Abstract
The PPAR gene pathway consists of interrelated genes that encode transcription factors, enzymes and downstream targets which coordinately act to regulate cellular processes central to glucose and lipid metabolism. The pathway includes the PPAR genes themselves, other class II nuclear hormone receptor transcription factors within the PPAR family, PPAR co-activators, PPAR co-repressors, and downstream metabolic gene targets. This review focuses on the transcription factors that comprise the PPAR transcriptional activator complex – the PPARs (PPARα, PPARβ, or PPARγ), PPAR heterodimeric partners, such as RXRα, and PPAR co-activators, such as PPARγ coactivator 1α (PGC-1α) and the estrogen related receptors (ERRα, ERRβ, and ERRγ). These transcription factors have been implicated in the development of myocardial hypertrophy and dilated cardiomyopathy as well as response to myocardial ischemia/infarction and, by association, ischemic cardiomyopathy. Human expression studies and animal data are presented as the background for a discussion of the emerging field of pharmacogenetics as it applies to these genes and the consequent implications for the individualization of therapy for patients with heart failure.
Keywords: Pharmacogenetics, Peroxisome Proliferator-activated Receptor, PPAR, PGC-1, ERR, Cardiovascular Disease
INTRODUCTION
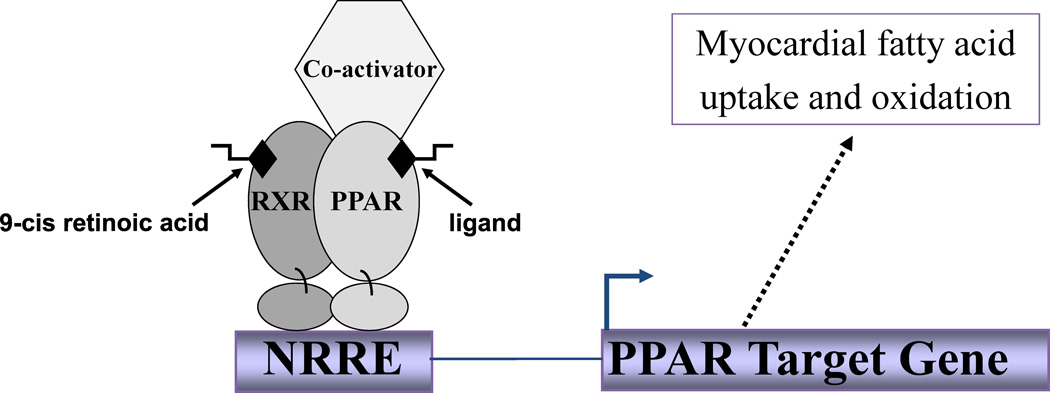
Peroxisome proliferator-activated receptors (PPARs) are members of the group II nuclear receptor superfamily. This family of transcription factors has been further subdivided into 3 groups distinguished by the affinity of ligand binding, the size of the ligand binding site, and the existence of known ligands (Table 1) (1–3). The PPARs belong to the group that has a relatively larger ligand-binding pocket with a relatively lower ligand binding affinity compared with the other two groups. These characteristics are thought to allow for binding of more diverse ligands, including metabolic intermediates (fatty acids) and xenobiotics (fibrates, thiazolidinediones) (1–4). In the presence of ligand, PPARα, β or γ binds to its cognate DNA regulatory element as a heterodimer with the retinoid X receptor α (RXRα; Figure 1) (5). Ligand binding results in a conformational change in the nuclear receptor complex that results in the association of co-activators, release of co-repressors and increased transcriptional activation of target genes (3;4;6–11).
Table 1.
Nuclear Hormone Receptors stratified by ligand
| Endocrine Receptors - High affinity binding sites; Ligands are hormonal lipids |
| Estrogen Receptor α,β (ERα,β) |
| Progesterone Receptor (PR) |
| Androgen Receptor (AR) |
| Glucocorticoid Receptor (GR) |
| Mineralocorticoid Receptor (MR) |
| Thyroid Receptor α, β (TRα, β) |
| Vitamin D Receptor (VDR) |
| Retinoic Acid Receptor α, β (RAR α, β) |
| Adopted Orphan Receptors – Low affinity binding sites; Ligands are dietary lipids |
| Peroxisome Proliferator Activated Receptor α, β/δ, γ (PPAR α, β/δ, γ) |
| Retinoid X Receptor α, β, γ (RXR α, β, γ) |
| Liver X Receptor α, β (LXR α, β) |
| Farnesoid X Receptor (FXR) |
| Pregnane X Receptor (PXR) |
| Constitutive Androstane Receptor (CAR) |
| Orphan Receptors – Unknown Ligands |
| Steroidogenic Factor 1 (SF-1) |
| Liver Receptor Homlogue 1 (LRH 1) |
| DAX-1 |
| Small Heterodimer Partner (SHP) |
| TLX |
| Photoreceptor Cell-specific Nuclear Receptor (PNR) |
| Nerve Growth Factor Inducible-B α, β,γ (NGFI-B α,β,γ) |
| RAR-related Orphan Receptor α,β,γ (ROR α,β,γ) |
| Estrogen Related Receptor α,β,γ (ERR α,β,γ) |
| RVR α,β,γ (RVR α,β,γ) |
| Germ Cell Nuclear Factor (GCNF) |
| Testicular Orphan Receptor (TR 2,4) Hepatocyte Nuclear Factor 4 (HNF4) |
| Chicken Ovalbumin Upstream Promoter Transcription Factor α, β, γ (COUP-TF α, β, γ) |
Figure 1.
Schematic of the PPAR transcriptional activator complex. PPAR-RE (PPAR response element) represents the DNA sequence that PPAR binds.
Several PPAR co-activators have been described. One such inducible transcriptional co-activator is PPARγ coactivator −1α (PGC-1α) (6;7). PGC-1α is highly expressed in both heart and the adipocyte and has been shown to exert physiologic control on glucose transport and gluconeogenesis as well as fatty acid oxidation and mitochondrial biogenesis (12;13). Another family of PPAR co-activators, known as the estrogen-related receptors (ERRα, ERRβ, and ERRγ), has recently been identified (13;14). ERRα and ERRγ are functional partners of PGC-1α(15–17) and gene expression profiling studies in cardiomyocytes demonstrate that ERRα and ERRγ up-regulate the expression of PPARα and PPARα-regulated genes.(18;19)
The PPARs, and more recently, PPAR-family members and transcription factors that comprise the PPAR transcriptional activator complex, have been the focus of intensive study as potential therapeutic targets in diseases such as diabetes, atherosclerosis and cardiomyopathy. Animal models and human expression data have implicated PPAR transcriptional activator complex genes in three general forms of cardiomyopathy: hypertrophic, diabetic and dilated cardiomyopathy. Additionally animal models have revealed that these genes are involved in the response to myocardial ischemia and infarction, thus likely affecting the development of ischemic cardiomyopathy. As these data represent the foundation for the investigation of genetic and pharmacogenetic associations with these genes, the data relevant to each gene and cardiomyopathic state are reviewed first. The genetic and pharmacogenetic association data with respect to these same phenotypes in patients are then discussed. The intention of this review is to provide the context for understanding the complexity of the PPAR-pharmacologic interaction as the field of PPAR pharmacogenetics, while currently in its infancy, is rapidly developing. Ultimately a better understanding of the genes described here will permit personalized medicine wherein medications are tailored to an individual patient’s genotype. Given the extensive data associating PPAR family members with cardiovascular diseases, outcomes, and response to drug therapy these genes offer much promise for further inquiry.
I. Peroxisome Proliferator-Activated Receptors (PPARs)
A. LEFT VENTRICULAR HYPERTROPHY
Both human and animal studies have demonstrated changes in PPARα expression during the development of left ventricular hypertrophy (LVH) (20–22). In a rat model of progressive LVH and heart failure, the expression of PPARα target genes was downregulated greater than 70% compared to controls (21). Similarly, in a transverse aortic constriction mouse model of pressure-overload induced LVH, PPARα mRNA levels were 39% lower in mice with aortic constriction compared with sham operated mice (22).
Recent supportive evidence for a key role of PPARβ/δ in the development of myocardial hypertrophy derives from the observation that mice with cardiomyocyte-restricted PPARδ deletion develop marked LVH in adulthood (23). In addition, the PPARβ/δ agonist L-165041 inhibits phenylephrine-induced hypertrophy in neonatal rat cardiomyocytes via increased interaction between PPARβ/δ and NF-κB (a transcription factor known to be important in the signaling pathway of myocardial hypertrophy) and decreased expression of NF-κB target genes (24).
Genetic and Pharmacogenetic Associations
The PPARA IVS7 2498 G>C polymorphism (located in intron 7 of PPARA; 36,354 nucleotides from the translation start site in genomic PPARA sequence; 2498 nucleotides downstream from the exon-intron junction) was identified by direct sequencing of a Taq1 restriction fragment length polymorphism generated in the initial cDNA cloning of human PPARA(25;26). Given its location within an intron, the functional consequences of this polymorphism remain unclear (25;26).
The PPARA IVS7 2498 polymorphism (designated “PPARA intron 7 G/C polymorphism” in the publication) has been associated with left ventricular hypertropy in response to exercise in 144 healthy British army recruits undergoing a 10-week intensive training program. This exercise regimen included upper and lower body strength and endurance training (25). At baseline, age, weight, BMI, blood pressures, and measurements of heart size (determined by cardiac MRI) were similar among all genotype groups (25). Cardiac MRI performed at the commencement and termination of training revealed an increase in LV mass that was largest in CC homozygotes (19.4 +/− 4.2g), intermediate in GC heterozygotes (11.8 +/− 1.9g), and lowest in GG homozygotes (6.7 +/− 1.5; p=0.009 between groups) (25). The genotype effect upon LV mass increase remained significant when adjusted for body surface area (P=.02) (25). No interaction between the effect of genotype and losartan (i.e. pharmacogenetic interaction) was noted in this study.
In a subset of the third MONitoring Trends and Determinants in CArdiovascular Disease (MONICA) Augsburg survey, 1142 German women and men ages 25–74 underwent echocardiographic analysis of LV mass (25;27). PPARA IVS7 2498 (designated “PPARA intron 7 G/C polymorphism” in the publication) CC homozygote men exhibited significantly higher left ventricular mass index (LVMI) and septal wall thickness than G allele carries (105.7 ± 4.8 g/m2 in CC; 92.2 ± 1.3 g/m2 in GC; 91.8 ± 1.0 g/m2 in GG; p=0.005) (25). PPARA IVS7 2498 CC homozygote women displayed a similar trend. In the combined sample of men and women, the hypertrophic effect of CC homozygosity, as measured by LVMI, was significant (25). Hypertension greatly increased the effect of PPARA IVS7 2498 genotype in males, with hypertensive C allele homozygote individuals 4 times as likely to have LVH as hypertensive G-allele carriers (25).
B. Dilated Cardiomyopathy
Several studies have implicated PPARα, PPARγ and PPARδ in the development of dilated cardiomyopathy. Sack and colleagues compared explanted left ventricular tissue from cardiac transplant recipients with Class III-IV heart failure to control hearts from patients who died of noncardiac etiologies and found significantly decreased mRNA and protein levels of PPARα target genes MCAD and LCAD in the cardiac tissue from patients with heart failure compared to controls. These findings were confirmed by Razeghi and colleagues who demonstrated a significant downregulation of PPARα target genes MCAD, LCAD, and mCPT-1 mRNA levels in 10 failing hearts of patients undergoing LVAD placement when compared to 9 normally functioning hearts that were not transplanted for technical reasons (28). Furthermore, several investigators have demonstrated that up-regulation of PPARα may be advantageous in heart failure, as administration of the PPARα agonist fenofibrate attenuated cardiac dysfunction in both pig and rat models of heart failure (29;30). While the preponderance of data suggests that PPARα and PPARα target gene expression is downregulated in dilated cardiomyopathy, one study has reported discordant results. When Schupp and colleagues studied explanted hearts from 16 patients with dilated cardiomyopathy undergoing transplant compared with 15 donor hearts (not used for transplantation due to technical reasons) PPARα mRNA expression was significantly increased in the explanted hearts compared to controls (136+/−25.4% vs. control, p<0.01)(31). mRNA levels of the PPAR-target gene CPT-1 were observed to have similarly increased levels (147+/−51% vs. control, p<0.05) (31). It is interesting to speculate that the differences in these findings may be related to different underlying etiologies of the dilated cardiomyopathic samples used in the studies or to the timing of transplantation with respect to the development of heart failure.
Cardiomyocyte restricted PPARγ knockout mice develop a dilated cardiomyopathy that is felt to be a consequence of reduced superoxide dismutase 2 (32), however when investigators compared the expression of PPARγ mRNA in ventricular samples from patients with dilated cardiomyopathy and controls, similar levels were observed (33). Mice with cardiomyocyte restricted PPARδ deletion develop a dilated cardiomyopathy after 4 months that is thought to be lipotoxic in nature (23) but no studies of PPARδ expression in human tissue have been performed to date.
C. Diabetic Cardiomyopathy
The PPAR genes have been implicated in the development of diabetic cardiomyopathy in animal models (34). The expression and activity of PPARα are increased in the hearts of both rat and mouse models of insulin-deficient and insulin-resistant diabetes. Mice with cardiac-restricted overexpression of PPARα have a cardiac phenotype, characterized by an increased dependence on fatty acid oxidation and a decreased dependence on glucose oxidation, similar to that seen in patients with diabetes(35;36). The hearts from mice with cardiac-restricted overexpression of PPARα also demonstrate features of diabetic cardiomyopathy including ventricular hypertrophy and systolic dysfunction compared to non-transgene controls (36).
Genetic and Pharmacogenetic Associations
While no genetic or pharmacogenetic associations of PPAR gene polymorphisms and development of diabetic cardiomyopathy have been reported to date, there are numerous reports of SNPs in these genes being associated with the onset, progression and severity of DM itself. For example, the common PPARG Pro12Ala SNP (located in exon B which is present only in adipocyte-specific isoforms of PPARγ) has been associated with the risk of developing DM in multiple populations of different ethnicity (37–42) and a SNP in the promoter of PPARA has been shown to influence the onset and progression of type 2 DM (43). Furthermore, associations have been observed between SNPs in these genes and differences in response to medications that are commonly used to treat type 2 DM. For example, the PPARG Pro12Ala SNP and PPARG haplotype have been reported to influence response to thiazolidinedione therapy in patients with DM (44;45) and the PPARG Pro12Ala SNP has been associated with the development of peripheral edema in patients with type 2 diabetes treated with the dual-acting PPARalpha/gamma agonist ragaglitazar (46).
D. Ischemic Cardiomyopathy
Both PPARα and PPARγ may have important roles in the myocardial response to ischemia. Experimental animal models suggest that if PPARα is upregulated during myocardial ischemia and reperfusion, left ventricular contractile function deteriorates, microinfarction and intramyocardial triglyceride deposition develop, and there is a significantly diminished recovery of cardiac function (20;47). Further supporting the deleterious role of PPARα during ischemia/reperfusion, PPARα-null mice have better recovery in ventricular function after ischemia/reperfusion compared to their wild-type littermates (48).
PPARγ also affects the myocardial response to ischemia and infarction, but its effects appear to be dependent on the timing of its expression. Upregulation of PPARγ in the post-infarction period appears to be deleterious. When the PPARγ agonist thiazolidinedione rosiglitazone was given to rats in the post infarction period (more than 6 hours after infarction), there was significantly increased mortality compared with controls (49). In contrast, there is evidence that upregulation of PPARγ may be protective in the pre-infarction period. When Zucker diabetic fatty rats were pretreated with rosiglitazone for 8 days prior to ischemia/reperfusion or myocardial infarction, the number of apoptotic cardiomyocytes was decreased by 58% and myocardial infarct size was decreased by 46% (50). Similar results were found when hypercholesterolemic rabbits were pretreated with rosiglitazone before being subjected to ischemia/reperfusion (51).
However, in randomized clinical trials, effects on cardiovascular outcomes for the 2 available TZDs are divergent. The PROactive trial, a randomized, double-blind outcome study in patients with type 2 DM and a history of macrovascular disease managed with diet and/or oral pioglitazone (52), demonstrated a reduction in the frequency of death, non-fatal MI, and cerebral infarction (53). Consistent with these observations, a meta-analysis of clinical trials demonstrated a significant reduction in death, MI, or stroke in patients with DM randomized to pioglitazone compared with placebo (54). In contrast, a meta-analysis including 42 clinical trials in DM patients demonstrated a significant 43% increased risk of MI and 64% increased risk of cardiovascular mortality in patients randomized to rosiglitazone compared with placebo (55). Additional data suggest that the effects of PPARα agonist drugs are equally complex and confusing. In contrast to the hyperglycemia seen in animal models where PPARα agonists are administered, in clinical trials administration of PPARα agonists to patients with DM results in a decrease in serum glucose levels and an apparent decrease in progression of coronary atherosclerosis (56).
Genetic and Pharmacogenetic Associations
Associations between the PPARA IVS7 2498 G>C polymorphism, atherosclerosis, and adverse cardiac events have been investigated. In the Lopid Coronary Angiography Trial (LOCAT - a study in 395 Finnish men less than 70 years old), PPARA IVS7 2498 C allele carriers had a significantly greater progression of coronary atherosclerosis compared with GG homozygotes with similar progression in both treated and untreated groups (no genotype-by-treatment interaction) (57). Among healthy middle aged men in the second Northwick Park Health Study (NPHS2), the PPARA IVS7 2498 CC genotype showed a trend toward greater incidence of ischemic events (myocardial infarction or coronary revascularization) (HR 1.83; 95%CI 0.96–3.51; p=0.07) (57).
We have recently investigated the influence ofPPARA IVS7 2498 genotypeon clinical outcomes in response to β-blocker therapy in patients admitted to the hospital with acute coronary syndromes(58). Mortality and cardiac rehospitalization through 1 year were assessed in 735 patients with acute coronary syndromes and significantly different outcomes associated with β-blocker therapy were observed according to PPARA IVS7 2498 genotype (p = 0.002 for interaction). Multivariable analysis, adjusting for propensity of BB therapy and all factors that differed by PPARA IVS7 2498 genotype, demonstrated that among ACS patients homozygous for the PPARA IVS7 2498 GG genotype, discharge on β-blocker therapy was associated with a 48% relative risk reduction in cardiac rehospitalization (HR 0.52, 95% CI 0.32–0.86; p=0.011). Discharge on β-blocker therapy in carriers of the PPARA IVS7 2498 C allele was associated with a nearly 3-fold relative increase in the risk of cardiac rehospitalization (HR 2.92, 95% CI 1.32–6.92; p=0.015; genotype interaction p=0.0005). We further demonstrated that PPARα mRNA expression in cardiac tissue from 34 hearts with normal left ventricular ejection fraction determined by echocardiography and no history of myocardial dysfunction donated for orthotopic cardiac transplantation but declined for reasons related to size or ABO blood type mismatch was significantly greater in the PPARA IVS7 2498 GG homozygote individuals compared to the PPARA IVS7 2498 GC heterozygote individuals (none of the individuals were CC homozygotes; 1.20 ± 0.30 vs. 0.94 ± 0.40 respectively; p = 0.04). To further explore the effect of increased PPARα expression on β-adrenergic responsiveness, we studied changes in heart rate and contractility in response to incremental doses of the β sympathomimetic dobutamine in transgenic mice with cardiac specific overexpression of PPARα (an animal model that mimicked the relative differences in cardiac PPARα expression demonstrated by our mRNA expression experiments in humans) (58). We found that, compared to non-transgene littermates, transgenic mice with cardiac specific overexpression of PPARα had a blunted response to incremental doses of dobutamine and proposed that PPARA IVS7 2498 C allele carriers have less of a general response to β-adrenergic stimulation compared to GG homozygote individuals, thereby reducing or eliminating the expected benefit derived from β-blocker therapy after acute coronary syndromes and potentially unmasking hazardous effects (58).
The PPARA Leu162Val SNP, located in the DNA binding region of PPARα and thought to confer differences in ligand-responsive activation of PPARα(59;60), has been associated with serum triglyceride, total cholesterol, LDL-cholesterol, and apolipoprotein B and C-III levels in small scale clinical studies (61–63). However when the association of the PPARA Leu162Val polymorphism with the risk of atherosclerosis was investigated in 3,012 healthy middle aged men in NPHS2, or in 395 Finnish men less than 70 years old from the LOCAT study, no association with variations in serum lipids were found (57). Nevertheless, the Val162 allele carriers had less progression of atherosclerosis in both treated and untreated groups in LOCAT (no interaction of drug treatment with genotype was noted) (57).
The PPARG Pro12Ala polymorphism is located in exon B of PPARG which is specific to PPARγ2, the PPARγ isoform restricted to adipose tissue (39). Compared with the PPARG Pro12 variant, in vitro DNA binding assays have demonstrated that the PPARG Ala12 variant has lower binding affinity for a PPAR responsive element and transient transfection experiments demonstrate that the PPARG Ala12 variant has decreased PPARγ-activation of a reporter construct in response to ligand (39). Several studies have suggested an association of this polymorphism with carotid atherosclerosis, coronary artery disease and/or myocardial ischemic events, however, not all studies have agreed on the direction of the association (64–66).
In individuals enrolled in the Physicians’ Health Study (14,916 men followed for a mean of 13.2 years) (67), the PPARG Pro12Ala polymorphism was genotyped in 523 individuals who developed a myocardial infarction and in 2092 controls obtained through a nested case-control design (66). Of those individuals who developed a myocardial infarction, the frequency of PPARG Ala12 allele carriers was significantly less than in the controls, with a decreased risk of subsequent MI (hazard ratio HR=0.77; 95% CI 0.60–0.98; p=0.034) (66). This relationship held even after controlling for traditional cardiac risk factors.
The association of this polymorphism with cardiovascular risk was also assessed in 2016 (all Caucasian) patients with type 2 DM from the genetic portion of the continually updated dataset known as the Diabetes Audit and Research in Tayside Scotland database (Go-DARTS) (65). A borderline, non-significant association of the Ala12 allele with non-fatal MI or revascularization (HR 0.54; 95%CI 0.27–1.08; p=0.08) was observed for the entire group. Subgroup analysis demonstrated a significant association if patients younger than 70 years old at time of enrollment were assessed separately (HR 0.43; CI 0.18–0.99; p=0.05) or if patients younger than 70 year old at time of enrollment with no prior history of stroke, MI or revascularization were evaluated for time to first event (HR 0.21; CI 0.06–0.69; p=0.01) (65).
More recently, association of PPARG Pro12Ala genotype with the risk of coronary artery disease was assessed prospectively in women enrolled in the Nurses’ Health Study (8 years mean follow-up) and in men (6 years mean follow-up) enrolled in the Health Professionals Follow-Up Study (64) 249 women and 266 men were identified who had a myocardial infarction. Nested case-controls, matched for age, smoking status and phlebotomy date, were used for comparison. This study demonstrated that, in contrast to the Physicians’ Health Study, men carriers of the Ala12 allele had an increased risk of MI or cardiac death (Relative Risk RR=1.44; CI 1.00–2.07; p=0.05). There was no statistical difference in nonfatal MI or fatal CHD in women carriers of the Ala12 allele (Relative Risk RR=1.17; CI 0.82–1.68; p=0.39) (64). When data were pooled for men and women, carriers of the Ala12 allele had an increased risk of MI or cardiac death (Relative Risk RR=130; CI 1.00–1.67; p=0.05) and, when stratified by body weight, men and women with a body mass index ≥ 25kg/m2 had a 1.68 fold increase in risk (CI 1.13–2.50; p=0.01) (64). While the results from these studies may seem contradictory, there are obvious differences in study design, patient cohorts, primary end-points and power. In addition, it is possible that geographic and ethnic differences in allele frequencies may contribute to variability in the study findings.
The PPARG 54,347 C>T (also referred to as PPARG 1431C>T and PPARG c.161C>T) polymorphism is a synonymous C>T substitution in nucleotide 161 of exon 6 (54,347 nucleotides from the translation start site in genomic PPARG sequence) (68). As exon 6 is found in all isoforms of PPARγ, it has been proposed that associations of this polymorphism may be more informative that those of isoform specifc polymorphisms (69). No functional information on this polymorphism is available to date. The PPARG 54,347 C>T polymorphism has been associated with the extent of coronary artery disease by angiography (69), carotid intima media thickness (70) and incidence of myocardial infarction among individuals younger than age 50 (71).
Several large-scale randomized trials conducted over the last two decades among patients with increased risk of cardiovascular disease, and perhaps especially those with type 2 DM, have demonstrated that statin treatment significantly reduces cardiovascular events (72–78). Although the physiologic mechanism of interaction between these pharmacologic agents and PPAR is not fully understood, it has been proposed that at least some of the cardiovascular effect of statins may result from increased expression and activation of PPARα and PPARγ(79;80).
Chen et al investigated whether a multi-locus haplotype association study combining information from all of the PPAR polymorphisms into a combined model would result in new insights into PPAR pharmacogenetics in the Lipoprotein and Coronary Atherosclerosis Study (LCAS; a randomized, placebo-controlled study of 429 subjects, 35–70 years old, with at least one 30–75% diameter stenosis on coronary angiography and LDL-cholesterol of 115–190 mg/dL despite diet) cohort assessing response to fluvastatin (81). 372 individuals were genotyped for seven PPAR polymorphisms (PPARA Leu162Val, PPARA −35,089 A>C, PPARG Pro12Ala, PPARG 54,347 C>T (designated “PPARG 161 C>T” in the publication), PPARG 25,506 C>T, PPARD −87 T>C (designated “PPARD 294 T>C” in the publication), PPARD −4401 C>T) and the change in serum lipids and progression of coronary artery disease (quantified by changes in lumen diameter and development of new coronary lesions) in response to fluvastatin was assessed (81). PPARD haplotypes were associated with significantly different changes in plasmid lipids and significantly different mean number of new coronary lesions in response to treatment with fluvastatin (81). PPARG haplotype was associated with significantly different changes in minimum lumen diameter (p=0.009) in response to fluvastatin treatment(81).
II. PPARγ Coactivator 1(PGC-1α)
PGC-1α acts as a coactivator to PPARs and other hormone receptors involved in the regulation of cellular energy metabolism and has a similar tissue expression pattern (expression of PGC-1α is enriched in tissues with high oxidative capacity including skeletal muscle, brown adipose tissue, and the heart). PGC-1α is thought to be the intersect point for the complex regulation of cellular energy balance, acting to “fine-tune” nuclear hormone receptor activity and orchestrating the response to multiple physiologic stimuli.
PGC-1α null mice have yielded interesting insights into the cardiac role of PGC-1α. PGC-1α null mice have been shown to have reduced systolic function (82) as well as diminished heart rate response to exercise and β-adrenergic stimulation. Thus PGC-1α regulation of cardiomyocyte metabolism implicates a role for this coactivator in cardiomyopathy.
A. Left Ventricular Hypertrophy
Given its integral role as a PPARα co-activator, it seems likely that PGC-1α would play a role in cardiac hypertrophy. Animal models have not directly addressed hypertrophy in relation to PGC-1α expression, however, in mouse models of chronic pressure overload (a model of LVH) PGC-1α levels are downregulated along with PPARα target genes (83). Human data suggests a role for PGC-1α in hypertrophic cardiomyopathy (HCM)(84).
Genetic and Pharmacogenetic Associations
Two PGC-1α gene (PPARGC1) polymorphisms have been associated with the development of HCM (84). The PPARGC1 Gly482Ser (G>A) and PPARGC1 Thr394Thr (C>T) SNPs were studied in 270 Chinese patients with hypertrophic cardiomyopathy (LV wall thickness >13 mm) and 894 healthy controls. Of note these SNPs are not in linkage dysequilibrium. PPARGC1 Ser482 allele frequency was significantly higher in HCM patients than in controls (84). Regression analysis controlling for age, sex, blood pressure, and BMI revealed an Odds Ratio for HCM of 1.52 (1.11–2.11) for PPARGC1 Ser482 carriers (84). The PPARGC1 Ser482 allele was also associated with increased maximum wall thickness in HCM patients (20.7 vs 19.1 mm; p <0.05) (84). The CC genotype of the PPARGC1 Thr394Thr SNP was more common in HCM patients than in controls (59.3 vs 49.7%; p <0.05), suggesting a recessive effect of the C allele (84). After multivariable regression analysis accounting for age, sex, BMI, and blood pressure, an odds ratio for HCM of 1.49 was noted for patients with CC homozygosity. The CC genotype was also associated with increased maximal wall thickness in HCM patients. Interestingly the PPARGC1 Ser482 and PPARGC1 Thr394Thr polymorphisms, when studied in 2486 hypertensive Chinese patients, were not associated with LVH (present in nearly 50% of these patients). The PPARGC1 Gly482Ser SNP was also studied in 2656 Danish patients, ages 41–71 as part of the DanMONICA study (85). Again no significant interaction between PPARGC1 Gly482Ser genotype and LV mass index was noted.
B. Dilated Cardiomyopathy
PGC-1α has recently been demonstrated to play a role in the development of dilated cardiomyopathy in animal models (12). Transgenic mice with cardiomyocyte-restricted constitutive overexpression of PGC-1α developed cardiomegaly with four chamber enlargement, severely reduced global contractile function and marked edema by 6 weeks of age (12). Histological section from left ventricular tissue revealed sarcomeric disruption, mild fibrosis and expansion of enlarged mitochondria (12). No genetic or pharmacogenetic associations with PGC-1α and the development of dilated cardiomyopathy have been reported to date.
C. Diabetic Cardiomyopathy
PGC-1α has also been implicated in the development of diabetic cardiomyopathy in animal models (34). The expression and activity of PGC-1α are increased in the hearts of two mouse models of diabetes (mice rendered diabetic by streptozotocin or leptin receptor deficiency) (36). Interestingly the association between PGC-1α and insulin resistance itself remains unclear. While studies focused on skeletal muscle would suggest a protective role, studies on pancreatic β cells suggest that PGC-1α reduces insulin secretion (86;87).
Genetic and Pharmacogenetic Associations
The Gly482Ser SNP of PPARGC1A (associated with hypertrophic cardiomyopathy, as discussed above) has been associated with the risk of developing type 2 DM and diabetic retinopathy in Caucasians (88;89).
III. Estrogen Related Receptors (ERRs)
ERRs were originally identified in a search for nuclear hormone receptors related to the estrogen receptors. Although downstream targets of ERR are incompletely understood to date, given that ERRs bind to the same nuclear hormone receptor binding site consensus sequence that the estrogen receptor binds they are thought to potentially have many of the same targets and effects as the estrogen receptors. As discussed above, ERRα and ERRγ have recently been recognized as important PPAR co-activators (13;14) as well as functional partners of PGC-1α(15–17). Gene expression profiling studies in cardiomyocytes demonstrate that ERRα and ERRγ up-regulate the expression of PPARα and PPARα-regulated genes(18;19).
A. Left Ventricular Hypertrophy
ERRα and ERRγ are thought to have a significant role in myocardial hypertrophy. ERRα mRNA levels are reduced in mice subjected to transient aortic constriction when compared to sham-operated controls, suggesting a role in the response to pressure overload (90). Mice null for ERRα have no significant difference in total body weight, but have significantly lower LV mass to body weight ratio when compared to wild-type mice (90). ERRγ is thought to have a similar role as ERRα in the myocardial hypertrophic response to pressure overload, however, direct binding to DNA may be required for ERRγ as mice with ERRγ lacking a DNA binding domain are characterized by lower LV mass to body weight ratio as well as conduction abnormalities (91).
Genetic and Pharmacogenetic Associations
To date, no genetic or pharmacogenetic associations of polymorphisms within the genes encoding ERRα and ERRγ (ESRRA and ESRRG, respectively) with ventricular hypertrophy have been reported. However, associations with hypertrophy have been described for two close relatives, the estrogen receptor α (ESR1) (92–95) and estrogen receptor β (ESR2) (96) genes. It seems likely that similar associations will be reported with the ERRs in the future.
B. Ischemic Cardiomyopathy
To date, there is no animal or human expression data that analyzes the role of ERRs in the myocardial response to ischemia. However, investigators have noted a genetic association of a polymorphism in the related gene ESR1 and the extent of coronary artery disease (92;93).
IV. Retinoid X Receptor α (RXRα)
As described earlier, PPARs heterodimerize with RXRα and bind to their cognate DNA regulatory elements. While studies relating RXRα to cardiomyopathic phenotypes do not currently exist, an RXRγ polymorphism has been associated with significantly higher indices of coronary stenosis in 105 patients with suspected coronary artery disease undergoing coronary angiography (97). This same polymorphism has been associated with familial combined hyperlipidemia(97).
DISCUSSION
The ultimate goal of pharmacogenetics is to define the genetic determinants of individual drug responsiveness and thereby provide personalized treatment to each individual. This review has discussed the growing body of data reporting the association of PPAR activator complex gene polymorphisms with cardiomyopathic phenotypes. To date, however, there are a limited number of important studies that have explored the interaction between PPAR activator complex gene variants and pharmacotherapy and even less that have explored the interaction between these variants and pharmacotherapy administered to treat heart failure. These studies, taken together, represent the beginning of a promising area of targeted genotype analysis as a potential guide to new drug development in the treatment of cardiomyopathy and heart failure.
Acknowledgments
Funding: (SC): NIH SCCOR in Cardiac Dysfunction and Disease P50 HL077113, 5 P60 DK20579 and 1R21HL089681
Reference List
- 1.Berkenstam A, Gustafsson JA. Nuclear receptors and their relevance to diseases related to lipid metabolism. Curr Opin Pharmacol. 2005;5:171–176. doi: 10.1016/j.coph.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Benoit G, Malewicz M, Perlmann T. Digging deep into the pockets of orphan nuclear receptors: insights from structural studies. Trends Cell Biol. 2004;14:369–376. doi: 10.1016/j.tcb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Y, Kan L, Qi C, Kanwar YS, Yeldandi AV, Rao MS, et al. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J Biol Chem. 2000;275:13510–13516. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]
- 5.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi C, Zhu Y, Reddy JK. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem Biophys. 2000 Spring;32:187–204. doi: 10.1385/cbb:32:1-3:187. [DOI] [PubMed] [Google Scholar]
- 7.Spiegelman BM, Puigserver P, Wu Z. Regulation of adipogenesis and energy balance by PPARgamma and PGC-1. Int J Obes Relat Metab Disord. 2000;24(4):S8–S10. doi: 10.1038/sj.ijo.0801492. [DOI] [PubMed] [Google Scholar]
- 8.Yu C, Markan K, Temple KA, Deplewski D, Brady MJ, Cohen RN. The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor gamma transcriptional activity and repress 3T3-L1 adipogenesis. J Biol Chem. 2005;280:13600–13605. doi: 10.1074/jbc.M409468200. [DOI] [PubMed] [Google Scholar]
- 9.Krogsdam AM, Nielsen CA, Neve S, Holst D, Helledie T, Thomsen B, et al. Nuclear receptor corepressor-dependent repression of peroxisome-proliferator-activated receptor delta-mediated transactivation. Biochem J. 2002;363:157–165. doi: 10.1042/0264-6021:3630157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, hado De OR, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westin S, Kurokawa R, Nolte RT, Wisely GB, McInerney EM, Rose DW, et al. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 12.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 14.Horard B, Vanacker JM. Estrogen receptor-related receptors: orphan receptors desperately seeking a ligand. J Mol Endocrinol. 2003;31:349–357. doi: 10.1677/jme.0.0310349. [DOI] [PubMed] [Google Scholar]
- 15.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 16.Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) J Biol Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 17.Ichida M, Nemoto S, Finkel T. Identification of a specific molecular repressor of the peroxisome proliferator-activated receptor gamma Coactivator-1 alpha (PGC-1alpha) J Biol Chem. 2002;277:50991–50995. doi: 10.1074/jbc.M210262200. [DOI] [PubMed] [Google Scholar]
- 18.Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, et al. Genome-wide Orchestration of Cardiac Functions by the Orphan Nuclear Receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Dewald O, Sharma S, Adrogue J, Salazar R, Duerr GD, Crapo JD, et al. Downregulation of peroxisome proliferator-activated receptor-alpha gene expression in a mouse model of ischemic cardiomyopathy is dependent on reactive oxygen species and prevents lipotoxicity. Circulation. 2005;112:407–415. doi: 10.1161/CIRCULATIONAHA.105.536318. [DOI] [PubMed] [Google Scholar]
- 21.Sack MN, Rader TA, Park S, Bastin J, McCune SA, Kelly DP. Fatty acid oxidation enzyme gene expression is downregulated in the failing heart. Circulation. 1996;94:2837–2842. doi: 10.1161/01.cir.94.11.2837. [DOI] [PubMed] [Google Scholar]
- 22.Barger PM, Brandt JM, Leone TC, Weinheimer CJ, Kelly DP. Deactivation of peroxisome proliferator-activated receptor-alpha during cardiac hypertrophic growth. J Clin Invest. 2000;105:1723–1730. doi: 10.1172/JCI9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng L, Ding G, Qin Q, Huang Y, Lewis W, He N, et al. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat Med. 2004;10:1245–1250. doi: 10.1038/nm1116. [DOI] [PubMed] [Google Scholar]
- 24.Planavila A, Rodriguez-Calvo R, Jove M, Michalik L, Wahli W, Laguna JC, et al. Peroxisome proliferator-activated receptor beta/delta activation inhibits hypertrophy in neonatal rat cardiomyocytes. Cardiovasc Res. 2005;65:832–841. doi: 10.1016/j.cardiores.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 25.Jamshidi Y, Montgomery HE, Hense HW, Myerson SG, Torra IP, Staels B, et al. Peroxisome proliferator--activated receptor alpha gene regulates left ventricular growth in response to exercise and hypertension. Circulation. 2002;105:950–955. doi: 10.1161/hc0802.104535. [DOI] [PubMed] [Google Scholar]
- 26.Sher T, Yi HF, McBride OW, Gonzalez FJ. cDNA cloning, chromosomal mapping, and functional characterization of the human peroxisome proliferator activated receptor. Biochemistry. 1993;32:5598–5604. doi: 10.1021/bi00072a015. [DOI] [PubMed] [Google Scholar]
- 27.Schunkert H, Hengstenberg C, Holmer SR, Broeckel U, Luchner A, Muscholl MW, et al. Lack of association between a polymorphism of the aldosterone synthase gene and left ventricular structure. Circulation. 1999;99:2255–2260. doi: 10.1161/01.cir.99.17.2255. [DOI] [PubMed] [Google Scholar]
- 28.Razeghi P, Young ME, Alcorn JL, Moravec CS, Frazier OH, Taegtmeyer H. Metabolic gene expression in fetal and failing human heart. Circulation. 2001;104:2923–2931. doi: 10.1161/hc4901.100526. [DOI] [PubMed] [Google Scholar]
- 29.Ichihara S, Obata K, Yamada Y, Nagata K, Noda A, Ichihara G, et al. Attenuation of cardiac dysfunction by a PPAR-alpha agonist is associated with down-regulation of redox-regulated transcription factors. J Mol Cell Cardiol. 2006;41:318–329. doi: 10.1016/j.yjmcc.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 30.Brigadeau F, Gele P, Wibaux M, Marquie C, Martin-Nizard F, Torpier G, et al. The PPARalpha activator fenofibrate slows down the progression of the left ventricular dysfunction in porcine tachycardia-induced cardiomyopathy. J Cardiovasc Pharmacol. 2007;49:408–415. doi: 10.1097/FJC.0b013e3180544540. [DOI] [PubMed] [Google Scholar]
- 31.Schupp M, Kintscher U, Fielitz J, Thomas J, Pregla R, Hetzer R, et al. Cardiac PPARalpha expression in patients with dilated cardiomyopathy. Eur J Heart Fail. 2006;8:290–294. doi: 10.1016/j.ejheart.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Ding G, Fu M, Qin Q, Lewis W, Kim HW, Fukai T, et al. Cardiac peroxisome proliferator-activated receptor gamma is essential in protecting cardiomyocytes from oxidative damage. Cardiovasc Res. 2007;76:269–279. doi: 10.1016/j.cardiores.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 33.Mehrabi MR, Haslmayer P, Humpeler S, Strauss-Blasche G, Marktl W, Tamaddon F, et al. Quantitative analysis of peroxisome proliferator-activated receptor gamma (PPARgamma) expression in arteries and hearts of patients with ischaemic or dilated cardiomyopathy. Eur J Heart Fail. 2003;5:733–739. doi: 10.1016/s1388-9842(03)00148-x. [DOI] [PubMed] [Google Scholar]
- 34.Finck BN, Kelly DP. Peroxisome proliferator-activated receptor alpha (PPARalpha) signaling in the gene regulatory control of energy metabolism in the normal and diseased heart. J Mol Cell Cardiol. 2002;34:1249–1257. doi: 10.1006/jmcc.2002.2061. [DOI] [PubMed] [Google Scholar]
- 35.Finck BN, Bernal-Mizrachi C, Han DH, Coleman T, Sambandam N, LaRiviere LL, et al. A potential link between muscle peroxisome proliferator- activated receptor-alpha signaling and obesity-related diabetes. Cell Metab. 2005;1:133–144. doi: 10.1016/j.cmet.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Finck BN, Lehman JJ, Leone TC, Welch MJ, Bennett MJ, Kovacs A, et al. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 38.Andrulionyte L, Zacharova J, Chiasson JL, Laakso M. Common polymorphisms of the PPAR-gamma2 (Pro12Ala) and PGC-1alpha (Gly482Ser) genes are associated with the conversion from impaired glucose tolerance to type 2 diabetes in the STOP-NIDDM trial. Diabetologia. 2004;47:2176–2184. doi: 10.1007/s00125-004-1577-2. [DOI] [PubMed] [Google Scholar]
- 39.Deeb SS, Fajas L, Nemoto M, Pihlajamaki J, Mykkanen L, Kuusisto J, et al. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 40.Meshkani R, Taghikhani M, Larijani B, Bahrami Y, Khatami S, Khoshbin E, et al. Pro12Ala polymorphism of the peroxisome proliferator-activated receptor-gamma2 (PPARgamma-2) gene is associated with greater insulin sensitivity and decreased risk of type 2 diabetes in an Iranian population. Clin Chem Lab Med. 2007;45:477–482. doi: 10.1515/CCLM.2007.095. [DOI] [PubMed] [Google Scholar]
- 41.Nemoto M, Sasaki T, Deeb SS, Fujimoto WY, Tajima N. Differential effect of PPARgamma2 variants in the development of type 2 diabetes between native Japanese and Japanese Americans. Diabetes Res Clin Pract. 2002;57:131–137. doi: 10.1016/s0168-8227(02)00027-x. [DOI] [PubMed] [Google Scholar]
- 42.Tai ES, Corella D, urenberg-Yap M, Adiconis X, Chew SK, Tan CE, et al. Differential effects of the C1431T and Pro12Ala PPARgamma gene variants on plasma lipids and diabetes risk in an Asian population. J Lipid Res. 2004;45:674–685. doi: 10.1194/jlr.M300363-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Flavell DM, Ireland H, Stephens JW, Hawe E, Acharya J, Mather H, et al. Peroxisome proliferator-activated receptor alpha gene variation influences age of onset and progression of type 2 diabetes. Diabetes. 2005;54:582–586. doi: 10.2337/diabetes.54.2.582. [DOI] [PubMed] [Google Scholar]
- 44.Kang ES, Park SY, Kim HJ, Kim CS, Ahn CW, Cha BS, et al. Effects of Pro12Ala polymorphism of peroxisome proliferator-activated receptor gamma2 gene on rosiglitazone response in type 2 diabetes. Clin Pharmacol Ther. 2005;78:202–208. doi: 10.1016/j.clpt.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 45.Wolford JK, Yeatts KA, Dhanjal SK, Black MH, Xiang AH, Buchanan TA, et al. Sequence variation in PPARG may underlie differential response to troglitazone. Diabetes. 2005;54:3319–3325. doi: 10.2337/diabetes.54.11.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen L, Ekstrom CT, Tabanera YP, Anant M, Wassermann K, Reinhardt RR. The Pro12Ala variant of the PPARG gene is a risk factor for peroxisome proliferator-activated receptor-gamma/alpha agonist-induced edema in type 2 diabetic patients. J Clin Endocrinol Metab. 2006;91:3446–3450. doi: 10.1210/jc.2006-0590. [DOI] [PubMed] [Google Scholar]
- 47.Sambandam N, Morabito D, Wagg C, Finck BN, Kelly DP, Lopaschuk GD. Chronic activation of PPARalpha is detrimental to cardiac recovery after ischemia. Am J Physiol Heart Circ Physiol. 2006;290:H87–H95. doi: 10.1152/ajpheart.00285.2005. [DOI] [PubMed] [Google Scholar]
- 48.Panagia M, Gibbons GF, Radda GK, Clarke K. PPAR-alpha activation required for decreased glucose uptake and increased susceptibility to injury during ischemia. Am J Physiol Heart Circ Physiol. 2005;288:H2677–H2683. doi: 10.1152/ajpheart.00200.2004. [DOI] [PubMed] [Google Scholar]
- 49.Lygate CA, Hulbert K, Monfared M, Cole MA, Clarke K, Neubauer S. The PPARgamma-activator rosiglitazone does not alter remodeling but increases mortality in rats post-myocardial infarction. Cardiovasc Res. 2003;58:632–637. doi: 10.1016/s0008-6363(03)00289-x. [DOI] [PubMed] [Google Scholar]
- 50.Yue TL, Bao W, Gu JL, Cui J, Tao L, Ma XL, et al. Rosiglitazone treatment in Zucker diabetic Fatty rats is associated with ameliorated cardiac insulin resistance and protection from ischemia/reperfusion-induced myocardial injury. Diabetes. 2005;54:554–562. doi: 10.2337/diabetes.54.2.554. [DOI] [PubMed] [Google Scholar]
- 51.Liu HR, Tao L, Gao E, Lopez BL, Christopher TA, Willette RN, et al. Anti-apoptotic effects of rosiglitazone in hypercholesterolemic rabbits subjected to myocardial ischemia and reperfusion. Cardiovasc Res. 2004;62:135–144. doi: 10.1016/j.cardiores.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 52.Charbonnel B, Dormandy J, Erdmann E, Massi-Benedetti M, Skene A. The prospective pioglitazone clinical trial in macrovascular events (PROactive): can pioglitazone reduce cardiovascular events in diabetes? Study design and baseline characteristics of 5238 patients. Diabetes Care. 2004;27:1647–1653. doi: 10.2337/diacare.27.7.1647. [DOI] [PubMed] [Google Scholar]
- 53.Dormandy JA, Charbonnel B, Eckland DJ, Erdmann E, Massi-Benedetti M, Moules IK, et al. Secondary prevention of macrovascular events in patients with type 2 diabetes in the PROactive Study (PROspective pioglitAzone Clinical Trial In macroVascular Events): a randomised controlled trial. Lancet. 2005;366:1279–1289. doi: 10.1016/S0140-6736(05)67528-9. [DOI] [PubMed] [Google Scholar]
- 54.Lincoff AM, Wolski K, Nicholls SJ, Nissen SE. Pioglitazone and risk of cardiovascular events in patients with type 2 diabetes mellitus: a meta-analysis of randomized trials. JAMA. 2007;298:1180–1188. doi: 10.1001/jama.298.10.1180. [DOI] [PubMed] [Google Scholar]
- 55.Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. N Engl J Med. 2007;356:2457–2471. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- 56.Koskinen P, Manttari M, Manninen V, Huttunen JK, Heinonen OP, Frick MH. Coronary heart disease incidence in NIDDM patients in the Helsinki Heart Study. Diabetes Care. 1992;15:820–825. doi: 10.2337/diacare.15.7.820. [DOI] [PubMed] [Google Scholar]
- 57.Flavell DM, Jamshidi Y, Hawe E, Pineda TI, Taskinen MR, Frick MH, et al. Peroxisome proliferator-activated receptor alpha gene variants influence progression of coronary atherosclerosis and risk of coronary artery disease. Circulation. 2002;105:1440–1445. doi: 10.1161/01.cir.0000012145.80593.25. [DOI] [PubMed] [Google Scholar]
- 58.Cresci S, Jones PG, Sucharov CC, Marsh S, Lanfear DE, Garsa A, et al. Interaction betweenPPARA genotype and β-blocker treatment influences clinical outcomes following acute coronary syndromes. Pharmacogenetics. 2008 doi: 10.2217/14622416.9.10.1403. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sapone A, Peters JM, Sakai S, Tomita S, Papiha SS, Dai R, et al. The human peroxisome proliferator-activated receptor alpha gene: identification and functional characterization of two natural allelic variants. Pharmacogenetics. 2000;10:321–333. doi: 10.1097/00008571-200006000-00006. [DOI] [PubMed] [Google Scholar]
- 60.Flavell DM, Pineda TI, Jamshidi Y, Evans D, Diamond JR, Elkeles RS, et al. Variation in the PPARalpha gene is associated with altered function in vitro and plasma lipid concentrations in Type II diabetic subjects. Diabetologia. 2000;43:673–680. doi: 10.1007/s001250051357. [DOI] [PubMed] [Google Scholar]
- 61.Vohl MC, Lepage P, Gaudet D, Brewer CG, Betard C, Perron P, et al. Molecular scanning of the human PPARa gene: association of the L162v mutation with hyperapobetalipoproteinemia. J Lipid Res. 2000;41:945–952. [PubMed] [Google Scholar]
- 62.Tai ES, Demissie S, Cupples LA, Corella D, Wilson PW, Schaefer EJ, et al. Association between the PPARA L162V polymorphism and plasma lipid levels: the Framingham Offspring Study. Arterioscler Thromb Vasc Biol. 2002;22:805–810. doi: 10.1161/01.atv.0000012302.11991.42. [DOI] [PubMed] [Google Scholar]
- 63.Tai ES, Corella D, Demissie S, Cupples LA, Coltell O, Schaefer EJ, et al. Polyunsaturated fatty acids interact with the PPARA-L162V polymorphism to affect plasma triglyceride and apolipoprotein C-III concentrations in the Framingham Heart Study. J Nutr. 2005;135:397–403. doi: 10.1093/jn/135.3.397. [DOI] [PubMed] [Google Scholar]
- 64.Pischon T, Pai JK, Manson JE, Hu FB, Rexrode KM, Hunter D, et al. Peroxisome proliferator-activated receptor-gamma2 P12A polymorphism and risk of coronary heart disease in US men and women. Arterioscler Thromb Vasc Biol. 2005;25:1654–1658. doi: 10.1161/01.ATV.0000171993.78135.7e. [DOI] [PubMed] [Google Scholar]
- 65.Doney AS, Fischer B, Leese G, Morris AD, Palmer CN. Cardiovascular risk in type 2 diabetes is associated with variation at the PPARG locus: a Go-DARTS study. Arterioscler Thromb Vasc Biol. 2004;24:2403–2407. doi: 10.1161/01.ATV.0000147897.57527.e4. [DOI] [PubMed] [Google Scholar]
- 66.Ridker PM, Cook NR, Cheng S, Erlich HA, Lindpaintner K, Plutzky J, et al. Alanine for proline substitution in the peroxisome proliferator-activated receptor gamma-2 (PPARG2) gene and the risk of incident myocardial infarction. Arterioscler Thromb Vasc Biol. 2003;23:859–863. doi: 10.1161/01.ATV.0000068680.19521.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Final report on the aspirin component of the ongoing Physicians’ Health Study. Steering Committee of the Physicians’ Health Study Research Group. N Engl J Med. 1989;321:129–135. doi: 10.1056/NEJM198907203210301. [DOI] [PubMed] [Google Scholar]
- 68.Meirhaeghe A, Fajas L, Helbecque N, Cottel D, Lebel P, Dallongeville J, et al. A genetic polymorphism of the peroxisome proliferator-activated receptor gamma gene influences plasma leptin levels in obese humans. Hum Mol Genet. 1998;7:435–440. doi: 10.1093/hmg/7.3.435. [DOI] [PubMed] [Google Scholar]
- 69.Wang XL, Oosterhof J, Duarte N. Peroxisome proliferator-activated receptor gamma C161-->T polymorphism and coronary artery disease. Cardiovasc Res. 1999;44:588–594. doi: 10.1016/s0008-6363(99)00256-4. [DOI] [PubMed] [Google Scholar]
- 70.Al-Shali KZ, House AA, Hanley AJ, Khan HM, Harris SB, Zinman B, et al. Genetic variation in PPARG encoding peroxisome proliferator-activated receptor gamma associated with carotid atherosclerosis. Stroke. 2004;35:2036–2040. doi: 10.1161/01.STR.0000138784.68159.a5. [DOI] [PubMed] [Google Scholar]
- 71.Chao TH, Li YH, Chen JH, Wu HL, Shi GY, Liu PY, et al. The 161TT genotype in the exon 6 of the peroxisome-proliferator-activated receptor gamma gene is associated with premature acute myocardial infarction and increased lipid peroxidation in habitual heavy smokers. Clin Sci (Lond) 2004;107:461–466. doi: 10.1042/CS20040014. [DOI] [PubMed] [Google Scholar]
- 72.Colhoun HM, Betteridge DJ, Durrington PN, Hitman GA, Neil HA, Livingstone SJ, et al. Rapid emergence of effect of atorvastatin on cardiovascular outcomes in the Collaborative Atorvastatin Diabetes Study (CARDS) Diabetologia. 2005;48:2482–2485. doi: 10.1007/s00125-005-0029-y. [DOI] [PubMed] [Google Scholar]
- 73.Colhoun HM, Betteridge DJ, Durrington PN, Hitman GA, Neil HA, Livingstone SJ, et al. Primary prevention of cardiovascular disease with atorvastatin in type 2 diabetes in the Collaborative Atorvastatin Diabetes Study (CARDS): multicentre randomised placebo-controlled trial. Lancet. 2004;364:685–696. doi: 10.1016/S0140-6736(04)16895-5. [DOI] [PubMed] [Google Scholar]
- 74.LaRosa JC, Grundy SM, Waters DD, Shear C, Barter P, Fruchart JC, et al. Intensive lipid lowering with atorvastatin in patients with stable coronary disease. N Engl J Med. 2005;352:1425–1435. doi: 10.1056/NEJMoa050461. [DOI] [PubMed] [Google Scholar]
- 75.Shepherd J, Barter P, Carmena R, Deedwania P, Fruchart JC, Haffner S, et al. Effect of lowering LDL cholesterol substantially below currently recommended levels in patients with coronary heart disease and diabetes: the Treating to New Targets (TNT) study. Diabetes Care. 2006;29:1220–1226. doi: 10.2337/dc05-2465. [DOI] [PubMed] [Google Scholar]
- 76.Waters DD, LaRosa JC, Barter P, Fruchart JC, Gotto AM, Jr, Carter R, et al. Effects of high-dose atorvastatin on cerebrovascular events in patients with stable coronary disease in the TNT (treating to new targets) study. J Am Coll Cardiol. 2006;48:1793–1799. doi: 10.1016/j.jacc.2006.07.041. [DOI] [PubMed] [Google Scholar]
- 77.Steiner G. A new perspective in the treatment of dyslipidemia : can fenofibrate offer unique benefits in the treatment of type 2 diabetes mellitus? Treat Endocrinol. 2005;4:311–317. doi: 10.2165/00024677-200504050-00004. [DOI] [PubMed] [Google Scholar]
- 78.Steiner G. Lipid intervention trials in diabetes. Diabetes Care. 2000;23(2):B49–B53. [PubMed] [Google Scholar]
- 79.Zelvyte I, Dominaitiene R, Crisby M, Janciauskiene S. Modulation of inflammatory mediators and PPARgamma and NFkappaB expression by pravastatin in response to lipoproteins in human monocytes in vitro. Pharmacol Res. 2002;45:147–154. doi: 10.1006/phrs.2001.0922. [DOI] [PubMed] [Google Scholar]
- 80.Inoue I, Goto S, Mizotani K, Awata T, Mastunaga T, Kawai S, et al. Lipophilic HMG-CoA reductase inhibitor has an anti-inflammatory effect: reduction of MRNA levels for interleukin-1beta, interleukin-6, cyclooxygenase-2, and p22phox by regulation of peroxisome proliferator-activated receptor alpha (PPARalpha) in primary endothelial cells. Life Sci. 2000;67:863–876. doi: 10.1016/s0024-3205(00)00680-9. [DOI] [PubMed] [Google Scholar]
- 81.Chen S, Tsybouleva N, Ballantyne CM, Gotto AM, Jr, Marian AJ. Effects of PPARalpha, gamma and delta haplotypes on plasma levels of lipids, severity and progression of coronary atherosclerosis and response to statin therapy in the lipoprotein coronary atherosclerosis study. Pharmacogenetics. 2004;14:61–71. doi: 10.1097/00008571-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 82.Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, et al. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 83.Lehman JJ, Kelly DP. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol. 2002;29:339–345. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- 84.Wang S, Fu C, Wang H, Shi Y, Xu X, Chen J, et al. Polymorphisms of the peroxisome proliferator-activated receptor-gamma coactivator-1alpha gene are associated with hypertrophic cardiomyopathy and not with hypertension hypertrophy. Clin Chem Lab Med. 2007 doi: 10.1515/CCLM.2007.189. [DOI] [PubMed] [Google Scholar]
- 85.Ambye L, Rasmussen S, Fenger M, Jorgensen T, Borch-Johnsen K, Madsbad S, et al. Studies of the Gly482Ser polymorphism of the peroxisome proliferator-activated receptor gamma coactivator 1alpha (PGC-1alpha) gene in Danish subjects with the metabolic syndrome. Diabetes Res Clin Pract. 2005;67:175–179. doi: 10.1016/j.diabres.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 86.Michael LF, Wu Z, Cheatham RB, Puigserver P, Adelmant G, Lehman JJ, et al. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc Natl Acad Sci U S A. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoon JC, Xu G, Deeney JT, Yang SN, Rhee J, Puigserver P, et al. Suppression of beta cell energy metabolism and insulin release by PGC-1alpha. Dev Cell. 2003;5:73–83. doi: 10.1016/s1534-5807(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 88.Petrovic MG, Kunej T, Peterlin B, Dovc P, Petrovic D. Gly482Ser polymorphism of the peroxisome proliferator-activated receptor-gamma coactivator-1 gene might be a risk factor for diabetic retinopathy in Slovene population (Caucasians) with type 2 diabetes and the Pro12Ala polymorphism of the PPARgamma gene is not. Diabetes Metab Res Rev. 2005;21:470–474. doi: 10.1002/dmrr.546. [DOI] [PubMed] [Google Scholar]
- 89.Kunej T, Globocnik PM, Dovc P, Peterlin B, Petrovic D. A Gly482Ser polymorphism of the peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) gene is associated with type 2 diabetes in Caucasians. Folia Biol (Praha) 2004;50:157–158. [PubMed] [Google Scholar]
- 90.Huss JM, Imahashi K, Dufour CR, Weinheimer CJ, Courtois M, Kovacs A, et al. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 91.Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 92.Pollak A, Rokach A, Blumenfeld A, Rosen LJ, Resnik L, Dresner PR. Association of oestrogen receptor alpha gene polymorphism with the angiographic extent of coronary artery disease. Eur Heart J. 2004;25:240–245. doi: 10.1016/j.ehj.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 93.Kunnas TA, Laippala P, Penttila A, Lehtimaki T, Karhunen PJ. Association of polymorphism of human alpha oestrogen receptor gene with coronary artery disease in men: a necropsy study. BMJ. 2000;321:273–274. doi: 10.1136/bmj.321.7256.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Figtree GA, Kindmark A, Lind L, Grundberg E, Speller B, Robinson BG, et al. Novel estrogen receptor alpha promoter polymorphism increases ventricular hypertrophic response to hypertension. J Steroid Biochem Mol Biol. 2007;103:110–118. doi: 10.1016/j.jsbmb.2006.09.035. [DOI] [PubMed] [Google Scholar]
- 95.Malmqvist K, Kahan T, Edner M, Held C, Hagg A, Lind L, et al. Regression of left ventricular hypertrophy in human hypertension with irbesartan. J Hypertens. 2001;19:1167–1176. doi: 10.1097/00004872-200106000-00023. [DOI] [PubMed] [Google Scholar]
- 96.Peter I, Shearman AM, Vasan RS, Zucker DR, Schmid CH, Demissie S, et al. Association of estrogen receptor beta gene polymorphisms with left ventricular mass and wall thickness in women. Am J Hypertens. 2005;18:1388–1395. doi: 10.1016/j.amjhyper.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 97.Nohara A, Kawashiri MA, Claudel T, Mizuno M, Tsuchida M, Takata M, et al. High frequency of a retinoid X receptor gamma gene variant in familial combined hyperlipidemia that associates with atherogenic dyslipidemia. Arterioscler Thromb Vasc Biol. 2007;27:923–928. doi: 10.1161/01.ATV.0000258945.76141.8a. [DOI] [PubMed] [Google Scholar]