Abstract
Purpose
The purpose of this article is to review the recent advances in the atomic-level understanding of the Epidermal Growth Factor Receptor Tyrosine Kinase (EGFR-TK). We aim to highlight the current and future importance of these studies for the understanding and treatment of malignancies where EGFR-TK is improperly activated.
Methods
The analysis was conducted on published crystal structures deposited in the Protein Data Bank (www.pdb.org) using the program O.
Results
In this review we emphasize how recent EGFR kinase domain crystal structures can explain the mechanisms of activation for L858R and other EGFR-TK mutations, and compare these distinct activating mechanisms with those recently described for the wild-type EGFR. We suggest an atomic-level mechanism for the poor efficacy of lapatinib against tumors with activating EGFR kinase domain point mutations compared to the efficacy of gefitinib and erlotinib, and demonstrate how structural insights help our understanding of acquired resistance to these agents. We also highlight how these new molecular-level structural data are expected to affect the development of EGFR-TK targeted small molecule kinase inhibitors.
Conclusion
There are now more crystal structures published for the EGFR-TK domain than for any other tyrosine kinase. This wealth of crystallographic information is beginning to describe the mechanisms by which proper regulation of EGFR-TK is lost in disease. These crystal structures are beginning to show how small molecules inhibit EGFR-TK activity and will aid development of EGFR-TK mutant targeted therapies.
Introduction
The Epidermal Growth Factor Receptor (EGFR) family of receptor tyrosine kinases includes four members: EGFR (HER1), HER2/neu, HER3 and HER41,2. This family has been shown to be important for proper regulation of many developmental, metabolic and physiological processes mediated by EGF, transforming growth factor-α and multiple other ligands. In numerous cancers, including glioblastomas, breast cancer and non-small-cell lung cancer (NSCLC), there is often a transforming deregulation of EGFR family kinase activity3–6. This deregulation can be caused by activating mutations, amplification or overexpression of EGFR or HER2, although HER2 mutations are very rare7–9. NSCLC is the leading cause of death by cancer in North America10, with only 30–40 % of metastatic non-small-cell lung cancer patients surviving for 12 months11 and EGFR overexpression seen in a large proportion of patients. The most prevalent and well studied EGFR-TK somatic mutations occur in NSCLC. While the exact signaling events that result from these somatic mutations are not completely understood, it seems clear that the ‘on-off’ equilibrium of EGFR-TK is altered. Two oral ATP-competitive EGFR tyrosine kinase targeted inhibitors, erlotinib (Tarceva, OSI/Genentech/Roche) and gefitinib (Iressa, AstraZeneca) lead to a significant clinical response in approximately 10 to 30% of NSCLC patients depending on ethnic origin, sex and smoking history12. Erlotinib has shown a modest overall survival benefit as compared to placebo in the second- and third-line treatment of patients with metastatic NSCLC and is currently approved by the US FDA for this indication13. Despite the overall modest efficacy, some patients respond dramatically to these agents. These “Lazarus phenomenon” responses became the subject of research for multiple groups and converged in primary tumor resequencing efforts of the EGFR tyrosine kinase domain that revealed multiple somatic mutations3–5,8,14–16. Correlation of these resequencing data with patient response showed that activating kinase domain mutations are present in the majority of the tumors that respond to the EGFR targeted ATP-competitive inhibitors, erlotinib and gefitinib. Multiple studies demonstrate that these EGFR mutations are oncogenic both in vitro and in vivo, and these mutations are believed to represent very early genetic events leading to lung cancer development17–19. There is an approximately 75% response rate to these agents in patients presenting with EGFR-mutant tumors, suggesting that where these mutations are present they, at least partially, drive the malignant transformation20,21. Overall the frequency of these mutations is approximately 5–20% depending on the population studied and is significantly more common in East Asians, women, non-smokers and patients with adenocarcinoma histology22. EGFR mutation testing is commercially available and current research efforts will define how best to incorporate EGFR mutational status into our current treatment paradigms23–26. Recent and ongoing clinical studies demonstrate the feasibility of mutation-driven patient selection. EGFR amplification as determined by EGFR-fluorescent in situ hybridization (FISH) and overexpression are other predictive and prognostic factors undergoing intense investigation27,28.
There are three broad classes of activating somatic mutations in EGFR-TK. These are categorized as: class (i) in-frame deletions in exon 19, class (ii) single-nucleotide substitutions that cause an amino acid alteration and class (iii) in-frame duplications and/or insertions in exon 208. The majority of the documented activating kinase domain mutations seen in tumors where EGFR-TK is deregulated can be classified as either class (i) or class (ii). Class (i) mutations are in-frame deletions that almost always include amino acid residues leucine-747 to glutamic acid-749 (LRE) (EGFR numbering throughout this review includes the 24 residue signal sequence) and are located at the N-terminus of the kinase domain C-helix (Figure 1). These deletions account for approximately 44% of the activating EGFR-TK domain mutations. Class (ii) mutations are dominated by a single point mutation in exon 21 that substitutes an arginine for a leucine at codon 858 (L858R). This point mutation has the highest prevalence of any single activating EGFR kinase domain point mutation and accounts for approximately 41% of EGFR-TK activating mutations (Figure 1A). An additional 4% of EGFR-TK activating mutations result in glycine-719 (G719) mutation to either serine, alanine or cysteine and a further 6% have been found to be other missense mutations. Class (iii) mutations account for the remaining 5% of EGFR-TK activating mutations. Although patients who present with “classical” activating EGFR tyrosine kinase domain mutations (with the exception of exon 20 insertions), often respond to anilinoquinazoline-based small molecule inhibitors, gefitinib and erlotinib3–5,29, evidence is accumulating that some of these activating mutations are more susceptible to treatment with certain kinase inhibitors than others20,29–32. The molecular-level reasons for these differences in response are not, however, clear. This review summarizes the recent crystallographic studies of EGFR-TK. We highlight how these structural data correlate with clinically seen effects of NSCLC treatment with small molecule kinase inhibitors and emphasize that these structural biology studies help describe distinct mechanisms of improper EGFR-TK activation.
Figure 1. EGFR tyrosine kinase crystal structures.
A. Cartoon illustrating the active state locations of the major structural regions of EGFR-TK discussed in this review. The position of an ATP analogue, AMP-PNP, in the catalytic cleft is shown (ATP) and the locations of the catalytic glutamic acid (Glu) and lysine (Lys) residues are shown (PDB accession code of structure used: 1ITW). B and C. Ribbon representation of the two crystallized conformations of Epidermal Growth Factor Receptor (EGFR) tyrosine kinase. Panel B shows the crystal structure of EGFR tyrosine kinase in complex with erlotinib34 (PDB accession code: 1M17). Panel C shows the crystal structure of EGFR tyrosine kinase in complex with lapatinib35 (PDB accession code: 1XKK). The kinase N- and C-lobes and C-helix are indicated, the activation loop is colored in gold and the glycine rich P-loop, blue. EGFR tyrosine kinase inhibitors erlotinib and lapatinib are shown as space-filling spheres. Locations of activating leucine-747 to glutamic acid-749 'LRE' deletion and leucine-858 to arginine (L858R) point mutation are shown in stick format in red and are labeled. The conformation of the two crystal structures differs with the activation loop of the erlotinib-bound structure seen in an active-like conformation and the activation loop of the lapatinib-bound crystal structure trapped in an inactive conformation. All structural figures made using the program PYMOL (http://www.pymol.org). Panel D shows a schematic of EGFR family activation based on crystal structures reviewed in Hubbard, 2005 69 and here. On extracellular ligand binding the receptor dimerizes allowing the cytoplasmic EGFR-TK to activate in a tail-to-head fashion36. The locations of regions within EGFR-TK that we discuss are indicated on the exon boundary map.
Methods
We downloaded the twenty crystal structures now available for the kinase domain of EGFR from the Protein Data Bank (PDB). The PDB is the repository of all published experimentally determined structures of macromolecules and can be accessed at www.pdb.org. We analyzed these crystal structures using the program O33.
Results
Crystal structures describe active and inactive “snapshots” of EGFR tyrosine kinase
Crystal structures of the tyrosine kinase domain of EGFR have now been published by four groups34–37 (Table 1). The structures provide crystallographic snapshots of EGFR-TK and illustrate the conformational flexibility important for proper catalytic activity. These crystal structures describe the active34 and inactive states35,36 of the kinase (Figure 1), show an activation mechanism for EGFR similar to cyclin-mediated activation of cyclin-dependent kinases36, and describe the binding of multiple EGFR-TK inhibitors to wild-type and mutant kinase37. The molecular reasons for improper activation of EGFR-TK and its targeted inhibition are being made increasingly clear by the work from these groups. The first crystal structures of the tyrosine kinase domain of EGFR, from Charles Eigenbrot and co-workers at Genentech (San Francisco, CA), show the enzyme trapped in an active-like conformation with and without the inhibitor erlotinib (PDB accession codes: 1M17 and 1M14 respectively)34. For this study the ligand-free protein was crystallized and then soaked with erlotinib once crystals had formed. The second EGFR tyrosine kinase crystal structure publication was by Lisa Shewchuk and co-workers at GlaxoSmithKline (Research Triangle Park, NC). They determined the EGFR-TK domain in complex with the inhibitor lapatinib (Tykerb, GlaxoSmithKline) (PDB accession code: 1XKK)35. This lapatinib-bound structure was seen in a conformation very similar to inactive cyclin-dependent kinases (CDK) and Src kinases and was suggested to be the inactive state of EGFR-TK by the authors. For most, but not all protein kinases, phosphorylation of the activation loop is a driving force for kinase activation38–40, the unphosphorylated active and inactive EGFR crystal structures therefore drew attention to a possible undescribed mechanism of EGFR-TK activation which does not require activation loop phosphorylation.
Table 1. Published crystal structures of EGFR-TK.
The table documents the EGFR-TK crystal structures currently published, including two recent EGFR-TK crystal structures in complex with anilinoquinazoline-based irreversible inhibitors 13-JAB and 34-JAB74.
| Protein Data Bank accession code |
Mutation | Inhibitor | Conformational State |
Space Group |
Resolution limit (Å2) |
Publication |
|---|---|---|---|---|---|---|
| 1M14 | Wild-Type | --- | active | I23 | 2.6 | Stamos (2002) |
| 1M17 | Wild-Type | erlotinib (Tarceva) | active | I23 | 2.6 | Stamos (2002) |
| 1XKK | Wild-Type | lapatinib (Tykerb) | inactive | P212121 | 2.4 | Wood (2004) |
| 2GS6 | Wild-Type | ATP analogue-peptide conjugate | active | I23 | 2.6 | Zhang (2006) |
| 2GS2 | Wild-Type | --- | active | I23 | 2.8 | Zhang (2006) |
| 2GS7 | V924R | AMP-PNP | inactive | P21 | 2.6 | Zhang (2006) |
| 2ITX | Wild-Type | AMP-PNP | active | I23 | 3.0 | Yun (2007) |
| 2J6M | Wild-Type | gefitinib (Iressa) | active | I23 | 3.1 | Yun (2007) |
| 2ITW | Wild-Type | AFN-941 | active | I23 | 2.9 | Yun (2007) |
| 2ITY | Wild-Type | gefitinib (Iressa) | active | I23 | 3.4 | Yun (2007) |
| 2ITN | G719S | AMP-PNP | active | I23 | 2.4 | Yun (2007) |
| 2ITO | G719S | gefitinib (Iressa) | active | I23 | 3.2 | Yun (2007) |
| 2ITP | G719S | AEE788 | active | I23 | 2.7 | Yun (2007) |
| 2ITQ | G719S | AFN-941 | active | I23 | 2.7 | Yun (2007) |
| 2ITT | L858R | AEE-788 | active | I23 | 2.7 | Yun (2007) |
| 2ITU | L858R | AFN-941 | active | I23 | 2.8 | Yun (2007) |
| 2ITV | L858R | AMP-PNP | active | I23 | 2.4 | Yun (2007) |
| 2ITZ | L858R | gefitinib (Iressa) | active | I23 | 2.8 | Yun (2007) |
| 2J5E | Wild-Type | 13-JAB | active | I23 | 3.1 | Blair (2007) |
| 2J5F | Wild-Type | 34-JAB | active | I23 | 2.95 | Blair (2007) |
Recently, two groups furthered the structural understanding of EGFR-TK and proposed mechanisms of activation and small molecule inhibition for wild-type and mutant EGFR-TK. The first of these studies, from the laboratory of John Kuriyan at the University of California, Berkeley, elegantly proposes an activation mechanism for EGFR (described below)36. In their paper wild-type EGFR-TK is documented in the active state bound to an ATP analogue-peptide conjugate (a substrate peptide conjugated to an ATP analogue) and also in the inactive state bound to a non-hydrolyzable ATP analogue, AMP-PNP. The conformation of the inactive form of EGFR-TK seen in this paper is similar to that seen for the lapatinib-bound structure and the study confirmed that this is indeed the inactive state. The second recent publication of EGFR-TK crystallographic data was from the laboratory of Michael Eck at Dana-Farber Cancer Institute and Harvard Medical School. In this study, 12 active state crystal structures of EGFR-TK were reported in complex with multiple inhibitors: gefitinib, a pyrrolopyrimidine AEE788 (Novartis), AMP-PNP (AMP-PNP can bind both the active and inactive state EGFR-TK as seen in the Eck and Kuriyan laboratory publications respectively) and AFN941 (Novartis) (an analogue of the pan-kinase inhibitor staurosporine)37. Four of the Eck laboratory structures were wild-type, four contained the leucine-858 to arginine (L858R) mutation and four contained the glycine-719 to serine (G719S) mutation. In this study the effect of the activating mutations on inhibitor binding to EGFR-TK was investigated.
Discussion
Activation mechanism of EGFR-TK is similar to cyclin-mediated activation of CDKs
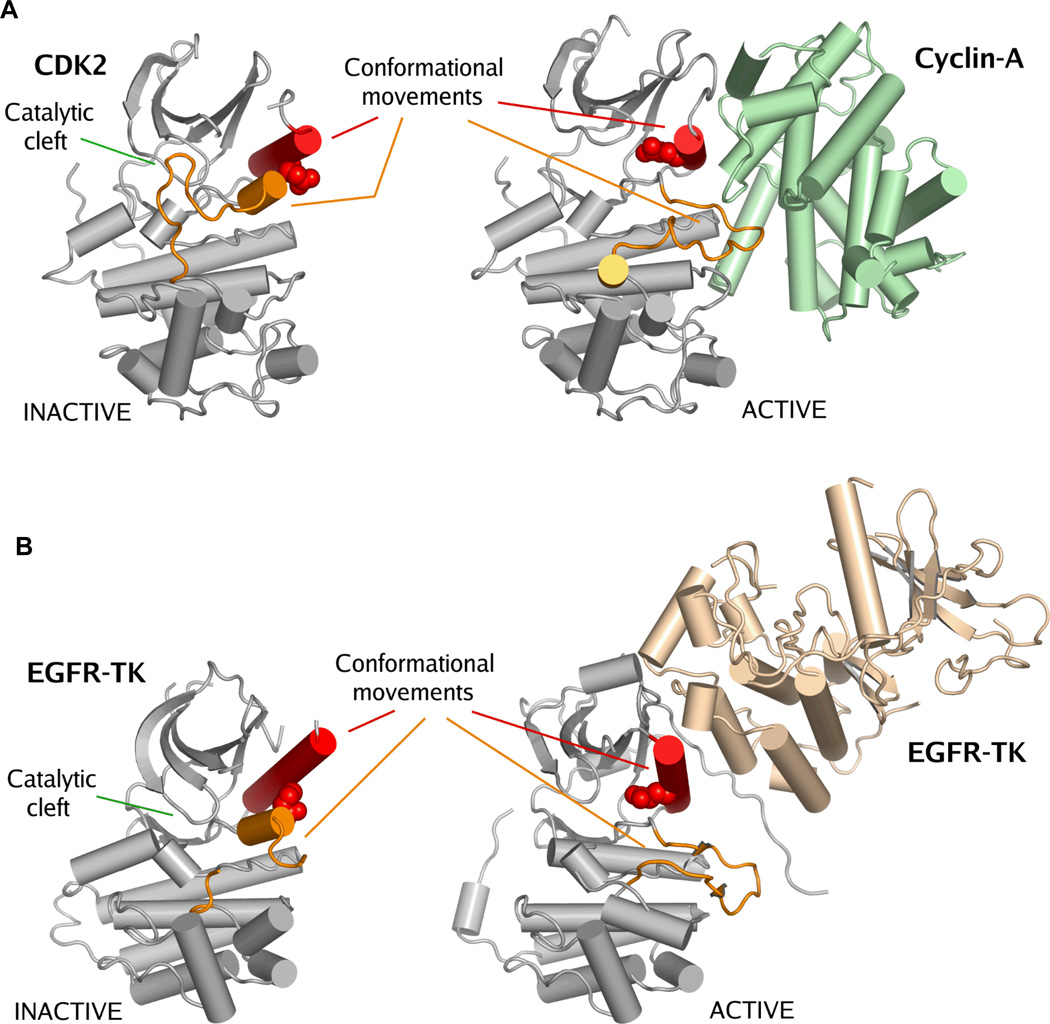
Protein kinase activity is regulated, in general, by the conformational state of the catalytic domain. The catalytic domain conformation, either active or inactive, governs the ability of the kinase to transfer a phosphate from ATP to peptide substrate and thus controls downstream signaling. Because of the critical cellular importance of phosphorylation there are several atomic-level mechanisms to regulate the ‘on-off’ equilibrium of protein kinases. These are comprehensively reviewed elsewhere38,39,41 but often include two components. Firstly, the correct amino acid residues must be oriented to facilitate phosphate transfer, and secondly, the peptide substrate binding site must not be occluded. Creation or removal of these conditions is often a critical step in the regulation of protein kinase activity. Two regions of kinase domains that are very frequently reoriented to meet, or break, these conditions are the activation loop and the C-helix. The activation loop in an active kinase is extended away from the cleft to allow peptide substrate binding while a “catalytic” glutamate residue (part of the C-helix) forms an ionic interaction with a lysine residue that coordinates the α and β phosphates of ATP38 (Figure 1A). In the inactive conformation, the activation loop often changes conformation dramatically to preclude the binding of peptide substrate, while the C-helix rotates away, pulling with it the critical “catalytic” glutamate residue38. There is a conformational equilibrium that exists between the active and inactive kinase states which is often modified by phosphorylation and dephosphorylation events, most frequently occurring on the activation loop of protein kinases. Usually in receptor tyrosine kinases ligand (e.g. cytokine, growth factor) binding to the extracellular portion of the receptor is followed by dimerization of the receptor. This allows transphosphorylation of the cytoplasmic kinase domains, on their activation loops and elsewhere, and results in consequent kinase activation and downstream signaling. In the EGFR family, however, ligand binding to the receptor does not result in immediate activation loop phosphorylation and activation loop phosphorylation is not required for kinase activity40, thus, the mechanism of activation for EGFR has remained elusive. Recent elegant studies by the Kuriyan laboratory propose the major EGFR activating mechanism to be driven by protein-protein interactions, similar to cyclin-mediated activation of CDKs36. For CDKs, the binding of cyclin to the N-lobe of the inactive state kinase forces a conformational rearrangement to the active state (Figure 2A). In inactive state CDKs the activation loop occludes the ATP-binding site and pushes the catalytically important C-helix residues out of position. On cyclin binding, structural rearrangements necessary for activation occur, including reorientation of the activation loop so that the peptide substrate binding site is exposed and rotation of the C-helix by approximately 90° orienting the correct position of the “catalytic” glutamic acid amino acid residue. A similar intermolecular activating complex for EGFR-TK has been hidden in plane sight for many years – it occurs in all of the active-state EGFR-TK crystal structures. In the inactive state, ordered parts of the activation loop fold into a helix similar to that seen in inactive cyclin-dependent kinase (CDK) family and Src family crystal structures35,36,42. In this conformation the activation loop prevents C-helix rotation towards the catalytic cleft35,36 thereby keeping the catalytically important lysine and glutamate residues distal from one another. On ligand-induced dimerization of the EGFR, the intracellular kinase domains are brought into close proximity allowing an asymmetrical kinase domain dimer to occur (Figures 1D and 2B). This tail-to-head interaction mediates an equilibrium shift to favor the active state. The Kuriyan laboratory study elegantly demonstrated how EGFR-TK becomes activated by this non-phosphorylation dependent mechanism. This knowledge may allow development of drugs to interrupt the asymmetric dimer formation and consequent activation, a potentially interesting avenue to explore as a therapeutic entry-point for tumors overexpressing EGFR.
Figure 2. Activation of CDKs and EGFRs occurs by a similar mechanism.
Panel A. Crystal structures of inactive CDK270 (PDB accession code: 1HCK) and cyclin-A in complex with CDK271 (PDB accession code: 1FIN). Panel B. Crystal structures of inactive EGFR tyrosine kinase domain35 (PDB accession code: 1XKK) and active state EGFR-TK36 (PDB accession code: 2GS6). There is a striking similarity between the inactive states of EGFR-TK and CDK2. Activation of these kinases is achieved by a protein-protein interaction that forces a structural rearrangement of the kinase towards the active state. In this figure kinase activation loops are colored orange and the C-helix is colored red. The catalytic glutamic acid is shown as space-filling spheres. The important conformational movements of the C-helix and the activation loop are indicated, but other conformational movements that occur between the active and inactive states are not shown38. The location of the catalytic cleft is indicated. It is important to note that this activating mechanism for wild-type EGFR is different from the disease-associated EGFR mutations.
It is also interesting to note that for CDKs there are a number of protein binding events that affect kinase activity and conformation by interfering with the interaction of cyclins with CDKs. Some of these interactions have been documented by X-ray crystallography (e.g. Cdk6-p16INK4a43 and p27Kip1-Cdk2-cyclin A44). Analogous protein-protein interactions that regulate activity by shielding or interruption of intermolecular binding sites may exist for EGFR family kinases and may be an interesting translational area to explore.
Somatic mutations improperly activate EGFR-TK by alternate mechanisms
For wild-type EGFR, asymmetric dimerization activates the kinase in a phosphorylation-independent manner. This asymmetric dimerization is probably not, however, the primary means of improper EGFR activation for many of the somatic EGFR mutations frequently seen in NSCLC. In these malignancies an equilibrium shift occurs between active and inactive states of EGFR-TK that favors the activated state, thereby leading to increased activation and consequent oncogenicity. An examination of the multiple EGFR-TK crystal structures now available suggests that these activations probably result from alternative structural mechanisms.
Class (ii) (single nucleotide substitution) mutations account for approximately 51% of EGFR-TK activating mutations. The study by the Eck laboratory37 solved multiple active state crystal structures of two of these mutants, leucine-858 to arginine (L858R) and glycine-719 to serine (G719S). Class (ii) mutations are dominated by a T➔G point mutation that results in a codon change and arginine expression at residue 858 instead of leucine. L858R accounts for approximately 41% of all EGFR-TK activating mutations in NSCLC8. Examination of the active and inactive conformation EGFR-TK crystal structures reveals the side chain of leucine-858 (L858) to be in two dramatically different local environments (Figure 1). In the active state, L858 is exposed on the protein’s surface, however, in the inactive state L858 is found in a closely packed hydrophobic pocket (Figure 3A and 3B). In the lapatinib-bound structure a hydrophobic portion of lapatinib packs against L858 (Figure 3A and 3B). It is expected from the active and inactive EGFR-TK crystal structures that replacement of small hydrophobic leucine with a large polar arginine will destabilize the inactive EGFR-TK conformation and stabilize the active conformation (Figure 3C). As the L858R mutation favors a solvent-exposed surface environment for residue 858, this mutation will push the conformational equilibrium that exists between the active and inactive states towards the active, catalytically competent conformation. Other class (ii) (single nucleotide substitution) mutations have also previously been discussed. These include the glycine rich P-loop mutation, G719S. The inactive state conformation is incompatible to non-glycine residues, therefore G719S is expected to favor the active state37. Further structural studies should also provide a clearer picture of how class (i) and class (iii) activating mutations function.
Figure 3. Crystal structures suggest mechanisms of activation and reduced inhibitor sensitivity.
A and B. In the crystal structure of the EGFR tyrosine kinase domain bound to lapatinib35 (PDB accession code: 1XKK) leucine-858 is located in a hydrophobic pocket. This pocket is defined by residues phenylalinine-723 (F723), leucine-747 (L747), methionine-766 (M766), leucine-788 (L788), leucine-861 (L861), leucine-862 (L862), a salt bridge between lysine-745 (K745) and aspartate-855 (D855), and the hydrophobic 3-fluorobenzyl-oxy group of EGFR tyrosine kinase-specific inhibitor. Panel A shows the residues which define this pocket in a stick-like format and Panel B depicts them as space-filling spheres. Leucine-858 is shown in stick format in red in both panels and lapatinib is shown as space-filling spheres in both panels. Secondary structure coloring as per Figure 1. Mutation of the small hydrophobic leucine-858 to a large charged arginine residue is expected to destabilize this conformation and push the kinase conformational equilibrium towards the active state. Pane C. The replacement of leucine-858 with an arginine residue is depicted. An arginine residue is shown in purple (with nitrogen atoms in blue) to illustrate that the relative size and charge of this residue are poorly compatible with this inactive conformation. This is panel is a model and not based on experimental diffraction data. Panel D. Difference in position of the C-helix from the erlotinib- and lapatinib-bound crystal structures of EGFR. The C-helices of the erlotinib- and lapatinib-bound structures are colored green and blue respectively. Residue methionine-742 (M742) is expected to sterically clash with lapatinib when the kinase is in the active conformation, with the expected result that lapatinib would bind poorly to the active conformation. The inset shows a close up of this clash. For this panel, crystal structures 1M17 and 1XKK were superimposed using the program TOPP72. Panel E. The most commonly seen resistance mutation to ATP-competitive inhibitors in EGFR-TK is threonine-790 to methionine. The location of this amino acid residue is deep in the catalytic cleft, but is predicted to deleteriously affect the binding of small molecule inhibitors gefitinib and erlotinib. Irreversible inhibitors to EGFR-TK have been shown to surmount this resistance and covalently bind to cysteine-797. Here we have illustrated these points by showing the molecular surface of active state EGFR in grey (PDB accession code: 1M17), the location of cysteine-797 as a yellow patch, and the location of the acquired methionine residue as red spheres (the orientation of this residue is modeled based on the crystal structure of insulin receptor kinase, 1IRK73, which is in a similar conformation and has a methionine at this location). The blue mesh indicates the location of erlotinib bound to active state EGFR-TK. Covalent inhibitor binding to cysteine-797 will occur proximal to the kinase catalytic cleft.
The atomic-level driving mechanisms for improper EGFR activation by somatic mutations will not be identical. This may result in differential responses to allosteric or ATP-competitive small molecule kinase inhibitors and may have significant implications for drug design and therapeutic use. For L858R or G719S this may include designing a direct interaction with R858 or S719, both of which are accessible from the ATP-cleft. We expect that further understanding and experimental evidence of the atomic-level structural consequences of EGFR mutations, alongside the recent L858R and G719S data, should aid development of drugs selective for one or another of these classes of mutation by allowing targeting to specific conformations of the mutant kinase. Indeed, to a certain degree, these mutant kinases can be regarded as novel targets. One potential clinical implication of the development of mutant-specific inhibitors could be that they improve the therapeutic window. These drugs could potentially lead to reduced off-target side effects since they would have less specificity for the wild-type EGF receptor. This might lead to both better tolerance of these drugs and improved efficacy since stronger target inhibition could be accomplished. Conceivably, this could lead to better and more durable responses. The development of such drugs could also assist in the selection of rational combination regimens that might limit the development of resistance further extending the clinical benefit of these compounds.
EGFR mutations can alter the binding of ATP-competitive inhibitors
EGFR-TK activating mutations respond to small molecule inhibitors differentially. For example, cells expressing the L858R mutant are significantly more sensitive to gefitinib than those that express the G719S mutant30, but NCSLC patients with an exon 19 (LRE) deletion mutation survive longer on erlotinib or gefitinib treatment than those with L858R20. These findings have important implications for treatment regimen design20,29,31. The Eck laboratory has begun to use a structural biology approach to discover the molecular reasons for these differing sensitivities to EGFR-targeted ATP-competitive kinase inhibitors. In their recent paper, crystal structures of wild-type, L858R and G719S EGFR-TK mutants were solved in complex with each of the four inhibitors, non-hydrolyzable ATP analogue AMP-PNP, gefitinib, AEE788 and a staurosporine analogue, AFN941. In all, a total of 12 crystal structures were published and enzyme kinetics for the wild-type and mutant constructs investigated37. An important finding from this report is that the mode of inhibitor binding can be altered by acquisition of a point mutation. This is seen in crystal structures of the staurosporine analogue AFN941 bound to wild-type and G719S mutant EGFR-TK. In these structures the orientation of AFN941 is changed, there is a ‘horizontal’ rotational difference of ∼30° in the plane of the catalytic cleft and a ‘vertical’ rotational difference of ∼15° perpendicular to the catalytic cleft. These are significant structural differences. A similar effect is seen in crystal structures of gefitinib bound to the EGFR-TK where rotation of the gefitinib alanine ring by ∼180° showed that in the L858R mutant the small molecule is able to adopt an alternate conformation37. Although the equilibrium shift towards the active state is thought to be of primary importance, it is possible that this alternate mode of gefitinib binding may be an additional factor in the 20-fold tighter affinity of gefitinib for the L858R mutant compared to wild-type EGFR-TK. Altogether, these crystal structures indicate that there may be structural reasons for some of the differences in sensitivity to EGFR-TK inhibitors. It should therefore be possible to rationally design inhibitors to differentially target mutant, over wild-type, EGFR-TK. Gefitinib and erlotinib seem to be serendipitous examples of differential EGFR-TK inhibitors. This might in part explain why these drugs lead to such a high likelihood of response in EGFR-mutant tumors, while their activity against tumors with EGFR overexpression is more modest. In essence, the therapeutic window of these drugs for an EGFR-mutant tumor is wide, while for an EGFR wild-type tumor it is quite narrow. The crystal structures of the L858R and G719S mutants in complex with gefitinib suggest that it should be possible to improve Kd/Km, ATP for mutant compared to wild-type by taking advantage of potential new interactions with the acquired point mutation or by making a better fit to the distorted ATP-cleft of the mutant kinase37. For patient populations as large as those harboring the L858R mutation or LRE deletion this seems to be a reasonable goal for drug discovery. The Eck laboratory crystal structures now facilitate rational in silico drug discovery efforts to target L858R and G719S EGFR mutants using experimentally derived crystal structures.
Structural analysis suggests why lapatinib has reduced efficacy towards activating mutants
The published crystal structure of EGFR-TK in complex with the dual EGFR and ErbB2 inhibitor, lapatinib, shows that this small molecule binds the kinase ATP-binding cleft when EGFR-TK is in the inactive conformation35. The active conformation EGFR-TK crystal structures, however, suggest that lapatinib may not bind as effectively to active state EGFR-TK. The crystal structures suggest that there will be steric clashes in the back pocket region, for example between methionine-742 and the 3-fluorobenzyl-oxy group of lapatinib (Figure 3D). Interestingly, in the study at GlaxoSmithKline35, EGFR-TK was soaked with inhibitor prior to crystallization trials, enabling the kinase to adopt the inactive conformation seen. It was noted, however, that soaking with lapatinib of the already formed active conformation ligand-free EGFR-TK crystals described by the Genentech group34 failed to produce acceptable co-crystals, providing some empirical evidence that lapatinib may not readily bind active state EGFR-TK. Structural analysis therefore appears to show that mutations which push the equilibrium of the kinase towards an active conformation (e.g. L858R) reduce the probability that the kinase will adopt the more lapatinib-accessible inactive state. Consequently, lapatinib may prove to be better used as an EGFR family kinase activity regulator for malignancies that do not contain kinase-activating point mutations, such as HER2/neu positive breast cancer, for which it was recently approved. In fact, limited clinical studies with lapatinib in patients with advanced NSCLC demonstrate a low response rate of 3% in unselected patients suggestive of the low utility of this compound for this disease45. On the other hand, the remarkable success of lapatinib in HER2/neu positive breast cancer in combination with chemotherapy and in patients with Herceptin-resistance does suggest that inhibition of the inactive state of ErbB2 is indeed an effective strategy46,47.
Acquired resistance to EGFR-TK inhibition
Despite the very high response rates to the reversible EGFR TKIs gefitinib and erlotinib in EGFR-mutant NSCLC, over time all patients unfortunately progress. Recent studies demonstrated that the mechanism of resistance, in over half of all patients, is secondary EGFR mutations that affect drug efficacy. The acquisition of resistance mutations to the targeted inhibition of kinases in cancer is by now a well-documented phenomenon48,49 but although the importance of the cancer stem cell is firmly established, the etiology of acquired resistance is still the subject of some debate49,50. In the case of EGFR-TK there are currently only two documented resistance point mutations to the anilinoquinazoline inhibitors gefitinib and erlotinib, threonine-790 to methionine (T790M)51,52 and aspartic acid-761 to tyrosine (D761Y)53
The most common secondary resistance mutation is by far T790M, a mutation which seems to occur in approximately half of all patients with secondary EGFR tyrosine kinase inhibitor resistance. The T790M mutation is located at the ‘gatekeeper’ position in the kinase ATP-binding cleft, a structural location often documented to interfere with inhibitor binding; imatinib resistance mutations threonine-315 to isoleucine (T315I) in BCR-Abl54 and threonine-670 to isoleucine (T670I) in Kit55 are both at the same structural location. Acquisition of T790M in EGFR results in an alteration of the topology of the ATP-binding pocket and is expected to sterically hinder inhibitor binding51,56 (Figure 3E). The irreversible inhibitors of EGFR-TK, Cl-387,785 (Wyeth), HKI-272 (Wyeth), EKB-569 (Wyeth) and Cl-1033 (Pfizer) seem to effectively inhibit T790M57 and are under clinical development for this indication. These drugs covalently bind to cysteine-797 of EGFR, however, atomic-level details of how they overcome the T790M mutation has not yet been described, we therefore eagerly await crystallographic descriptions of their modes of inhibition.
In the tail-to-head EGFR activation model (Figure 1D) the C-helix residue, D761, looks likely to form a salt bridge to lysine-949 in the activating (tail) molecule. This salt bridge would be disrupted by mutation to tyrosine, this potentially might have an impact on the catalytic cleft that could cause resistance.
Other mechanisms of secondary resistance to EGFR are expected to arise, including further secondary resistance point mutations and amplifications of EGFR itself and other downstream kinases. Recently, MET receptor tyrosine kinase amplification in lung cancer has been also shown to result in gefitinib resistance by activation of ERBB3, providing a novel mechanism of resistance and illustrating the potential for combination therapies in the treatment of this disease58.
Further secondary resistance point mutations for EGFR are beginning to be described59,60. As with BCR-Abl resistance mutations48 these are not predicted to be exclusively proximal to the catalytic cleft. Other potential secondary resistance mechanisms include further alteration of the active-inactive equilibrium, destabilization of the inhibitor-accessible conformation, alterations in the tail-to-head activating interface, mutations that alter the transmembrane helices61 and extracellular mutations.
The rational design of inhibitors to target acquired activating and resistance mutations in kinases has paradigms in the ongoing work for BCR-Abl and B-RAF62,63; small molecule inhibitors are being rationally designed to target oncogenic B-RAF64 and third generation BCR-Abl inhibitors against the T315I resistance mutation65.
Clinical implications of the EGFR-TK crystal structures
There are now twenty EGFR-TK crystal structures published in multiple conformational states and in complex with multiple inhibitors, substantially more than for any other tyrosine kinase. Differential effects on kinase activation have been seen for EGFR-TK mutations. These effects are beginning to be correlated with X-ray crystallographic data and significant conformational differences have been seen for inhibitor binding to mutant compared to wild-type kinase. The X-ray crystallographic, in vivo, in vitro and clinical data all seem to suggest that there is merit in regarding many of these different mutational EGFR-TK species as unique targets. The crystallographic studies also suggest that it may be possible to design inhibitors to selectively target mutant over wild-type kinase, therefore reducing the deleterious effects of off-target wild-type kinase inhibition while taking advantage of the oncogene addiction effect. These and other studies indicate that gefitinib and erlotinib may be fortuitous examples of mutant kinase-targeted inhibitors. The structural data reviewed here further suggests that in the future it will be possible to use knowledge of the mutational status of EGFR-TK to determine the optimal kinase inhibitor treatment regimen. While currently there are only two ATP-competitive EGFR-TK inhibitors approved in the US (erlotinib and lapatinib), in the future, the presence of specific mutations may indicate a specific kinase inhibitor therapy. All activating mutations do not have the same structural effects on the kinase, so while the presence of an activating mutation indicates the use of an EGFR-TK inhibitor, further work will be needed to determine the optimal inhibitor to use for each acquired activating mutation. The same should be true for acquired resistance mutations such as T790M and D761Y51,52. The ultimate hope should be that such crystallographic and molecular knowledge will lead to the development of more effective treatment strategies through the availability of better targeted drugs and/or rational combination strategies yielding a higher likelihood of responses, a better therapeutic window and through the prevention of the emergence of resistance, possibly long-term disease control.
The large-scale resequencing of tumor-expressed genes66 is continuing to discover activating, transforming and resistance mutations for many kinases with increasingly higher accuracies67,68. X-ray crystallographic analysis of these mutant kinases is a powerful tool to diagnose the spatial mechanisms of their deregulation and to aid discovery of improved strategies for inhibition. The study of EGFR-TK by multiple structural biology groups over recent years has provided a strong foundation for a comprehensive “Structural Pathology” approach to the description of improper EGFR-TK activation and targeted inhibition.
Acknowledgements
Michael J. Eck and Benjamin E. Turk are acknowledged for helpful discussions.
Research Support:
TJB is in receipt of an American Society of Hematology Junior Faculty Award Scholarship and is supported by the NIH/NIAID (1R01AI075133). ETP is supported by a NIH/National Cancer Institute T32 training grant (5T32CA009085). BH is a recipient of a Young Clinical Scientist Award from the Flight Attendant Medical Research Institute.
References
- 1.Schlessinger J. Common and distinct elements in cellular signaling via EGF and FGF receptors. Science. 2004;306:1506–1507. doi: 10.1126/science.1105396. [DOI] [PubMed] [Google Scholar]
- 2.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–21239. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 7.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 8.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118:257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 9.Roskoski R., Jr The ErbB/HER receptor protein-tyrosine kinases and cancer. Biochem Biophys Res Commun. 2004;319:1–11. doi: 10.1016/j.bbrc.2004.04.150. [DOI] [PubMed] [Google Scholar]
- 10.Jemal A, Murray T, Ward E, et al. Cancer statistics 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 11.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 12.Bailey R, Kris M, Wolf M, et al. Gefitinib (‘Iressa’ ZD1839) monotherapy for pretreated advanced non-small-cell lung cancer in IDEAL 1 and 2: tumor response is not clinically relevantly predictable from tumor EGFR membrane staining alone. Lung Cancer. 2003;41:S71. [Google Scholar]
- 13.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 14.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 15.Huang SF, Liu HP, Li LH, et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res. 2004;10:8195–8203. doi: 10.1158/1078-0432.CCR-04-1245. [DOI] [PubMed] [Google Scholar]
- 16.Kosaka T, Yatabe Y, Endoh H, et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 17.Politi K, Zakowski MF, Fan PD, et al. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji H, Li D, Chen L. et al: The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–495. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 19.Sordella R, Bell DW, Haber DA, et al. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 20.Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 21.Jackman DM, Yeap BY, Sequist LV, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- 22.Riely GJ, Politi KA, Miller VA, et al. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res. 2006;12:7232–7241. doi: 10.1158/1078-0432.CCR-06-0658. [DOI] [PubMed] [Google Scholar]
- 23.Janne PA, Engelman JA. Johnson BE,Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol. 2005;23:3227–3234. doi: 10.1200/JCO.2005.09.985. [DOI] [PubMed] [Google Scholar]
- 24.Asahina H, Yamazaki K, Kinoshita I, et al. A phase II trial of gefitinib as first-line therapy for advanced non-small cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer. 2006;95:998–1004. doi: 10.1038/sj.bjc.6603393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue A, Suzuki T, Fukuhara T, et al. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24:3340–3346. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- 26.Sutani A, Nagai Y, Udagawa K, et al. Gefitinib for non-small-cell lung cancer patients with epidermal growth factor receptor gene mutations screened by peptide nucleic acid-locked nucleic acid PCR clamp. Br J Cancer. 2006;95:1483–1489. doi: 10.1038/sj.bjc.6603466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch FR, Witta S. Biomarkers for prediction of sensitivity to EGFR inhibitors in non-small cell lung cancer. Curr Opin Oncol. 2005;17:118–122. doi: 10.1097/01.cco.0000155059.39733.9d. [DOI] [PubMed] [Google Scholar]
- 28.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 29.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang J, Greulich H, Janne PA, et al. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res. 2005;65:8968–8974. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]
- 31.Thomas RK, Greulich H, Yuza Y, et al. Detection of oncogenic mutations in the EGFR gene in lung adenocarcinoma with differential sensitivity to EGFR tyrosine kinase inhibitors. Cold Spring Harb Symp Quant Biol. 2005;70:73–81. doi: 10.1101/sqb.2005.70.056. [DOI] [PubMed] [Google Scholar]
- 32.Choong NW, Dietrich S, Seiwert TY, et al. Gefitinib response of erlotinib-refractory lung cancer involving meninges--role of EGFR mutation. Nat Clin Pract Oncol. 2006;3:50–57. doi: 10.1038/ncponc0400. quiz 1 p following 57. [DOI] [PubMed] [Google Scholar]
- 33.Jones TA, Zhou JY, Cowan SW, et al. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Stamos J, Sliwkowski MX. Eigenbrot C,Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–16272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 35.Wood ER, Truesdale AT, McDonald OB, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Gureasko J, Shen K, et al. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Yun CH, Boggon TJ, Li Y, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 39.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 40.Tice DA, Biscardi JS, Nickles AL, et al. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc Natl Acad Sci U S A. 1999;96:1415–1420. doi: 10.1073/pnas.96.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison SC. Variation on an Src-like theme. Cell. 2003;112:737–740. doi: 10.1016/s0092-8674(03)00196-x. [DOI] [PubMed] [Google Scholar]
- 42.Xu W, Doshi A, Lei M, et al. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 43.Russo AA, Tong L, Lee JO, et al. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 44.Russo AA, Jeffrey PD, Patten AK, et al. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 45.Ross HJ, Blumenschein GR, Dowlati A, et al. Preliminary safety results of a phase II trial comparing two schedules of lapatinib (GW572016) as first line therapy for advanced or metastatic non-small cell lung cancer. ASCO Annual Meeting Proceedings. 2005;23:7099. [Google Scholar]
- 46.Geyer CE, Forster J, Lindquist D, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 47.Konecny GE, Pegram MD, Venkatesan N, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 48.Shah NP, Nicoll JM, Nagar B, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 49.O'Hare T, Corbin AS, Druker BJ. Targeted CML therapy: controlling drug resistance, seeking cure. Curr Opin Genet Dev. 2006;16:92–99. doi: 10.1016/j.gde.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Graham SM, Jorgensen HG, Allan E, et al. Primitive, quiescent Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 51.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–7892. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 52.Pao W, Miller VA, Politi KA, et al. Acquired Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib Is Associated with a Second Mutation in the EGFR Kinase Domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–64501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 54.Branford S, Rudzki Z, Walsh S, et al. High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood. 2002;99:3472–3475. doi: 10.1182/blood.v99.9.3472. [DOI] [PubMed] [Google Scholar]
- 55.Tamborini E, Bonadiman L, Greco A, et al. A new mutation in the KIT ATP pocket causes acquired resistance to imatinib in a gastrointestinal stromal tumor patient. Gastroenterology. 2004;127:294–299. doi: 10.1053/j.gastro.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 56.Clark J, Cools J, Gilliland DG. EGFR inhibition in non-small cell lung cancer: resistance, once again rears its ugly head. PLoS Med. 2005;2:e75. doi: 10.1371/journal.pmed.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 58.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 59.Yu Z, Jin C, Kobayashi S, et al. Predictive model of resistance mechanisms against irreversible EGFR inhibitors reveals novel treatment strategies. Abstract number 2555, AACR 2007 Meeting. 2007 [Google Scholar]
- 60.Costa DB, Kobayashi S, Halmos B, et al. Bim mediates EGFR tyrosine kinase inhibitor induced apoptosis in non-small cell lung cancers and is linked to the resistance conferred by secondary EGFR mutations, T790M and the novel L747S. Abstract number LB-61. AACR 2007 meeting. 2007 [Google Scholar]
- 61.Mendrola JM, Berger MB, King MC, et al. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J Biol Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 62.Weisberg E, Manley PW, Cowan-Jacob SW, et al. Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukaemia. Nat Rev Cancer. 2007;7:345–356. doi: 10.1038/nrc2126. [DOI] [PubMed] [Google Scholar]
- 63.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6:313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 64.King AJ, Patrick DR, Batorsky RS, et al. Demonstration of a genetic therapeutic index for tumors expressing oncogenic BRAF by the kinase inhibitor SB-590885. Cancer Res. 2006;66:11100–11105. doi: 10.1158/0008-5472.CAN-06-2554. [DOI] [PubMed] [Google Scholar]
- 65.Burley SK. Cancer and kinases: reports from the front line. Genome Biol. 2006;7:314. doi: 10.1186/gb-2006-7-4-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weir B, Zhao X, Meyerson M. Somatic alterations in the human cancer genome. Cancer Cell. 2004;6:433–438. doi: 10.1016/j.ccr.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 67.Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas RK, Nickerson E, Simons JF, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 69.Hubbard SR. EGF receptor inhibition: attacks on multiple fronts. Cancer Cell. 2005;7:287–288. doi: 10.1016/j.ccr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 70.De Bondt HL, Rosenblatt J, Jancarik J, et al. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 71.Jeffrey PD, Russo AA, Polyak K, et al. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 72.CCP4: The CCP4 suite: Programs for protein crystallography. Acta Cryst. 1994;D50:760–776. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 73.Hubbard SR, Wei L, Ellis L, et al. Crystal structure of the tyrosine kinase domain of the human insulin receptor [see comments] Nature. 1994;372:746–754. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- 74.Blair JA, Rauh D, Kung C, et al. Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat Chem Biol. 2007;3:229–238. doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]