Abstract
Phospholamban (PLB) is a 52 amino acid integral membrane protein that interacts with the sarcoplasmic reticulum Ca2+ ATPase (SERCA) and helps to regulate Ca2+ flow. PLB inhibits SERCA impairing Ca2+ translocation. The inhibition can be relieved upon phosphorylation of PLB. The Arg9 to Cys (R9C) mutation is a loss of function mutation with reduced inhibitory potency. The effect R9C PLB has on the membrane surface and the hydrophobic region dynamics was investigated by 31P and 2H solid-state NMR spectroscopy in multilamellar vesicles (MLVs). The 31P NMR spectra indicate that, like the phosphorylated PLB (P-PLB), the mutated R9C-PLB protein has significantly less interaction with the lipid bilayer headgroup when compared to wild-type PLB (WT-PLB). Similar to P-PLB, R9C-PLB slightly decreases 31P T1 values in the lipid headgroup region. 2H SCD order parameters of 2H nuclei along the lipid acyl chain decrease less dramatically for R9C-PLB and P-PLB when compared to WT-PLB. The results suggest that R9C-PLB interacts less with the membrane surface and hydrophobic region than WT-PLB. Detachment of the cytoplasmic domain of R9C-PLB from the membrane surface could be related to its loss of function.
Keywords: Phospholamban, Multilamellar vesicles, Membrane interaction, Solid-state NMR spectroscopy
Introduction
Phospholamban is a 52 amino acid membrane protein located in the sarcoplasmic reticulum (SR). PLB interacts with and inhibits SERCA which pumps Ca2+ from the cytoplasm into the SR [1-3]. Ca2+ translocation into the SR induces heart muscle relaxation, while Ca2+ translocation back to the cytoplasm induces heart muscle contraction [1]. The inhibition of SERCA by PLB can be relieved either upon phosphorylation of PLB at position 16 or 17 by protein kinase A or by elevated Ca2+ concentrations in the cytoplasm [4, 5]. PLB has been shown to consist of four domains: the cytoplasmic domain Ia (1-16), loop (17-22), domain Ib (23-30) and the hydrophobic transmembrane domain (31-52) [6]. There has long been a debate on the cytoplasmic domain (domain Ia) of PLB and whether it exists as an α-helix [7] or random coil [8], and if it is anchored on the membrane surface [7], or sticking out into the cytoplasm as a bell-flower assembly [9].
Current research indicates that an equilibrium exists between two conformations of PLB [10-12]. The two conformations are the T and R states. The T state exists at a low energy level with domain Ia folded as an α-helix and attached to the membrane surface. The R state exists at a higher energy level with domain Ia unfolded and dissociated from the membrane surface [10, 13]. Excellent work by the Veglia group using solid-state NMR indicates that most of WT-PLB exist as the T-state pentamer with the α-helical cytoplasmic domain laying on the membrane surface [6, 7]. Phosphorylation of PLB shifts the PLB population from the T state to the R state or an intermediate T’ state with a partially unfolded cytoplasmic domain [7, 13-15], Domain Ib has a larger dynamic population than in the cytoplasmic domain [16]. It inserts into the membrane surface as an α-helix with unknown function [17]. The transmembrane domain inserts into the membrane as an α-helix forming a 13° angle with respect to the bilayer normal [7, 18], Wild-type PLB forms a pentamer with a Leu-zipper and three cysteines in the transmembrane domain stabilizing the complex [7], Small populations of PLB exist in a monomeric form, which is believed to interact with and inhibit SERCA [1]. Thus, the pentamer form of PLB acts like an inactive storage form, while the monomer is actively involved in SERCA inhibition [6, 7]. However, in vivo studies demonstrate that the monomeric form of PLB can inhibit SERCA to the same extent as WT-PLB by reducing SERCA-Ca2+ affinity, but the monomer cannot regulate heart muscle relaxation rates as the pentamer form does [19].
The R9C-PLB mutation relieves inhibition function on SERCA as P-PLB [20], Individuals that carry this mutation tend to exhibit an enlarged heart chamber and heart muscle contraction dysfunction which can lead to heart failure [20], In vivo studies using transgenic mice indicate that the R9C-PLB mutation induces fibrosis and enlargement of the heart and nuclei in heart muscle cells [20], Previous studies have shown that the population of the R9C-PLB pentamer form is slightly more than that of WT-PLB and R9C-PLB and cannot be phosphorylated efficiently [21, 22], R9C-PLB has a similar affinity to SERCA as WT-PLB and P-PLB [21, 23]. However, it is not clear why the R9C mutation induces loss of function. Multiple mechanisms including oligomerization state, structural conformations, affinity to SERCA and membrane interactions could be involved.
Membrane proteins are difficult to study due to their relatively large size, hydrophobicity and loss of stability or function or changes of conformation in a non-native lipid bilayer environment [24, 25]. Solid-state NMR spectroscopy is a powerful and robust way to probe the dynamic and structural properties of membrane proteins [25-29], By incorporating membrane proteins into lipids, the proteins are stabilized in their native state retaining their functionality. Multilamellar vesicles, which are composed of multi-layers of lipid bilayers, are commonly used to study membrane proteins [30-32]. Membrane structure and dynamic changes induced by the incorporation of proteins into the phospholipid bilayer can be revealed using 31P solid-state NMR spectroscopy [33-35]. Also, 2H solid-state NMR spectroscopy of 2H labels along the lipid acyl chain can be used to study the dynamics of the hydrophobic region of the membrane [34-42], In this study, interactions between lipid bilayers and WT-PLB, P-PLB and R9C-PLB are investigated from the membrane perspective using 31P and 2H solid-state NMR spectroscopy. These experiments shed light onto the loss of functionality of the R9C mutation from the membrane perspective.
Materials and Methods
Materials
Phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL). l-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) was dissolved in chloroform and stored at −20°C before use. Trifluoroethanol (TFE) and N-[2-hydroxyethyl]piperazine-N’-2-ethane sulfonic acid (HEPES) were obtained from Sigma-Aldrich (St. Louis, MO). Ethylenediaminetetraacetic acid (EDTA) and sodium chloride (NaCl) were obtained from Fisher Scientific (Pittsburgh, PA). Fmoc amino acids were purchased from Applied Biosystems (Carlsbad, CA). All other peptide synthesis and HPLC reagents and solvents were purchased from VWR (San Dimas, CA).
Synthesis, Purification and Characterization of WT-PLB and R9C-PLB
WT-PLB and R9C-PLB (sequences identified in humans) were synthesized via Fmoc-based solid-phase peptide synthesis using a CEM solid-phase peptide synthesizer. A General Electric (GE) AKTA purifier HPLC was utilized to purify the peptides via reverse phase chromatography on a CI8 column. The purified peptides were lyophilized and characterized by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. The peptides were at least 95% pure.
NMR Sample Preparation
MLVs were prepared according to a protocol established by Rigby and coworkers [40], Lyophilized WT-PLB and R9C-PLB were dissolved in a minimal amount of TFE. 35 mg of POPC was mixed with dissolved peptides (4 mol% with respect to lipid) in a 12×75 mm test tube. The solvent was removed by a steady stream of N2 gas and the tube was placed in a vacuum dessicator overnight. The lipid peptide mixture was rehydrated with 95 μL HEPES buffer (5 mM EDTA, 20 mM NaCl and 30mM HEPES at pH 7.0). Vesicles were formed in a warm water bath at 45 °C for about 30 min using a vortexer and bath sonication. The sample was transferred into a 4 mm NMR rotor. 2H NMR samples were prepared with a lipid mixture consisting of 8 mg of 1-palmitoyl(d31)-2-oleoyl-sn-glycero-3-phosphocholine (P0PC-d31) and 27 mg of POPC.
Solid-State NMR Spectroscopy
31P and 2H solid-state NMR spectra were acquired using a 500 MHz WB Bruker Avance solid-state NMR spectrometer with a Bruker 4-mm double resonance CP-MAS probe. For 31P data, the NMR spectrometer was operated at 202.4 MHz and 1024 transients were averaged with 100 Hz line broadening. Static powder pattern NMR spectra were collected using the spin echo (90°-τ1-180°-τ1-acq) pulse sequence with 1H decoupling [35], The recycle delay was set to 5 s. All spectra were acquired at 25 °C. 31P T1 relaxation was detected using an inversion recovery (180°-τ-90°-acq) pulse sequence [43], τ was set from 10 ms to 6 s. The sample was spun at 4 kHz at the magic angle. For T1 measurements, the spectra were acquired at 25 °C, 35 °C, 45 °C and 55 °C for each sample and the samples were equilibrated at these temperatures for at least 10 min before spectral acquisition.
For 2H NMR experiments, the NMR spectrometer was operated at 76.77 MHz. The quadrupolar echo pulse sequence was employed using quadrature detection with complete phase cycling of the pulse pairs [40], The 90° pulse length was 3 μs, the interpulse delay was 40 μs, and the recycle delay was 0.3 s. All spectra were acquired at 25 °C. The spectral width was 100 kHz and line broadening was set to 200 Hz. A total of 40960 transients were averaged for each spectrum.
NMR Data Analysis
For 31P NMR simulations, the DMFIT software package was used [44], The spectra were fit to a sum of a minimal number of lineshapes. The 31P chemical shift anisotropy (CSA) was obtained from the simulated spectra. For T1 calculations, the data were fit to a single-exponential function: I(t)=I(0)-Aexp(−t/T1).
2H NMR spectra were deconvoluted (dePaked) using the procedures of McCabe and Wassall [45, 46]. The order parameters of each 2H on the lipids were calculated from the quadrupolar splittings of the dePaked 2H NMR spectra according to the following expression [38,47]:
| (Eq. 1) |
ΔνiQ is the quadrupolar splitting of a deuteron attached to the ith carbon. e2qQ/h is the quadrupolar splitting constant (168 kHz for the deuterons in C-2H bonds). SiCD is the chain order parameter of a deuteron attached to the ith carbon of the acyl chain of POPC. SCD of CD3 was calculated by multiplying by 3 to obtain the lipid CD3 terminal order parameters according to the literature [30, 48], 2H nuclei attached to the terminal methyl carbon was assigned as carbon number 15. The remaining 2H atoms were assigned in decreasing order along the lipid acyl chain.
Results
31P NMR Spectroscopy of WT-PLB, P-PLB andR9C-PLB in Phospholipid Bilayers
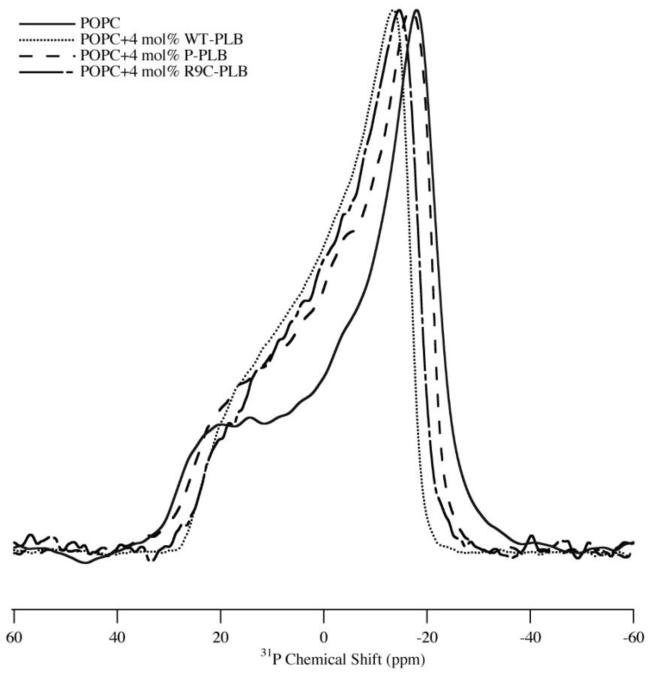
31P NMR spectra can provide unique structural and dynamic information on the phospholipid headgroup region. Figure 1 shows 31P NMR spectra of MLVs of POPC with and without 4 mol% WT-PLB, P-PLB and R9C-PLB. All static 31P NMR spectra show a characteristic axially symmetric powder pattern lineshape both with and without WT-PLB, P-PLB and R9C-PLB. In Figure 1, the addition of 4 mol% WT-PLB decreases the 31P CSA line width dramatically from 48(±0.2) ppm to 41 (±0.2) ppm indicating an increase in 31P lipid membrane surface dynamics. The addition of P-PLB and R9C-PLB have relatively moderate effects on the 31P CSA width by decreasing it to 44(±0.2) ppm and 43(±0.2) ppm (Figure 1). The results suggest that WT-PLB perturbs the membrane surface head group region more when compared to P-PLB or R9C-PLB. Also, the 31P NMR data suggest that P-PLB and R9C-PLB may interact with the membrane surface in a similar way and associate less with the membrane surface than WT-PLB. The 31P NMR data were collected in the liquid-crystalline phase at 25 °C, and the 31P chemical shift anisotropy (CSA) was obtained from the simulated fit of the spectra.
Figure 1.
1H-decoupled 31P NMR powder spectra of MLV samples of POPC control (solid line), POPC with 4 mol% WT-PLB (dotted line), POPC with 4 mol% P-PLB (dashed line) and POPC with 4 mol% R9C-PLB (dotted-dashed line).
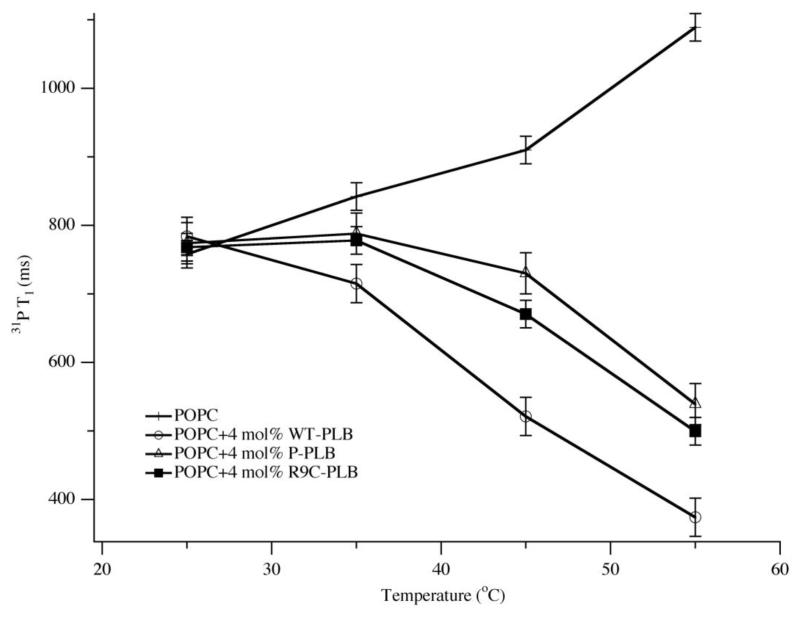
31P T1 spin-lattice relaxation times can reveal changes in lipid motions including the long axis of rotation and lipid diffusion upon protein interaction [49]. Figure 2 shows the 31P T1 values of MLVs of POPC with and without 4 mol% WT-PLB, P-PLB and R9C-PLB as a function of temperature. At 25 °C, the 31P T1 values of the lipids with and without WT-PLB, P-PLB and R9C-PLB are similar. As the temperature increases from 25 °C to 55 °C, the 31P T1 values of the POPC lipid control increases (Figure 2, crosses). The addition of 4 mol% WT-PLB decreases the 31P T1 values dramatically and the reduction is more pronounced as the temperature increases. P-PLB and R9C-PLB also decrease the 31P T1 values, but to a lesser extent when compared to WT-PLB.
Figure 2.
31P T1 of MLV samples of POPC control (crosses), POPC with 4 mol% WT-PLB (open circles), POPC with 4 mol% P-PLB (open triangles) and POPC with 4 mol% R9C-PLB (solid squares) at 25 °C, 35 °C, 45 °C and 55 °C. The error bars were obtained by averaging 31P T1 values from multiple different sample sets.
2HNMR Spectroscopy of WT-PLB, P-PLB and R9C-PLB in Phospholipid Bilayers
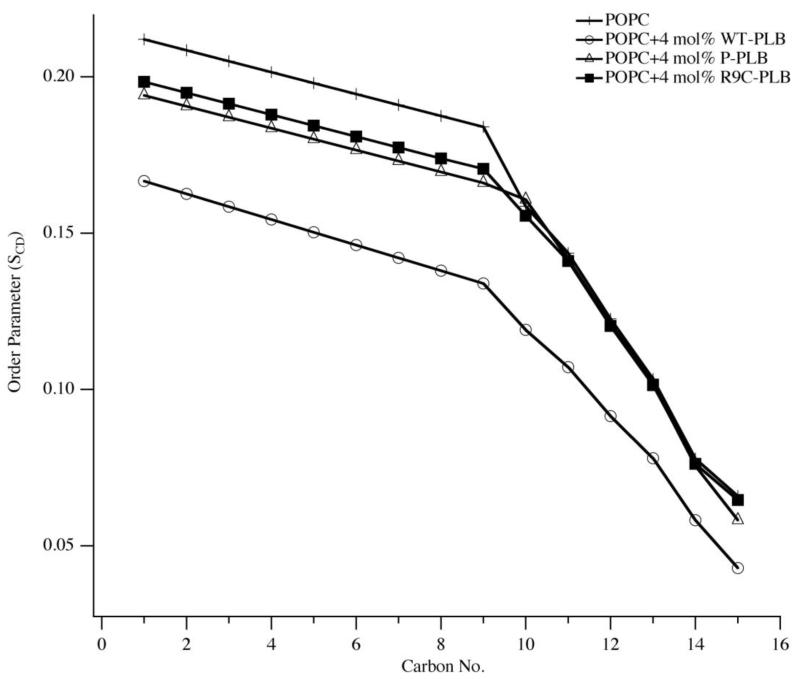
Figure 3 shows 2H NMR spectra of MLVs of POPC with and without 4 mol% WT-PLB, P-PLB and R9C-PLB. From Figure 3, the control POPC lipids give the largest quadrupolar splittings (Figure 3, solid line) and the addition of WT-PLB, P-PLB and R9C-PLB decreases the quadrupolar splittings (Figure 3). P-PLB and R9C-PLB have a smaller and similar reduction in quadrupolar splittings than WT-PLB. Figure 4 shows the calculated 2H SCD order parameters of the POPC acyl chains with and without 4 mol% WT-PLB, P-PLB and R9C-PLB. Quadrupolar splittings of dePaked 2H NMR spectra reveal the corresponding order parameters of the C-D methylene groups and the terminal methyl groups of the lipid acyl chain [39-41]. The quadrupolar splitting of the CD3 methyl groups at the end of the acyl chains is the smallest and closest to 0 kHz since they are less restricted than the ones near the membrane surface [39-41]. The next smallest quadrupolar splitting is the 2H attached to C-14 and so forth along the acyl chain. The POPC control sample has the largest order parameters among the four samples along the entire acyl chain (Figure 4). Decreasing order parameters indicate increased dynamic motion [39-41], The 4 mol% WT-PLB decreases the SCD order parameters of 2H along the lipid acyl chain dramatically (Figure 4, open circles). This indicates a strong interaction between WT-PLB and the membrane hydrophobic region. P-PLB (Figure 4, open triangles) and R9C-PLB (Figure 4, solid squares) induce less order parameter reductions than WT-PLB and the order parameters are similar for both of them along the acyl chain. For all three types of PLB, a more pronounced reduction in SCD order parameters is observed for the 2H near the membrane surface than the ones in the hydrophobic core. Both P-PLB and R9C-PLB have subtle effects on the 2H SCD order parameters in the lipid hydrophobic core region. The data suggest that the membrane core is less susceptible to protein interaction than the membrane surface.
Figure 3.
2H NMR spectra of MLV samples of POPC control (solid line), POPC with 4 mol% WT-PLB (dotted line), POPC with 4 mol% P-PLB (dashed line) and POPC with 4 mol% R9C-PLB (dotted-dashed line).
Figure 4.
Smoothed acyl chain (DMPC-d54) order parameter (SCD) calculated from the dePaked spectra of MLV samples of POPC control (crosses), POPC with 4 mol% WT-PLB (open circles), POPC with 4 mol% P-PLB (open triangles) and POPC with 4 mol% R9C-PLB (solid squares).
Discussion
It is important to study the structural and dynamic properties of membrane proteins in their native environment. Lipid bilayers help stabilize hydrophobic membrane proteins [50, 51], Different forms of PLB including WT-PLB, P-PLB and mutated PLB result in various effects on SERCA. WT-PLB inhibits SERCA while P-PLB relieves the inhibition [1], Different mutated forms of PLB can either enhance or impair the inhibition and are called gain of function mutations and loss of function mutations [52, 53], Gain of function PLB mutants, like N27A and N30C, reveals a super-inhibition effect on SERCA when compared with WT-PLB [52, 54, 55]. Loss of function PLB mutants, like R9C, cannot inhibit SERCA function as efficiently as WT-PLB [20].
Direct effects of mutation or phosphorylation on PLB can be investigated from several different perspectives. These effects, including changes in PLB oligomerization, conformations, interaction with membrane and affinity to SERCA, may lead to PLB function variations on SERCA. Previous oligomerization studies indicate that loss of functions mutants exist predominantly as pentamers, while some gain of function PLB mutants exist mostly in the monomeric form [49, 50], R9C-PLB has a slightly larger pentamer population than WT-PLB [53, 54]. Oligomerization change could partially contribute to the alterations of PLB mutant function on SERCA. SERCA-PLB interaction studies suggest that all WT-PLB, P-PLB and several PLB mutants have similar binding affinity to SERCA, so PLB affinity to SERCA is distinct from its inhibition potency [53], Lipid interaction with PLB may partially contribute to function variation according to WT-PLB and P-PLB studies. Most WT-PLB is in the pentameric T-state with the cytoplasmic domain anchored on the membrane surface and perturbing it significantly [7, 35], Upon phosphorylation, the PLB population shifts toward an intermediate T’ state or R state with less perturbations on the membrane surface [17]. This indicates a possible dissociation process of the P-PLB cytoplasmic domain from the membrane surface [35, 50, 56]. Thus, membrane associated WT-PLB can inhibit SERCA, while the membrane dissociated P-PLB relieves the inhibition. The membrane bilayer is essential here to serve as both a native system and a possible mediation being actively involved in regulating PLB functionality. Studies focused on the interaction between phospholamban and the membrane can provide insight into the functional variation of different forms of PLB.
The interaction of R9C-PLB with the membrane is examined in this work. 31P solid-state NMR spectra provide evidence for dynamic changes on the membrane surface with the addition of PLB. WT-PLB dramatically decreases the 31P CSA width, which indicates a dynamic increase in the 31P headgroup region on the membrane surface [35]. R9C-PLB induces a moderate decrease in the CSA width, which is very similar to the effect of P-PLB (Figure 1). The data reveal that both R9C-PLB and P-PLB have less association with the membrane surface when compared with WT-PLB. Phosphorylation shifts the PLB population from the T state to the R state or the intermediate T’ state with the cytoplasmic domain detached or partially detached from the membrane surface [13]. Like phosphorylation, the R9C mutation may also shift PLB toward a conformation in which the cytoplasmic domain tends to dissociate from the lipid bilayer headgroup region. This hypothesis is further supported by the 31P NMR T1 relaxation data. The 31P T1 value reflects membrane surface and acyl chain conformation changes, long axis rotation, and lipid diffusion upon protein interaction [49]. At room temperature, the 31P T1 values of the POPC lipids with and without WT-PLB, P-PLB and R9C-PLB have no significant difference (Figure 2). As the temperature increases from 25 °C to 55 °C, the T1 values of the POPC control increases. The addition of WT-PLB, P-PLB and R9C-PLB decreases the 31P T1 values. The WT-PLB induces the most dramatic reduction in T1 while the P-PLB and R9C-PLB produce a similar and moderate reduction (Figure 2). When the T1 value reaches its smallest value, relaxation is more efficient [57, 58], For the POPC lipid control sample, the temperature with the lowest T1 value is lower than 25 °C, which is typical for phospholipids [49, 57, 58], With the addition of WT-PLB, P-PLB or R9C-PLB, the T1 value decreases as temperature increases. This indicates that the temperature at the minimal T1 value has been shifted to above 55 °C by WT-PLB, P-PLB and R9C-PLB with WT-PLB yielding the most pronounced effect on T1. The membrane surface is perturbed mostly by the cytoplasmic domain of PLB [6, 7]. As various conformations of this region interact differently with the lipid bilayer surface, it can potentially tune the PLB inhibitory effect [7, 59], The similarity in the data of R9C-PLB and P-PLB effect on membrane surface suggests there is less association with the membrane surface when compared to WT-PLB. Phosphorylation on the cytoplasmic domain could enhance the hydrophillicity of this region resulting in less interaction with the membrane surface. For the R9C-PLB, the dissociation could be due to the mutation of positively charged Arg into neutral Cys eliminating the electrostatic interaction between Arg and the POPC head group or the changes of the hydrophobicity of residue 9 [60], The lipid-mediated dissociation observed in this study could facilitate them adopting an inhibition removal favorable conformation.
2H quadrupolar splittings of 2H labels on the entire acyl chain of the POPC-d31 lipids reflect dynamics of the lipid bilayer hydrophobic region [34, 35, 42], SCD order parameters calculated from the 2H quadrupolar splittings yield individual 2HH dynamics along the acyl chain. 2H nuclei near the membrane surface have larger order parameters than the ones near the membrane core since 2H nuclei near the membrane surface are more restricted [41]. WT-PLB decreases SCD order parameters to a greater extent than P-PLB and R9C-PLB on the entire acyl chain (Figure 4). The SCD order parameter values indicate additional interaction of the membrane hydrophobic region with WT-PLB when compared with P-PLB and R9C-PLB. Moderate effects of R9C-PLB on the lipid hydrophobic region similar to P-PLB may relate to their inhibition relief effect on SERCA. 2H nuclei on the acyl chain near the membrane surface region are disturbed more significantly by PLB than the ones located in the membrane center. In this region, the more distinct SCD values of the lipid with the addition of WT-PLB compared with P-PLB and R9C-PLB may result from a domain Ib conformation change upon phosphorylation or R9C mutation [61], Interestingly, the region in the membrane center is not significantly disturbed by either R9C-PLB or P-PLB, while WT-PLB does alter this region. The data suggest that acyl chain dynamics in the membrane core is less susceptible to membrane protein perturbation than the region near the surface.
Conclusions
In conclusion, R9C-PLB function loss could be due to its oligomerization states, structural conformations and interaction with membrane. From a membrane perspective, similar to P-PLB, R9C-PLB has a less significant interaction with both the surface and hydrophobic regions of the lipid bilayer when compared with WT-PLB. The less significant disturbance of R9C-PLB and P-PLB on the membrane surface may result from the detachment or partial detachment of the cytoplasmic domain from the lipid bilayer. The membrane bilayer may act as a mediation to regulate PLB function. Future studies can directly focus on the R9C-PLB conformations that relate to its function.
Highlights.
-
>
Phospholamban was incorporated into lipid bilayers and studied by solid-state NMR spectroscopy.
-
>
WT-PLB dramatically perturbs the membrane surface and the hydrophobic regions of the bilayer.
-
>
R9C-PLB and P-PLB have similar moderate effects on the membrane surface and hydrophobic region.
-
>
The R9C-PLB cytoplasmic domain associates less tightly with the membrane surface than WT-PLB.
Acknowledgement
We thank Dr. Sergey Maltsev for technical assistance and Dr. Shadi Abu-Baker and Dan Mayo for helpful discussion. This work was supported by NIH grant (R01GM080542 and GM080542Z) and AHA grant (0755602B).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].MacLennan DH, Kranias EG. Phospholamban: A crucial regulator of cardiac contractility. Nature Rev. Mol. Cell Biol. 2003;4:566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- [2].Mueller B, Karim CB, Negrashov IV, Kutchai H, Thomas DD. Direct detection of phospholamban and sarcoplasmic reticulum Ca-ATPase interaction in membranes using fluorescence resonance energy transfers. Biochemistry. 2004;43:8754–8765. doi: 10.1021/bi049732k. [DOI] [PubMed] [Google Scholar]
- [3].Toyoshima C, Asahi M, Sugita Y, Khanna R, Tsuda T, MacLennan DH. Modeling of the inhibitory interaction of phospholamban with the Ca2+ ATPase. Proc. Natl. Acad. Sci. U. S. A. 2003;100:467–472. doi: 10.1073/pnas.0237326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chen Z, Akin BL, Jones LR. Ca(2+) binding to site I of the cardiac Ca(2+) pump is sufficient to dissociate phospholamban. J. Biol. Chem. 2010;285:3253–3260. doi: 10.1074/jbc.M109.080820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Catalucci D, Latronico MVG, Ceci M, Rusconi F, Young HS, Gallo P, Santonastasi M, Bellacosa A, Brown JH, Condorelli G. At increases sarcoplasmic reticulum Ca(2+) cycling by direct phosphorylation of phospholamban at Thr(17) J. Biol. Chem. 2009;284:28180–28187. doi: 10.1074/jbc.M109.036566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10165–10170. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9101–9106. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J. Amer. Chem. Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- [9].Oxenoid K, Chou JJ. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Traaseth NJ, Veglia G. Probing excited states and activation energy for the integral membrane protein phospholamban by NMR CPMG relaxation dispersion experiments. Biochim. Biophys. Acta. 2010;1798:77–81. doi: 10.1016/j.bbamem.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lian P, Wei D-Q, Wang J-F, Chou K-C. An allosteric mechanism inferred from molecular dynamics simulations on phospholamban pentamer in lipid membranes. Plos One. 2011;6 doi: 10.1371/journal.pone.0018587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Seidel K, Andronesi OC, Krebs J, Griesinger C, Young HS, Becker S, Baldus M. Structural characterization of Ca2+-ATPase-bound phospholarnban in lipid bilayers by solid-state nuclear magnetic resonance (NMR) Spectroscopy. Biochemistry. 2008;47:4369–4376. doi: 10.1021/bi7024194. [DOI] [PubMed] [Google Scholar]
- [13].Masterson LR, Yu T, Shi L, Wang Y, Gustavsson M, Mueller MM, Veglia G. cAMP-dependent protein kinase A selects the excited state of the membrane substrate phospholamban. J. Mol. Biol. 2011;412:155–164. doi: 10.1016/j.jmb.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Paterlini MG, Thomas DD. The alpha-helical propensity of the cytoplasmic domain of phospholamban: A molecular dynamics simulation of the effect of phosphorylation and mutation. Biophys. J. 2005;88:3243–3251. doi: 10.1529/biophysj.104.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sayadi M, Feig M. Role of conformational sampling of Serl6 and Thrl7-phosphorylated phospholamban in interactions with SERCA. Biochim. Biophys. Acta-Biomembranes. 2013;1828:577–585. doi: 10.1016/j.bbamem.2012.08.017. [DOI] [PubMed] [Google Scholar]
- [16].Ghimire H, Abu-Baker S, Sahu ID, Zhou AD, Mayo DJ, Lee RT, Lorigan GA. Probing the helical tilt and dynamic properties of membrane-bound phospholamban in magnetically aligned bicelles using electron paramagnetic resonance spectroscopy. Biochim. Biophys. Acta-Biomembranes. 2012;1818:645–650. doi: 10.1016/j.bbamem.2011.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Karim CB, Zhang ZW, Howard EC, Torgersen KD, Thomas DD. Phosphorylation-dependent conformational switch in spin-labeled phospholamban bound to SERCA. J. Mol. Biol. 2006;358:1032–1040. doi: 10.1016/j.jmb.2006.02.051. [DOI] [PubMed] [Google Scholar]
- [18].Abu-Baker S, Lu J-X, Chu S, Shetty KK, Gor’kov PL, Lorigan GA. The structural topology of wild-type phospholamban in oriented lipid bilayers using N-15 solid-state NMR spectroscopy. Prot. Sci. 2007;16:2345–2349. doi: 10.1110/ps.072977707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chu GX, Li L, Sato Y, Harrer JM, Kadambi VJ, Hoit BD, Bers DM, Kranias EG. Pentameric assembly of phospholamban facilitates inhibition of cardiac function in vivo. J. Biol. Chem. 1998;273:33674–33680. doi: 10.1074/jbc.273.50.33674. [DOI] [PubMed] [Google Scholar]
- [20].Schmitt JP, Kamisago M, Asahi M, Li GH, Ahmad F, Mende U, Kranias EG, MacLennan DH, Seidman JG, Seidman CE. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- [21].Ha KN, Masterson LR, Hou Z, Verardi R, Walsh N, Veglia G, Robia SL. Lethal Arg9Cys phospholamban mutation hinders Ca(2+)-ATPase regulation and phosphorylation by protein kinase A. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2735–2740. doi: 10.1073/pnas.1013987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ceholski DK, Trieber CA, Holmes CFB, Young HS. Lethal, hereditary mutants of phospholamban elude phosphorylation by protein kinase A. J. Biol. Chem. 2012;287:2659626605. doi: 10.1074/jbc.M112.382713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gruber SJ, Haydon S, Thomas DD. Phospholamban mutants compete with wild type for SERCA binding in living cells. Biochem. Biophys. Res. Commun. 2012;420:236–240. doi: 10.1016/j.bbrc.2012.02.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kang C, Li Q. Solution NMR study of integral membrane proteins. Curr. Opin. Chem. Biol. 2011;15:560–569. doi: 10.1016/j.cbpa.2011.05.025. [DOI] [PubMed] [Google Scholar]
- [25].Saito H, Naito A. NMR studies on fully hydrated membrane proteins, with emphasis on bacteriorhodopsin as a typical and prototype membrane protein. Biochim. Biophys. Acta. 2007;1768:3145–3161. doi: 10.1016/j.bbamem.2007.08.026. [DOI] [PubMed] [Google Scholar]
- [26].Brown MF, Heyn MP, Job C, Kim S, Moltke S, Nakanishi K, Nevzorov AA, Struts AV, Salgado GFJ, Wallat I. Solid-State H-2 NMR spectroscopy of retinal proteins in aligned membranes. Biochim. Biophys. Acta. 2007;1768:2979–3000. doi: 10.1016/j.bbamem.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mahalakshmi R, Marassi FM. Orientation of the Escherichia coli outer membrane protein OmpX in phospholipid bilayer membranes determined by solid-state NMR. Biochemistry. 2008;47:6531–6538. doi: 10.1021/bi800362b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schneider R, Ader C, Lange A, Giller K, Hornig S, Pongs O, Becker S, Baldus M. Solid-state NMR spectroscopy applied to a chimeric potassium channel in lipid bilayers. J. Amer. Chem. Soc. 2008;130:7427–7435. doi: 10.1021/ja800190c. [DOI] [PubMed] [Google Scholar]
- [29].Lorigan GA. Magnetic resonance spectroscopic studies of the integral membrane protein phospholamban. In: Webb GA, editor. Modern Magnetic Resonance: Applications in Chemistry, Biological abd Marine Sciences. Vol. 1. Springer; 2008. pp. 313–318. [Google Scholar]
- [30].Stockton GW, Polnaszek CF, Tulloch AP, Hasan F, Smith ICP. Molecular-motion and order in single-bilayer vesicles and multilamellar dispersions of egg lecithin and lecithin-cholesterol mixtures-deuterium nuclear magnetic-resonance study of specifically labeled lipids. Biochemistry. 1976;15:954–966. doi: 10.1021/bi00650a003. [DOI] [PubMed] [Google Scholar]
- [31].Traikia M, Langlais DB, Cannarozzi GM, Devaux PF. High-resolution spectra of liposomes using MAS NMR. The case of intermediate-size vesicles. J. Magn. Reson. 1997;125:140–144. doi: 10.1006/jmre.1996.1068. [DOI] [PubMed] [Google Scholar]
- [32].Jesorka A, Orwar O. Liposomes: Technologies and analytical applications. Annu. Rev. Anal. Chem. 2008;1:801–832. doi: 10.1146/annurev.anchem.1.031207.112747. [DOI] [PubMed] [Google Scholar]
- [33].Dave PC, Tiburu EK, Damodaran K, Lorigan GA. Investigating structural changes in the lipid bilayer upon insertion of the transmembrane domain of the membrane-bound protein phospholamban utilizing P-31 and H-2 solid-state NMR spectroscopy. Biophys. J. 2004;86:1564–1573. doi: 10.1016/S0006-3495(04)74224-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chu SD, Hawes JW, Lorigan GA. Solid-state NMR spectroscopic studies on the interaction of sorbic acid with phospholipid membranes at different pH levels. Magn. Reson. Chem. 2009;47:651–657. doi: 10.1002/mrc.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Abu-Baker S, Lorigan GA. Phospholamban and its phosphorylated form interact differently with lipid bilayers: A (31)P, (2)H, and (13)C solid-state NMR spectroscopic study. Biochemistry. 2006;45:13312–13322. doi: 10.1021/bi0614028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Abu-Baker S, Lu JX, Chu SD, Brinn CC, Makaroff CA, Lorigan GA. Side chain and backbone dynamics of phospholamban in phospholipid bilayers utilizing H-2 and N-15 solid-state NMR spectroscopy. Biochemistry. 2007;46:11695–11706. doi: 10.1021/bi700749q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Huster D, Arnold K, Gawrisch K. Influence of docosahexaenoic acid and cholesterol on lateral lipid organization in phospholipid mixtures. Biochemistry. 1998;37:17299–17308. doi: 10.1021/bi980078g. [DOI] [PubMed] [Google Scholar]
- [38].Huster D, Yao YL, Jakes K, Hong M. Conformational changes of colicin la channel-forming domain upon membrane binding: a solid-state NMR study. Biochim. Biophys. Acta. 2002;1561:159–170. doi: 10.1016/s0005-2736(02)00340-1. [DOI] [PubMed] [Google Scholar]
- [39].Lafleur M, Fine B, Sternin E, Cullis PR, Bloom M. Smoothed orientational order profile lipid bilayers by H-2 Nuclear Magnetic Resonance. Biophys. J. 1989;56:1037–1041. doi: 10.1016/S0006-3495(89)82749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rigby AC, Barber KR, Shaw GS, Grant CWM. Transmembrane region of the epidermal growth factor receptor: Behavior and interactions via H-2 NMR. Biochemistry. 1996;35:12591–12601. doi: 10.1021/bi9611063. [DOI] [PubMed] [Google Scholar]
- [41].Seelig A, Seelig J. Dynamic structure of fatty acyl chains in a phospholipid bilayer measured by deuterium magnetic-resonance. Biochemistry. 1974;13:4839–4845. doi: 10.1021/bi00720a024. [DOI] [PubMed] [Google Scholar]
- [42].Yu X, Chu S, Hagerman AE, Lorigan GA. Probing the Interaction of Polyphenols with Lipid Bilayers by Solid-State NMR Spectroscopy. Journal of Agricultural and Food Chemistry. 2011;59:6783–6789. doi: 10.1021/jf200200h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Aussenac F, Laguerre M, Schmitter JM, Dufourc EJ. Detailed structure and dynamics of bicelle phospholipids using selectively deuterated and perdeuterated labels. H-2 NMR and molecular mechanics study. Langmuir. 2003;19:10468–10479. [Google Scholar]
- [44].Massiot D, Fayon F, Capron M, King I, Le Calve S, Alonso B, Durand JO, Bujoli B, Gan ZH, Hoatson G. Modelling one- and two-dimensional solid-state NMR spectra. Magn. Reson. Chem. 2002;40:70–76. [Google Scholar]
- [45].McCabe MA, Wassall SR. Fast-Fourier-Transform DePaking. J. Magn. Reson. Series B. 1995;106:80–82. [Google Scholar]
- [46].McCabe MA, Wassall SR. Rapid deconvolution of NMR powder spectra by weighted fast Fourier transformation. Solid State Nucl. Magn. Reson. 1997;10:53–61. doi: 10.1016/s0926-2040(97)00024-6. [DOI] [PubMed] [Google Scholar]
- [47].Dave PC, Tiburu EK, Nusair NA, Lorigan GA. Calculating order parameter profiles utilizing magnetically aligned phospholipid bilayers for H-2 solid-state NMR studies. Solid State Nucl. Magn. Reson. 2003;24:137–149. doi: 10.1016/S0926-2040(03)00052-3. [DOI] [PubMed] [Google Scholar]
- [48].Dufourc EJ, Parish EJ, Chitrakorn S, Smith ICP. Structural and dynamical details of cholesterol lipid interaction as revealed by deuterium NMR. Biochemistry. 1984;23:6062–6071. [Google Scholar]
- [49].Lu J.-x., Blazyk J, Lorigan GA. Exploring membrane selectivity of the antimicrobial peptide KIGAKI using solid-state NMR spectroscopy. Biochim. Biophys. Acta. 2006;1758:1303–1313. doi: 10.1016/j.bbamem.2006.02.002. [DOI] [PubMed] [Google Scholar]
- [50].Gustavsson M, Traaseth NJ, Karim CB, Lockamy EL, Thomas DD, Veglia G. Lipid-mediated folding/unfolding of phospholamban as a regulatory mechanism for the sarcoplasmic reticulum Ca(2+)-ATPase. J. Mol. Biol. 2011;408:755–765. doi: 10.1016/j.jmb.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gustavsson M, Traaseth NJ, Veglia G. Activating and deactivating roles of lipid bilayers on the Ca(2+)-ATPase/phospholamban complex. Biochemistry. 2011;50:10367–10374. doi: 10.1021/bi200759y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cornea RL, Jones LR, Autry JM, Thomas DD. Mutation and phosphorylation change the oligomeric structure of phospholamban in lipid bilayers. Biochemistry. 1997;36:2960–2967. doi: 10.1021/bi961955q. [DOI] [PubMed] [Google Scholar]
- [53].Lockamy EL, Cornea RL, Karim CB, Thomas DD. Functional and physical competition between phospholamban and its mutants provides insight into the molecular mechanism of gene therapy for heart failure. Biochem. Biophys. Res. Commun. 2011;408:388392. doi: 10.1016/j.bbrc.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chu S, Coey AT, Lorigan GA. Solid-state (2)H and (15)N NMR studies of side-chain and backbone dynamics of phospholamban in lipid bilayers: Investigation of the N27A mutation. Biochim. Biophys. Acta. 2010;1798:210–215. doi: 10.1016/j.bbamem.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Akin BL, Chen Z, Jones LR. Superinhibitory phospholamban mutants compete with Ca(2+) for binding to SERCA2a by stabilizing a unique nucleotide-dependent conformational state. J. Biol. Chem. 2010;285:28540–28552. doi: 10.1074/jbc.M110.151779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zamoon J, Nitu F, Karim C, Thomas DD, Veglia G. Mapping the interaction surface of a membrane protein: Unveiling the conformational switch of phospholamban in calcium pump regulation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4747–4752. doi: 10.1073/pnas.0406039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Pinheiro TJT, Watts A. Resolution of individual lipids in mixed phospholipid-membranes and specific lipid cytochrome-c interactions by magic-angle spinning solid-state P-31 NMR. Biochemistry. 1994;33:2459–2467. doi: 10.1021/bi00175a014. [DOI] [PubMed] [Google Scholar]
- [58].Pinheiro TJT, Watts A. Lipid specificity in the interaction of cytochrome-c with anionic phospholipid-bilayers revealed by solid-state P-31 NMR. Biochemistry. 1994;33:2451–2458. doi: 10.1021/bi00175a013. [DOI] [PubMed] [Google Scholar]
- [59].Gustavsson M, Traaseth NJ, Veglia G. Probing ground and excited states of phospholamban in model and native lipid membranes by magic angle spinning NMR spectroscopy. Biochim. Biophys. Acta-Biomembranes. 2012;1818:146–153. doi: 10.1016/j.bbamem.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ceholski DK, Trieber CA, Young HS. Hydrophobic imbalance in the cytoplasmic domain of phospholamban is a determinant for lethal dilated cardiomyopathy. J. Biol. Chem. 2012;287:16521–16529. doi: 10.1074/jbc.M112.360859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yu X, Lorigan GA. Secondary structure, backbone dynamics and structural topology of phospholamban and its phosphorylated and Arg9Cys mutated forms in phospholipid bilayers utilizing (13)C and (15)N solid-state NMR spectroscopy. 2012. in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]