Summary
Somites are embryonic precursors of the axial skeleton and skeletal muscles, and establish the segmental vertebrate body plan. Somitogenesis is controlled in part by a segmentation clock that requires oscillatory expression of genes including Lunatic fringe (Lfng). Oscillatory genes must be tightly regulated both at the transcriptional and post-transcriptional levels for proper clock function. Here we demonstrate that microRNA-mediated regulation of Lfng is essential for proper segmentation during chick somitogenesis. We find that mir-125a-5p targets evolutionarily conserved sequences in the Lfng 3′UTR, and that preventing interactions between mir-125a-5p and Lfng transcripts in vivo causes abnormal segmentation and perturbs clock activity. This provides strong evidence that miRNAs function in the post-transcriptional regulation of oscillatory genes in the segmentation clock. Further, this demonstrates that the relatively subtle effects of miRNAs on target genes can have broad effects in developmental situations that have critical requirements for tight post-transcriptional regulation.
Introduction
Somites are cohorts of cells that bud from the anterior end of the presomitic mesoderm (PSM) and give rise to the axial skeleton and other structures (reviewed in Hirsinger et al., 2000). During somitogenesis, the expression levels of numerous genes oscillate in the PSM as part of a segmentation clock that controls the timing of somite formation. The Notch target c-hairy1 was the first gene found expressed in this pattern (Palmeirim et al., 1997). In mouse and chick, a key oscillatory gene is Lunatic fringe (Lfng), which encodes a glycosyltransferase that modulates Notch signaling (Moloney et al., 2000). During vertebrate segmentation, both Lfng transcript levels and LFNG protein levels oscillate with a period that matches the rate of somite formation (2 hours in the mouse, 90 minutes in the chick) (Dale et al., 2003; Pourquie, 2001).
Either loss of Lfng expression, or sustained, non-oscillatory Lfng activity perturbs somite formation and patterning, presumably by altering its oscillatory expression (Dale et al., 2003; Evrard et al., 1998; Serth et al., 2003; Zhang and Gridley, 1998). It is known that cyclic Lfng expression is regulated at the transcriptional level (Cole et al., 2002), but little is known about the post-transcriptional mechanisms that contribute to the rapid oscillations. Stable oscillatory expression patterns have been proposed to be regulated by feedback inhibition mechanisms coupled with transcriptional time delays (Lewis, 2003; Monk, 2003). Some mathematical models of the segmentation clock invoke delayed feedback loops involving regulation of Notch1, Lfng and Hes7 (or c-hairy1 in chick). In these models, mRNA and protein half-lives of oscillatory genes must be tightly regulated to ensure proper clock function (Feng and Navaratna, 2007; Gonzalez and Kageyama, 2009). The Lfng 3′UTR is evolutionarily conserved, and has been proposed to regulate RNA half-life (Chen et al., 2005; Hilgers et al., 2005). One possible source of such regulation would be miRNAs, non-coding RNA molecules that direct post-transcriptional repression of protein-coding genes by promoting RNA turnover and/or by decreasing translational efficiency of their target transcripts (reviewed in Bartel, 2004), and one model of oscillatory gene expression has proposed miRNA functions in the clock (Xie et al., 2007).
We hypothesized that the oscillatory expression of Lfng in the segmentation clock could require post-transcriptional regulation by miRNAs. Here we identify an miRNA (mir-125a-5p) that is enriched in the PSM, and targets evolutionarily conserved sequences in the Lfng 3′UTR. Inhibiting mir-125a-5p function or preventing interactions between mir-125a-5p and endogenous Lfng transcripts in vivo perturbs somitogenesis and disrupts clock function in the PSM of developing chick embryos. These findings support the hypothesis that regulation of oscillatory genes by miRNAs may provide a mechanism for post-transcriptional control of the segmentation clock.
Results
mir-125a-5p is expressed in the PSM and targets the Lfng 3′UTR
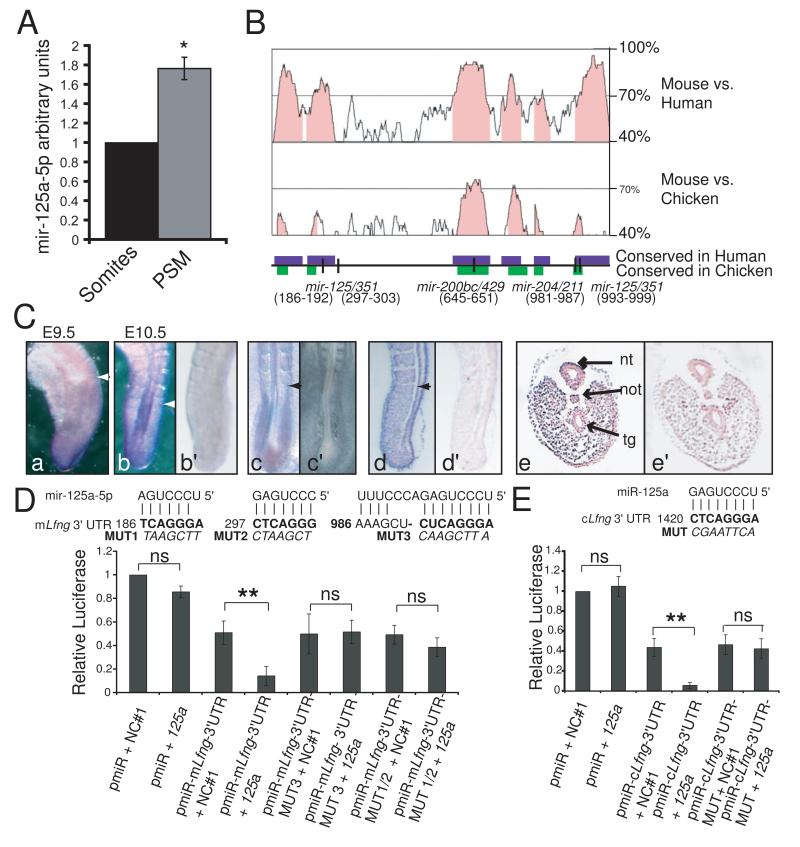
To examine the possibility that Lfng oscillations might be regulated by miRNAs, we assessed the expression of candidate miRNAs in the PSM, where the clock is active. By QRT-PCR (Fig 1A) and miRNA microarray (data not shown) we found that mir-125a-5p levels are higher in the mouse PSM, than in the mature somites. Thus, its expression is enriched in the PSM where Lfng is predicted to require a short RNA half life. mir-125a-5p is proposed to target three sites in the mouse Lfng 3′UTR, and one of the sites is conserved in chicken (Fig. 1B). Whole mount in-situ hybridization confirmed specific expression of mir-125a-5p in the PSM of mouse and chicken embryos (Fig 1C, panels a - c). Futher, mir-125a-5p expression was observed in mouse embryos in the ectoderm and mesoderm, but was largely excluded from the neural tube, notochord, and tailgut (Fig 1C, panels d and e).
Figure 1.
The Lfng 3′UTR is an evolutionarily conserved target of mir-125a-5p A. By QRT-PCR mir-125a-5p is significantly enriched in the PSM compared to the mature somites of E9.5 mouse embryos (*= p<0.05, Student’s T-Test. Error bars = SD). B. Lfng 3′UTR schematic showing high conservation among mouse, human, and chicken Lfng 3′UTRs (mVista Bray et al., 2003). Conserved regions shown as colored boxes. Positions of TargetScan predicted miRNA binding sites define the first nt of the 3′UTR as 1.C. RNA in situ analysis of mir-125a-5p in whole mount mouse embryos at E9.5 (a) and E10.5 (b), and in HH10 chick embryos (c). Section in situ of E10.5 mouse embryos (d= saggital, e=transverse) demonstrate mir-125a-5p expression in the ectoderm and mesoderm, but not in the neural tube, notochord, and tail gut. Arrows indicate most recent somite boundary, nt = neural tube, not=notochord, tg= tailgut. In situs with negative control probe on adjacent sections did not exhibit staining (panels b’, c’, d’, e’). D. Transfection of premir-125a-5p (125a) significantly reduces luciferase expression from pmir-mLfng3′UTR (pMIR-REPORT + mouse Lfng 3′UTR) compared to transfection of a negative control pre-mir (NC#1). Mutations of the mir-125a-5p binding sites at either end of the 3′UTR (MUT1-3) abrogate this effect. E. Transfection of premir-125a-5p significantly reduces luciferase expression from pmir-cLfng3′UTR (pMIR-REPORT + chicken Lfng 3′UTR). Mutations in the mir-125a-5p (MUT) binding site abrogate this effect. Two way ANOVA, Bonferroni post hoc; *=p<.05, **=p<.01, error bar = SD. See Fig. S1 for related analyses.
The Lfng 3′UTR can be directly targeted by mir-125a-5p
To test whether Lfng is a direct target of mir-125a-5p, we examined the effects of the miRNA on transcripts containing the 3′UTR of Lfng. Vectors containing either mouse or chick Lfng 3′UTR sequence exhibit lower luciferase expression than control vectors in these cells due to the effects of endogenous miRNAs (Fig. S1A). However, expression of exogenous mir-125a-5p causes a further significant reduction in luciferase expression only from vectors containing the mouse or chicken Lfng 3′UTR (Fig. 1D-E). In contrast, mir-125a-5p binding sites were not identified in the 3′UTRs of other oscillatory genes, and mir-125a-5p expression had no effect on expression of transcripts containing the Hes7 3′UTR (Fig. S1B). Mutation of predicted mir-125a-5p binding sites at either end of the mouse 3′UTR, or of the single site in the chicken 3′UTR abrogates the effect of exogenous mir-125a-5p, indicating that mir-125a-5p directly interacts with the Lfng 3′UTR, and that these interactions are conserved among organisms that utilize Lfng in the segmentation clock.
Inhibition of mir-125a-5p activity perturbs segmentation
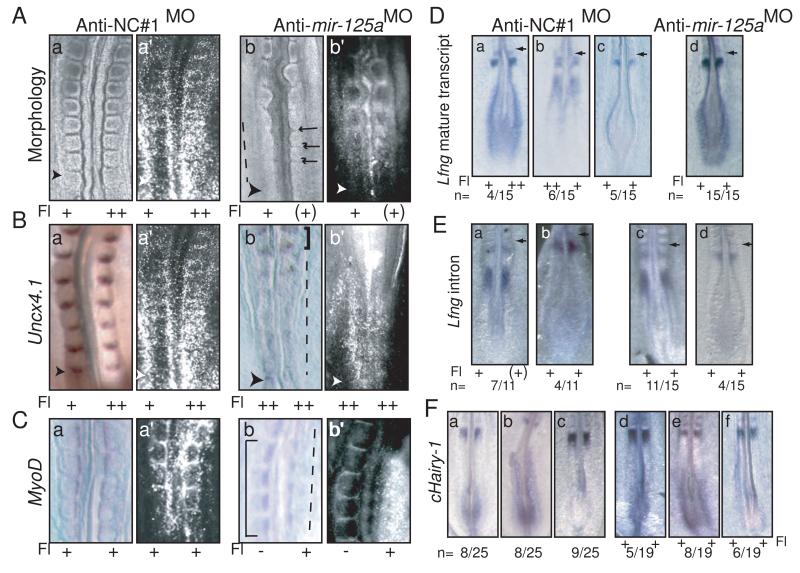
To dissect the function of mir-125a-5p in segmentation, we performed loss-of-function experiments in chick embryos using in ovo electroporation of antisense morpholinos to inhibit mir-125a-5p activity. Morpholinos complementary to mir-125a-5p (anti-mir-125aMO, Supplemental Experimental Methods) bind to endogenous mir-125a-5p, and inhibit interactions with its target transcripts, as shown by our inability to detect mir-125a-5p by in situ hybridization in electroporated chick embryos (Fig S2A). 24 hours post-electroporation, the targeted region has undergone segmentation. Inhibition of mir-125a-5p activity perturbs formation and patterning of mature somites (n=18/18; Fig. 2). Intersomitic boundaries were absent or disorganized in the electroporated regions of the embryo (Fig. 2A). Somite patterning was also disorganized ,with reduced and diffuse expression of Uncx4.1, a marker of the caudal somite compartment (Fig. 2B). Inhibition of mir-125a-5p also leads to formation of disorganized and irregular myotome compartments, as evidenced by weak and diffuse MyoD expression in the electroporated region (Fig. 2C). Interestingly, phenotypes were observed even in cases where electroporations levels were comparably low. This is unlikely to be due to non-cell autonomous effects of the morpholino. Instead, it reflects one of the roles of the segmentation clock which is to synchronize the oscillations of neighboring cells. Mosaic regions of the embryo containing wild type cells mixed with cells that have altered clock function are predicted to exhibit phenotypes at the tissue level due to lack of cell:cell synchronization. This effect has recently been confirmed in mouse embryos that are chimeric for wild type and Lfng null cells (Okubo et al., 2012). Together, these findings indicate that mir-125a-5p activity is required for normal formation and patterning of epithelial somites.
Figure 2.
Inhibition of mir-125a-5p perturbs somitogenesis in chicken embryos, and stabilizes Lfng transcripts in the PSM A. Electroporation of anti-NC#1MO has no effect on somite morphology (panel a), while electroporation of anti-mir-125aMO results in abnormal somite morphology with absent (dashed line) or disorganized (arrows) intersomitic boundaries in electroporated regions (panel b).B. Uncx4.1 staining is reduced and diffuse in embryos electroporated with anti-mir-125aMO (dashed line, panel b) compared to anti-NC#1MO (panel a). Note relatively normal somites in the older region of the anti-mir-125aMO embryo in the region that is less positive for the morpholino (square bracket). The same embryo is pictured in part A panel a and part B panel a C. MyoD expression is disorganized in somites electroporated with anti-mir-125aMO (dashed line, panel b) compared to embryos electroporated with anti-NC#1MO (panel a). Note normal myotomes in the unelectroporated regions of the anti-mir-125aMO embryo (square brackets). D 2h post-electroporation with anti-NC#1MO, endogenous Lfng is observed in the three described phases (Pourquie and Tam, 2001). In contrast, in anti-mir-125aMO positive embryos, robust, non-cyclic Lfng expression is observed in the caudal PSM of all embryos (panel d, n=15/15). E In situ analysis with a probe specific for the Lfng intron demonstrates that both control embryos (a,b) and embryos electroporated with anti-mir-125aMO (c,d) exhibit dynamic patterns of newly transcribed Lfng RNA. F. c-hairy1 expression is cyclic in control embryos (a-c), or in embryos electroporated with anti-mir-125aMO (d-f)Right hand panels reflect the fluorescein signal demarcating the electroporated regions of each embryo. Fl: fluorescein. Degree of electroporation efficiency designated as “++ ” = strong, “+” = moderate, “(+)” = weak,“ −” = negative. Arrowheads indicate the most recent somite boundary. See figure S2 for fluorescein images as well as analysis of mir-125a-5p by in situ in electroporated embryos.
mir-125a-5p activity is required for normal cyclic Lfng expression
We next examined the effect of mir-125a-5p inhibition on the expression of endogenous Lfng. Lfng expression was examined 2 hours post-electroporation with anti-mir-125MO, when the targeted region of the embryo is confined to the PSM. In embryos electroporated with control morpholinos, Lfng expression is cyclic (Fig. 2D panels a-c). In contrast, all embryos electroporated with anti-mir-125aMO exhibit stable Lfng expression in the caudal PSM as well as a band in the rostral PSM (n=15/15; Fig. 2D, panel d). The expression of a stable Lfng stripe in the anterior PSM may reflect the distinct control mechanisms found in the anterior and posterior PSM (Cole et al., 2002). The sustained, non-oscillatory expression of Lfng in the posterior PSM, where the clock is active, suggests that loss of mir-125a-5p activity stabilizes the Lfng transcript, preventing its rapid turnover.
To assess whether the change in the Lfng expression reflects a direct effect on endogenous Lfng RNA transcripts, as opposed to an indirect effect on Lfng transcription, we examined the expression of newly transcribed Lfng using an in situ probe for the first intron of the gene (Morales et al., 2002). In control embryos, dynamic expression of intron-containing Lfng transcripts is observed, with embryos exhibiting a single anterior band or an anterior band and a posterior band of varying width (Fig. 2E, panels a-b). This dynamic pattern is maintained in embryos electroporated with anti-mir-125aMO (Fig. 2E, panels c-d), suggesting that in the short time frame of these experiments, loss of mir-125a-5p activity directly affects turnover of mature Lfng transcripts. Further, we find that the c-hairy1 expression pattern appears oscillatory two hours post-transfection with anti-mir-125aMO (Fig. 2F). This is expected, as c-hairy1 oscillations have been shown to persist for one or two cycles even in the presence of cycloheximide (Palmeirim et al., 1997), thus we do not predict an overt effect on c-hairy1 oscillations within the short time frame of this experiment, unless there are direct effects of chairy transcript stability. Together, these data suggest that the short term effects of mir-125a-5p act through effects on Lfng transcript stability.
Direct interactions between endogenous mir-125a-5p and the Lfng 3′UTR are required for normal somitogenesis and clock function
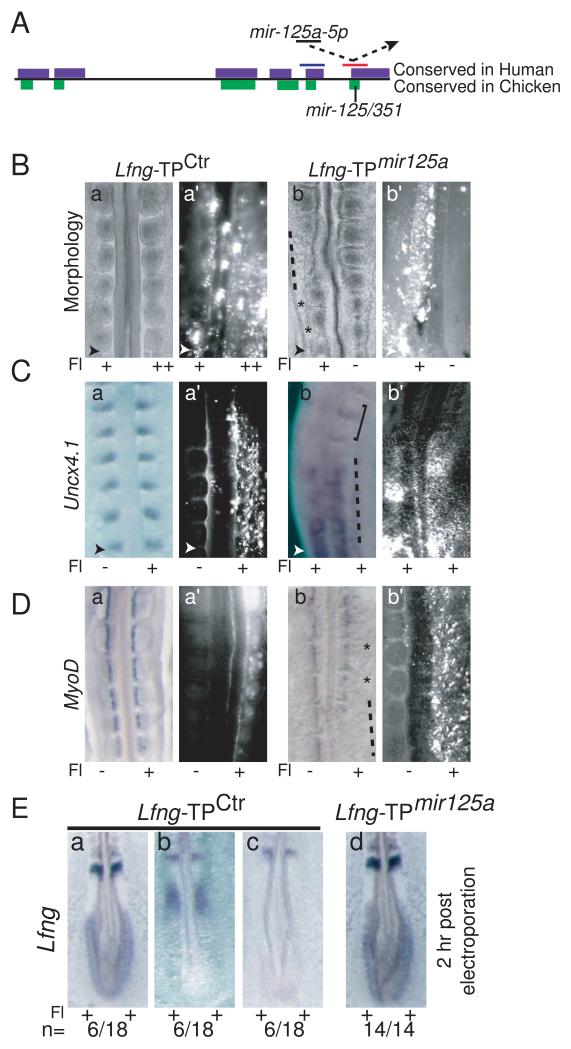
The segmentation phenotypes observed when mir-125a-5p activity is inhibited are reminiscent of those observed when Lfng is ubiquitously expressed in the chick PSM (Dale et al., 2003), supporting the hypothesis that mir-125a-5p inhibition affects Lfng expression. To further investigate the specificity of this effect, we directly examined the functional relevance of the mir-125a-5p:Lfng interaction, using Target Protectors (TP) (Choi et al., 2007) to specifically disrupt the binding of endogenous mir-125a-5p to endogenous Lfng transcripts in chick embryos. Target Protectors bind to miRNA recognition sites in mRNAs, physically preventing interactions between endogenous transcripts and the miRNA (Fig. 3A). A TP that binds to and blocks the mir-125a-5p binding site in the chick Lfng 3′UTR (Lfng-TPmir125a), or a control TP complementary to a nearby conserved site in the chick 3′UTR (Lfng-TPCtrl) were used (Fig 3A, Supplementary Experimental Methods). In cell culture, these TPs do not themselves affect expression from transcripts containing the Lfng 3′UTR, but Lfng-TPmir-125a protects transcripts from the effects of exogenous mir-125a-5p (Fig S3), indicating that Lfng-TPmir-125a blocks binding of mir-125a-5p to the 3′UTR. Specifically blocking interactions between endogenous mir-125a-5p and Lfng in the chick PSM with Lfng-TPmir-125a severely perturbed segmentation. Somites were disorganized (n=21/21, Fig. 3B), with diffuse and reduced Uncx4.1 expression (Fig. 3C). Myotome formation occurred, but compartments exhibited abnormal size and spacing reflecting underlying defects in somite morphogenesis (Fig. 3D). To examine the effects of mir-125a-5p on cyclic Lfng expression, we examined the expression of endogenous Lfng mRNA 2h after electroporation of target protectors. All embryos positive for Lfng-TPmir-125a exhibit strong, non-oscillatory Lfng expression in the caudal PSM (n= 14/14, Fig. 3E). Thus, LfngTPmir-125a recapitulates the phenotypes seen after mir-125a-5p inhibition, indicating that interactions between mir-125a-5p and Lfng are essential for proper somite formation and clock function in the developing chick embryo.
Figure 3.
Blocking interactions between mir-125a-5p and Lfng perturbs somitogenesis and segmentation clock function. A. Schematic of the Lfng 3′UTR and the TPs used, with the single mir-125a-5p binding site in chicken. Approximate positions of Lfng-TPCtrl (blue) and Lfng-TPmir- 125a (red) are shown (not to scale). B. Electroporation of Lfng-TPctr has no effect on somite morphology (panels a, a’), while electroporation of Lfng-TPmir-125a results in abnormal somite morphology with absent (dashed lines) or disorganized (*) intersomitic boundaries (panels b, b’). C. Uncx4.1 expression is disorganized and sometimes reduced in Lfng-TPmir-125a positive embryos (dashed line, panels b, b’) compared to embryos electroporated with Lfng-TPctr (panels a, a’). Note relatively normal somites in the less positive region of panel b (square bracket) D. MyoD expression in the somites of Lfng-TPmir-125a positive embryos (panels b, b’) indicates that myotomes are formed, but somite compartments are of irregular sizes (*) compared to embryos electroporated with Lfng-TPctr (panel a, a’). In some regions of the embryo, MyoD expression is strongly downregulated or delayed (dashed line, panel b). E. 2h post-electroporation, cyclic expression of endogenous Lfng is observed in Lfng-TPCtrl embryos (panels a-c), while robust, non-cyclic Lfng expression is observed in the caudal PSM of Lfng-TPmir-125a positive embryos (panel d, n=14/14). Fl: fluorescein as described in Fig. 2. Arrowheads indicate the most recent somite boundary. See Fig. S3 for analysis of target protector activity in cell lines, and Fig. S4 for fluorescein images.
Stabilization of Lfng transcripts affects clock function via a feedback loop
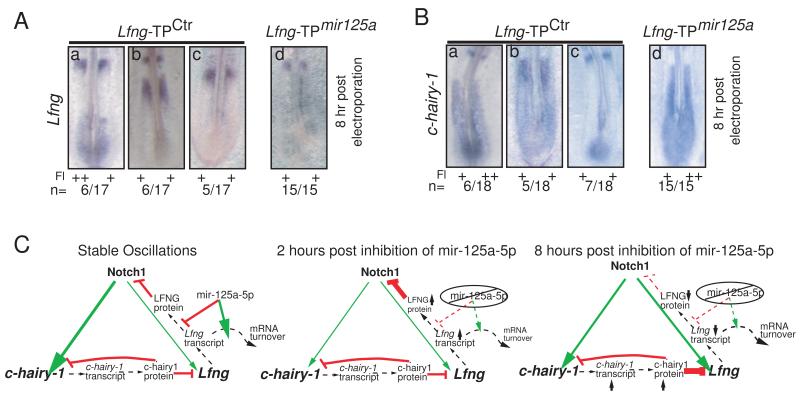
Models of the segmentation clock predict that altering the mRNA half-life of individual clock components will eventually perturb oscillations of other clock-linked genes, as the feedback loop affects the transcription of other clock components like chairy-1 (Feng and Navaratna, 2007; Gonzalez and Kageyama, 2009; Hirata et al., 2004). To address this issue, we examined endogenous Lfng and chairy-1 expression 8h (approximately 5 cycles) after electroporation. At this timepoint, all embryos positive for Lfng-TPmir-125a express Lfng as a single band in the anterior PSM, with no expression observed in the caudal PSM. This suggests that Lfng expression stabilizes at a level below our limit of detection after long term inhibition of the mir-125a:Lfng interaction (n=15/15, Fig. 4A). Disrupting interactions between mir-125a-5p and Lfng also perturbs chairy1 oscillations, with all Lfng-TPmir-125a positive embryos exhibiting constitutive chairy1 expression in the caudal PSM (n=15/15; Fig. 4B). This suggests that the segmentation defects observed after inhibiting mir-125a-5p:Lfng interactions result from disruption of segmentation clock function, and that altering Lfng transcript stability affects other clock components via feedback loops.
Figure 4.
Long term inhibition of interactions between mir-125a-5p and Lfng perturbs the oscillatory expression of genes linked to the segmentation clock via feedback. A. 8h post-electroporation, cyclic expression of endogenous Lfng is observed in Lfng-TPCtrl embryos (a-c) while endogenous Lfng expression is non-cyclic, with no expression detected in the caudal PSM of Lfng-TPmir-125a positive embryos (panel d, n=15/15). B. 8h post-electroporation, cyclic expression of endogenous chairy1 is observed in Lfng-TPCtrl embryos (panels a-c), but non-cyclic cHairy expression is observed in the caudal PSM of Lfng-TPmir-125a positive embryos (panel d, n=15/15). Fl: Fluorescein; ‘+’ and ‘++’ indicate relative levels of fluorescein signal, images available in Fig S4. C Model for effects of mir-125a-5p in the clock. Stable clock oscillations are governed in part by interlocking feedback loops where Notch activates Lfng and c-hairy-1 (green arrows), while LFNG protein inhibits Notch signalling, and c-hairy-1 protein inhibits its own transcription and that of Lfng (red lines). The lengths of delays imposed by transcription rate and translational efficiency (dashed lines), as well as the half-lives of transcripts and proteins are critical for maintenance of stable oscillations. mir-125a-5p is proposed to increase the rate of Lfng transcript turnover and/or decrease the efficiency of translation. In the absence of mir-125a-5p, (middle) levels of Lfng transcript and LFNG protein increase. In the long term (right), the effect of increased Lfng transcript stability is loss of robust oscillations, with stable, increased levels of c-hairy-1 transcript, and stable decreased transcription of Lfng. See also Fig. S4 for fluorescein images
Discussion
Our results suggest that mir-125a-5p functions in post-transcriptional regulation of the chick segmentation clock by destabilizing Lfng transcripts. Although miRNA-based regulation in animal systems has been suggested to act largely via translational efficiency, it is clear that transcript turnover is frequently accelerated by miRNAs (Baek et al., 2008; Guo et al., 2010). Overall, our findings are consistent with a model wherein blocking interactions between mir-125a-5p and Lfng in chick embryos initially results in stabilization of Lfng mRNA, although we cannot rule out that this interaction may also affect translational efficiency of the transcripts (Fig. 4C).
Because Lfng is a component of the oscillating clock machinery, functioning in a negative feedback loop along with Notch1 and c-hairy-1, the increase in Lfng transcript stability is predicted to have a long term effect on the expression of other clock components (Fig. 4C). Consistent with this, we observe a loss of robust oscillation of clock genes after long term inhibition of mir-125a-5p, with very low expression of Lfng in the caudal PSM (below our levels of detection by in situ), and a stable increase in the expression of chairy-1 (Fig. 4C). Therefore, we propose a model where miRNAs function to regulate transcript turnover and/or translational delays in the chicken segmentation clock, facilitating the oscillatory dynamics generated by the delayed negative feedback loop during the rapid period of the clock. During the revision of this manuscript, it was reported that mir-9 expression can influence the oscillatory expression of a Hes1 luciferase reporter in tissue culture, supporting the idea that miRNA:transcript interactions can be important in the regulation of cyclically expressed genes (Bonev et al., 2012). The work here extends this finding by altering endogenous miRNA:transcript interactions in vivo and revealing a robust and dramatic phenotype, supporting the hypothesis that regulation of oscillatory transcripts by miRNAs play a critical, functional role in the segmentation clock.
The conservation of this precise mechanism in other vertebrates remains unclear. Recent findings suggest that conditional inactivation of Dicer in the mesoderm of developing mouse embryos may not affect clock function in the short term (Zhang et al., 2011). However, inactivation of Dicer prevents miRNA maturation, and significant data suggests that mature miRNAs have long half-lives, ranging from 28-211 hours (Gantier et al., 2011). Thus, even after cre-mediated excision of Dicer, it is possible that cells in the PSM during early embryogenesis will still contain mature miRNAs, and that the relatively normal segmentation that was observed through E11.5 in this study could perhaps rely on residual mature miRNAs that are present in the caudal PSM cells.
We find that interfering with interactions between mir-125a-5p and Lfng transcripts in vivo in chick embryos stabilizes those transcripts in the PSM, suggesting an effect on RNA turnover. However, transgenic analysis examining the function of the Lfng 3′UTR in GFP reporter transgenes suggests that the mir-125a-5p binding sites may not have a dramatic effect on the 3′UTR’s ability to destabilize an mRNA stability in the mouse PSM (data not shown). The possibility that the clock function of mir-125a-5p might not be conserved between mouse and chicken would not be surprising given that different organisms can utilize completely distinct sets of protein components in their segmentation clocks (Krol et al., 2011). Thus, it is possible that different miRNA:transcript pairs are important in the mouse segmentation clock, or that regulation occurs via translational efficiency rather than through effects on transcript stability. Testing of these models will require targeted mutation of the mir-125a-5p binding sites in the Lfng 3′UTR at the endogenous locus, to examine whether these binding sites are required for normal mouse segmentation. However, it is attractive to hypothesize that different mechanisms of post-transcriptional clock regulation could contribute to the differences in clock period observed among distinct vertebrate species.
Experimental Procedures (Supplemental Methods online)
miRNA QRT-PCR
Total RNA was extracted from the PSM and mature somites of E 9.5 embryos and QRT-PCR was performed using Taqman primers specific for mmu-mir-125a-5p, 2198, PN4395309 in triplicate on at least three biologically independent replicates. Results show mean +/− SD after normalizing expression levels of the somite samples to 1. Significance calculated by Student’s T test.
In-situ Hybridization
RNA in-situ hybridization was performed essentially as described with mRNA probes(Shifley et al., 2008), or with DIG labeled miRCURY LNA probes (Exiqon) either in whole mount embryos(Sweetman, 2011) or in section in situs(Nuovo, 2011). Section in situs were performed essentially as described (Nuovo, 2011). Details of mRNA probes and in situ protocols are found in the online supplement.
Luciferase Assays
The mouse or chick Lfng 3′UTR was amplified and cloned into pMIR-REPORT™ (Ambion). PCR mutagenesis of the mir-125a-5p seed regions using primers described in the supplemental experimental methods was confirmed by sequencing. Luciferase assays were performed in NIH3T3 cells transfected with reporter, pSVRenilla, and precursor miRNA. Cells were assayed for luciferase activity 40h post-transfection (Promega). All values reflect at least three independent experiments. Statistical analysis was performed using two way ANOVA, with Bonferroni post hoc.
In ovo electroporation
Fluorescein-tagged Target Protectors and Anti-mir-morpholinos (Supplemental Experimental Methods) were ordered from Gene Tools, LLC. To reduce the chance of off target interference with other miRNAs, an antisense morpholino corresponding to the NC#1 control sequence (Ambion) was ordered for use as a control morpholino. As the NC#1 sequence does not have predicted targets in vertebrate genomes, this morpholino is unlikely to affect the activity other miRNAs, and was used in preference to a scrambled sequence morpholino that might have exhibited unexpected effects. In ovo electroporation was performed essentially as described (Dubrulle et al., 2001). Embryos of stage 7-8HH were used for in ovo electroporation. Target Protectors or anti-miR morpholinos were laid on the anterior primitive streak using a glass capillary. An electric pulse of 6V, 25 mseconds was charged three times. Embryos were incubated for 2, 8 or 24 hours prior to removal and analysis. Only embryos exhibiting robust fluorescein expression and normal morphology outside of the electroporated region were used for analysis. For these analyses, 981 embryos were electroporated, with between 15 and 60% of embryos being analyzed for any particular electroporation. A subset of embryos exhibited fluorescein positive somites on only one side of the embryo. Further protocol details are in the supplemental online methods.
Supplementary Material
Highlights.
mir-125a-5p affects the stability of Lfng transcripts in the chick PSM
Inhibiting mir-125a-5p:Lfng binding perturbs the segmentation clock in chickens
Oscillatory Lfng expression in the clock requires mir-125a-5p activity
Loss of mir-125a-5p activity causes abnomal segmentation in chick embryos
Acknowledgments
We thank R. Wharton and A. Hopper for comments, A. Fischer for help with electroporation, and the OSUCCC Nucleic Acids core facility for QRT-PCR. This work was supported by NSF grant # IOS-0919649 and NIH grant # R03HD062722 to SEC and a Pelotonia Predoctoral Fellowship to MFR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baek D, J. V, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bonev B, Stanley P, Papalopulu N. MicroRNA-9 Modulates Hes1 Ultradian Oscillations by Forming a Double-Negative Feedback Loop. Cell Rep. 2012;2:10–18. doi: 10.1016/j.celrep.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N, Dubchak I, Pachter L. AVID: A global alignment program. Genome Res. 2003;13:97–102. doi: 10.1101/gr.789803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kang L, Zhang N. Negative feedback loop formed by Lunatic fringe and Hes7 controls their oscillatory expression during somitogenesis. Genesis. 2005;43:196–204. doi: 10.1002/gene.20171. [DOI] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Cole SE, Levorse JM, Tilghman SM, Vogt TF. Clock regulatory elements control cyclic expression of Lunatic fringe during somitogenesis. Dev Cell. 2002;3:75–84. doi: 10.1016/s1534-5807(02)00212-5. [DOI] [PubMed] [Google Scholar]
- Dale JK, M. M, Dequeant ML, Malapert P, McGrew M, O. P. Periodic notch inhibition by lunatic fringe underlies the chick segmentation clock. Nature. 2003;421:275–278. doi: 10.1038/nature01244. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, McGrew MJ, Pourquié O. FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell. 2001;106:219–232. doi: 10.1016/s0092-8674(01)00437-8. [DOI] [PubMed] [Google Scholar]
- Evrard YA, Lun Y, Aulehla A, Gan L, Johnson RL. Lunatic fringe is an essential mediator of somite segmentation and patterning. Nature. 1998;394:377–381. doi: 10.1038/28632. [DOI] [PubMed] [Google Scholar]
- Feng P, Navaratna M. Modelling periodic oscillations during somitogenesis. Math Biosci Eng. 2007;4:661–673. doi: 10.3934/mbe.2007.4.661. [DOI] [PubMed] [Google Scholar]
- Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, Williams BR. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Kageyama R. Hopf bifurcation in the presomitic mesoderm during the mouse segmentation. J Theor Biol. 2009;259:176–189. doi: 10.1016/j.jtbi.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Pourquié O, Dubrulle J. In vivo analysis of mRNA stability using the Tet-Off system in the chicken embryo. Dev Biol. 2005;284:292–300. doi: 10.1016/j.ydbio.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Hirata H, Bessho Y, Kokubu H, Masamizu Y, S. Y, Lewis J, Kageyama R. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- Hirsinger E, Jouve C, Dubrulle J, O. P. Somite formation and patterning. Int Rev Cytol. 2000;198:1–65. doi: 10.1016/s0074-7696(00)98002-1. [DOI] [PubMed] [Google Scholar]
- Krol AJ, Roellig D, Dequeant ML, Tassy O, Glynn E, Hattem G, Mushegian A, Oates AC, Pourquie O. Evolutionary plasticity of segmentation clock networks. Development. 2011;138:2783–2792. doi: 10.1242/dev.063834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, Vogt TF. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- Monk N. Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays. Curr Biol. 2003;13:1409–1413. doi: 10.1016/s0960-9822(03)00494-9. [DOI] [PubMed] [Google Scholar]
- Morales AV, Yasuda Y, Ish-Horowicz D. Periodic Lunatic fringe expression is controlled during segmentation by a cyclic transcriptional enhancer responsive to notch signaling. Dev Cell. 2002;3:63–74. doi: 10.1016/s1534-5807(02)00211-3. [DOI] [PubMed] [Google Scholar]
- Nuovo GJ. In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. Methods. 2011;52:307–315. doi: 10.1016/j.ymeth.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Okubo Y, Sugawara T, Abe-Koduka N, Kanno J, Kimura A, Saga Y. Lfng regulates the synchronized oscillation of the mouse segmentation clock via trans-repression of notch signalling. Nat Commun. 2012;3:1141. doi: 10.1038/ncomms2133. [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Pourquie O. The vertebrate segmentation clock. J Anat. 2001;199:169–175. doi: 10.1046/j.1469-7580.2001.19910169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquie O, Tam PP. A nomenclature for prospective somites and phases of cyclic gene expression in the presomitic mesoderm. Dev Cell. 2001;1:619–620. doi: 10.1016/s1534-5807(01)00082-x. [DOI] [PubMed] [Google Scholar]
- Serth K, Schuster-Gossler K, Cordes R, Gossler A. Transcriptional oscillation of lunatic fringe is essential for somitogenesis. Genes Dev. 2003;17:912–925. doi: 10.1101/gad.250603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifley ET, Vanhorn KM, Perez-Balaguer A, Franklin JD, Weinstein M, Cole SE. Oscillatory lunatic fringe activity is crucial for segmentation of the anterior but not posterior skeleton. Development. 2008;135:899–908. doi: 10.1242/dev.006742. [DOI] [PubMed] [Google Scholar]
- Sweetman D. In Situ Detection of microRNAs in Animals. Methods Mol Biol. 2011;732:1–8. doi: 10.1007/978-1-61779-083-6_1. [DOI] [PubMed] [Google Scholar]
- Xie ZR, Yang HT, Liu WC, Hwang MJ. The role of microRNA in the delayed negative feedback regulation of gene expression. Biochem Biophys Res Commun. 2007;358:722–726. doi: 10.1016/j.bbrc.2007.04.207. [DOI] [PubMed] [Google Scholar]
- Zhang N, Gridley T. Defects in somite formation in Lunatic fringe deficient mice. Nature. 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]
- Zhang Z, O’Rourke JR, McManus MT, Lewandoski M, Harfe BD, Sun X. The microRNA-processing enzyme Dicer is dispensable for somite segmentation but essential for limb bud positioning. Dev Biol. 2011;351:254–265. doi: 10.1016/j.ydbio.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.