Abstract
The avalanche of genomic information in the past decade has revealed that natural product biosynthesis using the ribosomal machinery is much more widespread than originally anticipated. Nearly all of these compounds are crafted through posttranslational modifications of a larger precursor peptide that often contains the marching orders for the biosynthetic enzymes. We review here the available information for how the peptide sequences in the precursors govern the posttranslational tailoring processes for several classes of natural products. In addition, we highlight the great potential these leader peptide directed biosynthetic systems offer for engineering conformationally restrained and pharmacophore-rich products with structural diversity that greatly expands the proteinogenic repertoire.
Introduction
The past decade has seen an upsurge in the available genomic information with about 800 fully sequenced bacterial genomes and around 700 unfinished genomes as of August 2009. This explosion of genomic information has provided unparalleled new insights into the genetic capacity of organisms to generate secondary metabolites1, including a rapid rise in the discovery of natural products that are ribosomally synthesized and posttranslationally modified2-13. These tailoring processes release the peptides from the structural and functional constraints imposed on natural ribosomal peptides, while at the same time restricting conformational flexibility to allow better target recognition and increase metabolic and chemical stability. For the vast majority of natural products of ribosomal origin, the initial precursor peptide is much larger than the final product. These precursors typically contain N-terminal leader peptides, and in some cases, C-terminal extensions that are removed in the last step of the maturation process. Interestingly, a recent comprehensive analysis of the structural motifs generated using these pathways concluded that the types of structures accessible through the ribosomal route are remarkably similar to those produced via nonribosomal biosynthesis14. This review will discuss the currently available information regarding the roles of these leader peptides as well as the prospects that leader peptide directed biosynthesis (LDB) offers for natural product engineering.
Proposed Roles for the Leader Peptides
For almost all natural products produced via the ribosome, the precursor genes encode a peptide that contains an N-terminal leader extension in addition to the C-terminal core peptide that is processed to the mature compound (Fig. 1). Many hypotheses have been offered for the function of the leader peptides, but before discussing these proposed roles, we will outline the nomenclature used in this review because the terminology in the literature differs greatly for the various compound classes (see Supplementary Fig. 1 online). The initial ribosomally produced peptides will be referred to as precursor peptides, with the peptide segment that is converted into the mature natural product denoted as the core peptide (Fig. 1). The peptide sequence that is appended to the N-terminus of the core peptide will be termed the leader sequence, and any sequence that is attached to the C-terminus of a core peptide will be referred to as a recognition sequence. For some natural products from higher organisms, an N-terminal signal peptide may also be present that directs the subcellular localization of the peptide.
Figure 1. General Scheme and Examples of Leader Peptide Directed Biosynthesis (LDB).
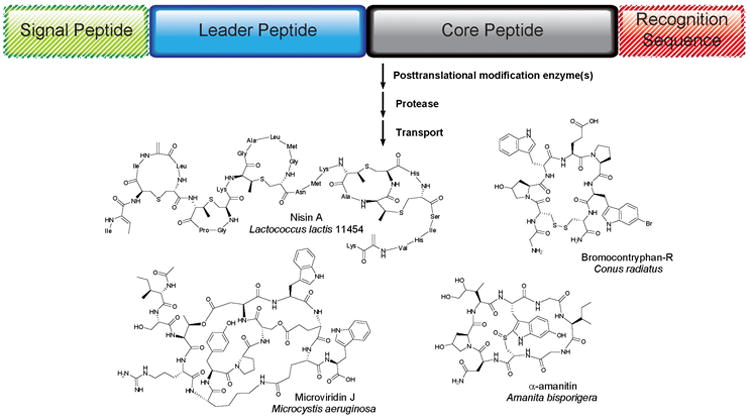
The precursor peptides typically consist of an N-terminal leader and a C-terminal core peptide. A signal peptide governing subcellular localization may be attached to the N-terminus of the leader peptide, and recognition sequences are sometimes found to the C-terminus of the core peptide. The precursor peptides are ribosomally synthesized and posttranslationally modified to their active structures.
The role most commonly proposed for the leader peptides is that of a secretion signal. However, the vast majority of leader peptides of natural product biosynthesis have no homology with the peptides of the typical Sec and twin-arginine translocation pathways that are used in bacteria, archaea and plants to transport proteins across membranes. A second role that is frequently postulated is that of a recognition motif for the posttranslational modification (PTM) enzymes. It is this role that is most enticing from a natural product engineering perspective as it may allow generation of analogs by attachment of core peptide variants or even very different peptides to the leader peptides. A related proposed task is that of a cis-acting chaperone in which the leader actively assists during the PTM process. Taking clues from the roles of leader peptides attached to enzymes15, they may also assist in folding of the precursor peptide, stabilizing the precursor against degradation, or keeping the precursor peptide inactive during biosynthesis inside the host until the appropriate time for secretion and proteolysis. Indeed, many of the proteases involved in ribosomal natural product biosynthesis are localized on the outside of the producing cells and in some cases the protease is part of the transporter. As discussed in this review, support for almost all of these roles has been reported, but the function of the leader peptide differs for different classes of compounds and sometimes even within a certain natural product group. Further delineating the function of these peptide sequences will offer exciting insights into basic biological processes as well as provide improved opportunities to reprogram natural product structures by utilizing the leader peptide directed biosynthetic enzymes.
Lantibiotics
Lantibiotics are distinguished by the presence of thioether crosslinks called lanthionines and methyllanthionines (Fig. 2a). These thioether linkages are introduced by dehydration of a serine or threonine to generate a dehydroalanine or dehydrobutyrine, respectively, followed by addition of a cysteine thiol as illustrated in Fig. 2b for lacticin 481. Including the thioether crosslinks and dehydro amino acids, an impressive 15 different PTMs have been documented in various family members (Fig. 2a)16. Currently, lantibiotics are divided into two classes on the basis of the sequences of their leader peptides and biosynthetic enzymes. The role of the leader peptide has been investigated for both groups using in vivo mutagenesis studies and in vitro reconstitution of the posttranslational modification reactions.
Figure 2. Posttranslational modifications in lantibiotics.
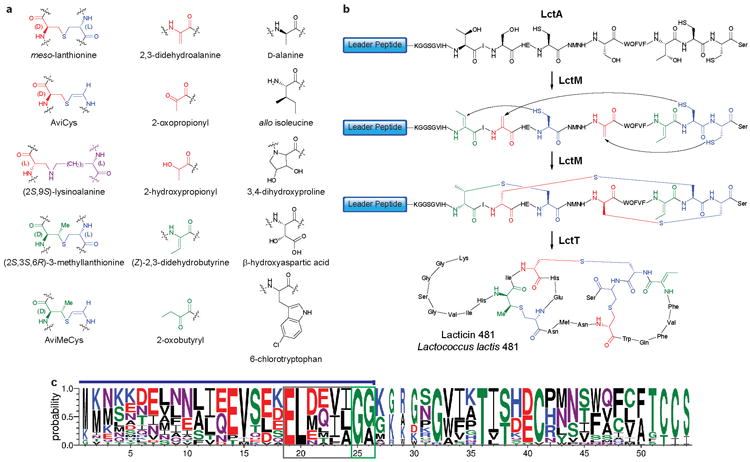
a. Different PTMs found in lantibiotics. Colors denote the amino acid origins: red structures are derived from Ser, green from Thr, blue from Cys, and violet from Lys. b. Biosynthesis of lacticin 481, a class II lantibiotic. The lacticin 481 precursor peptide (LctA) containing an N-terminal leader and C-terminal core peptide is transformed into a polycyclic thioether product through the action of a bifunctional enzyme (LctM) that dehydrates Ser to dehydroalanine and Thr to dehydrobutyrine, and subsequently catalyzes the Michael-type addition of Cys residues to these unsaturated amino acids. The leader is proteolytically removed from the modified core peptide by a bifunctional protease/transporter enzyme (LctT). Coloring scheme as in Figure 2a. Although the process is drawn as complete dehydration before the commencement of cyclization, recent studies suggest the dehydration and cyclization events may be alternating30,105. c. Sequence logo representing sequence alignments of selected class II lantibiotic precursor peptides (for alignments, see Supplementary Fig. 3 online). The probability of each amino acid is depicted by the height of the letter and is scaled (width of the letter) according to how many sequences contributed to that position (i.e. narrow letters were generated from a smaller number of sequences than wider letters)106. Blue line above the sequence logo indicates the leader peptide, the ELXXBX motif is boxed in grey, and the double Gly motif is boxed in green.
Class I lantibiotics include nisin (Fig. 1), a compound used for more than 40 years in more than 80 countries as an effective agent to combat food-borne pathogens17. The leader peptides of class I lantibiotics are about 25 amino acids in length and are rich in Asp residues (Supplementary Fig. 2 online). NMR studies on fully processed nisin attached to its leader peptide did not reveal any interactions between the leader and the nisin molecule, both in solution and in micelles18. Moreover, none of the investigated lantibiotic leader peptides displayed secondary structure in aqueous solution, although they attain α-helical conformations in trifluoroethanol19 and structure prediction tools anticipate helical character. These observations do not support the hypothesis that the leader peptide acts as a chaperone for folding. However, leader peptides attached to the mature lantibiotics greatly reduce or abolish their antimicrobial activity, consistent with a protective mechanism20,21. In addition, ATP-binding cassette transporters clearly use the class I leader peptides for recognition because various engineered non-lantibiotic cargo attached to these leader sequences are secreted22,23. Similarly, the leader peptide of nisin is also recognized by the dehydratase and cyclase enzymes as demonstrated by in vivo processing of non-lantibiotic therapeutic peptides attached to the nisin leader peptide24,25, and by in vitro studies on the cyclase enzyme21. The nisin cyclase, NisC, is made up of an α,α-barrel that contains the active site and a separate domain proposed to contain the leader peptide binding site21.
The leader peptides of class II lantibiotics are typically also rich in Asp and Glu residues, contain an ELXXBXG motif (B = V, L, or I), and usually end in a double Gly motif (Fig. 2c and Supplementary Fig. 3 online). Originally defined as consecutive Gly residues preceding the proteolytic cleavage site of non-lantibiotic bacteriocins26, this motif now also includes GlyAla and GlySer/Thr sequences. Like the leader peptides of class I lantibiotics, the double Gly leader peptides play several roles that include keeping the modified core peptide inactive when still attached to the leader peptide27. In addition, in vitro studies on lacticin 481 have shown that the leader peptide is important for efficient dehydration and cyclization (Fig. 2b), but not essential. When the leader peptide and the core region of the lacticin 481 precursor peptide were presented in trans to lacticin 481 synthetase, dehydration still occurred, albeit with much decreased efficiency28. Unexpectedly, even incubation of the synthetase with just the core peptide resulted in dehydration. Hence, the leader peptide is not a compulsory allosteric element for dehydratase activity. Instead, the enzyme has a low level basal activity in the absence of the leader peptide, suggesting that the leader peptide may influence the equilibrium between an inactive and an active form of the enzyme by binding to the latter29. Upon binding, lacticin 481 synthetase processes its substrate peptide distributively and directionally, moving from N- to C-terminus30, whereas in the absence of the leader peptide, no such directionality was observed28.
Site-directed mutagenesis studies have shown that the double Gly motif is essential for proteolytic processing31,32, but not for installation of lanthionine rings29. Hence, the recognition of the leader peptides is different for the protease and synthetase. It has proven difficult to delineate the exact factors that are essential for recognition by the synthetase as nearly all single point mutants of the lacticin 481 leader peptide were still processed29. A similar plasticity was observed for microcin B17 as discussed below. The only clear disruption of processing by the synthetase was observed with a series of mutants that introduced Pro residues, supporting the hypothesis that the leader peptide attains an α-helical conformation when bound to the synthetase29. Interestingly, leader peptides of the double Gly type with clear homology with lantibiotic leader peptides are also found in many bacteriocins that do not undergo posttranslational modifications (Supplementary Fig. 4 online). Therefore, these leader peptides are thought to only be important for secretion and/or for reducing the biological activity of the bacteriocin33. Hence, the leader peptides of class II lantibiotics may have evolved from a role in secretion to include additional roles in guiding the enzymes involved in lantibiotic biosynthesis. However, the hypothesis that non-conserved amino acids in these leader peptides would convey recognition by the PTM enzymes for each specific lantibiotic is not supported by mutagenesis studies29.
Microcins
Enterobacteria produce a structurally diverse group of ribosomally produced small peptides (<10 kDa) called the microcins34. At present, four kinds of PTMs have been reported for the microcin family. The potent inhibitor of DNA gyrase, microcin B17, contains four oxazole and four thiazole heterocycles (Fig. 3a)35. A different type of cyclization is found in microcin J25, the best-studied member of the lasso peptides that inhibit RNA polymerase (Fig. 3a)36-38. Microcin C7 (also known as C51)34 and microcin E49239 are also posttranslationally modified but without the need of a leader peptide40,41 and will therefore not be discussed here.
Figure 3. Posttranslational modifications in microcin and cytolysin biosynthesis.
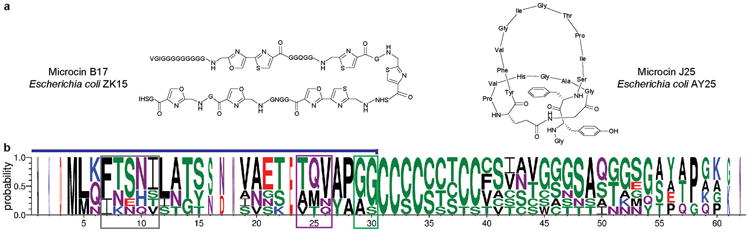
a. Structures of microcin B17 and J25. b. Sequence logo representing sequence alignments of selected cytolysin precursor peptides (for alignments, see Supplementary Fig. 6 online). See the legend to Fig. 2b for further information about the logo format. The blue line above the sequence logo indicates the leader peptide, the FXXXB motif is boxed in grey, the TQV motif is boxed in violet, and the double Gly motif is boxed in green.
Microcin B17 was the first posttranslationally modified peptide antibiotic for which the biosynthesis was reconstituted in vitro42. The precursor peptide contains a 24-amino acid leader peptide that does not carry a net charge and does not show homology with the leader peptides of bacteriocins, lantibiotics, or other microcins. Both in vivo and in vitro studies have shown the requirement of the leader peptide for the installation of the oxazole and thiazole heterocycles42,43. On the other hand, the leader peptide appears not to be involved in transport43,44, an unusual exception to this general role for leader peptides. Furthermore, unlike the observations for the lantibiotic lacticin 481, the leader peptide of microcin B17 cannot act in trans45. The installation of the heterocycles requires a three-protein complex composed of a cyclodehydratase that generates oxazoline and thiazoline structures from GlySer and GlyCys sequences, respectively, a flavin-dependent dehydrogenase that oxidizes these intermediates to oxazoles and thiazoles, and a third scaffolding protein that is required for catalysis. Photocrosslinking experiments suggest that the leader peptide binds to this scaffolding protein46. The first modified residue in the core region of microcin B17 is separated from the end of the leader peptide by an essential47 Gly-rich spacer peptide that is proposed to ensure the correct register between the two segments of the precursor peptide and allow the core region to reach the active sites for modification. Like the maturation process for the lantibiotic lacticin 481, heterocyclization of the microcin B17 precursor takes place by a distributive and directional process, with the synthetase moving from N- to C-terminus of the core region48. Other similarities with the lantibiotic systems are the propensity of the microcin B17 leader peptide to form an amphipathic α-helix45, and the observation that the modified core peptide of microcin B17 with its leader still attached has greatly reduced antimicrobial activity46. A final common theme between lantibiotics and microcin B17 is that many residues in the leader peptide can be mutated without abolishing the ability of the biosynthetic enzymes to process the precursor peptide. However, two residues, Phe8 and Leu12 in a FXXXL motif are critical for binding of the leader peptide to the biosynthetic enzymes45. These observations further support an α-helical conformation as these residues are located on one face of the α-helix structure determined by NMR analysis in aqueous trifluoroethanol. In addition, incorporation of proline residues to disrupt the α-helical nature precluded processing of the core peptide.
A structurally intriguing collection of compounds are the lasso peptides, such as microcin J25, in which the N-terminal amino group is engaged in a lactam bond with a carboxylate side chain of an Asp/Glu residue at position 8 in a manner that traps the C-terminal peptide chain threaded through the ring (Fig. 3a). Bulky, often aromatic, side chains of the amino acids that flank the ring prevent the tail from slipping out of the lasso. The precursor peptide to microcin J25 contains a 37-amino acid leader peptide and a 21-residue core peptide (Supplementary Fig. 5 online). In vitro reconstitution of microcin J25 biosynthesis demonstrated that the leader peptide is required for posttranslational modification by two enzymes49. One protein catalyzes the adenylation of the carboxylate side chain of Glu/Asp8. A second protein cleaves off the leader peptide and installs the new lactam amide bond. The leader peptide cleavage site for the lasso peptides found by database mining is not of the double Gly type49. Because synthetic efforts to construct the lasso peptides have not resulted in the threaded structure37,38, the tail must be held in a near-mature orientation before the lactam is formed. It has been suggested that the leader peptide might be involved in this orienting of the tail50, but it is equally plausible that the biosynthetic enzymes position the C-terminal segment while the ring is formed around it.
Cytolysins
In the past few years, many systems related to microcin B17 have been discovered in sequenced genomes, demonstrating that the ribosomal pathway to thiazole and oxazole containing natural products is widespread and not limited to Gram-negative enterobacteria6,7. Notably, gene clusters encoding the required biosynthetic machinery are found in various human pathogens including Clostridium botulinum, Listeria monocytogenes, Staphylococcus aureus, and Streptococcus pyogenes, as well as in cyanobacteria, archaea, and others. Many of these clusters contain genes that suggest these compounds undergo additional posttranslational tailoring processes6 in much the same way that lantibiotics contain a myriad of structural modifications. Hence, these clusters will be a rich source of novel natural products of great structural diversity. The two systems that have been investigated experimentally are streptolysin S6 and listeriolysin S7 produced by similar routes in S. pyogenes and L. monocytogenes, respectively.
Streptolysin S (SLS) is responsible for the characteristic lysis of red blood cells (β-hemolysis) induced by group A Streptococcus. The structure of the compound has not yet been determined, in part because its activity is rapidly lost in the absence of bacterial cells or other stabilizing factors. It is made from a 53-amino acid precursor peptide containing a 23-residue leader peptide of the double Gly type and a 30-residue core region51 (Fig. 3b and Supplementary Fig. 6 online). Its biosynthetic gene cluster contains orthologs of the three enzymes required for microcin B17 maturation. Heterologous expression of these proteins and the precursor peptide and in vitro reconstitution of its biosynthesis6 has further confirmed that SLS contains oxazoles/thiazoles. In contrast to the systems discussed so far, SLS displays hemolytic activity even when the leader peptide is still attached. The importance of residues in the leader peptide for SLS maturation has been investigated by alanine scanning mutagenesis revealing TQV and FXXXB (B = I, V, Fig. 3b) sequences that are key for recognition, with the latter resembling the FXXXL motif discussed above for microcin B17. Although these motifs are important for SLS maturation, they are not uniformly conserved in the cytolysin leader peptides (Fig. 3b), suggesting that substrate recognition is different for the biosynthetic enzymes of different family members. Another interesting observation is that the leader sequence inhibits microcin B17 biosynthesis in vitro with a Ki in the micromolar range, similar to the Km of the precursor peptide45, but that the leader peptide of the SLS precursor has much higher affinity (KD = 6.7 nM) for its synthetase52. Furthermore, the SLS leader sequence binds to the cyclodehydratase, not the scaffold protein. The relative affinities of the leader peptides of microcin B17 and SLS for their respective biosynthetic enzymes may be a consequence of microcin B17 being inactive with its leader peptide still attached, whereas SLS is very active even before leader peptide removal. Therefore, the tight complex with the cyclodehydratase may protect the producing organism.
Goadsporin is another linear thiazole and oxazole containing natural product that is biosynthesized using similar logic as microcin B1753. Like the thiopeptides discussed below, goadsporin contains dehydroamino acids in addition to the five-membered heterocycles. Its biosynthetic cluster contains enzymes with homology to the microcin B17 and lantibiotic tailoring enzymes, but at present, the role of the leader peptide has not yet been investigated.
Cyanobactins, Amatoxins, and Cyclotides
The patellamides (Fig. 4) belong to the cyanobactin family, which includes more than 100 unique cyclic peptides2. Members of this group are produced by diverse cyanobacteria including symbionts of marine invertebrates2,54,55, and several have antitumor activity. In addition to sharing similar structural features such as thiazoles with the microcins and cytolysins, cyanobactins also share similar biosynthetic pathways2,42,56,57. The heterocyclic structures are the result of posttranslational tailoring of Ser, Thr, and Cys residues by two enzymes (Fig. 4), a cyclodehydratase (PatD), and a bifunctional enzyme with an oxidase domain (PatG). The final step in cyanobactin maturation is the release of the bioactive product via cleavage of the leader and recognition sequences from the core peptides and concomitant cyclization (Fig. 4).
Figure 4. Proposed biosynthesis of patellamides C and A.
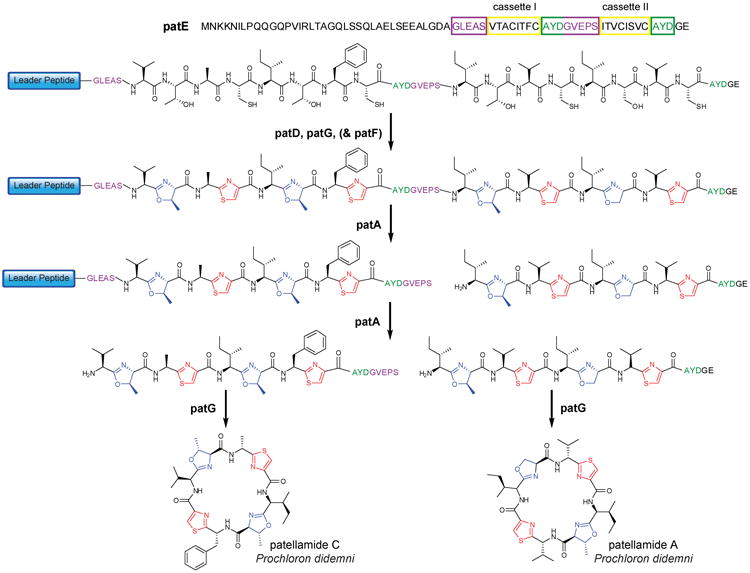
The precursor peptide (PatE) consists of an N-terminal leader peptide and two core peptide cassettes, each sandwiched between two recognition sequences (green/violet). It is hypothesized that PatD is responsible for heterocyclization and PatG may be responsible for oxidation of the heterocycles. PatA sequentially removes cassette I and II from the leader peptide and the N-terminal recognition sequences (violet) and PatG removes the remaining recognition sequences (green) and catalyzes cyclization. PatF is essential for patellamide biosynthesis, but its precise role has not been identified2.
Most cyanobactin precursors possess a leader peptide, two core peptides, and flanking recognition sequences that direct posttranslational modification (Fig. 4). Interestingly, the precursor peptides contain highly conserved leader peptides and recognition sequence regions, but have highly variable core peptides (Supplementary Fig. 7 online). The high conservation of the leader and recognition sequences may reflect conserved recognition elements for the biosynthetic enzymes, while the hypervariable core peptides enable high chemical diversity in the products. The role of the leader peptide in directing the PTMs has not been extensively studied in vitro, but the demonstration that patellamides can be produced heterologously in E. coli54,57 suggests such studies can be carried out.
The C-terminal segment of the cyanobactin precursor peptides harbors two core peptide cassettes (Fig. 4), sandwiched between flanking recognition sequences2. The immediate N-terminal flanking sequences typically consist of G(L/V)E(A/P)S, whereas AYDG(E) is observed C-terminal to the cassettes. Both of these recognition sequences play critical roles in the excision and cyclization of the core peptides. A subtilisin-like serine protease (PatA) catalyzes two amide bond hydrolysis reactions in a sequential manner58, first proteolytically removing cassette II and its C-terminally attached recognition sequence from the full-length precursor, followed by the removal of cassette I from the previously trimmed precursor (Fig. 4). Recent in vitro studies showed that the heterocycles are not required for proteolysis58, further suggesting the protease recognizes the recognition sequences and not the core peptides. Both cleavage reactions result in free N-termini for subsequent cyclization catalyzed by a second subtilisin-like serine protease (PatG) that simultaneously removes the C-terminal recognition sequences through a transamidation mechanism58. Cyanobactins do not have the double Gly protease recognition motif.
Like the cyanobactins, the precursor peptides for amatoxins (e.g. α-amanitin, Fig. 1) and phallotoxins - the cause of most fatal mushroom poisonings – also display conserved leader and recognition sequences and a variable core region that is excised and cyclized (Supplementary Fig. 8 online)5. Proteolytic cleavage at both ends of the core peptide is performed by a Pro oligopeptidase59. Cyclotides produced by plants are cyclized in a head-to-tail fashion and contain a threaded knot of three disulfide bonds60. Like cyanobactins, their precursor peptides contain core peptides with recognition sequences preceding and following each core region (Supplementary Fig. 9 online)61. The presumed helical structure of the precursor peptides62 and the identification of an Asn peptidase that may be involved in their processing63 offer hints for further exploration.
Thiopeptides
Another large group of thiazole containing compounds are the thiopeptides comprising more than 80 members, most of which inhibit protein biosynthesis64. Known for over 60 years, their ribosomal assembly was only revealed by four independent studies in early 200910-13. Most thiopeptides consist of a macrocylic core made up of a string of thiazoles/thiazolines that are joined together by a 6-membered heterocycle that can be a pyridine, a hydroxypyridine or a dehydropiperidine (e.g. thiostrepton, Fig. 5). Aided by fully and partially sequenced genomes, the biosynthetic gene clusters were recently reported for thiostrepton11, thiocillin10, siomycin12, thiomuracin13, and nosiheptide65. The common modifications include dehydrations of Ser and Thr by enzymes with homology to lantibiotic dehydratases, thiazole and thiazoline formation by enzymes with homology with the cyanobactin, microcin B17, and SLS biosynthetic enzymes, and proposed cyclizations of two dehydroalanines to produce the central 6-membered heterocycle by as of yet unidentified proteins (Fig. 5). Additional compound-specific modifications complete the maturation process. As in many of the systems discussed thus far, the leader sequences (34-48 residues) are rich in Asp and Glu and contain some highly conserved hydrophobic residues. Several of the leader peptides end in the typical double Gly motif (Supplementary Fig. 10 online). Currently, the role of the leader peptide has not yet been investigated in vivo or in vitro, but it is likely that it directs at least part of the posttranslational modifications.
Figure 5. Biosynthesis of thiostrepton.
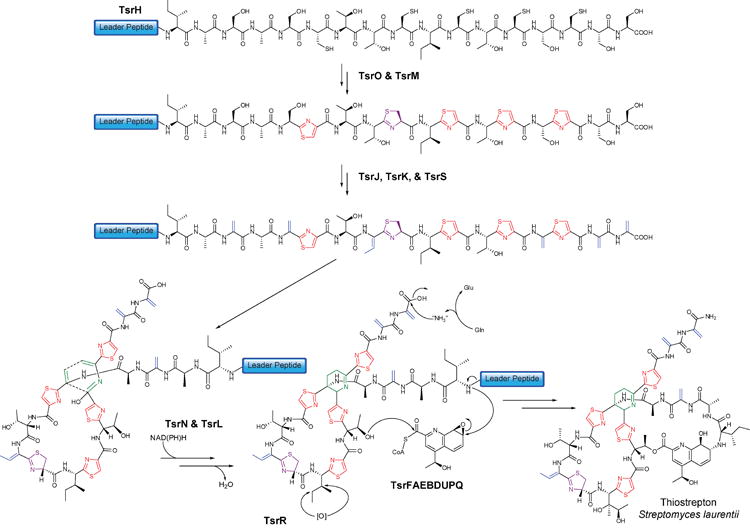
TsrH is ribosomally synthesized as a precursor peptide consisting of an N-terminal leader and C-terminal core peptide. Heterocyclization and dehydration of hydroxyl-amino acids results in a conformationally constrained core peptide, which is further tailored to include a central dehydropiperidine ring, a quinaldic acid moiety, and oxidative modifications. Although the transformations are shown in a particular order, the actual sequence of the modifications is not known. Alphabetical gene nomenclature is used as in 12.
Microviridins
Like most natural products discussed in this review, microviridins have a polycyclic architecture. The crosslinks in these tricyclic compounds are formed by lactone and lactam linkages between the carboxyl groups of Asp/Glu and the side chains of Lys and Ser/Thr residues (Fig. 1). They are potent inhibitors of proteases. Depsipeptides such as the microviridins are typically biosynthesized by non-ribosomal peptide synthetases, but using the fully sequenced genome of an Anabaena strain and the raw genome of a Planktothrix agardhii strain, two groups recently discovered that the microviridins are ribosomally produced8,9. The precursor peptides are made up of a 13-17 residue core peptide and a leader peptide of 34-37 amino acids that does not have a double Gly motif but does harbor several very conserved residues. Interestingly, in the Anabaena strain, a cryptic cluster contains a precursor peptide that appears to contain a double Gly leader peptide, three core regions, and intervening recognition sequences8 in a similar architecture as discussed above for the cyanobactins (Supplementary Fig. 11 online). Microviridin B and J from Microcystis strains were heterologously produced in E. coli8, and microviridin K from P. agardhii was produced in vitro using purified heterologously expressed proteins. The amide and ester bonds are installed by two ATP-grasp ligases with the two ester bonds formed first in a strictly ordered process9,66. The in vitro studies also showed that the leader peptide is required for the tailoring reactions.
Conopeptides
Conopeptides comprise a group of structurally diverse peptides produced by predatory cone snails that target a broad range of ion channels, membrane receptors, and transporters67. A single cone snail species can produce up to 200 different toxins in its venom68 (e.g. bromocontryphan-R, Fig. 1). It is estimated that collectively, 50,000-100,000 different conopeptides may be produced by this genus67,68. They have been classified into different gene superfamilies67,68, and they contain a wide variety of PTMs (Fig. 6a). Conopeptides are translated as precursor peptides of 80-100 residues that are subsequently processed to the mature compound69. The N-termini contain a hydrophobic signal peptide of ∼20 residues that is highly conserved within each gene superfamily (Supplementary Fig. 12 online) and that directs the peptide to the endoplasmic reticulum (ER) where the peptide is modified70,71. This N-terminal signal peptide is followed by a 20-60 residue leader sequence that is conserved within each gene superfamily; little or no sequence homology is found for the leader peptides from different families (Supplementary Fig. 12 online). Interestingly the leader peptides of the contryphans (Fig. 6b), the M-superfamily, and the T-superfamily (Supplementary Fig. 12 online) have several conserved prolines arguing against the helical conformation proposed for most leader peptides discussed in this review. The C-terminal core peptide is typically 11-30 residues in length and, aside from highly conserved Cys residues, shows a remarkably high degree of polymorphism72 (Supplementary Fig. 12 online).
Figure 6. Posttranslational modifications in conopeptides.
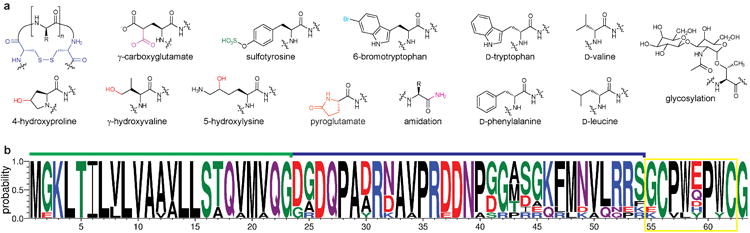
a. Structures of the PTMs found in conopeptides. b. Sequence logo representing sequence alignments of selected contryphan precursor peptides (for alignments, see Supplementary Fig. 12 online). See the legend to Fig. 2b for further information about the logo format. The blue line above the sequence logo indicates the leader peptide, the green line above the sequence logo indicates the signal peptide, and the hypervariable core peptide region is boxed in yellow.
A range of PTM enzymes have been identified67,73,74, but at present, the role(s) of the leader peptides have not been fully investigated. Some conopeptide precursors have a leader peptide containing a γ-carboxylation recognition sequence (γ-CRS) defined by the (K/R)XXZXXXX(K/R) motif where Z is a hydrophobic amino acid71,75-77. γ –Carboxylation can also be achieved with the leader peptide provided in trans75. Protein disulfide isomerase (PDI) assisted disulfide formation is also promoted by the leader peptide compared to oxidative folding of just the core peptide alone77,78. However, not all modifications are guided by the leader peptide as amidation of the C-terminus of the core peptide is governed by the presence of XG-(K/R)n (n = 0-4) motifs in the C-terminal recognition sequences resulting in amidation of residue X67. On the other hand, the sequence (L/X)X(K/X)RX in the leader sequence governs cleavage after the Arg residue in some conopeptides to remove the signal and leader peptide from the modified core region. Proteolysis will not occur when this recognition sequence is found within the core peptide67,79. The exact timing and sub-cellular location of proteolytic removal of the N-terminal signal and leader peptide (and C-terminal recognition sequence) is unknown, but this step is essential to yield the biologically active toxin67. A major protease proposed to catalyze such activity has been identified in Conus textile79. Importantly, the molecular recognition for all these transformations may involve both leader peptide binding as well as recognition of initial PTMs by subsequent modification enzymes80,81. For instance, γ-carboxylation of glutamate residues77 or hydroxylation of proline residues82 can have a dramatic influence on the oxidative folding of the core peptide.
Other ribosomally synthesized natural products
The past decade has also seen the discovery of several head-to-tail backbone cyclized bacteriocins83. They are produced with leader peptides of various lengths, but very little is currently known about their processing. This group of peptides includes subtilosin A that contains unusual thioether bridges in which Cys residues are crosslinked to the α-carbons of Thr and Phe residues84. The leader peptide for subtilosin A formation is unusually short (7 amino acids)85 compared to the typical leader length found for all other systems discussed herein. Interestingly, the gene cluster for the spore killing factor (skf) in bacilli encodes a similar precursor peptide as subtilosin86, suggesting skf may contain similar modifications, but currently its structure is not known. Bacilli also produce a series of structurally related ComX pheromones that are important for inducing competence. They are produced as 55-amino acid precursors that are posttranslationally modified to introduce a prenylated and cyclized Trp87 into a 10-residue final product. Similarly, the thiolactone quorum sensing peptides produced by staphylococci that function as important virulence factors are ribosomally synthesized. For both groups, the precise role of the leader peptide has not yet been investigated, but for the thiolactones, it appears to be involved in localizing the precursor peptide to the membrane88. They have recently been expressed heterologously in E. coli89, setting up future detailed studies.
Reprogramming Leader Peptide Directed Biosynthesis
Although the molecular details of leader peptide recognition by the biosynthetic enzymes are still largely unknown, researchers have already explored the potential of LDB for engineering of natural products as illustrated below.
Lantibiotics
A large number of site-directed mutants of various lantibiotic precursor peptides are correctly processed by the biosynthetic machinery in whole cell studies with homologous and heterologous hosts resulting in lantibiotic variants (for reviews, see90-92). The most extensive examples are full alanine scanning of both peptides of the two-peptide lantibiotic lacticin 314793 as well as the investigation of all possible single amino acid mutations for mersacidin94. Collectively, these in vivo studies have illustrated the remarkably promiscuous nature of the biosynthetic enzymes and in some instances, these investigations have resulted in compounds with improved properties95-97. In vitro engineering studies have also demonstrated that core peptides containing nonproteinogenic amino acids can be modified to lantibiotic analogs with improved antimicrobial activities98. Intriguingly, the lantibiotic biosynthetic enzymes have not only been used to prepare analogs, but also to install dehydro amino acids and thioether rings in a large variety of other peptides attached to the leader peptides24,25,99-101, further highlighting the potential of LDB. In these studies, the nisin dehydratase dehydrated Ser/Thr residues that were located as far as 42 amino acids C-terminal to the end of the leader peptide24. Finally, the biosynthetic enzymes involved in haloduracin biosynthesis were still active when a recognition sequence for a commercial protease was engineered at the junction between the leader peptide and the core peptide, allowing leader peptide removal even when the endogenous protease was not available3.
Microcins and Cytolysins
The microcin B17 biosynthetic pathway has been explored both in vivo and in vitro for its potential to generate analogs for structure-function relationships102,103. These studies have demonstrated that oxazoles can be interchanged for thiazoles and vice versa, or can be eliminated or introduced at non-native positions. On the other hand, the Ser residues involved in oxazole formation could not be replaced by Thr, and the Gly residues that take part in both oxazole and thiazole formation could not be substituted by other amino acids47, showing some limitations to the substrate promiscuity. Interestingly, in vitro the streptolysin S synthetase partially installed oxazoles/thiazoles into the microcin B17 precursor as well as into a related precursor peptide from C. botulinum despite the leader peptides of the latter two having only very limited sequence identity with the SLS leader52. Moreover, an entirely artificial core peptide was designed and attached to the SLS leader peptide and after in vitro synthetase action, a novel cytolytic product was obtained.
Engineering of the lasso peptides has focused so far on in vivo whole cell experiments in which the gene for the precursor peptide was mutated. No less than 381 mutants of microcin J25 have been probed in the native E. coli host, resulting in 242 mutants that were correctly procesed104. Only the residues that form the noose of the lasso (Gly1 and Glu8) and Gly2 were intolerant to substitution, while only conserved substitutions were compatible at the second aromatic amino acid (Tyr20) that prevents slippage of the tail out of the lactam ring.
Patellamides, Microviridins, and Conopeptides
The portable nature of leader peptide directed biosynthesis has also been demonstrated for the cyanobactins. Genetic engineering of the patellamide hypervariable region and heterologous expression in E. coli resulted in the production of a novel cyclic peptide57. Similarly, co-expression of the machinery involved in production of the hexapeptide patellin 2 and the octapeptide patellin 3 with a precursor peptide for the heptapeptide trunkamide resulted in the production of the latter56. The components of the conopeptide biosynthetic machinery that have been investigated for engineering studies have also shown great potential. For instance, appending the leader peptide with the γ-CRS sequence to either N- or C-terminus of a non-native synthetic peptide resulted in γ-carboxylation of glutamate residues in the synthetic peptide71,75. In contrast, the microviridin biosynthetic machinery so far has proven less amenable to leader peptide directed engineering. Whereas substitution of Thr with Ser in the lactone generating step was tolerated, efforts to increase or decrease the size of the ring were unsuccessful66.
Summary and Outlook
Given the remarkable pace at which natural products of ribosomal origin are being discovered, it is likely that new classes of such compounds will be reported in the near future. Furthermore, the number of members of existing classes will rapidly increase through genome database mining exercises that focus on conserved sequences of either the leader peptides or the PTM enzymes. The biosynthetic enzymes for most of the systems discussed here are inherently promiscuous as they perform posttranslational modifications on multiple residues in the core peptide(s) that are in very different sequence contexts. The strategy to bind a leader peptide/recognition sequence in order to modify core peptides with great sequence tolerance offers a very efficient approach for rapid evolution of structural and functional diversity through hypermutation of the core sequences. It may therefore come as no surprise that the leader peptide directed biosynthetic strategy appears to be widespread in nature. As a corollary, most of these systems will likely lend themselves to leader peptide directed bioengineering to produce natural product analogs, and ultimately to provide a toolbox to design and create tailor-made pharmacophore-rich products with structural diversity that greatly exceeds the proteinogenic repertoire.
For these goals to be reached, the role of the leader peptide needs to be investigated in greater detail for all systems discussed here and especially structural information of the leader peptides bound to the posttranslational modification machinery would be extremely valuable. In all investigated cases, the leader peptides adopt an unstructured conformation in aqueous solution but addition of trifluoroethanol readily stabilizes an α-helical conformation, Pro mutants abolish correct processing, and key residues identified by mutagenesis tend to cluster on one face of a helix. It is anticipated that the biosynthetic enzymes may also stabilize and bind the helical conformation, but no direct evidence is available for this model. Nevertheless, even in the absence of detailed knowledge of the molecular recognition between the leader peptides and the biosynthetic enzymes, the power of leader peptide directed engineering has already been demonstrated for generating natural product analogs.
Supplementary Material
Acknowledgments
Our work on LDB has been supported by the National Institutes of Health (GM58822). We thank Prof. D. Mitchell (UIUC) for helpful discussions.
Footnotes
Competing Interests Statement: The authors declare that they have no competing financial interests
References
- 1.Bode HB, Muller R. The impact of bacterial genomics on natural product research. Angew Chem, Int Ed Engl. 2005;44:6828–46. doi: 10.1002/anie.200501080. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt EW, et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102:7315–20. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McClerren AL, et al. Discovery and in vitro biosynthesis of haloduracin, a new two-component lantibiotic. Proc Natl Acad Sci USA. 2006;103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawton EM, Cotter PD, Hill C, Ross RP. Identification of a novel two-peptide lantibiotic, Haloduracin, produced by the alkaliphile Bacillus halodurans C-125. FEMS Microbiol Lett. 2007;267:64–71. doi: 10.1111/j.1574-6968.2006.00539.x. [DOI] [PubMed] [Google Scholar]
- 5.Hallen HE, Luo H, Scott-Craig JS, Walton JD. Gene family encoding the major toxins of lethal Amanita mushrooms. Proc Natl Acad Sci USA. 2007;104:19097–101. doi: 10.1073/pnas.0707340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SW, et al. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad Sci USA. 2008;105:5879–84. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter PD, et al. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008;4:e1000144. doi: 10.1371/journal.ppat.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziemert N, Ishida K, Liaimer A, Hertweck C, Dittmann E. Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angew Chem, Int Ed Engl. 2008;47:7756–9. doi: 10.1002/anie.200802730. [DOI] [PubMed] [Google Scholar]
- 9.Philmus B, Christiansen G, Yoshida WY, Hemscheidt TK. Post-translational modification in microviridin biosynthesis. ChemBioChem. 2008;9:3066–73. doi: 10.1002/cbic.200800560. [DOI] [PubMed] [Google Scholar]
- 10.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc Natl Acad Sci USA. 2009;106:2549–53. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J Am Chem Soc. 2009;131:4327–34. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 12.Liao R, et al. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol. 2009;16:141–7. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris RP, et al. Ribosomally Synthesized Thiopeptide Antibiotics Targeting Elongation Factor Tu. J Am Chem Soc. 2009:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 14.McIntosh JA, Donia MS, Schmidt EW. Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat Prod Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun P, Tommassen J. Function of bacterial propeptides. Trends Microbiol. 1998;6:6–8. doi: 10.1016/S0966-842X(97)01188-8. [DOI] [PubMed] [Google Scholar]
- 16.Willey JM, van der Donk WA. Lantibiotics: Peptides of Diverse Structure and Function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 17.Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol Life Sci. 2008;65:455–476. doi: 10.1007/s00018-007-7171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Hooven HW, Rollema HS, Siezen RJ, Hilbers CW, Kuipers OP. Structural features of the final intermediate in the biosynthesis of the lantibiotic nisin. Influence of the leader peptide. Biochemistry. 1997;36:14137–45. doi: 10.1021/bi9713106. [DOI] [PubMed] [Google Scholar]
- 19.Beck-Sickinger AG, Jung G. In: Nisin and Novel Lantibiotics. Jung G, Sahl HG, editors. ESCOM; Leiden: 1991. pp. 218–230. [Google Scholar]
- 20.van der Meer JR, et al. Influence of amino acid substitutions in the nisin leader peptide on biosynthesis and secretion of nisin by Lactococcus lactis. J Biol Chem. 1994;269:3555–62. [PubMed] [Google Scholar]
- 21.Li B, et al. Structure and Mechanism of the Lantibiotic Cyclase Involved in Nisin Biosynthesis. Science. 2006;311:1464–1467. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- 22.Izaguirre G, Hansen JN. Use of alkaline phosphatase as a reporter polypeptide to study the role of the subtilin leader segment and the SpaT transporter in the posttranslational modifications and secretion of subtilin in Bacillus subtilis 168. Appl Environ Microbiol. 1997;63:3965–71. doi: 10.1128/aem.63.10.3965-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuipers A, et al. NisT, the transporter of the lantibiotic nisin, can transport fully modified, dehydrated and unmodified prenisin and fusions of the leader peptide with non-lantibiotic peptides. J Biol Chem. 2004;279:22176–82. doi: 10.1074/jbc.M312789200. [DOI] [PubMed] [Google Scholar]
- 24.Rink R, et al. Production of dehydroamino acid-containing peptides by Lactococcus lactis. Appl Environ Microbiol. 2007;73:1792–1796. doi: 10.1128/AEM.02350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kluskens LD, et al. Post-translational Modification of Therapeutic Peptides By NisB, the Dehydratase of the Lantibiotic Nisin. Biochemistry. 2005;44:12827–12834. doi: 10.1021/bi050805p. [DOI] [PubMed] [Google Scholar]
- 26.Håvarstein LS, Holo H, Nes IF. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by gram-positive bacteria. Microbiology. 1994;140(Pt 9):2383–9. doi: 10.1099/13500872-140-9-2383. [DOI] [PubMed] [Google Scholar]
- 27.Xie L, et al. Lacticin 481: in vitro reconstitution of lantibiotic synthetase activity. Science. 2004;303:679–81. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 28.Levengood MR, Patton GC, van der Donk WA. The Leader Peptide is not Required for Post-translational Modification by Lacticin 481 Synthetase. J Am Chem Soc. 2007;129:10314–15. doi: 10.1021/ja072967+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patton GC, Paul M, Cooper LE, Chatterjee C, van der Donk WA. The Importance of the Leader Sequence for Directing Lanthionine Formation in Lacticin 481. Biochemistry. 2008;47:7342–51. doi: 10.1021/bi800277d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee MV, et al. Distributive and directional behavior of lantibiotic synthetases revealed by high-resolution tandem mass spectrometry. J Am Chem Soc. 2009;131:12258–64. doi: 10.1021/ja9033507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen P, Qi FX, Novak J, Krull RE, Caufield PW. Effect of amino acid substitutions in conserved residues in the leader peptide on biosynthesis of the lantibiotic mutacin II. FEMS Microbiol Lett. 2001;195:139–44. doi: 10.1111/j.1574-6968.2001.tb10511.x. [DOI] [PubMed] [Google Scholar]
- 32.Furgerson Ihnken LA, Chatterjee C, van der Donk WA. In vitro Reconstitution and Substrate Specificity of a Lantibiotic Protease. Biochemistry. 2008;47:7352–63. doi: 10.1021/bi800278n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sprules T, Kawulka KE, Gibbs AC, Wishart DS, Vederas JC. NMR solution structure of the precursor for carnobacteriocin B2, an antimicrobial peptide from Carnobacterium piscicola. Eur J Biochem. 2004;271:1748–56. doi: 10.1111/j.1432-1033.2004.04085.x. [DOI] [PubMed] [Google Scholar]
- 34.Duquesne S, Destoumieux-Garzon D, Peduzzi J, Rebuffat S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep. 2007;24:708–34. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 35.Bayer A, Freund S, Jung G. Posttranslational Backbone Modifications in the Ribosomal Biosynthesis of the Glycine-Rich Antibiotic Microcin B17. Angew Chem, Int Ed Engl. 1993;32:1336–1339. [Google Scholar]
- 36.Wilson KA, et al. Structure of microcin J25, a peptide inhibitor of bacterial RNA polymerase, is a lassoed tail. J Am Chem Soc. 2003;125:12475–83. doi: 10.1021/ja036756q. [DOI] [PubMed] [Google Scholar]
- 37.Bayro MJ, et al. Structure of antibacterial peptide microcin J25: a 21-residue lariat protoknot. J Am Chem Soc. 2003;125:12382–3. doi: 10.1021/ja036677e. [DOI] [PubMed] [Google Scholar]
- 38.Rosengren KJ, et al. Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone. J Am Chem Soc. 2003;125:12464–74. doi: 10.1021/ja0367703. [DOI] [PubMed] [Google Scholar]
- 39.Thomas X, et al. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J Biol Chem. 2004;279:28233–42. doi: 10.1074/jbc.M400228200. [DOI] [PubMed] [Google Scholar]
- 40.Guijarro JI, et al. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J Biol Chem. 1995;270:23520–32. doi: 10.1074/jbc.270.40.23520. [DOI] [PubMed] [Google Scholar]
- 41.Nolan EM, Fischbach MA, Koglin A, Walsh CT. Biosynthetic tailoring of microcin E492m: post-translational modification affords an antibacterial siderophore-peptide conjugate. J Am Chem Soc. 2007;129:14336–47. doi: 10.1021/ja074650f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science. 1996;274:1188–93. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 43.Madison LL, Vivas EI, Li YM, Walsh CT, Kolter R. The leader peptide is essential for the post-translational modification of the DNA-gyrase inhibitor microcin B17. Mol Microbiol. 1997;23:161–8. doi: 10.1046/j.1365-2958.1997.2041565.x. [DOI] [PubMed] [Google Scholar]
- 44.Yorgey P, Davagnino J, Kolter R. The maturation pathway of microcin B17, a peptide inhibitor of DNA gyrase. Mol Microbiol. 1993;9:897–905. doi: 10.1111/j.1365-2958.1993.tb01747.x. [DOI] [PubMed] [Google Scholar]
- 45.Roy RS, Kim S, Baleja JD, Walsh CT. Role of the microcin B17 propeptide in substrate recognition: solution structure and mutational analysis of McbA1-26. Chem Biol. 1998;5:217–28. doi: 10.1016/s1074-5521(98)90635-4. [DOI] [PubMed] [Google Scholar]
- 46.Milne JC, et al. Cofactor requirements and reconstitution of microcin B17 synthetase: a multienzyme complex that catalyzes the formation of oxazoles and thiazoles in the antibiotic microcin B17. Biochemistry. 1999;38:4768–81. doi: 10.1021/bi982975q. [DOI] [PubMed] [Google Scholar]
- 47.Sinha Roy R, Belshaw PJ, Walsh CT. Mutational analysis of posttranslational heterocycle biosynthesis in the gyrase inhibitor microcin B17: distance dependence from propeptide and tolerance for substitution in a GSCG cyclizable sequence. Biochemistry. 1998;37:4125–36. doi: 10.1021/bi9728250. [DOI] [PubMed] [Google Scholar]
- 48.Kelleher NL, Hendrickson CL, Walsh CT. Posttranslational Heterocyclization of Cysteine and Serine Residues in the Antibiotic Microcin B17: Distributivity and Directionality. Biochemistry. 1999;38:15623–15630. doi: 10.1021/bi9913698. [DOI] [PubMed] [Google Scholar]
- 49.Duquesne S, et al. Two Enzymes Catalyze the Maturation of a Lasso Peptide in Escherichia coli. Chem Biol. 2007;14:793–803. doi: 10.1016/j.chembiol.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 50.Tsai CJ, Ma B, Nussinov R. Intra-molecular chaperone: the role of the N-terminal in conformational selection and kinetic control. Phys Biol. 2009;6:13001. doi: 10.1088/1478-3975/6/1/013001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nizet V, et al. Genetic locus for streptolysin S production by group A streptococcus. Infect Immun. 2000;68:4245–54. doi: 10.1128/iai.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitchell DA, et al. Structural and functional dissection of the heterocyclic peptide cytotoxin streptolysin S. J Biol Chem. 2009;284:13004–12. doi: 10.1074/jbc.M900802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Onaka H, Nakaho M, Hayashi K, Igarashi Y, Furumai T. Cloning and characterization of the goadsporin biosynthetic gene cluster from Streptomyces sp TP-A0584. Microbiology. 2005;151:3923–33. doi: 10.1099/mic.0.28420-0. [DOI] [PubMed] [Google Scholar]
- 54.Long PF, Dunlap WC, Battershill CN, Jaspars M. Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. ChemBioChem. 2005;6:1760–5. doi: 10.1002/cbic.200500210. [DOI] [PubMed] [Google Scholar]
- 55.Ziemert N, et al. Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa. Appl Environ Microbiol. 2008;74:1791–7. doi: 10.1128/AEM.02392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4:341–3. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donia MS, et al. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2:729–35. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 58.Lee J, McIntosh J, Hathaway BJ, Schmidt EW. Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates. J Am Chem Soc. 2009;131:2122–4. doi: 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo H, Hallen-Adams HE, Walton JD. Processing of the Phalloidin Proprotein by Prolyl Oligopeptidase from the Mushroom Conocybe albipes. J Biol Chem. 2009;284:18070–7. doi: 10.1074/jbc.M109.006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gruber CW, et al. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell. 2008;20:2471–83. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jennings C, West J, Waine C, Craik D, Anderson M. Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Oldenlandia affinis. Proc Natl Acad Sci USA. 2001;98:10614–9. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dutton JL, et al. Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J Biol Chem. 2004;279:46858–67. doi: 10.1074/jbc.M407421200. [DOI] [PubMed] [Google Scholar]
- 63.Saska I, et al. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J Biol Chem. 2007;282:29721–8. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- 64.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 65.Yu Y, et al. Nosiheptide Biosynthesis Featuring a Unique Indole Side Ring Formation on the Characteristic Thiopeptide Framework. ACS Chem Biol. 2009 doi: 10.1021/cb900133x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Philmus B, Guerrette JP, Hemscheidt TK. Substrate specificity and scope of MvdD, a GRASP-like ligase from the microviridin biosynthetic gene cluster. ACS Chem Biol. 2009;4:429–34. doi: 10.1021/cb900088r. [DOI] [PubMed] [Google Scholar]
- 67.Buczek O, Bulaj G, Olivera BM. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 2005;62:3067–79. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Halai R, Craik DJ. Conotoxins: natural product drug leads. Nat Prod Rep. 2009;26:526–36. doi: 10.1039/b819311h. [DOI] [PubMed] [Google Scholar]
- 69.Woodward SR, Cruz LJ, Olivera BM, Hillyard DR. Constant and hypervariable regions in conotoxin propeptides. Embo J. 1990;9:1015–1020. doi: 10.1002/j.1460-2075.1990.tb08204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Craig AG, Pradip B, Baldomero MO. Post-translationally modified neuropeptides from Conus venoms. Eur J Biochem. 1999;264:271–275. doi: 10.1046/j.1432-1327.1999.00624.x. [DOI] [PubMed] [Google Scholar]
- 71.Brown MA, et al. Precursors of Novel Gla-Containing Conotoxins Contain a Carboxy-Terminal Recognition Site That Directs gamma-Carboxylation. Biochemistry. 2005;44:9150–9159. doi: 10.1021/bi0503293. [DOI] [PubMed] [Google Scholar]
- 72.Olivera BM, et al. Speciation of cone snails and interspecific hyperdivergence of their venom peptides. Potential evolutionary significance of introns. Ann N Y Acad Sci. 1999;870:223–37. doi: 10.1111/j.1749-6632.1999.tb08883.x. [DOI] [PubMed] [Google Scholar]
- 73.McIntosh JM, Olivera BM, Cruz LJ, Gray WR. Gamma-carboxyglutamate in a neuroactive toxin. J Biol Chem. 1984;259:14343–14346. [PubMed] [Google Scholar]
- 74.Bulaj G. Formation of disulfide bonds in proteins and peptides. Biotechnol Adv. 2005;23:87–92. doi: 10.1016/j.biotechadv.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 75.Bandyopadhyay PK, et al. Conantokin-G Precursor and Its Role in gamma-Carboxylation by a Vitamin K-dependent Carboxylase from a Conus Snail. J Biol Chem. 1998;273:5447–5450. doi: 10.1074/jbc.273.10.5447. [DOI] [PubMed] [Google Scholar]
- 76.Eva C, et al. Novel gamma-carboxyglutamic acid-containing peptides from the venom of Conus textile. FEBS J. 2006;273:2779–2788. doi: 10.1111/j.1742-4658.2006.05294.x. [DOI] [PubMed] [Google Scholar]
- 77.Bulaj G, et al. Efficient oxidative folding of conotoxins and the radiation of venomous cone snails. Proc Natl Acad Sci U S A. 2003;100:14562–14568. doi: 10.1073/pnas.2335845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buczek O, Olivera BM, Bulaj G. Propeptide Does Not Act as an Intramolecular Chaperone but Facilitates Protein Disulfide Isomerase-Assisted Folding of a Conotoxin Precursor. Biochemistry. 2004;43:1093–1101. doi: 10.1021/bi0354233. [DOI] [PubMed] [Google Scholar]
- 79.Milne TJ, Abbenante G, Tyndall JDA, Halliday J, Lewis RJ. Isolation and Characterization of a Cone Snail Protease with Homology to CRISP Proteins of the Pathogenesis-related Protein Superfamily. J Biol Chem. 2003;278:31105–31110. doi: 10.1074/jbc.M304843200. [DOI] [PubMed] [Google Scholar]
- 80.Craig AG, et al. A Novel Post-translational Modification Involving Bromination of Tryptophan. Identification of the residues, L-6-bromotryptophan, in peptides from Conus imperialis and Conus radiatus venom. J Biol Chem. 1997;272:4689–4698. doi: 10.1074/jbc.272.8.4689. [DOI] [PubMed] [Google Scholar]
- 81.Pisarewicz K, Mora D, Pflueger FC, Fields GB, Mari F. Polypeptide Chains Containing D-gamma-Hydroxyvaline. J Am Chem Soc. 2005;127:6207–6215. doi: 10.1021/ja050088m. [DOI] [PubMed] [Google Scholar]
- 82.Lopez-Vera E, Walewska A, Skalicky JJ, Olivera BM, Bulaj G. Role of Hydroxyprolines in the in Vitro Oxidative Folding and Biological Activity of Conotoxins. Biochemistry. 2008;47:1741–1751. doi: 10.1021/bi701934m. [DOI] [PubMed] [Google Scholar]
- 83.Maqueda M, et al. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol Rev. 2008;32:2–22. doi: 10.1111/j.1574-6976.2007.00087.x. [DOI] [PubMed] [Google Scholar]
- 84.Kawulka K, et al. Structure of Subtilosin A, an Antimicrobial Peptide from Bacillus subtilis with Unusual Posttranslational Modifications Linking Cysteine Sulfurs to a-Carbons of Phenylalanine and Threonine. J Am Chem Soc. 2003;125:4726–4727. doi: 10.1021/ja029654t. [DOI] [PubMed] [Google Scholar]
- 85.Zheng G, Yan LZ, Vederas JC, Zuber P. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J Bacteriol. 1999;181:7346–55. doi: 10.1128/jb.181.23.7346-7355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Molle V, et al. The Spo0A regulon of Bacillus subtilis. Mol Microbiol. 2003;50:1683–701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 87.Okada M, et al. Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat Chem Biol. 2005;1:23–4. doi: 10.1038/nchembio709. [DOI] [PubMed] [Google Scholar]
- 88.Zhang L, Lin J, Ji G. Membrane anchoring of the AgrD N-terminal amphipathic region is required for its processing to produce a quorum-sensing pheromone in Staphylococcus aureus. J Biol Chem. 2004;279:19448–56. doi: 10.1074/jbc.M311349200. [DOI] [PubMed] [Google Scholar]
- 89.Thoendel M, Horswill AR. Identification of Staphylococcus aureus AgrD residues required for autoinducing peptide biosynthesis. J Biol Chem. 2009;284:21828–38. doi: 10.1074/jbc.M109.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuipers OP, et al. Protein engineering of lantibiotics. Antonie van Leeuwenhoek. 1996;69:161–169. doi: 10.1007/BF00399421. [DOI] [PubMed] [Google Scholar]
- 91.Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and Mode of Action of Lantibiotics. Chem Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 92.Cortés J, Appleyard AN, Dawson MJ. Whole-cell generation of lantibiotic variants. Methods Enzymol. 2009;458:559–74. doi: 10.1016/S0076-6879(09)04822-8. [DOI] [PubMed] [Google Scholar]
- 93.Cotter PD, et al. Complete alanine scanning of the two-component lantibiotic lacticin 3147: generating a blueprint for rational drug design. Mol Microbiol. 2006;62:735–47. doi: 10.1111/j.1365-2958.2006.05398.x. [DOI] [PubMed] [Google Scholar]
- 94.Appleyard AN, et al. Dissecting structural and functional diversity of the lantibiotic mersacidin. Chem Biol. 2009;16:490–8. doi: 10.1016/j.chembiol.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu W, Hansen JN. Enhancement of the chemical and antimicrobial properties of subtilin by site-directed mutagenesis. J Biol Chem. 1992;267:25078–85. [PubMed] [Google Scholar]
- 96.Field D, Connor PM, Cotter PD, Hill C, Ross RP. The generation of nisin variants with enhanced activity against specific gram-positive pathogens. Mol Microbiol. 2008;69:218–30. doi: 10.1111/j.1365-2958.2008.06279.x. [DOI] [PubMed] [Google Scholar]
- 97.Rollema HS, Kuipers OP, Both P, de Vos WM, Siezen RJ. Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl Environ Microbiol. 1995;61:2873–8. doi: 10.1128/aem.61.8.2873-2878.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Levengood MR, Knerr PJ, Oman TJ, van der Donk WA. In vitro mutasynthesis of lantibiotic analogues containing nonproteinogenic amino acids. J Am Chem Soc. 2009;131:12024–5. doi: 10.1021/ja903239s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chatterjee C, Patton GC, Cooper L, Paul M, van der Donk WA. Engineering Dehydro Amino Acids and Thioethers into Peptides using Lacticin 481 Synthetase. Chem Biol. 2006;13:1109–1117. doi: 10.1016/j.chembiol.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 100.Rink R, et al. NisC, the Cyclase of the Lantibiotic Nisin, Can Catalyze Cyclization of Designed Nonlantibiotic Peptides. Biochemistry. 2007;46:13179–13189. doi: 10.1021/bi700106z. [DOI] [PubMed] [Google Scholar]
- 101.Levengood MR, van der Donk WA. Use of Lantibiotic Synthetases for the Preparation of Bioactive Constrained Peptides. Bioorg Med Chem Lett. 2008;18:3025–8. doi: 10.1016/j.bmcl.2008.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sinha Roy R, Kelleher NL, Milne JC, Walsh CT. In vivo processing and antibiotic activity of microcin B17 analogs with varying ring content and altered bisheterocyclic sites. Chem Biol. 1999;6:305–18. doi: 10.1016/s1074-5521(99)80076-3. [DOI] [PubMed] [Google Scholar]
- 103.Zamble DB, et al. In vitro characterization of DNA gyrase inhibition by microcin B17 analogs with altered bisheterocyclic sites. Proc Natl Acad Sci USA. 2001;98:7712–7. doi: 10.1073/pnas.141225698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pavlova O, Mukhopadhyay J, Sineva E, Ebright RH, Severinov K. Systematic structure-activity analysis of microcin J25. J Biol Chem. 2008;283:25589–95. doi: 10.1074/jbc.M803995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lubelski J, Khusainov R, Kuipers OP. Directionality and Coordination of Dehydration and Ring Formation during Biosynthesis of the Lantibiotic Nisin. J Biol Chem. 2009;284:25962–72. doi: 10.1074/jbc.M109.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


