Abstract
Accurate measurements of the thermodynamic stability of folded membrane proteins require methods for monitoring their conformation that are free of experimental artifacts. For tryptophan fluorescence emission experiments with membrane proteins folded into liposomes, there are two significant sources of artifacts: the first is light scattering by the liposomes; the second is the nonlinear relationship of some tryptophan spectral parameters with changes in protein conformation. Both of these sources of error can interfere with the method of determining the reversible equilibrium thermodynamic stability of proteins using titrations of chemical denaturants. Here, we present methods to manage light scattering by liposomes for tryptophan emission experiments and to properly monitor tryptophan spectra as a function of protein conformation. Our methods are tailored to the titrations of membrane proteins using common chemical denaturants. One of our recommendations is to collect and analyze the right-angle light scattering peak that occurs around the excitation wave- length in a fluorescence experiment. Another recommendation is to use only those tryptophan spectral parameters that are linearly proportional to the protein conformational population. We show that other commonly used spectral commonly used parameters lead to errors in protein stability measurements.
1 Introduction
The physical forces governing the thermodynamic stability of a membrane protein are important cues to the relationship of the protein’s structure and its function. The most relevant measurements of a membrane protein’s stability will come from studies that use thermodynamically stable mimetics of biological lipid bilayers. Stable mimetics include large unilamellar vesicles (LUVs) composed of glycerophospholipids. However, LUVs can pose challenges to membrane protein experiments with luminescent spectroscopy because of their tendency to scatter light.
One example of luminescent spectroscopy that is popular for membrane protein studies is tryptophan fluorescence emission spectroscopy. All three research groups that have so far reported reversible equilibrium thermodynamic information for the stability of membrane proteins in liposomes chose tryptophan fluorescence emission as their primary source of data on protein conformational states (Hong and Tamm, 2004; Sanchez et al., 2008; Huysmans et al., 2010). These three groups also employed titrations of chemical denaturants as a technique to perturb equilibrium between the folded and unfolded conformations of their respective membrane proteins. Denaturant titrations pose two problems that need to be solved for thermodynamic studies of membrane proteins: (a) the light scattering by LUVs depends on the denaturant concentration, and (b) a mixture of protein conformations in a sample has a non-linear relationship with most tryptophan spectral parameters. In this paper, we propose solutions to overcome both of these problems that enable accurate measurements of tryptophan emission spectra and membrane protein stabilities.
2 Issues with managing light scattering from liposomes
Light scattering by liposomes can be ruinous to spectroscopic experiments if it is not appropriately managed. Ladokhin and coworkers have provided several extremely useful procedures to minimize or correct for the effects of light scattering on tryptophan spectrofluorometry (Ladokhin et al., 2000). Here, we expand upon some of those techniques to tailor them for the study of membrane protein folding, especially folding perturbed by the titration of chemical denaturants, such as urea or guanidine HCl.
2.1 The contribution of light scattering to a tryptophan fluorescence emission scan can be divested from true tryptophan emission
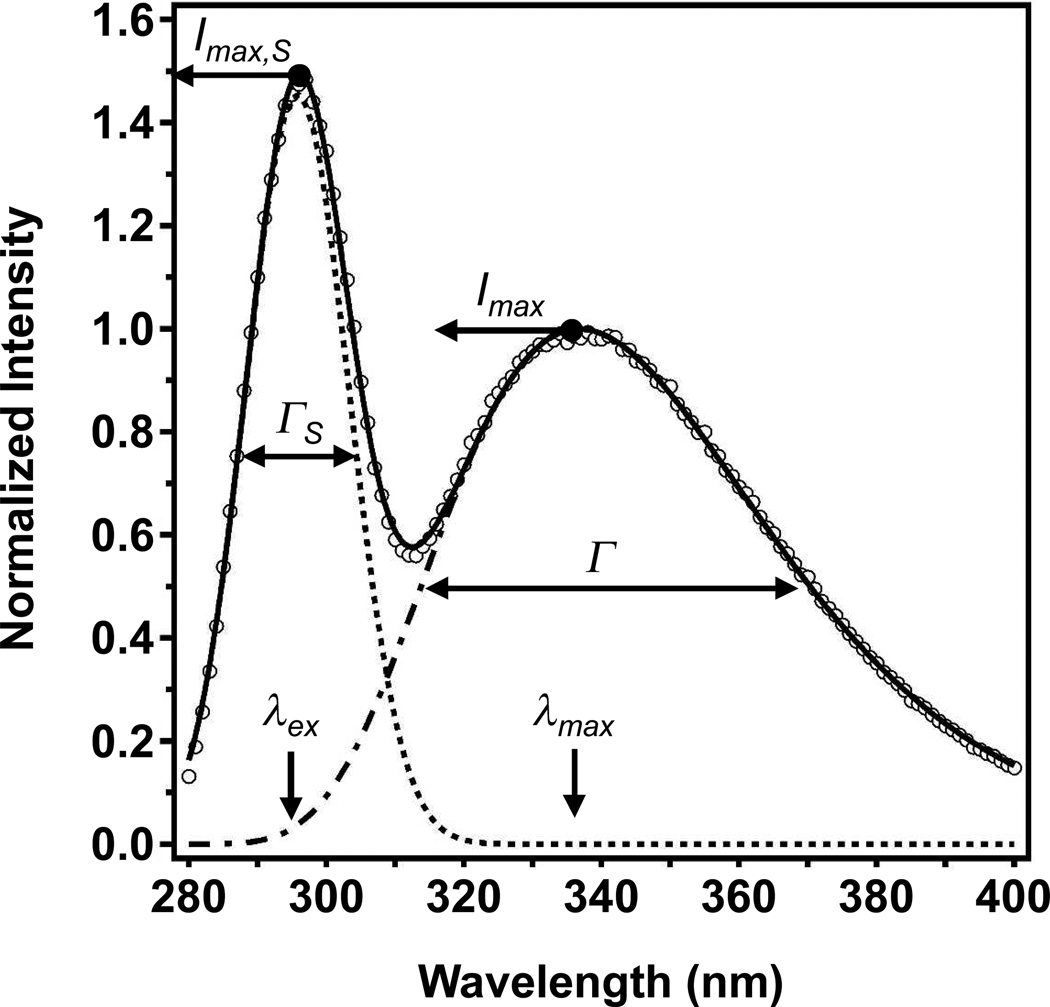
We show in Fig. 1 how light scattering usually manifests in a typical fluorescence experiment. In this experiment, we folded the fatty acid transporter FadL from the outer membrane of E. coli into LUVs of 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC). We excited the folded protein sample with light at 295 nm and then collected its fluorescence emission data from 280–400 nm. By collecting emission data all the way down to 280 nm, we could observe the full peak caused by light scattering that was distributed around the excitation wavelength. The light scattering peak is easily discernable (Fig. 1) from the emission peak that came from the tryptophans in FadL.
Fig. 1.
Relative contributions of light scattering and tryptophan fluorescence emission to the observed data from a typical fluorescence experiment. The observed data (open circles) came from a sample of 400 nM FadL folded into LUVs of DLPC at a lipid-to-protein ratio of 2000-1 and in 1.5 M guanidine HCl, 2mM EDTA, and 100mM citrate, pH 3.8. The sample was excited at 295 nm, the excitation polarizer was set at 90°, and the emission polarizer was set at 0°. The sample pathlength was 10 mm, the excitation slits were 2.4 mm wide, and the emission slits were 2.0 mm wide. The solid line represents a fit to the observed data by a sum of a normal distribution and a log normal distribution as described by Eq. (3). Fit parameters from Eq. (3) are labeled next to the parts of the data that they describe. The dotted line is the normal distribution component of Eq. (2), which represents light scattering at the excitation wavelength. The dot/dashed line represents the log normal distribution component of Eq. (1), which describes the fluorescence emission of the tryptophans in FadL.
The most common procedure to account for light scattering in an emission scan is to eliminate it by subtracting a scan of a blank sample of liposomes in buffer from the protein scan. However, we have chosen to keep the light scattering peaks in our emission data to understand how they change from one experiment to another. We can fully divest the light scattering and protein emission peaks and consider them separately if we fit both peaks with an expression expanded from the single log-normal distribution used by Permyakov (Permyakov, 1993; Ladokhin et al., 2000).
The single log-normal distribution describes the asymmetric shape of a tryptophan emission peak (Fig. 1). It can give the tryptophan emission intensity at a wavelength λ by:
| (1) |
where Imax is the maximum emission intensity of the tryptophan peak, λmax is the emission wavelength at Imax, Γ is the width of the peak at half of Imax, and ρ is a factor that describes the degree of asymmetry in the peak.
The shape of the light scattering peak can be adequately described by a normal distribution. The right-angle (90°) light scattering intensity at wavelength λ is given by:
| (2) |
where Imax,S is the maximum emission intensity of the light scattering peak, λex is the excitation wavelength, and ΓS is the width of the peak at half of Imax,S.
We use the sum of Eq. (1) and Eq. (2) to describe our observed data from emission scans of tryptophans and liposomes:
| (3) |
It does not take much extra time to include the light scattering peak in a tryptophan fluorescence scan. Once collected, it can be removed directly from an existing set of data by using Eq. (3) to determine the log-normal parameters coming only from tryptophan emission.
2.2 Effects of light scattering on the tryptophan fluorescence from membrane proteins
Ladokhin and coworkers noted four problems that light scattering has for tryptophan fluorescence experiments: (1) it directly contributes to the observed emission signal, (2) it causes less of the excitation light to reach the tryptophans, (3) it causes less of the light emitted by the tryptophans to reach the detector, (4) it causes the observed emission spectrum to be red-shifted because it asymmetrically affects tryptophan’s excitation band (Ladokhin et al., 2000).
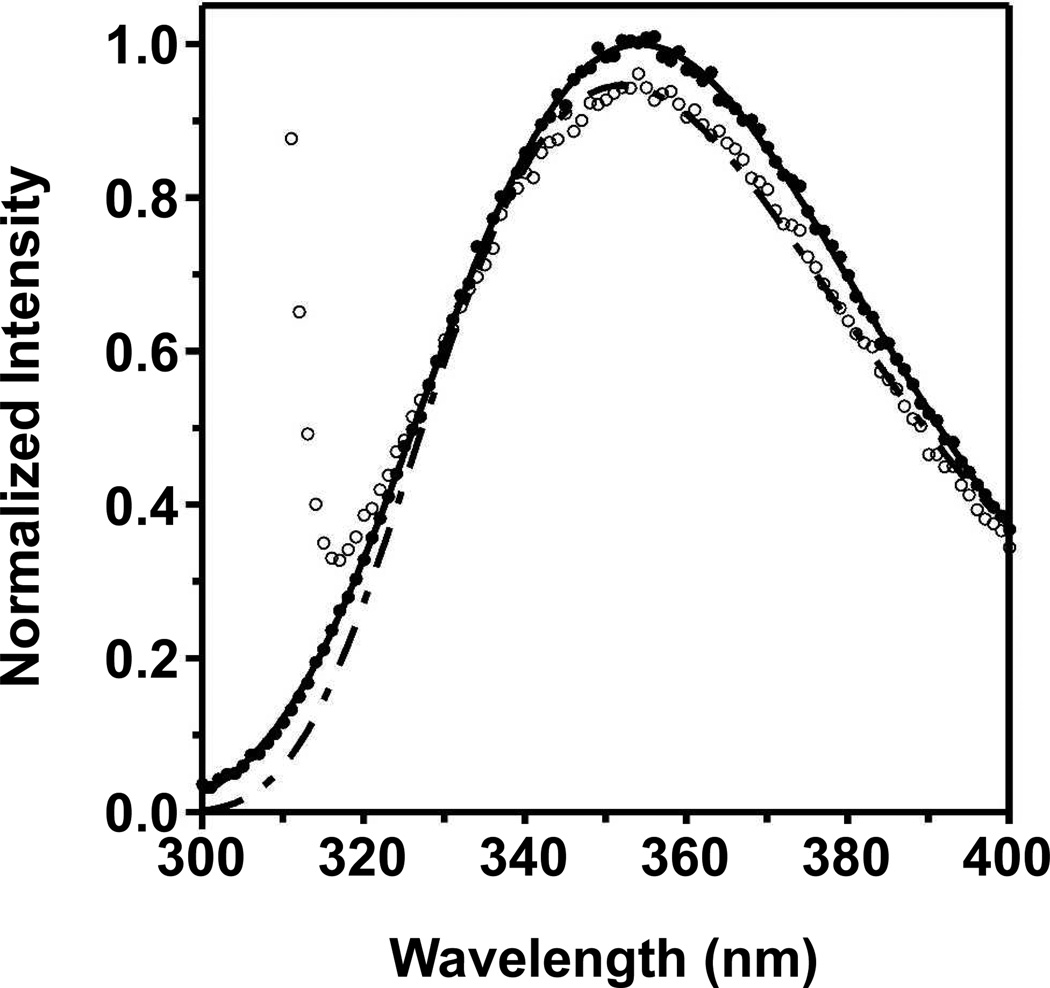
These problems are demonstrated in Fig. 2. We dissolved one sample of L-tryptophan zwitterion into a blank buffer solution and a second sample into the same buffer solution that also contained LUVs of DLPC. The tryptophan zwitterion should not have partitioned onto the LUVs (Ladokhin et al., 2000). In principle then, the fluorescence emission should have been the same from both samples. However, Fig. 2 shows that there were spectral changes due to light scattering from the LUVs.
Fig. 2.
Light scattering changes the spectral properties of tryptophan fluorescence. Two samples of L-tryptophan zwitterion were dissolved at a concentration of 6.0 µM in a background buffer of 2mM EDTA, and 100mM citrate, pH 3.8. One sample included LUVs of DLPC (open circles) at a lipid concentration of 800 µM. The solid line represents a fit of Eq. (1) to the open circles. The other sample had no LUVs (filled circles). The dot/dashed line represents the log normal distribution component of Eq. (3) fit to the filled circles.
The first problem with light scattering (Ladokhin et al., 2000) is apparent in both Figs. 1 and 2 where the experimentally observed emission at low wavelengths (295–325 nm) from a sample containing LUVs is higher than the true tryptophan emission component that resulted from a fit of Eq. (3) to the observed data. The light scattering directly added to the tryptophan signal to increase the observed emission at those low wavelengths. Eliminating the contribution from the light scattering, either by subtracting a blank scan or by using Eq. (3), can remove the first problem. Problems 2 and/or 3 are to blame for the peak emission intensity (Imax) of the sample with LUVs being lower than the peak intensity of the sample not containing LUVs. Problem 4 can be observed in Fig. 2 as a very subtle increase in the wavelength position of the peak emission (λmax) going from the sample without LUVs to the sample with LUVs. Altogether, these observations suggest that inaccuracies in protein conformation measurements will arise if tryptophan emission data that were troubled by these problems were used to measure the thermodynamics of membrane protein folding. To prevent problems 2–4, Ladokhin and coworkers advocated for the use of a simple method to correct protein spectra using their L-tryptophan spectra as a reference (Ladokhin et al., 2000).
2.3 Reducing light scattering with a spectrofluorometer
Besides correcting protein spectra or removing the direct contribution from light scattering, Ladokhin and coworkers also proposed several means of reducing light scattering to begin with by changing the experimental set-up with the spectrofluorometer itself (Ladokhin et al., 2000). These means included: using an excitation polarizer at 90° and an emission polarizer at 0°, using emission slits at or narrower than 5 mm and excitation slits at or narrower than 10 mm, and using an excitation wavelength of 295 nm. The emission polarizer also eliminates the Wood’s anomaly that is an artifact of many monochromators and that would disrupt proper fitting of the spectra with Eq. (3) (Ladokhin et al., 2000). The relatively long excitation wavelength at 295 nm (compared to 280 nm) avoids energy transfer from tyrosines to tryptophans and additionally prevents intertryptophyl transfer (Burstein et al., 1973). Not having either of these energy transfer events allows the position-width analysis of tryptophan spectra that we discuss below. Ladokhin and coworkers finally recommend using a cuvette with a pathlength ≤ 4 mm to reduce light scattering (Ladokhin et al., 2000). However, to prevent protein aggregation, we used protein concentrations so low that a 10 mm square cuvette was necessary to increase our signal-to-noise ratio. Otherwise, we followed all of the advice from Ladokhin and coworkers in all of our experiments presented here (See Materials and Methods).
2.4 Reducing light scattering by refractive index matching
Light scattering by liposomes can also be reduced by matching their refractive index with solutes. As we describe more fully below, the ratio between the refractive index of a lipid bilayer and the refractive index of the background solution is one of the factors that influence how much light is scattered by liposomes (Matsuzaki et al., 2000). If the refractive index of the background solution is raised by the addition of solutes, then the liposomes will scatter less light. Virtually any solute, including buffers, can raise the refractive index of the background solution. However, some solutes in high concentrations could affect the structure of lipid bilayers or the structure of membrane proteins. Therefore, solutes with high refractive indices coupled with high solubility would be the best candidates for refractive index matching. One group has used sucrose to make liposomes invisible to linear dichroism spectroscopy for the study of membrane pore forming peptides (Ardhammar et al., 2002).
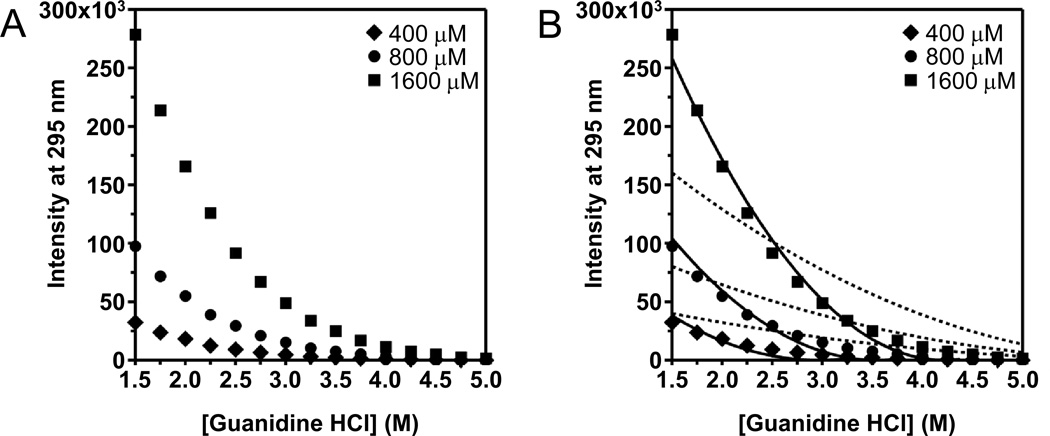
Importantly for membrane protein folding experiments, the most frequently used chemical denaturants (urea and guanidine HCl) are in fact good solutes for refractive index matching. In Fig. 3A, we show the effects of guanidine HCl on light scattering at three different concentrations of DLPC where it can be observed that the peak intensity of scattered light measured as right-angle emission in our spectrofluorimeter decays with increasing concentrations of guanidine HCl. At a lipid concentration of 400 µM, the LUVs are essentially invisible in solutions having greater than 3.5 M guanidine. At higher lipid concentrations, it takes more guanidine to make the LUVs invisible because there are more LUVs scattering light. The data in Fig. 3A are fully reversible and we recover the same amount of light scattering whether the LUVs are first put in concentrated guanidine and then diluted or whether they are first put in buffer and then titrated into guanidine. Also, if LUVs of DLPC are prepared by extrusion in 8.0 M guanidine, they do not scatter light until the guanidine is diluted.
Fig. 3.
Denaturants can reduce light scattering from LUVs by matching the refractive index of the lipid bilayers. Peak intensities of RGD light scattering by LUVs of DLPC at an excitation wavelength of 295 nm are plotted. Dotted lines represent fits to Eq. (9). Solid lines represent fits to Eq. (11). The background buffer for all samples was 2mM EDTA and 100 mM citrate, pH 3.8. (A) Effect of the number of LUVs on their light scattering at different concentrations of the solute guanidine HCl. (B) Effect of the solute sucrose on the light scattering at a lipid concentration of 800 µM.
If we had included a membrane protein in the titrations shown in Fig. 3A, we would also have needed a full set of blank samples of liposomes without protein for each concentration of guanidine HCl in order to adequately remove the direct contribution of scattering from the tryptophan emission signal from each protein sample. There are three key reasons why we do not favor this approach. First, preparing a full set of blanks for each protein titration is inefficient for a titration with a large number of points and it doubles the data collection time. Second, since light scattering is sensitive to the output of a spectrofluorimeter’s excitation light source, one day’s data may not align with another day’s data. A typical xenon arc lamp, for example, can loose brightness over time as it ages, quickly making a set of data from blank samples obsolete. Third, since the peaks are well distinguished from each other, collecting the light scattering and explicitly divesting its contribution from that of the true fluorescence in the exact same sample works well. Therefore, we suggest using Eq. (3) as the primary method to remove the contribution of light scattering from observed fluorescence emission data when doing experiments with membrane proteins in different concentrations of denaturants. At the very least, we suggest collecting the full light scattering peak as a second stream of information about each sample. This information could reveal issues, such as the formation of membrane protein aggregates, which would not be readily apparent from just the tryptophan emission peak.
2.5 Rayleigh-Gans-Debye theory describes light scattering by liposomes
The right-angle light scattering by liposomes seen in fluorescence emission experiments can be described by the Rayleigh-Gans-Debye (RGD) theory of light scattering by particles around the same size as the wavelength of the incident light (Matsuzaki et al., 2000). The RGD scattering is not the same as Rayleigh light scattering, which comes from particles much smaller than the wavelength of light.
Matsuzaki and coworkers found that RGD scattering by LUVs is proportional to the number of LUVs in the pathlength (Np), the radius of the LUVs (R), the excitation wavelength (λex), and the refractive indices of both the LUVs (nluv) and the background solution (nback) (Matsuzaki et al., 2000). For our experiments, we assumed Np is directly proportional to the lipid concentration. We also assumed that all of our LUVs that were extruded through membranes with 100 nm pores had a radius of 50 nm. These parameters can be combined in the Rayleigh RGD equation for the intensity of right-angle light scattering (I(90°)). If the LUVs are treated as optically homogenous spheres, then the RGD equation is:
| (4) |
where the size parameter (α) is:
| (5) |
and
| (6) |
(Matsuzaki et al., 2000). The exact value of I(90°) in a given experiment would also be governed by the intensity of the excitation light, and the orientation of any polarizers or filters that are in line with the incident beam or the detector.
To account for changing refractive index matching across a titration of guanidine HCl, we used the following polynomial, which relates the concentration of guanidine in a solution to the difference in the measured refractive indices of the guanidine solution (nGdnHCl) and the blank buffer (nbuffer):
| (7) |
The guanidine becomes part of the background solution for liposomes. Our particular buffer (2mM EDTA, 100 mM citrate, pH 3.8) has a refractive index (nbuffer) of 1.337 measured with an Abbe-3L refractometer at a wavelength of 589.3 nm. Refractive indices vary with wavelength according to the Sellmeir equation, but for a first approximation, we used the difference in refractive indices measured with our refractometer for Eq. (7). We used nbuffer and Eq. (7) to express the refractive index of the background solution (nback) containing both buffer and guanidine as:
| (8) |
Combining Eq. (4) and Eq. (7) gives:
| (9) |
In Fig. 3B, we show global fits of Eq. (9) to the same three sets of data that are shown in Fig. 3A where we held nluv to be the same value for each of the three lipid concentrations. The fits are shown as dotted lines. The fits gave the value of nluv as 1.445, but they do not describe the data well. Nevertheless, the fits do accurately represent the general decay in light scattering intensity 14 with increasing concentrations of guanidine. We then fit the data in Fig. 3B with a refined model that assumed that the guanidine had an effect on the lipid bilayer structure of LUVs of DLPC. This new model included the following simple linear decrease in the refractive index of the lipids with increasing concentrations of guanidine:
| (10) |
where is the refractive index of the LUVs in the absence of guanidine and mluv describes the linear decrease in the refractive index. The refractive index of a lipid bilayer depends on its thickness and on the spacing between the lipid headgroups (Ohki, 1968). As a chaotrope and a Hofmeister ion, guanidine HCl may affect both bilayer thickness and headgroup spacing (Feng et al., 2002; Aroti et al., 2007). Combining Eq. (9) and Eq. (10) gives:
| (11) |
The solid lines in Fig. 3B are global fits of Eq. (11) to the three sets of titration data where we held to be the same value for each of the three lipid concentrations. The refined model does reasonably describe the data considering we made such a simple assumption about the behavior of lipid bilayers in guanidine. The global fit value for was 1.5008, which is very consistent with the expected refractive index of bilayers of phosphatidylcholine lipids at a wavelength of 295 nm (Chong and Colbow, 1976; Huang et al., 1991).
3 Using tryptophan spectral properties to monitor membrane protein folding into liposomes
It is well understood that the spectral parameters of tryptophan fluorescence emission are sensitive to the environment of tryptophans and that a protein’s tryptophans can report on environmental changes during events such as folding or unfolding. For example, the λmax of many proteins’ emission scans will red-shift upon unfolding as the tryptophans become more exposed to polar solvent. What is less understood is which spectral parameter is the best one to observe for a given experiment. We focus here on the choice for the best parameter to use for measurements of the equilibrium thermodynamic stability of a membrane protein when it is folded in lipid bilayers.
3.1 Position-width analysis
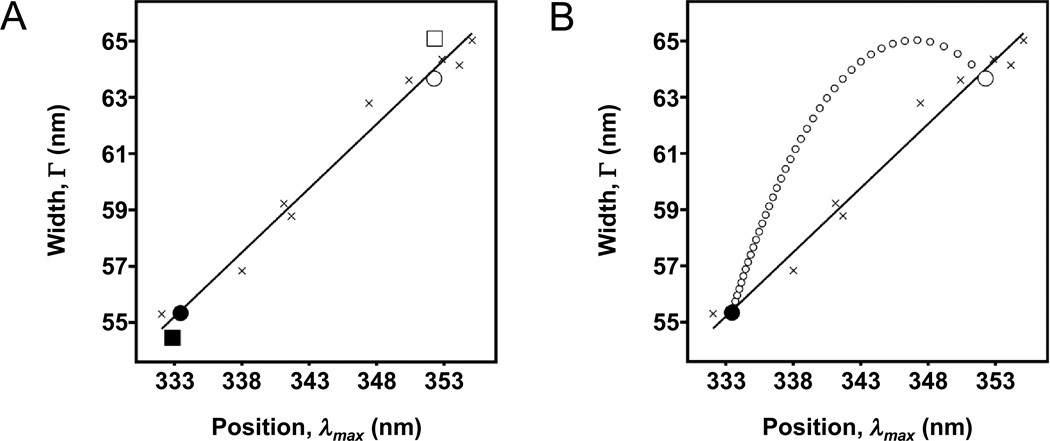
In 1973, Burstein and coworkers showed that a plot of peak spectral position (λmax) versus width (Γ) can reveal the degree of heterogeneity of tryptophan microenvironments in a protein (Burstein et al., 1973). The group compared the position-width data of proteins against position-width data for N-Acetyl-L-tryptophanamide (NATA) and other indole derivatives that were dissolved in various solvents. We also prepared a similar set of NATA samples (see Materials and Methods) and plotted their position-width values in Fig. 4A. The position-width data for NATA varied linearly with the polarity of the solvent solutions. When NATA was in apolar solutions, it had a blue-shifted λmax and a smaller Γ. When it was in water or other polar solutions, it had a red-shifted λmax and a larger Γ. A line fit to our NATA data (Fig. 4A) can be expressed by Γ(λmax) = 0.457λmax −96.8. This line is characteristic of our ISS PC1 spectrofluorometer set-up. Ladokhin and coworkers also produced a similar position-width plot of NATA and found a line expressed by Γ(λmax) = 0.624λmax −156.7 (Ladokhin et al., 2000). Burstein and coworkers found an even steeper line fit to their indole data (Burstein et al., 1973). The variation in these three lines suggests that some leeway is needed when comparing spectral parameters reported from different groups or from different spectrofluorometer set-ups.
Fig. 4.
Position-width analysis of tryptophan fluorescence from folded and unfolded outer membrane proteins. Positions (λmax) and widths (Γ) come from fits to Eq. (3). The solid line is a linear fit to data from spectra of N-acetyl-tryptophan-amide dissolved in different solvents or mixtures of solvents (crosses). (A) Two proteins, FadL (filled square) and OmpW (filled circle), were folded into LUVs of DLPC in 1.5 M guanidine HCl. The two proteins were also unfolded in the presence of the same LUVs but in 5.5 M guanidine HCl (open symbols of the same shapes). (B) Simulated data (small open circles) showing the expected heterogeneity of tryptophan environments when different fractions of an ensemble of OmpW molecules are folded versus unfolded. These different fractions could be obtained, for example, when a sample of OmpW is titrated into different concentrations of guanidine HCl. Each data point along the arc comes from Eq. (12) and represents a 2.5% step in a mixture of the folded and unfolded states. The endpoints of the arc (large circles) were taken from the actual data for OmpW shown in (A).
These lines for indole behavior have significance for tryptophan fluorescence spectra of proteins. Burstein and coworkers concluded that protein emission spectra having a position–width pair falling on the indole line indicated that all of the protein’s tryptophans in the sample were in equivalent environments (Burstein et al., 1973). If more than one tryptophan environment were sampled, the position-width data would fall above the indole line. Multiple environments could occur in a single protein conformation for proteins that have more than one tryptophan. Multiple environments could also occur from a mixture of more than one conformation of the same protein being present at the same time. For example, Ladokhin and coworkers showed that a mixture of peptide oligomeric states could have a mixture of tryptophan environments and yield position-width data above their NATA line (Ladokhin et al., 2000). Position-width analysis also allows the identification of classes of tryptophan environments (Burstein et al., 1973). On our NATA line, class I tryptophans in apolar environments would have a λmax around 333 nm and a Γ around 55 nm. Class III tryptophans in polar environments would have a λmax around 352 nm and a Γ around 64 nm.
Along with the NATA data in Fig. 4A, we show position-width values for two outer membrane proteins from E coli, FadL and Outer Membrane Protein W (OmpW), in both their folded and unfolded conformations. The folded conformations were with LUVs of DLPC in 1.5 M guanidine HCl. The unfolded conformations were also with the LUVs but were in 5.5 M guanidine HCl. FadL has eleven tryptophans and OmpW has five. According to their crystal structures (van den Berg et al., 2004; Hong et al., 2006), if FadL and OmpW were folded, all of their tryptophans would either be buried inside their barrel structures or inside the membrane. The position-width data points for both proteins were consistent with that expectation since they fell on our NATA line in the class I region when the proteins were folded. When the proteins were unfolded, they produced position-width data points that fell on the NATA line in the class III region. We concluded that the unfolded proteins were not embedded in membranes and had all of their tryptophans fully accessible to water.
3.2 Variation of tryptophan spectral properties with fractional populations of folded protein
We considered what the position-width data for OmpW would look like if there were a heterogeneous mixture of its folded and unfolded conformations. In Fig. 4B, we show a simulated transition between the actual data for the folded and unfolded states of OmpW shown in Fig. 4A. To create this transition, we generated a set of spectra for discrete steps in the fraction of folded protein (ffold) in the mixture. In Fig. 5A, we show an example of the generated spectra where ffold is 0.5. The generated spectra were calculated from the equation:
| (12) |
where Istep(λ) is the tryptophan emission intensity at a given wavelength at each step in ffold, Ifold(λ) is the emission intensity taken from a fit of Eq. (3) to the data from the folded sample of OmpW in 1.5 M guanidine HCl, and Iunfold(λ) is the emission intensity taken from a fit of Eq. (3) to the data from the unfolded sample of OmpW in 5.5 M guanidine HCl. We fit all of our generated spectra to Eq. (3) to extract the simulated values for λmax and Γ for each step. In Fig. 4B, we show position-width values for 0.025 steps in ffold as small circles. The small circles form an arc above the NATA line. This arc implies that mixtures of folded and unfolded OmpW contain mixtures of tryptophan environments and would produce spectral widths that are broader than the widths from either conformation alone.
Fig. 5.
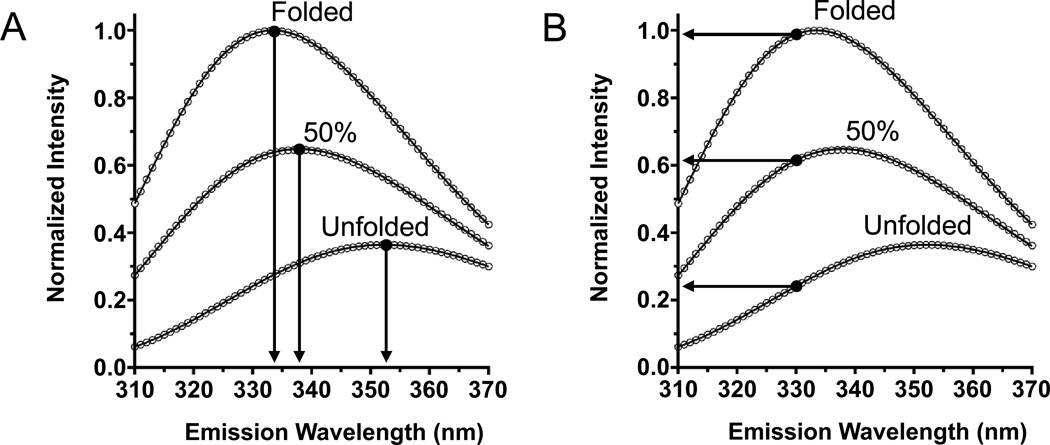
Comparison of the dependence of two spectral parameters to the fraction of folded OmpW in a sample. Tryptophan fluorescence emission intensity is normalized to the peak intensity (Imax) of a folded sample of OmpW in LUVs of DLPC in 1.5 M guanidine HCl. The unfolded sample of OmpW is in the same LUVs but instead is in 5.5 M guanidine HCl. The curve for 50% folded protein was generated using Eq. (12) and the spectra from folded and unfolded protein samples. (A) Emission intensity at a fixed wavelength (e.g., 330 nm) linearly depends on the fraction of folded protein. (B) The position of maximum emission (λmax) does not linearly depend on the fraction of folded protein.
It is also notable to observe that the small circles in Fig. 4B are also more closely spaced along the left-side of the arc for lower values of ffold than they are on the right-side of the arc for higher values of ffold. This result is because of a characteristic of tryptophan spectra resulting from mixtures of tryptophan environments already noted by Ladokhin and coworkers—that the parameters λmax and Γ do not vary linearly with ffold (Ladokhin et al., 2000). In Fig. 5A, we show λmax for the spectra from folded and unfolded samples of OmpW and for the spectra generated from Eq. (12) when ffold is 0.5. It can be observed that their values are not spaced evenly λmax. In fact, the only spectral parameter from tryptophan fluorescence emission that does vary linearly with ffold is the emission intensity at a specific wavelength (I(λ)) (Ladokhin et al., 2000). The parameter I(330) is highlighted in Fig. 5B and it is spaced evenly among the three spectra. The intensity at 330 nm is a good choice to monitor for membrane protein folding experiments with liposomes because the wavelength is long enough to avoid direct contribution from light scattering (Figs. 1 and 2).
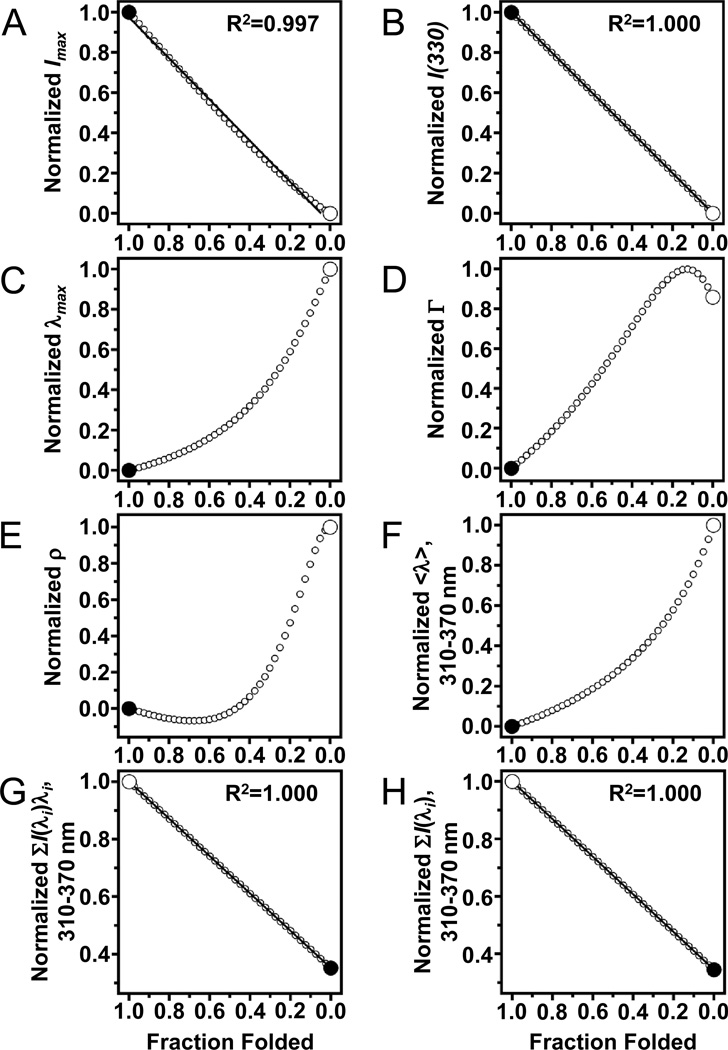
Figure 6 is a more thorough representation of the relationship between spectral parameters determined from the same generated spectra as depicted in Fig. 4B and ffold. The parameter Imax (Fig. 6A) does not vary linearly with ffold. The non-linearity of Imax with ffold is subtle, but the correlation coefficient of a linear fit to the relationship of the two values in this generated data is still not unity, whereas Fig. 6B shows that that the correlation coefficient of I(330) versus ffold is in fact unity. The additional spectral parameters shown in Figs. 6C-6F all significantly deviate from linear relationships with ffold. Figure 6C shows the nonlinearity of the λmax parameter that was used by Sanchez and coworkers to measure the thermodynamic stability of folded OmpA (Sanchez et al., 2008). Figure 6F shows the relationship of the average emission wavelength parameter (<λ>) with ffold. The magnitude of <λ> depends on the window of wavelengths over which the parameter is calculated. An expression for calculating <λ> is given by:
| (13) |
where λi is the ith emission wavelength sampled and I(λi) is the emission intensity at that wavelength (Hong and Tamm, 2004). The <λ> from a window between 300–400 nm was used by Hong and Tamm to measure the thermodynamic stability of folded OmpA (Hong and Tamm, 2004). The <λ> from a window between 310–370 nm shown in Fig. 6F was used by Huysmans and coworkers to measure the thermodynamic stability of folded PagP (Huysmans et al., 2010).
Fig. 6.
Not every spectral parameter of tryptophan fluorescence linearly depends on fraction folded. The endpoints of each plot (large circles) were taken from actual data for folded and unfolded samples of OmpW with LUVs of DLPC in 1.5 M and 5.5 M guanidine HCl, respectively. Other data points (small circles) come from Eq. (12) and represent 2.5% steps of simulated mixtures of the folded and unfolded samples. (A) Peak intensity (Imax). Solid line represents a linear fit to the data points. (B) Emission intensity at 330 nm. Solid line represents a linear fit to the data points. (C) Position of maximum emission (λmax). (D) Spectral width (Γ) at half the peak intensity. (E) Log-normal asymmetry parameter (ρ). (F) Average emission wavelength <λ> using a window of emission from 310–370 nm. (G) Numerator of Eq. (13) using a window of emission from 310–370 nm. Solid line represents a linear fit to the data points and has a slope of −0.648. (H) Denominator of Eq. (13) using a window of emission from 310– 370 nm. Solid line represents a linear fit to the data points and has a slope of −0.655.
Both the numerator and denominator of Eq. (6.13) are linear functions of the emission intensity at specific wavelengths. Since intensity at a specific wavelength varies linearly with ffold, both the numerator and denominator also vary linearly with ffold. This conclusion is shown in Figs. 6.6G and 6.6H, which depict the linearity of ffold with the numerator and denominator of Eq. (6.13), respectively. Equation (6.13) is then a rational function being that it is a linear function divided by another linear function. Therefore, as Fig. 6.6F shows, <λ> does not vary linearly with ffold. However, as Hong and Tamm show, <λ> can be normalized to convert it back to its linear components during further analysis to measure thermodynamic values from data (see below) (Hong and Tamm, 2004).
4 Choosing an appropriate tryptophan spectral property to measure the thermodynamic stabilities of folded membrane proteins
How does the complex relationship of tryptophan spectral parameters to ffold affect the measurements of stabilities of membrane proteins? In order to measure the stabilities of OmpA and PagP, the three groups we discussed above each used a two-state linear extrapolation model that compares the relationship of observed spectral parameters to chemical titrations of denaturant (Yobs([D])) to find the standard state free energy of their proteins in the absence of denaturant (). An example of such a model is given by:
| (14) |
where Yunf,w and Yfold,w are the values of the chosen spectral parameter in the absence of denaturant for the unfolded and folded conformations, respectively; Sunf and Sfold are the slopes of linear baselines in the unfolded and folded regions of the data, respectively; m is a constant that describes how steeply the protein’s free energy depends on [D]; R is the gas constant; and T is the temperature in Kelvin (Street et al., 2008).
The model in Eq. (14) assumes that a protein’s equilibrium constant (Keq) in a given denaturant concentration can be determined by:
| (15) |
(Street et al., 2008). This assumption can only be true if Yobs([D]) varies linearly with ffold. Therefore, the parameter λmax will give an erroneous value for if used in Eq. (14). So will the <λ> parameter unless it is normalized in Eq. (14). The normalization can be done with a ratio of the denominator of Eq. (13) calculated for each of the folded and unfolded conformations of the protein being studied (Hong and Tamm, 2004). This ratio (QR) is given by:
| (16) |
for the same window of emission intensities as used for calculating <λ>. Equation (16) can then be used with Eq. (14) to give the following two-state linear extrapolation model to be used when Yobs is <λ> (Hong and Tamm, 2004):
| (17) |
Of all popular tryptophan spectral parameters, only two will give correct values of : emission intensity and <λ>, as long as it is normalized to a linear function of emission intensity as in Eq. (6.17).
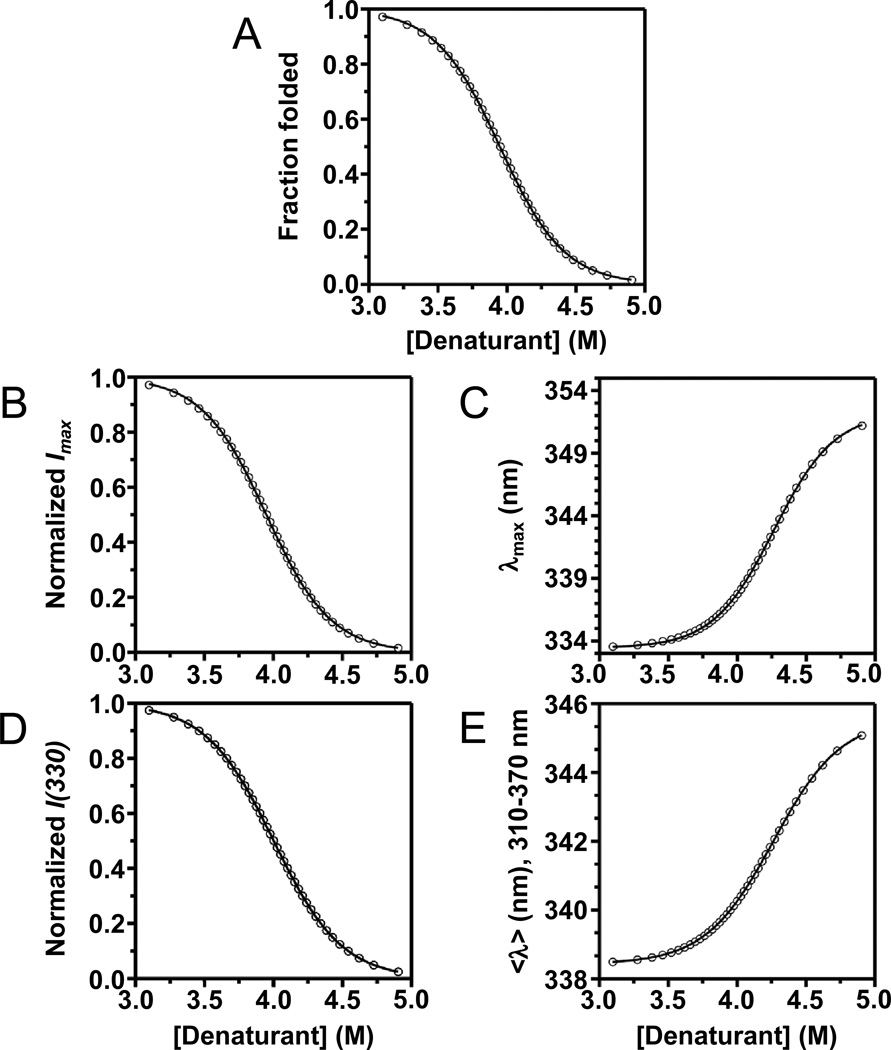
To demonstrate the validity of that conclusion, we simulated a two-state reversible equilibrium folding transition of OmpW that might occur with a denaturant titration. For the simulation, we assumed the protein had an unfolding free energy of equal to 10.0 kcal mol−1 in the absence of denaturant and an m-value of 2.50 kcal mol−1 M−1. We also assumed that Sunf and Sfold from Eq. (14) would both be zero for each spectral parameter. In Fig. 7A, we show the plot of ffold versus [D] that would be needed to produce our assumed values of , m, Sunf, and Sfold. The other panels of Fig. 7 show plots generated for Imax, λmax, I(330), and <λ> (for the emission window 310–370 nm) that replace the ffold values from Fig. 7A with the corresponding values for each spectral parameter as calculated using Eq. (12). We then fit Eq. (14) to each generated curve in Figs. 7B-E and determined the values of and m that would result from the respective spectral parameters. We also fit Eq. (17) to the generated curve in Fig. 7E and determined the same thermodynamic values. The results of these fits are shown in Table I. The only parameter that reproduced our assumed values for and m were those that were linear with intensity, i.e., I(330) and the normalized <λ>. The other spectral parameters misrepresented the thermodynamic values, primarily because they incorrectly positioned the denaturant midpoint of the conformational transition of the protein (Cm). These incorrect midpoints can be easily seen in Fig. 6. For example, when is ffold 0.50, the λmax would be only 23 percent of the way through its transition (Fig. 6C). We also note that the misrepresentations of the thermodynamic parameters shown in Table I would be even worse if the spectral parameters had non-zero values of Sunf and Sfold.
Fig. 7.
Relationship of spectral parameters of tryptophan fluorescence to isotherms from chemical denaturation. The vertical-axis positions of the data points in each plot come from Eq. (12) and represent 2.5% steps of simulated mixtures of folded and unfolded samples of OmpW. The horizontal-axis positions of the data points come from Eq. (14) and represent a simulated equilibrium, two-state denaturant titration of OmpW at 37° C. For the simulation, was assumed to be 10 kcal mol−1, the m-value was assumed to be 5 kcal mol−1 M−1, and the baseline regions were assumed to have zero slope. Solid lines represent a best fit of Eq. (14) to the simulated data. (A) Fraction folded. (B) Peak intensity (Imax). (C) Position of maximum emission (λmax). (D) Intensity emitted at 330 nm. (E) Average emission intensity, <λ>, using a window of emission from 310–370 nm.
Table I.
Thermodynamic parameters recovered from the response of tryptophan spectral parameters to denaturant concentration.
| Assumed | Imax | I(330) | λmax | <λ>310–370 nm | <λ>normalized | ||
|---|---|---|---|---|---|---|---|
|
|
10.0 | 10.4 | 10.0 | 11.9 | 10.7 | 10.0 | |
| m2 | 2.5 | 2.6 | 2.5 | 2.8 | 2.5 | 2.5 | |
| Cm3 | 4.0 | 3.9 | 4.0 | 4.3 | 4.3 | 4.0 |
values are in kcal mol−1
values are in kcal mol−1 M−1
values are in M
5 Conclusions
Measuring the thermodynamic stability of membrane proteins folded in liposomes is approachable by monitoring the tryptophan fluorescence emission by the proteins. We have suggested appropriate procedures to improve the accuracy of these measurements by managing the light scattering from the liposomes and by selecting the appropriate spectral parameter to relate to the fraction of folded protein in experimental samples. Our suggestions are tailored to the use of denaturant titrations. We conclude that the direct contribution of light scattering to tryptophan spectra should be removed by fitting emission intensities to a sum of a normal distribution describing light scattering and a log-normal distribution describing tryptophan emission. We further demonstrate that the emission intensity at a specific wavelength (e.g., 330 nm) should be used for the determination of thermodynamic parameters.
6 Materials and Methods
6.1 Protein folding reactions
Both FadL and OmpW were expressed to inclusion bodies and purified in a manner previously described (Burgess et al., 2008). We made fresh unfolded protein stocks prior to each experiment by dissolving inclusion body pellets in a buffer of 8 M guanidine HCl. Stock guanidine solutions were prepared from UltraPure powder (Invitrogen). The concentration of guanidine was checked by refractometry. The background buffer for all experiments was 100 mM citrate (Sigma) and 2 mM EDTA (Sigma), pH 3.8. We used lipid stocks in chloroform (Avanti Polar Lipids) and dried them briefly under nitrogen followed by at least 8 hours of dehydration under vacuum. We then wetted the lipids with the background buffer to a lipid concentration of 20 mg/mL for one hour with occasional vortexing and then prepared LUVs by extruding the lipid suspensions 21 times through two stacked 0.1 µM filters.
We prepared the protein folding/unfolding reactions in three steps. The first step was a dilution of the unfolded protein stocks to a final guanidine concentration of 3.0 M and a final protein concentration of 6.0 µM. Also present was 1.4 mM 3-(N,N-Dimethylmyristyl-ammonio) propanesulfonate detergent (Sigma), which is just above its critical micelle concentration (CMC) in 3.0 M guanidine. Without detergent the proteins visibly precipitated, even in 3.0 M guanidine and the presence of liposomes. The second reaction step was a threefold dilution of the solvated proteins into the presence of LUVs of 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC) at a 2000-1 lipid-protein ratio. For folding reactions, the final guanidine concentration was 1.5 M. For unfolding reactions, the final guanidine concentration was 5.5 M. In either concentration of guanidine, the three-fold dilution was enough to take the detergent below its CMC. If we kept the detergent above its CMC, then the light scattering due to the LUVs disappeared as the LUVs were solubilized by the detergent. Below the detergent CMC, the amount of light scattering by LUVs was the same as for samples of LUVs that had never been exposed to detergent. The third reaction step was a five-fold dilution, which kept the same guanidine concentrations from the second step. The final protein concentration was 400 nM and the final lipid concentration was 800 µM. After the dilutions, we incubated all samples with gentle mixing at 37°C for 40–50 hours before fluorescence experiments.
6.2 L-tryptophan, blank LUVs, and NATA reactions
We prepared samples of L-tryptophan (Sigma) in the same manner as the protein folding experiments, except that we did not include any detergent and the final L-tryptophan concentration was 6.0 µM. The final lipid concentration was 800 µM. We prepared blank samples of LUVs at the different final lipid concentrations shown in Fig. 3 in the same manner as the protein folding experiments, except that we did not include any detergent or protein. We prepared samples of N-Acetyl-L-tryptophanamide (NATA) by dissolving powder (Sigma) in the following solvents: dichloromethane, acetonitrile, isopropyl alcohol, propanol, water, various percentages of methanol in water, and 10 M urea.
6.3 Spectrofluorometery
All protein samples were excited at 295 nm in an ISS PC1 spectrofluorometer (Champaign, IL). We used an excitation polarizer at 90° and an emission polarizer at 0°. Excitation slits were 2.4 mm and emission slits were 2.0 mm. The pathlength of our cuvettes was 10 mm. We collected a minimum of four emission scans from 280–400 nm for each sample and then averaged the data before fitting. We used Igor Pro v6.12 (www.wavemetrics.com) for all least-squares fitting routines.
Acknowledgements
The authors thank the National Science Foundation (MCB 0423)807 and 0919868) and the National Institutes of Health (R01 GM079440, T32 GM008403) for financial support.
References
- Ardhammar M, Lincoln P, Nordén B. Invisible liposomes: refractive index matching with sucrose enables flow dichroism assessment of peptide orientation in lipid vesicle membrane. Proc. Natl. Acad. Sci. USA. 2002;24:15313–15317. doi: 10.1073/pnas.192583499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroti A, Leontidis E, Dubois M, Zemb T. Effects of monovalent anions of the Hofmeister series on DPPC lipid bilayers Part I: swelling and in-plane equations of state. Biophys J. 2007;93:1580–1590. doi: 10.1529/biophysj.106.094482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess NK, Dao TP, Stanley AM, Fleming KG. β-Barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behaviorin vitro. J. Biol. Chem. 2008;39:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein EA, Vedenkina NS, Ivkova MN. Fluorescence and the location of tryptophan residues in protein molecules. Photochem. Photobiol. 1973;18:263–279. doi: 10.1111/j.1751-1097.1973.tb06422.x. [DOI] [PubMed] [Google Scholar]
- Chong CS, Colbow K. Light scattering and turbidity measurements on lipid vesicles. Biochim. Biophys. Acta. 1976;436:260–282. doi: 10.1016/0005-2736(76)90192-9. [DOI] [PubMed] [Google Scholar]
- Feng Y, Yu Z, Quinn PJ. Effect of urea, dimethylurea, and tetramethylurea on the phase behavior of dioleoylphosphatidylethanolamine. Chem. Phys. Lipids. 2002;114:149–157. doi: 10.1016/s0009-3084(01)00198-0. [DOI] [PubMed] [Google Scholar]
- Hong H, Tamm LK. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Natl. Acad. Sci. USA. 2004;101:4065–4070. doi: 10.1073/pnas.0400358101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Patel DR, Tamm LK, van den Berg B. The outer membrane protein OmpW forms an eight-stranded β-barrel with a hydrophobic channel. J. Biol. Chem. 2006;281:7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- Huang HW, Liu W, Olah GA, Wu Y. Physical techniques of membrane studies – study of membrane active peptides in bilayers. Prog. Surf. Sci. 1991;38:145–139. [Google Scholar]
- Huysmans GHM, Baldwin SA, Brockwell DJ, Radford SE. The transition state for folding of an outer membrane protein. Proc. Natl. Acad. Sci. USA. 2010;107:4099–4104. doi: 10.1073/pnas.0911904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladokhin AS, Jayasinghe S, White SH. How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Anal. Biochem. 2000;285:235–245. doi: 10.1006/abio.2000.4773. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K, Murase O, Sugishita K, Yoneyama S, Akada K, Ueha M, Nakamura A, Kobayashi S. Optical characterization of liposomes by right angle light scattering and turbidity measurement. Biochim. Biophys. Acta. 2000;1467:219–226. doi: 10.1016/s0005-2736(00)00223-6. [DOI] [PubMed] [Google Scholar]
- Ohki S. Dielectric constant and refractive index of lipid bilayers. J. Theoret. Biol. 1968;19:97–115. doi: 10.1016/0022-5193(68)90008-8. [DOI] [PubMed] [Google Scholar]
- Permyakov EA. Luminescent Spectroscopy of Proteins. Boca Raton: CRC Press; 1993. pp. 40–42. [Google Scholar]
- Sanchez KM, Gable JE, Schlamadinger DE, Kim JE. Effects of tryptophan microenvironment, soluble domain, and vesicle size on the thermodynamics of membrane protein folding: lessons from the transmembrane protein OmpA. Biochemistry. 2008;47:12844–12852. doi: 10.1021/bi800860k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street TO, Courtemanche N, Barrick D. Protein folding and stability using denaturants. Methods Cell Biol. 2008;84:295–325. doi: 10.1016/S0091-679X(07)84011-8. [DOI] [PubMed] [Google Scholar]
- van den Berg B, Black PN, Clemons WM, Rapoport TA. Crystal structure of the long-chain fatty acid transporter FadL. Science. 2004;304:1506–1509. doi: 10.1126/science.1097524. [DOI] [PubMed] [Google Scholar]