and that the possession of the CTE conferred upon the human and the fusion enzymes two features absent from eCysRS. First, both hCysRS and the fusion enzyme have the flexibility to also recognize A37 in the context of the E. coli core. While both strongly discriminated against G37A in htRNACys (mutant htRNA-hM1), both also acylated the A37-containing etRNACys (etRNA-wt) with high efficiency, indicating an ability to recognize G37 in the context of the human core and to recognize A37 in the context of the E. coli core. In contrast, the CTE-lacking eCysRS is unable to recognize G37 when associated with the human core. Efficient aminoacylation of htRNACys by eCysRS occurred only after introduction of G37A and extensive remodeling of the human core (mutant htRNA-hM2), the latter of which includes G15-G48 and specific substitutions in the D and variable loops8. Second, the CTE in the human and fusion enzymes renders the coordinated recognition of G37 with the human core much stronger than the coordinated recognition of A37 with the E. coli core. While both the human and fusion enzymes discriminated against G37A in the context of the human core by 105–106-fold (htRNA-wt versus htRNA-hM1), they discriminated against A37G in the context of the E. coli core by only 103-fold (etRNA-wt versus etRNA-eM1). The coordinated recognition is also evident in eCysRS, albeit weaker with only a 103-fold discrimination against A37G in the context of the E. coli core (etRNA-wt versus etRNA-eM1). Together, these results show that the cross-species barrier is associated with coordinated changes of the core and nucleotide 37 in the transition from etRNACys to htRNACys, and that the CTE is required to accompany this transition in the development from eCysRS to hCysRS.
Higher codon selectivity of htRNACys than etRNACys
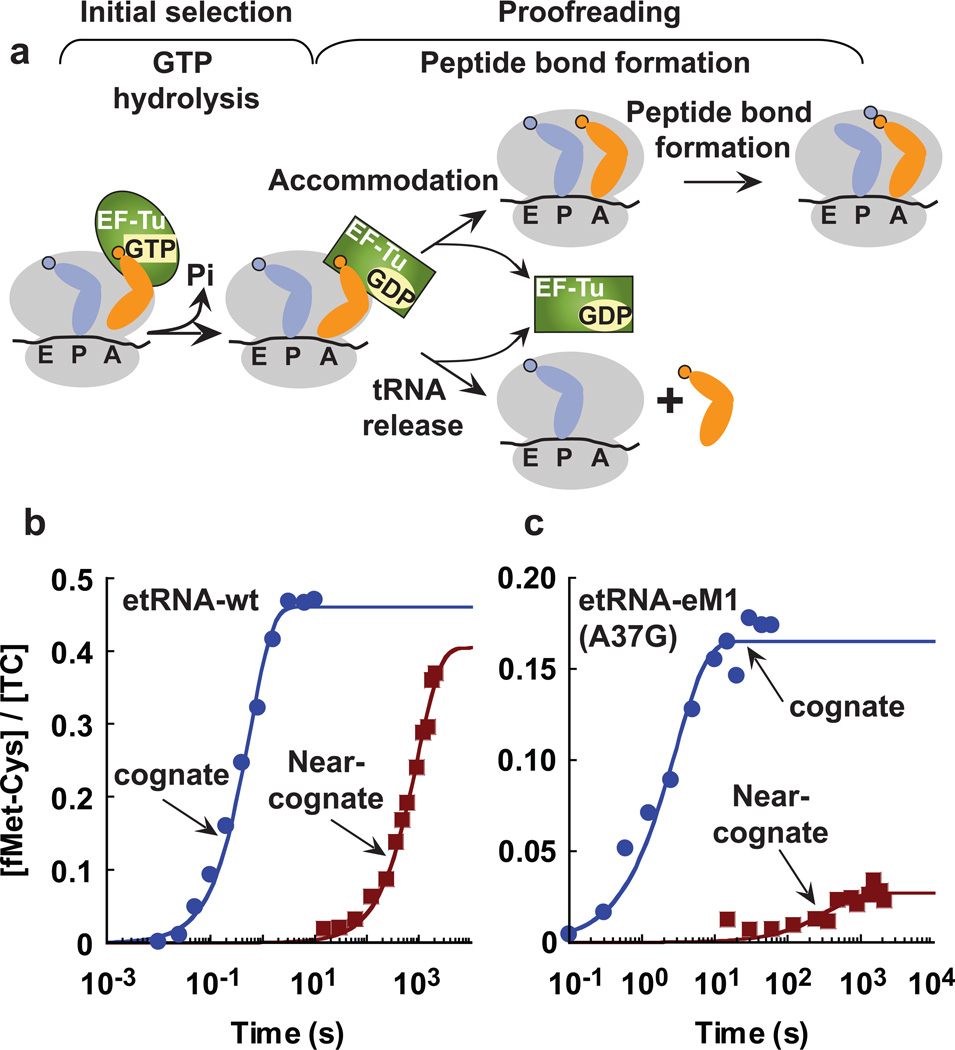
We next examined the codon selectivity of etRNACys and htRNACys on the ribosome. The accuracy of codon recognition is governed in two stages (Fig. 3a): initial selection and proofreading22–25. Initial selection begins with the entry of an aa-tRNA in a ternary complex with EF-Tu and GTP to the ribosome, allowing the sorting of the tRNA by codon-anticodon sampling. Correct pairing leads to rapid GTP hydrolysis26–28, whereas near-cognate pairings hinder GTP hydrolysis and non-cognate pairings are rejected from the ribosome29. GTP hydrolysis is irreversible and sets the stage for proofreading, where cognate aa-tRNA is accommodated for rapid peptide bond formation, whereas near-cognate ones are rejected. Because selectivity in both stages is largely controlled by kinetics22–24, we used kinetic assays with the E. coli ribosome22–24. Mammalian ribosomes have not been developed with such assays.
Figure 3.
Codon recognition on the ribosome. (a) A scheme of codon recognition, showing initial selection of tRNA in the first stage, which is assayed by GTP hydrolysis, followed by proofreading in the second stage, which is assayed by peptide bond formation. (b) Time courses of dipeptide formation on etRNA-wt on UGC (cognate) or CGC (near-cognate) codons measured with a limiting concentration of the ternary complex (0.25 µM). (c) Time courses of dipeptide formation on etRNA-eM1 harboring the A37G mutation on UGC or CGC codon measured with a limiting concentration of the ternary complex (0.25 µM). Data in (b) and (c) were obtained in single turnover conditions and were well fit to a single exponential equation and plotted on a semi-log scale.
Selectivity at the initial stage was assayed by the release of Pi from hydrolysis of GTP upon recognition of a cognate codon-anticodon pairing. Each tRNA was prepared by in vitro transcription and used in the form of Cys-tRNACys in complex with Tu and GTP. A 70S ribosome with fMet-tRNAfMet in the P site was programmed with a synthetic mRNA to place at the A site the cognate codon for cysteine (UGC) or one of three near-cognate (CGC, UAC, UGG) codons. Each complex was rapidly mixed with a purified ternary complex to allow measurement of GTP hydrolysis at 3 µM ribosome (Table 1), which was ~2-fold of the Km of etRNACys or htRNACys for the ribosome (Table 2). Discrimination of etRNA at the cognate codon (kapp of 13 ± 1 s−1) against the 2nd-position and 3rd-position near-cognate codons (kapps of 0.013 ± 0.003 and 0.033 ± 0.006 s−1, respectively) was apparent at a level similar to the discrimination reported for other tRNA30. However, discrimination against the 1st-position near cognate codon (4.5 ± 0.4 s−1) was relatively weak. A similar pattern was observed with htRNA, in which the kapp for the cognate codon (2.9 ± 0.3 s−1) was significantly faster than the kapps for the 2nd-position and 3rd-position near-cognate codons (0.0022 ± 0.0004 and 0.010 ± 0.003 s−1, respectively), but only slightly faster than the kapp for the 1st-position near-cognate (0.06 ± 0.01 s−1). Notably, while both tRNAs were weak at discriminating against the 1st-postiion near-cognate codon, htRNA was 17-fold more discriminative.
Table 1.
Discrimination against codon-anticodon mismatches
| A-site codon (5' NNN 3') | kapp (s−1) (3 µM ribosomes) |
|
Ratio (htRNA/etRNA) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| etRNA | htRNA | etRNA | htRNA | ||||||
| Cognate | UGC | 13 ± 1 | 2.9 ± 0.3 | ||||||
| Near- cognate |
1st position | CGC | 4.5 ± 0.4 | 0.06 ± 0.01 | 2.9 | 48 | 17 | ||
| 2nd position | UAC | 0.013 ± 0.003 | 0.0022 ± 0.0004 | 1000 | 1300 | 1.3 | |||
| 3rd position | UGG | 0.033 ± 0.006 | 0.010 ± 0.003 | 390 | 290 | 0.7 | |||
Bold face letters underlined indicate mismatches.
Table 2.
Kinetic parameters of GTP hydrolysis
| tRNACys | Cognate (UGC) | Near-cognate (CGC) | Selectivity | Selectivity relative to etRNA (wt) |
||||
|---|---|---|---|---|---|---|---|---|
|
Km (µM) |
kGTP (s−1) |
kGTP/Km (µM−1s−1) |
Km (µM) |
kGTP (s−1) |
kGTP/ Km (µM−1s−1) |
|||
| etRNA (wt) | 1.7 ± 0.1 | 20 ± 1 | 12 ± 1 | 2.1 ± 0.4 | 4.5 ± 0.4 | 2.1 ± 0.4 | 5.7 | 1 |
| htRNA (wt) | 1.1 ± 0.3 | 3.9 ± 0.4 | 3.5 ± 0.4 | 1.0 ± 0.1 | 0.08 ± 0.01 | 0.08 ± 0.01 | 44 | 7.7 |
| etRNA (native) | 2.8 ± 0.3 | 49 ± 3 | 18 ± 2 | 2.4 ± 0.2 | 4.9 ± 0.2 | 2 ± 0.2 | 9 | 1.6 |
| eM1 | 1.7 ± 0.1 | 29 ± 1 | 17 ± 1 | 1.9 ± 0.2 | 0.6 ± 0.1 | 0.30 ± 0.06 | 57 | 10 |
| eM5 | 2.5 ± 0.6 | 25 ± 3 | 10 ± 3 | 2.5 ± 0.9 | 0.5 ± 0.1 | 0.2 ± 0.1 | 50 | 8.8 |
Mutations: eM1 (A37G); eM5 (A13U, U21A, A37G).
Selectivity = [kGTP/Km](Cognate)/ [kGTP/Km](Near-cognate).
The selectivity of cognate against the 1st-position near-cognate codon was determined by analysis of kapp at increasing ribosome concentrations (Table 2, Supplementary Fig. S3). This revealed that the Km of each tRNA on the ribosome was similar, whereas the saturating rate constant kGTP varied. Analysis of the specificity constant kGTP/Km between cognate and near-cognate codons showed that the selectivity of htRNACys (ratio of 44) was 7.7-fold higher than the selectivity of etRNACys (ratio of 5.7). The lower selectivity of etRNACys was not due to the lack of nucleotide modifications in the transcript, because native etRNACys increased the selectivity from a ratio of 5.7 to 9.0, still 5-fold lower than the selectivity of htRNACys. The modest gain in selectivity with native etRNACys may be attributable to the natural modification 2-methylthio-N6-isopentenyl (ms2i6) of the A37 base31.
Higher selectivity of htRNACys conferred by G37
To determine whether it was the human core or the G37 being responsible for the higher selectivity of htRNACys, two mutants of etRNACys were created (Table 2, Supplementary Fig. S2, Supplementary Figure S4): eM1 harbored the single A37G substitution and eM5 harbored additional substitutions to recapitulate features of the human core. The A37G substitution alone elevated the selectivity of etRNACys to 57, similar to that of htRNACys, while the additional substitutions in the eM5 mutant made no further improvement (selectivity of 50). Thus, G37 alone is sufficient to elevate the selectivity of etRNACys to that of htRNACys. The selectivity exhibited by the A37G substitution in etRNACys is 10-fold higher than that of etRNACys-wt, an effect similar to others induced by a single nucleotide substitution of tRNA (Supplementary Table S4).
We tested the role of G37 in the proofreading stage of decoding by monitoring peptide bond formation. The etRNACys-eM1 mutant was chosen for comparison with etRNACys-wt, because the mutant had the same sequence context as the etRNACys-wt except for the A37G substitution, while showing the same selectivity as htRNACys-wt in the GTPase assay. A 70S ribosome with fMet-tRNAfMet in the P site was programmed with a synthetic mRNA to place at the A site the cognate (UGC) or near-cognate (CGC) codon. Each complex was rapidly mixed with a ternary complex to allow measurement of fMet-Cys formation (Fig. 3b, c, Table 3). At the cognate codon, the apparent rate constant (kapp) of etRNA-wt (1.6 ± 0.2 s−1) was similar to the values measured by others24,32, while the kapp of the eM1 mutant (0.31 ± 0.06 s−1) was slower. At the near-cognate codon, the kapp of etRNA-wt (0.0011 ± 0.0001 s−1) was substantially decreased and the value of the mutant was too low to be precisely determined.
Table 3.
Kinetics of peptide bond formation
| Cys-tRNACys | kapp (s−1) | [fMet-Cys]/[TC] | Fidelity of proofreading |
||
|---|---|---|---|---|---|
| UGC codon | CGC codon | UGC codon | CGC codon | ||
| etRNA-WT | 1.6 ± 0.2 | 0.0011 ± 0.0001 | 0.46 | 0.40 | 1.2 |
| etRNA-eM1 | 0.31 ± 0.06 | Not determined | 0.17 | 0.03 | 5.7 |
Summary of the rate constant kapp of dipeptide formation and the amplitude [fMet-Cys]/[TC] for calculation of fidelity. TC = ternary complex.
Selectivity of proofreading was determined from the amplitude of dipeptide formation per ternary complex loaded to the ribosome23,24,33,34. The selectivity of etRNA-wt at the cognate codon (0.46) and near-cognate codon (0.40) was similar, indicating that discrimination did not occur at the peptide formation step for the wt tRNA. In contrast, the selectivity of the etRNA-eM1 mutant at the cognate codon (0.17) was more than 5-fold greater than at the near-cognate codon (0.03). Thus, the mutant exhibited discrimination at a step where the wt tRNA was incapable of discrimination. From the ratio between the cognate and near-cognate values, the selectivity of proofreading was 1.2 for etRNA-wt and 5.7 for etRNA-eM1, demonstrating a 4.8-fold increase in fidelity of the mutant. The overall selectivity improvement of the eM1 mutant relative to etRNA-wt, calculated as the product of the improvement in the initial selection (10-fold) and in the proofreading (~5-fold), was ~50-fold.
Discussion
Using the cross-species barrier of cysteinylation of tRNA as a framework, we have provided a first line of evidence supporting the hypothesis that, due to the expansion of the translational apparatus that accompanies organismal development, the tRNA component of the apparatus has developed discrete determinants interdependent on aminoacylation and decoding. The cross-species barrier in htRNACys is localized to the G15-C48 tertiary core and to the mammal-specific G37 in the anticodon loop. Fusion of eCysRS with the human CTE overcomes the barrier at both regions and, more importantly, it re-directs the landscape of tRNA recognition such that G37 has become a major determinant for aminoacylation, as important as the conserved U73. The significance of G37 to the fusion protein far exceeds the significance of A37 to the bacterial enzyme. Using the E. coli ribosome, we then showed that the single A37-to-G37 transition in etRNACys confers a 10-fold increase in initial selectivity and an additional 5-fold increase in proofreading, indicating the capacity for a 50-fold increase in the overall accuracy of decoding.
The increase in initial selectivity by the A37G mutation is demonstrated in tRNA transcripts. In their native forms, A37 in etRNACys is modified to ms2t6 A37 and G37 in htRNACys is modified to m1 G37. Both modifications can further increase tRNA selectivity on the ribosome35, suggesting that the higher accuracy displayed by htRNACys relative to etRNACys in the transcript form should maintain in the native form. We were unable to assess decoding accuracy of the two tRNAs using rabbit reticulocyte ribosome, and we considered yeast ribosome unsuitable, because it presumably co-adapted with yeast tRNACys, which harbors A37. However, because the nucleotides that control accuracy of decoding in the ribosome are strictly conserved, the results with the E. coli ribosome strongly suggest that the A37-to-G37 transition will confer higher selectivity on the human ribosome.
Crystal structures of bacterial ribosomes reveal that the accuracy of initial selection is controlled by an induced-fit process of the 30S, involving three strictly conserved nucleotides in the 16S rRNA to closely monitor the geometry of codon-anticodon interactions26,36,37. The three conserved nucleotides are also implicated in the proofreading stage by monitoring the entire anticodon loop structure of tRNA38. Nucleotide 37 is localized in a dense network of interactions between the three conserved nucleotides, suggesting that any change in and around the network can result in rearrangement of the contacts and affect the overall accuracy of decoding. The nature of how the A37-to-G37 transition in etRNACys increases accuracy of decoding is unknown, due to the lack of crystal structures of ribosome-tRNACys complexes. The A37-to-G37 transition may increase accuracy by limiting the rearrangement of the contacts to respond only to cognate codon-anticodon interactions. Although the effect of the A37-to-G37 transition may depend on the sequence context of the anticodon loop, examples of this transition are abundant in tRNA databases: the frequency of G37 in tRNA increases from 12% in bacteria to 29% in humans, while that of A37 decreases from 88% to 70% (Supplementary Table S5), indicating a shift of preference from A37 to G37.
Recognition of the A37-to-G37 transition for aminoacylation requires the CTE, which coordinates the transition with changes of the tertiary core. Without the CTE, the coordination still exists but is 103-fold weaker for eCysRS relative to the fusion protein and hCysRS. Structural modeling suggests that the CTE is well suited to perform the coordination, as it emanates from the terminus of the anticodon-binding domain and can reach up to a solvent-accessible channel between the stem-contact fold and the inner corner of the core. Coordinated recognition by the CTE is mediated by indirect readout and is likely secured by the extensive and complementary enzyme-tRNA interface already present in the eCysRS-tRNA structure18. The CTE is unique to eukaryotic CysRS, present from yeast to humans, with no sequence homology to any other protein domains. The specificity of the CTE to CysRS is illustrated by the analysis of a CTE-fusion to eMetRS, a close structural homolog of eCysRS. While the native eMetRS is able to acylate the bacterial but not the eukaryotic tRNAMet, illustrating the existence of a cross-species barrier; the CTE fusion to eMetRS does not overcome the barrier but instead impairs the enzyme activity with bacterial tRNAMet (Supplementary Fig. S5).
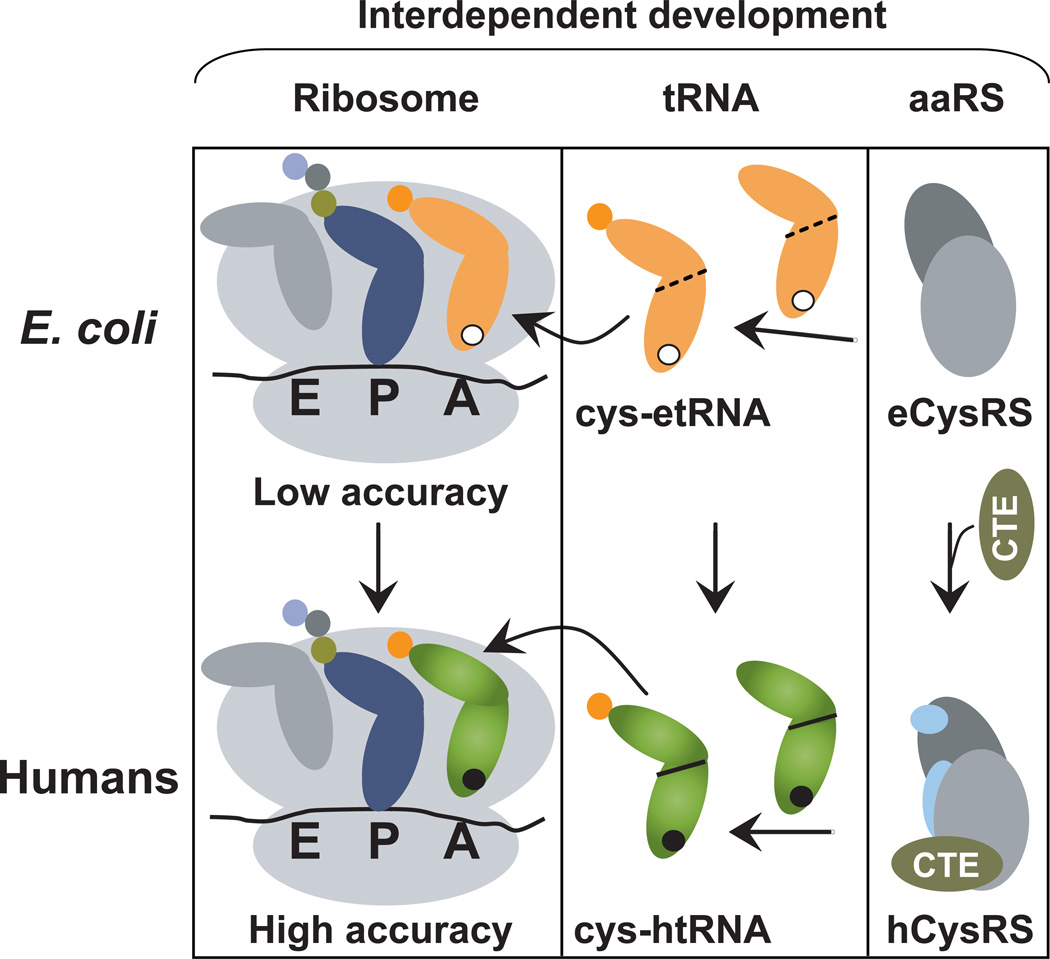
Our work suggests a model consistent with the A37-to-G37 transition being interdependent on both aminoacylation and ribosome decoding (Fig. 4). This transition would occur only concomitantly with the development of the CTE in human CysRS as an appendage to the core domains in bacterial CysRS. Using an indirect readout mechanism, the CTE would reach around from the anticodon loop all the way to the top of the tertiary core, where it enforces recognition of the human core. To secure the presence of G37 in humans, the CTE would redirect the human CysRS to adopt G37 as a major determinant for aminoacylation. The absence of the CTE from bacterial CysRSs explains their inability to recognize the A37-to-G37 transition.
Figure 4.
A model of interdependent development of tRNACys nucleotides. Center: The A37-to-G37 transition (an open circle to a black circle) from etRNACys (orange) to htRNACys (green) is proposed to couple with the transition from the G15-G48 tertiary core (a dashed line) in E. coli to the G15-C48 tertiary core (a solid line) in humans. Right: This coupled development is concomitant with the CTE fusion to the core domains of eCysRS (dark grey: activation domain; light grey: tRNA docking domains) to enable recognition of the transition for aminoacylation. The development process also recruits two other eukaryote-specific domains (light blue) to form hCysRS. Left: The A37-to-G37 transition in the coupled development of tRNA nucleotides confers high codon selectivity on the A site of the ribosome to satisfy the demand for accuracy of protein synthesis in humans.
The conservation of A37 in bacterial tRNACys has important implications. First, calculation of the error frequency of the A37-bearing etRNACys from the kapp of dipeptide synthesis at near-cognate (0.001 s−1) and cognate (1.6 s−1) codons reveals a value of 0.6 × 10−3, consistent with the error frequency reported for other bacterial tRNA24. At this error frequency, the fidelity of etRNACys is sufficient for faithful synthesis of an average protein in bacteria, suggesting that higher accuracy is not necessary. Second, even if the A37-to-G37 transition had occurred by a single-site nucleotide substitution among bacterial species over time, the resulting tRNA mutant would be unrecognizable to bacterial CysRS, due to the lack of the CTE. Development of the CTE, with more than 100 amino acids in specific structural arrangements, is arguably more difficult than the single nucleotide substitution in tRNA. Third, addition of the CTE to bacterial CysRS is not beneficial, because it would slow down the rate of aminoacylation and incur disadvantage to bacteria. Note that the CTE fusion of eCysRS delays kchem (9.5 ± 0.4 s−1) by 2-fold in single turnover assays relative to the unfused enzyme (15.2 ± 0.5 s−1), and also delays kcat (0.44 ± 0.02 s−1) by 2.5-fold in steady state relative to the control (1.0 ± 0.1 s−1). Reduced rates of aminoacylation are harmful to bacteria particularly during rapid cell growth. In the case of hCysRS, while the native enzyme exhibits a slower kchem (20 ± 1 s−1) than a °C mutant lacking the CTE (~120 s−1), there is an advantage in having the CTE, because the specificity of tRNA aminoacylation of the native enzyme is higher than that of the mutant7. Thus, hCysRS would benefit from a trade-off between speed and accuracy by having the CTE.
In mitochondria, tRNACys is strictly conserved with A37 from yeast to humans and the occurrence of A37 remains stable (25.93% in yeast and 22.22% in humans, Supplementary Table S6). In addition, mitochondrial CysRSs consistently lack the CTE, suggesting that aminoacylation of tRNACys by these enzymes is operated by a bacteria-like mechanism. Because mitochondrial CysRSs are nuclear-encoded, this bacteria-like mechanism would prevent cross-acylation of the cytosolic G37-bearing tRNACys during their transient residence in the cytoplasm before import into mitochondria. The compartment-specific acylation should prevent two enzymes from acylating the same tRNA, a situation that would promote errors and challenge the stability of the genetic code39.
Cross-species barriers to aminoacylation are widespread among aaRSs. At least for the CysRS enzymes, an interdependent relationship exists between tRNA aminoacylation and tRNA decoding on the ribosome, involving the development of a specific domain in the evolution of CysRS from bacteria to humans. Certainly analysis of other members of the aaRS family for interdependent relationships of tRNA aminoacylation and decoding will be necessary to gain a broader insight into the evolution of these enzymes and their role in the maintenance of the genetic code stability.
Methods
Reagents
The gene for the eCysRS+CTE fusion protein was generated by inserting the coding sequence of the CTE of hCysRS7 (from V642 to Q748) into the eCysRS clone40 at the BamHI site, such that expression of the fusion extended the C-terminal K461 of eCysRS with the sequence of the CTE. Recombinant His-tagged eCysRS, hCysRS, and eCysRS+CTE were each expressed at 37 °C in BL21(DE3) upon induction with 0.4 mM IPTG and were purified using Talon resin, followed by chromatography through a Mono Q column on an Akta FPLC. Enzyme concentrations were measured by Bradford assay with BSA as the standard, and corrected by active site burst assay41. Unmodified transcripts of tRNAs were prepared by in vitro transcription with T7 RNA polymerase from DNA templates generated by primer extension of overlapping olignucleotides. After denaturing gel purification and ethanol precipitation, tRNA transcripts were determined for concentration by absorption at 260 nm and by plateau charging. Tight-coupled ribosomes were prepared from E. coli MRE600 cells, while E. coli His-tagged EF-Tu, IF1, IF2, and IF3 were affinity purified42. The mRNA sequence for translation was the 022 sequence, encoding 5'-GGG-AAG-GAG-GUA-AAA-AUG-XXX-GUU-CTA-UAC-AAG-ACU-CAC-CAC-CAC-CAC-CAC-3', where the underline and bold face letters indicate the Shine-Delgarno sequence and the initiation codon, respectively, and XXX indicates the triplet sequence of the cognate and near-cognate codons for cysteine. This sequence was chosen because its initiation region confers high expression levels in E. coli43. Each mRNA was synthesized by in vitro transcription from a T7 promoter.
Aminoacylation
Steady-state assays40 were performed at 37 °C in a buffer containing 20 mM Tris-HCl, pH 7.5, 20 mM KCl, 10 mM MgCl2, 25 mM DTT, 2 mM ATP, 50 µM 35S-cysteine with 0–120 µM tRNA, and 0–4 nM of CysRS. The Km (tRNA), kcat, and kcat/Km values for each tRNA and enzyme were derived from fitting the data to Michaelis-Menten equation. The kcat/Km data for the G37A mutant was estimated from initial rate of aminoacylation under sub-Km concentrations of the tRNA substrate (40 µM) with 10 µM enzyme. Single-turnover assays were performed similarly but with CysRS in excess of tRNA44 on the RQF-3 KinTek instrument.
GTP hydrolysis
GTPase reactions were performed with 7 mM MgCl2 intermediate between 3.5 mM MgCl2 of HiFi buffer and 20 mM MgCl2 of LoFi buffer23. This buffer was chosen because it conferred higher stoichiometry in complex formation and faster rates compared with the HiFi buffer. Ternary complex was formed by activating 2 µM EF-Tu with 8 nM γ-32P-GTP (6,000 Ci/mmole) for 15 min at 37 °C after which Cys-tRNACys was added to 1.0 µM and the incubation continued for 15 min at 4 °C. Free GTP was removed from the ternary complex by a Centrispin-20 spin. The 70S Initiation complex (70SIC) was formed by incubating 1–6 µM 70S ribosome with a 1.5-fold molar excess of IF1, IF2 and IF3 and a 2-fold excess of native fMet-tRNAfMet and mRNA with 1 mM GTP for 25 min at 37 °C. Titration of initiation factors relative to the ribosome showed a consistent stoichiometry of 0.7 for fMet-tRNAfMet bound per ribosome. Titration of a ternary complex relative to the 70SIC also showed a stoichiometry of 0.7 for each complex per 70SIC. Aliquots of the mixture of a ternary complex and 70SIC were quenched in acid, followed by reaction with sodium molybdate, and extraction with ethyl acetate42. Analysis of kapp versus ribosome concentration fit to a hyperbola (R2 > 0.98) to yield Km and kGTP. Assays with native etRNACys were performed with a crude mixture of E. coli tRNAs isolated from an over-producer strain that generated Cys-etRNACys as 5–7% of total tRNA. The strong affinity of EF-Tu for aa-tRNA over tRNA provided the selectivity for loading Cys-etRNACys to the ribosome. Native fMet-tRNAfMet was prepared similarly, except that aminoacylation with MetRS was done in the presence of methionyl formyl transferase and N10-formyltetrahydrofolate.
Peptide bond formation
Assays were performed by mixing 70SIC (1.0 µM 70S ribosome, 1.5 µM IF1, IF2 and IF3, 1.0 µM 35S-fMet-tRNAfMet, 2 µM mRNA and 1 mM GTP in Buffer A (50 mM Tris-HCl, pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, and 10 mM DTT) and ternary complex (1 µM EF-Tu, 0.5 µM Cys-tRNACys and 1 mM GTP in Buffer A) at 25 °C. Aliquots were quenched in acid and analyzed by electrophoretic TLC45. The fraction of 35S-fMet converted to dipeptide was plotted as a function of time. Single exponential fitting of the plots yielded apparent rate constants for peptide bond formation. If reactions were in multiple turnover conditions, the data would not fit to a single exponential equation. The slow reactions at the near-cognate codon were not due to recycling of Cys-tRNACys, because once Cys-tRNACys left EF-Tu-GTP it could not re-bind to EF-Tu, which would have hydrolyzed GTP to GDP in the absence of a bound aa-tRNA46. Neither were the slow reactions at the near-cognate codon due to hydrolysis of Cys-tRNACys, which would have shown the same plateau level for both the wt and eM1 mutant tRNA.
Supplementary Material
Acknowledgements
We thank Professor Uttam RajBhandary for the expression clone of E. coli methionyl-tRNA formyl transferase and Dr. Takao Igarashi for help with purification of methionyl-tRNA synthetase and methionyl-tRNA formyl transferase. We thank Professors Paul Schimmel and William Merrick for discussion. This work was supported by NIH grants GM081601 (Y.M.H.) and GM071014 (B.S.C).
Footnotes
Author contributions:
C. L., H. G., H. L., and Y. M. H. designed research; C. L. and H. G. performed research; C. L., H. G., B. C., and Y. M. H. analyzed data. All authors discussed and agreed with the content of the manuscript. Y. M. H. wrote the paper.
Competing financial interests:
All of the authors claimed no competing financial interests of the work.
References
- 1.Reynolds NM, Lazazzera BA, Ibba M. Cellular mechanisms that control mistranslation. Nat Rev Microbiol. 2010;8:849–856. doi: 10.1038/nrmicro2472. [DOI] [PubMed] [Google Scholar]
- 2.Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- 3.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakharkar MK, Kangueane P, Sakharkar KR, Zhong Z. Huge proteins in the human proteome and their participation in hereditary diseases. In Silico Biol. 2006;6:275–279. [PubMed] [Google Scholar]
- 5.Kramer EB, Vallabhaneni H, Mayer LM, Farabaugh PJ. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA. 2010;16:1797–1808. doi: 10.1261/rna.2201210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, et al. Kinetic Quality Control of Anticodon Recognition by a Eukaryotic Aminoacyl-tRNA Synthetase. J Mol Biol. 2007;367:1063–1078. doi: 10.1016/j.jmb.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipman RS, Hou YM. Aminoacylation of tRNA in the evolution of an aminoacyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1998;95:13495–13500. doi: 10.1073/pnas.95.23.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinn CL, Tao N, Schimmel P. Species-specific microhelix aminoacylation by a eukaryotic pathogen tRNA synthetase dependent on a single base pair. Biochemistry. 1995;34:12489–12495. doi: 10.1021/bi00039a001. [DOI] [PubMed] [Google Scholar]
- 10.Shiba K, et al. Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescues escherichia coli double-defective mutant. J Biol Chem. 1997;272:22809–22816. doi: 10.1074/jbc.272.36.22809. [DOI] [PubMed] [Google Scholar]
- 11.Wakasugi K, Quinn CL, Tao N, Schimmel P. Genetic code in evolution: switching species-specific aminoacylation with a peptide transplant. Embo J. 1998;17:297–305. doi: 10.1093/emboj/17.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pallanck L, Li S, Schulman LH. The anticodon and discriminator base are major determinants of cysteine tRNA identity in vivo. J Biol Chem. 1992;267:7221–7223. [PubMed] [Google Scholar]
- 13.Hamann CS, Hou YM. Enzymatic aminoacylation of tRNA acceptor stem helices with cysteine is dependent on a single nucleotide. Biochemistry. 1995;34:6527–6532. doi: 10.1021/bi00019a034. [DOI] [PubMed] [Google Scholar]
- 14.Zhang CM, Liu C, Slater S, Hou YM. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys) Nat Struct Mol Biol. 2008;15:507–514. doi: 10.1038/nsmb.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauenstein SI, Hou YM, Perona JJ. The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity. J Biol Chem. 2008;283:21997–22006. doi: 10.1074/jbc.M801838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levitt M. Detailed molecular model for transfer ribonucleic acid. Nature. 1969;224:759–63. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- 17.Nissen P, Thirup S, Kjeldgaard M, Nyborg J. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure. 1999;7:143–156. doi: 10.1016/s0969-2126(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 18.Hauenstein S, Zhang CM, Hou YM, Perona JJ. Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat Struct Mol Biol. 2004;11:1134–1141. doi: 10.1038/nsmb849. [DOI] [PubMed] [Google Scholar]
- 19.Hou YM, Motegi H, Lipman RS, Hamann CS, Shiba K. Conservation of a tRNA core for aminoacylation. Nucleic Acids Res. 1999;27:4743–4750. doi: 10.1093/nar/27.24.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou YM, Westhof E, Giege R. An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc Natl Acad Sci U S A. 1993;90:6776–6780. doi: 10.1073/pnas.90.14.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou YM. Structural elements that contribute to an unusual tertiary interaction in a transfer RNA. Biochemistry. 1994;33:4677–4681. doi: 10.1021/bi00181a603. [DOI] [PubMed] [Google Scholar]
- 22.Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. Embo J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- 24.Wohlgemuth I, Pohl C, Rodnina MV. Optimization of speed and accuracy of decoding in translation. EMBO J. 2010;29:3701–3709. doi: 10.1038/emboj.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruusala T, Ehrenberg M, Kurland CG. Is there proofreading during polypeptide synthesis? EMBO J. 1982;1:741–745. doi: 10.1002/j.1460-2075.1982.tb01240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyltRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villa E, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci U S A. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuette JC, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 30.Gromadski KB, Daviter T, Rodnina MV. A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol Cell. 2006;21:369–377. doi: 10.1016/j.molcel.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Czerwoniec A, et al. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ledoux S, Olejniczak M, Uhlenbeck OC. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat Struct Mol Biol. 2009;16:359–364. doi: 10.1038/nsmb.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fersht A. Structure and Mechanism in Protein Science. New York: W.H. Freeman and Company; 1998. [Google Scholar]
- 34.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Esberg B, Curran JF, Bjork GR. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J Mol Biol. 1997;271:209–221. doi: 10.1006/jmbi.1997.1176. [DOI] [PubMed] [Google Scholar]
- 36.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 37.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 38.Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17:1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 39.Ribas de Pouplana L, Schimmel P. Operational RNA code for amino acids in relation to genetic code in evolution. J Biol Chem. 2001;276:6881–6884. doi: 10.1074/jbc.R000032200. [DOI] [PubMed] [Google Scholar]
- 40.Zhang CM, Christian T, Newberry KJ, Perona JJ, Hou YM. Zinc-mediated Amino Acid Discrimination in Cysteinyl-tRNA Synthetase. J Mol Biol. 2003;327:911–917. doi: 10.1016/s0022-2836(03)00241-9. [DOI] [PubMed] [Google Scholar]
- 41.Fersht AR, et al. Active site titration and aminoacyl adenylate binding stoichiometry of aminoacyl-tRNA synthetases. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 42.Pan D, Zhang CM, Kirillov S, Hou YM, Cooperman BS. Perturbation of the tRNA Tertiary Core Differentially Affects Specific Steps of the Elongation Cycle. J Biol Chem. 2008;283:18431–18440. doi: 10.1074/jbc.M801560200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calogero RA, Pon CL, Canonaco MA, Gualerzi CO. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1988;85:6427–6431. doi: 10.1073/pnas.85.17.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang CM, Perona JJ, Ryu K, Francklyn C, Hou YM. Distinct Kinetic Mechanisms of the Two Classes of Aminoacyl-tRNA Synthetases. J Mol Biol. 2006;361:300–311. doi: 10.1016/j.jmb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 45.Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 46.Fasano O, De Vendittis E, Parmeggiani A. Hydrolysis of GTP by elongation factor Tu can be induced by monovalent cations in the absence of other effectors. J Biol Chem. 1982;257:3145–3150. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.